Abstract
CysK (O-acetylserine sulfhydrylase) is a pyridoxal-5′ phosphate-dependent enzyme which catalyzes the second step of the de novo cysteine biosynthesis pathway by converting O-acetyl serine (OAS) into l-cysteine in the presence of sulfide. The first step of the cysteine biosynthesis involves formation of OAS from serine and acetyl CoA by CysE (serine acetyltransferase). Apart from role of CysK in cysteine biosynthesis, recent studies have revealed various additional roles of this enzyme in bacterial physiology. Other than the suggested regulatory role in cysteine production, other activities of CysK include involvement of CysK-in contact-dependent toxin activation in Gram-negative pathogens, as a transcriptional regulator of CymR by stabilizing the CymR-DNA interactions, in biofilm formation by providing cysteine and via another mechanism not yet understood, in ofloxacin and tellurite resistance as well as in cysteine desulfurization. Some of these activities involve binding of CysK to another cellular partner, where the complex is regulated by the availability of OAS and/or sulfide (H2S). The aim of this study is to present an overview of current knowledge of multiple functions performed by CysK and identifying structural features involved in alternate functions. Due to possible role in disease, promoting or inhibiting a “moonlighting” function of CysK could be a target for developing novel therapeutic interventions.
Keywords: CysK, Cysteine biosynthesis pathway, Cysteine synthase complex, Multifaceted roles, Contact-dependent growth inhibition, Ofloxacin resistance, Tellurite resistance, CymR regulation, Biofilm formation, Cysteine desulfurization, CysK inhibitors, Moonlighting functions
Introduction
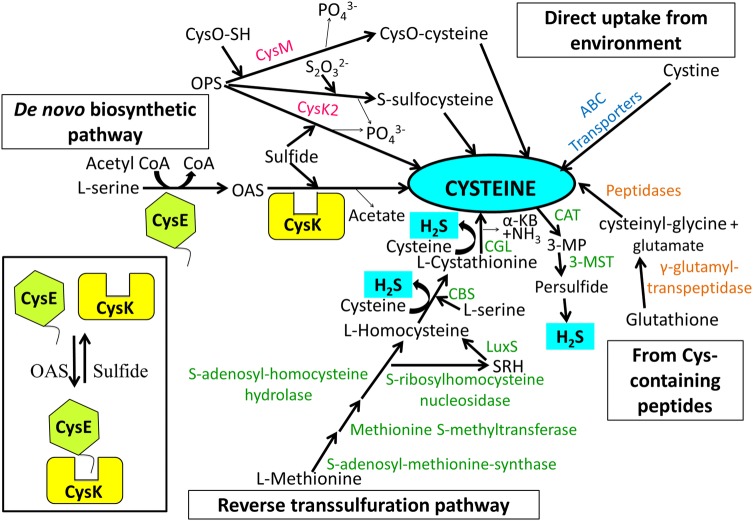
The rise in clinical incidence of antibiotic-resistant bacteria is a major global health care issue. It is especially true when the infections with multidrug-resistant (MDR) pathogens impose a significant and increasing burden on both patients and healthcare providers. Therefore, identification of novel drug targets and subsequent development of drugs against these targets is a dire need for improving current treatment. In Mycobacterium tuberculosis (Mtb) essentiality of cysteine biosynthesis pathway is well-documented in its persistent phase (Schnell et al. 2015). Similarly in Entamoeba histolytica (Ehi) and Salmonella typhimurium (Sty), cysteine, generated through the de novo cysteine biosynthesis pathway is vital for the pathogenic antioxidative defense mechanism (Chinthalapudi et al. 2008; Turnbull et al. 2008). These cases serve as a paragon to explore cysteine biosynthesis pathway enzymes in other pathogenic organisms also (Gerdes et al. 2003; Baba et al. 2006; Akerley et al. 2002; Chaudhuri et al. 2009; Kobayashi et al. 2003; Sassetti et al. 2003; Sassetti and Rubin 2003; Rengarajan et al. 2005; Schnell et al. 2015; Chinthalapudi et al. 2008; Turnbull et al. 2008). Cysteine can be produced in bacteria (i) from L-serine via a two-step de novo biosynthetic pathway (Kredich et al. 1966 and discussed in the ensuing section), (ii) by conversion of methionine-to-cysteine via reverse transsulfuration pathway (Seiflein and Lawrence 2001; Sekowska and Danchin 1999; Thomas and Surdin-Kerjan 1997; Vermeij and Kertesz 1999; Wheeler et al. 2005) (iii) by hydrolysis of Glutathione (GSH) or cysteine-containing peptides by proteolytic enzymes (Suzuki et al. 1993, 2001) and (iv) direct uptake as cysteine dimer from the environment through transporters or symporters (reviewed by Guédon et al. 2006 and references therein). The major biosynthetic pathways and additional sources of cysteine in bacteria are presented in Fig. 1.
Fig. 1.
Sources of cysteine and cysteine-generated H2S in bacteria. Cysteine in bacteria is produced from—(i) l-serine via classical de novo biosynthetic pathway (ii) l-methionine via reverse transsulfuration pathway that involves conversion of Met to S-adenosyl-methionine, followed by generation of S-adenosyl-homocysteine (SAH) which is cleaved to l-homocysteine either by SAH-hydrolase or by successive action of SAH-nucleosidase and S-ribosyl-homocysteinase (LuxS). Subsequently, PLP-dependent enzyme, cystathionine β-synthase (CBS) catalyses the condensation of homocysteine and serine to form l-cystathionine which is converted by cystathionine γ-lyase (CGL) to cysteine, α-ketobutyrate (α-KB) and ammonia (Thomas and Surdin-Kerjan 1997; Guédon et al. 2006) (iii) peptides (cystathione) and (iv) direct intake of extracellular cystine via ABC transporters. Enzymatic biogenesis of H2S mainly involves CBS, CGL, and 3-MGT/CAT routes. The schematic figure depicts all enzymes as monomers without any relevance to the biologically active oligomeric form. Thin arrows in the figure represent release of a by-product. Inset depicts formation of CSC under varying cellular availability of OAS. In absence, CysK and CysE form CSC where the activity of CysK is inhibited and there is no production of cysteine. Once OAS starts accumulating, the complex dissociates to release CysK for cysteine production
The de novo biosynthesis of cysteine in bacteria requires two enzymes; serine acetyltransferase (SAT/CysE) and O-acetylserine sulfhydrylase (OASS/CysK) that are encoded by cysE and cysK genes, respectively. CysE catalyzes the transfer of acetate from acetyl-CoA to serine, thereby, generating O-acetylserine (OAS). CysK, the next enzyme of the pathway, uses pyridoxal 5′-phosphate (PLP) as a cofactor to convert OAS and sulfide into cysteine and acetate. There are other OASS present as well, designated CysM and CysK2, which are 43% and 26% identical to CysK, respectively, but catalyzes the formation of preferred O-phospho-l-serine (OPS) with either CysO-SH sulfur donor in case of CysM or sulfide/sulphate donor in case CysK2 to produce cysteine via CysO-cysteine or S-sulfocysteine intermediates, respectively (Kredich 1996; Steiner et al. 2014; Schnell et al. 2015; Fig. 1). Though cysK and cysM double mutant of S. typhimurium is a cysteine auxotrophs but mutant of either one is able to catalyze the second step of de novo biosynthesis pathway, demonstrating that even if one of them is functional, cysteine is produced (Hulanicka et al. 1979). Evolution of these isozymes for redundancy in the pathway step may have arisen because of the need of the microbe to survive under varying habitats (sulfide or thiosulfate rich) or for stress adaptation in some pathogens. For instance CysM appears to be the key enzyme for catalyzing the production of l-cysteine during dormant M. tuberculosis and maintaining intracellular redox homeostasis inside host macrophages (Steiner et al. 2014). Similarly, it is reported that S. typhimurium lacking cysK and cysM is less virulent and exhibits reduced antibiotic resistance (Turnbull et al. 2008, 2010).
Intriguingly, additional roles of the CysK are now being discovered apart from its original role of cysteine biosynthesis. These include the involvement of CysK- (1) in Cysteine Synthase Complex (CSC) formation with CysE (Kredich et al. 1969; Droux et al. 1998; Mino et al. 2000a, b; Wirtz et al. 2004; Campanini et al. 2015a, b), (2) in activation of contact-dependent growth inhibition (CDI) toxin (Diner et al. 2012; Johnson et al. 2016), (3) as a transcriptional promoter by stabilizing the interaction between transcriptional regulator CymR and DNA (Tanous et al. 2008), (4) in biofilm formation in Vibrio fischeri (Vfi) (Singh et al. 2015), (5) in ofloxacin (Frávega et al. 2016) and tellurite resistance (Vásquez et al. 2001; Ramìrez et al. 2006) and (6) in cysteine desulfurization (Mino et al. 2003; Hullo et al. 2007).Therefore, inhibition of CysK may downregulate not only the cysteine biosynthesis pathway but may also influence the above-listed functions.
Considering the importance of CysK in bacterial physiology, attempts are ongoing to develop its inhibitors mainly against CysK from Haemophilus influenzae (Hin), Sty and Mtb. Based on the structural determinants identified from interaction between CysK and CysE C-terminal tail, the peptide inhibitors and the first synthetic inhibitor, 2-substituted-cyclopropane-1-carboxylic acids were developed by the Mozzarelli’s group (Amori et al. 2012). Subsequently, optimization strategies to improve both potency and drug-like properties were carried out in many laboratories. The aim of this review is to offer a better understanding of multifaceted roles of CysK and to cover the progress and challenges associated with discovery of CysK inhibitors with therapeutic potential.
Structural insights into CysK
Oligomeric state and overall topology
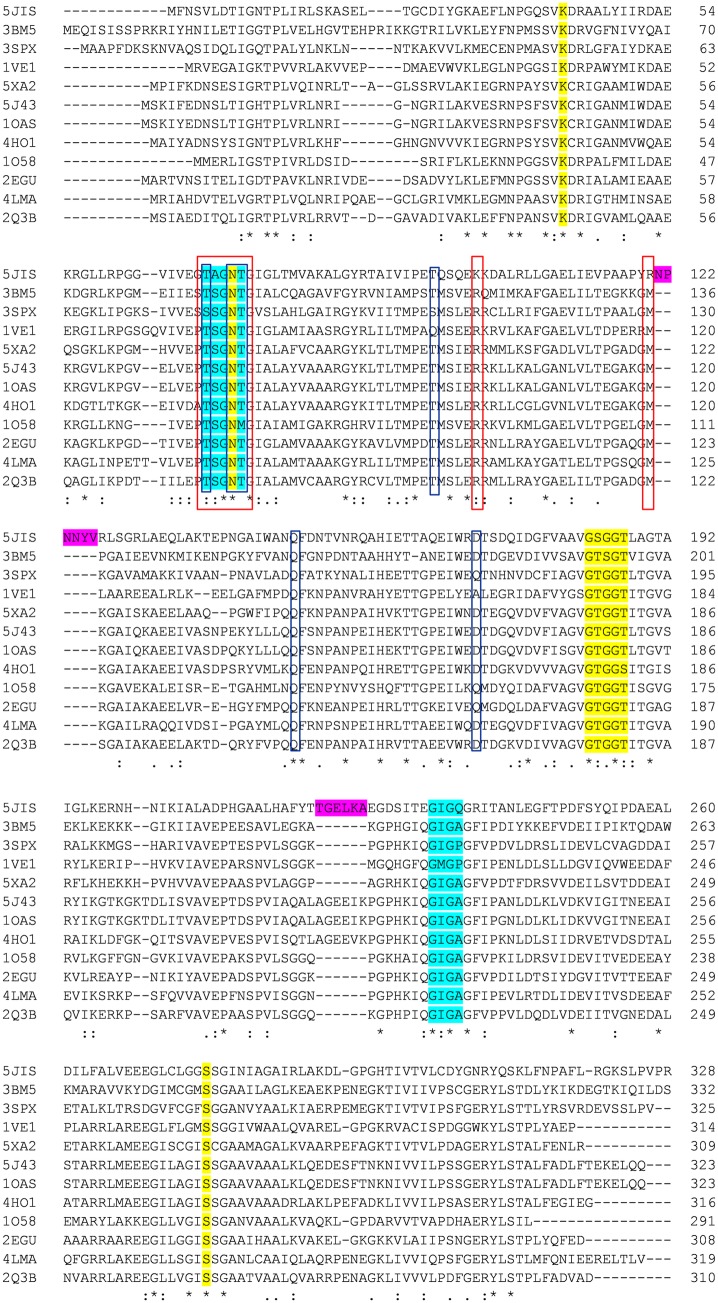
Although most studies predominantly report CysK to be a dimer, tetrameric form of CysK has been reported in the case of Thermotoga maritime (Heine et al. 2004), Sty (Cook et al. 1978), Saccharomyces cerevisiae (Yamagata et al. 1976) and Arabidopsis thaliana (Droux et al. 1998) with molecular weight of monomer unit being around 34.5 kDa. In recent years, a number of crystal structures of CysK and its isomers as well as mutants have been solved. Table 1 compiles the list of structures of CysK available in protein data bank (PDB) for ready reference. Comparison of amino acid sequence (Fig. 2) and 3D structure reveals similarity on both fronts. Overall topology of CysK resembles that of other type II PLP-dependent enzymes such as tryptophan synthase-β and threonine deaminase (Burkhard et al. 1998; Gallagher et al.1998; Grishin et al. 1995; Schneider et al. 2000) which consists of two domains (one large and the other small), each comprising of α/β fold. A cleft between these two domains harbors the binding site for the PLP moiety.
Table 1.
Source and PDB code for available structures of CysK Enzyme either as apo or in ligand-bound form
| Domain | Organism | PDB ID (References) | CysK-apo or ligand-bound |
|---|---|---|---|
| BACTERIA | Microcystis aeruginosa | 4LMA (Lu et al. 2014) | Complexed with MPD [(4 s)-2-methyl-2,4-pentanediol] and plp |
| Escherichia coli | 5J43; 5J5V (Johnson et al. 2016) | Complexed with tRNA nuclease CdiA and PLP; complexed with tRNA nuclease CdiA, immunity protein CdiI, MSE and PLP | |
| Haemophilus influenzae | 1Y7L (Huang et al. 2005) | Complexed with decameric fragment of CysE, SO4 and PLP | |
| 3IQG; 3IQH; 3IQI (Salsi et al. 2010) | Complexed with MNWNI pentapeptide and PLP; complexed with MNYDI pentapeptide, SO4and PLP; complexed with MNENI pentapeptide and PLP | ||
| 4HO1; 4LI3;5DBH; 4NU8; 4ORE; 4ZU1; 4ZU6; 5DBE; 5XCW; 5XCN; 5XCP (no available references) | Complexed with GOL (glycerol), EDO (1,2-ethanediol) and PLP; complexed with peptide from CysE, GOL and PLP; complexed with GOL and 0JO (2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid); complexed with peptide from CysE, GOL and PLP; complexed with peptide inhibitor and OAS; complexed with C-terminal peptide from CysE, GOL and OAS; complexed with C-terminal peptide from CysE, OAS and PLP; complexed with C-terminal peptide from CysE, OAS, GOL and 0JO; covalently linked to PLP; covalently linked to PLP; covalently linked to PLP | ||
| Mycobacterium tuberculosis | 2Q3B; 2Q3C; 2Q3D (Schnell et al. 2007) | Complexed with CL (chloride ion) and MPD, covalently linked to PLP; complexed with DFSI inhibitory peptide and MPD, covalently linked to PLP; complexed with PDA and MPD | |
| 3ZEI (Poyraz et al. 2013) | Complexed with AWH (3-[(Z)-[(5Z)-5-[[2-(2-hydroxy-2-oxoethyloxy)phenyl]methylidene]-3-methyl-4-oxidanylidene-1,3-thiazolidin-2-ylidene]amino]benzoic acid), MPD and PLP | ||
| Salmonella typhimurium | 1OAS (Burkhard et al. 1998) | Complexed with PLP | |
| 1FCJ (Burkhard et al. 2000) | Complexed with SO4, CL and PLP | ||
| 1D6S (Burkhard et al. 1999) | Complexed with PLP and MET (methionine) | ||
| Thermus thermophiles | 1VE1; 2EFY; 2ECO; 2ECQ (no available references) | Complexed with PLP; complexed with 4AT (5-oxohexanoic acid) and PLP; complexed with 4MV (4-methyl valeric acid) and PLP; complexed with PLP and 3HL ((3 s)-3-hydroxybutanoic acid) | |
| Planctomyces limnophila | 5XA2; 5XOQ (no available references) | Covalently linked to PLP; complexed with GLY-PHE-SER-GLY-GLY-ASP-GLY-ILE and PEG, covalently linked to PLP | |
| Leishmania donovani | 3SPX; 3T4P; 3TBH (Raj et al. 2012) | Complexed with NA (sodium ion), CL and PEG (di(hydroxyethyl)ether), covalently linked to PLP; complexed with CL and PEG, covalently linked to PLP; complexed with octapeptide from CysE and NA, covalently linked to PLP | |
| Brucella abortus | 5JIS; 5JJC (Dharavath et al. 2017) | PLP covalently linked to K42 residue of CysK | |
| Thermotoga maritima | 1O58 (Heine et al. 2004) | Complexed with PO4 | |
| Geobacillus kaustophilus | 2EGU (no available references) | Apo | |
| Mycobacterium marinum | 3RR2 (Baugh et al. 2015) | Apo | |
| EUKARYOTA | Entamoeba histolytica | 3BM5; 2PQM (Chinthalapudi et al. 2008) | Complexed with SO4, PLP and CYS (cysteine); complexed with SO4 and PLP |
| 4JBL; 4JBN; 4IL5 (Raj et al. 2013) | Complexed with SO4 and MET, covalently linked to PLP; complexed with tetrapeptide from CysE, and SO4, covalently linked to PLP; complexed with SO4 and ILE (isoleucine), covalently linked to PLP | ||
| Arabidopsis thaliana | 2ISQ (Francois et al. 2006) | Complexed with octapeptide from CysE, SO4 and PLP | |
| 4AEC (Feldman-Salit et al. 2012) | Complexed with ACT (acetate ion), GOL and PLP | ||
| 1Z7W; 1Z7Y (Bonner et al. 2005) | Complexed with SO4 and PLP; complexed with AA5(n-[(3-hydroxy-2-methyl-5-{[(trihydroxyphosphoranyl)oxy]methyl}pyridin-4-yl)methylene]methionine) |
Fig. 2.
Structure-based sequence alignment of CysK. Sequence alignment is generated through Clustal Omega using the sequences from Brucella abortus (5JIS), Entamoeba histolytica (3BM5), Leishmania donovani (3SPX), Thermus thermophiles (1VE1), Planctomyces limnophila (5XA2), Escherichia coli (5J43), Salmonella typhimurium (1OAS), Haemophilus influenzae (4HO1), Thermotoga maritima (1O58), Geobacillus kaustophilus (2EGU), Microcystis aeruginosa (4LMA), and Mycobacterium tuberculosis (2Q3B). OAS- and PLP-binding residues are highlighted in blue and yellow colors, respectively. The two insertions 121N–126V and 218Y–223L in the active site of BabCysK are emphasized in magenta color. Residues involved in interacting with CysE and CdiA-CT toxin are boxed in red and blue, respectively. Completely conserved residues are indicated by the asterisks at the bottom line of the alignment figure while highly and moderately conserved residues are shown with double and single dots, respectively
PLP-binding site
From the insights gained from the crystal structures of S. typhimurium (Burkhard et al. 1998), M. tuberculosis (Schnell et al. 2007), E. histolytica (Chinthalapudi et al. 2008), H. influenzae (Salsi et al. 2010), L. donovani(Raj et al. 2012), M. aeruginosa (Lu et al. 2014), E. coli (Johnson et al. 2016), and B. abortus (Bab) (Dharavat et al. 2017), the PLP-binding site is located at the interface between N- and C-terminal domains. It forms an internal aldimine (Schiff-base linkage) with the Lys41 (residue number is of Sty; Figs. 2 and 3). The glycine-rich residues—Gly176, Thr177, Gly178, Gly179, and Thr180 loops around the phosphate group of PLP making hydrogen bonds to the non-ester phosphate oxygen atoms. The unprotonated 3′-hydroxyl group of PLP which is in hydrogen bonding with the side-chain amide nitrogen of the Asn71, forms the final attachment point of the coenzyme. The N1 nitrogen of pyridine ring of PLP is in H-bonding with Ser272 (Fig. 3).
Fig. 3.
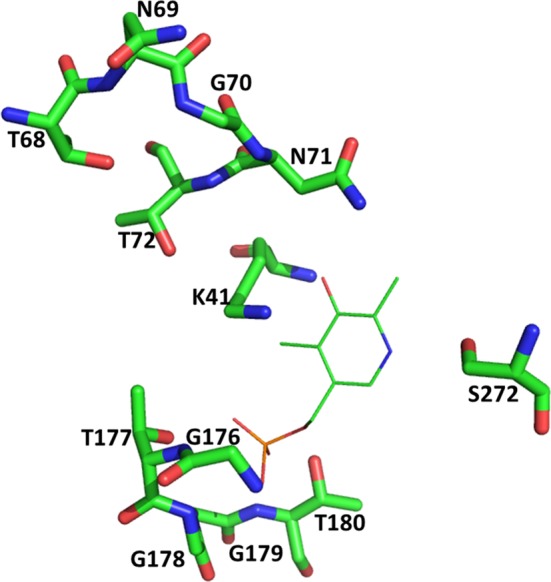
PLP-binding site of CysK based on the crystal structure of StyCysK (1OAS). PLP-binding residues are depicted with stick representation whereas PLP is shown as line
Active site
The binding site of OAS is predominantly reported through crystal structures of CysK in complex with either C-terminal end of CysE or l-methionine that mimics the substrate OAS (Salsi et al. 2010; Burkhard et al. 1999). The α-carboxylate-binding subsite is formed by four amide N-atoms of highly conserved Asn loop (69TSGNT73; color coded blue in Fig. 2) in the N-terminal domain of the CysK subunit (Burkhard et al. 1998). Upon formation of the reaction intermediate, N-terminal domain encompassing the Asn loop folds over the active site, thereby restricting the solvent access to the hydrophobic active site and protecting the reaction intermediate. Another well-conserved Gly loop (229GIGA232; color-coded blue in Fig. 2) in the C-terminal domain also plays an important role in substrate and active site interaction (Raj et al. 2013; Fig. 4).
Fig. 4.
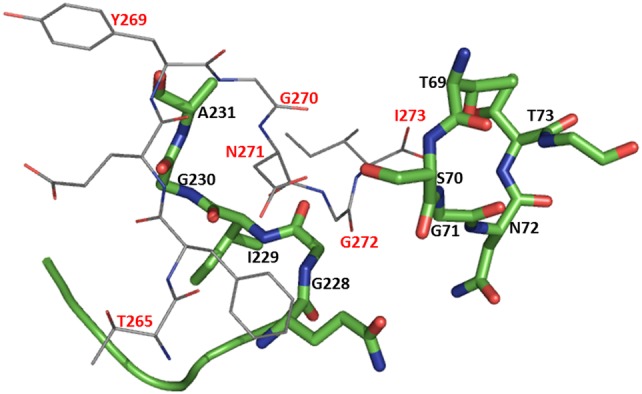
Active site of CysK bound to CysE C-terminal peptide based on the crystal structure of HinCysK (4NU8): CysK active-site residues (69TSGNT73; 228GIGA231) are represented by green sticks with residue labeling in black; CysE C-terminal peptide residues (265TFEYGDGI273) are shown as grey lines with labeling in red
Insights into mechanism of CysK
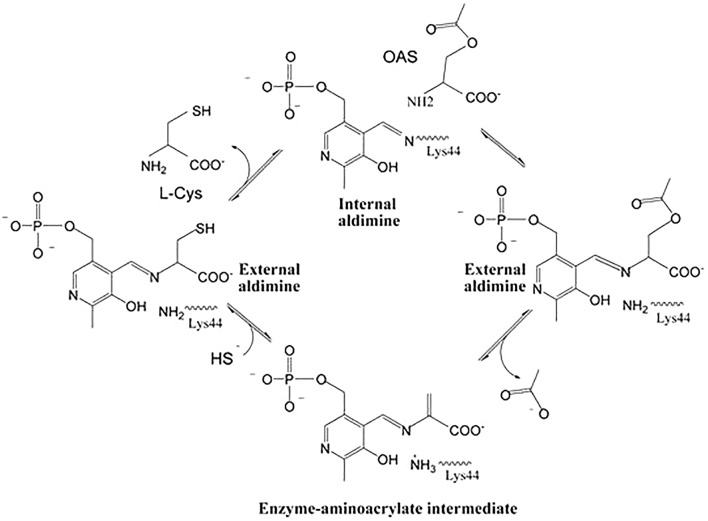
CysK follows a Ping Pong Bi Bi reaction kinetic that starts with the diffusion of OAS into the active site (Burkhard et al. 1999). The geminal di-amines are formed when the α-amine of the substrate attacks C4ʹ of the internal aldimine between PLP and Lys41 (StyCysK numbering). These geminal di-amines intermediates are converted to an external aldimine intermediate in which the carboxylate of the substrate interacts with the asparagine-loop adjacent to the cofactor and the positive end of the α-helix 2, triggering the closure of the active site. Formation of external aldimine results in the release of Lys41 which then acts as a general base in the α,β-elimination of acetate resulting in the formation of α-aminoacrylate intermediate. Once the acetate has been eliminated, the nucleophilic attack of sulfide onto this α-aminoacrylate intermediate, results in the formation of cysteine (Fig. 5) (Schnell et al. 2015; Burkhard et al. 1999).
Fig. 5.
Catalytic mechanism of CysK from M. tuberculosis (Scheme 1 from Schnell et al. 2015, reprinted with permission from Elsevier)
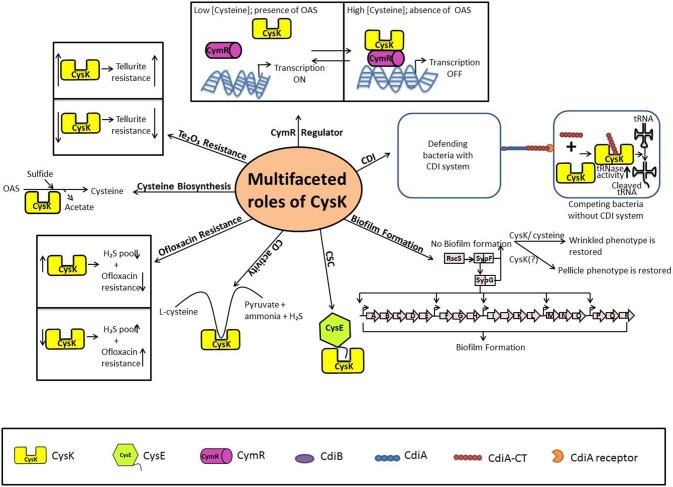
Multifaceted roles of CysK
The role of CysK in cysteine biosynthesis pathway is well established. In addition, recent studies provide significant evidences that CysK plays an integral role in other cellular activities in bacteria. These activities are listed in Table 2 and Fig. 6. The multiple functions of CysK are discussed below.
Table 2.
Multiple documented roles of CysK as compiled from various studies
| S. No. | Activity | Mechanism | Binding partners | Organisms | Reference |
|---|---|---|---|---|---|
| 1 | Cysteine synthase | Conversion of OAS to cysteine in presence of sulfide | – | Plants and microorganisms | Kredich et al. (1966) |
| 2 | Regulation of Cysteine biosynthesis pathway | CysK forms complex with CysE in which its activity is inhibited and subsequently, cysteine production is inhibited | CysE | Plants and microorganisms | Kredich et al. (1969) |
| 3 | CdiA-CT toxin activation | CysK present in the target cell binds to CDI-CT and activates it | CdiA-CT | E. coli and B. subtilis (Diner, result unpublished) | Diner et al. (2012), Johnson et al. (2016), Benoni et al. (2017), Kaundal et al. (2016) |
| 4 | Regulation of CymR | CysK binds to CymR and stabilizes the CymR–DNA interaction and increases the half-life of the complex | CymR | B. subtilis | Tanous et al. (2008) |
| 5 | Biofilm formation | Exact mechanism is unknown | – | V. fischeri and C. albicans | Singh et al. (2015), Murillo et al. (2005) |
| 6 | Ofloxacin resistance | Inhibiton of CysK results in incresase in H2S-mediated defense system in bacteria | – | S. typhimurium | Frávega et al. (2016) |
| 7 | Tellurite resistance | Biomechanism is unknown | – | S. typhimurium, A. brasilense, and B. stearothermophilus | Vásquez et al. (2001), Ramìrez et al. (2006) |
| 8 | Cysteine desulfurization | CysK catalyzes desulfurization of cysteine | – | L. casei, B. subtilis, A. pernix and E.coli | Bogicevic et al.(2012); Hullo et al. (2007), Mino et al. (2003), Awano et al. (2005) |
Fig. 6.
Multifaceted roles of CysK involving multiple binding partner and variable cellular environment
Role in regulation of cysteine biosynthesis pathway
Cysteine biosynthetic pathway is well regulated at transcriptional level by CysB transcriptional activator (Kredich et al. 1992) and through the coordinated activities of CysE and CysK at post-transcriptional level. Cysteine is a feedback inhibitor of CysE activity (Kredich et al. 1966). A recent study where a natural A241V variant of S. flexneri CysE has unstable quaternary assembly and reduced activity, suggests transient dissociation of quaternary structure of CysE to be another regulatory mechanism for cysteine synthesis (Verma et al. 2018). CysE and CysK together form a bi-enzyme complex called cysteine synthase or cysteine regulatory complex (CRC) as it regulates cysteine production. In this complex, the C-terminal of CysE binds to the active site of CysK via the key residue Ile-273 (numbering of StyCysE), competitively dissociates the substrate OAS and inhibits CysK enzymatic activity (Fig. 1b; Wirtz et al. 2001; Mino et al. 2000a, b). Until recently, how a low-affinity OAS substrate (KM in mM) could competitively dissociate a high-affinity C-terminal CysE peptide inhibitor (Kd in nM or µM) from the CysK active site was unfathomable. However, a study conducted by Singh and coworkers illustrates that “competitive allosteric inhibition” otherwise “substrate-induced facilitated dissociation” is responsible for expulsion of competitive inhibitor from the active site of CysK. Their work clearly demonstrates that OAS upon entering the active site of CysK (already occupied with the C-terminal CysE peptide), triggers conformational changes that alter an open-binding competent state to a closed-binding incompetent state, leading to inhibitor peptide moving out of the active site (Singh et al. 2017). It has been observed that the activity of CysK when complexed with CysE is reduced, with twofold decrease in kcat and fivefold increase in KM compared to the free enzyme values (Mino et al. 2000a, b). Recent systematic study on assembly of Sty reveals that the complex exists in two states-a low molecular weight species composed of one CysE hexamer and two CysK dimers (~ 310 kDa) and a high molecular weight species comprising of one CysE hexamer and four CysK dimers (~ 490 kDa). These two forms are in dynamic equilibrium and sensitive to the physiological levels CysK, OAS, and sulfide. Increased levels of OAS, under sulfur-limiting conditions, favor dissociation of CSC complex and continuous cysteine formation (Kaushik et al. 2017). The CSC complex appears to have no effect on the activity of CysE but it is suggested that one of the roles of CysK is to prevent the aggregation of CysE, therefore, enhancing its stability in the solution (Droux et al. 1998).
Recent studies indicate that some microorganisms do not form this CSC complex. This has been attributed to the active site cleft and peptide-binding properties of CysK. For example, the active site opening of CysK from Thermotoga maritima is only about 8 Å wide, as opposed to 11 Å for L. donovani CysK and 12 Å for MtbCysK that are able to form the complex (Raj et al 2012; Schnell et al. 2007; Heine et al. 2004). Interestingly, the active site of CysK from Brucella abortus (Bab) is ~ 13 Å wide but there is no complex formation in this organism. The answer to this anomaly lies in the two insertions 121N-126V and 218Y-223L (residue numbering are of BabCysK; color-coded magenta in Fig. 2), unique to the organism, which are responsible for differences in the vicinity of the active site. In fact, the researchers prepared a double mutant (Q96A-Y125A) of BabCysK with smaller amino acids that made the active site more accessible and as expected, the mutant exhibited higher affinity for BabCysE. The residues of active site of CysK that interact with the C-terminal peptide of CysE in Bab are shown inside the red box in Fig. 2 (Dharavath et al. 2017). Similarly, Ehi CysK and CysE cannot form the bi-enzyme complex because of differences in the sequence of the C-terminal tail (Kumar et al. 2011).
Role in CDI
Two general strategies employed by Gram-negative bacteria to inhibit competing bacteria are (i) secretion of bacteriocins and other soluble factors which diffuse through the environment and kill the bacteria at a distance and, (ii) contact-dependent growth inhibition (CDI) mediated by CdiA and CdiB, where the latter aids in exporting CdiA from cytosol to the outer membrane surface, which in turn forms a β-helical filament that extends from the defending bacterial cell to the CdiA receptor present on the surface of the competing bacteria. After the attachment of the β-helical filament to the CdiA receptor, the defending bacteria then release C-terminal toxin domain (CdiA-CT), which is a tRNase that cleaves tRNA of the competing target and arrests its growth. This CdiA-CT toxin tRNase is activated only when its C-terminal tail is inserted into the active site of CysK present in the target cell. The C-terminal Ile227 of the tail interacts with the Thr69, Asn72, Thr73, and Gln143 of CysK to form H–bonds; while Tyr225 of the tail forms H-bonds with Thr95 and Asp164 (E.coli CysK numbering; Fig. 2; Johnson et al. 2016). Both, CdiA-CT as well as CysE have been reported to bind to CysK with nanomolar affinities (Diner et al. 2012). It is intriguing how CdiA-CT secreting cells prevent autoinhibition. The presence of an immunity protein known as CdiI is responsible for the phenomenon (Aoki et al. 2011). CdiI forms a complex with CdiA-CT to neutralize its tRNase activity but apparently this complex dissociates rapidly (Benoni et al. 2017; Kaundal et al. 2016). A study by Kaundal et al. categorically demonstrates the role of CysK in stabilizing the binary complex CdiI/CdiA-CT with ~ 130 fold decrease in dissociation rate therefore, proving CysK to be a modulator of CdiA-CT/CdiI activity (Kaundal et al. 2016; Johnson et al. 2016).
Role in regulation of CymR
Cysteine is considered as a crucial and very significant amino acid in the survival of all microorganisms because of its thiol group. This is why all the genes involved in the biosynthesis, transport, and degradation of cysteine are tightly regulated. It has been observed that CymR regulator, which belongs to the family of Rrf2 family of transcription factors, is the central repressor of cysteine metabolism. It directly binds to a 27 bp sequence of the promoter region of the genes involved and regulates their expression. Intriguingly, it has been observed in B. subtilis, CymR binds to CysK with affinity which is slightly lower than between CysE and CysK. CymR, alone, is able to bind with the DNA, but CysK stabilizes the CymR–DNA interaction and increases the half-life of the complex for about seven times and positively regulates the gene repression by CymR. The cellular availability of cysteine is communicated to CymR by CysK via sensing the availability of its own substrate, OAS. Under cysteine-rich conditions, concentration of OAS is low and CymR–CysK complex is formed which transcriptionally represses the cysteine regulon. With low cysteine availability, OAS is abundant which prevents the CymR–CysK complex formation and may also dissociate the preformed complex either (i) by directly competing with CymR for the CysK active site or (ii) by altering the conformation of CysK, so that it cannot make complex with CymR. It is suggested that in B. subtilis, the intracellular concentrations of metabolites govern the interaction between different binding partners of CysK (Tanous et al. 2008).
Role in biofilm formation
The ability of bacteria to grow in biofilms facilitates them to survive in unfavorable conditions and to colonize a number of biotic and abiotic surfaces. The luminous marine bacterium V. fischeri is one such example, where the microbe establishes symbiosis with its host Euprymna scolopes (squid), when it forms a transient biofilm on the light organ of the juvenile squid (Nyholm et al. 2000). After 2–4 h, Vfi migrates through this biofilm towards the pores present on surface of the host leading to the internal organs, where it multiplies to a high cell density and establishes a long-term association with the squid. The formation of biofilm in Vfi is dependent on an 18-gene symbiosis polysaccharide locus-syp, which encodes the proteins that are involved in the production and export of the polysaccharides (a major component of biofilm matrix), and expression of matrix proteins BmpA, BmpB and BmpC (genes bmpA, bmpB, bmpC, respectively). Additional colonization factors include RscS and SypF (sensor kinases) and downstream response regulator SypG (DNA binding protein) that control syp locus. The visualization of the biofilm in the laboratory is performed with the overexpression of a sensor kinase called RscS which results in the wrinkled colonies-a biofilm phenotype. Another kind of biofilm phenotype is pellicle which forms at the liquid–air interface in static culture. Mutants with disrupted syp genes fail to form biofilms, expressing the wrinkled and pellicle phenotype, and colonize the squid (Fig. 6; mutated sypG and sypF genes are colored white). Further mutations in the cysH, cysJ, cysK, and cysN genes that are involved in cysteine metabolism also failed in Biofilm development in vitro, although supplementing the growth media with cysteine and/or overexpressing cysK could rescue the growth defect. Surprisingly, while cysteine could complement some defect (restoration of pellicle phenotype), CysK could overcome all (restoration of pellicle and wrinkled phenotype) suggesting the latter has additional role in biofilm formation other than providing cysteine (Singh et al. 2015).
Role in ofloxacin resistance
Emergence of drug-resistant strains of Sty to commonly used ampicillin or chloramphenicol led to shifting to second-generation flouroquinolones (ofloxacin, ciprofloxacin) for treating uncomplicated enteric fever as these drugs were found to be more effective and safer too (Fu et al. 1990; Chandey et al. 2012). However, reports of resistance of Sty strains to these drugs led (Joshi and Amarnath 2007; Crump et al. 2008) Gil and coworkers to explore fluoroquinolone resistance mechanism (Frávega et al. 2016). Based on an elegant study which established that endogenously produced H2S is cytoprotective for some bacteria by virtue of its ability to suppress oxidative stress generated by antibiotics (Shatalin et al. 2011), the investigators decided to focus on pathways responsible for accumulation of this signaling molecule. With the help of real-time reverse transcription (qRT) PCR analysis of Sty treated with bactericidal antibiotics they observed that cysK gene was slightly downregulated and also cysteine level was repressed. Further experiments with ΔcysK knockout Sty mutant corroborated reduction in l-cysteine synthesis and accumulation of H2S that accounted for the mutant exhibiting eightfold higher resistance to ofloxacin as opposed to wild type Sty. Therefore, inhibition of bacteria derived H2S-generating pathways in bacteria should make them sensitive to antibiotics. H2S is produced in bacteria from methionine or cysteine by cystathionine β-synthase (CBS) and Cystathionine γ-lyase (CGL) enzymes of transsulfuration pathway (with l-cysteine as substrate instead of l-serine). Additional path involves deamination between cysteine and α-ketobutyrate (α-KB) by aspartate/cysteine aminotransferase (CAT) to produce 3-mercaptopyruvate (3-MP) which under reducing conditions gets converted to H2S by 3-mercaptopyruvate sulfurtransferase (3-MST) as depicted in Fig. 1 (Pal et al. 2018). Interestingly, recent studies indicate that CBS from Bacillus subtilis (Hullo et al. 2007), Lactobacillus plantarum (Matoba et al. 2017), and Bacillus anthracis (Devi et al. 2017) unlike eukaryotic homologs, exhibit OASS activity as well as CBS activity (L-OAS dependent) and is able to generate H2S from a mixture of l-cysteine and l-homocysteine. A phylogenetic study by Gourinath and coworkers shows that these l-OAS-dependent CBS present in bacteria compared to CBS and CysK form a different branch of the phylogenetic tree suggesting their evolution as a new member in the PLP-bound enzyme family (Devi et al. 2017).
Role in tellurite resistance
Metal ions are necessary for the survival of the bacteria when present in lower concentration. However, if the concentration of these metal ions is too high, they generate oxidative stress leading to toxicity and eventually death of the bacteria. One such metal ion which is highly toxic for bacteria is tellurite, which possess strong oxidizing properties. Tellurites employ their toxicity through replacement of sulfur and selenium in various cellular functions (Garberg et al. 1999). It has been demonstrated that the cellular reductases convert tellurite to non-toxic tellurium at the expense of NAD(P)H (Chiong et al. 1988; Moscoso et al. 1998). Studies on identifying the cellular determinants in tellurite resistance have shown that CysK encoded by cysK gene plays an essential role in mediating tellurite resistance in Sty LT2, E. coli and Azospirillum brasilense. Disruption of cysK gene has been shown to decrease the tellurite resistance, while, introduction of cysK gene from Bacillus stearothermophilusV conferred tellurite resistance to E. coli. However, the exact biomechanism behind tellurite resistance mediated by CysK is still unknown (Vásquez et al. 2001; Ramìrez et al. 2006).
Role in cysteine desulfurization
Analogous to cysteine desulfurization (CD) activity of PLP-dependent CBS for producing pyruvate, ammonia and sulfide in vitro (Dwivedi 1982; Fig. 1), CysK in E. coli is reported to possess the CD activity (Awano et al. 2005). Similar findings are reported in Lactobacillus casei, (Bogicevic et al. 2012), B. subtilis (Hullo et al. 2007) and A. pernix (Mino et al. 2003). Given the existence of other CD proteins in bacterial cell, implication of CD activity of CysK is not clear, but based on gene disruption, a role in l-cysteine degradation and production is suggested (Awano et al. 2005).
Current status of inhibitors of CysK
Due to multifaceted roles of CysK and its absence in humans, efforts to identify a potent inhibitor are underway in many laboratories. The rational of design is mostly based on interactions between the active site residues of CysK- and CysE-derived peptide. Promising leads with strength and limitations are summarized in Table 3 and significant findings are discussed below.
Table 3.
Overview of existing CysK inhibitors with strength and/or limitations
| S. no. | Scaffold | Inhibitor | Organisms |
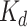 (µM) (µM) |
IC50 (µM) | Strengths and/or weaknesses | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Peptide | MNLNI | HinCysK | 24.9 ± 3.6 | – | Low in vivo stability, low bioavailability and poor pharmacokinetics | Salsi et al. (2010) |
| 2 | MNWNI | HinCysK | 24.9 ± 3.6 | – | |||
| 3 | MNYDI | HinCysK | 109 ± 10 | -– | Benoni et al. (2016) | ||
| 4 | MNYDI | StyCysK | 4.4 ± 0.3 | – | |||
| 5 | Compound 1 | StyCysK | 3.7 ± 0.4 | – | Needs further lead optimization | Spyrakis et al. (2013) | |
| 6 | 2-Nitro-5H-thiazolo[3,2-a]pyrimidine-5,7(6H)-dione | 8-Nitro-4-(2-(trifluoromethyl)phenyl)-4,4a-dihydro-2H-pyrimido[5,4-e]thiazolo[3,2-a]pyrimidine-2,5(3H)-dione (4n) | MtbCysK | – | 17.7 | Only inhibitor available with no cytotoxicity and MIC study (MIC = 7.6 µM) | Jean Kumar et al. (2013) |
| 7 | 4-Hydroxy-2-[2-(1H-indol-3-yl)-2-oxoethyl]sulfanyl-1H-pyrimidin-6-one | EhiCysK | ~ 8.0 | – | Needs further lead optimization to develop a strong inhibitor | Nagpal et al. (2012) | |
| 8 | Cyclopropane | (±)-trans-2-[(1E)-prop-1-en-1-yl]cyclopropanecarboxylic acid (7) | HinCysK | 1.46 ± 0.26 | 600 | Harsh to synthesize, prone to decomposition in the stock and highly volatile | Amori et al. (2012) |
| 9 | (1S,2S)-1-(4-Chlorobenzyl)-2-phenylcyclopropanecarboxylic acid (15b) | StyCysK | 0.054 ± 0.008 | 0.099 ± 0.004 | It is the only inhibitor with cyclopropane scaffold to bind CysK in nanomolar concentrations | Pieroni et al. (2016) | |
| 10 | Cis (±)-1-methyl-2-phenylcyclopropane-1,2-dicarboxylic acid (23) | StyCysK | 9 ± 1 | – | It appears as a valuable pharmacophore which represents the solid base for the synthesis of improved, drug-like and more efficient CysK inhibitors | Annunziato et al. (2016) | |
| 11 | Heterocyclic carboxylic acids | 1H-pyrazole-5-carboxylic acid (4c) | StyCysK | 122 ± 5 | – | The physicochemical properties indicate potential for penetration through the Gram-negative bacterial cell wall and possess drug-like properties | Magalhāes et al. (2018) |
| 12 | Thiazolidine | 3-((Z)-((Z)-5-(4-Fluorobenzylidene)-3-methyl-4-oxothiazolidin-2-ylidene)amino)benzoic acid (87) | MtbCysK | – | 0.019 ± 0.001 | Most potent inhibitor reported | Poyraz et al. (2013) |
| 13 | Fluoroalanine | Trifluoroalanine | StyCysK | – | 130 ± 10 | Non-specific and inefficient | Franko et al. (2018) |
Inhibition by the endogenous CysE peptide
In their quest to find inhibitor peptides against HinCysK, group of A. Mozzarelli computed the binding free energy of 400 pentapeptides in silico docked into the active site of HinCysK. The origin of this database was permutation combinations in the sequence motif MNX1X2I (derived on the basis of consensus sequence in the C-terminal CysE peptide from other organisms) where X1 corresponds to an aromatic amino acid and X2 to a strong hydrogen bond donor (Benoni et al. 2015; Campanini 2015a, b). Subsequently, 14 of these pentapeptides were selected for validating the computational predictions. The crystallized peptide MNWNI was found to be the best binder among the tested peptides followed by MNYDI, MNENI, MNETI and MNKGI in decreasing order. The terminal Ile of the CysE C-terminal tail is conserved and essential for interaction with CysK active site since its mutation to Ala resulted in disruption of complex (Zhao et al. 2006). The carboxylate group of Ile is secured in the interior of the CysK active site by hydrogen bonds with Asn72, Thr73, Thr79 and Gln143. These CysK residues also interact with the carboxylate of the OAS substrate (Salsi et al. 2010).In addition to Ile, Asn is also a significant amino acid that interacts with Ser70 in the case of HinCysK (Huang et al. 2005).Similarly, a comprehensive study on MNYDI peptide and StyCysK reveals that the aromatic residue at X1 is essential for hydrophobic interaction with Phe144 in the apolar active site of HinCysK (Salsi et al. 2010; Benoni et al. 2016). These structural determinants extracted from crystal structures of various peptide-enzyme complexes formed the basis for developing peptide/non-peptide inhibitors. Since then several studies have reported inhibitors against open conformation of CysK from different organisms with affinities for CysK generally ranging from millimolar to micromolar concentrations (Jean Kumar et al. 2013; Spyrakis et al. 2013; Yadava et al. 2015; Kant et al. 2018). Natural compounds as potential inhibitors of CysK have also been reported, however, the leads from these studies require rational modifications to achieve better efficacy (Nagpal et al. 2012; Mori et al. 2015, 2018). Comprehensive overviews of all these peptide and synthetic inhibitors (discussed below) are provided in the studies by Campanini (Campanini et al. 2015a, b) and Mazumder (Mazumder and Gourinath 2016).
Synthetic inhibitors
To avoid the problems of low stability and bioavailability associated with peptide inhibitors, battery of novel synthetic compounds were screened in silico for their inhibitor potency. Some of these molecules displayed better stability and pharmacokinetics than the natural peptide inhibitors in vitro and are listed below:
UPAR40
It is a class of 2-substituted-cyclopropane-1-carboxylic acids which was synthesized inspired by knowledge gained by analysing pentapeptides that were efficiently bound to HinCysK. The carboxylic moiety and the C2 lipophilic side chain of Ile267 which are molecular determinants of these pentapeptides formed the scaffold of the new chemical structures. Further, a cyclopropane ring was chosen as spacer due its rigid structure which provides the ligand a restrained conformation. Compound 7 with more lipophilic properties had the highest affinity for the HinCysK active site (Amori et al. 2012). A subsequent study by the same group illustrated that UPAR40 inhibits CysK activity by stabilizing closed conformation of the enzyme and formulated a hypothesis on the compound’s ability to additionally inhibit CysK or its isoforms from other species (Bruno et al. 2013).
α-Substituted-2-phenylcyclopropane carboxylic acids
The inhibitors developed by Amori had their own limitations such as hard to synthesize, highly volatile and prone to decomposition. A further attempt to optimize the inhibitors for better stability and binding efficacy led to the replacement of C2 alkenyl chain to phenyl ring maintaining the trans stereo-geometry with carboxylic moiety. A substituted chlorine at the para position of benzyl at the C1 position of the cyclopropane yielded a potent inhibitor (compound 15b) with dissociation constant in nanomolar range (Pieroni et al. 2016).
Cyclopropane-1,2-dicarboxylic acids
Next step of lead improvement included a substitution at C2 or di-substitution at C1 and C2 with carboxylic acid in the basic 2-phenylcyclopropane carboxylic acid. This led to better penetration of the outer membrane of Gram-negative bacteria and improved drug likeness. The most potent inhibitor in this series, Cis (±)-1-methyl-2-phenylcyclopropane-1,2-dicarboxylic acid (compound 23) which bear a phenyl moiety at C2 showed comparable activity with (1S,2S)-1-(4-Chlorobenzyl)-2-phenylcyclopropanecarboxylic acid (15b) (Annunziato et al. 2016).
Novel fragments inhibiting CysK
In quest to develop novel inhibitors against CysK, Barbara Campanini and co-workers performed scaffold hopping using the previously identified hits (Amori et al. 2012; Pieroni et al. 2016; Annunziato et al. 2016). They found 1H-pyrazole-5-carboxylic acid (compound 4c) in the series with the best inhibitory activity, low ClogP, high total polar surface area (TPSA) and reduced size for efficient penetration across Gram-negative cell membrane (Magalhāes et al. (2018). This study gave a rough idea about the structure–activity relationships (SAR) of these inhibitors where pyrazole substituent is probably binding in the lipophilic pocket near active site. Along with the lipophilic pocket, the size of the molecule was also found to have some influence in binding.
Thiazolidine inhibitors
Group of Schneider developed a rational inhibitor of nanomolar potency (IC50 of 103 nM) based on the crystal structure of CysK-peptide complex. This lead compound with thiazolidine scaffold was further optimized by employing protein crystallography, e-pharmacophore modeling, synthetic chemistry and in vitro testing. A library of 98 compounds was synthesized to obtain a comprehensive SAR data to understand the importance of substitutions in the phenyl ring system and the nitrogen of the thiazolidine nucleus. The results revealed that the carboxyl moiety of the secondary phenyl ring of the scaffold and the para position in the benzylidene ring are crucial for strong binding and inhibition of CysK. These findings were used to develop inhibitor 3-((Z)-((Z)-5-(4-Fluorobenzylidene)-3-methyl-4-oxothiazolidin-2-ylidene) amino) benzoic acid with IC50 19 nM and appear to be the most potent inhibitor till date (Poyraz et al. 2013; Table 3).
Fluoroalanine derivatives
Fluoroalanine derivatives are irreversible inhibitors that exploit the intrinsic reactivity of protein residues. Among these, mono-/di-/tri-halogenated (chloro/flouro based) alanine have been used as inhibitors of PLP-based enzymes. A study conducted by Franko and coworkers to find irreversible inhibitors for CysK concluded that F-ala competes with the substrate and slowly degrades with time whereas TF-ala is comparatively better owing to its stability and irreversible inactivation of CysK. However, in the case of CysK, even TF-ala is inefficient compared to that of the reversible inhibitors (Table 3). Therefore, this inhibitor requires efforts to be further understood and optimized in the context of the CysK (Franko et al. 2018).
The development of CysK inhibitors is still in the nascent stage. Crystal structures of the ternary complex of CysK with inhibitors provide some useful insights into the SAR between the inhibitor and the active site of the enzyme. From the investigations, it is clear that an efficient inhibitor should occupy as much space as possible of the active site. In addition, one must exercise caution and keep the other roles and properties of CysK in mind while designing inhibitors. For instance, ∆cysK sty mutant is more resistant to Ofloxacin as compared to wild-type strain indicating that inhibiting this enzyme will be a paradox for tackling MDR problem. In fact, in this context as CBS or CSE are more valuable drug targets for combating drug resistance menace, search for inhibitors for the Lactobacillus plantarum CBS is in progress (Devi et al. 2017; Matoba et al. 2017).
Conclusion
In recent years, the incidents of emerging MDR strains of the pathogens are increasing at an alarming rate which makes it imperative that the scientific community identifies novel targets for drug development purposes. Due to its multiple activities that may influence bacteria’s survival, CysK appears to be one such target. But development of drugs against a particular function entails better understanding of the complex interplay between moonlighting activities of CysK and its diverse binding partners. Dissociating the interface with the binding partner may be a promising approach. Current ongoing efforts through in silico and in vitro analysis to develop a non-peptidyl inhibitor in nano-molar efficacy against CysK have identified is 3-((Z)-((Z)-5-(4-Fluorobenzylidene)-3-methyl-4-oxothiazolidin-2-ylidene) amino) benzoic acid as the most potent inhibitor so far with IC50 19 nM. This review is an attempt to summarize the growing list of diverse activities and inhibitors of CysK for development of improved and specific therapeutics.
Acknowledgements
Authors are grateful to Indian Council for Medical Research (ICMR), Government of India, for the research grant (BIC/12(16)/2012). We sincerely thank Department of Biotechnology, Jaypee Institute of Information Technology for providing an opportunity to present this work at International Conference on Advances in Biosciences and Biotechnology—2018.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Contributor Information
Pallavi Joshi, Email: joshipallavi5795@gmail.com.
Abhinal Gupta, Email: abhinalgupta34@gmail.com.
Vibha Gupta, Phone: 91-120-2594207, Email: vibha.gupta@jiit.ac.in.
References
- Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci USA. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amori L, Katkevica S, Bruno A, Campanini B, Felici P, Mozzarelli A, Costantino G. Design and synthesis of trans-2-substituted-cyclopropane-1-carboxylic acids as the first non-natural small molecule inhibitors of O-acetylserine sulfhydrylase. Med Chem Commun. 2012;3:1111–1116. [Google Scholar]
- Annunziato G, Pieroni M, Benoni R, Campanini B, Pertinhez TA, Pecchini C, Bruno A, Magalhães J, Bettati S, Franko N, Mozzarelli A, Costantino G. Cyclopropane-1,2-dicarboxylic acids as new tools for the biophysical investigation of O-acetylserine sulfhydrylases by fluorimetric methods and saturation transfer difference (STD) NMR. J Enzyme Inhib Med Chem. 2016;31:78–87. doi: 10.1080/14756366.2016.1218486. [DOI] [PubMed] [Google Scholar]
- Aoki SK, Poole SJ, Hayes CS, Low DA. Toxin on a stick: modular CDI toxin delivery systems play roles in bacterial competition. Virulence. 2011;2:356–359. doi: 10.4161/viru.2.4.16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awano N, Wada M, Mori H, Nakamori S, Takagi H. Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl Environ Microbiol. 2005;71:4149–4152. doi: 10.1128/AEM.71.7.4149-4152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L, et al. Increasing the structure coverage of tuberculosis drug targets. Tuberculosis (Edinb) 2015;95:142–148. doi: 10.1016/j.tube.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoni R, Pertinhez TA, Spyrakis F, Davalli S, Pellegrino S, Peredi G, Pezzotti A, Bettati S, Campanini B, Mozzarelli A. Structural Insights into the Interaction of O-acetylserine sulfhydrylase with competitive, peptidic inhibitors by saturation transfer difference-NMR. FEBS Lett. 2016;590:943–953. doi: 10.1002/1873-3468.12126. [DOI] [PubMed] [Google Scholar]
- Benoni R, Beck CM, Garza-Sanchez F, Bettati S, Mozzarelli A, Hayes CS, Campanini B. Activation of an anti-bacterial toxin by the biosynthetic enzyme CysK: mechanism of binding, interaction specificity and competition with cysteine synthase. Nature. 2017;7:146–157. doi: 10.1038/s41598-017-09022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogicevic B, Berthoud H, Portmann R, Meile L, Irmler S. CysK from Lactobacillus casei encodes a protein with O-acetylserine sulfhydrylase and cysteine desulfurization activity. Appl Microbiol Biotechnol. 2012;94:1209–1220. doi: 10.1007/s00253-011-3677-5. [DOI] [PubMed] [Google Scholar]
- Bonner ER, Cahoon RE, Knapke SM, Jez JM. Molecular basis of cysteine biosynthesis in plants: structural and functional analysis of O-acetylserine sulfhydrylase from Arabidopsis thaliana. J Biol Chem. 2005;280(46):38803–38813. doi: 10.1074/jbc.M505313200. [DOI] [PubMed] [Google Scholar]
- Bruno A, Amori L, Constantino G. Computational insights into the mechanism of inhibition of OASS-A by a small molecule inhibitor. Mol Inform. 2013;32:447–457. doi: 10.1002/minf.201200174. [DOI] [PubMed] [Google Scholar]
- Burkhard P, Jagannatha Rao GS, Hohenester E, Schnackerz KD, Cook PF, Jansonius JN. Three- dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium. J Mol Biol. 1998;283:121–133. doi: 10.1006/jmbi.1998.2037. [DOI] [PubMed] [Google Scholar]
- Burkhard P, Tai CH, Ristroph CM, Cook PF, Jansonius JN. Ligand binding induce a large conformational change in O-acetylserine sulfhydrylase from Salmonella typhimurium. J Mol Biol. 1999;291:941–953. doi: 10.1006/jmbi.1999.3002. [DOI] [PubMed] [Google Scholar]
- Burkhard P, Tai CH, Jansonius JN, Cook PF. Identification of an allosteric anion-binding site on O-acetylserine sulfhydrylase: structure of the enzyme with chloride bound. J Mol Biol. 2000;303:279–286. doi: 10.1006/jmbi.2000.4109. [DOI] [PubMed] [Google Scholar]
- Campanini B, Pieroni M, Raboni S, Bettati S, Benoni R, Pecchini C, Costantino G, Mozzarelli A. Inhibitors of the sulfur assimilation pathway in bacterial pathogens as enhancers of antibiotic therapy. Curr Med Chem. 2015;22:187–213. doi: 10.2174/0929867321666141112122553. [DOI] [PubMed] [Google Scholar]
- Campanini B, Benoni R, Bettati S, Beck CM, Hayes CS, Mozzarelli A. Moonlighting O-acetylserine sulfhydrylase: new functions for an old protein. Biochim Biophys Acta. 2015;1854:1184–1193. doi: 10.1016/j.bbapap.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandey M, Multani AS. Comparative study of efficacy and safety of azithromycin and ofloxacin in uncomplicated typhoid fever: a randomised, open labelled study. J Clin Diagn Res. 2012;6:1736–1739. doi: 10.7860/JCDR/2012/4702.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH) BMC Genom. 2009;10:291. doi: 10.1186/1471-2164-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinthalapudi K, Kumar M, Kumar S, Jain S, Alam N, Gourinath S. Crystal structure of native O-acetylserine sulfhydrylase from Entamoeba histolytica and its complex with cysteine: structural evidence for cysteine binding and lack of interactions with serine acetyl transferase. Proteins. 2008;72:1222–1232. doi: 10.1002/prot.22013. [DOI] [PubMed] [Google Scholar]
- Chiong M, González E, Barra R, Vásquez C. Purification and biochemical characterization of tellurite-reducing activitiesfromThermustherrmophilusHB8. J Bacteriol. 1988;170:3269–3273. doi: 10.1128/jb.170.7.3269-3273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PF, Wedding RT. Cysteine synthetase from Salmonella typhimurium LT-2. Aggregation, kinetic behavior, and effect of modifiers. J Biol Chem. 1978;253:7874–7879. [PubMed] [Google Scholar]
- Crump JA, Kretsinger K, Gay K, Hoekstra RM, Vugia DJ, Hurd S, Segler SD, Megginson M, Luedeman LJ, Shiferaw B, Hanna SS, Joyce KW, Mintz ED, Angulo FJ. Clinical response and outcome of infection with Salmonella enterica serotype Typhi with decreased susceptibility to fluoroquinolones: a United States foodnet multicenter retrospective cohort study. Antimicrob Agents Chemother. 2008;52:1278–1284. doi: 10.1128/AAC.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi S, Abdul Rehman SA, Tarique KF, Gourinath S. Structural characterization and functional analysis of cystathionine β-synthase: an enzyme involved in the reverse transsulfuration pathway of Bacillus anthracis. FEBS J. 2017;284:3862–3880. doi: 10.1111/febs.14273. [DOI] [PubMed] [Google Scholar]
- Dharavath S, Raj I, Gourinath S. Structure-based mutational studies of O-acetylserine sulfhydrylase reveal the reason for the loss of cysteine synthase complex formation in Brucella abortus. Biochem J. 2017;474:1221–1239. doi: 10.1042/BCJ20161062. [DOI] [PubMed] [Google Scholar]
- Diner EJ, Beck CM, Webb JS, Low DA, Hayes CS. Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI) Genes Dev. 2012;26:515–525. doi: 10.1101/gad.182345.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droux M, Ruffet ML, Douce R, Job D. Interactions between Serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants—structural and kinetic properties of the free and bound enzymes. Eur J Biochem. 1998;255:235–245. doi: 10.1046/j.1432-1327.1998.2550235.x. [DOI] [PubMed] [Google Scholar]
- Dwivedi CM, Ragin RC, Uren JR. Cloning, purification, and characterization of beta-cystathionase from Escherichia coli. Biochem. 1982;21:3064–3069. doi: 10.1021/bi00256a005. [DOI] [PubMed] [Google Scholar]
- Feldman-Salit A, Wirtz M, Lenherr ED, Throm C, Hothorn M, Scheffzek K, Hell R, Wade RC. Allosterically gated enzyme dynamics in the cysteine synthase complex regulate cysteine biosynthesis in arabidopsis thaliana. Structure. 2012;20(2):292–302. doi: 10.1016/j.str.2011.11.019. [DOI] [PubMed] [Google Scholar]
- Francois JA, Kumaran S, Jez JM. Structural basis for interaction of O-acetylserine sulfhydrylase and serine acetyltransferase in the arabidopsis cysteine synthase complex. Plant Cell. 2006;18(12):3647–3655. doi: 10.1105/tpc.106.047316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franko N, Grammatoglou K, Campanini B, Costantino G, Jirgensons A, Mozzarelli A. Inhibition of O-acetylserine sulfhydrylase by fluoroalanine derivatives. J Enzyme Inhib Med Chem. 2018;33:1343–1351. doi: 10.1080/14756366.2018.1504040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frávega J, Álvarez R, Dìaz F, Inostroza O, Tejìas C, Rodas PI, Paredes-Sabja D, Fuentes JA, Calderòn IL, Gil F. Salmonella Typhimurium exhibits fluoroquinolone resistance mediated by the accumulation of the antioxidant moleculeH2S in a CysK-dependent manner. J Antimicrob Chemother. 2016;71:3409–3415. doi: 10.1093/jac/dkw311. [DOI] [PubMed] [Google Scholar]
- Fu KP, Hilliard J, Isaacson D, et al. In-vivo evaluation of ofloxacin in Salmonella typhimurium infection in mice. J Antimicrob Chemother. 1990;25:263–268. doi: 10.1093/jac/25.2.263. [DOI] [PubMed] [Google Scholar]
- Gallagher DT, Gilliland GL, Xiao G, Zondlo J, Fisher KE, Chinchilla D, Eisenstein E. Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase. Structure (Lond) 1998;6:465–475. doi: 10.1016/s0969-2126(98)00048-3. [DOI] [PubMed] [Google Scholar]
- Garberg P, Engman L, Tolmachev V, Lundqvist H, Gerdes R, Cotgreave I. Binding of tellurium to hepatocellular selenoproteins during incubation with inorganic tellurite: consequences for the activity of selenium-dependent glutathione peroxidase. Int J Biochem Cell Biol. 1999;31:291–301. doi: 10.1016/s1357-2725(98)00113-7. [DOI] [PubMed] [Google Scholar]
- Gerdes SY, Scholle MD, Campbell JW, Balázsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D’Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabási AL, Oltvai ZN, Osterman AL. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin NV, Phillips MA, Goldsmith EJ. Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci. 1995;6:465–475. doi: 10.1002/pro.5560040705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guédon E, Martin-Verstraete I. Cysteine Metabolism and Its Regulation in Bacteria. Amino Acid Biosynth Pathw Regulat Metab Eng. 2006;5:195–218. [Google Scholar]
- Heine A, et al. Crystal Structure of O-Acetylserine sulfhydrylase (TM0665) from Thermotoga maritima at 1.8 Å Resolution. Proteins. 2004;56:387–391. doi: 10.1002/prot.20003. [DOI] [PubMed] [Google Scholar]
- Huang B, Vetting MW, Roderick SL. The active site of O-acetylserine sulfhydrylase is the anchor point for bienzyme complex formation with serine acetyltransferase. J Bacteriol. 2005;187:3201–3205. doi: 10.1128/JB.187.9.3201-3205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulanicka MD, Hallquist SG, Kredich NM, Mojica-A T. Regulation of O-acetylserine sulfhydrylase B by l-cysteine in Salmonella typhimurium. J Bacteriol. 1979;140:141–146. doi: 10.1128/jb.140.1.141-146.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullo MF, Auger S, Soutourina O, Barzu O, Yvon M, Danchin A, Martin-Verstraete I. Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J Bacteriol. 2007;189:187–197. doi: 10.1128/JB.01273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean kumar VU, Poyraz O, Saxena S, Schnell R, Yogeeswari P, Schneider G, Sriram D. Discovery of novel inhibitors targeting the Mycobacterium tuberculosis O-acetylserine sulfhydrylase (CysK1) using virtual high-throughput screening. Bioorg Med Chem Lett. 2013;23:1182–1186. doi: 10.1016/j.bmcl.2013.01.031. [DOI] [PubMed] [Google Scholar]
- Johnson PM, Beck CM, Morse RP, Garza-Sanchez F, Low DA, Hayes CS, Goulding CW. Unraveling essential role of CysK in CDI toxin activation. Proc Natl Acad Sci USA. 2016;113:9792–9797. doi: 10.1073/pnas.1607112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Amarnath SK. Fluoroquinolone resistance in Salmonella typhi and S. paratyphi A in Bangalore, India. Trans R Soc Trop Med Hyg. 2007;101:308–310. doi: 10.1016/j.trstmh.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Kant V, Vijayakumar S, Sahoo GC, Chaudhery SS, Das P. In-silico screening and validation of high-affinity tetra-peptide inhibitor of Leishmania donovani O-acetyl serine sulfhydrylase (OASS) J Biomol Struct Dyn. 2018;36:1–15. doi: 10.1080/07391102.2018.1429315. [DOI] [PubMed] [Google Scholar]
- Kaundal S, Uttam M, Thakur KG. Dual Role of a biosynthetic enzyme, CysK, in contact dependent growth inhibition in bacteria. Plos One. 2016;10:1–18. doi: 10.1371/journal.pone.0159844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik A, Ekka MK, Kumaran S. Two distinct assembly states of cysteine regulatory complex of salmonella typhimurium are regulated by enzyme-substrate cognate pairs. Biochem. 2017;56:1–43. doi: 10.1021/acs.biochem.6b01204. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredich NM. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol Microbiol. 1992;6:2747–2753. doi: 10.1111/j.1365-2958.1992.tb01453.x. [DOI] [PubMed] [Google Scholar]
- Kredich NM, Tomkins GM. The enzymic synthesis of l-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem. 1966;241:4955–4965. [PubMed] [Google Scholar]
- Kredich NM, Becker MA, Tomkins GM. Purification and characterization of cysteine synthetase, a bifunctional protein complex, from Salmonella typhimurium. J Biol Chem. 1969;244:2428–2439. [PubMed] [Google Scholar]
- Kumar S, Raj I, Nagpal I, Subbarao N, Gourinath S. Structural and biochemical studies of serine acetyltransferase reveal why the parasite Entamoeba histolytica cannot form a cysteine synthase complex. J Biol Chem. 2011;286:12533–12541. doi: 10.1074/jbc.M110.197376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Xu BY, Zhou K, Cheng W, Jiang YL, Chen Y, Zhou CZ. Structural and biochemical Analysis of Microcystis aeruginosa O-acetylserine sulfhydrylases reveal a negative feedback regulation of cysteine biosynthesis. Biochim Biophys Acta. 2014;1844:308–315. doi: 10.1016/j.bbapap.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Magalhães J, Franko N, Annunziato G, Welch M, Dolan SK, Bruno A, Mozzarelli A, Armao S, Jirgensons A, Pieroni M, Costantino G, Campanini B. Discovery of novel fragments inhibiting O-acetylserine sulphhydrylase by combining scaffold hopping and ligand-based drug design. J Enzyme Inhib Med Chem. 2018;33:1444–1452. doi: 10.1080/14756366.2018.1512596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba Y, Yoshida T, Izuhara-Kihara H, Noda M, Sugiyama M. Crystallographic and mutational analyses of cystathionine b-synthase in the H2S synthetic gene cluster in Lactobacillus plantarum. Protein Sci. 2017;26:763–783. doi: 10.1002/pro.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder M, Gourinath S. Structure-based design of inhibitors of the crucial cysteine biosynthetic pathway enzyme O-acetyl serine sulfhydrylase. Curr Top Med Chem. 2016;16:948–959. doi: 10.2174/1568026615666150825142422. [DOI] [PubMed] [Google Scholar]
- Mino K, Ishikawa K. Characterization of a novel thermostable O-acetylserine sulfhydrylase from Aeropyrum pernix K1. J Bacteriol. 2003;185:2277–2284. doi: 10.1128/JB.185.7.2277-2284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino K, Hiraoka K, Imamura K, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K. Characteristics of serine acetyltransferase from Escherichia coli deleting different lengths of amino acid residues from the C-terminus. Biosci Biotechnol Biochem. 2000;64:1874–1880. doi: 10.1271/bbb.64.1874. [DOI] [PubMed] [Google Scholar]
- Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishin K. Effects of bienzyme complex formation of cysteine synthetase from Escherichia coli on some properties and kinetics. J Stage. 2000;64:1628–1640. doi: 10.1271/bbb.64.1628. [DOI] [PubMed] [Google Scholar]
- Mori M, Jeelani G, Masuda Y, Sakai K, Tsukui K, Waluyo D, Tarwadi WY, Nonaka K, Matsumoto A, Ōmura S, Nozaki T, Shiomi K. Identification of natural inhibitors of Entamoeba histolytica cysteine synthase from microbial secondary metabolites. Front Microbiol. 2015;6:962. doi: 10.3389/fmicb.2015.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Tsuge S, Fukasawa W, Jeelani G, Nakada-Tsukui K, Nonaka K, Matsumoto A, Ōmura S, Nozaki T, Shiomi K. Discovery of antiamebic compounds that inhibit cysteine synthase from the enteric parasitic protist Entamoeba histolytica by screening of microbial secondary metabolites. Front Cell Infect Microbiol. 2018;8:409. doi: 10.3389/fcimb.2018.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso H, Saavedra C, Loyola C, Pichuantes S, Vásquez C. Biochemical characterization of tellurite reducing activities from Bacillus stearothermophilus V. Res Microbiol. 1998;49:389–397. doi: 10.1016/s0923-2508(98)80321-5. [DOI] [PubMed] [Google Scholar]
- Murillo LA, Newport G, Lan CY, Habelitz S, Dungan J, Agabian NM. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot Cell. 2005;4:1562–1573. doi: 10.1128/EC.4.9.1562-1573.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal I, Raj I, Subbarao N, Gourinath S. Virtual screening, identification and in vitro testing of novel inhibitors of O-Acetyl-l-Serine sulfhydrylase of Entamoeba histolytica. PLoS One. 2012;7:e30305. doi: 10.1371/journal.pone.0030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterium association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci USA. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieroni M, Annunziato G, Beato C, Wouters R, Benoni R, Campanini B, Pertinhez TA, Bettati S, Mozzarelli A, Costantino G. Rational design, synthesis, and preliminary structure-activity relationships of α–substituted-2-phenylcyclopropane carboxylic acids as inhibitors of Salmonella typhimuriumO–acetylserine sulfhydrylase. J Med Chem. 2016;59:2567–2578. doi: 10.1021/acs.jmedchem.5b01775. [DOI] [PubMed] [Google Scholar]
- Poyraz O, Jeankumar VU, Saxena S, Schnell R, Haraldsson M, Yogeeswari P, Sriram D, Schneider G. Structure-guided design of novel thiazolidine inhibitors of O-acetylserine sulfhydrylase from Mycobacterium tuberculosis. J Med Chem. 2013;56:6457–6466. doi: 10.1021/jm400710k. [DOI] [PubMed] [Google Scholar]
- Raj I, Kumar S, Gourinath S. The narrow active-site cleft of O-acetylserine Sulfhydrylase from Leishmania donovani allows complex formation with serine acetyltransferases with a range of C-termini sequences. Acta Crystallogr. 2012;68:909–919. doi: 10.1107/S0907444912016459. [DOI] [PubMed] [Google Scholar]
- Raj I, Mazumder M, Gourinath S. Molecular basis of ligand recognition by OASS from E. histolytica: insights from structural and molecular dynamics simulation studies. Biochim Biophys Acta. 2013;1830:4573–4583. doi: 10.1016/j.bbagen.2013.05.041. [DOI] [PubMed] [Google Scholar]
- Ramìrez A, Castañeda M, Xiqui ML, Sosa A, Baca BE. Identification, cloning and characterization of cysK, the gene encoding O-acetylserine (thiol)-lyase from Azospirillum brasilense, which is involved in tellurite resistance. FEMS Microbiol Lett. 2006;261:272–279. doi: 10.1111/j.1574-6968.2006.00369.x. [DOI] [PubMed] [Google Scholar]
- Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci USA. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsi E, Bayden AS, Spyrakis F, Amadasi A, Campanini B, Bettati S, Dodatko T, Cozzini P, Kellogg GE, Cook PF, Roderick SL, Mozzarelli A. Design of O-acetylserine sulfhydrylase by mimicking nature. J Med Chem. 2010;53:345–356. doi: 10.1021/jm901325e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Schneider G, Käck H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000;8:R1–R6. doi: 10.1016/s0969-2126(00)00085-x. [DOI] [PubMed] [Google Scholar]
- Schnell R, Oehlmann W, Singh M, Schneider G. Structural insights into catalysis and inhibition of O-acetylserine sulfhydrylase from Mycobacterium tuberculosis. J Biol Chem. 2007;282:23473–23481. doi: 10.1074/jbc.M703518200. [DOI] [PubMed] [Google Scholar]
- Schnell R, Sriram D, Schneider G. Pyridoxal-dependent mycobacterium cysteine synthases: structure, mechanism and potential as a drug targets. Biochim Biophys Acta. 2015;1854:1175–1183. doi: 10.1016/j.bbapap.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Seiflein TA, Lawrence JG. Methionine-to-cysteine recycling in Klebsiella aerogenes. J Bacteriol. 2001;183:336–346. doi: 10.1128/JB.183.1.336-346.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekowska A, Danchin A. Identification of yrrU as the methylthioadenosine nucleosidase gene in Bacillus subtilis. DNA Res. 1999;6:255–264. doi: 10.1093/dnares/6.5.255. [DOI] [PubMed] [Google Scholar]
- Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;18:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- Singh P, Brooks IIJF, Ray VA, Mandel MJ, Visick KL. CysK plays a role in biofilm formation and colonization by Vibrio fischeri. Appl Environ Microbiol. 2015;81:5223–5234. doi: 10.1128/AEM.00157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Ekka MK, Kaushik A, Pandya V, Singh RP, Banerjee S, Mittal M, Singh V, Kumaran S. Substrate-induced facilitated dissociation of the competitive inhibitor from the active site of O-acetyl serine sulfhydrylase reveals a competitive-allostery mechanism. Biochemistry. 2017;56:5011–5025. doi: 10.1021/acs.biochem.7b00500. [DOI] [PubMed] [Google Scholar]
- Spyrakis F, Singh R, Cozzini P, Campanini B, Salsi E, Felici P, Raboni S, Benedetti P, Cruciani G, Kellogg GE, Cook PF, Mozzarelli A. Isozyme specific ligands for O-acetylserine sulfhydrylase, a novel antibiotic target. Plos One. 2013;8:1–13. doi: 10.1371/journal.pone.0077558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner EM, Böth D, et al. CysK2 from mycobacterium tuberculosis is an ophospho-l-serine-dependent S-sulfocysteine synthase. J bacteriol. 2014;196:3410–3420. doi: 10.1128/JB.01851-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Hashimoto W, Kumagai H. Escherichia coli K-12 can utilize an exogenous gamma-glutamyl peptide as an amino acid source, for which gammaglutamyltranspeptidase is essential. J Bacteriol. 1993;175:6038–6040. doi: 10.1128/jb.175.18.6038-6040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kamatani S, Kim ES, Kumagai H. Aminopeptidases A, B, and N and dipeptidase D are the four cysteinylglycinases of Escherichia coli K-12. J Bacteriol. 2001;183:1489–1490. doi: 10.1128/JB.183.4.1489-1490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanous C, Soutourina O, Raynal B, Francoise M, Mervelet P, Gilles AM, Noirot P, Danchin A, England P, Verstraete IM. The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis. J Biol Chem. 2008;283:35551–35562. doi: 10.1074/jbc.M805951200. [DOI] [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull AL, Surette MG. l-Cysteine is required for induced antibiotic resistance in actively swarming Salmonella enterica serovar Typhimurium. Microbiology. 2008;154:3410–3419. doi: 10.1099/mic.0.2008/020347-0. [DOI] [PubMed] [Google Scholar]
- Turnbull AL, Surette MG. Cysteine biosynthesis, oxidative stress and antibiotic resistance in Salmonella typhimurium. Res Microbiol. 2010;161:643–650. doi: 10.1016/j.resmic.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Vásquez CC, Saavedra CP, Loyola CA, Araya MA, Pichuantes S. The product of the cysK Gene of Bacillus stearothermophilus V mediates potassium tellurite resistance in Escherichia coli. Curr Microbiol. 2001;43:418–423. doi: 10.1007/s002840010331. [DOI] [PubMed] [Google Scholar]
- Verma D, Gupta S, Kaur KJ, Gupta V. Is perturbation in the quaternary structure of bacterial CysE, another regulatory mechanism for cysteine synthesis? Int J Biol Macromol. 2018;111:1010–1018. doi: 10.1016/j.ijbiomac.2018.01.076. [DOI] [PubMed] [Google Scholar]
- Vermeij P, Kertesz MA. Pathways of assimilative sulfur metabolism in Pseudomonas putida. J Bacteriol. 1999;181:5833–5837. doi: 10.1128/jb.181.18.5833-5837.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler PR, Coldham NG, Keating L, Gordon SV, Wooff EE, Parish T, Hewinson RG. Functional demonstration of reverse transsulfuration in the Mycobacterium tuberculosis complex reveals that methionine is the preferred sulfur source for pathogenic mycobacteria. J Biol Chem. 2005;280:8069–8078. doi: 10.1074/jbc.M412540200. [DOI] [PubMed] [Google Scholar]
- Wirtz M, Berkowitz O, Droux M, Hell R. The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein–protein interaction. Eur J Biochem. 2001;268:686–693. doi: 10.1046/j.1432-1327.2001.01920.x. [DOI] [PubMed] [Google Scholar]
- Wirtz M, Droux M, Hell R. O-acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot. 2004;55:1785–1798. doi: 10.1093/jxb/erh201. [DOI] [PubMed] [Google Scholar]
- Yadava U, Shukla BK, Roychoudhury M, Kumar D. Pyrazolo[3,4-d]pyrimidines as novel inhibitors of O-acetyl-l-serine sulfhydrylase of Entamoeba histolytica: an in silico study. J Mol Model. 2015;21:96. doi: 10.1007/s00894-015-2631-3. [DOI] [PubMed] [Google Scholar]
- Yamagata S. O-Acetylserine and O-acetylhomoserine sulfhydrylase of yeast. Subunit structure. J Biochem. 1976;80:787–797. doi: 10.1093/oxfordjournals.jbchem.a131339. [DOI] [PubMed] [Google Scholar]
- Zhao C, Moriga Y, Feng B, Kumada Y, Imanaka H, Imamura K, Nakanishi K. On the interaction site of serine acetyltransferase in the cysteine synthase complex from Escherichia coli. Biochem Biophys Res Commun. 2006;341:911–916. doi: 10.1016/j.bbrc.2006.01.054. [DOI] [PubMed] [Google Scholar]