Abstract
Heavy metal lead, which enters the human body through food intake, endangers human health. Microbe has the ability of adsorbing heavy metal, among which lactic acid bacteria are promising microbes to adsorb and remove Pb2+. The purpose of this study was to screen lactic acid bacteria from Ya’an pickle water to effectively remove Pb2+. The 7 strains having strong ability to effectively remove Pb2+ were detected. These strains were identified by microscopic examination and 16S rDNA sequencing, 4 strains of Lactobacillus plantarum and 3 strains of Lactobacillus brevis were obtained. Then the bacteria had a blind adsorption effect on Pb2+. After microwave digestion, the Pb2+ concentration was measured by flame atomic absorption spectrometry. The highest removal reached 82.25%. The adsorption mechanism of lactic acid bacteria was mainly divided into biosorption and bioaccumulation. The 7 strains of lactic acid bacteria could provide potential for detoxification of contaminated foods and reduction of the Pb2+ accumulation in the human diet and animal feed. At the same time, this study was helpful to further understand the mechanism of Pb2+ being adsorbed by lactic acid bacteria.
Electronic supplementary material
The online version of this article (10.1186/s13568-018-0724-y) contains supplementary material, which is available to authorized users.
Keywords: Pickle water, Lactic acid bacteria, Pb2+, Adsorption
Introduction
Currently, pesticide residue, veterinary drugs residue and heavy metal pollution are blamed for the food safety incident. The pesticide and veterinary drugs residues in food may be reduced by pretreatments, such as heat blanching, brine rinsing and peeling. But heavy metal pollution is difficult to handle. Once the food is contaminated by heavy metal, it means it can only be destroyed because of the nonbiodegradability and accumulation of heavy metals (Ekere et al. 2016). Lead, as an important industrial raw material, has been widely applied to modern industry including metallurgy, printing, military, medicine, electronics, ceramics, pigment and transportation industry (Özcan et al. 2009), but it is also toxic heavy metal. The lead ion (Pb2+) is one of the sources of environmental pollution. It has various sources and high cumulative toxicity, which can cause serious harm to human nervous system, hematopoietic function of bone marrow and so on, and environmental problems of ecosystems such as air pollution, brine pollution and soil pollution (Cui et al. 2017). Consequently, removing Pb2+ has aroused more people’s attention.
The traditional approaches for removing Pb2+ are precipitation, coagulation, adsorption and ion exchange. The ion exchange was applied for the selective removal of Pb2+ from waste brine samples. The polyacrylamide zirconium (IV) hybrid cation exchanger has a high selectivity to Pb2+ in comparison to other metal ions (Rahman et al. 2013). With the development of the technology, the use of ceramsite (Wang et al. 2018), ferric oxide (Saha et al. 2014) and other substances to adsorb heavy metals has also appeared. Ferric oxide has been widely used in the treatment of Pb2+ in waste brine (Zhang and Li 2016), but it was not suitable to use the iron oxide to fill in the pipe column directly, which was poor permeability and low efficiency. In general, most of these approaches required special equipment and rigorous experimental condition, which greatly limited their practically applicable value (Kobya et al. 2005). Recently, biosorption, a new method to eliminate heavy metal pollution, has been researched as a popular topic because of the advantages of low investment, high efficiency and safety with no side effects (Tural et al. 2017). Therefore, biosorption was important to remove Pb2+ from the aqueous solution selectively and efficiently.
Many reports commonly showed the biosorption of heavy metals by bacteria and fungi. Iskandar et al. reported the biosorption by filamentous fungi isolated from a fresh brine ecosystem (Iskandar et al. 2012), and there were some reports showed that pretreating Aspergillus niger would significantly improve the biosorption of Pb2+ (Kapoor and Viraraghavan 1998). While, some bacteria used for biosorption may be pathogenicity, which was easy to cause secondary pollution. More reports showed that lactic acid bacteria (LAB) strains were probiotic bacteria which were acknowledged as the safety level microorganisms which were widely used to produce fermented food (Teusink and Molenaar 2017; Jahromi et al. 2017), and were also applied into heavy metal biosorption. Schut et al. found that copper could be removed by biosorption of wine-relevant lactobacilli (Schut et al. 2011). Besides, some LAB strains were evaluated and selected through biosorption of cadmium, arsenic and mercury by Kinoshita et al. (Kinoshita et al. 2013). However, researches on the adsorption of LAB are still relatively rare.
In our previous studies, we have reviewed that the progress in mechanism and influence of biosorption between LAB and Pb2+ (Lin et al. 2017). Due to the probiotic role of the LAB, the security of adsorption with LAB would be higher than other methods of removing Pb2+. Considering the fact that LAB were extensive in pickle brine (Zafar et al. 2018), high-quality pickle brine was selected from Ya’an, Sichuan (Xia et al. 2017), and expected to screen and identify LAB with strong adsorption capacity for Pb2+, and provided a microbiological method for further solving contamination in food.
Materials and methods
Materials and reagents
Pb2+ solutions were prepared from PbNO3 (Hangfeng Chemical Co., Inc., Chengdu, China) for using in different experiments. 0.85% physiological saline, 6% nitric acid (HNO3), hydrogen peroxide (H2O2), 25% potassium iodide solution (KI), ascorbic acid solution (AA), and methyl isobutyl ketone (MIBK) were purchased from Sigma–Aldrich (Shanghai, China). All solutions were prepared using analytical-grade reagents and water was distilled and deionized with a Milli-Q® system (Millipore, Billerica, MA, USA).
Sampling
Three samples of locally ripened Ya’an pickle brine were collected in July of 2015 from three different local farmers in Ya’an, Sichuan, China (every farmer provided 1 sample with 10 mL brine). The containers used for storing pickle brine were shaken prior to sampling. The samples were collected at ambient temperatures and kept on ice during the 3 h transport to our laboratory. The pickle brine from each sample was aseptically collected for further analysis.
Isolation and screening of resistant bacteria
3 samples were exerted appropriate tenfold serial dilutions (10−1 to 10−4) with sterile PS, respectively. Every dilution of 50 μL was inoculated in MRS agar plate by spread plate method. After the incubation for 24 h at 37 °C, by the flat colony counting method, the plates colony number of 30–300 were selected out as the mother plates (TMP). Colonies were randomly selected from TMP. After purification and isolation, some suspected LAB colonies were inoculated respectively on the selective MRS plates (TSP) containing 2 mL 200 g/L Pb2+ by the plate streak. After then, TSP were incubated for 24 h at 37 °C. Some colonies of the TSP were selected out for acclimatization of bacteria.
They were inoculated in the De Man, Rogosa, and Sharpe (MRS) agar plate (specific for LAB) with 2 mL solution containing Pb2+ respectively (500 mg/L, 1000 mg/L, 1500 mg/L, 2000 mg/L) (Pb2+ medium) and to be isolated the strains which were capable of tolerating high concentration of Pb2+. After culture for 48 h at 37 °C,the dominant Pb2+-resistant strains were isolated from Pb2+ medium, and preserved with 30% glycerin at − 20 °C. These strains would be marked for subsequent identification.
Identification of Pb2+ resistant bacteria
The morphological characteristics of the isolated strains of LAB were observed and recorded before and after Gram staining (Michael 1983).
The genotypic characterization of strains was performed by sequencing the gene 16S rDNA (Sontakke et al. 2009). Omega’s DNA extraction kit was used for total genomic DNA extraction, when strains grew to their late log phase. Fragments of bacterial 16S rDNA were amplified by polymerase chain reaction (PCR) using the primers 27F (5′-GCCTGTGCGGGGTGCTATAC-3′) and 1492R (5′-CGCCGTTGGCGGCGTGCTA-3′) (Bioengineering Co., Inc., Shanghai, China) with the thermocycler, whose accession number is MH681598, MH681599, MH681600, MH681601, MH681602, MH681603 and MH681604 respectively. Lactobacillus plantarum (L. plantarum) (MH681599) GDMCC 11516 is a L. plantarum strain that has been deposited in the culture collection ‘Guangdong Microbial Culture Center’, acronym GDMCC, the strain’s registration number is GDMCC 11516. PCR amplification products were sequenced by Sanger chain termination method and spliced by Contig Express splicing program.
The reaction parameters of PCR included five min of denaturation at 95 °C, followed by 35 cycles of 95 °C, for 30 s, 58 °C, for 30 s, 72 °C, for 90 s, and a final extension at 72 °C, for 7 min. After the reaction, the PCR products were subjected to 1% agarose gel electrophoresis to confirm the PCR amplification fragments. The PCR products were recovered by using the Omega’s DNA extraction kit (Omega Bio-tek, Co., Inc. USA). The purified PCR products were sent to Shanghai Bioengineering Limited by Share Ltd to sequence. Finally, the spliced sequence file was compared with the data in the NCBI ribosomal DNA sequence (Bacteria and Archaea) database by using the NCBI Blast program, and the strain information with the largest similarity to the sequence of the tested bacteria was obtained (Emel et al. 2017). The phylogenetic tree was analyzed and constructed with the neighbor-joining method (Gitzendanner et al. 2018).
Bacterial adsorption assay
Those resistant strains were activated 2 times in 5 mL MRS broth at 37 °C for 24 h. Then amplification culture was in 10 mL MRS broth at 37 °C for 24 h. The cultured stains were centrifuged (4000 rpm, 4 °C, 15 min) and then washed with sterile PS for 2 times to get the wet bacteria.
Adsorption experiments were carried out on the basis of 3 g/L (the weight of wet bacteria/the volume of 200 mg/L Pb2+ solution). All the samples were exerted shake cultivation for 1 h at 37 °C, pH close to neutral. After incubation, the suspension of samples was centrifuged (4000 rpm, 4 °C, 15 min). A sample was taken from the supernatant to be digested.
All glass instruments and polytetrafluoroethylene tank (digestion tank) were soaked with 6% HNO3 for 24 h at room temperature, and then washed by ultrapure water for 2–3 times, then dried. The solvent was added to every sample and two reagent blanks, meanwhile every sample need a parallel experiment. All the digestion experiments were set at 600 watts (W). Microwave digestion instrument (MDI, CEM Co., Inc., USA) was started in strict accordance with the operating instructions (Acar et al. 2016).
According to microwave digestion program (Additional file 1: Table S1) and the setting of solvent proportion (Additional file 1: Table S2), the sample was digested and the optimum proportion of solvent was determined. After the temperature of digestion tanks reducing to room temperature, tanks were slowly unscrewed to drive away the acid, then all the solution was transferred into the volumetric flask (50 mL). All the volumetric flasks were titrated to 50 mL with deionized water, and then homogenized for being measured.
Determination by Flame Atomic Absorption Spectrometry (FAAS)
A fast sequential FAAS (Jena Analytical Instruments Co., Ltd, Germany) was employed to carry out all measurements. Pb2+ hollow cathode lamps were used as radiation sources. An air/acetylene flame was used for Pb2+ element determinations. The FAAS operating conditions were 0.4 nm Slit width, 2.0 mA, Applied current, 1.0 nm Spectral resolution, 1700 L min−1 Acetylene flow rate.
Pb2+ standard solution samples of 0 mg/L, 0.05 mg/L, 0.1 mg/L, 0.2 mg/L, 0.4 mg/L, 0.8 mg/L (the standard series) was prepared to draw the Pb2+ standard curve, after determination by FAAS.
KI, AA and MIBK were used as extractants. 1.5 mL KI, 1 mL AA and 5 mL MIBK in sequence was added to the sample digestion solution, the reagent blank and the standard series. Each time an extractant was added, the sample was oscillated for 2 min.
After the extraction of 10 min, the sample was 200 times diluted, and then the organic phase was introduced into the FAAS. Determination of absorbance at 283.3 nm wavelengths, a standard curve or calculate a linear regression equation was drew with the standard series of absorbance. The absorbance of the sample was brought into the equation to obtain the residual Pb2+ concentration.
The adsorption capacity of bacteria
The adsorption capacity of the strains towards Pb2+ was expressed as the removed rate and bacterial adsorption capacity (Baig et al. 2010).
Co is the initial Pb2+ concentration, Ce is the residual Pb2+ concentration after removal.
Qe: Bacterial adsorption capacity, Co represents the initial Pb2+ concentration, Ce represents the equilibrium solution of Pb2+ concentration. V: the volume of Pb2+ solution, m: weight of wet cell, m/V = 3 g/L.
Statistical analysis
All the experiments were carried out in triplicate, and the results are provided as the mean ± SD (standard deviation) values. One-way analysis of variance (ANOVA) was performed, and the significance of each mean property value was determined (p < 0.05) with Duncan’s multiple range test with SPSS software (SPSS Inc., IL, USA).
Result
Colony morphology
As could be seen from Fig. 1, most of the bacteria were stalked, single or chain. However, the cell size of bacteria shrank obviously and the morphology atrophied with the rod structure absent, which could be seen in Fig. 1b. The Pb2+ on TSP inhibited the accumulation of bacterial components (Goswami et al. 2017) or changed the synthesis of cell walls (Xu et al. 2016), leading to the change in the morphology of the bacteria (Li et al. 2017).
Fig. 1.
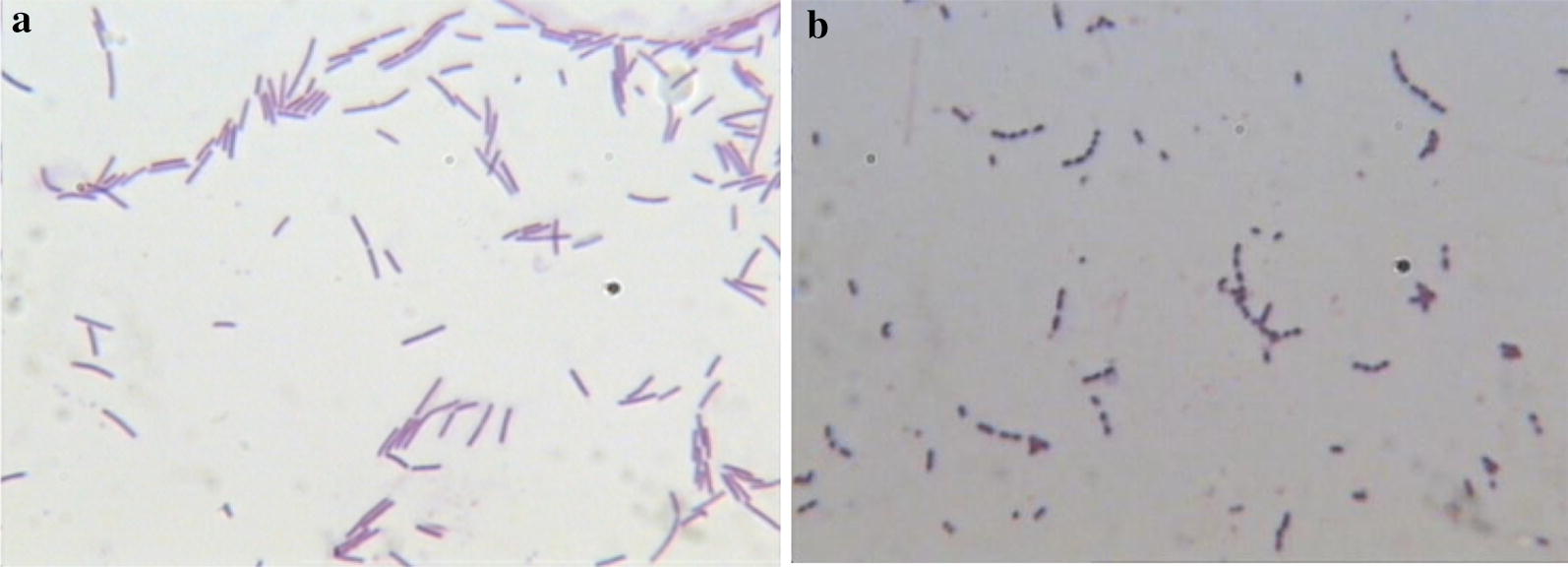
Gram staining and microscopy of colonies. a Represents TMP and b represents TSP
Morphological characteristics were compared between TMP and TSP (Table 1). The colony on TMP was generally larger than TSP’s. The colony morphologies of TMP and TSP both were white, smooth surface, tidy edge and round colony with different size. The colony on TMP grew well, showing a normal growth state of bacteria, while the colonies on TSP became very small and transparent, which showed that Pb2+ has a great effect on the growth of bacteria. The colony on TMP was generally larger than TSP’s. Combined with Fig. 1, the mutation of bacteria was further determined.
Table 1.
Colony morphology comparison of TMP and TSP
| TMP | TSP | |
|---|---|---|
| Image |

|
|
| Size | Medium-sized, 1–5 mm diameter | Small, compact arrangement |
| Form | Round, regular edges | Round, regular edges |
| Protruding | Slightly bulging | Raised |
| Surface state | Smooth | Smooth |
| Surface gloss | Glossy | Little glossy |
| Texture | Wet and sticky, easy to pick | Sticky, difficult to pick |
| Color and transparency | Thick and white | White and transparent |
According to Bergey’ s manual of systematic bacteriology (Peladan and Monteil 1984), some suspected LAB strains were inoculated on TSP for screening Pb2+ resistant strains. In the end, 32 strains were selected from TSP and marked from LAB-01 to LAB-32.
Acclimatization of bacteria
All the 32 strains could grow on the Pb2+ medium, but with the increase of Pb2+ concentration, the number of colonies with morphological variation increased (Table 2). At the same time, the culture time of the strains in MRS ager medium was about 32 h, but when Pb2+ was added into the medium, the strains had a prolonged adaptation period and a significant decrease number of colonies. Pb2+ could decrease the growth rate of bacteria by regulating or inhibiting some enzymatic reactions or by binding with DNA, membranes and cell walls (Kurniawan et al. 2018). In present experiment, the stable growth time of the colony was 48 h.
Table 2.
Colony morphology of Pb2+ medium
| 500 mg/L | 1000 mg/L | 1500 mg/L | 2000 mg/L |
|---|---|---|---|
|

|
|

|
| Well growth | Slow growth | Slow growth | Slow growth and some variation |
Finally, 7 strains of Pb2+-resistance strains (LAB-05, LAB-08, LAB-13 LAB-32 LAB-10, LAB-23, LAB-27) were preserved for identification.
Identification of strains
According to the contrast of PCR products detected by 1% agarose gel electrophoresis between Marker and 7 strains (Fig. 2), specific bands of 7 strains were found at 1500 bp–2000 bp, which conformed to expectation, indicating that the PCR amplification was successful.
Fig. 2.
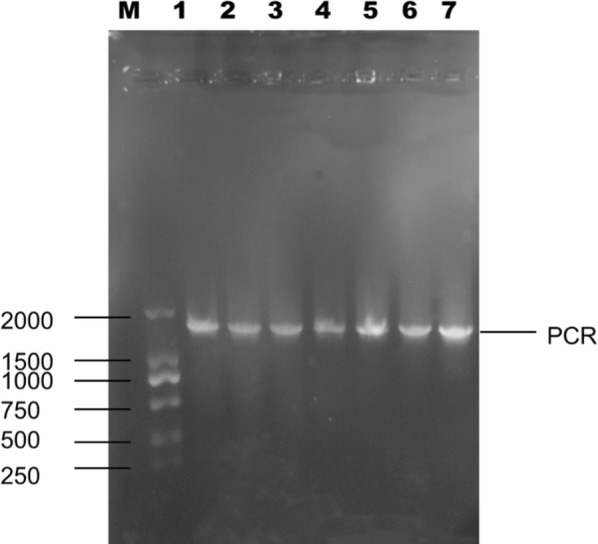
Electrophoresis of PCR-amplified 16 S rDNA gene fragments from 7 strains (left to right: Marker, 05, 08, 13, 23, 27, 10, 32)
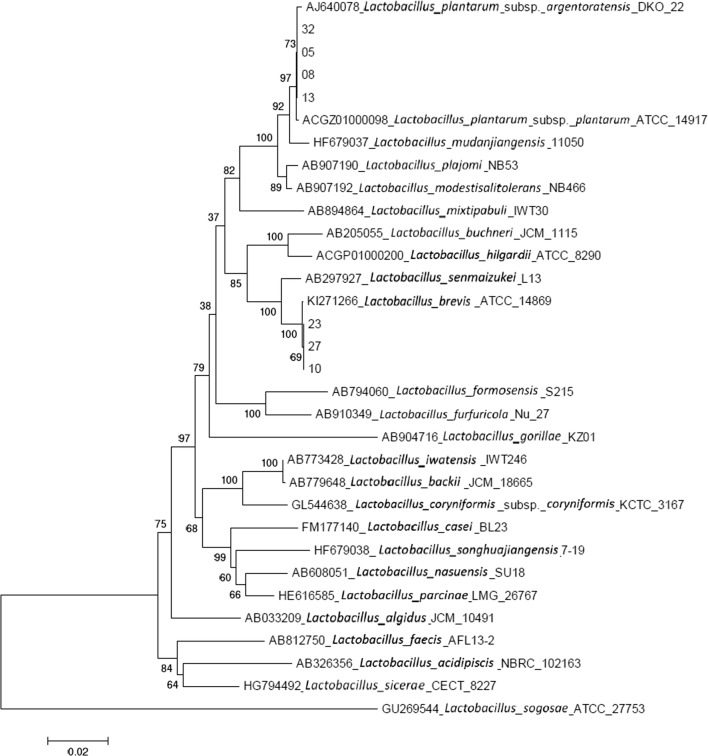
After sequencing, the 16S-rDNA gene sequence of each strain was carried out BLAST analysis in GenBank. To show the relationship between strains, the phylogenetic tree (Fig. 3) was constructed by using MEGA 5.1 software based on 16 s rDNA gene sequence extracted from those LAB and GenBank database (Alexander et al. 2014).
Fig. 3.
Phylogenetic tree based on 16 S-rDNA sequences of 7 strains
Computational integration of genomic traits into 16S rDNA microbiota sequencing was studied. The homologies of the 7 strains and the reference bacteria were more than 97%. When sequence homologies of 16S rDNA were over 97%, it could be considered to belong to the same genus (Auch et al. 2010). Thereafter, 05, 08, 13 and 32 belong to a same genus, 10, 23 and 27 belong to a same genus. As Fig. 3 shown, 05, 08, 13 and 32 were closely related to L. plantarum, and 10, 23 and 27 were closely related to Lactobacillus (L. brevis). The similarity rate of two groups to their similar bacteria was over 97%. The genus of the 7 strains of LAB were shown in Table 3.
Table 3.
The genus of 7 strains
| LAB | Genus |
|---|---|
| 05 | Lactobacillus plantarum |
| 08 | Lactobacillus plantarum |
| 10 | Lactobacillus brevis |
| 13 | Lactobacillus plantarum |
| 23 | Lactobacillus brevis |
| 27 | Lactobacillus brevis |
| 32 | Lactobacillus plantarum |
Optimization of digestion conditions
As could be seen from Table 4 that the second method was digested fully and sufficiently. HNO3 had strong oxidation property, and was easily vaporized acid after digestion. Therefore, 0.5 mL of sample, 6 mL of HNO3 and 2 mL of H2O2 were taken.
Table 4.
Digestion degree of different proportion of solvent
| No. | Degree of digestion |
|---|---|
| One | + + + + + |
| Two | + + + + |
| Three | + + + |
Adsorption capacity of LAB
As could be seen from Fig. 4, the removal rate of the 7 LAB strains, of which LAB-32 had the highest removal rate and its average adsorption capacity reached 54.03 mg/g, LAB-23 had the lowest removal rate and its average adsorption capacity reached 43.26 mg/g. Besides, LAB-08, LAB-10 and LAB-13 had similar adsorption capacity.
Fig. 4.
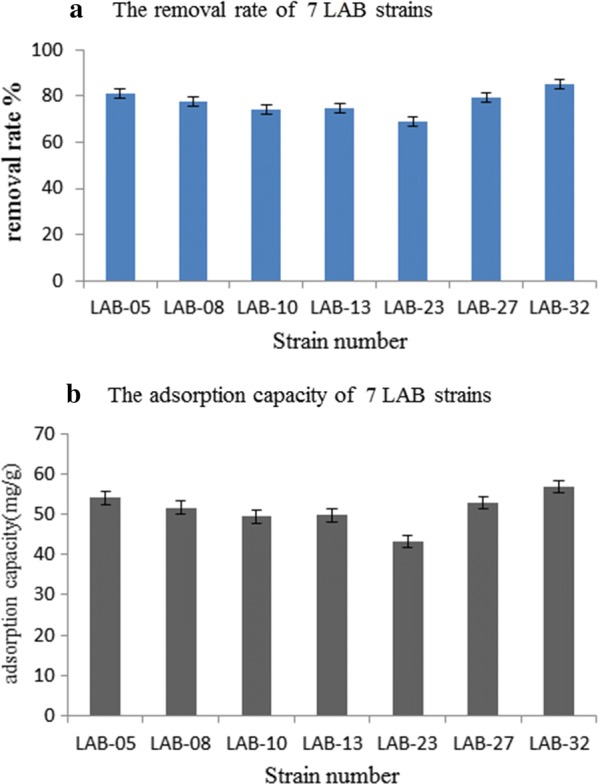
The removal rate (a) and adsorption capacity (b) of 7 LAB strains. The blue bar chart represents the removal rate and the gray bar chart represents the adsorption amount
At present, there were few reports on the adsorption of LAB towards Pb2+, especially for L. plantarum and L. brevis, which were almost not reported. The adsorbents of Pb2+ were divided into non-biological adsorbent and biological adsorbent, different kinds of adsorbents had different removal rates (Table 5).
Table 5.
The removal rate of different material towards Pb2+
| Type of adsorbent | Adsorbent | Maximum adsorption (mg/g) | References |
|---|---|---|---|
| Non-biological adsorbent | Coconut-shell carbon (dry) freshbrine algae | 26.50 | Sekar et al. (2004) |
| Original diatomite (DO) | 3.1101 | Sögüt and Caliskana (2017) | |
| Manganese oxide modified Diatomite (DMn) | 12.6322 | ||
| Sawdust | 4.59 | Li et al. (2007) | |
| Modified peanut husk | 4.66 | ||
| Acidified multi-walled CNTs | 49.71 | Wang et al. (2007) | |
| Original bentonite | 19.19 | Kul and Koyuncu (2010) | |
| Cicer arientinum biomass | 27.79 | Nadeem et al. (2009) | |
| Coontail or hornwort | 44.8 | Keskinkan et al. (2004) | |
| Pine wood char | 4.13 | Mohan et al. (2007) | |
| ACF-400 | 30.11 | ||
| Biological adsorbent | Oedogonium hatei | 40.9–44.2 | Gupta et al. (2010) |
| Derived vermicompost (CV) | 38.11 | Zhu et al. (2017) | |
| Cow manure (CM) | 43.01 | ||
| L. rhamnosus- GG | 46.8 | Halttunen et al. (2007) | |
| B. longum 2C | 45.4 | ||
| L. rhamnosus LC-705 | 29.1 | Ibrahim et al. (2006) | |
| LAB-05 (Lactobacillus plantarum) |
55.31 | This work | |
| LAB-08 (Lactobacillus plantarum) |
51.83 | This work | |
| LAB-10 (Lactobacillus brevis) |
50.31 | This work | |
| LAB-13 (Lactobacillus plantarum) |
50.94 | This work | |
| LAB-23 (Lactobacillus brevis) |
44.17 | This work | |
| LAB-27 (Lactobacillus brevis) |
53.71 | This work | |
| LAB-32 (Lactobacillus plantarum) |
57.31 | This work |
As can be seen from the comparison in Table 5, in general, the adsorption capacity of 7 strains in different adsorbents was at a high level. In the non-biological adsorbent, the maximum adsorption capacity was 49.71 mg/g, while in the biological adsorbent, the maximum adsorption capacity reached 55.72 mg/g.
Discussion
Due to the high adsorption capacities of 7 strains to Pb2+, we referred to existing research results to speculate on the process of adsorption of Pb2+ by LAB in this experiment (Fig. 5).
Fig. 5.
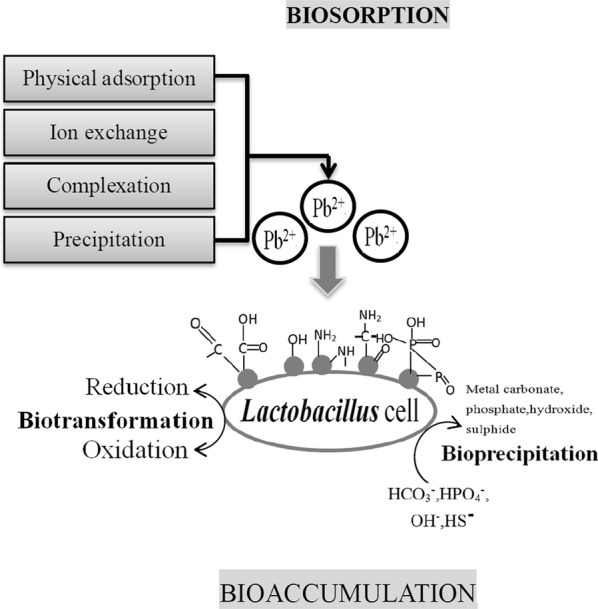
The mechanism of biosorption and bioaccumulation of a microbial cell
Metal ions enter the living LAB usually through extracellular biosorption and intracellular bioaccumulation (Huang et al. 2018). Biological adsorption mainly refers to the adsorption of metal ions onto biological matrix by one or more combination of includes physical adsorption, precipitation, complexation, coordination, chelation, ion exchange, electrostatic interaction and microprecipitation (Vijayaraghavan and Yun 2008; Diep et al. 2018). Bioaccumulation includes cytoplasmic sequestration, enzymatic detoxification and trafficking to efflux systems (Huang et al. 2018). The former was a fast process, which took only dozens of minutes without consuming energy. But bioaccumulation was a metabolic process that requires energy. Microorganisms were really sensitive to heavy metal stress. Heavy metals show the highest toxicity to cell, exhibiting an increase in the production of apoptosis and cell death. The damage of the DNA of bacteria were negatively correlated with adsorption ability of bacteria (Zhou et al. 2008; Giller et al. 1998). Therefore, the abnormal situation in this experiment was closely related to the toxicity and stress of heavy metals. The deeper reason may be the damage to bacteria DNA and other substances caused by heavy metals.
Biological materials had good biosorption capacity for metal ions (Vijayaraghavan and Yun 2008). Compared with abiotic adsorbent, the effect of biological adsorbents was more stable, more widely used and cheaper (Vinod et al. 2015). The bacterial cell wall was the first component in contact with metal ions, which were deposited first on the cell surface or in the cell wall structure. The chemical functional groups of the cell wall played a vital role in biosorption (Vijayaraghavan and Yun 2008), such as carboxylate, hydroxyl, sulfate, phosphate and amino, etc (François, et al. 2012). Sintuprapa et al. (2000) suggested that the functional groups of cell wall containing negative charges in living cells of Penicillium sp., such as phosphate groups, hydroxyl groups and carboxyl groups, reacted with Zn2+ to adsorb Zn2+. Then Zn2+ bound to polyphosphate granules and accumulated and precipitated in cells. Feng et al. (2012) isolated Pb2+-resistant train Lb. plantarum 70810 EPS from traditional Chinese pickled cabbage, they observed a large number of Pb2+ particles adsorbed on the surface of cell by scanning electron microscopy, which showed that the functional groups joining in adsorption involve the hydroxyl group, carboxyl group, sulfate group, amino group and amide group and so on. In this study, LAB mainly relied on cell surface adsorption to blind free Pb2+ through positive and negative charges to the surface of cell wall. The cell wall of LAB was mainly composed of mannan, glucan, chitin and protein. The hydroxyl group, carboxyl group, sulfate group, amino group and amide group in these components can be complex with Pb2+, then retained by mineral nucleation (Murthy et al. 2012). In addition, cell wall peptidoglycans and/or surface extracellular polymers with cell wall dissociation could also form effective biosorption matrices to adsorb metal ions (Vijayaraghavan and Yun 2008). Gram-positive microorganisms had a large adsorption capacity because they have a thick peptidoglycan layer and contain a large number of adsorption sites (Timková et al. 2018). François et al. (2012) also confirmed that the biosorption of mercury by Bacillus sp. CM111 depends on the surface extracellular polymeric substances. Similarly, the extracellular polymeric substances of LAB could be used as metal ion binders to adsorb Pb2+. Metal ions and bacterial cell surface could be combined by electrostatic interaction, van der Waals force, covalent bond, redox interaction and precipitation (Murthy et al. 2012).
Due to all the samples in this study were exerted shake cultivation for 1 h, which was a relatively short time. A large number of Pb2+ could be adsorbed in a short time, and extracellular biosorption played the most important role, followed by bioaccumulation.
Bioaccumulation is a metabolic process in which metal ions are absorbed into the intracellular space by the import complexes which create a translocation pathway through the lipid bilayer and then sequestered by the protein and peptide ligands (Diep et al. 2018). Bioaccumulation is a process after biosorption (Christoforidis et al. 2015). When most of places on the cell wall that can be used for metal ion binding are occupied, living microbial cells initiated intracellular bioaccumulation (Mrvčić et al. 2012). After Pb2+ was transported inside LAB, there were two possible ways to explain bioaccumulation. For one thing, Pb2+ was transported to the cytosol and isolated in certain areas to prevent damage to important organelles and functional molecules. For another, Pb2+ bound to the Pb2+-binding protein, which reduced the toxicity of Pb2+ and kept the cells growing normally. It was obvious that the reaction of Pb2+ and binding proteins was irreversible. Bioaccumulation of Pb2+ in cells depended on the growth of LAB (Podder and Majumder 2018).
In addition, pH, temperature, pretreatment, initial metal ion concentration, contact time, interfering ions, biomass concentration and other factors also had different impact on the adsorption capacity of LAB (Lin et al. 2017).
Adsorption capacity of L. brevis and L. plantarum was studied under the optimum growth conditions of LAB, but not under their own optimum growth conditions. If under those conditions, the adsorption capacity of them should be improved. So, next, we would continue to study the optimal growth conditions of these two strains of LAB and used them separately or jointly to solve the problem of Pb2+ contamination in food.
In general, this study was to screen the Pb2+ resistant LAB from Ya’an pickle brine. 7 strains with strong tolerance to Pb2+ were isolated for identification. The results showed that 4 strains of L. plantarum and 3 strains of L.s brevis were obtained in the 7 strains of LAB. Through the blinding experiment, their adsorption capacities towards Pb2+ were at 43.26–56.83 mg/g, which were at a higher level. Through the explanation of the adsorption mechanism of LAB, the adsorption of LAB towards Pb2+ mainly involved biosorption and bioaccumulation. The 7 strains of LAB mainly relied on biosorption to remove Pb2+, at the same time, the complexation on the bacterial surface played a decisive impact. The 7 strains of LAB could be great potential as a safe adsorbent to solve heavy metal pollution in the food and the accumulation of heavy metals in human body.
Additional file
Additional file 1: Table S1. Microwave digestion program. Table S2. The proportion of solvent.
Authors’ contributions
DL initiated the writing of this manuscript. WQ designed the structure of this manuscript, ZW gave some valuable advices about the structure of the manuscript, DW interpreted results, YH drafted the manuscript, HC compiled information, QH, HC, JZ and QZ made contribution to the revision of the manuscript, YZ, RJ, TQ, YC and DW performed the experiments. All authors read and approved the final manuscript.
Acknowledgements
The authors are grateful for financial supports from the National Natural Science Foundation of China (41807364), “211 Engineering Double Support Plan”, Sichuan Agricultural University (03572195), and the education department of Sichuan Province major project for financial support (17ZB0338).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Data and material are authentic and believable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Research ethical approval was waived by the China the Ministry of Health’s National Biomedical Research Ethics Committee.
Funding
Source of fund: Natural Science Foundation of China; Fund Number: 41807364.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Derong Lin, Phone: +86-835-288-2187, Email: lindr2018@sicau.edu.cn.
Hongfu Cao, Email: 943941556@qq.com.
Yixin Zhong, Email: 844929149@qq.com.
Yichen Huang, Email: 1187207651@qq.com.
Jinpeng Zou, Email: 1270107884@qq.com.
Qi He, Email: 635420623@qq.com.
Ran Ji, Email: 137249060@qq.com.
Tao Qin, Email: 2912859717@qq.com.
Yuan Chen, Email: 1628072829@qq.com.
Dan Wang, Email: 1825296531@qq.com.
Zhijun Wu, Email: wzj@sicau.edu.cn.
Wen Qin, Email: qinwen@sicau.edu.cn.
Dingtao Wu, Email: DT_Wu@sicau.edu.cn.
Hong Chen, Email: chenhong945@sicau.edu.cn.
Qing Zhang, Email: zhangqing@sicau.edu.cn.
References
- Acar O, Tunçeli A, Türker AR. Comparison of wet and microwave digestion methods for the determination of copper, iron and zinc in some food samples by FAAS. Food Anal Method. 2016;9(11):01–08. [Google Scholar]
- Alexander K, Hannes H, Frank F, Jörg S. Computational integration of genomic traits into 16S rDNA microbiota sequencing studies. Gene. 2014;549(1):186–191. doi: 10.1016/j.gene.2014.07.066. [DOI] [PubMed] [Google Scholar]
- Auch AF, Jan MV, Klenk HP, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2(1):117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig JA, Kazi TG, Shah AQ, Kandhro GA, Afridi HI, Khan S, Kolachi NF. Biosorption studies on powder of stem of Acacia nilotica: removal of arsenic from surface water. J Hazard Mater. 2010;178(1–3):941. doi: 10.1016/j.jhazmat.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Christoforidis AK, Orfanidis S, Papageorgiou SK, Lazaridou AN, Favvas EP, Mitropoulos ACh. Study of Cu(II) removal by Cystoseira crinitophylla biomass in batch and continuous flow biosorption. Chem Eng J. 2015;277:334–340. [Google Scholar]
- Cui Z, Zhang X, Yang H, Sun L. Bioremediation of heavy metal pollution utilizing composite microbial agent of Mucor circinelloides, Actinomucor sp. and Mortierella sp. J Environ Chem Eng. 2017;5(4):3616–3621. [Google Scholar]
- Diep P, Mahadevan R, Yakunin A. Heavy Metal Removal by Bioaccumulation Using Genetically Engineered Microorganisms. Front Bioeng Biotechnol. 2018;6:157. doi: 10.3389/fbioe.2018.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekere NR, Agwogin AB, Ihedioha JN. Studies of biosorption of Pb(2+), Cd(2+) and Cu(2+) from aqueous solutions using Adansonia digitata root powders. Int J Phytoremediat. 2016;18(2):116–125. doi: 10.1080/15226514.2015.1058329. [DOI] [PubMed] [Google Scholar]
- Emel Öz, Güzin K, Özlem B, Mükerrem K. Isolation and identification of lactic acid bacteria from pastırma. Food Control. 2017;77:158–162. [Google Scholar]
- Feng M, Chen X, Li C, Nurgul R, Dong M. Isolation and identification of an Exopolysaccharide-Producing lactic acid bacterium strain from chinese paocai and biosorption of Pb(II) by its exopolysaccharide. J Food Sci. 2012;77(6):T111–T117. doi: 10.1111/j.1750-3841.2012.02734.x. [DOI] [PubMed] [Google Scholar]
- François F, Lombard C, Guigner JM, Soreau P, Brian-Jaisson F, Martino G, Vandervennet M, Garcia D, Molinier AL, Pignol D, Peduzzi J, Zirah S, Rebuffat S. Isolation and characterization of environmental bacteria capable of extracellular biosorption of mercury. Appl Environ Microbiol. 2012;78(4):1097–1106. doi: 10.1128/AEM.06522-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giller KE, Witter E, Mcgrath SP. Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol Biochem. 1998;30(10–11):1389–1414. [Google Scholar]
- Gitzendanner MA, Soltis PS, Yi TS, Li DZ, Soltis DE. Plastome phylogenetics: 30 years of inferences into plant evolution. Adv Bot Res. 2018;85:293–313. [Google Scholar]
- Goswami L, Manikandan NA, Pakshirajan K, Pugazhenthi G. Simultaneous heavy metal removal and anthracene biodegradation by the oleaginous bacteria Rhodococcus opacus. 3. Biotech. 2017;7(1):37. doi: 10.1007/s13205-016-0597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta VK, Rastogi A, Nayak A. Biosorption of nickel onto treated alga (Oedogonium hatei): application of isotherm and kinetic models. J Colloid Interf Sci. 2010;342(2):533–539. doi: 10.1016/j.jcis.2009.10.074. [DOI] [PubMed] [Google Scholar]
- Halttunen T, Finell M, Salminen S. Arsenic removal by native and chemically modified lactic acid bacteria. Int J Food Microbiol. 2007;120:173–178. doi: 10.1016/j.ijfoodmicro.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Huang F, Wang ZH, Cai YX, Chen SH, Tian JH, Cai KZ. Heavy metal bioaccumulation and cation release by growing Bacillus cereus RC-1 under culture conditions. Ecotox Environ Safe. 2018;157:216–226. doi: 10.1016/j.ecoenv.2018.03.077. [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Halttunen T, Tahvonen R, Salminen S. Probiotic bacteria as potential detoxification tools: assessing their heavy metal binding isotherms. Can J Microbiol. 2006;52(9):877–885. doi: 10.1139/w06-043. [DOI] [PubMed] [Google Scholar]
- Iskandar I, Koike K, Sendjaj P. Identifying groundwater arsenic contamination mechanisms in relation to arsenic concentrations in water and host rocks. Environ Earth Sci. 2012;65(7):2015–2026. [Google Scholar]
- Jahromi MF, Liang JB, Ebrahimi R, Soleimani AF, Rezaeizadeh A, Abdullah N, Shokryazdan P. Protective potential of Lactobacillus species in lead toxicity model in broiler chickens. Animal. 2017;11(5):1–7. doi: 10.1017/S175173111600224X. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Viraraghavan T. Biosorption of heavy metals on Aspergillus niger: effect of pretreatment. Bioresour Technol. 1998;63(2):109–113. [Google Scholar]
- Keskinkan O, Goksu MZ, Basiuyuk M, Forster CF. Heavy metal adsorption properties of a submerged aquatic plant (Ceratophyllum demersum) Bioresour Technol. 2004;92(2):197–200. doi: 10.1016/j.biortech.2003.07.011. [DOI] [PubMed] [Google Scholar]
- Kinoshita H, Sohma Y, Ohtake F, Ishida M, Kawai Y, Kitazawa H, Saito T, Kimura K. Biosorption of heavy metals by lactic acid bacteria and identification of mercury binding protein. Res Microbiol. 2013;164(7):701–709. doi: 10.1016/j.resmic.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Kobya M, Demirbas E, Senturk E, Ince E. Adsorption of heavy metal ions from aqueous solutions by activated carbon prepared from apricot stone. Bioresour Technol. 2005;96(13):1518–1521. doi: 10.1016/j.biortech.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Kul AR, Koyuncu H. Adsorption of Pb(II) ions from aqueous solution by native and activated bentonite: kinetic, equilibrium and thermodynamic study. J Hazard Mater. 2010;179(1):332–339. doi: 10.1016/j.jhazmat.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Kurniawan SB, Purwanti IF, Titah HS. The Effect of pH and Aluminium to Bacteria Isolated from Aluminium Recycling Industry. J Ecol Eng. 2018;19(3):154–161. [Google Scholar]
- Li Q, Zhai J, Wang M, Zhou J. Kinetic studies of adsorption of Pb(II), Cr(III) and Cu(II) from aqueous solution by sawdust and modified peanut husk. J Hazard Mater. 2007;141(1):163–167. doi: 10.1016/j.jhazmat.2006.06.109. [DOI] [PubMed] [Google Scholar]
- Li X, Meng D, Li J, Yin H, Liu H, Liu X, Cheng C, Xiao Y, Liu Z, Yan M. Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ Pollut. 2017;231(1):908–917. doi: 10.1016/j.envpol.2017.08.057. [DOI] [PubMed] [Google Scholar]
- Lin D, Ji R, Wang D, Xiao M, Zhao J, Zou J, Li Y, Qin T, Xing B, Chen Y, Liu P, Wu Z, Wang L, Zhang Q, Chen H, Qin W, Wu D, Liu Y, Liu Y, Li S. The research progress in mechanism and influence of biosorption between lactic acid bacteria and Pb(II): a review. Crit: Rev Food Sci; 2017. [DOI] [PubMed] [Google Scholar]
- Michael F. B4-gram staining technique. Method Mycoplasmol. 1983;1:39–41. [Google Scholar]
- Mohan D, Pittman CU, Bricka JM, Smith F, Yancey B, Mohammad J, Steele PH, Alexandre-Franco MF, Gómez-Serrano V, Gong H. Sorption of arsenic, cadmium, and lead by chars produced from fast pyrolysis of wood and bark during bio-oil production. J Colloid Interf Sci. 2007;310(1):57–73. doi: 10.1016/j.jcis.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Murthy S, Bali G, Sarangi SK. Lead biosorption by a bacterium isolated from industrial effluents. Int J Microbiol Res. 2012;4(3):192. [Google Scholar]
- Mrvčić J, Stanzer D, Šolić E, Stehlik-Tomas V. Interaction of lactic acid bacteria with metal ions: opportunities for improving food safety and quality. World J Microbiol Biotechnol. 2012;28(9):2771–2782. doi: 10.1007/s11274-012-1094-2. [DOI] [PubMed] [Google Scholar]
- Nadeem R, Nasir MH, Hanif MS. Pb (II) sorption by acidically modified Cicer arientinum biomass. Chem Eng J. 2009;150(1):40–48. [Google Scholar]
- Özcan AS, Tunali̇ S, Akar T, Özcan A. Biosorption of lead(II) ions onto waste biomass of Phaseolus vulgaris L.: estimation of the equilibrium, kinetic and thermodynamic parameters. Desalination. 2009;244(1):188–198. [Google Scholar]
- Peladan F, Monteil H. Intérêt taxonomique de la caractérisation des Pseudomonas par l’analyse des acides gras volatils produits en culture. Annales de l’Institut Pasteur/Microbiologie. 1984;135:411–425. [PubMed] [Google Scholar]
- Podder MS, Majumder CB. Optimization of environmental conditions for the growth and bioaccumulation of As (III) and As (V) ions by growing Corynebacterium glutamicum MTCC 2745: inhibition kinetic study. Sustain Water Resour Manage. 2018;4(1):23–44. [Google Scholar]
- Rahman N, Haseen U, Rashid M. Synthesis and characterization of polyacrylamide zirconium (IV) iodate ion-exchanger: its application for selective removal of lead (II) from wastewater. Arab. J. Chem. 2013;10:S1765–S1773. [Google Scholar]
- Saha I, Gupta K, Chakraborty S, Chatterjee D, Ghosh UC. Synthesis, characterization and As(III) adsorption behavior of β-cyclodextrin modified hydrous ferric oxide. J Ind Eng Chem. 2014;20(4):1741–1751. [Google Scholar]
- Schut S, Zauner S, Hampel G, König H, Claus H. Biosorption of copper by wine-relevant lactobacilli. Int J Food Microbiol. 2011;145(1):126–131. doi: 10.1016/j.ijfoodmicro.2010.11.039. [DOI] [PubMed] [Google Scholar]
- Sekar M, Sakthi V, Rengaraj S. Kinetics and equilibrium adsorption study of lead(II) onto activated carbon prepared from coconut shell. J Colloid Interf Sci. 2004;279(2):307–313. doi: 10.1016/j.jcis.2004.06.042. [DOI] [PubMed] [Google Scholar]
- Sintuprapa W, Thiravetyan P, Tanticharoen M. A possible mechanism of Zn2+ uptake by living cells of Penicillium sp. Biotechnol Lett. 2000;22(21):1709–1712. [Google Scholar]
- Söğüt EG, Caliskana N. Isotherm and kinetic studies of Pb(II) adsorption on raw and modified diatomite by using non-linear regression method. Fresenius Environ Bull. 2017;26(4):2720–2728. [Google Scholar]
- Sontakke S, Cadenas MB, Maggi RG, Diniz PPVP, Breitschwerdt EB. Use of broad range16S rDNA PCR in clinical microbiology. J Microbiol Method. 2009;76(3):217–225. doi: 10.1016/j.mimet.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Timková I, Sedláková-Kaduková J, Pristaš P. Biosorption and Bioaccumulation Abilities of Actinomycetes/Streptomycetes Isolated from Metal Contaminated Sites. Separations. 2018;5(4):54. [Google Scholar]
- Teusink B, Molenaar D. Systems biology of lactic acid bacteria: for food and thought. Curr Opin Syst Biol. 2017;6:7–13. doi: 10.1016/j.coisb.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tural B, Ertaş E, Enez B, Fincan SA, Tural S. Preparation and characterization of a novel magnetic biosorbent functionalized with biomass of Bacillus subtilis: kinetic and isotherm studies of biosorption processes in the removal of Methylene Blue. J Environ Chem Eng. 2017;5(5):4795–4802. [Google Scholar]
- Vijayaraghavan K, Yun YS. Bacterial biosorbents and biosorption. Biotechnol Adv. 2008;26(3):266–291. doi: 10.1016/j.biotechadv.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Vinod KG, Arunima N, Shilpi A. Bioadsorbents for remediation of heavy metals: current status and their future prospects. Environ Eng Res. 2015;20(1):1–18. [Google Scholar]
- Wang HJ, Zhou AL, Peng F, Yu H, Chen LF. Adsorption characteristic of acidified carbon nanotubes for heavy metal Pb(II) in aqueous solution. Mater Sci Eng, A. 2007;466(1):201–206. [Google Scholar]
- Wang J, Zhao Y, Zhang P, Yang L, Xu H, Xi G. Adsorption characteristics of a novel ceramsite for heavy metal removal from stormwater runoff. Chin J Chem Eng. 2018;26:96–103. [Google Scholar]
- Xia Y, Liu X, Wang G, Zhang H, Xiong Z, Sun Y, Ai L. Characterization and selection of Lactobacillus brevis starter for nitrite degradation of Chinese pickle. Food Control. 2017;78:126–131. [Google Scholar]
- Xu YB, He XL, Huang JX, Wen LH, Zheng L, Qiao QX, Du QP. Antagonistic effects of the combination of both zinc and cefradine on the growth and morphology of the opportunistic pathogen Pseudomonas fluorescens YZ2. Int Biodeter Biodegr. 2016;111:85–92. [Google Scholar]
- Zafar SB, Siddiqui NN, Shahid F, Qader SAU, Aman A. Bioprospecting of indigenous resources for the exploration of exopolysaccharide producing lactic acid bacteria. J Genet Eng Biotechnol. 2018;16(1):17–22. doi: 10.1016/j.jgeb.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li Z. Heavy metals removal using hydrogel-supported nanosized hydrous ferric oxide: synthesis, characterization, and mechanism. Sci Total Environ. 2016;580:776–786. doi: 10.1016/j.scitotenv.2016.12.024. [DOI] [PubMed] [Google Scholar]
- Zhou S, Wei C, Liao C, Wu H. Damage to DNA of effective microorganisms by heavy metals: impact on wastewater treatment. J Environ Sci. 2008;20(12):1514–1518. [Google Scholar]
- Zhu W, Du W, Shen X, Zhang H, Ding Y. Comparative adsorption of Pb2+ and Cd2+ by cow manure and its vermicompost. Environ Pollut. 2017;227:89–97. doi: 10.1016/j.envpol.2017.04.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Microwave digestion program. Table S2. The proportion of solvent.
Data Availability Statement
Data and material are authentic and believable.