Abstract
During early palate development, gene expression and regulation exhibit heterogeneity along the anterior-posterior axis. Transforming growth factor-β (TGF-β) and bone morphogenetic protein (BMP) signaling pathways play essential roles in secondary palatal formation but the exact relationship between the TGF-β and BMP pathways in palate development remains unknown. Here, we demonstrate that, during early secondary palate development, phospho-(p)Smad1/5/8 is highly expressed in the anterior palate but relatively lowly expressed in the posterior palate. Conversely, pSmad2/3 has a lower expression level in the anterior palate than in the posterior palate. With the BRE-Gal reporter, we found that the canonical BMP signaling pathway was not activated in the anterior palate but exhibited a moderate level in the posterior palate. Co-immunoprecipitation revealed that Smad4 bound to pSmad1/5/8 only in the posterior palate and not in the anterior palate. Knocking-out of Tgfbr2 (Wnt1-Cre;Tgfbr2f/f;BRE) in the palatal mesenchyme enhanced canonical BMP activity in the posterior palate but not in the anterior palate, because of decreased pSmad2/3. pSmad1/5/8-Smad4 complexes were found to be dramatically increased in posterior palatal mesenchymal cells at embryonic day 13.5 cultured in the presence of SB525334. Proximity ligation assay also showed pSmad1/5/8-Smad4 complexes were increased in the posterior palate of Wnt1-Cre;Tgfbr2f/f mice. Therefore, the reduction of pSmad2/3 level in the palatal mesenchyme of Wnt1-Cre;Tgfbr2f/f;BRE mice contributes primarily to the increase of pSmad1/5/8-Smad4 complexes leading to enhanced canonical BMP activity in the posterior palate. Moreover, during early development, canonical BMP signaling operates in the posterior palate but is completely absent in the anterior palate. Canonical TGF-β signaling suppresses canonical BMP signaling activity in the posterior palate by competing limited Smad4.
Keywords: Bone morphogenetic protein, Smad1/5/8, Smad2/3, Transforming growth factor-β, Palate development
Introduction
Signaling by the transforming growth factor β (TGF-β) superfamily, including bone morphogenetic proteins (BMP), is transduced into cells by the binding of ligands to the type I and type II transmembrane serine/threonine kinase complex. With the binding of the ligand, the type II receptor activates the type I receptor by phosphorylation of the type I receptor. The activated type I receptor phosphorylates receptor-activated Smads (R-Smads, including Smad-1, −2, −3, −5, and −8) in the cytoplasm. Canonical (or Smad-dependent) TGF-β or BMP signaling occurs via the phosphorylation of Smad-2/3 or Smad1/5/8, respectively. The phosphorylated Smad-2/3 (pSmad2/3) or Smad1/5/8 (pSmad1/5/8) binds to common Smad (Smad4) and enters the nucleus to interact with other transcription factors and regulates downstream gene expression. In addition to the canonical Smad pathway, TGF-β/BMP receptors can also activate members of the mitogen-activated protein kinase (MAPK) pathway, known as the noncanonical pathway, including the TGF-β activated kinase 1 (TAK1; Yamaguchi et al. 1995). TAK1, a member of the MAPKKK family, was originally identified as a key regulator of MAPK kinase activation in TGF-β signaling pathways (Yamaguchi et al. 1995). Upon activation, TAK1 phosphorylates MKK3/6 directly, leading to the activation of a number of downstream kinases including P38 and JNK kinases (Zhang 2009).
The development of the palate requires a series of interactions between pharyngeal ectoderm-derived epithelium and the cranial neural crest (CNC)-derived mesenchyme. The palate comprises two structures: the primary and the secondary palates. In mice, the development of the secondary palate begins at embryonic day 11.5 (E11.5) with the emergence of bilateral outgrowths, called palatal shelves, on the oral side of the maxillary process. At E12.5-E13.5, the palatal shelves initially grow vertically flanking the developing tongue. Around E14.0, the bilateral palatal shelves reorient to the horizontal positon above the tongue. Subsequently, two palatal shelves grow towards and finally fuse with each other at the midline to form the intact roof of the oral cavity (Liu et al. 2008).
The mammalian secondary palate is divided anatomically into the anterior bony region (hard palate) and the posterior muscular region (soft palate), each of which have specialized functions in occlusion, speech and swallowing. Consistent with the morphological differences in the anterior and posterior palate, previous studies have clearly demonstrated the molecular heterogeneity along the anterior-posterior (AP) axis of the developing secondary palate (Hilliard et al. 2005; Liu et al. 2008). During early palate development, several critical signaling molecules and transcription factors exhibit regional differential expression along the AP axis. For example, the expression of the Shox2, Msx1 and Fgf10 genes is restricted to the anterior palatal mesenchyme, whereas the Bmp3, Barx1, Mn1, Meox2 and Tbx22 genes are preferentially expressed in the posterior palatal mesenchyme (Alappat et al. 2005; Barlow et al. 1999; Li and Ding 2007; Liu et al. 2008; Nie 2005; Rice et al. 2004; Yu et al. 2005; Zhang et al. 2002). In addition, anterior and posterior palatal mesenchymal cells exhibit distinct responses in terms of gene expression and cellular behavior to growth factors. For example, exogenous BMP is able to induce Msx1 and Bmp4 expression in the anterior palate but not in the posterior palate, whereas FGF8 induces Pax9 expression in the posterior but not in the anterior palatal mesenchyme (Hilliard et al. 2005; Zhang et al. 2002). All these investigations indicate that the regulation of palate development is the result of distinct signaling and genetic networks in the anterior and the posterior regions of the palate.
Gene inactivation studies in mice have demonstrated that the TGF-β superfamily plays essential roles in secondary palatal formation, including Bmp4, Bmpr1a, Tgfb2, Tgfb3 and Tgfbr2, indicating that the TGF-β and BMP pathways interact to regulate palate development (Baek et al. 2011; Ito et al. 2003; Kaartinen et al. 1995; Li et al. 2011; Liu et al. 2005; Rice et al. 2004; Zhang et al. 2002). However, the exact relationship between the TGF-β and BMP pathways during secondary palate development remains unknown.
Here, we provide evidence that, during early secondary palate development, the canonical BMP signaling pathway is not activated in the anterior palate but is involved in posterior palate development. TGF-β inhibits the canonical BMP signaling pathway in the posterior palate region but not in the anterior region.
Materials and methods
Animals and embryo collection
The generation of BRE-Gal (BRE), Wnt1-Cre, Tgfbr2f/+ and Tak1f/+ lines have been described previously (Chytil et al. 2002; Danielian et al. 1998; Javier et al. 2012; Liu et al. 2006). Wnt1-Cre;Tgfbr2f/f;BRE mice and Wnt1-Cre;Tak1f/f;BRE mice were generated by mating Wnt1-Cre;Tgfbr2f/+ with Tgfbr2f/+;BRE mice and by mating Wnt1-Cre;Tak1f/+ with Tak1f/+;BRE, respectively. All wild-type mice were on a CD-1 or C57BL6/J background. Embryos were collected from timed-mated females and subjected to genotyping based on the polymerase chain reaction.
Immunofluorescence and X-gal staining
For immunofluorescence, samples were fixed with 4% paraformaldehyde at 4 °C overnight, dehydrated through a graded ethanol series and then processed for paraffin sectioning. Sections were subjected to standard immunofluorescence staining as described previously (He et al. 2010). The following primary antibodies were used: anti-pSmad1/5/8 (Cell Signaling), anti-pSmad2/3 (Santa Cruz Biotechnology), anti-Smad4 (Abcam) and anti-phosphorylated P38 (pP38; R&D Systems). Alexa Fluor 568 or Alexa Fluor 488 (Invitrogen) acted as labels on secondary antibodies. Nuclei were counter-stained with 4,6-diamidino-2-phenylindole (Invitrogen). For X-gal staining, samples were fixed in 4% paraformaldehyde, washed in ice-cold phosphate-buffered saline (PBS), subsequently dehydrated with 30% sucrose/PBS, embedded in O.C.T. (Tissue-Tek) and cryo-sectioned. Standard X-gal staining was conducted as described previously (Ito et al. 2003). The tissue anterior or posterior to the first molar tooth bud was considered as the anterior or posterior palate, respectively.
Co-immunoprecipitation
Primary cultured palatal mesenchymal cells were lysed in RIPA buffer (Cell Signaling Technology) with protease inhibitor for 15 min at 4 °C. For organs, tissues were digested with 0.25% trypsin (Life Technologies) at 37 °C for 10 min prior to lysis. Lysates were centrifuged at 12,000 rpm at 4 °C for 20 min to remove cellular debris. The supernatant was precleared with 40 μl Dynabeads protein A/G (Life Technologies) at 4 °C for 30 min. Anti-Smad4 antibody or negative control IgG (Santa Cruz Biotechnology, 3 μg each) was added to the precleared lysates following incubation at 4 °C overnight and then with 50 μl Dynabeads protein A/G for 2 h to precipitate immunocomplexes. Beads were washed three times with wash buffer (10 mM TRIS-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 1% Nonidet P-40 and 2 mM EDTA). Of the sample buffer, 50 μl were added to the beads and samples were heated to 95 °C for 10 min. pSmad1/5/8 or pSmad2/3 were detected in the protein complex by Western blot.
Western blot
Proteins were separated with SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was blocked in 5% nonfat milk for 30 min at room temperature and then incubated with antibodies against pSmad1/5/8 or aD-glyceraldehyde-3-phosphate dehydrogenase (Santa Cruz Biotechnology) overnight at 4 °C with constant rocking. The membrane was then washed three times and incubated with IRDye 800 secondary antibody (Li-COR) for 1 h. Immunoreactive bands were visualized with the Odyssey imaging system (Li-COR).
Primary palatal mesenchymal cell culture
E13.5 embryonic posterior palate was dissected out and incubated with 2 units/ml Dispase (Life Technologies) in PBS for 30 min at room temperature. Palatal epithelium was removed from mesenchyme with the aid of fine forceps. The isolated mesenchyme was then digested with 0.25% trypsin at 37 °C for 10 min and dissociated into a single-cell suspension by gentle pipetting. Cells were plated onto a 6-well plate and cultured in DMEM (Life Technologies) supplemented with 20% FBS (Life Technologies) at 37 °C. For the blocking of TGF-β/pSmad2/3 signaling, cells were treated with SB525334 (Tocris Bioscience) at a final concentration of 10 μM and were harvested after 24 h after culture.
In situ proximity ligation assay
Endogenous pSmad1/5/8-Smad4 and pSmad2/3-Smad4 complexes in the posterior palate were detected by using an in situ proximity ligation assay (PLA) kit (Duolink kit, Sigma-Aldrich) as described previously (Yang et al. 2014). Briefly, tissue sections were blocked with blocking reagent and incubated with primary antibodies (anti-Smad4 and anti-pSmad1/5/8 or anti-pSmad2/3 antibodies) overnight at 4 °C. Then, sections were incubated with secondary antibodies conjugated with oligonucleotides (anti-mouse PLA probe Minus and Anti-rabbit PLA probe Plus). Ligation and amplification were performed according to the protocol provided by the kit. PLA signals were detected under a fluorescence microscope.
Data quantification and statistical analysis
All experiments were repeated at least three times and at least three samples were collected for each transgenic or mutant mouse embryo. Histological immunofluorescence intensity was analyzed from three continuous sections of each sample with Image J software. Statistical analysis was performed by using Student’s t-test.
Results
Expression pattern of down-stream mediators of TGF-β/BMP signaling pathway in early developing palate
To investigate the differential engagement of TGF and BMP signaling in palate development, we first examined the expression of TGF-β/BMP signaling down-stream mediators, including pSmad1/5/8, pSmad2/3, Smad4 (Smad-dependent) and pP38 (Smad-independent), in early developing palatal shelves along the AP axis. Immunofluorescent results showed that pSmad1/5/8 and pP38 were highly expressed in the anterior palate but were expressed at a relatively low level in the posterior palate (Fig. 1a–h). Conversely, pSmad2/3 showed a lower expression level in the anterior palate but a higher level of expression in the posterior palate (Fig. 1c, d). For Smad4, comparable expression levels were detected in the anterior and the posterior palatal shelves (Fig. 1e, f).
Fig. 1.
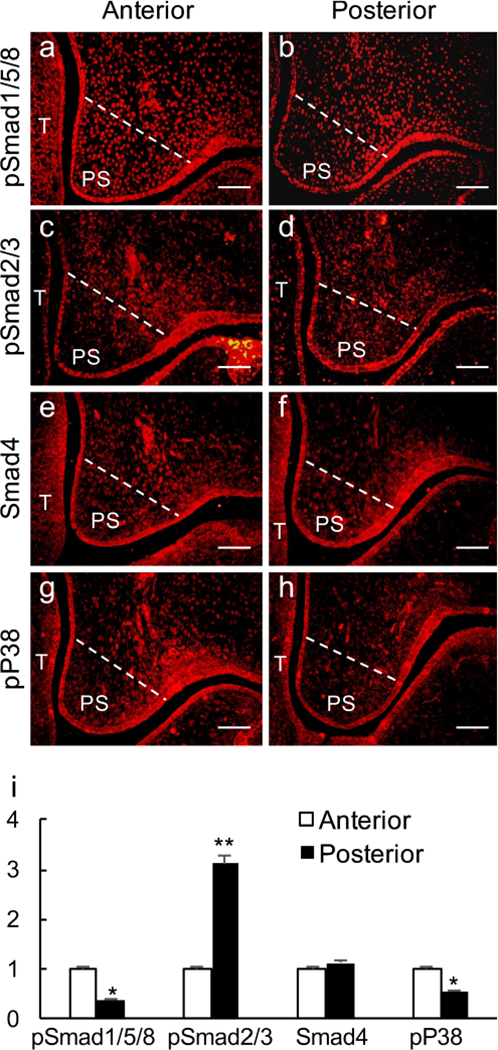
Immunofluorescent staining of down-stream mediators of transforming growth factor α (TGF-β)/bone morphogenetic protein (BMP) signaling pathway in the developing palate at E13.5. a, b pSmad1/5/8; c, d pSmad2/3; e, f Smad4; g, h pP38. a, c, e, g Anterior palate; b, d, f, h posterior palate. White lines demarcate the palatal region for quantifying the immunofluorescence intensity (T tongue, PS palatal shelf). Bars 50 μm. h Statistical comparison of immunofluorescence intensity. *P < 0.05, **P < 0.01
Canonical BMP signaling is operating in developing posterior palate but not in anterior palate
We next attempted to map the exact activity of the canonical BMP signaling pathway in the developing palate. We took advantage of a reporter mouse line harboring seven copies of BRE and the Id3 minimal promoter with LacZ reporter (BRE-Gal), which has been shown to serve as a faithful indicator of BMP canonical signaling activity in developing mouse embryo (Javier et al. 2012). To our surprise, this BRE-Gal reporter failed to show canonical BMP activity in the anterior palate but revealed a moderate level of canonical BMP activity in the posterior palate (Fig. 2a–d), suggesting that canonical BMP signaling is not operating in the anterior palate.
Fig. 2.
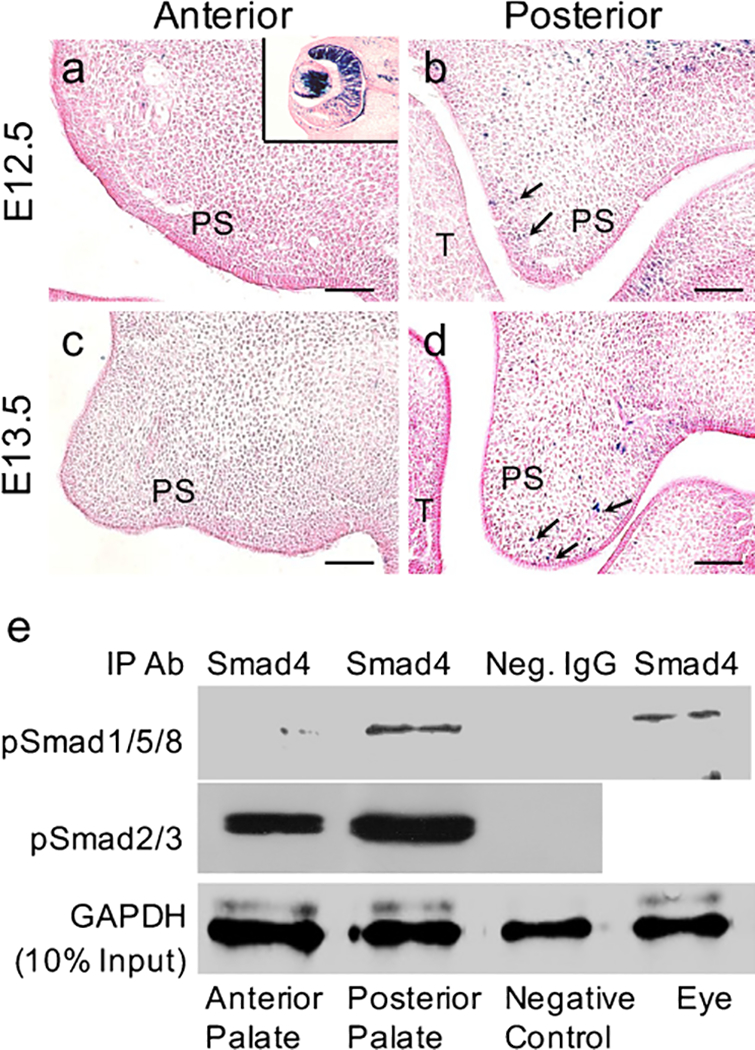
Canonical BMP signaling is operating in the posterior but not in anterior portion of the developing palatal shelves. a-d X-gal staining of the anterior (a, c) and posterior (b, d) palate from BRE-Gal reporter mice at embryyonic day 12.5 (E12.5; a, b) and E13.5 (c, d). Black arrows indicate positive X-gal Staining (T tongue, PS palatal shelf). Insert in a Eye tissue as a positive control for X-gal staining. Bars 50 μm. e Western blot showing pSmad1/5/8-Samd4 and pSmad2/3-Samd4 complexes in palatal shelves. Eye tissues were included as positive controls for pSmad1/5/8-Samd4 complexes (IP Ab immunoprecipitation antibody, Neg. IgG negative control IgG, GAPDH D-glyceraldehyde-3-phosphate dehydrogenase)
Because activation of the BRE reporter requires the formation of the pSmad1/5/8-Smad4 complex (Javier et al. 2012), we then set out to determine whether the failed activation of BRE-Gal in the anterior palate was attributed to the lack of pSmad1/5/8-Smad4 complex formation by conducting a co immunoprecipitation assay. Eye tissue was used as a positive control, as it exhibited a high level of canonical BMP activity (Yang et al. 2014). Indeed, using Smad4 antibody, we were able to precipitate abundant pSmad1/5/8 in posterior palate and eye tissue but pSmad1/5/8 precititated from the anterior palate were barely detectable by Western blot (Fig. 2e). These results indicate that Smad4 binds to pSmad1/5/8 only in the posterior palate but not in the anterior palate. Furthermore, we found amounts of pSmad2/3-Smad4 complex both in the anterior and in the posterior palate (Fig. 2e).
TGF-β signaling inhibits canonical BMP signaling activity in posterior palate
We reported previously that TGF-β signaling suppressed canonical BMP signaling activity in the early developing tooth germ (Yang et al. 2014). In view of the expression of pSmad2/3 and the abundance of pSmad2/3-Smad4 complex in both the anterior and posterior palates, we tested whether the same mechanism exists in the developing palate. To do this, we generated mice carrying compounded BRE-Gal and Wnt1-Cre;Tgfbr2f/f alleles. We examined BRE-Gal activity in the palatal mesenchyme in the absence of Tgfbr2. Surprisingly, although canonical BMP activity remained undetectable in the anterior palatal shelves, significantly enhanced canonical BMP activity was indeed found in the posterior palate (Fig. 3a, b), indicating that TGF-β signaling attenuates canonical BMP signaling activities. To confirm further that the enhanced canonical BMP activity in the posterior palatal shelves of Wnt1-Cre;Tgfbr2f/f;BRE mice was correlated with the absence of TGF-β signaling, the expression levels of pSmad1/5/8, pSmad2/3, Smad4 and pP38 were examined in the mutant mice by immunofluorescent staining. pSmad1/5/8 and Smad4 showed no obvious changes either in the anterior or in the posterior palate as compared with wild-type controls (Fig. 3c–h). However, as expected, the level of pSmad2/3 was remarkably downregulated in the mesenchyme of both the anterior and the posterior palate (Fig. 3e, f). The expression level of pP38, a downstream target of TGF-β signaling, was also dramatically decreased in both the anterior and the posterior palates (Fig. 3i, j).
Fig. 3.
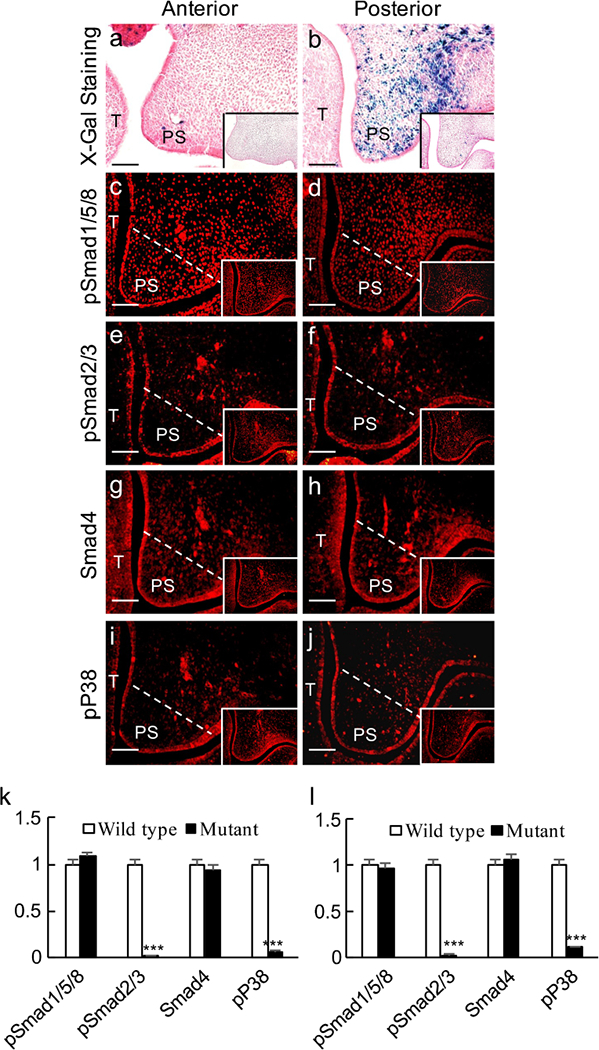
Inactivation of Tgfbr2 in the palatal mesenchyme enhances canonical BMP activity. a, b X-gal staining of the anterior (a) and posterior (b) palate from an E13.5 Wnt1-Cre;Tgfbr2f/f;BRE embryo. c-j Immunofluorescence staining of pSmad1/5/8 (c, d), pSmad2/3 (e, f), Smad4 (g, h) and pP38 (i, j) in the anterior (a, c, e, g, i) and posterior (b, d, f, h, j) palate from E13.5 Wnt1-Cre;Tgfbr2f/f;BRE embryos. White lines demarcate the palatal region for quantifying the immunofluorescence intensity (T tongue, PS palatal shelf). Inserts Wild-type controls. Bars 50 μm. k, l Statistical comparison of immunofluorescence intensity. ***P < 0.001
TAK1, a key kinase activator of P38, is recognized as an important regulator of Smad-independent TGF-β signaling in palate development and is expressed in the anterior and posterior domains of E13.5 palatal shelves (Rice et al. 2004; Song et al. 2013). To exclude the possibility that the elevated canonical BMP activity in the posterior palate was the consequence of a dramatically reduced pP38 level in Wnt1-Cre;Tgfbr2f/f;BRE mice, we generated Wnt1-Cre;Tak1f/f;BRE mice and examined BRE-Gal activity in the palatal shelves. As shown in Fig. 4, the deletion of Tak1 in the palatal mesenchyme did not result in an increased canonical BMP activity such as that observed in Wnt1-Cre;Tgfbr2f/f;BRE mice (Fig. 4a–d).
Fig. 4.
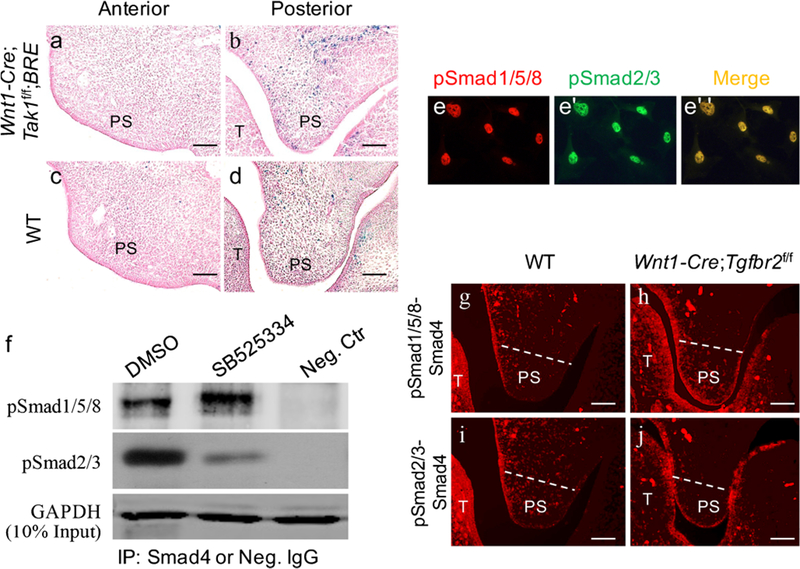
Inhibition of TGF-β/Smad2/3 signaling induces pSmad1/5/8-Smad4 complex formation. a-d X-gal staining of the anterior (a, c) and posterior (b, d) palatal shelves from E12.5 Wnt1-Cre;Tak1f/f;BRE (a, b) and wild-type (c, d) embryos (T tongue, PS palatal shelf). e–e” Immunofluorescence showing the co-localization of pSmad1/5/8 and pSmad2/3 in cultured primary posterior palatal mesenchymal cells from E13.5 wild-type embryo. f Western blot showing a significantly increased level of pSmad1/5/8-Smad4 complexes after treatment with SB525334 (Neg. Ctr negative control, IP immunoprecipitation, Neg. IgG negative control IgG). g-j In situ proximity ligation assay shows pSmad1/5/8-Smad4 (g, h) and pSmad2/3-Smad4 (i, j) complexes in the posterior palate of E13.5 wild-type mice (g, i) and Wnt1-Cre;Tgfbr2f/f mice (h, j). Bars 50 μm
We thus assumed that reduced/absent pSmad2/3 in the palatal mesenchyme of Wnt1-Cre;Tgfbr2f/f;BRE mice contributed primarily to increased pSmad1/5/8-Smad4 complexes leading to enhanced canonical BMP activity in the posterior palate. To test this hypothesis, we first examined and detected the co-localization of pSmad15/8 and pSmad4 in primary posterior palatal mesenchymal cells (Fig. 4e–e”). Then, we used in vitro primary palatal cell culture with SB525334, a widely used selective inhibitor of TGF-β receptor I (ALK5) that can specifically block phosphorylation of Smad2/3 induced by TGF-β1 (Grygielko et al. 2005). E13.5 posterior palatal mesenchymal cells were isolated and placed in cell culture in the presence/absence of SB525334. The inhibition efficiency was assessed by detecting the expression of pSmad2/3 (data not show). As determined by the co-immunoprecipitation assay shown in Fig. 4f, with the decreased pSmad2/3-Smad4 complex, the amount of pSmad1/5/8-Smad4 complex dramatically increased in the cells treated with SB525334 as compared with the control cells, demonstrating that the enhanced canonical BMP activity in the posterior palate of Wnt1-Cre;Tgfbr2f/f;BRE mice is indeed attributable to increased pSmad1/5/8-Smad4 complexes. To confirm this deduction further, we detected the pSmad1/5/8-Smad4 and pSmad2/3-Smad4 complexes in a tissue section by using in situ PLA. As expected, no pSmad2/3-Smad4 complexes but an increased level of pSmad1/5/8-Smad4 complexes was detected in the posterior palate of Wnt1-Cre;Tgfbr2f/f mice compared with wild-type mice (Fig. 4g–j). Taken together, our results demonstrate that canonical TGF-β signaling suppresses canonical BMP signaling activity in the posterior palate by competing limited Smad4.
Discussion
In the present study, we revealed the differential regulatory mechanisms of TGF-β/BMP signaling in the anterior and the posterior portions of the developing palate. Canonical BMP signaling operates in the posterior palate, although at a moderate level but is completely absent in the anterior palate during early development. Inactivation of TGF-β signaling is able to enhance canonical BMP signaling in the posterior palate but surprisingly not in the anterior palate.
In the developing palate, several Bmp genes are expressed in dynamic and differential patterns along the AP axis and BMP signaling has been shown to regulate cell proliferation in the anterior palatal mesenchyme and to maintain palatal epithelial integrity in the posterior portion (Bush and Jiang 2012; He et al. 2010; Hilliard et al. 2005; Zhang et al. 2002). In the early developing palatal shelves, both Bmp2 and Bmp4 transcripts are present and Bmp3 is moderately expressed at the posterior palate at E13.5 (Nie 2005). At E12.5 and E 13.5, Bmpr1a is highly expressed in the anterior palate and weakly expressed in the posterior palate, whereas Bmpr1b is only expressed in the anterior palate but not in the posterior palate (Baek et al. 2011; Li et al. 2011). The presence of BMP ligand or BMP signaling components is not an unequivocal indicator of BMP signaling activity (Javier et al. 2012). BRE has been identified as a regulatory sequence activated by canonical BMP signaling in various BMP target genes, including the Xeno-pus id3 and ventx2 genes and the Drosophila brk gene (von Bubnoff et al. 2005; Yao et al. 2006). BRE is recognized by Smad1/5-Smad4 complex in response to the activated BMP type I receptor (Katagiri et al. 2002), which makes BRE a faithful indicator for the transcriptional response of cells to the canonical BMP signaling. In the present investigation, we found no BRE activation in the anterior palate because of the absence of pSmad1/5/8-Smad4 complexes. However, abundant pSmad1/5/8-Smad4 complexes have been detected in the posterior palate, which is capable of activating BRE. The reason for these different responses to BMP signaling might be the different expression levels of BMP and pSmad1/5/8 in the anterior and the posterior palates. High levels of BMP and pSmad1/5/8 have been determined in the anterior palate as compared with those in the posterior palate (Nie 2005; Parada and Chai 2012; this study). In some cellular processes that depend on low levels of BMP signaling, Smad4 is essential for R-Smads to maximize their full function, probably by forming an obligate heterotrimer with R-Smads. However, in other cellular processes that have high levels of BMP signaling, abundant phosphorylated R-Smads can be formed. Under these conditions, R-Smads can still activate their downstream targets and Smad4 appears to be less important (Tong and Kwan 2013). In the posterior palate, pSmad1/5/8 are at a lower expression level and need the help of Smad4 to transduce BMP signaling, which activates the canonical BMP signaling. Furthermore, the lower level of pSmad1/5/8 makes this canonical BMP signaling more sensitive to TGF-β signaling activity. Inactivation of TGF-β signaling reduces the pSmad2/3 level and releases Smad4 from pSmad2/3-Smad4 complexes facilitating the formation of Smad1/5/8-Smad4 complexes. This explains the enhanced X-gal staining in the posterior palate of Wnt1-Cre;Tgfbr2f/f;BRE mice. In the present investigation, we observed that canonical TGF-β signaling suppresses canonical BMP signaling activity in the mesenchymal cells of the posterior palate by competing limited Smad4. The antagonism between TGF-β and BMP signaling has also been reported in several other cells, such as dermal papilla cells, cardiomyocytes and breast cancer cells (Bin et al. 2013; Grönroos et al. 2012; Wang et al. 2012).
We have shown previously that an atypical canonical BMP signaling (Smad1/5/8-dependent but Smad4-independent) pathway operates in the dental mesenchyme during early craniofacial development (Yang et al. 2014). Indeed, this atypical canonical BMP signaling pathway has been implicated in several other developing organs, such as nervous system, lens, bone and heart (Chu et al. 2004; Rajagopal et al. 2009; Tan et al. 2007; Tong and Kwan 2013); it might also operate in the anterior palatal shelves, as evidenced by the presence of high levels of pSmad1/5/8 and the absence of BRE activity and Smad1/5/8-Smad4 complexes. The Msx1 homeobox gene, a well-established downstream target of BMP signaling, is expressed in developing facial primordia, particularly at the sites where epithelial-mesenchymal interactions occur during organogenesis, including of the tooth and palate (Zhang et al. 2002). Msx1-gene-deficient mice exhibit cleft secondary palate and an arrest of tooth development at the bud stage (Houzelstein et al. 1997; Satokata and Maas 1994). In the early developing dental mesenchyme, Msx1 is activated by BMP via the atypical canonical BMP (Smad4-independent) signaling pathway (Yang et al. 2014). Our preliminary data also show that the formation of pSmad1/5/8-Smad4 complexes by the overexpression of Smad4 suppresses Msx1 expression in dental mesenchymal cells (unpublished data). This might explain the observation that exogenous BMP4 is able to induce Msx1 expression in the anterior palatal mesenchyme but not in the posterior palatal mesenchyme (Zhang et al. 2002). Thus, in the anterior palate, BMP probably transduces signals through the atypical canonical pathway, which is able to induce Msx1 transcription. However, in the posterior palate, BMP signaling is transduced by the canonical pathway via the pSmad1/5/8-Smad4 complex, which suppresses the Msx1 expression in the posterior palate.
The present investigation thus reveals different regulation mechanisms of BMP signaling in the anterior and posterior portions of the early developing palate. In the anterior palate, an atypical canonical BMP signaling pathway may control palate development, because of the absence of the pSmad1/5/8-Smad4 complex. In the posterior palate, the canonical BMP signaling pathway operates and TGF-β signaling attenuates and modulates canonical BMP signaling activity by competing Smad4.
Acknowledgments
Funding
This study was supported by grants from the National Natural Science Foundation of China (81371105, 81570942 to G.B.Y. and 81470708 to G.H.Y.) and the grants from National Institutes of Health (R01DE024152 and R01DE026482 to Y.P.C.).
Abbreviations
- AP
Anterior-posterior
- BMP
Bone morphogenetic protein
- CNC
Cranial neural crest
- E
Embryonic day
- MAPK
Mitogen-activated protein kinase
- p
Phospho
- PLA
Proximity ligation assay
- R
Receptor-activated
- TAK-1
TGF-β activated kinase 1
- TGF-β
Transforming growth factor-β
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
References
- Alappat SR, Zhang Z, Suzuki K, Zhang X, Liu H, Jiang R, Yamada G, Chen Y (2005) The cellular and molecular etiology of the cleft secondary palate in Fgf10 mutant mice. Dev Biol 277:102–113 [DOI] [PubMed] [Google Scholar]
- Baek JA, Lan Y, Liu H, Maltby KM, Mishina Y, Jiang R (2011) Bmpr1a signaling plays critical roles in palatal shelf growth and palatal bone formation. Dev Biol 350:520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow AJ, Bogardi JP, Ladher R, Francis-West PH (1999) Expression of chick Barx-1 and its differential regulation by FGF-8 and BMP signaling in the maxillary primordia. Dev Dyn 214:291–302 [DOI] [PubMed] [Google Scholar]
- Bin S, Li HD, Xu YB, Qi SH, Li TA, Liu XS, Tang JM, Xie JL (2013) BMP-7 attenuates TGF-β1-induced fibroblast-like differentiation of rat dermal papilla cells. Wound Repair Regen 21:275–281 [DOI] [PubMed] [Google Scholar]
- Avon Bubnoff, Peiffer DA Blitz IL, Hayata T, Ogata S, Zeng Q, Trunnell M, Cho KW (2005) Phylogenetic footprinting and genome scanning identify vertebrate BMP response elements and new target genes. Dev Biol 281:210–226 [DOI] [PubMed] [Google Scholar]
- Bush JO, Jiang R (2012) Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development 139:231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ (2004) Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development 131:3501–3512 [DOI] [PubMed] [Google Scholar]
- Chytil A, Magnuson MA, Wright CV, Moses HL (2002) Conditional inactivation of the TGF-beta type II receptor using Cre:lox. Genesis 32:73–75 [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP (1998) Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol 8: 1323–1326 [DOI] [PubMed] [Google Scholar]
- Grönroos E, Kingston IJ, Ramachandran A, Randall RA, Vizán P, Hill CS (2012) Transforming growth factor β inhibits bone morphogenetic protein-induced transcription through novel phosphorylated Smad1/5-Smad3 complexes. Mol Cell Biol 32:2904–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grygielko ET, Martin WM, Tweed C, Thornton P, Harling J, Brooks DP, Laping NJ (2005) Inhibition of gene markers of fibrosis with a novel inhibitor of transforming growth factor-beta type I receptor kinase in puromycin-induced nephritis. J Pharmacol Exp Ther 313:943–951 [DOI] [PubMed] [Google Scholar]
- He F, Xiong W, Wang Y, Matsui M, Yu X, Chai Y, Klingensmith J, Chen Y (2010) Modulation of bmp signaling by noggin is required for the maintenance of palatal epithelial integrity during palatogenesis. Dev Biol 347:109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP (2005) Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J Anat 207:655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzelstein D, Cohen A, Buckingham ME, Robert B (1997) Insertional mutation of the mouse Msx1 homeobox gene by an nlacZ reporter gene. Mech Dev 65:123–133 [DOI] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P Jr, Nakajima A, Shuler CF, Moses HL, Chai Y (2003) Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development 130:5269–5280 [DOI] [PubMed] [Google Scholar]
- Javier AL, Doan LT, Luong M, Reyes de Mochel NS, Sun A, Monuki ES, Cho KW (2012) Bmp indicator mice reveal dynamic regulation of transcriptional response. PLoS One 7:e42566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J (1995) Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelialmesenchymal interaction. Nat Genet 11:415–421 [DOI] [PubMed] [Google Scholar]
- Katagiri T, Imada M, Yanai T, Suda T, Takahashi N, Kamijo R (2002) Identification of a bmp-responsive element in Id1, the gene for inhibition of myogenesis. Genes Cells 7:949–960 [DOI] [PubMed] [Google Scholar]
- Li L, Lin M, Wang Y, Cserjesi P, Chen Z, Chen Y (2011) Bmpr1a is required in mesenchymal tissue and has limited redundant function with BmprIb in tooth and palate development. Dev Biol 349:451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ding J (2007) Gene expression analysis reveals that formation of the mouse anterior secondary palate involves recruitment of cells from the posterior side. Int J Dev Biol 51:167–172 [DOI] [PubMed] [Google Scholar]
- Liu HH, Xie M, Schneider MD, Chen ZJ (2006) Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci U S A 103:11677–11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF (2005) Distinct functions for bmp signaling in lip and palate fusion in mice. Development 132:1453–1461 [DOI] [PubMed] [Google Scholar]
- Liu W, Lan Y, Pauws E, Meester-Smoor MA, Stanier P, Zwarthoff EC, Jiang R (2008) The Mn1 transcription factor acts upstream of Tbx22 and preferentially regulates posterior palate growth in mice. Development 135:3959–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie XG (2005) Differential expression of Bmp2, Bmp4 and Bmp3 in embryonic development of mouse anterior and posterior palate. Chin Med J 118:1710–1716 [PubMed] [Google Scholar]
- Parada C, Chai Y (2012) Roles of bmp signaling pathway in lip and palate development. Front Oral Biol 16:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal R, Huang J, Dattilo LK, Kaartinen V, Mishina Y, Deng CX, Umans L, Zwijsen A, Roberts AB, Beebe DC (2009) The type I BMP receptors, Bmpr1a and Acvr1, activate multiple signaling pathways to regulate lens formation. Dev Biol 335:305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DP (2004) Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest 113:1692–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I, Maas R (1994) Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet 6: 348–356 [DOI] [PubMed] [Google Scholar]
- Song Z, Liu C, Iwata J, Gu S, Suzuki A, Sun C, He W, Shu R, Li L, Chai Y, Chen Y (2013) Mice with Tak1 deficiency in neural crest lineage exhibit cleft palate associated with abnormal tongue development. J Biol Chem 288:10440–10450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Weng T, Zhang J, Wang J, Li W, Wan H, Lan Y, Cheng X, Hou N, Liu H, Ding J, Lin F, Yang R, Gao X, Chen D, Yang X (2007) Smad4 is required for maintaining normal murine postnatal bone homeostasis. J Cell Sci 120:2162–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KK, Kwan KM (2013) Common partner Smad-independent canonical bone morphogenetic protein signaling in the specification process of the anterior rhombic lip during cerebellum development. Mol Cell Biol 33:1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sun A, Li L, Zhao G, Jia J, Wang K, Ge J, Zou Y (2012) Up-regulation of BMP-2 antagonizes TGF TGF-β1/ROCK-enhanced cardiac fibrotic signalling through activation of Smurf1/Smad6 complex. J Cell Mol Med 16:2301–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K (1995) Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science 270:2008–2011 [DOI] [PubMed] [Google Scholar]
- Yang G, Yuan G, Ye W, Cho KW, Chen Y (2014) An atypical canonical bone morphogenetic protein (BMP) signaling pathway regulates Msh homeobox 1 (Msx1) expression during odontogenesis. J Biol Chem 289:31492–31502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao LC, Blitz IL, Peiffer DA, Phin S, Wang Y, Ogata S, Cho KW, Arora K, Warrior R (2006) Schnurri transcription factors from Drosophila and vertebrates can mediate bmp signaling through a phylogenetically conserved mechanism. Development 133:4025–4034 [DOI] [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y, Chen Y (2005) Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development 132:4397–4406 [DOI] [PubMed] [Google Scholar]
- Zhang YE (2009) Non-Smad pathways in TGF-beta signaling. Cell Res 19:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y (2002) Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development 129:4135–4146 [DOI] [PubMed] [Google Scholar]


