Abstract
Self-assembly of amelogenin plays a key role in controlling enamel biomineralization. Recently, we generated self-aligning nanoribbons of amelogenin in water-in-oil emulsions stabilized by the full-length protein (rH174). Here, we tested the hypothesis that the hydrophilic C-terminus is critical for self-assembly of amelogenin into nanoribbons. The self-assembled structures of two amelogenin cleavage products, rH163 and rH146, were compared with structures of rH174 at different pH values and degrees of saturation using atomic force microscopy, electron microscopy, and dynamic light scattering. We observed that the number density of rH174 nanoribbons increased significantly when the initial pH was raised from 4.5 to 5.6. Nanoribbons, as well as unique helical nanostructures, were also readily observed when amelogenin rH146 was used, but showed little tendency for parallel alignment and did not bundle into fibrils like rH174. In contrast, rH163 rarely formed nanoribbons but predominantly assembled into nanospheres under the same conditions. We conclude that the presence of a hydrophilic C-terminus may not be a prerequisite for nanoribbon formation but may be critical for ribbon alignment and subsequent fibril formation. These results highlight the contribution of the hydrophobic domain in the self-assembly of elongated structures of amelogenins. Molecular mechanisms governing these processes based on the formation of reverse micelles are discussed.
Keywords: amelogenin, atomic force microscopy, electron microscopy, nanoribbons, self-assembly
Self-assembly guides exquisite bio-organization during the secretory stage of enamel formation. In vitro, amelogenin proteins, the main component of the enamel matrix, self-assemble into nanospheres and affect nucleation and crystal growth of calcium phosphate mineral (1–4) by molecular mechanisms not well understood. Previous studies on the role of amelogenin self-assembly in enamel formation (4, 5) concluded that the formation of amelogenin nanospheres was dominated by hydro-phobic driving forces (6, 7). Severe aggregation of the protein is prevented through the formation of a hydro-phobic central core of about 20–40 amelogenin molecules, exposing the hydrophilic C-terminus to the surface of the nanosphere (7). Other studies reported on the prior formation of nanospheres as a nucleator of chain-like assemblies (4, 6–9), which aligned themselves parallel to the C-axis of fluoroapatite FAP crystals (10). Amelogenin lacking the hydrophilic C-terminus showed abnormal nanosphere aggregation (7) and a low tendency to form chain-like structures (11). In contrast, our recent studies showed that the full-length amelogenin protein can form nanoribbons of specific width without prior nanosphere formation. Nanoribbons formed in metastable water–oil emulsions with the participation of calcium and phosphate ions. This finding supported the role of calcium ions as an initiator for amelogenin self-assembly (12–14) that may be key for the formation of more complex assemblies of amelogenin and its interaction with hydroxyapatite (HA) crystals during enamel formation. In the developing enamel, full-length amelogenin has been reported to be associated with the forming mineral (15).
The amino acid sequence of amelogenin is highly conserved among different species (16) and provides the amphiphilic character of this molecule. It is composed of a large, mainly hydrophobic, core of about 160 amino acids and a small hydrophilic and charged C-terminal domain. In our previous work (17), we developed a procedure that allowed the formation of water–oil emulsions that were stabilized by amelogenin. Micro-meter- to nanometer-sized droplets formed and became temporarily stable when rH174 molecules aligned themselves at the water–oil interface (17), leading to the formation of amelogenin reverse micelles, in which the hydrophilic C-terminus of the protein is oriented towards the water compartment of the droplet. The exposure of the hydrophobic core on the surface of the reverse micelles permitted intermolecular interactions between residues in the hydrophobic domain that were not readily accessible in aqueous solutions as a result of the strong tendency of the protein to form nanospheres from a hydrophobic collapse. The use of a water–oil emulsion system ultimately resulted in the supramolecular assembly of amelogenin nanoribbons of 16.7 nm in width and several micrometers in length (17). The nanoribbons had a strong tendency to align themselves and to form bundles of several hundreds of nanometers in width and several micrometers in length.
The aim of this study was to elucidate the role of the hydrophilic C-terminus of the protein in self-assembly at the water–oil interface. We compared the controlled modulation and elongation of rH174 assemblies in the water–oil system with the behavior of the amelogenin cleavage products rH163 and rH146, which lack the hydrophilic C-terminus. The putative role of the hydrophilic C-terminus of the protein as a crucial element for the ability of amelogenin proteins to form reverse micelles was tested.
Material and methods
Recombinant human amelogenin rH174 and cleavage products rH163 and rH146 were produced in Escherichia coli and purified according to a procedure previously described (18). The matrix metalloproteinase 20 (MMP20) proteolytic products amelogenin rH163 and rH146 lack 11 and 28 amino acids at the C-terminus, respectively.
Preparation of emulsions
Protein, calcium, and phosphate stock solutions were prepared in double-deionized water using reagent-grade chemicals. All buffer stock solutions were filtered (through a 0.22-μm filter) prior to use. Amelogenin emulsions were prepared from a mixture of ethyl acetate/octanol [3:7, volume by volume (v/v)] and oil/water [4:1 (v/v)], according to a procedure previously described (17). Software developed by Larsen (Ion products and solubility of calcium phosphates. Aarhus, Royal Dental College, 2001) was applied for calculating the degree of saturation (DS). The final concentrations of CaCl2 and KH2PO4 were 33.4 mM and 20.9 mM at pH 4.5 [DS of hydroxyapatite (DSHA) = 3.6]. A final concentration of 0.37 or 3.7 mg ml−1 of protein was used. The pH was measured using a calibrated micro glass-electrode and adjusted to 4.5 by adding small amounts of KOH or HCl. Emulsions were incubated at 37°C for up to 7 d. After 4–7 d, mineralization was induced by raising the pH to 5.6, resulting in an increase of the DSHA to 12.
Atomic force microscopy (AFM), transmission electron microscopy (TEM), and dynamic light scattering (DLS) were used as previously described (14). Atomic force microscopy analysis of the immobilized products was performed in dry conditions using tapping mode. Transmission electron microscopy analysis was performed on protein absorbed onto glow-discharged carbon-coated copper grids (Ted Pella, Redding, CA, USA) negatively stained with a 2% methylamine tungstate solution (NANO-W; Nanoprobes, Yaphank, NY, USA). Specimens were analyzed with a Tecnai 12 transmission electron microscope at 120 kV (FEI, Hillsboro, OR, USA). Dynamic light scattering studies on the water phase of emulsions were carried out on a dynamic light-scattering analyzer (Zetasizer Nano-ZS; Malvern Instruments, Worcestershire, UK). Dynamic light scattering analysis was calibrated using a silica standard and the refractive index of water. Samples (n > 3, including various batches) of 1.5 ml (total volume) were analyzed for particle size in multiple runs, at 37°C. The particle sizes represent hydrodynamic diameters derived from the time-correlation function of the particle number density averaged over 50 acquisitions (10 s each).
Results
Metastable amelogenin water–oil emulsions were prepared as previously reported (17). Upon vortexing the water–oil mixtures, emulsions formed, as indicated by the white appearance. The emulsions gradually phase separated into oil and water phases, forming a whitish interface in which protein accumulated. The time to phase separate differed for each protein type. Emulsions containing 0.37 mg ml−1 of rH174 at pH 4.5 separated visibly within 24–36 h, while rH163 and rH146 emulsions were less stable and separated in 1 or 12 h, respectively. Use of amelogenin concentrations of 3.7 mg ml−1 almost doubled the time to phase separate.
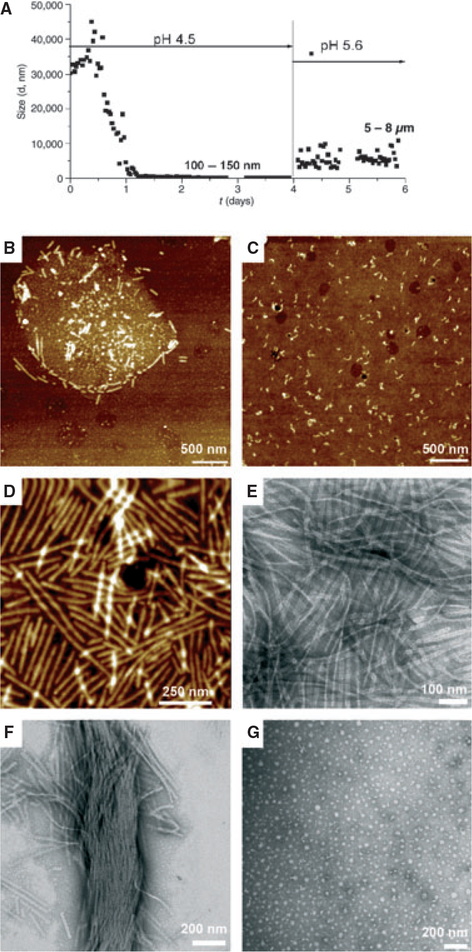
Particle size analysis of the water portion of the emulsion formed by rH174 at pH 4.5 (DSHA = 3.7) was carried out by DLS over a period of 6 d (Fig. 1A). Initially, large particles, in the range of > 10 lm, were detected. The particle size decreased rapidly over time, reaching about 5 μm in 12 h and plateaued at 100–150 nm in 1–2 d. After day 4, samples were treated with mineralizing conditions by raising the pH to 5.6. As the pH was raised, the formation of a white precipitate, presumably calcium phosphate mineral, was observed. The pH measured at day 6 decreased by an average of 0.5 units, presumably as a result of apatite formation. The particle size showed a large increase, to 5–8 μm, and remained in this range for the rest of the experiment. Samples were extracted at different time points and analyzed. Atomic force microscopy analysis of the emulsion at time zero revealed the presence of sphere-like particles (1–2 μm in diameter), comprising spherical structures of about 30 nm (Fig. 1B). High-resolution AFM of these structures showed specific accumulation of short and long ribbon-like structures running along the micelle circumference in an emulsion at 5 min. When the solution was completely phase separated, short, ribbon-like structures of 100–200 nm were observed in the water portion (Fig. 1C). The length and the number of ribbons increased abruptly when the pH was raised to 5.6 (Fig. 1D,E). Ribbons were oriented in a parallel manner and often formed bundles (Fig. 1F). Atomic force microscopy analysis from flat areas coated with homogeneous distributions of nanoribbons showed a length of up to 500 nm. The average ribbon width of 17 nm, measured by TEM, was consistent over incubation time, different pH values or different water–oil ratios (17). Self-assembly experiments carried out in the absence of calcium showed characteristic nanospheres of full-length amelogenin (Fig. 1G).
Fig. 1.
Amelogenin rH174: time-course of assembly and ribbon elongation at the water–oil interface as a function of pH and the water–oil ratio. Metastable water-in-oil emulsions of amelogenin (0.37 mg ml−1) were prepared by mixing with octanol/ethyl acetate (water/oil ratio = 1:4) in the presence of high concentrations of calcium and phosphate ions at pH 4.5 [degree of saturation of hydroxyapatite (DSHA) = 3.7]. (A) Particle size analysis by time-resolved dynamic light scattering. The particle size decreased over time from micrometer-size to nanometer-size particles of 100–150 nm. At day 4, the pH was increased to 5.6 (DSHA = 11–12) and the particle size increased to 5–8 μm. Atomic force microscopy topographic images (tapping mode) and transmission electron microscopy images of supramolecular amelogenin microstructures and nanostructures were obtained. Micelle structures and ribbon-like structures were observed within the first minute of emulsion preparation and immobilization on a glass substrate (B); short ribbons appeared in the water phase after 4 d (C). Abundant elongated amelogenin nanoribbons (D, E) and bundles (F) formed at pH 5.6. In the absence of calcium, amelogenin assembled into characteristic 20-nm nanospheres (G).
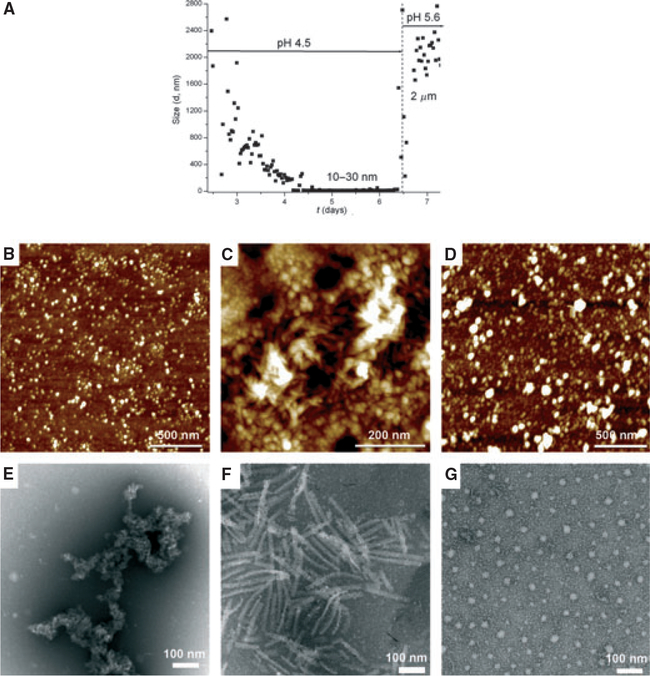
Amelogenin rH163, lacking 11 C-terminal amino acids, formed emulsions that were very unstable and phase separated within 1 h. The particle size (Fig. 2A) decreased slowly over time, taking 4–5 d to reach a plateau of 10–30 nm in diameter. Atomic force microscopy and TEM analysis showed the presence of nano-spheres or aggregates of 40–50 nm in size at day 6 (Fig. 2B,E). These nanostructures remained largely unaltered after the pH was raised to 5.6. In general, nanoribbons were not observed for rH163. Only when using high concentrations of rH163 (3.7 mg ml−1) were scarce numbers of short ribbons, of about 100–200 nm in length, observed at pH 5.6 (Fig. 2C,F). BSA was selected as a control protein to eventually demonstrate the specificity of the protein self-assembly process occurring through the water–oil system. In this case, spherical particles (25–40 nm) co-existed with aggregates of about 70 nm (Fig. 2D,G). Spherical structural motifs of BSA remained unaltered independently of the pH, incubation time, or when a water-only system was used.
Fig. 2.
Amelogenin rH163. Metastable water-in-oil emulsions of amelogenin (0.37 mg ml−1) were prepared by mixing protein suspension in the presence of high concentrations of calcium and phosphate ions at pH 4.5 [degree of saturation of hydroxyapatite (DSHA) = 3.7] with octanol/ethyl acetate. (A) Particle size analysis by time-resolved dynamic light scattering (DLS): the particle size decreased over time from micrometer-size particles to particles of 10–30 nm. The pH was raised to 5.6 (DS = 12) inducing particles to increase in size to around 2 μm. Atomic force microscopy topographic images (tapping mode) and transmission electron microscopy images of amelogenin nanostructures formed under these conditions were recorded. (B, E) Nanospheres of 40–50 nm or aggregates were observed after 6 d of incubation. (C, F) Short nanostrings (100–200 nm long and about 20 nm wide) formed at pH 5.6. BSA was used as a control. BSA samples, collected after the pH was increased to 5.6 (D, G), showed spherical particles of 25–40 nm and large particles or clusters of 50–70 nm.
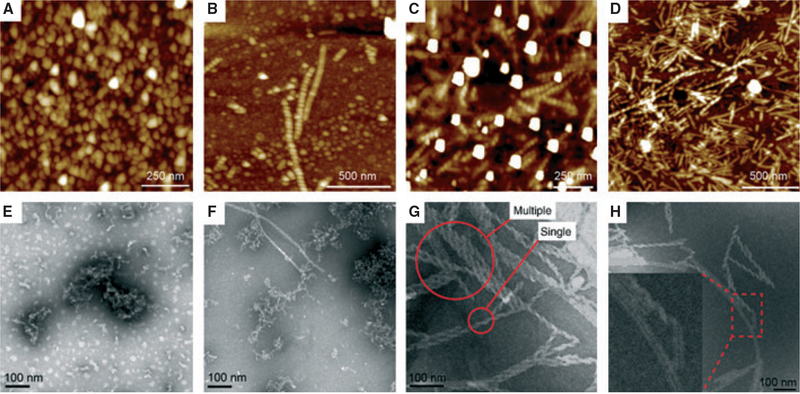
Amelogenin rH146 formed emulsions at pH 4.5, which took at least 12 h to completely phase separate. Atomic force microscopy and TEM analysis showed the formation predominantly of nanospheres with diameters of between 10 and 50 nm in the first 24 h (Fig. 3A,E). After 7 d at pH 4.5, protein assemblies formed and nanoribbons co-existed with a very low number of helices or twisted ribbons (Fig. 3B,F). When the pH was increased to 5.6 at day 7, nanohelices and nanoribbons became abundant (Fig. 3C,G). Atomic force microscopy and TEM specimens showed the presence of numerous helices mixed with nanoribbons without alignment (Fig. 3 D,H). Occasionally, two helices would align and form pairs. The nanoribbons formed by rH146 were about 17 nm wide and approximately the same width as ribbons from rH174 (Fig. 1D,E). Large particles of 100 nm average diameters, possibly apatite crystals, located close to rH146 helices, were also observed (Fig. 3A). The periodicity of the helices varied among different strings, and a mixture of left-handed and right-handed spirals was observed. Additionally, some ribbons appear partially twisted in some images (Fig. 3H inset), suggesting a two-step process: ribbon formation followed by twisting.
Fig. 3.
Amelogenin rH146: metastable water–oil emulsions of amelogenin (3.7 mg ml−1) were prepared by mixing with octanol/ethyl acetate (water/oil ratio = 1:4) in the presence of high concentrations of calcium and phosphate ions at pH 4.5 [degree of saturation of hydroxyapatite (DSHA) = 3.7]. On days 6–7, the pH was increased to 5.6 (DSHA = 11–12). Atomic force microscopy topographic images (A–D) and the corresponding transmission electron microscopy images (E–H) of amelogenin nanostructures. At day 1, at an initial pH of 4.5, nanosphere particles of 30–40 nm and aggregates are observed (A, E). At day 7 and at a pH of 4.5, nanospheres and aggregates co-existed with a very low number of helices ranging from 100 to 500 nm (B, F). At pH 5.6, numerous helices of different lengths (300–500 nm), and of randomly organization, appeared (C, D, G, H). Samples consisted of mixtures of flat ribbons and helices.
Discussion
In this study we validated the methodology for ribbon formation and elongation of the full-length amelogenin protein rH174 at the water–oil interface, as reported earlier (17). Owing to its amphiphilic character, amelogenin was able to play the role of a surfactant in this water–oil system and temporarily stabilized water droplets in oil. We proposed that interactions of oriented amelogenin molecules at the water–oil interface resulted in dimerization of amelogenin molecules, which subsequently self-assembled into nanoribbons in the presence of calcium and phosphate, while the emulsion phase separated (17). Raising the pH and simultaneously inducing mineralization resulted in a very significant increase in the numbers of ribbons of rH174 and rH146, suggesting a cooperative interaction between these two components (Fig. 1D,E) (19). While calcium and phosphate ions are present in the assembled structures, as shown previously by Energy Dispersive X-ray espectroscopy, EDX (17), we were not able to identify a solid mineral phase directly associated with the protein structures.
Much has been reported on the role of the hydrophilic C-terminus in controlling amelogenin assembly and protein–mineral interactions (20, 21). Based on Small angel X-ray scattering, SAXS and DLS, a model was proposed in which the N-terminal and central domains of amelogenin molecules would form a dense core, while the hydrophilic C-terminus would be exposed to the aqueous environment (7). On the contrary, during reverse micelle formation, the hydrophilic C-terminus of rH174 is oriented towards the inside of the micelle, while the hydrophobic portions of rH174 are exposed to the oil phase. Hence, when micelles approach each other, intermolecular interactions, such as hydrogen bonds, van der Waals forces, or electrostatic forces between the hydrophobic portions, can form (17). Considering the requirements for the formation of reverse micelles, cleavage products of amelogenin, rH163, and rH146, lacking the hydrophilic C-terminus, were initially considered as negative controls. While the current conditions of the water–oil system did not allow for rH163 to form somewhat stable emulsions, the addition of rH146 allowed for temporary emulsification, and nanoribbons and nanohelices were readily observed (Fig. 3).
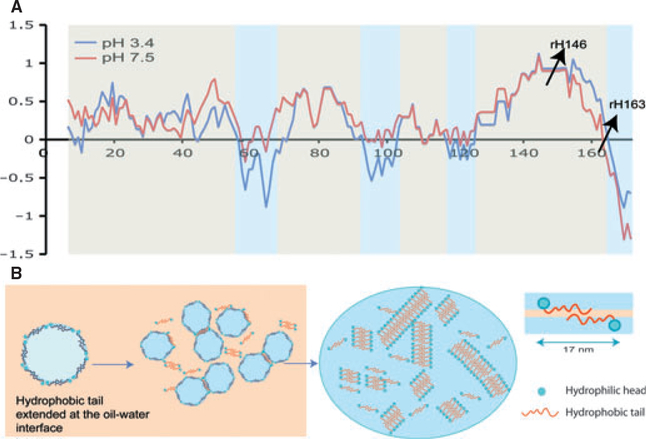
The current view of considering amelogenin as a hydrophobic protein may not be correct when the protein is exposed to an acidic environment. Amelogenin is rich in histidine, which has a pKa of 6.0, and other acidic residues, for example glutamic acid and aspartic acid, have a pKa of around 4. Figure 4A, shows a hydrophilicity plot of rH174 at pH 7.4 and pH 3.4. With a decrease in pH, the protein becomes more charged, mainly by protonation of histidine residues, and several domains of the protein become hydrophilic. Our self-assembly experiments are initially performed at pH 4.5. We hypothesize that at pH 4.5, hydrophilic portions, which are present, not only at the C-terminus but also around residues 60, 95, and 120, will position themselves towards the water phase. This leads to an alignment of the amelogenin molecules along the water–oil interface following the circumference of the micelles (Fig. 4B). This alignment is also supported by AFM in Fig. 1B and appears to be a prerequisite for amelogenin self-assembly into nanoribbons because it prevents a hydrophobic collapse upon pH increase and allows for intermolecular bonds to form in the hydrophobic portion of amelogenin, as suggested in Fig. 4B. It could be said that this hypothesis does not fit completely with amelogenin rH163, which did not readily form elongated supramolecular structures. Additionally, as illustrated by the inability of the rH163 molecule to stabilize an emulsion, we attribute the lack of formation of elongated nano-structures to the water–oil conditions in the study, which do not promote intermolecular interaction between rH163 molecules.
Fig. 4.
Model for amelogenin ribbons formed at the oil/water interface. (A) Hydrophobicity plots for full-length amelogenin molecule at pH 3.4 and pH 7.4 were obtained from the Cowan hydrophobicity scale using the ExPASy server http://web.expasy.org/ protscale. Normalized values show alternate patterns of hydrophobic (+) and hydrophilic (−) domains across the molecule. Cleavage sites for rH163 and rH146 are indicated with arrows. (B) Based on the alternate hydrophilic-phobic domain configuration in the amelogenin molecule, the model proposes that during reverse micelle formation the protein will extend itself along the micelle circumference, facing out hydrophobic domains, while hydrophilic domains will orient themselves towards the water phase. Amelogenin dimers form when micelles approach each other through intermolecular interactions and formation of secondary bonds along the core and the N-terminus of the molecules. Self-assembly is initiated when dimers interact with each other to form ribbons. This reaction only occurs in the presence of both calcium and phosphate ions.
The formation of helices and twisted ribbons by rH146 leads to a change in our model of amelogenin self-assembly at the water–oil interface, as our initial hypothesis required a bipolar or amphiphilic nature of the molecule for the formation of reverse micelles. As shown in our model in Fig. 4, we hypothesize that the molecule over its entire length stretches along the water–oil interface, with the hydrophilic domains, which develop under acidic conditions, oriented towards the water phase. Similarly to our previous model described by He et al. (17), the alignment of amelogenin along the interface prevents the molecule from hydrophobic collapse and facilitates intermolecular interaction along the entire backbone of the molecule, which drive the self-assembly into nanoribbons.
Indeed, self-assembly that leads to the formation of flat, twisted, and helical ribbons has been reported for amphiphilic peptides or proteins by others (22–25). In these studies, the pH played a critical role in determining the structure that evolved. An example of similar self-assembly behavior resembling some of the findings of this study is an investigation on bolaamphiphiles by cryo-TEM. Depending on the length of the hydrophobic spacer, bolaamphiphile self-assembled into nanofibres or helical nanoribbons at pH 3, while nanoparticles or vesicles developed at pH 12 (22).
For this study, and in general for amelogenin molecules, the challenge is to prevent the hydrophobic domains from collapsing to allow ribbon formation. We hypothesize that as a critical first step, alignment of the amelogenin molecule at the water–oil interface is required to prevent a hydrophobic collapse and to facilitate hydrophobic interactions between amelogenin molecules that stabilize the N termini. Second, intermolecular bonds form, possibly among the numerous pro-line and Glc residues within the hydrophobic portion of the protein. Third, a self-assembly process is triggered, possibly by the formation of bridges between amelogenin dimers through calcium and phosphate ions, leading to the formation of nanoribbons.
Surface charges are greatly diminished at the isoelectric point of amelogenin but strongly increase in the acidic and basic pH regions (26). Increased surface charges prevent aggregation of the protein and lower the average particle size when measured by DLS (27). An acidic pH value of 4.5 has been shown to enhance self-assembly of amelogenin into ribbons, indicating that electrostatic repulsion is required initially to stabilize an extended hydrophobic core of the molecule. Nanoribbons formed at this pH, but the numbers of ribbons drastically increased when the pH was raised to 5.6, suggesting that surface charges need to be reduced to permit self-assembly. The role of pH in secretory pathways plays a critical role in the post-translational modification and folding of proteins (28). Proteins destined for secretion are initially synthesized in the endoplasmic reticulum, at a pH of about 7.2, then undergo further post-translational modifications in the Golgi and trans-Golgi network at pH 6, and finally proteins are delivered or accumulated in secretory granules at a pH of 5.7–5.2 (28). Although the pH of the secretory pathway for ameloblasts has not been reported, there is no reason to assume that proteins differs much from other cell’s pathways. Hence our in vitro biomimetic approach may not differ much from the in vivo conditions where amelogenin assembly could occur at pH values of between 4.5 and 6 after exocytosis through vesicles.
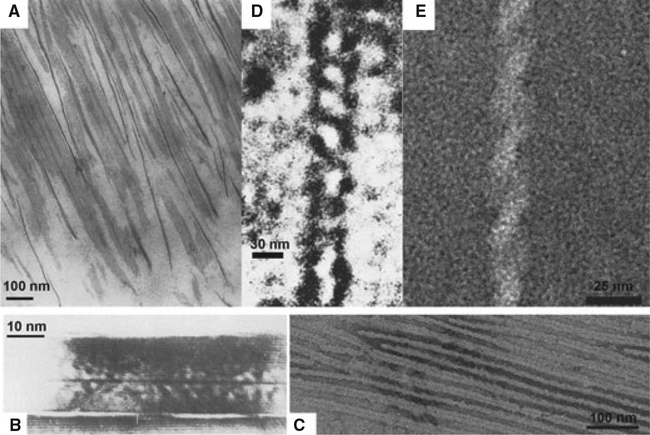
Further supporting the biological relevance of the amelogenin supramolecular structures developed by our water–oil emulsion system, are earlier studies in developing enamel that reported remarkably similar amelogenin ribbons and helical structures. Travis & Glimcher (29) described the organic matrix of developing bovine enamel in the following way: “The organic matrix in decalcified sections of enamel is strikingly similar in its over-all organization to that of the fully mineralized tissue. When viewed in longitudinal prism profiles, the intraprismatic organic matrix is composed of relatively thin dense lines, approximately 48Å wide, which are relatively parallel to each other and have their fiber axes parallel to the long axes of the prisms within which they are located. Many of these dense lines, which have the appearance of thin filaments, are organized into doublets…” (Fig. 5A). The TEM images of ribbons observed in that study are juxtapositioned to ribbons observed in this study in Fig. 5C.
Fig. 5.
Amelogenin ribbons and helices observed in developing enamel and reported in the literature in comparison with the structures observed in this study. (A) Matrix of developing bovine enamel showed ribbon-like structures [Travis & Glimcher (29); reprint with permission of J Cell Biol]; (B) Marshall’s crushed enamel-milled crystallites from human mature teeth exhibiting a dark central line [Marshall & Lawless (33); reprint with permission of J Dent Res]; (C) Amelogenin rH174 ribbons from this study; (D) Smales’s helical structures found in developing matrix of rat enamel [Smales (34); reprint with permission of Nature]; (E) Characteristic amelogenin rH146 helices from this study.
In 1981, Marshall et al., in agreement with previous observations (30–32), published a TEM study of the central dark line in enamel crystallites from human mature teeth (33) (Fig. 5B) that may correspond to original ribbon-like organic templates upon which apatite crystals grew. Furthermore, Smales found helical structures (Fig. 5D), with widths of 15–30 nm and variable pitches, from the developing matrix of rat enamel, which res-embed the helical structures of rH146 observed in this study (Fig. 5E) (34). Based on TEM observations, Smales proposed the idea that helices are an integral part of the enamel structure, at least during some stages in enamel development.
The aforementioned ribbons and helices observed by others in developing enamel support our in vitro results and the hypothesis that complex supramolecular amelogenin assemblies as ribbons or helices may be participating to some extent in enamel development. These results add new insights into amelogenin biology that may lead us to decode how nature produces a highly functional mineralized tissue such as dental enamel. Ongoing self-assembly and template mineralization studies in aqueous medium will provide further insights into the roles of the different segments and their important implications for enamel development.
Acknowledgements –
The authors wish to thank Dr Wu Li at UCSF for the support in protein synthesis and purification. These studies were supported by NIH/NIDCR grants DERO1–017529 and RO1–017529S2.
Footnotes
Conflicts of interest – The authors declare no conflicts of interest.
References
- 1.Fincham AG, Moradian-Oldak J, Simmer JP. The structural biology of the developing dental enamel matrix. J Struct Biol 1999; 126: 270–299. [DOI] [PubMed] [Google Scholar]
- 2.Moradian-Oldak J Amelogenins: assembly, processing and control of crystal morphology. Matrix Biol 2001; 20: 293–305. [DOI] [PubMed] [Google Scholar]
- 3.Iijima M, Moradian-Oldak J. Control of apatite crystal growth in a fluoride containing amelogenin-rich matrix. Biomaterials 2005; 26: 1595–1603. [DOI] [PubMed] [Google Scholar]
- 4.Beniash E, Simmer JP, Margolis HC. The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J Struct Biol 2005; 149: 182–190. [DOI] [PubMed] [Google Scholar]
- 5.Fincham AG, Moradian-Oldak J, Diekwisch TG, Lyaruu DM, Wright JT, Bringas P Jr, Slavkin HC. Evidence for amelogenin “nanospheres” as functional components of secre-tory-stage enamel matrix. J Struct Biol 1995; 115: 50–59. [DOI] [PubMed] [Google Scholar]
- 6.Aichmayer B, Wiedemann-Bidlack FB, Gilow C, Simmer JP, Yamakoshi Y, Emmerling F, Margolis HC, Fratzl P. Amelogenin nanoparticles in suspension: deviations from spherical shape and pH-dependent aggregation. Biomacromolecules 2010; 11: 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aichmayer B, Margolis HC, Sigel R, Yamakoshi Y, Simmer JP, Fratzl P. The onset of amelogenin nanosphere aggregation studied by small-angle X-ray scattering and dynamic light scattering. J Struct Biol 2005; 151: 239–249. [DOI] [PubMed] [Google Scholar]
- 8.Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. pH triggered self-assembly of native and recombinant amelogenins under physiological pH and temperature in vitro. J Struct Biol 2007; 160: 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science 2005; 307: 1450–1454. Erratum in: Science 2005; 309: 2166. [DOI] [PubMed] [Google Scholar]
- 10.Habelitz S, Kullar A, Marshall SJ, Denbesten PK, Balooch M, Marshall GW, Li W. Amelogenin-guided crystal growth on fluoroapatite glass-ceramics. J Dent Res 2004; 83: 698–702. [DOI] [PubMed] [Google Scholar]
- 11.Kwak SY, Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Litman A, Margolis HC. Role of 20-kDa amelogenin (P148) phosphorylation in calcium phosphate formation in vitro. J Biol Chem 2009; 284: 18972–18979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan Y, Sun Z, Wang R, Abbott C, Moradian-Oldak J. Enamel inspired nanocomposite fabrication through amelogenin supramolecular assembly. Biomaterials 2007; 28: 3034–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchko GW, Tarasevich BJ, Bekhazi J, Snead ML, Shaw WJ. A solution NMR investigation into the early events of amelogenin nanosphere self-assembly initiated with sodium chloride or calcium chloride. Biochemistry 2008; 47: 13215–13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X, Li W, Habelitz S. The cooperative self-assembly of 25 and 23kDa amelogenins. J Struct Biol 2008; 164: 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchida T, Tanabe T, Fukae M, Shimizu M, Yamada M, Miake K, Kobayashi S. Immunochemical and immunohisto-chemical studies, using antisera against porcine 25 kDa amelogenin, 89 kDa enamelin and the 13–17 kDa nonamelogenins, on immature enamel of the pig and rat. Histochemistry 1991; 96: 129–138. [DOI] [PubMed] [Google Scholar]
- 16.Paine ML, White SN, Luo W, Fong H, Sarikaya M, Snead ML. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol 2001; 20: 273–292. [DOI] [PubMed] [Google Scholar]
- 17.He X, Wu S, Martinez-Avila O, Cheng Y, Habelitz S. Self-aligning amelogenin nanoribbons in oil-water system. J Struct Biol 2011; 174: 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Gao C, Yan Y, Denbesten P. X-linked amelogenesis imperfecta may result from decreased formation of tyrosine rich amelogenin peptide (TRAP). Arch Oral Biol 2003; 48: 177–183. [DOI] [PubMed] [Google Scholar]
- 19.Fowler CE, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. Co-operative mineralization and protein self-assembly in amelogenesis: silica mineralization and assembly of recombinant amelogenins in vitro. Eur J Oral Sci 2006; 114: 297–303. [DOI] [PubMed] [Google Scholar]
- 20.Moradian-Oldak J, Paine ML, Lei YP, Fincham AG, Snead ML. Self-assembly properties of recombinant engineered amelogenin proteins analyzed by dynamic light scattering and atomic force microscopy. J Struct Biol 2000; 131: 27–37. [DOI] [PubMed] [Google Scholar]
- 21.Paine ML, Lei YP, Dickerson K, Snead ML. Altered amelogenin self-assembly based on mutations observed in human X-linked amelogenesis imperfecta (AIH1). J Biol Chem 2002; 277: 17112–17116. [DOI] [PubMed] [Google Scholar]
- 22.Wang T, Jiang J, Liu Y, Li Z, Liu M. Hierarchical self-assembly of bolaamphiphiles with a hybrid spacer and L-glutamic acid headgroup: pH- and surface-triggered hydrogels, vesicles, nanofibers, and nanotubes. Langmuir 2010; 26: 18694–18700. [DOI] [PubMed] [Google Scholar]
- 23.Mizuno N, Baxa U, Steven AC. Structural dependence of HET-s amyloid fibril infectivity assessed by cryoelectron microscopy. Proc Natl Acad Sci U S A 2011; 108: 3252–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, Marini DM, Hwang W, Santoso S. Design of nanostructured biological materials through self-assembly of peptides and proteins. Curr Opin Chem Biol 2002; 6: 865–871. [DOI] [PubMed] [Google Scholar]
- 25.Kulkarni VS, Boggs JM, Brown RE. Modulation of nano-tube formation by structural modifications of sphingolipids. Biophys J 1999; 77: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uskokovic V, Castiglione Z, Cubas P, Zhu L, Li W, Habelitz S. Zeta-potential and particle size analysis of human amelogenins. J Dent Res 2010; 89: 149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moradian-Oldak J, Leung W, Fincham AG. Temperature and pH-dependent supramolecular self-assembly of amelogenin molecules: a dynamic light-scattering analysis. J Struct Biol 1998; 122: 320–327. [DOI] [PubMed] [Google Scholar]
- 28.Paroutis P, Touret N, Grinstein S. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology (Bethesda) 2004; 19: 207–215. [DOI] [PubMed] [Google Scholar]
- 29.Travis DF, Glimcher MJ. The structure and organization of, and the relationship between the organic matrix and the inorganic crystals of embryonic bovine enamel. J Cell Biol 1964; 23: 447–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nylen MU, Eanes ED, Omnell KA. Crystal growth in rat enamel. J Cell Biol 1963; 18: 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frazier PD. Adult human enamel: an electron microscopic study of crystallite size and morphology. J Ultrastruct Res 1968; 22: 1–11. [DOI] [PubMed] [Google Scholar]
- 32.Ronnholm E The amelogenesis of human teeth as revealed by electron microscopy. II. The development of the enamel crystallites. J Ultrastruct Res 1962; 6: 249–303. [DOI] [PubMed] [Google Scholar]
- 33.Marshall AF, Lawless KR. TEM study of the central dark line in enamel crystallites. J Dent Res 1981; 60: 1773–1782. [DOI] [PubMed] [Google Scholar]
- 34.Smales FC. Structural subunit in prisms of immature rat enamel. Nature 1975; 258: 772–774. [DOI] [PubMed] [Google Scholar]