Abstract
Alternative splicing of pre-messenger RNA increases genetic diversity and recent studies estimate that most human multi-exon genes are alternatively spliced. If this process is not highly regulated and accurate, it leads to mis-splicing events which may result in proteins with altered function. A growing body of work has implicated mis-splicing events as contributing to a wide range of diseases including cancer, neurodegenerative diseases, and muscular dystrophies. Understanding the mechanisms that cause aberrant splicing events and how this leads to disease is vital for designing effective therapeutic strategies. This review focuses on advances in therapies targeting splicing and highlights the animal models developed to recapitulate disease phenotypes as a model for testing these therapies.
Keywords: RNA splicing, disease, animal models
Disrupted splicing in disease
Our knowledge about pre-mRNA maturation has greatly expanded since the discovery of “split” genes by Richard Roberts and Phillip Sharp in 1977, for which they were jointly awarded the Nobel Prize in 1993 for their independent discoveries [1, 2]. We now know that eukaryotic genes contain a series of exons and introns, the latter of which have to be removed during RNA maturation in order for the mature mRNA to be translated into protein (Box 1). Splicing is a precise process by necessity as mistakes in splicing can also lead to unintended effects causing coding or frameshift mutations. In order to direct splicing effectively with high fidelity, the process relies on signals within the transcript itself, such the splice sites and associated sequences, as well as factors that recognize them to facilitate molecular reorganization, recruitment of cofactors, and catalysis of the reaction. Hence, genetic mutations that lead to sequence alterations within the transcript can create new splice sites or enhancer sequences that lead to the recognition of new exons, also known as cryptic exons, or disrupt splicing sequences required for the recognition of exons that lead to the exclusion of constitutive exons from the transcript. These types of splicing mistakes create aberrant transcripts and contribute to disease. Similarly, defects in splicing machinery can lead to aberrant splicing of multiple transcripts and also contribute to the disease state.
Box 1: Mechanisms of RNA Splicing.
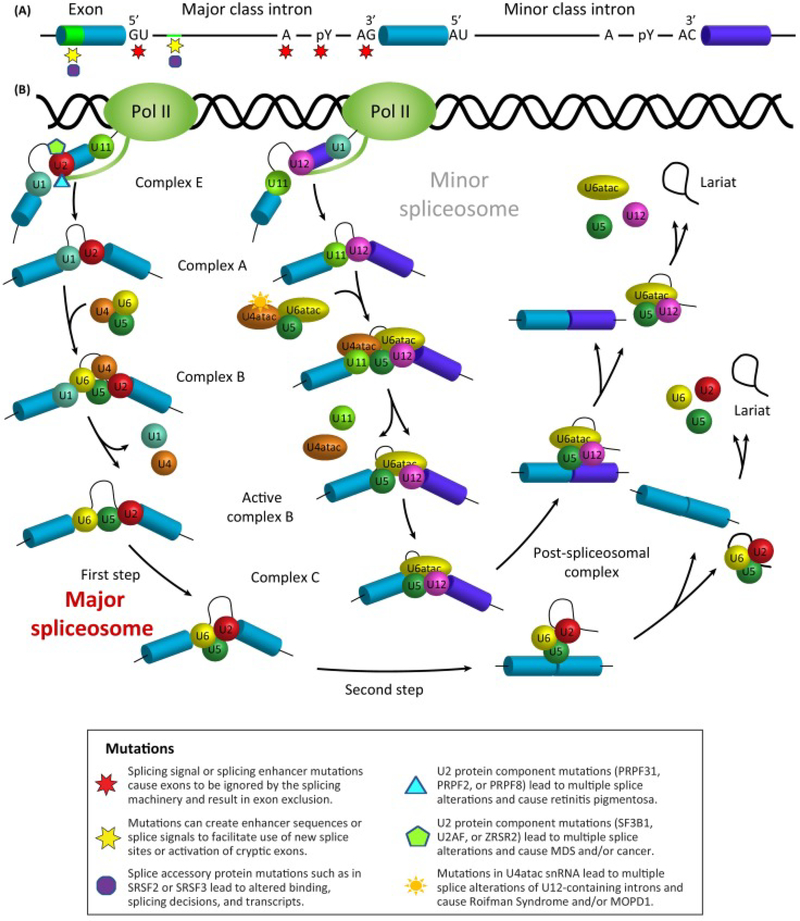
Pre-mRNA splicing involves the removal of regions of the transcript called introns and joining together the exons that will make up the mature RNA transcript. While most of the mature transcript is responsible for coding the resultant protein, some exons are also regulatory in nature making up the untranslated region (UTR) of the mRNA. Splicing is a highly-ordered process performed by a macromolecular machine called the spliceosome that rivals the ribosome in its complexity and sheer number of proteins and RNAs involved [64, 65]. The spliceosomal small nuclear RNAs and proteins or snRNP (small nuclear ribonucleoprotein) complexes are essential for the recognition of splicing sites and the catalysis of the splicing reaction (Fig. 1). The location of splice sites at both the 5’ and 3’ ends of the intron, are demarcated by degenerate canonical sequences, which are recognized by core elements of the spliceosome. The composition of the 5’ splice site, or donor splice site, includes a nearly invariant “GU” dinucleotide sequence along with less conserved residues downstream. In rare cases, a “GC” dinucleotide has been documented to serve as the 5’ splice site when the remaining nucleotides match the “consensus” sequence [66]. The 3’ splice site, or acceptor splice site, is composed of three conserved elements: a branch point, polypyrimidine or Py tract, and an “AG” at the 3’ end. The branch point is typically an adenosine located about 18 to 40 nucleotides from the 3’ splice site, and this is followed by a track of 15–20 polypyrimidine residues, particularly uracil. Atypical branch points also exist that can be more distant from the 3’ss as well as those that utilize a non-adenosine base [67].
The splicing process is accomplished by a two-step trans-esterification reaction which is carried out by either the major spliceosome, for approximately 99% of transcripts, (Figure 1B left, labeled in bold red) or much less frequently by the minor spliceosome (Figure 1B, labeled in gray). The first step is the lariat formation via a nucleophilic attack of the 5’ splice site phosphate group by the 3’ hydroxyl group of the branch point adenosine. In the second step the free hydroxyl of the detached exon attacks the 3’ splice site, leading to the generation of two ligated exons and a lariat intron. [68]. In the first step of spliceosome assembly, the U1 snRNP associates in a pairwise manner with the 5’ splice site, and weakly with the U2 snRNP, forming Complex E [69]. When the U2 snRNP tightly binds the branch point by base-pairing, the complex A is said to be assembled; this is then followed by the recruitment of the tri-snRNP U4/U5/U6, to generate the pre-catalytic complex B. After several conformational rearrangements facilitated by RNA helicases, the complex B adopts a conformation that catalyzes the first of the transesterification reactions. This process generates complex C, which results in the free 5’ exon and lariat intron. Complex C undergoes rearrangement to perform the second step of splicing that allows for the ligation of adjacent exons and release of the lariat intron. At this point the snRNPs are released in an ATP-dependent manner to catalyze additional rounds of splicing [69, 70].
The minor spliceosome works in much the same way; however, the intron boundaries are characterized by divergent “AU” and “AC” dinucleotide sequences at the 5’ and 3’ termini respectively. These sequence variants require U11 and U12 snRNPs for the recognition of splice sites and the subsequent catalysis. Additionally, U4atac and U6atac replace U4 and U6, with U5 as the only snRNP component in common between the two spliceosomes.
Figure 1A. Mutations in the RNA transcript itself can lead to splicing alterations. It should be noted that similar mutations are also possible in the sequences controlling splicing of the minor introns, though not depicted. Figure 1B. Similarly, mutations in core spliceosome machinery of both the major and minor spliceosome have been shown to lead to altered splicing networks. Both of these scenarios can manifest in a disease phenotype.
Further adding to the complexity of splicing is the fact that genes are not always spliced in the same manner; alternative splicing produces different proteins from a single gene (Box 2). It is currently estimated that greater than 95% of human genes are alternatively spliced [3, 4]. While alternative splicing expands the genetic diversity of an organism and presents the opportunity for specific discreet transcripts to be expressed in developmentally or tissue specific ways, there has been a flourish of research defining how inappropriately controlled alternative splicing events can contribute to disease onset and severity. Likewise, there has been new research investigating how alternative splicing events can be targeted for therapeutic development.
Box 2: Alternative splicing.
In addition to constitutive splicing, most eukaryotic cells are able to perform alternative splicing; the process wherein different coding regions of the pre-mRNA are included in the mature mRNA (Fig.2A). Some exons of a gene may be included or excluded from the final transcript. Alternative splicing is one of the leading drivers of proteomic diversity, as one gene can generate many different mRNA messages that translate into several proteins with divergent sizes and/or functions. One of the contributing factors to alternative splicing is the length of introns in higher vertebrates, as long introns have a high likelihood of undergoing alternative splicing [71]. Alternative splicing can occur co-transcriptionally with many of the protein factors that participate in regulated alternative splicing residing on the CTD tail of RNA polymerase II [72].
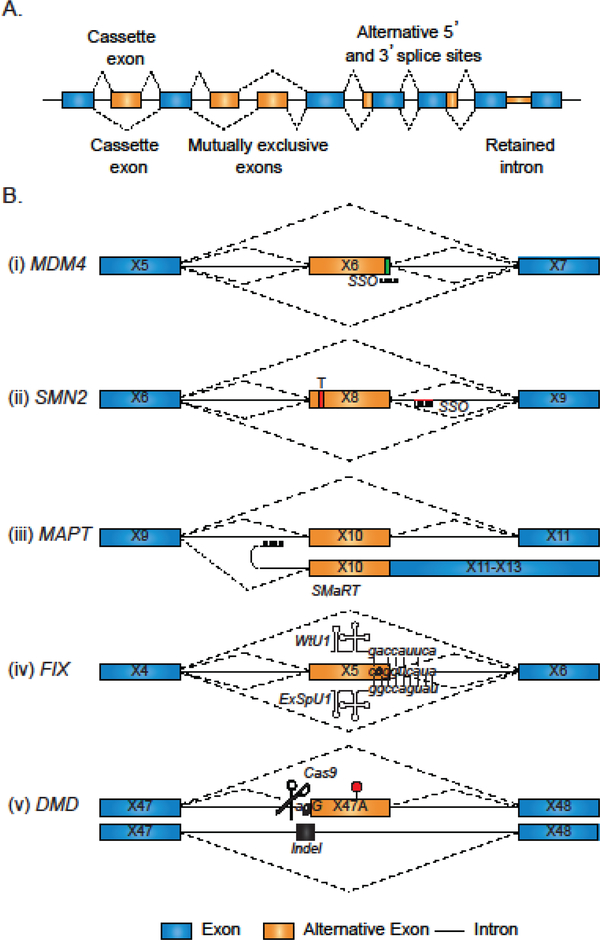
Alternative splicing is a highly regulated process and happens in response to developmental cues and external stimuli, including stress. Moreover, alternative splicing can result in one of several outcomes in splice site selection: cassette exons – the preferential exclusion of an exon under certain conditions; alternative 3’ and 5’ splice site selection – the selection of a distal splice site instead of the normally utilized splice site, mutually exclusive exons – sets of exons that are never included in the same transcript; and intron retention – a normally excluded intron is retained in the final mRNA transcript.
Alternative splicing is regulated by transcriptional elongation, core spliceosomal proteins, RNARNA basepairing, chromatin structure, and well as a host of RNA-binding proteins that interact with splicing elements on the pre-mRNA. Aside from the canonical splicing signals, the 5’ splice site, branch point, and 3’ splice site, there are interactions between cis elements on the pre-mRNA and trans-acting protein factors. These cis-acting elements are known as splicing regulatory elements (SREs), and are generally classified as either being enhancers, which promote the recognition of an exon, or silencers, whose proteins facilitate the masking of splice signals to result in exon exclusion events. SREs can occur both in introns and exons and have been characterized in many organisms from the fly to human. SREs can be classified as an exonic splicing enhancer (ESE), intronic splicing enhancer (ISE), exonic splicing silencer (ESS), or an intronic splicing silencer (ISS).
Numerous avenues have been pursued to accomplish splice alterations for therapeutic benefit (Fig.2B). Examples of splice-altering therapies are: (i) ASOs targeting an exon/intron boundary to induce exon exclusion; (ii) ASOs targeting a splicing silencer to promote exon inclusion; (iii) trans splicing to an exogenous construct to increase full-length splicing; (iv) exonic-specific U1 to improve the recognition of a mutant splice site; (v) CRIPR-Cas9 genome editing to delete a mutant cryptic spice site and restore constitutive splicing. Exons are depicted graphically by boxes, introns by horizontal lines, and splice choices by angled dashed lines. For 2B, reported splicing enhancer elements are depicted by a green box within the exon while splicing silencer elements are depicted by a red line (intronic) or a red box (exonic). Splice choices in the absence of therapeutic intervention are depicted by dashed angled lines above each transcript and the therapeutic intervention below. The dark angled lines are the preferred splicing choice, while the light dashed lines occur less frequently.
This review will highlight the role that animal models mimicking splicing dysregulation have played in both understanding the disease etiology as well as providing a platform for developing and testing novel treatment strategies that target the deleterious splicing event.
Splicing regulation
As alluded to above, differential splicing choices, are regulated through the coordinated interactions between cis elements (splicing regulatory elements-SREs) on the pre-mRNA and trans binding protein factors. The trans-factors are RNA-binding proteins that form multiprotein complexes and facilitate recruitment (for positive regulation) or inhibition (for negative regulation) of the spliceosome components. Though there are numerous types of RNA binding proteins that interface with splicing regulation, the splicing regulatory proteins that are rich in serine/arginine (SR) residues, also known as SR proteins, and the heterogeneous ribonucleoprotein (hRNPs) are two of the most common. Some splice regulatory proteins are tissue specific and help to guide and delineate the tissue specific profiles of the transcripts. The combinatorial binding of these proteins is dynamic and consequently can give rise to various permutations of exons and introns being included or excluded (Box 2). Given the complex regulatory patterns of alternative splicing regulation, there are many functional links between disrupted alternative splicing and disease (Supplemental Table and reviewed in [5]). From mutations in cis SREs on the pre-mRNA, to overexpression or mutation of splicing factors, there are myriad diseases that result from perturbed alternative splicing. In fact, of roughly 80,000 mutations reported in the human gene mutation database, 10% affect splice sites [6, 7]. The number of mutations affecting splicing indirectly by their association with SREs, cis elements, or their associated trans factors has not been tracked and therefore mutations affecting splice choices is much greater than quantitating splice site mutations alone predicts.
Laboratory animals to model and study splicing defects
Scientists first began to associate defects in RNA splicing with disease in the early 1980s with the discovery of a retained intron in patients with β-thalassemia [8] (Box 3). Since then, hundreds of different splicing mutations have been identified as the underlying cause of diseases [9, 10]. With this growing list of diseases related to splicing defects, it is essential to investigate the factors that contribute to mis-splicing events, study how they lead to disease, and devise ways to correct the splicing for therapy. Cellular and animal models have been instrumental in augmenting our understanding of splicing in disease, as well as serving as important tools for pre-clinical therapy testing. From models in Drosophila and zebrafish to mammalian systems such as mice and dogs, many laboratory models have been designed or fortuitously characterized for the purpose of understanding and treating splicing defects (Table 1, Key Table).
Box 3. Evolution of therapies for splicing targets: β-thalassemia.
β-thalassemias are a group of diseases affecting hemoglobin synthesis. There are over 200 identified mutations leading this genetic disorder, with splicing mutations among the most common [73, 74]. Many of the mutations are clear examples of how single nucleotide changes can lead to the activation of what are known as cryptic splice sites, or novel 5’-donor or 3’-aceptor sequences that generate aberrantly spliced transcripts. IVS2–654 β-thalassemia is one of the most common of these splicing mutations, causing a C to T change in the position 654 of the IVS2 intron in the β-hemoglobin gene (HBB) to generate an aberrant 5′-donor splice site at position 652 and activates a cryptic 3′acceptor splice site at position 579. This allows for the aberrant inclusion of an intron and the translation of a non-functional β-globin protein [75]. Expression of this non-functional protein leads to low oxygen levels and a shortage of red blood cells, which can lead to life threatening anemia.
Lewis and colleagues were the first to successfully generate a humanized mouse model of βthalassemia that expresses an aberrant splice variant by carrying common IVSII-654 β-thalassemia splicing mutation [76]. More recently, subsequent models were generated harboring additional splice-disrupting mutations [77, 78]. Importantly, not only were these models useful for gaining further understanding of the molecular mechanisms involved with the disease, but also as a platform for testing splice switching oligonucleotides [79]. Furthermore, these studies have led to new advances for SSO delivery including the lentiviral vectors that stably express U7 snRNAs carrying splice-switching SSO sequences targeted to the cryptic 5’ splice site recently tested in erythroid cells from patients [80].
Genome editing techniques are a new avenue of therapy likewise being applied to the correction of splicing defects in the β-thalassemias. Using CRISPR-Cas9 and a piggyBac transposon Yuet Wai Kan’s lab was able to correct HBB mutations in an induced pluripotent stem cells (iSPCs) derived from a β-thalassemia patient. These corrected clones were differentiated into hematopoietic progenitors expressing a 16-fold increase in HBB [81]. Remarkably, the genome editing therapy yielded no detectable off-target effects. Thus, from its earliest association as a splice-dependent disease, to the development of animal models, and to the testing of splice-correcting therapies, the βthalassemia field has paved the way for understanding splicing defects in disease and to finding therapies to correct them.
Table 1.
Key Table. Commonly used animals to model splicing changes and test splice-altering therapies in disease.
| Disease | Animal Model | Therapies | Outcome | References |
|---|---|---|---|---|
| Alzheimer’s Disease and other Tauopathies | Mouse model expressing N296H mutant Tau isoform | PNA, SMaRT | PNA treatment induces MAPT exon 10 skipping in vitro, Transfection of a MAPT trans-splicing donor plasmid carrying exons 10 to 13 induced exon 10 inclusion in vivo. | [86–88] [89] |
| Knock-in mouse model expressing the Apoer2 gene with deleted exon 19. | SSO | SSO treatment increase inclusion of ApoER2 exon 19 in vitro and in vivo. Treatment improves hippocampal basal synaptic transmission, learning and memory in AD mice. | ||
| Amyotrophic Lateral Sclerosis | Mouse model expressing Q331K mutant TDP-43, | SSO | SSO treatment induces FUS exon 7 skipping in vitro. | [32, 37, 41, 90] |
| Mouse expressing R521C mutant FUS | ||||
| Mouse model expressing a Q101X mutation in the Tardbp gene | ||||
| Autism Spectrum Disorder | Mouse model expressing a nSR100/Srrm4 isoform with deleted exon 7 and 8 | N/A | N/A | [91] |
| Autoimmune Diseases | Knock-in mouse model overexpressing the 1/4Ctla4 splice variant | SSO | Treatment with SSO induced skipping of CTLA4 exon 2 in vitro and in vivo increasing expression of the liCTLA-4 isoform. Induction of the splice isoform delayed the onset of diabetes and decreased its incidence in NOD mice. | [92–94] |
| Breast Cancer | Knock-in mouse model expressing human Δ16HER2, | Small molecule (Dacomitinib) | Dacomitinib treatment completely suppressed Δ16HER2-driven carcinogenesis in vitro and in vivo. In Δ16HER2 mice, treatment completely abolished autochthonous mammary tumors formation. | [95–98] |
| Knock-in mice expressing ERBB4 isoforms CYT-1 and CYT-2 | SSO | SSO treatment induced skipping of ERBB4 exon 26, thus increasing CYT-2 isoform expression in vitro. | ||
| Cystic Fibrosis | Transgenic mouse model carrying a YAC with a mutated CFTR gene in exon 9 (TG12T5 insertion) | SSO, ExSpeU1, CRISPR/Cas9 | Treatment with SSO or an exon-specific U1 snRNA induced correct inclusion of CFTR exon 16 in vitro. Treatment with CRISPR/Cas9 induced skipping of cryptic exons in the CFTR introns 12, 21, and 22 in vitro. | [99–102] |
| Deafness | Mouse model expressing nSR100/Srrm4 isoform with deletion on intron 12 and exon 13 | N/A | N/A | [103] |
| Duchenne Muscular Dystrophy | Multiple models as reviewed in [11]; including a dog model carrying an A>G mutation in the intron 6 of the canine DMD gene | SSO, SMaRT, CRISPR-Cas9 | Transfection of a transsplicing AAV vector containing Dmd intron 22 and exon 23 induced expression of WT protein in vivo and in vitro. SSO treatment induced skipping of Dmd exon 51 producing an in-frame mRNA in vivo. CRISPR/Cas9 induced skipping of exon 51 in the Dmd gene in vivo.
Treatments and restoration of the WT protein induced muscle normalization both structurally and functionally. In patients SSO treatment lowered the incidence of loss of ambulation. |
[11, 12, 15, 84, 104–106] |
| Mouse models carrying a C>T mutation in exon 23 of the Dmd gene or a deletion of the exon 52 | ||||
| Mouse model with the Dmd exon 50 deleted | ||||
| Epilepsy | Knock-out mouse model with a deletion of the exon E8a in the gene Kdm1a/Lsd1 (neuroLSD1 null) | N/A | N/A | [107] |
| Familial dysautonomia | Knock-in mouse model carrying a mutated human IKBKAP gene in intron 20 (T>C) | Small molecule (Kinetin) | Kinetin treatment induces IKBKAP exon 20 inclusion in patients. | [108, 109] |
| Mouse model with Ikbkap knock-out | ||||
| Fukuyama Muscular Dystrophy | Knock-in mouse model carrying a mutant Fktn with exon 10 replaced by a SINEVNTR-Alu-containing human exon 10 | SSO | SSO treatment abolishes inclusion of fukutin aberrant exon 11 in vivo. Treatment restored normal function of Fukutin. | [110, 111] |
| Hemophilia B | Mouse model expressing human Factor 9 with mutant exon 5 | ExSpeU1 | Treatment with an exonspecific U1 snRNA induces F9 exon 5 inclusion in vitro. | [101, 112, 113] |
| Hereditary Myopathy with Lactic Acidosis | Mouse model carrying a G>C mutation in intron 4 of the ISCU gene | SSO | SSO treatment induces skipping of the cryptic ISCU exon 4a in vitro. | [114, 115] |
| Hereditary Tyrosinemia type I | Mouse model carrying a G>A mutation in exon 8 of the Fah gene | CRISPR/Cas9 | CRISPR/Cas9 treatment induced inclusion of exon 8 in Fah in vivo. Treatment ameliorated liver damage of Fahmut/mut mice. | [116] |
| Huntington’s Disease | Knock-in mouse model carrying an insertion of 150 CAG repeats into exon 1 of the HTT gene | SMaRT | Transfection of an HTT exon 1 trans-splicing donor plasmid with 21 CAG repeats abolished exon 1 skipping in vitro. | [117, 118] |
| Hutchinson-Gilford Progeria syndrome | Knock-in mouse model carrying a C>T mutation in exon 11 of the Lmna gene | SSO | Treatment with SSO induced correct inclusion of Lmna exon 11 in vitro. | [119, 120] |
| Hypertrophic Cardiomyopathy | Knock-in mouse model carrying a G>A mutation in exon 6 of the Mybpc3 gene | SMaRT | Transfection of a transsplicing AAV vector containing Mybpc3 exons 1–6 induced inclusion of exon 6 in vivo. Although treatment repaired cMyBP-C protein, it was still not enough to ameliorate the cardiac phenotype in KI mice. | [121, 122] |
| Infantile Hypophosphatasia | Mouse model carrying a T>G mutation in intron 9 of the Alpl gene | N/A | N/A | [123] |
| Leber’s Congenital Amaurosis | Knock-in mouse model carrying a A>G mutation in intron 26 of the CEP290 gene | SSO, CRISPR/Cas9 | SSO and CRISPR/Cas9 treatment induced skipping of a cryptic exon in CEP290 intron 26 in vitro. | [124, 125] |
| Melanoma | Knock-out mouse expressing Mdm4-S (exon 6 deleted) | SSO | Treatment with SSO induced skipping of Mdm4 exon 6 in vitro and in vivo. In patient-derived mouse xenographs, treatment decreased tumor growth and sensitized them to BRAFV600E inhibition. | [126, 127] |
| Muscular dystrophy type 1A | Mouse model carrying a G>A mutation in the intron 2 of the Lama2 gene | CRISPR/Cas9 | Treatment with AAV containing the CRISPR/Cas9 components induced inclusion of exon 8 in the Lama2 gene in vivo. Genome editing of dy2J/dy2J mice showed improved muscle architecture and decreased fibrosis with concomitant improvement on paralysis of the hind limbs and mobility. | [52] |
| Myelodysplastic Syndromes and Acute Myeloid Leukemia | Knock-in mouse model expressing K700E mutant Sf3b1 | Small molecules (E7107, H3B-8800) | Treatment with E7107 resulted in widespread intron retention and reduced leukemic burden in Srsf2 mutant mice. Treatment with H3B-8800 produced splicing modulation and inhibited tumor growth in mouse xenografts. | [21, 22, 24, 128–130] |
| Knock-in mouse model expressing P95H mutant Srsf2 | ||||
| Heterozygous knock-out mouse model with the Hnrnpk gene deleted | ||||
| Mouse model expressing S34F mutant U2AF1 isoform | ||||
| Myotonic Dystrophy type 1 | Knock-in mouse model expressing a mutant HSA gene with 250 CTG repeats in its 3’ UTR | Small molecule (Manumycin A) | Treatment with manumycin A induced inclusion of Clcn1 exon 7a in vivo. | [131, 132] |
| Niemann-Pick Type C disease | Mouse model carrying a G>A mutation in intron 9 of the Npc1 gene | SSO | Treatment with the SSO inhibited inclusion of a cryptic exon in the Npc1 intron 9 in patient cells. | [133, 134] |
| Pompe Disease | Knock-out mouse models with a disrupted/deleted exon 6 on the Gaa gene | SSO | Treatment with the SSO inhibited inclusion of a cryptic exon in the Gaa intron 15 in patient cells. | [135] |
| Prader-Willi syndrome | Knock-out mouse model with the Snord115 gene deleted | SSO | Treatment with the SSO induces HTR2C exon 5b inclusion in vitro an in vivo. In both fasted mice and in genetically altered, hyperphagic mice, treatment reduced food intake. | [136, 137] |
| Prostate Cancer | Knock-in mouse expressing human AR3/AR-V7 | Small Molecule (Niclosamide) | Niclosamide treatment induced AR-V7 proteasomal degradation both in vitro and in vivo. In mouse xenographs, treatment decreased tumor growth of enzalutamideresistant prostate cancer. | [138, 139] |
| Retinitis Pigmentosa | Zebra fish and mouse models expressing mutant Prpf31 | SMaRT | Transfection of a transsplicing AAV vector containing Rho exons 2–5 and an artificial intron 2 induced expression of wild type protein in vivo. Nevertheless, in severely affected transgenic mice, treatment was not sufficient to delay or stop photoreceptor degeneration. | [140–143] |
| Knock-in mice expressing mutant Prpf3 or Prfpf8 | ||||
| Rett Syndrome | Knock-out mouse model with the Mecp2 gene deleted | N/A | N/A | [144] |
| Rhabdomyosarcoma | Transgenic mouse model expressing human MDM2ALT1 | SSO | SSO treatment induced inclusion of MDM2 exons 4 to 11 thus increasing MDM2 full length protein expression in vitro. | [145, 146] |
| Spinal Muscular Atrophy | Multiple models as reviewed in [147]; including a knock-in mouse model carrying a mutated human Smn gene in exon 7 (C>T) | SSO, Small molecules (7-piperazinylcou marins, RG7800), ExSpeU1 | Treatment with SSO, small molecules or an exon-specific U1 snRNA induced retention of SMN2 exon 7 in vivo and patient cells. SSO treatment in mouse studies showed marked improvement in muscle morphology and motor function as well as extended life expectancy. Treatment with SSO RG7800 has entered clinical trials. | [12, 148–153] |
| Heterozygous Smn exon 2 knock-out mice | ||||
| Knock-in mouse model carrying a mutated human Smn gene in exon 7 (TTT insertion) | ||||
| Usher Syndrome | Knock-in mouse model carrying a G>A mutation in exon 3 of the Ush1c gene | SSO | Treatment with SSO induced correct inclusion of Ush1c exon 3 in vitro and in vivo. Transgenic 216AA mice treated with the SSO showed therapeutic correction of deafness. | [154, 155] |
| X-linked agammaglobulinemia | Knock-in mouse model carrying a mutated human BTK gene in intron 4 (A>T) and | SSO | Treatment with SSO induced skipping of a cryptic exon in the BTK intron 3 in patient cells. | [156, 157] |
| Mouse model with a BTK knock-out | ||||
| β-Thalassemias | Knock-in mouse models carrying either a mutated human HBB gene in intron 2 (C>T), in intron 1 (G>A) or (T>C) | PNA, SSO | Treatment with PNA induced skipping of cryptic exon in the HBB intron 2 in vitro. Treatment with ASO induced skipping of cryptic exon in the HBB intron 2 in vivo. In IVS2–654 mice, treatment improved red blood cellmorphology and hemoglobin levels. | [158–160] |
N/A = Not available
The vast majority of animal models were designed and generated by engineering mouse embryonic cells to harbor the splice affecting mutations via homologous recombination techniques. These models allow the splicing defects to be verified in the affected tissues of the animal. Furthermore, the models enable researchers to prove the causative nature of the defect for the designated disease by characterizing the pathology in the mouse and provide an appropriate genetic background for preclinical trials in animals.
Additionally, a few models were identified that occurred naturally in nature. For example, the widely used mdx mouse model of Duchenne Muscular Dystrophy is derived from a mouse line with a naturally occurring nonsense mutation in exon 23 of the mouse dystrophin gene. While there is no splicing defect in the mdx mouse, it is a useful backdrop in which to investigate therapies that can induce removal of the mutated exon by interfering with its splicing (see Table 1 and Box 4). Similarly, multiple dog breeds with a Duchenne-like phenotype have been identified that carry a variety of DMD mutations [11]. Of these, the affected Golden Retriever DMD model, carries an A-to-G nucleotide base change in the intron 6 of the canine DMD gene is now referred to as the GRDMD. The intronic mutation in the GRDMD model causes the exclusion of exon 7 from the DMD transcript and an out-of-frame protein. Colonies of this dog model have been established to allow disease characterization and therapeutic testing in a mammalian system other than mouse. In fact, this model has been utilized to show the efficacy of SSOs for systemic multiexon-skipping therapy ([11] and see below).
Box 4. Splicing induction as therapy in non-splice mediated diseases.
Duchenne muscular dystrophy (DMD) is a genetic disorder characterized by progressive muscle wasting and weakness caused by the loss of a functional dystrophin protein. The disease is caused by hundreds of different known mutations in the DMD gene. These mutations encompass a wide variety of genetic alterations from exon deletions or duplications and even frameshift mutations, with just one thing in common: they all lead to a non-functional dystrophin protein.
One of the ways that researchers have approached the therapeutic design against this disease is taking advantage of alternative splicing and using it to produce a partially functional dystrophin protein. The use of antisense oligonucleotides has been one strategy to do this (Fig. 2B), both by inducing skipping of a single mutated exon [82] and by inducing skipping of multiple exons using a cocktail of SSOs [83]. The goal of these therapies is to delete the exons carrying the deleterious mutations and restore the reading frame of the DMD protein. Although the DMD protein is lacking the sequence encoded by the exon that is induced to be skipped, its function is conserved sufficiently to restore muscle physiology and to alleviate disease symptoms. Using splice-switching oligomers in the clinic, though, has therapeutic limitations related to delivery and molecule stability.
In an effort to design new therapies for DMD, researchers have explored using novel genome editing CRISPR-Cas9 technology to correct DMD functionality by inducing splicing changes to delete “bad” exons. The lab created a new mouse model of DMD by using CRISPR-Cas9 to delete exon 50 of the mouse Dmd gene (ΔEx50), which caused a deleterious frameshift in the gene and resulted in mice with severe muscle dysfunction [84]. The DMD phenotype could be rescued by systemic delivery of adeno-associated virus encoding the CRISPR/Cas9 genome editing components to create reframing mutations and allowed skipping of exon 51 [84]. Armed with this powerful proof of principle, the lab went on to target 12 exons of the human DMD gene, for which reframing mutations and exon skipping could result in therapeutic benefit. They were able to correct multiple different mutations in human engineered heart muscle cells using the CRISPR-Cas9 technique [85].
Controlling splicing for therapeutic benefit
One of the challenges of designing therapies that target alternative splicing is ensuring target specificity. There are tens of thousands of mRNA messages in the cell and modulating the splicing behavior of one particular gene, perhaps in one specific tissue, without having undesirable off-target effects is challenging. To this end, targeting specific transcripts and affecting their RNA processing is emerging as a viable option to ensure target specificity and to minimize potentially deleterious off-target effects (See Figure 2B). In diseases that arise from a splicing defect, a common therapeutic strategy is to use of splice switching oligonucleotides (SSOs) to restore the normal splice variant (Reviewed in [12]). One type of SSO is a single-stranded RNA molecule that binds to complementary SRE targets to occlude binding of a specific RNA binding factor, thereby impacting the levels of splicing. If the SSO is targeting a splice site or enhancer signal, the resultant mature RNA will exclude the exon targeted. In contrast, if the SSO is targeting a splicing silencer, the strategy can be used to force the inclusion of an exon and restore full-length protein expression.
Fig 2.
Alternative Splicing and Therapeutic Strategies.
Small molecule compounds that target splicing factors are another means of changing alternative splicing that have been gaining popularity for treatment of diseases arising from splice defects. Frequently, libraries of compounds are used to screen splicing reporter constructs to discover synthetic and natural compounds that alter splicing of a particular exon. Additionally, some small molecules have shown efficacy in inhibiting splicing in general. Given that mutations in general splicing factors have recently been identified (discussed in more detail below), small molecules affecting the core spliceosome are a viable option for disease treatment. Specifically, Spliceostatin A and pladienolide bind the SF3B spliceosomal protein, which is an essential component of the U2 snRNP [13]. While these compounds do not target splicing of a particular gene, they are useful in disrupting splicing networks and cellular homeostasis in cells that are metabolically active in diseases such as cancer. AIDS, a systemic disease caused by a viral infection, has shown some benefit from small molecule splicing inhibitors is HIV. IDC16, a small indole derivative, was similarly identified in an in vitro screen. IDC16 interferes with exonic splicing enhancers for SRSF1 and suppresses the expression of key viral proteins, inhibiting the replication of several strains, including those that are resistant to viral protease and reverse transcriptase inhibitors [14]. This is an important example highlighting the relevance of splicing inhibition to both infectious and genetic diseases. Finally, another avenue of investigation in correcting splicing mutations is genome editing, where specific mutations can be corrected or circumvented (discussed comprehensively below).
It is important to note that there are multiple instances in which splicing therapies have been designed, tested in animal models, undergone clinical trials and are now FDA approved drugs. The evolution of two of these therapies for the treatment of Spinal Muscular Atrophy (SMA) and Duchenne Muscular Dystrophy (DMD) have been chronicled and discussed thoroughly previously [12, 15]. For both of these diseases, SSO are used to modify splicing, for exon inclusion in the case of SMA, and for exon exclusion for the case of DMD. These two success stories highlight the power of splice altering therapies and the importance of testing them in animal models for real, clinical use. While these and additional therapeutics targeting splicing defects are summarized in Table 1, we also detail the use of animal models in the evolution and testing of some of these therapies in the remainder of the review. In three vignettes described below, we discuss how developing technologies and animal models have impacted the RNA splicing field in the recent past and provide new insights into disease and the therapies to treat them.
Mutations in splicing core components as the source of disease
Given the prevalence of splicing alterations in disease, it is not surprising that recent studies point towards components of the spicing machinery as the basis of numerous diseases. This has been reported in myelodysplastic syndromes (MDS), a group of rare clonal hematopoietic stem cell disorders characterized by ineffective hematopoiesis, which can progress into acute myeloid leukemia (AML), as well as in retinitis pigmentosa, a breakdown and loss retinal cells that can lead to blindness (See supplemental table 1, Key table 1 and [16, 17]).
In MDS and AML, several mutations in splicing factors have been identified; with SRSF2, U2AF and SF3B1 being the most common ones [18]. SF3B1 is mutated in approximately one quarter of MDS patients and is component of the U2 snRNP [19]. Therefore, mutations in the SF3B1 splicing factor affect binding of the U2 snRNP to the branch-point of pre-mRNAs, and consequently, leads to alterations in the splicing of several genes [20]. The A-to-G transition in exon 15 of SF3B1, one of the most common mutations in MDS, leads to the K700E substitution and also affects 3’ splice site selection fidelity. A novel mouse model containing this mutation was recently developed. This mouse develops progressive macrocytic anemia and myelodysplasia and induces increased alternative 3’ splice site selection in several genes. Notably, a subset of the splicing changes seen in this mouse model are also affected in the human condition [21]. Other MDS patient mutations have likewise been modeled in mice. For example, mice with the P95H mutation in splicing factor SRSF2 also replicates the MDS phenotype in mice [22]. Similarly, a mouse model expressing a U2AF S34F mutant splicing factor that showed increased alternative 3’ splice site usage with an increased frequency of myeloid progenitors in both spleen and bone marrow was generated [23]. Each of these models reinforces how important pre-mRNA splicing and its core components are for the development of pathogenesis.
The generation of novel animal models that accurately reflect the molecular mechanisms and phenotype of the disease is a critical step for the development of treatments. For example, in MDS clinical trials treatment with E7107, a pladienolide analog, in patient-derived xenografts (PDX) decreased leukemic burden in mice with spliceosomal gene mutations [24]. This drug was also tested in vivo in the mouse model designed by Obeng and collaborators, where administration of E7107 reduced chimerism after bone marrow transplant of a mixture of Sf3b1 mutant and WT cells [21]. Moreover, this drug entered phase I clinical trial, but it was stopped shortly after due to two reported cases of vision loss [25]. H3B-8000, another modulator of the SF3b complex has recently entered clinical trials after showing antitumor effects in PDX mouse models with tumor cells from patients harboring SF3B1 or SRSF2 mutations [26]. Therefore, splice modulators emerge as a clear alternative in the treatment of diseases arising from mutations in the core splicing components with animal models harboring conserved disease-causing mutations being key to early testing in the pipeline to the clinic.
Phase separation and alternative splicing in neurodegenerative diseases: new players in an old game
Eukaryotic cells are known to be compartmentalized into specific membrane-bound compartments called organelles. However, membrane-less organelles have emerged as important functional compartments whose cellular organization is less well understood. Moreover, these membrane-less organelles are composed of a mix of proteins and nucleic acids. It had become clear recently that these membrane-less organelles are brought together and maintained as a result of phase separation phenomenon happening within the cells. The nucleolus, Cajal bodies, nuclear speckles and nuclear stress granules are all examples of membrane-less organelles, where several splicing factors and ribonucleoprotein complexes and/or miRNAs reside. Hence, it is not surprising that RNA metabolism is intricately influenced by phase separation and the formation, reorganization, or disruption of these membrane-less organelles [27].
Phase separation is facilitated in part by proteins with low complexity domains. When these proteins are mutated it can hinder their ability to coalesce and disrupt the normal physiological liquid-liquid phase separation. When physiological phase separation is disrupted, pathogenic protein aggregates may occur and, in some cases, lead to neurodegenerative disease. Recent studies have shown that some of these aggregating and disease-causing proteins are in fact splicing factors due to low complexity regions commonly found in RNA binding proteins [28, 29]. Simply put, specific mutations in some splicing factors alter the biophysical properties of the protein to contribute to pathological aggregation and potentially lead to their sequestration and functional inhibition. One such example is trans-activating response region DNA binding protein 43 (TDP-43) encoded by the TARDBP gene. More than 40 different mutations have been identified in the C-terminal domain, which can cause TDP43 to form toxic aggregates by phase separation [30, 31]. These aggregates can also alter normal splicing of the cells by promoting inclusion of cassette exons or modifying the usage of mutually exclusive exons [32, 33]. Moreover, this protein has been shown to affect the splicing of another splicing factor, hnRNP A1, by increasing the inclusion of exon 7B, causing it to also form aggregates [34]. Hence, it is still a matter of debate whether the aggregate accumulation in the cytoplasm is toxic itself, or whether the altered splicing is responsible for the disease phenotype [32, 33, 35].
Animal models carrying mutant forms of TDP-43 have been generated that replicate some of the abnormal alternative splicing seen in ALS. For example, mice harboring a mutant Q331K TDP-43 transgene leads to a loss-of-function in direct splicing targets like Kcnip2 or Atp2b1 as well as age-dependent motor neuron loss [36]. Additionally, the Tardbp (Tdp-43) gene was modified to produce a truncated protein by the change of a glutamine to a stop codon in position 110 in another mouse model. These mice exhibited a loss of splicing fidelity in Tdp-43 target genes in the brain as well as functional defects manifesting as impaired hindlimb strength when measured by a classical clasping test and abnormal body tone [37]. These animal models reinforce TDP-43 as a key splicing factor involved in ALS pathogenesis.
The protein FUS (FUsed in Sarcoma), is another example of a splicing factor affected in ALS. Mutations in the SYGQ low-complexity domain situated in the N-terminal end of the protein, such as the G156E, can induce the disruption of the normal liquid-liquid phase separation in favor of protein aggregates and RNP granules [38, 39]. Furthermore, FUS has been shown to interact with the U11 snRNP, a component of the minor spliceosome, thus regulating the splicing of minor intron containing mRNAs. In the case of neurodegenerative diseases like ALS, cytoplasmic aggregates of FUS sequester and trap U11 and U12 snRNAs, affecting minor intron splicing [40]. A mouse model harboring the mutant R521C FUS protein was able to recapitulate the molecular mechanisms affected in ALS related to alternative splicing defects. In these transgenic mice, several genes had increased intron retention compared to the WT mice, especially in genes related to the extracellular matrix such as collagens and cadherins, which are key in neurite outgrowth and synapsis specificity [41].
Alzheimer’s disease (AD) is another neurodegenerative pathology where the abnormal liquid-liquid phase separation phenomenon is involved; in this case, the microtubule-associated protein Tau forms aggregates. While Tau is not a splicing factor itself, in this disease (and other neurodegenerative diseases categorized as tauopathies) the abnormal alternative splicing of Microtubule Associated Protein Tau (MAPT) gene is what causes an imbalance between Tau splice isoforms, leading to protein aggregates [42]. A total of 14 different mutations have been identified that affect different splicing enhancers or silencers in exon 10 and intron 11, thus altering the inclusion or exclusion of exon 10 of MAPT [43, 44]. The resultant isoforms 4R-Tau and 3R-Tau contain different numbers of the imperfect repeat sequences and are normally represented in the adult brain at a ratio of 1:1. However, the splicing changes affecting exon 10 disrupt this normal ratio and are crucial for the pathogenic accumulation of Tau. Furthermore, the altered ratio of these two isoforms has different clinical manifestations leading to different tauopathies. For example, in Pick’s disease, also known as frontotemporal dementia (FTD), there is relative change in the Tau isoform ratio to more 3R-Tau than 4R-Tau [45]. This example, once again, reinforces the importance of alternative splicing changes leading to alterations in cellular phase separation and further highlights how harnessing these processes could lead to the therapeutic avenues for multiple diseases.
In an effort to modulate alternative splicing as a means of therapy for tauopathies and AD, a hTau transgenic mouse model that expresses all six splice isoforms simultaneously from the human MAPT gene in a mouse Mapt null background was generated. Interestingly, this model shows Tau altered splicing where exon 10 exclusion is increased, thus producing more 3R-Tau isoform and modeling what is observed in Pick’s disease [46]. The hTau transgenic mouse model was additionally used to test the therapeutic capabilities of the spliceosome-mediated RNA trans-splicing (SMaRT) technology (Fig. 2Biii). hTau mice infected with a lentivirus carrying the pre-trans-splicing RNA molecule (PTM) that includes the 3’ end of a Mapt cDNA including exon 10 and its associated 5’ss and splicing signals showed increased exon 10 inclusion in the brain [47]. In another instance, hTau mice were treated with SSOs that targeted regulatory elements within intron 12 of the MAPT gene. The SSO treatment successfully shifted the ratio of Tau isoforms by increasing the inclusion of exon 10. The resultant changes produced higher levels of the isoform 4R-Tau [48]. Surprisingly, this shift in Tau isoforms also caused increased protein aggregate formation and increased seizures in mouse models, reflecting the importance of the titration of isoform products for alternative splicing in disease manifestation and therapy.
As summarized above, it is well known that in neurodegenerative diseases like the tauopathies, AD and ALS, different protein aggregates are underlying cause. Moreover, recent research has shown that some splicing factors can, in fact, also form aggregates by the phase separation phenomenon[28, 29]. These events lead to altered pre-mRNA splicing, which can be detected in the different pathologies. Therefore, the generation of novel and improved animal models, which can replicate not only the outcome of the disease, but also the alterations in splicing is crucial. Animal models such as these allow for the design of splicing modulating therapies to combat the effects of pathological aggregation and splicing defects neurodegenerative diseases.
Using new technologies as therapeutic modulators of splicing: targeted genome editing with CRISPR-Cas9
In order to ensure target specificity of RNA therapeutics, there is a growing interest in developing new and improved ways to regulate alternative splicing or better delivery methods for existing ones. The rise of new technologies in molecular biology and genetics emerges as a fundamental answer to these problems. Examples include next-generation sequencing (NGS), nanoparticles, single-molecule studies and targeted genome editing with the clustered regularly interspaced short palindromic repeats-Cas9 (CRISPR-Cas9) technology. This powerful system, which is part of the innate immune response in bacteria, has been co-opted by molecular biologists to induce precise genome editing in a number of organisms. For instance, CRISPR-Cas9 has been used to generate genetic knockouts, tag endogenous proteins, induce disease-causing mutations, and also therapeutically correct toxic gene products in in vitro systems and animal models [49, 50]. In the recent years the use of the CRISPR-Cas9 technology has grown exponentially and applying them to diseases related to alternative splicing is not the exception.
Genome editing technologies have been harnessed for the study of new treatment modalities for multiple diseases arising from mutations that disrupt splicing fidelity, such as the β-thalassemia group of diseases (Box 3). Other studies using this technology have likewise been pursued, as highlighted by recent studies of hereditary tyrosinemia type I (HTI), a fatal genetic disease caused by mutation of fumarylacetoacetate hydrolase (FAH). This disease has been modeled in mice by inducing the same mutation that is observed in humans: a homozygous G>A point mutation that causes exclusion of exon 8 during splicing and creates a truncated, unstable FAH protein. Using CRISPR-Cas9, researchers have been able to correct the FAH mutation in the livers of mice, generating full-length protein sufficient to restore the weight loss of a mouse model of HTI [51].
Another example of the CRISPR-Cas9 technology being used in a splicing-related disease is the work published in congenital muscular dystrophy type 1A (MDC1A). This disease is characterized by severe muscle wasting as is seen in many other neuromuscular diseases (NMD). The dy2j mouse model harbors a 5’ splice site mutation in intron 2 of the LAMA2 gene that causes aberrant skipping of exon 2, leading to murine dystrophia muscularis-2J. This group was able to restore the production of the full-length protein in the dy2j/dy2j mouse model by using the CRISPR-Cas9 technology in conjunction with an adeno-associated virus (AAV) for delivery into the mouse. In this model, the authors used genome editing to delete the mutation in intron 2 and therefore promote exon 2 inclusion by creating a new functional donor splice site [52].
Similarly, genome editing has been utilized to create new mouse models and treatment options for Duchene muscular dystrophy (DMD, Box 4). While therapies aimed at splicing in DMD are not correcting mis-splicing in the disease, splicing changes are being utilized as a powerful way to manipulate the DMD gene to remove disease causing frameshift mutations.
While the genome editing technologies, we have discussed thus far target the DNA, and hence, the genetic code, new systems have been developed to redirect the CRISPR/Cas 9 toolbox to target RNA directly. Microsatellite repeat expansions in the genome can give rise to pathogenic RNA molecules that cause several diseases such as myotonic dystrophy, Huntington’s disease or ALS (when associated with the accumulation of the C9orf72 mRNA). These diseases lend themselves well to the application of the new RNA-targeting Cas9 enzyme to eliminate the toxic RNAs. In a proof of principle experiment, the RNA-targeting CRISPR-Cas9 system was used to successfully eradicate repeat-containing RNAs in myotonic dystrophy patient cells. Encouragingly, decreasing the amount of toxic RNA also correlated with the correction of some of the validated disease-specific splicing defects[53].
The successes described above are examples of what the future holds in the design of new therapies against diseases related to mis-splicing. Nevertheless, several barriers will have to be overcome before these technologies can be used in the clinic. For the CRISPR-Cas9 system specifically, there are potential problems with possible off-target effects that will need to be carefully addressed, as the correction of a specific mutation may lead to other genome editing events that could have deleterious effects. Additionally, a long discussion about the ethical implications of using this technology is just starting and regulations need to be in order before this emerging technology can be used to treat human diseases.
Conclusions and future perspectives
Even though research has taken important steps into understanding how the processes of post-transcriptional regulation are involved in the generation of diseases, there remain many open opportunities (see Outstanding Questions). For example, recent studies showing that in several cancers splicing factors, in fact, act as oncogenes or tumor suppressors (Table 1 and extensively reviewed in [54]). Consequently, current efforts are focused on developing new cancer therapies that target splice regulatory factors, or even alter their post-translational regulation, as is the case for the SRPK inhibitors that target the kinases that phosphorylate many of the SR proteins [55].
Outstanding Questions.
Will RNA modifications affect the splicing of a subset of RNAs? Does mis-regulation of these modifications lead to disease and thus new therapeutic opportunities?
Is there overlap in chromatin remodeling drugs and splice altering therapy? Since transcription and splicing are intricately linked, is it possible to repurpose drugs targeting transcription for RNA splicing therapy? On the other hand, could drugs targeting chromatin also cause unintended deleterious consequences on global RNA splicing profiles?
As with all therapies, delivery and dosing will be important for therapies targeting the RNA splicing pathway. How will cellular uptake be optimized? Will it be possible to limit delivery to specific tissues and/or cell types?
Given the success of SSO therapies, can RNA molecules be designed and delivered to make up a “designer” spliceosome? This is especially appealing for treatment of diseases caused by mutations in core splicing components.
New regulatory methods that impact splicing will also drive the future of therapeutic design. Recent published work indicates that N6-methyl-adenosine, or m6A RNA methylation, directly impacts pre-mRNA splicing and may be regulated to impact splicing upon certain types of stress [56]. Though this topic is hotly debated and there is strong evidence that this method of regulation is not broadly applicable to the majority of RNA transcripts, the role of m6A in RNA metabolism and regulation of stability is broadly accepted [57–59]. Similarly, there have been reports of long non-coding RNAs, sisRNAs and snoRNAs regulating splicing and, in some cases, impacting disease [60–62]. Thus, there is still much to learn about the regulatory intricacies of RNA and it’s splicing via new modifications, RNA-RNA interactions and RNA-protein complexes, all paving the way for novel therapeutic targets.
The implementation of new technologies as well as the generation of new knowledge will move us one step closer to the development of new and improved therapies to fight disease. As has been done in DMD, splicing therapy can also be used more broadly to repair a damaged or mutated gene by inducing splicing changes to exclude a bad exon. Conversely, splicing may be harnessed to cause a non-functional transcript to be generated for any identified disease-causing gene by deleting an essential exon to induce a deleterious deletions or frameshift. Thus, therapeutics targeting the splicing process may be applicable to a multitude of genetic diseases with gain-of-function mutations or inappropriately expressed proteins as was accomplished with SSOs targeting the amyloid beta precursor protein as a therapeutic target for Alzheimer’s disease [63].
Supplementary Material
Fig 1.
Disease Causing Mutations in the pre-mRNA and Major and Minor Spliceosomes. (A) Mutations in the RNA transcript itself can lead to splicing alterations. Similar mutations are also possible in the sequences controlling splicing of the minor introns, although are not depicted here. (B) Similarly, mutations in core spliceosome machinery of both the major and minor spliceosome have been shown to lead to altered splicing networks. Both of these scenarios can manifest in a disease phenotype. Abbreviations: MDS, myelodysplastic syndromes; MOPD1, microcephalic osteodysplastic primordial dwarfism type 1; SRSF, serine/arginine-rich splicing factor.
Highlights.
Recent studies have shown that approximately one third of all disease-causing mutations are related to RNA splicing.
Research on new and/or improved animal models that can replicate in a better way diseases caused by aberrant splicing has not only helped to increase the understanding of these diseases, but also to design new therapies against them.
The appearance of new antisense oligonucleotides and small molecules, together with novel techniques such as the SMaRT or the ExSpeU1, gives a light of hope in the fight against splicing-related diseases as they present as promising therapy options.
The development of new technologies in molecular biology such as next-generation sequencing or the CRISPR-Cas9 genome editing technology has opened the door for the research on RNA mis-splicing events and how they relate to disease.
Acknowledgements
We apologize that space limitations did not allow us to cite all the important studies that have contributed to this field. We also thank the funding sources for this work: grants from the Elsa U. Pardee Foundation, the Wolfe Foundation, the Sunbeam Foundation, Pelotonia, and the National Institutes of Health NIH CA195324 and NS084187 to DSC as well as NS077984 to BLS, trainee.
Glossary Terms
- Adeno-associated viral (AAV) vector
Gene therapy technology used for the delivery of genetic material. In this case, an infection with a harmless virus is performed, the virus can persist inside the cells as episomal, thus not integrating in the host genome. For example, this technology can be used to deliver components of the CRISPR-Cas9 gene editing system, as discussed within this review.
- Cassette exon
A complete exon that may be excluded or included in alternative splicing events.
- Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 System
Gene editing technique based on bacterial adaptive immunity, where a guide RNA (gRNA) is used to target the gene of interest to be cleaved by the bacterially derived nuclease, Cas9. This can be used to induce mutations taking advantage of the cellular repair machinery.
- Cryptic splice site
Splice sites that are not normally used in wild-type pre-mRNA splicing, but that are activated as a result of mutations and can be used instead of the actual splice sites, hence affecting the normal splicing pattern of a transcript.
- Exon-specific U1 snRNA (ExSpeU1)
Technology that takes advantage of the degenerate nature of the U1 snRNA base complementarity with the 5’ splice site of a target pre-mRNA. Due to the presence of 3 to 4 mismatches in this binding, improving the base complementarity with an artificial exon-specific U1 snRNA can target the splice site more specifically and may be used to correct aberrant splicing (Fig 2Biv).
- Minor spliceosome
A U12-dependent ribonucleoprotein responsible for catalyzing removal of U12type introns, present in most eukaryotes. U12-type introns are defined by the strong conservation of the 5’ splice site and branch point sequences. In contrast to the major spliceosome, the 5’ splice site and branch point are cooperatively recognized by the U11 and U12 snRNPs, respectively, and the U4atac, U5, and U6atac tri-snRNP complex is utilized for complex B formation.
- Peptide nucleic acid (PNA)
Synthetic polymers based on modified nucleic acids, where the sugar-phosphate backbone is replaced by repeating units of N-(2-aminoethyl)-glycine linked through peptide bonds. The lack of charged phosphate groups makes PNA have strong binding affinities. They can be used in antisense technology to modulate gene expression and splicing.
- Phase separation
Physicochemical phenomenon occurring inside eukaryotic cells in dynamic structures called membrane-less organelles, formed by proteins and nucleic acids. Some examples of this organelles are the nucleoli, the nuclear speckles and the stress granules. They are thought to be involved in different aspects such as gene regulation, mRNA metabolism and processing.
- Pladienolide analog E7107
A derivative of pladienolide B with improved stability which binds to the SF3B component of U2 snRNP to interrupt spliceosome assembly.
- Splice Switching Oligonucleotide (SSO)
Synthetic polymers based on modified nucleic acids, normally designed as single stranded nucleic acids which are complementary (antisense) to a specific mRNA molecule (also called Antisense Oligonucleotides, ASOs or AONs). They can be used to modify gene expression by promoting mRNA degradation or altering RNA splicing.
- Spliceosome-mediated RNA trans-splicing (SMaRT)
Technology that takes advantage of the uncommon splicing between 2 separate pre-mRNA molecules or trans-splicing. Using an artificial pre-mRNA trans-splicing molecule (PTM), containing a binding domain, a splicing domain and a coding domain; it is possible to trigger a trans-splicing event between this PTM and a target pre-mRNA. The resultant artificially spliced transcript is able to correct an error in the target pre-mRNA by replacing the mutated exon and all downstream exons (encoded by the PTM) via the trans-splicing event (see also Fig 2Biii).
- SF3B
A heptameric protein complex in the U2 snRNP (in humans – SF3b155, SF3b130, SF3b145, SF3b49, SF3b14b, p14SF3b14a, and SF3b10), in which mutations have been shown to lead to alternative branch point selection and has been linked to cancer.
- sisRNA
Introns are generally degraded upon splicing; however, stable intronic sequence RNAs have been suggested to have other roles in the cell including regulation of gene expression, acting as a molecular sponge, and regulating protein translation.
- snoRNA
Small nucleolar RNAs generally exist in two classes and primarily serve as guides for chemical modification of ribosomal RNA including methylation (Box C/D class) and pseudouridylation (Box H/ACA).
- SRSF1
Serine/arginine-rich splicing factor 1 (previously known as ASF/SF2) is a splicing factor that contains an arginine- and serine-rich domain and two RNA recognition motifs and is required for 5’ splice site selection and cleavage. SRSF1 is also reported to be a proto-oncogene.
- SRSF2
Much like SRSF1, serine/arginine-rich splicing factor 2 (previously known as SC35) is important for splice site selection and spliceosome assembly. Mutation of SRSF2 is implicated in MDS and AML.
- Stress granule
Dense aggregates which form in eukaryotic cells under stress conditions like heat-shock, they are composed of RNA and pre-mRNA processing factors such as splicing-related proteins. They are thought to be a cellular mechanism of rapid reprograming of gene expression.
- U2AF
U2 auxiliary factor is a heterodimer consisting of a large and a small subunit, U2AF65 and U2AF35 respectively. The complex is required for definition of 3’ splice sites. Mutations in U2AF have been implicated in blood diseases and cancers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Berget SM et al. (1977) Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A 74 (8), 3171–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow LT et al. (1977) An amazing sequence arrangement at the 5’ ends of adenovirus 2 messenger RNA. Cell 12 (1), 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Pan Q et al. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40 (12), 1413–1415. [DOI] [PubMed] [Google Scholar]
- 4.Wang ET et al. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456 (7221),470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scotti MM and Swanson MS (2015) RNA mis-splicing in disease. Nature Reviews Genetics 17, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper DN et al. (2006) The Human Gene Mutation Database (HGMD) and its exploitation in the study of mutational mechanisms. Curr Protoc Bioinformatics Chapter 1, Unit 1.13. [DOI] [PubMed] [Google Scholar]
- 7.Tazi J et al. (2009) Alternative splicing and disease. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1792 (1), 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley TJ et al. (1982) RNA processing errors in patients with beta-thalassemia. Proceedings of the National Academy of Sciences of the United States of America 79 (15), 4775–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorman SN et al. (2014) Splicing mutation analysis reveals previously unrecognized pathways in lymph node-invasive breast cancer. Scientific Reports 4, 7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y et al. (2017) Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett 393, 40–51. [DOI] [PubMed] [Google Scholar]
- 11.McGreevy JW et al. (2015) Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Dis Model Mech 8 (3), 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havens MA and Hastings ML (2016) Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Research 44 (14), 6549–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaida D et al. (2007) Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3 (9), 576–83. [DOI] [PubMed] [Google Scholar]
- 14.Bakkour N et al. (2007) Small-molecule inhibition of HIV pre-mRNA splicing as a novel antiretroviral therapy to overcome drug resistance. PLoS Pathog 3 (10), 1530–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havens MA et al. (2013) Targeting RNA splicing for disease therapy. Wiley Interdiscip Rev RNA 4 (3), 247–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L et al. (2017) Mutation screening in genes known to be responsible for Retinitis Pigmentosa in 98 Small Han Chinese Families. Scientific Reports 7 (1), 1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezquerra-Inchausti M et al. (2017) High prevalence of mutations affecting the splicing process in a Spanish cohort with autosomal dominant retinitis pigmentosa. Scientific Reports 7, 39652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue D and Abdel-Wahab O (2016) Modeling SF3B1 Mutations in Cancer: Advances, Challenges, and Opportunities. Cancer Cell 30 (3), 371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yip BH et al. (2016) Impact of Splicing Factor Mutations on Pre-mRNA Splicing in the Myelodysplastic Syndromes. Curr Pharm Des 22 (16), 2333–44. [DOI] [PubMed] [Google Scholar]
- 20.Qiu J et al. (2016) Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. Rna 22 (10), 1535–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obeng EA et al. (2016) Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell 30 (3), 404–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim E et al. (2015) SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 27 (5), 617–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirai CL et al. (2015) Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer cell 27 (5), 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SC et al. (2016) Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med 22 (6), 672–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong DS et al. (2014) A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs 32 (3), 436–44. [DOI] [PubMed] [Google Scholar]
- 26.Seiler M et al. (2018) H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat Med 24 (4), 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uversky VN (2017) Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol 44, 18–30. [DOI] [PubMed] [Google Scholar]
- 28.Molliex A et al. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163 (1), 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conlon EG and Manley JL (2017) RNA-binding proteins in neurodegeneration: mechanisms in aggregate. Genes & Development 31 (15), 1509–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H-R et al. (2018) TAR DNA-binding protein 43 (TDP-43) liquid-liquid phase separation is mediated by just a few aromatic residues. Journal of Biological Chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conicella AE et al. (2016) ALS Mutations Disrupt Phase Separation Mediated by alpha-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure 24 (9), 1537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold ES et al. (2013) ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proceedings of the National Academy of Sciences 110 (8), E736–E745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang A et al. (2018) A single N‐terminal phosphomimic disrupts TDP‐43 polymerization, phase separation, and RNA splicing. The EMBO Journal 37 (5), e97452.29438978 [Google Scholar]
- 34.Deshaies JE et al. (2018) TDP-43 regulates the alternative splicing of hnRNP A1 to yield an aggregation-prone variant in amyotrophic lateral sclerosis. Brain 141 (5), 1320–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Conti L et al. (2015) TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic and mitotic cellular pathways. Nucleic Acids Research 43 (18), 8990–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ditsworth D et al. (2017) Mutant TDP-43 within motor neurons drives disease onset but not progression in amyotrophic lateral sclerosis. Acta Neuropathologica 133 (6), 907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricketts T et al. (2014) A nonsense mutation in mouse Tardbp affects TDP43 alternative splicing activity and causes limb-clasping and body tone defects. PLoS One 9 (1), e85962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel A et al. (2015) A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162 (5), 1066–1077. [DOI] [PubMed] [Google Scholar]
- 39.Nomura T et al. (2014) Intranuclear Aggregation of Mutant FUS/TLS as a Molecular Pathomechanism of Amyotrophic Lateral Sclerosis. Journal of Biological Chemistry 289 (2), 1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reber S et al. (2016) Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants. Embo j 35 (14), 1504–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu H et al. (2014) ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. The Journal of Clinical Investigation 124 (3), 981–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park SA et al. (2016) Tau mis-splicing in the pathogenesis of neurodegenerative disorders. BMB Reports 49 (8), 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu F and Gong C-X (2008) Tau exon 10 alternative splicing and tauopathies. Molecular Neurodegeneration 3, 8–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lisowiec J et al. (2015) Structural determinants for alternative splicing regulation of the MAPT pre-mRNA. RNA Biology 12 (3), 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falcon B et al. (2018) Structures of filaments from Pick’s disease reveal a novel tau protein fold. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cathy A et al. (2003) Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. Journal of Neurochemistry 86 (3), 582–590. [DOI] [PubMed] [Google Scholar]
- 47.Avale ME et al. (2013) Trans-splicing correction of tau isoform imbalance in a mouse model of tau mis-splicing. Human Molecular Genetics 22 (13), 2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoch KM et al. (2016) Increased 4R-Tau Induces Pathological Changes in a Human-Tau Mouse Model. Neuron 90 (5), 941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu PD et al. (2014) Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157 (6), 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrangou R and Doudna JA (2016) Applications of CRISPR technologies in research and beyond. Nature Biotechnology 34, 933. [DOI] [PubMed] [Google Scholar]
- 51.Shao Y et al. (2018) Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats. J Biol Chem 293 (18), 6883–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kemaladewi DU et al. (2017) Correction of a splicing defect in a mouse model of congenital muscular dystrophy type 1A using a homology-directed-repair-independent mechanism. Nat Med 23 (8), 984–989. [DOI] [PubMed] [Google Scholar]
- 53.Batra R et al. (2017) Elimination of Toxic Microsatellite Repeat Expansion RNA by RNA-Targeting Cas9. Cell 170 (5), 899–912.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dvinge H et al. (2016) RNA splicing factors as oncoproteins and tumor suppressors. Nature reviews. Cancer 16 (7), 413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siqueira RP et al. (2015) Potential Antileukemia Effect and Structural Analyses of SRPK Inhibition by N-(2(Piperidin-1-yl)-5-(Trifluoromethyl)Phenyl)Isonicotinamide (SRPIN340). PLOS ONE 10 (8), e0134882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao W et al. (2016) Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Molecular Cell 61 (6), 925. [DOI] [PubMed] [Google Scholar]
- 57.Darnell RB et al. (2018) Pre-mRNA processing includes N(6) methylation of adenosine residues that are retained in mRNA exons and the fallacy of “RNA epigenetics”. RNA 24 (3), 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao BS et al. (2018) Our views of dynamic N(6)-methyladenosine RNA methylation. Rna 24 (3), 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ke S et al. (2018) Saturation mutagenesis reveals manifold determinants of exon definition. Genome Research 28 (1), 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dey BK et al. (2014) Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription 5 (4), e944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osman I et al. (2016) Stable intronic sequence RNAs (sisRNAs): a new layer of gene regulation. Cellular and molecular life sciences : CMLS 73 (18), 3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Falaleeva M et al. (2016) Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proceedings of the National Academy of Sciences 113 (12), E1625–E1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang JL et al. (2018) Targeting Amyloid-beta Precursor Protein, APP, Splicing with Antisense Oligonucleotides Reduces Toxic Amyloid-beta Production. Mol Ther 26 (6), 1539–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cvitkovic I and Jurica MS (2013) Spliceosome Database: a tool for tracking components of the spliceosome. Nucleic Acids Research 41 (Database issue), D132–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmidt C et al. (2014) Mass spectrometry–based relative quantification of proteins in precatalytic and catalytically active spliceosomes by metabolic labeling (SILAC), chemical labeling (iTRAQ), and label-free spectral count. RNA 20 (3), 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan J et al. (2016) Noncanonical registers and base pairs in human 5′ splice-site selection. Nucleic Acids Research 44 (8), 3908–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mercer TR et al. (2015) Genome-wide discovery of human splicing branchpoints. Genome Research 25 (2), 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72, 291–336. [DOI] [PubMed] [Google Scholar]
- 69.Will CL and Luhrmann R (1997) Protein functions in pre-mRNA splicing. Curr Opin Cell Biol 9 (3), 320–8. [DOI] [PubMed] [Google Scholar]
- 70.Matera AG and Wang Z (2014) A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15 (2), 108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fox-Walsh KL et al. (2005) The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci U S A 102 (45), 16176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Misteli T and Spector DL (1999) RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell 3 (6), 697–705. [DOI] [PubMed] [Google Scholar]
- 73.Thein SL (2013) The molecular basis of beta-thalassemia. Cold Spring Harb Perspect Med 3 (5), a011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao A and Galanello R (2010) Beta-thalassemia. Genet Med 12 (2), 61–76. [DOI] [PubMed] [Google Scholar]
- 75.Phanthong P et al. (2017) Enhancement of beta-Globin Gene Expression in Thalassemic IVS2–654 Induced Pluripotent Stem Cell-Derived Erythroid Cells by Modified U7 snRNA. Stem Cells Transl Med 6 (4), 1059–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lewis J et al. (1998) A Common Human β Globin Splicing Mutation Modeled in Mice. Blood 91 (6), 2152–2156. [PubMed] [Google Scholar]
- 77.Breveglieri G et al. (2015) Generation and Characterization of a Transgenic Mouse Carrying a Functional Human beta -Globin Gene with the IVSI-6 Thalassemia Mutation. Biomed Res Int 2015, 687635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vadolas J et al. (2006) Humanized beta-thalassemia mouse model containing the common IVSI-110 splicing mutation. J Biol Chem 281 (11), 7399–405. [DOI] [PubMed] [Google Scholar]
- 79.Xie SY et al. (2011) Correction of beta654-thalassaemia mice using direct intravenous injection of siRNA and antisense RNA vectors. Int J Hematol 93 (3), 301–310. [DOI] [PubMed] [Google Scholar]
- 80.Preedagasamzin S et al. (2018) Engineered U7 snRNA mediates sustained splicing correction in erythroid cells from beta-thalassemia/HbE patients. Biochem Biophys Res Commun 499 (1), 86–92. [DOI] [PubMed] [Google Scholar]
- 81.Xie F et al. (2014) Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Research 24 (9), 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.R. MJ et al. (2013) Eteplirsen for the treatment of Duchenne muscular dystrophy. Annals of Neurology 74 (5), 637–647. [DOI] [PubMed] [Google Scholar]
- 83.Echigoya Y and Yokota T (2014) Skipping multiple exons of dystrophin transcripts using cocktail antisense oligonucleotides. Nucleic Acid Ther 24 (1), 57–68. [DOI] [PubMed] [Google Scholar]
- 84.Amoasii L et al. (2017) Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Science Translational Medicine 9 (418). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Long C et al. (2018) Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Science Advances 4 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wobst HJ et al. (2017) Increased 4R tau expression and behavioural changes in a novel MAPT-N296Hgenomic mouse model of tauopathy. Sci Rep 7, 43198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peacey E et al. (2012) Targeting a pre-mRNA structure with bipartite antisense molecules modulates tau alternative splicing. Nucleic Acids Research 40 (19), 9836–9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Avale ME et al. (2013) Trans-splicing correction of tau isoform imbalance in a mouse model of tau mis-splicing. Hum Mol Genet 22 (13), 2603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hinrich AJ et al. (2016) Therapeutic correction of ApoER2 splicing in Alzheimer’s disease mice using antisense oligonucleotides. EMBO Mol Med 8 (4), 328–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou Y et al. (2013) ALS-Associated FUS Mutations Result in Compromised FUS Alternative Splicing and Autoregulation. PLOS Genetics 9 (10), e1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quesnel-Vallieres M et al. (2016) Misregulation of an Activity-Dependent Splicing Network as a Common Mechanism Underlying Autism Spectrum Disorders. Mol Cell 64 (6), 1023–1034. [DOI] [PubMed] [Google Scholar]
- 92.Liu SM et al. (2012) Overexpression of the CTLA-4 isoform lacking exons 2 and 3 causes autoimmunity(). Journal of immunology (Baltimore, Md. : 1950) 188 (1), 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ichinose K et al. (2013) Brief report: increased expression of a short splice variant of CTLA-4 exacerbates lupus in MRL/lpr mice. Arthritis Rheum 65 (3), 764–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mourich DV et al. (2014) Alternative Splice Forms of CTLA-4 Induced by Antisense Mediated Splice-Switching Influences Autoimmune Diabetes Susceptibility in NOD Mice. Nucleic Acid Therapeutics 24 (2), 114126. [DOI] [PubMed] [Google Scholar]
- 95.Tilio M et al. (2016) Irreversible inhibition of Δ16HER2 is necessary to suppress Δ16HER2-positive breast carcinomas resistant to Lapatinib. Cancer Letters 381 (1), 76–84. [DOI] [PubMed] [Google Scholar]
- 96.Marchini C et al. (2011) The Human Splice Variant Δ16HER2 Induces Rapid Tumor Onset in a Reporter Transgenic Mouse. PLOS ONE 6 (4), e18727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wali VB et al. (2014) Convergent and divergent cellular responses by ErbB4 isoforms in mammary epithelial cells. Mol Cancer Res 12 (8), 1140–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nielsen TO et al. (2013) Directing HER4 mRNA expression towards the CYT2 isoform by antisense oligonucleotide decreases growth of breast cancer cells in vitro and in vivo. Br J Cancer 108 (11), 2291–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Manson AL et al. (1997) Complementation of null CF mice with a human CFTR YAC transgene. The EMBO Journal 16 (14), 4238–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Igreja S et al. (2016) Correction of a Cystic Fibrosis Splicing Mutation by Antisense Oligonucleotides. Hum Mutat 37 (2), 209–15. [DOI] [PubMed] [Google Scholar]
- 101.Fernandez Alanis E et al. (2012) An exon-specific U1 small nuclear RNA (snRNA) strategy to correct splicing defects. Human Molecular Genetics 21 (11), 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sanz DJ et al. (2017) Cas9/gRNA targeted excision of cystic fibrosis-causing deep-intronic splicing mutations restores normal splicing of CFTR mRNA. PLOS ONE 12 (9), e0184009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakano Y et al. (2012) A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse. PLoS Genet 8 (10), e1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lorain S et al. (2013) Dystrophin rescue by trans-splicing: a strategy for DMD genotypes not eligible for exon skipping approaches. Nucleic Acids Res 41 (17), 8391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bish LT et al. (2012) Long-term restoration of cardiac dystrophin expression in golden retriever muscular dystrophy following rAAV6-mediated exon skipping. Mol Ther 20 (3), 580–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miyatake S et al. (2018) Exon Skipping Therapy Using Phosphorodiamidate Morpholino Oligomers in the mdx52 Mouse Model of Duchenne Muscular Dystrophy In Duchenne Muscular Dystrophy: Methods and Protocols (C. Bernardini ed), pp. 123–141, Springer; New York. [DOI] [PubMed] [Google Scholar]
- 107.Rusconi F et al. (2016) LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proc Natl Acad Sci U S A 113 (13), 3651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Axelrod FB et al. (2011) Kinetin improves IKBKAP mRNA splicing in patients with familial dysautonomia. Pediatr Res 70 (5), 480–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morini E et al. (2016) Sensory and autonomic deficits in a new humanized mouse model of familial dysautonomia. Hum Mol Genet 25 (6), 1116–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kanagawa M et al. (2009) Residual laminin-binding activity and enhanced dystroglycan glycosylation by LARGE in novel model mice to dystroglycanopathy. Human Molecular Genetics 18 (4), 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Taniguchi-Ikeda M et al. (2011) Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature 478 (7367), 127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barbon E et al. (2016) Transposon-mediated Generation of Cellular and Mouse Models of Splicing Mutations to Assess the Efficacy of snRNA-based Therapeutics. Mol Ther Nucleic Acids 5 (11), e392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Balestra D et al. (2016) An Exon-Specific U1snRNA Induces a Robust Factor IX Activity in Mice Expressing Multiple Human FIX Splicing Mutants. Mol Ther Nucleic Acids 5 (10), e370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rawcliffe DFR et al. (2016) The High Level of Aberrant Splicing of ISCU in Slow-Twitch Muscle May Involve the Splicing Factor SRSF3. PLOS ONE 11 (10), e0165453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sanaker PS et al. (2012) Antisense oligonucleotide corrects splice abnormality in hereditary myopathy with lactic acidosis. Gene 494 (2), 231–6. [DOI] [PubMed] [Google Scholar]
- 116.Yin H et al. (2014) Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32 (6), 551–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sathasivam K et al. (2013) Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci U S A 110 (6), 2366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rindt H et al. (2012) Replacement of huntingtin exon 1 by trans-splicing. Cell Mol Life Sci 69 (24), 4191204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Osorio FG et al. (2011) Splicing-directed therapy in a new mouse model of human accelerated aging. Sci Transl Med 3 (106), 106ra107. [DOI] [PubMed] [Google Scholar]
- 120.Scaffidi P and Misteli T (2005) Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med 11 (4), 440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vignier N et al. (2009) Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ Res 105 (3), 239–48. [DOI] [PubMed] [Google Scholar]
- 122.Mearini G et al. (2013) Repair of Mybpc3 mRNA by 5’-trans-splicing in a Mouse Model of Hypertrophic Cardiomyopathy. Mol Ther Nucleic Acids 2, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sabrautzki S et al. (2012) New mouse models for metabolic bone diseases generated by genome-wide ENU mutagenesis. Mammalian Genome 23 (7–8), 416–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Garanto A et al. (2016) In vitro and in vivo rescue of aberrant splicing in CEP290-associated LCA by antisense oligonucleotide delivery. Hum Mol Genet 25 (12), 2552–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ruan GX et al. (2017) CRISPR/Cas9-Mediated Genome Editing as a Therapeutic Approach for Leber Congenital Amaurosis 10. Mol Ther 25 (2), 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dewaele M et al. (2016) Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest 126 (1), 68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bardot B et al. (2015) Mice engineered for an obligatory Mdm4 exon skipping express higher levels of the Mdm4-S isoform but exhibit increased p53 activity. Oncogene 34 (22), 2943–2948. [DOI] [PubMed] [Google Scholar]
- 128.Buonamici S et al. (2016) H3B-8800, an Orally Bioavailable Modulator of the SF3b Complex, Shows Efficacy in Spliceosome-Mutant Myeloid Malignancies. Blood 128 (22), 966. [Google Scholar]
- 129.Gallardo M et al. hnRNP K Is a Haploinsufficient Tumor Suppressor that Regulates Proliferation and Differentiation Programs in Hematologic Malignancies. Cancer Cell 28 (4), 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shirai CL et al. (2015) Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer Cell 27 (5), 631–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mankodi A et al. (2002) Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell 10 (1), 35–44. [DOI] [PubMed] [Google Scholar]
- 132.Oana K et al. (2013) Manumycin A corrects aberrant splicing of Clcn1 in myotonic dystrophy type 1 (DM1) mice. Scientific Reports 3, 2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gómez-Grau M et al. (2017) New murine Niemann-Pick type C models bearing a pseudoexon-generating mutation recapitulate the main neurobehavioural and molecular features of the disease. 7, 41931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rodriguez-Pascau L et al. (2009) Antisense oligonucleotide treatment for a pseudoexon-generating mutation in the NPC1 gene causing Niemann-Pick type C disease. Hum Mutat 30 (11), E993–e1001. [DOI] [PubMed] [Google Scholar]
- 135.Bergsma AJ et al. (2016) From Cryptic Toward Canonical Pre-mRNA Splicing in Pompe Disease: a Pipeline for the Development of Antisense Oligonucleotides. Molecular Therapy. Nucleic Acids 5 (9), e361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Garfield AS et al. (2016) Increased alternate splicing of Htr2c in a mouse model for Prader-Willi syndrome leads disruption of 5HT2C receptor mediated appetite. Molecular Brain 9 (1), 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang Z et al. (2016) Oligonucleotide-induced alternative splicing of serotonin 2C receptor reduces food intake. EMBO Molecular Medicine 8 (8), 878–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sun F et al. (2014) Androgen receptor splice variant AR3 promotes prostate cancer via modulating expression of autocrine/paracrine factors. J Biol Chem 289 (3), 1529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu C et al. (2014) Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration resistant prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 20 (12), 3198–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bujakowska K et al. (2009) Study of gene-targeted mouse models of splicing factor gene Prpf31 implicated in human autosomal dominant retinitis pigmentosa (RP). Invest Ophthalmol Vis Sci 50 (12), 592733. [DOI] [PubMed] [Google Scholar]
- 141.Yin J et al. (2011) Mutant Prpf31 causes pre-mRNA splicing defects and rod photoreceptor cell degeneration in a zebrafish model for Retinitis pigmentosa. Molecular Neurodegeneration 6 (1), 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Graziotto JJ et al. (2011) Three gene-targeted mouse models of RNA splicing factor RP show late-onset RPE and retinal degeneration. Invest Ophthalmol Vis Sci 52 (1), 190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Berger A et al. (2015) Repair of rhodopsin mRNA by spliceosome-mediated RNA trans-splicing: a new approach for autosomal dominant retinitis pigmentosa. Mol Ther 23 (5), 918–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li R et al. (2016) Misregulation of Alternative Splicing in a Mouse Model of Rett Syndrome. PLOS Genetics 12 (6), e1006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Comiskey DF Jr. et al. (2017) A novel mouse model of rhabdomyosarcoma underscores the dichotomy of MDM2-ALT1 function in vivo. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Comiskey DF Jr. et al. (2015) Splicing factor SRSF1 negatively regulates alternative splicing of MDM2 under damage. Nucleic Acids Res 43 (8), 4202–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bebee TW et al. (2012) Mouse models of SMA: tools for disease characterization and therapeutic development. Hum Genet 131 (8), 1277–93. [DOI] [PubMed] [Google Scholar]
- 148.Gladman JT et al. (2010) A humanized Smn gene containing the SMN2 nucleotide alteration in exon 7 mimics SMN2 splicing and the SMA disease phenotype. Human Molecular Genetics 19 (21), 4239–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bowerman M et al. (2012) Fasudil improves survival and promotes skeletal muscle development in a mouse model of spinal muscular atrophy. BMC Medicine 10 (1), 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Passini MA et al. (2011) Antisense Oligonucleotides Delivered to the Mouse CNS Ameliorate Symptoms of Severe Spinal Muscular Atrophy. Science Translational Medicine 3 (72), 72ra18–72ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Woll MG et al. (2016) Discovery and Optimization of Small Molecule Splicing Modifiers of Survival Motor Neuron 2 as a Treatment for Spinal Muscular Atrophy. Journal of Medicinal Chemistry 59 (13), 60706085. [DOI] [PubMed] [Google Scholar]
- 152.Ratni H et al. (2016) Specific Correction of Alternative Survival Motor Neuron 2 Splicing by Small Molecules: Discovery of a Potential Novel Medicine To Treat Spinal Muscular Atrophy. Journal of Medicinal Chemistry 59 (13), 6086–6100. [DOI] [PubMed] [Google Scholar]
- 153.Nizzardo M et al. (2015) Spinal muscular atrophy phenotype is ameliorated in human motor neurons by SMN increase via different novel RNA therapeutic approaches. 5, 11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pan B et al. (2017) Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c. Nat Biotechnol 35 (3), 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lentz JJ et al. (2013) Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat Med 19 (3), 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bestas B et al. (2015) Splice-Correction Strategies for Treatment of X-Linked Agammaglobulinemia. Current Allergy and Asthma Reports 15 (3), 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rattanachartnarong N et al. (2014) In vitro correction of a novel splicing alteration in the BTK gene by using antisense morpholino oligonucleotides. Arch Immunol Ther Exp (Warsz) 62 (5), 431–6. [DOI] [PubMed] [Google Scholar]
- 158.Breveglieri G et al. (2015) Generation and Characterization of a Transgenic Mouse Carrying a Functional Human β-Globin Gene with the IVSI-6 Thalassemia Mutation. BioMed Research International 2015, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chin JY et al. (2008) Correction of a splice-site mutation in the beta-globin gene stimulated by triplex-forming peptide nucleic acids. Proceedings of the National Academy of Sciences 105 (36), 13514–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Svasti S et al. (2009) RNA repair restores hemoglobin expression in IVS2–654 thalassemic mice. Proc Natl Acad Sci U S A 106 (4), 1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.