Abstract
DNA methylation is a fundamental means of epigenetic gene regulation that occurs in virtually all cell types. In many higher organisms, including humans, it plays vital roles in cell differentiation and homeostatic maintenance of cell phenotype. The control of DNA methylation has traditionally been attributed to a highly coordinated, linear process, whose dysregulation has been associated with numerous pathologies including cancer, where it occurs early in, and even prior to, the development of neoplastic tissues. Recent experimental evidence has demonstrated that, contrary to prevailing paradigms, methylation patterns are actually maintained through inexact, dynamic processes. These processes normally result in minor stochastic differences between cells that accumulate with age. However, various factors, including cancer itself, can lead to substantial differences in intercellular methylation patterns, viz. methylation heterogeneity. Advancements in molecular biology techniques are just now beginning allow insight into how this heterogeneity contributes to clonal evolution and overall cancer heterogeneity. In the current review, we begin by presenting a didactic overview of how the basal bimodal methylome is established and maintained. We then provide a synopsis of some of the factors that lead to the accrual of heterogeneous methylation and how this heterogeneity may lead to gene silencing and impact the development of cancerous phenotypes. Lastly, we highlight currently available methylation assessment techniques and their suitability to the study of heterogeneous methylation.
1. Introduction
DNA methylation, and cytosine methylation in particular, has been studied as a means of epigenetic gene regulation since the 1970’s [1, 2]. Its dysregulation was soon linked to cancer [3–5] and by 2000, significant experimental evidence had confirmed aberrant DNA methylation to be a primary contributor to carcinogenesis [6]. Since that time, significant advances in sequencing and other molecular biology techniques have begun to allow researchers to gain insight into how the various patterns of DNA methylation influence cellular phenotype and tumorigenesis. In the current review, we aim to provide a brief overview of how the highly organized, bimodal methylome established during embryogenesis can become altered by various endogenous and exogenous factors to yield the highly heterogeneous patterns of methylation often seen in cancer. We then seek to summarize recent progress in elucidating how these patterns affect, and are affected by, cancer-associated phenomena. Lastly, we review currently available methylation analysis techniques and discuss their suitability for the analysis of heterogeneous methylation.
Biochemically speaking, cytosine methylation occurs by donation of a methyl (CH3) group from S-adenosylmethionine to the fifth position in the cytosine pyrimidine ring resulting in the formation of 5–methylcytosine (5mC). The reaction occurs immediately following DNA synthesis and is catalyzed by a family of enzymes called DNA methyltransferases (DNMTs). 5mC is predominantly found within the genome in the context of 5’-cytosine-phosphate-guanine-3’ (CpG) dinucleotides. CpG dinucleotides are notable in that they form symmetrical CpG dinucleotides on the complementary strand, providing an ideal scaffold for DNMTs (predominantly DNMT1) to propagate DNA methylation patterns from parent strands to both daughter strands during DNA replication. This mechanism confers methylation with the definitive epigenetic attribute of mitotic heritability. As will be discussed later, while heritable, these methylation patterns are maintained and modified via dynamic regulation leading to minor stochastic differences in methylation patterns.
CpG sites are globally rare throughout the genome [7] and predominantly (70–80%) constitutively methylated in healthy cells [8]. Approximately 10% of CpG sites, however, are densely concentrated into “CpG islands” (CGIs), among which more than half are located in the promoter regions near the transcription start sites of over 60% of human genes [9, 10]. Unlike the majority of CpG sites throughout the genome, the sites located within CGIs are often constitutively hypomethylated in healthy cells. The reason for this hypomethylation lies in one of the primary putative functions of cytosine methylation in mammals, the regulation of genetic expression by the reduction of, or “silencing,” of gene transcription.
While other functions of DNA methylation also exist, these appear to be heavily dependent upon the genomic context in which the methylation occurs [11, 12], as well as cell phenotype [13]. For example, DNA methylation within promoter CGIs has considerably different implications for gene expression than methylation within gene bodies or regulatory elements [14, 15]. A detailed discussion of the various nuanced roles of DNA methylation is beyond the current scope, however a number of excellent reviews can be found elsewhere [11, 16].
The current review is primarily focused upon the role of heterogeneous methylation in the context of promoter-related gene silencing. Nonetheless, in order to approach the etiology of heterogeneous methylation, it is useful to begin with an overview of how the default methylation patterns are established and maintained within mammalian cells. A list of some of the key terms and biomolecules involved in these processes are detailed in Table 1.
Table 1.
Key Terms
| Term | Description |
|---|---|
| Chromatin | The overall nucleoprotein complex that acts as both a functional and structural scaffold to coordinate access to and control of DNA. |
| Heterochromatin | Chromatin that is tightly condensed into higher order structures to disallow access to specific genomic regions and the genetic material within. |
| Euchromatin | Chromatin that is open and uncondensed, enabling access to the genomic region, binding of transcription factors and gene transcription. |
| Histones | Core proteins that assemble to form an eight protein complex upon which genomic DNA is wound at 147 base pairs to form a single nucleosome. Each octameric histone protein is composed of two H3–H4 dimers bridged together to form a tetramer coordinated with two H2A-H2B dimers. |
| Histone Modifications | Chemical moieties on histone tails that function as editable epigenetic instructions for directing and coordinating cellular machinery to achieve a particular chromatic state. |
| DNA Methyltransferases (DNMTs) | Family of enzymes that catalyze the reaction transferring a methyl group from methionine and cytosine to form 5mC. |
| • DNMT1 | Classically considered to be solely responsible for maintenance methylation, ensuring daughter DNA strands maintain the same methylation pattern as the parent strands. DNMT1 has also recently been shown to play a role in de novo methylation [17, 18]. |
| • DNMT3A and DNMT3B | Primarily responsible for so-called de novo methylation, viz. methylation of previously unmethylated cytosine bases. Recent studies have also shown the de novo DNMTs to be important for maintenance methylation of repeat elements [19]. |
| • DNMTL | A closely related homolog to the DNMT proteins that lack methyltransferase activity. DNMTL binds to H3K4 and actively recruits de novo DNMTs to chromatin. |
| Methylcytosine Binding Proteins (MBDs) | A family of proteins whose putative function is to recognize and bind 5mC in order to translate DNA methylation into functionally silent chromatin [20]. |
| Polycomb Repressive Complexes (PRC1 and PRC2) | Coordinated multiprotein complexes of the Polycomb group (PcG) of proteins that typically act to repress gene expression. |
| Ten-Eleven Translocation (TET) Enzymes | A family of enzyme that are thought to be involved in active cytosine demethylation by catalyzing the iterative oxidation of 5mC to yield 5-Hydroxymethylcytosine (5hmC), 5-formylcytosine, and 5-carboxylcytosine [21, 22]. |
2. Generation of the Basal, Bimodal Methylome
Roughly speaking, the methylome of mammals is not heterogeneous by default, but bimodal, with the majority of CpG dinucleotides being methylated, with the notable exception of those located within the aforementioned CpG islands. This distribution is established early on in embryogenesis where, prior to implantation, parental DNA methylation is rapidly erased from the DNA of the fertilizing sperm and the primordial germ cells of the developing embryo [23]. Both of these global demethylation processes are preceded by a number of epigenetic remodeling events that are at least partially mediated by genome-wide oxidation of 5mC by TET1, a member of the larger TET family of demethylating enzymes [24–27].
Following these global demethylation events, CpG sites remain unmethylated roughly until the time of implantation [28, 29], when most CpG-poor regions become indiscriminately remethylated by the de novo DNMTs, DNMT3A and DNMT3B. Promoter CGIs are protected from arbitrary methylation events such as these by a number of mechanisms, including: nearby cis-regulatory regions such as transcription factor binding sites [30, 31], so-called R-loops that arise due to asymmetric cytosine/guanine distributions within CGIs [32], the persistent presence of TET1 and, notably, mutually exclusive histone modifications. These protections result in notable “CGI-shore” boundaries between unmethylated CpG-rich CGIs and methylated CpG-poor regions such as gene bodies [33].
In some instances, promoters CGIs are required to be temporarily inactive, but must still remain quiescent for later use. In these cases, repression can be accomplished through histone modifications, namely trimethylation of H3K27 via the polycomb repressive complex, PRC2, which is directed to CGIs by a number of different mechanisms [34]. Interestingly, it also appears that, H3K27me3 likely protects silenced promoter CGIs from unwanted spurious cytosine methylation, as the two are normally mutually exclusive in healthy cells [35, 36]. Many of these CGIs lose their protection by H3K27me3 with age or during carcinogenesis, potentially leading to aberrant hypermethylation and potential silencing [37, 38].
A minority of promoters, particularly those with lower CpG densities, do become targets of DNA methylation. The genes associated with these promoters require long-term, stable repression and include inactive X-chromosome genes, imprinted genes and those expressed exclusively in the germ line. These genes are typically silenced by a combination of H3K9 methylation and DNA methylation that ensures long-term silencing through formation of stable heterochromatic structures [39].
Lastly, some genes that provide embryonic stem cells with the attributes of self-renewal and pluripotency [40] are bivalently marked with both active (H3K4me3) and repressive (H3K27me3) histone modifications [41, 42], resulting in basal low-level expression during embryogenesis. During differentiation, this bivalency is lost and many of these pluripotency genes are targeted for DNA methylation, preventing dedifferentiation [43, 44]. In fact, demethylation of bivalent genes is thought to be a necessary step in somatic cell reprogramming, a key process that occurs during carcinogenesis that allows differentiated cells to revert back to a pluripotent state [45].
3. Defining Maintenance Stochasticism and Aberrant Methylation
According to the classical theory of DNA methylation, the bimodal methylation patterns established during embryogenesis are maintained during cell division by DNMT1 in a tightly controlled, linear process that ostensibly results in the propagation of precise methylation patterns. This classical view continues to be reiterated in the literature to this day [46], despite its inability to account for a number of experimental observations, particularly the considerable evidence demonstrating widespread methylation heterogeneity even amongst nominally healthy cells [38, 47, 48].
There are a number of other consistently-observed phenomena for which the classic model cannot account. For one, the DNMT1 enzyme is highly error-prone, showing only a 10- to 40-fold preference for hemimethylated (methylated on only one strand) over unmethylated CpG sites. This belies DNMT1’s role as the sole enzyme responsible for faithful maintenance methylation and further cannot account for the relatively high degree of transgenerational fidelity between mother and daughter cells [49]. Similarly, the classic concept of methylation being faithfully copied to the daughter strands on a site-by-site basis has also been invalidated by numerous experiments [38, 47, 48, 50]. Evidence from these experiments has strongly indicated that it is the methylation density, rather than precise patterns, that pass from mother to daughter cells [48, 50–52]. The classic model also fails to account for counteracting phenomena that prevent or remove methylation, such as mutually-exclusive histone marks and passive or active demethylation, e.g. through the TET family of enzymes. Lastly, in the classic model, the DNMT enzymes have separate and distinct roles. However, recent studies have indicated that in the processes of both maintenance and de novo methylation [53], methylation is accomplished through a coordinated effort between DNMT1 and DNMT3A/B enzymes [19, 54].
In light of these discrepancies, several new candidate models have been developed to account for experimental data that are discordant with the classic theory [53, 55–57]. While differing on some specifics, each of these second generation models utilize a dynamic stochastic theory (herein referred to as maintenance stochasticism) of both de novo and maintenance methylation. In these models, methylation is maintained through the local activity of methyltransferases, DNA demethylases and the DNA replication rate [53].
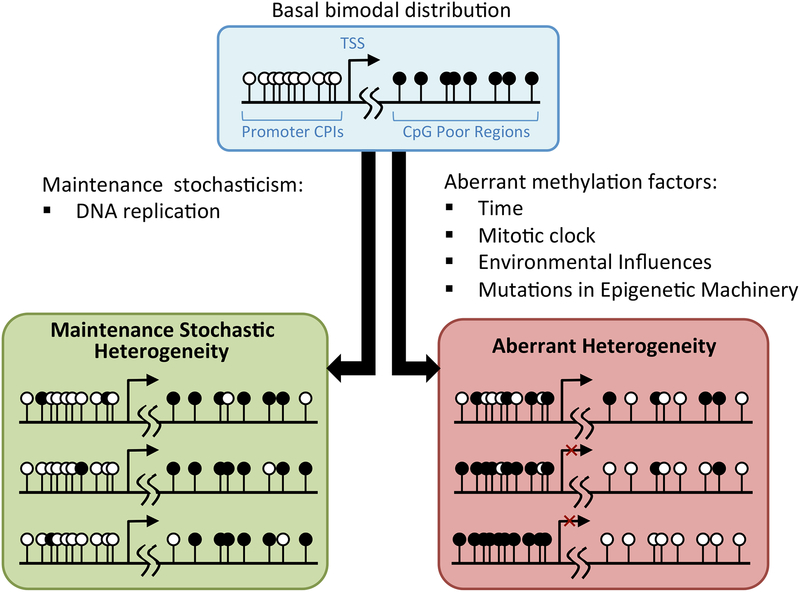
The concept of DNA methylation as a metastable dynamic process has considerable implications in the interpretation of heterogeneous methylation vis-à-vis cancer-specific aberrant methylation. Specifically, it predicts that the slight differences in methylation patterns observed in fully differentiated cells of the same tissue are not only tolerated, but expected and minor differences in pattern are unlikely to significantly affect cell phenotype or, in the case of cancer, yield neoplastic clones. This prediction has been supported in experiments, for example in the case of promoter CGIs, changes in methylation density, not specific methylation patterns, have been shown to determine whether or not a gene is silenced and to what extent [58–61]. Consequently, in many cases aberrant methylation might only be considered as such if it results in a notable alteration in methylation density or in the erosion of the boundary between CGIs and CpG-poor regions [33, 47, 62]. Indeed, as illustrated in Figure 1, this paradigm implies that all aberrant methylation could be considered heterogeneous (with respect to “normal” methylation patterns), however the converse would not necessarily be true. Dynamic methylation also provides viable mechanism that can at least partially account for the considerable change in methylation patterns that occur as humans age [63, 64].
Figure 1. Aberrant methylation vs maintenance stochasticism.
Recently-developed dynamic models of methylation propose that the basal methylation patterns established during embryogenesis (top) are propagated from parent to daughter DNA by cellular machinery that is dynamically regulated. This results in minor stochastic differences (left) in methylation patterns in subsequent cellular generations. Over time, these differences can be greatly exacerbated by various factors to produce aberrant methylation (right) that can result in inappropriate gene expression contributing to carcinogenesis.
4. The Accrual of Heterogeneous Methylation
Upwards of several thousand promoter regions can become heterogeneously and/or aberrantly methylated during the transformation of otherwise healthy cells into a neoplastic state [65]. How these loci become inappropriately methylated prior to and during carcinogenesis are open questions of active interest. To date, there are a number of known factors that researchers have determined to be contributors to the development of aberrant heterogeneous methylation, including age, number of cellular divisions, environmental exposures, and numerous cancer-associated phenomena, including genetic mutations in epigenetic regulators and hyperproliferation.
4.1. Age
Humans gradually accrue spurious changes in their methylome (epimutations) throughout their lifetime in what is referred to as epigenetic drift [64, 66]. This accumulation of spurious methylation (resulting in heterogeneous methylation patterns) is relatively consistent between individuals and can even be used as an “epigenetic clock” to predict the age of an individual within five years [67]. Studies have shown that the majority of this aberrant methylation does not arise in a random manner. Rather, errors preferentially accumulate at sites that carry PRC2 repressive [68] or bivalent [69] marks such as occur in multipotent cell-types, including embryonic and adult stem cells. Notably, hypermethylation of these loci appear to be independent of tissue type and have further been shown to be particularly associated with adult cancers and even preneoplastic populations [68, 70].
4.2. Mitotic Clock
A separate but related concept is that of the mitotic clock, which is a measure of the aberrant methylation that is due exclusively to the aforementioned lack of fidelity of the DNA methylation maintenance machinery. A number of studies have attempted to isolate the contribution from this source, with most estimates demonstrating a fidelity of roughly 99.9% for CpG sites located within promoter CGIs, and even higher in other contexts [38, 51]. This amounts to an error rate of one error per division for every 1000 CpG sites on average, which is six to seven orders of magnitude higher than the rate for DNA replication [71]. Nonetheless, it stands to reason that the mitotic clock is not the primary driver of the epigenetic clock as there is little variation in epigenetic drift between tissues that differ widely in turnover rate [64].
4.3. Environment
One area of intensive research is the question of how exogenous influences from the environment affect the methylome and epigenetic phenomena [72–75]. There is a growing list of environmental factors that have been shown to “speed up” the epigenetic clock and result in an overall increase in heterogeneous methylation. Among these are: environmental pollutants [72], acute and chronic chemical exposure [76, 77], smoking [78], alcohol use [79], diet [80, 81], UV exposure [82] and viral infection [83, 84], to name a few. Many of these studies have further shown that, similar to age-related methylation, most of these environmental influences also result in preferential aberrant methylation of PRC2 and bivalent promoter loci [64].
4.4. Mutations Affecting Epigenetic Regulators
Recent progress in next-generation sequencing technologies have enabled performance of large-scale genomic studies that have identified mutations affecting epigenetic regulation that frequently appear in a number of cancer types. Given the strong reliance upon properly functioning epigenetic machinery for the maintenance of epigenetic marks, including DNA methylation, it is no surprise that these mutations might result in an increase in aberrant methylation.
While a detailed list of these genes is beyond the current scope, they can be broadly categorized into three groups: epigenetic modulators, epigenetic modifiers and epigenetic mediators [85]. Epigenetic modulators are genes that activate or repress epigenetic machinery in cancer. Examples of modulators include IDH1/2, KRAS, APC and TP53, among others. Epigenetic modifiers are genes responsible for modifying DNA methylation or chromatin structure, and can be further subclassified into writers, editors and readers [86]. Of these three subcategories, only the writers and editors actually directly alter DNA methylation. Writer-modifiers include the DNMT enzymes, while editor-modifiers include the TET enzymes and reader-modifiers include the MBD proteins. Some other examples of epigenetic modifiers are MLL 1/2/3, SETD2, EZH2 and BRD4. Lastly, epigenetic mediators are genes that are regulated by modifiers and result in an increase in pluripotency or survival, such as those which are epigenetically poised in stem cell populations. These genes include OCT4, NANOG and SOX2 as well as others.
The preponderant co-occurrence of aberrantly methylated DNA along with genetic mutations in cancer strongly suggests that genetic and epigenetic stability are inextricably linked. Some recent investigations have begun to uncover how genetic and epigenetic factors can interact in shaping the methylome in cancer. Among these are notable examples of genetic-epigenetic interplay in certain forms of the so-called “CpG island methylator phenotype (CIMP).” CIMP was first described shortly after the initial gold rush for genetic loci with cancer-associated methylation, when it became clear that some cases of colorectal cancer were characterized by significantly more CGI methylation than others of the same type [87]. And while CIMP has been identified in numerous other tumor types, including bladder, breast, ovarian, prostate, endometrial, gastric, glioblastomas, hepatocellular, lung and pancreatic cancers [88], the molecular underpinnings have for the most part remained elusive. However, more recent studies, particularly of CIMP in glioma and leukemia, have identified mutations involved in an axis of pathways that lead to aberrant methylation. Specifically, somatic mutations were found in isocitrate dehydrogenase-1 (IDH1) in many cases of glioblastoma CIMP [89], while somatic mutations in both IDH1 and IDH2, as well as TET2 [90] were shown to occur in leukemic forms of CIMP [91]. Each of these mutations were ultimately found to lead to in an increase in the oncometabolite, 2-hydroxyglutarate, which acts as an inhibitor of the TET proteins, yielding an increase in aberrant hypermethylation, particularly within promoters targeted by PRC2.
Another example of genetic-epigenetic interplay occurs in activating mutations in the KRAS gene. These have been shown to increase ZNF304, a zinc-finger DNA-binding protein, which recruits a corepressor complex containing DNMT1, resulting in targeted methylation of promoter CGIs of a number of tumor suppressor genes associated with CIMP [92]. Similarly, mutations in the retinoblastoma tumor suppressor gene (RB) result in an increase in histone acetyltransferase-dependent activation of the DNA damage response gene, ataxia telangiectasia mutated (ATM), which associates with DNMT1, ultimately resulting in aberrant hypermethylation of various promoter CGIs [93].
As might be expected, the factors influencing the rate of aberrant methylation accrual are not mutually exclusive. For example, a recent study demonstrated that the same loci that become aberrantly methylated in buccal cells in response to smoking in vitro, are not only predictive of lung cancer, but numerous other, non-smoking-related, cancers as well [94]. The authors proposed that these results might be explained by the existence of a common mechanism between methylation that occurs as a result of acute environmental influences and cancerous tissue at large. For example, in both of these cases there is an overall increase in both the number and frequency of cell divisions, as experienced by the buccal cells exposed to smoke and cancer cells with shortened cell cycles. This hypothesis is supported by recent evidence indicating that that variations in cell cycle length may greatly perturb epigenetic memory, leading to significant increases in heterogeneous methylation [47, 95].
5. Distinguishing the Contribution of Heterogeneous Methylation to Cancer Heterogeneity
The heterogeneous methylation that builds up as a result of these various factors can lead to the development of loci with significant changes in methylation density and erosion of the boundaries between hypomethylated and hypermethylated regions [62, 96]. Experimental evidence indicates that these differentially methylated regions (DMRs) positively correlate with the probability of developing neoplasia [62, 68]. In fact, it has recently been shown that methylation heterogeneity actually peaks immediately prior to cancer initiation and often declines somewhat thereafter [97, 98]. But correlation is not causation and while it seems clear that some of this methylation contributes to inappropriate long-term silencing of key “driver” genes such as tumor suppressors [11], experimental evidence strongly suggests that a significant portion of these and other methylation events are so-called “passenger” events with little to no functional effect on cell phenotype [63, 99, 100]. For example, age-associated hypermethylation is known to preferentially accumulate in regions that would normally not be expressed by the tissue of origin [63, 101, 102] or are already silenced by other mechanisms, such as loci associated with PRCs and H3K9 methylation [33, 39, 103, 104]. Likewise, the majority of age- and cancer-associated hypomethylation yields no functional effect [63]. Said another way, it appears that heterogeneous methylation often acts to reinforce existing chromatin states, as opposed to creating them [11].
That is not to say that heterogeneous methylation does not directly contribute to carcinogenesis. In fact, even if the majority of silencing and unsilencing is established by other mechanisms, it is currently thought that it is changes in methylation that are mainly responsible for propagating these states to daughter cells as a means to the development of epigenetic clonal populations [105–108]. An example of this can be seen in somatic cell reprogramming, which recent evidence suggests may act as a prerequisite for tumorigenesis [45]. In this case, despite the considerable amount of epigenetic reprogramming that occurs via histone marks, full reprogramming of somatic cells cannot be accomplished without alterations in methylation. In particular, TET-mediated demethylation of the promoters associated with OCT4 and NANOG is required in order to achieve the fully pluripotent state thought to underlie much of the epigenetic and overall tumor cell plasticity that is a common characteristic of many cancerous phenotypes [107]. Similarly, during the well-known phenomenon of epithelial-mesenchymal transition (EMT), methylation acts to reinforce the H3K9me3-mediated silencing of promoters of genes associated with epithelial traits, thereby providing directional transition and development of clones with a mesenchymal phenotype [108]. This paradigm is also supported by clinical studies. For example, it has recently been shown that when comparing methylation patterns between patients suffering from metastatic cancer of the same type, there is often considerable heterogeneity in methylation between individuals (intertumor heterogeneity), but minimal heterogeneity between metastases within the same individual (intratumor heterogeneity) [105]. This indicates that by the time cancer has reached the point of metastasis, the high methylation heterogeneity seen in the initial stages of neoplasia has resolved into stably propagated methylation profiles producing distinct, phenotypically-advantageous epigenetic clones [97].
However, overall, despite tremendous effort in the elucidation of the effects of heterogeneous methylation, it still remains difficult to posit definitive rules to determine a priori whether heterogeneous methylation of a given locus controls gene expression, stabilizes it or even exerts any functional effect at all [109]. Consequently, any discussion regarding the effects of heterogeneous methylation upon a particular locus typically requires reference to its potential functionality within the precise context in which it occurs. Nonetheless, when taken as a whole, current experimental data does allow assertion of a number of general themes regarding the effects of heterogeneous methylation vis-à-vis clonal development and cancer heterogeneity. Furthermore, by noting the covariation of these themes, a picture of the potential interplay between heterogeneous methylation and cancer heterogeneity begins to begin to emerge [97].
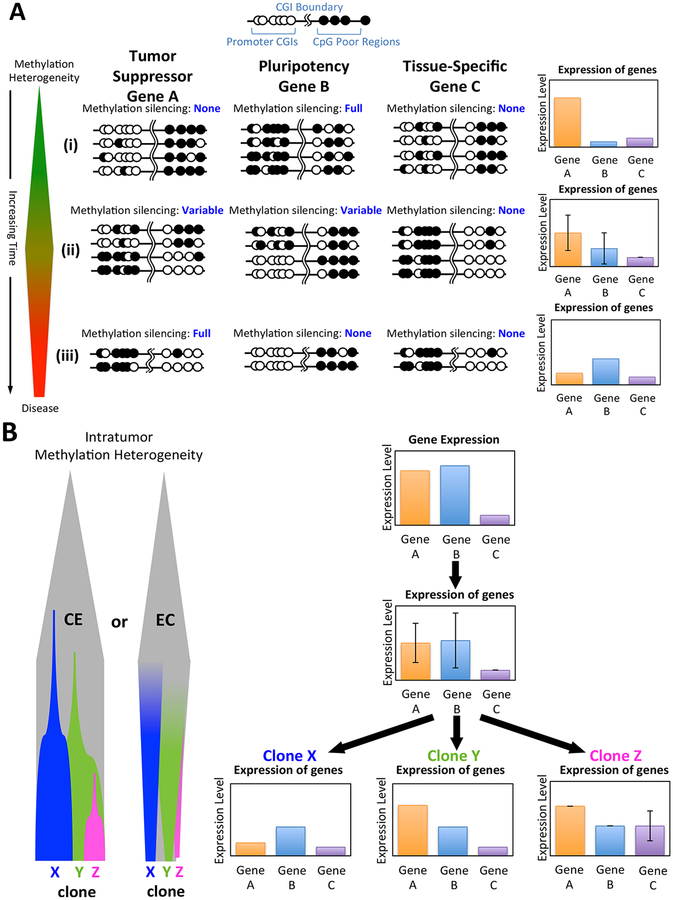
The potential interplay between various experimentally observed themes can perhaps best be exemplified by a prototype schema, such as the one illustrated in Figure 3A. Shown is a simplified hypothetical example how this interplay might result in the emergence of a particular clone. At the top are methylation patterns representing CpG-rich (e.g., promoter CGIs) and CpG-poor loci associated with three types of hypothetical genes, Genes A, B and C. The bar graphs represent the respective expression levels of these genes prior to and throughout the course of carcinogenesis. Tumor Suppressor Gene A represents tumor suppressor or other “driver” genes that act as key regulators of cell maintenance, such as CDKN2A [110], SFRP genes [111], SOCS1, SHP1 [112], RASSF1A [113], CHFR [114], et al.. On the other hand, Tissue-Specific Gene C represents any of the many genes that are not expressed or required by the respective tissue [63, 101, 102]. Lastly, Pluripotency Gene B represents genes important for the establishment of pluripotency that are typically bivalently marked during embryogenesis, but later silenced by methylation during differentiation, examples include: NANOG and OCT4 [115, 116] ,ESRRB [117] and SOX2 [118].
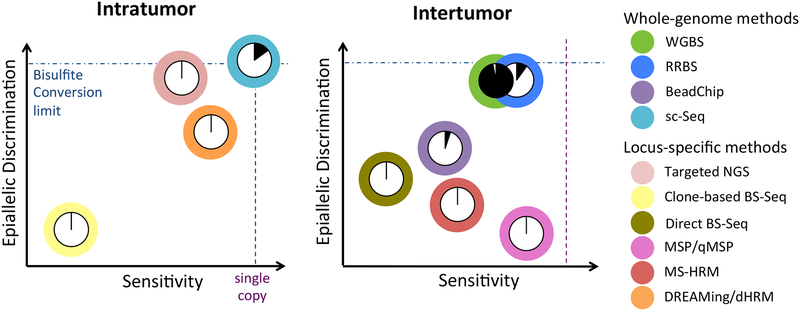
Figure 3. Comparison of DNA methylation detection techniques.
Methylation detection methods are shown categorized according to whether they are best suited to the assessment of intratumor heterogeneity (left) or intertumor heterogeneity (right). Each technique is represented by a color-coded circle with a pie chart indicating its relative methylome coverage and plotted according to its relative ability to resolve different methylation patterns (epiallelic discrimination) versus its relative ability to detect those patterns within a mixed population (sensitivity).
The schema begins with the bimodal patterns of methylation established during embryogenesis. Over time (progressing from top to bottom) and in response to other exogenous factors, cells within the tissue develop heterogeneous methylation patterns at the three loci. Some patterns develop that contain only minor methylation differences as predicted by a dynamic theory of methylation maintenance (maintenance stochasticism). Others emerge that differ in density and/or position beyond maintenance stochasticism and are termed aberrant. Early on and prior to the development of disease, only minor pattern variations with little heterogeneity are present within the promoters of both Gene A and Gene B, which remain expressed and unexpressed, respectively. In the promoters of tissue-specific genes, such as Gene C, methylation preferentially accumulates but has no functional effect, as they are already silenced by other mechanisms.
As time progresses toward the initial stages of carcinogenesis, aberrant methylation stochastically accrues resulting in a high degree of methylation heterogeneity throughout the entire genome as a result of, and resulting in, widespread epigenetic instability [97]. The patterns of methylation within the promoters of Gene A and Gene B begin to exhibit a high levels of heterogeneity leading to significant variability in gene expression between different cells. The cells exhibiting significant hypomethylation in the Gene B promoter start to develop a pluripotent phenotype at this time. By this point, the promoter of Gene C has already accumulated large amounts of aberrant hypermethylation and remains unexpressed.
As the cancer advances, some cells begin to establish definitive expression patterns and develop into a quasi-stable clone, Clone X, with particular phenotypic characteristics. In this case, Clone X emerges only from cells that also show hypomethylation in Gene B. Evolutionary pressures select only those cells that also exhibit hypermethylation-associated silencing of Tumor Suppressor A. Lastly, the promoter of Gene C remains fully methylated with little expression. The combination of these final methylation patterns along with the numerous others in the clonal genome ultimately combine to create the cancerous cellular phenotype of Clone X.
It’s important to note that the evolution of Clone X is not predetermined. In fact, immediately prior to cancer initiation, the high level of epigenetic instability and associated methylation heterogeneity may have the potential to result in the generation of numerous different phenotypes in time and space [85, 119, 120]. Whether these clones form via expansion from a single cell (e.g., cancer stem cells) [121] or through evolutionary convergence [122] remains an important open question [99]. These points are illustrated in Figure 3B, which shows how other clones may emerge simultaneously or later on during disease progression. For example, Clone Y might emerge from cells that retain expression of Gene A and some of the cells of Clone Z might have undergone EMT as a result of demethylation and re-expression of Gene C, as has been experimentally observed [123]. The particular phenotype of the clones that ultimately develop will be largely dependent on the precise evolutionary pressures and microenvironment within the tissue of origin. Somewhat counterintuitively, methylation heterogeneity often decreases as the clonal populations within an individual stabilize and expand while interclonal/interindividual heterogeneity increases due to the generation of clones with different phenotypes arising from the selective pressures particular to each individual [105].
In short, while the phenomena of methylation heterogeneity and epigenetic instability may be common to almost all cancer types [33], it can result in the development of cancers containing clones with considerably disparate characteristics. This has clear implications for cancer therapeutics. In fact, experimental evidence indicates that cancers exhibiting higher degrees of methylation heterogeneity are more difficult to treat [124, 125]. Consequently, the development of technologies capable of adequately detecting and treating polyclonal cancers will inevitably require a more thorough understanding of the correlations between heterogeneous methylation and therapeutic resistance [107].
Evaluating the complex relationships between the large numbers of aberrantly-methylated promoters and carcinogenesis is an extremely daunting task that will involve a broad collective effort between epigenetic researchers using numerous different means of analysis. In the last section of our review we present a summary of the techniques for assessing heterogeneous methylation that have been and continue to be developed to accomplish this gargantuan task.
6. Detecting Heterogeneous Methylation
While there exist numerous methods for the assessment of DNA methylation, they greatly vary in their respective abilities to detect and differentiate patterns of methylation, as well in their coverage of the genome. Consequently, the choice of which technique to use for the assessment of heterogeneous methylation can be largely dependent upon the question being asked. Here we seek to discuss the relative attributes of each technique in order to direct the reader to the most appropriate one for his or her application. Toward this end, Figure 3 shows two plots, one of technologies deemed most suitable for the assessment of intratumor heterogeneity and another of those better suited for intertumor assessment. The techniques are shown as pie charts indicating their relative methylome coverage, and plotted according to their relative abilities to resolve different methylation patterns (epiallelic discrimination) versus their relative abilities to detect those patterns within a mixed population (sensitivity).
Current methods for assessing DNA methylation typically involve bisulfite conversion of genomic DNA followed by downstream analysis methods. In bisulfite conversion (BSC), target DNA is treated with sodium bisulfite, which chemically converts unmethylated cytosine into uracil, while methylated cytosine residues are nominally preserved. During PCR amplification, uracil is replicated as thymine, thereby effectively translating changes in methylation to changes in primary sequence that can be readily analyzed by traditional genetic analysis techniques. It’s useful to note up front that the BSC process is not completely efficient, as 0.1% or more unmethylated cytosine residues typically fail to convert. Thus reliance upon BSC ultimately limits the ability of most methods to detect rare epialleles with minor differences in methylation patterns.
For clarity, we have divided the techniques into three categories: locus-specific, genome-wide and “in development” approaches.
6.1. Locus-specific approaches
6.1.1. Direct Bisulfite Sequencing
Traditional bisulfite sequencing (BS-Seq) relies upon PCR amplification of a particular locus of interest (LOI) within bisulfite-treated (BST) DNA, followed by Sanger sequencing [126]. The sequence obtained from the resulting amplicons can be used to obtain the average methylation level of each CpG dinucleotide in the original sample. The primary drawback to this method is amplification bias, which limits detection to methylation events that are present in 10–20% of the original template strands and prevents accurate quantitative analysis [127]. Today, BS-Seq is more commonly employed with pyrosequencing, which improves the sensitivity to ~5% [128]. Nonetheless, use of this method with 1st-generation sequencing technology precludes the ability to resolve different methylation patterns (epialleles) and is thus not suitable for the study of intratumor heterogeneity. Alternatively, next-generation sequencing technologies can be employed to somewhat overcome this limitation by offering the ability to detect and differentiate epiallelic fractions over 0.1–1% [129].
6.1.2. Clone-based Bisulfite Sequencing
Amplicons produced as above can alternatively be cloned into vectors and transfected into competent cells prior to sequencing (clone-based bisulfite sequencing). While capable of distinguishing individual epialleles, this method has become largely obsolete due to high costs and long processing times. The sensitivity of this method is also limited by PCR bias, as well as the impracticality of sequencing large numbers of clones.
6.1.3. Methylation-Specific PCR (MSP)-Based Methods
MSP is a methylation detection method designed to specifically amplify only fully-methylated epialleles from BST DNA [130]. This method is highly sensitive, cost-effective and especially useful when the targeted region contains well-validated biomarkers. However, some of the major disadvantages of MSP are that it is not quantitative and can be prone to false positives due to incomplete BSC. Real-time PCR analogs of MSP, Quantitative Methylation Specific PCR (qMSP) [131] or MethyLight [132], were created to partially address these problems, but nonetheless remain fundamentally limited by the inability to detect more than one [typically fully-methylated] pattern at a time. Furthermore, due to the fact that the presence of partially-methylated sequences or mixed populations can complicate MSP results, the technique is ill-suited for the assessment of intratumor heterogeneity.
6.1.4. Methylation-Sensitive High Resolution Melting (MS-HRM)
MS-HRM [133] is a post-PCR analysis method that identifies variations in nucleic acid sequences based on differences in their respective responses to thermal denaturation. An intercalating dye is incorporated during PCR and used to observe the denaturation [melt] profile and melt temperature (Tm) of the resulting amplicons. MS-HRM takes advantage of the higher binding energies of C-G versus A-T base pairs that result in an increase in Tm proportional to the total number of methylated cytosine in the template strands and thus allows discernment of the relative methylation density of the target strands. Despite its reasonable sensitivity (0.1% epiallelic fractions) and ability to assess methylation density, it is not well-suited to the evaluation of heterogeneous samples due to the inability to resolve complex-composite melt profiles [134]. This problem can in principle be addressed through sample digitization, however this remains impractical due to limitations in the number of reactions that can be digitized and simultaneously monitored [135].
6.1.5. Discrimination of Rare EpiAlleles by Melt (DREAMing)
DREAMing [136] represents a significant adaptation of MS-HRM that provides the capability to distinguish and enumerate individual methylated epialleles at single-copy sensitivity and single-CpG-site resolution. The method employs semi-limiting dilution of BST DNA targets followed by PCR and HRM analysis. By diluting such that no more than one epiallelic variant is presented in a reaction, this technique can detect individual epialleles and identify their respective methylation densities based on Tm. The methylation heterogeneity within the region of interest of each sample can then be visualized by plotting a histogram of the Tm within each reaction. This technique is ultrasensitive (0.005%), cost-effective and can be supplemented with sequencing of any amplicons of interest. Its primary drawback is lack of throughput as each sample must be quasi-digitized into a format compatible with existing HRM technology.
6.2. Genome-wide approaches
The advent of high-throughput sequencing (next-generation sequencing - NGS) effectively ushered in a new era of an affordable means for the assessment of DNA sequences at single-base resolution across the entire genome. While clearly a tremendous advancement, it is not without its caveats, particularly with respect to highly heterogeneous samples. The overall sensitivity of NGS-based approaches is largely dependent on the total number of reads, but fundamental limitations due to errors and biases prevent the achievement of sensitivities below 0.1–1% epiallelic fractions. A number of clever strategies, including SAFE-Seqs [137] and duplex sequencing [138], have been developed to overcome this barrier, but these require significantly more labor and cost. Furthermore, these have not yet been applied to the assessment of methylation, but ostensibly would remain fundamentally limited by the efficiency of the BSC process.
6.2.1. Whole-Genome Bisulfite-Sequencing (WGBS)
WGBS has been considered to be the gold standard in genome-wide methylation profiling [139]. The major advantage of WGBS is the comprehensive evaluation of methylation within almost all CpG sites of the entire genome at single-base resolution and without selective bias. However, up to now, only a handful of studies have successfully employed WGBS in the assessment of cancer methylomes [62, 140–142]. This is largely due to the extremely high cost of the assay and its technical downstream complexity. An adaptation of this method, called Reduced Representation Bisulfite Sequencing (RRBS) [143], involves the same sequencing technologies as WGBS, but utilizes a key pre-enrichment step to selectively enrich for CpG-dense regions. This reduced representation lowers the number of reads necessary to yield accurate results, thus decreasing overall costs and time compared to WGBS. RRBS typically covers only ~4% of all CpG dinucleotides (as opposed to ~95% by WGBS) due to the notable exclusion of intergenic and distal regulatory elements.
6.2.2. Single-cell sequencing (sc-Seq)
Until recently, all genome-wide DNA methylation profiling methods utilized epigenetically-mixed populations of cells as a starting material. As mentioned above, limitations in the technology preclude the assessment of epiallelic fractions below 0.1% and thus prevent the assessment of individual epialleles. However, exciting advancements have recently been made that now allow DNA derived from individual cells to be extracted, amplified and analyzed. Likewise, a number of genome bisulfite sequencing techniques have been amended to single-cell sequencing technologies for the assessment of methylation within individual cells. These techniques include scRRBS [144], scBS-Seq [145] and scWGBS [146]. While, in principle, scBS–Seq technologies offer the ability to compare the methylation states of different individuals, considerable hurdles remain that prevent their utility for the assessment of methylation heterogeneity. Specifically, the preparation of DNA from each cell involves a relatively complicated and inherently lossy process that introduces significant technical artifacts such as DNA loss/degradation, coverage nonuniformities, allelic dropouts, as well as high false-positive and false-negative rates. These issues greatly limit overall mapping efficiencies, making it necessary to perform >10 million reads to achieve typical coverages of 5–10% of all CpG sites. Consequently, scBS-Seq studies are currently limited to the assessment to relatively homogenous populations of cells, as heterogeneous populations would require sequencing of inordinate numbers of cells to achieve conclusive results.
6.2.3. Microarrays: Illumina Infinium BeadChips
The Infinium BeadChip arrays are microarray platforms that incorporate large numbers of microbeads, each containing a high density of oligonucleotides designed to probe for a single CpG site. The most widely-used version of this platform is the 450K array that provides the average methylation status of over 480,000 individual CpG-sites (~1.7% of all CpG sites) within a given sample. Moderate running costs and fairly rapid data processing times make the BeadChip platform a popular choice for the assessment of intertumor methylation heterogeneity, including numerous studies that have been incorporated into The Cancer Genome Atlas [147]. However, there are some notable precautions to bear in mind when using this method, including relatively high levels of false-positives due to probe cross-reactivity and overlap of CpGs with known SNPs [148]. An improved version (MethylationEPIC BeadChip microarray) has recently been introduced that increases the coverage to over 850,000 CpG sites or roughly 3.2% of all genomic CpG sites [149],
6.3. Methods in Development
Along with the continuous improvement of NGS, a new generation of single-molecule sequencing technologies (sometimes referred to as third generation sequencing - TGS) is emerging. These new platforms are in principle capable of detecting DNA modifications directly without bisulfite conversion or enzymatic modifications. The analysis of methylation patterns on a molecule-by-molecule basis without the need for BSC is the proverbial “holy grail” for the assessment of methylation heterogeneity. However, third generation sequencing remains under active development and requires significant improvements in reliability and/or throughput before primetime use.
6.3.1. Single-Molecule, Real-Time (SMRT) Sequencing
The SMRT sequencing method, currently marketed by Pacific Biosciences, allows direct, real-time detection of single nucleotide modifications through the use of zero-mode waveguides [150]. Detection is accomplished by monitoring the incorporation of fluorescently-labeled nucleotides into the complementary strand during synthesis by DNA polymerase. The method has been demonstrated to provide, not only the primary sequence, but also information regarding structural variants and epigenetic modifications (e.g., N6-methyladenine, 5mC and 5hmC) at a single-base pair resolution [151]. This platform seems to be a possible solution for limitations imposed by bisulfite-conversion-based techniques, since it holds the potential to assess methylation directly from unmodified DNA. However, the main drawbacks of this developing technology are the significant running cost and high error rate, estimated at upwards of 20% [152]. So far, detection of methylation by this method has only been demonstrated with bacterial DNA. The considerable technical hurdles associated with this technology will likely need to be overcome and its performance validated before genome-wide methylome studies can be attempted.
6.3.2. Nanopore Sequencing
Nanopore sequencing is a TGS technology that assesses single molecules of unmodified DNA by sensing alterations in electrical current that occur as different bases pass through a nanopore. The nanopore itself can be a biological protein [153] or synthetic solid-state construct [154]. Oxford Nanopore Technologies has recently released the first commercially-available sequencer based on this technology. The instrument, named MinION, is sold as a moderate-cost, hand-held and portable sequencing device that offers read lengths of hundreds of kilobases. It accepts samples as small as 10pg and does not require PCR amplification prior to analysis. Nanopores are also capable of distinguishing between cytosine, 5mC, 5hmC, albeit with considerable high error rates (1.7–12.2%) [155], currently limiting this technology to proof-of-concept research only [156, 157]. Studies involving genome-wide DNA methylation assessment have yet to be reported with this technology.
6.4. DNA Methylation Detection Techniques Summary
In general, genome-wide approaches are best suited for the assessment of intertumor methylation heterogeneity and biomarker discovery. NGS-based techniques can also be used to evaluate intratumor heterogeneity, but are limited by their inability to readily detect minor epiallelic fractions under 1%. On the other hand, locus-specific techniques provide a cost-effective means of assessing large sample cohorts for intra- and inter-individual methylation heterogeneity within pre-identified LOIs. Furthermore, a number of locus-specific techniques, such as targeted NGS and DREAMing, are lower in throughput, but provide the requisite sensitivity for the detection of minor epiallelic variants. A number of other particularly exciting technologies are under active development. Single-cell sequencing methods offer the potential to evaluate the methylomes of individual cells to provide insight into the relationship between heterogeneous methylation and cell phenotype. Third-generation sequencing technologies are particularly promising for the assessment of methylation heterogeneity as they hold the unique potential for the assessment of genome-wide methylation at single-molecule sensitivity without the need for BSC of the template strands.
7. Conclusions
In this review, we sought summarize the processes that contribute to the alteration of the basic bimodal patterns of methylation that are established early on in embryogenesis and how these alterations contribute to, and are affected by, carcinogenesis. Although investigations into these relationships remain in their relative infancy, a number of key generalizations were extracted and assimilated into a working model of how different clones might arise from the epigenetic instability associated with methylation heterogeneity. Lastly, we briefly discussed some of the available methods for assessing heterogeneous methylation and how these methods may be used to shed light on the many unanswered questions that remain in the elucidation of the complex relationship between methylation and cancer heterogeneity.
Figure 2. The contribution of heterogeneous methylation to the emergence of cancerous clones.
(A) At the earliest stage (i), minor stochastic differences in methylation patterns accumulate in the promoters of Tumor Suppressor Gene A, Pluripotency Gene B and Tissue-Specific Gene C. Immediately prior to the development of neoplasia (ii), significant methylation heterogeneity has developed in Gene A, resulting in considerable variance in expression between cells. Some select cells become hypomethylated in Gene B, resulting in a pluripotent phenotype. Gene C is not normally expressed and preferentially accrues high levels of CGI methylation in its promoter. As the cancer develops (iii), methylation heterogeneity is somewhat reduced as quasi stable clone X emerges via widespread methylation-mediated silencing of Gene A and hypomethylation of Gene B. Gene C remains unexpressed with heavy methylation in its promoter. (B) The phenotype of Clone X might emerge from methylation/epigenetic heterogeneity through clonal expansion (CE) of a cell that develops a particular phenotypic advantage or through evolutionary convergence (EC), where shared evolutionary pressures yield groups of cells with similar methylation profiles. These same phenomena can yield other phenotypic clones (Clones Y and Z) throughout the course of carcinogenesis, contributing to overall intratumor heterogeneity.
Acknowledgments
Funding
This work was supported by the National Institutes of Health [R01CA155305 and R21CA186809 U54CA151838].
References
- [1].Holliday R, Pugh J, DNA modification mechanisms and gene activity during development, Science 187(4173) (1975) 226–232. [PubMed] [Google Scholar]
- [2].Riggs AD, X inactivation, differentiation, and DNA methylation, Cytogenet. Cell Genet 14(1) (1975) 9–25. [DOI] [PubMed] [Google Scholar]
- [3].Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, et al. , The 5-methylcytosine content of DNA from human tumors, Nucleic Acids Research 11(19) (1983) 6883–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Feinberg AP, Vogelstein B, Hypomethylation distinguishes genes of some human cancers from their normal counterparts, Nature 301(5895) (1983) 89–92. [DOI] [PubMed] [Google Scholar]
- [5].Baylin SB, Höppener JWM, de Bustros A, Steenbergh PH, Lips CJM, Nelkin BD, DNA Methylation Patterns of the Calcitonin Gene in Human Lung Cancers and Lymphomas, Cancer Res 46(6) (1986) 2917–2922. [PubMed] [Google Scholar]
- [6].Baylin SB, Herman JG, DNA hypermethylation in tumorigenesis: epigenetics joins genetics, Trends in Genetics 16(4) (2000) 168–174. [DOI] [PubMed] [Google Scholar]
- [7].Swartz MN, Trautner TA, Kornberg A, Enzymatic Synthesis of Deoxyribonucleic Acid: XI. Further Studies On Nearest Neighbor Base Sequences In Deoxyribonucleic Acids, J. Biol. Chem 237(6) (1962) 1961–1967. [PubMed] [Google Scholar]
- [8].Ehrlich M, Gama-Sosa MA, Huang L-H, Midgett RM, Kuo KC, McCune RA, et al. , Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells, Nucleic Acids Research 10(8) (1982) 2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Antequera F, Bird A, Number of CpG islands and genes in human and mouse, Proceedings of the National Academy of Sciences of the United States of America 90(24) (1993) 11995–11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bird AP, CpG islands as gene markers in the vertebrate nucleus, Trends in Genetics 3 (1987) 342–347. [Google Scholar]
- [11].Jones PA, Functions of DNA methylation: islands, start sites, gene bodies and beyond, Nat Rev Genet 13(7) (2012) 484–492. [DOI] [PubMed] [Google Scholar]
- [12].Schubeler D, Function and information content of DNA methylation, Nature 517(7534) (2015) 321–326. [DOI] [PubMed] [Google Scholar]
- [13].Spruijt CG, Vermeulen M, DNA methylation: old dog, new tricks?, Nat. Struct. Mol. Biol 21(11) (2014) 949–954. [DOI] [PubMed] [Google Scholar]
- [14].Hwang W, Oliver VF, Merbs SL, Zhu H, Qian J, Prediction of promoters and enhancers using multiple DNA methylation-associated features, BMC Genomics 16(Suppl 7) (2015) S11–S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Martino D, Saffery R, Characteristics of DNA methylation and gene expression in regulatory features on the Infinium 450k Beadchip, bioRxiv (2015). [Google Scholar]
- [16].Reddington JP, Pennings S, Meehan RR, Non-canonical functions of the DNA methylome in gene regulation, Biochem. J 451 (2013) 13–23. [DOI] [PubMed] [Google Scholar]
- [17].Fatemi M, Hermann A, Gowher H, Jeltsch A, Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA, European Journal of Biochemistry 269(20) (2002) 4981–4984. [DOI] [PubMed] [Google Scholar]
- [18].Kim G-D, Ni J, Kelesoglu N, Roberts RJ, Pradhan S, Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases, The EMBO Journal 21(15) (2002) 4183–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, et al. , Cooperativity between DNA Methyltransferases in the Maintenance Methylation of Repetitive Elements, Molecular and Cellular Biology 22(2) (2002) 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Clouaire T, Stancheva I, Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin?, Cellular and molecular life sciences: CMLS 65(10) (2008) 1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hill PWS, Amouroux R, Hajkova P, DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: An emerging complex story, Genomics 104(5) (2014) 324–333. [DOI] [PubMed] [Google Scholar]
- [22].Tahiliani M, Koh KP, Shen YH, Pastor WA, Bandukwala H, Brudno Y, et al. , Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1, Science 324(5929) (2009) 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, et al. , Chromatin dynamics during epigenetic reprogramming in the mouse germ line, Nature 452(7189) (2008) 877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopina J, Chen PY, et al. , DNA Demethylation Dynamics in the Human Prenatal Germline, Cell 161(6) (2015) 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, et al. , Germline DNA Demethylation Dynamics and Imprint Erasure Through 5-Hydroxymethylcytosine, Science 339(6118) (2013) 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, et al. , The Dynamics of Genome-wide DNA Methylation Reprogramming in Mouse Primordial Germ Cells, Molecular Cell 48(6) (2012) 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sabag O, Zamir A, Keshet I, Hecht M, Ludwig G, Tabib A, et al. , Establishment of methylation patterns in ES cells, Nat Struct Mol Biol 21(1) (2014) 110–112. [DOI] [PubMed] [Google Scholar]
- [28].Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, et al. , Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line, Genes & Development 6(5) (1992) 705–714. [DOI] [PubMed] [Google Scholar]
- [29].Monk M, Boubelik M, Lehnert S, Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development, Development 99(3) (1987) 371–382. [DOI] [PubMed] [Google Scholar]
- [30].Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Names A, et al. , Spl elements protect a CpG island from de novo methylation, Nature 371(6496) (1994) 435–438. [DOI] [PubMed] [Google Scholar]
- [31].Macleod D, Charlton J, Mullins J, Bird AP, Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island, Genes & Development 8(19) (1994) 2282–2292. [DOI] [PubMed] [Google Scholar]
- [32].Ginno Paul A., Lott Paul L., Christensen Holly C., Korf I, Chédin F, R-Loop Formation Is a Distinctive Characteristic of Unmethylated Human CpG Island Promoters, Molecular Cell 45(6) (2012) 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Timp W, Feinberg AP, Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host, Nat Rev Cancer 13(7) (2013) 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Blackledge NP, Rose NR, Klose RJ, Targeting Polycomb systems to regulate gene expression: modifications to a complex story, Nat Rev Mol Cell Biol 16(11) (2015) 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Brinkman AB, Gu H, Bartels SJJ, Zhang Y, Matarese F, Simmer F, et al. , Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk, Genome Research 22(6) (2012) 1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Murphy PJ, Cipriany BR, Wallin CB, Ju CY, Szeto K, Hagarman JA, et al. , Single-molecule analysis of combinatorial epigenomic states in normal and tumor cells, Proceedings of the National Academy of Sciences 110(19) (2013) 7772–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. , Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer, Nat Genet 39(2) (2007) 232–236. [DOI] [PubMed] [Google Scholar]
- [38].Landan G, Cohen NM, Mukamel Z, Bar A, Molchadsky A, Brosh R, et al. , Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues, Nat Genet 44(11) (2012) 1207–1214. [DOI] [PubMed] [Google Scholar]
- [39].Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, et al. , Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation, Genes & Development 17(15) (2003) 1855–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. , 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming, Nat. Commun 2 (2011) 8. [DOI] [PubMed] [Google Scholar]
- [41].Voigt P, Tee W-W, Reinberg D, A double take on bivalent promoters, Genes & Development 27(12) (2013) 1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bernstein BE, Mikkelsen TS, Xie XH, Kamal M, Huebert DJ, Cuff J, et al. , A bivalent chromatin structure marks key developmental genes in embryonic stem cells, Cell 125(2) (2006) 315–326. [DOI] [PubMed] [Google Scholar]
- [43].Hagarman JA, Motley MP, Kristjansdottir K, Soloway PD, Coordinate Regulation of DNA Methylation and H3K27me3 in Mouse Embryonic Stem Cells, PLoS ONE 8(1) (2013) e53880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, et al. , G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis, Nat Cell Biol 8(2) (2006) 188–194. [DOI] [PubMed] [Google Scholar]
- [45].Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, Ablain J, et al. , A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation, Science 351(6272) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Law JA, Jacobsen SE, Establishing, maintaining and modifying DNA methylation patterns in plants and animals, Nat Rev Genet 11(3) (2010) 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shipony Z, Mukamel Z, Cohen NM, Landan G, Chomsky E, Zeliger SR, et al. , Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells, Nature 513(7516) (2014) 115–119. [DOI] [PubMed] [Google Scholar]
- [48].Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, et al. , DNA Methylation Analysis of Chromosome 21 Gene Promoters at Single Base Pair and Single Allele Resolution, PLoS Genet 5(3) (2009) e1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bird A, DNA methylation patterns and epigenetic memory, Genes & Development 16(1) (2002) 6–21. [DOI] [PubMed] [Google Scholar]
- [50].Silva AJ, Ward K, White R, Mosaic Methylation in Clonal Tissue, Developmental Biology 156(2) (1993) 391–398. [DOI] [PubMed] [Google Scholar]
- [51].Ushijima T, Watanabe N, Okochi E, Kaneda A, Sugimura T, Miyamoto K, Fidelity of the Methylation Pattern and Its Variation in the Genome, Genome Research 13(5) (2003) 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Laird CD, Pleasant ND, Clark AD, Sneeden JL, Hassan KMA, Manley NC, et al. , Hairpin-bisulfite PCR: Assessing epigenetic methylation patterns on complementary strands of individual DNA molecules, Proceedings of the National Academy of Sciences of the United States of America 101(1) (2004) 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jeltsch A, Jurkowska RZ, New concepts in DNA methylation, Trends in Biochemical Sciences 39(7) (2014) 310–318. [DOI] [PubMed] [Google Scholar]
- [54].Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, et al. , In Vivo Control of CpG and Non-CpG DNA Methylation by DNA Methyltransferases, PLoS Genet 8(6) (2012) e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Riggs AD, Xiong Z, Methylation and epigenetic fidelity, Proceedings of the National Academy of Sciences of the United States of America 101(1) (2004) 4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sneppen K, Dodd IB, Nucleosome dynamics and maintenance of epigenetic states of CpG islands, Phys. Rev. E 93(6) (2016) 8. [DOI] [PubMed] [Google Scholar]
- [57].Sormani G, Haerter JO, Lovkvist C, Sneppen K, Stabilization of epigenetic states of CpG islands by local cooperation, Mol. Biosyst 12(7) (2016) 2142–2146. [DOI] [PubMed] [Google Scholar]
- [58].Kass SU, Pruss D, Wolffe AP, How does DNA methylation repress transcription?, Trends in Genetics 13(11) (1997) 444–449. [DOI] [PubMed] [Google Scholar]
- [59].Hsieh CL, Dependence of transcriptional repression on CpG methylation density, Molecular and Cellular Biology 14(8) (1994) 5487–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Boyes J, Bird A, Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein, The EMBO Journal 11(1) (1992) 327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, et al. , Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome, Nat Genet 39(4) (2007) 457–466. [DOI] [PubMed] [Google Scholar]
- [62].Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, et al. , Increased methylation variation in epigenetic domains across cancer types, Nature Genetics 43(8) (2011) 768–U77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yuan T, Jiao YM, de Jong S, Ophoff RA, Beck S, Teschendorff AE, An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging, Plos Genetics 11(2) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zheng SC, Widschwendter M, Teschendorff AE, Epigenetic drift, epigenetic clocks and cancer risk, Epigenomics 8(5) (2016) 705–719. [DOI] [PubMed] [Google Scholar]
- [65].Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, et al. , Aberrant CpG-island methylation has non-random and tumour-type-specific patterns, Nat Genet 24(2) (2000) 132–138. [DOI] [PubMed] [Google Scholar]
- [66].Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. , Aging and Environmental Exposures Alter Tissue-Specific DNA Methylation Dependent upon CpG Island Context, PLoS Genet 5(8) (2009) e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Horvath S, DNA methylation age of human tissues and cell types, Genome Biology 14(10) (2013) 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, et al. , Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer, Genome Research 20(4) (2010) 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rakyan VK, Down TA, Maslau S, Andrew T, Yang T-P, Beyan H, et al. , Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains, Genome Research 20(4) (2010) 434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. , A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing, Nat Genet 39(2) (2007) 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Loeb LA, Monnat RJ, DNA polymerases and human disease, Nat Rev Genet 9(8) (2008) 594–604. [DOI] [PubMed] [Google Scholar]
- [72].Cao Y, Environmental pollution and DNA methylation: carcinogenesis, clinical significance, and practical applications, Front. Med 9(3) (2015) 261–274. [DOI] [PubMed] [Google Scholar]
- [73].Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, et al. , Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships, Hum. Genet 131(10) (2012) 1565–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Feil R, Fraga MF, Epigenetics and the environment: emerging patterns and implications, Nat Rev Genet 13(2) (2012) 97–109. [DOI] [PubMed] [Google Scholar]
- [75].Barrow TM, Michels KB, Epigenetic epidemiology of cancer, Biochem. Biophys. Res. Commun 455(1–2) (2014) 70–83. [DOI] [PubMed] [Google Scholar]
- [76].Chappell G, Pogribny IP, Guyton KZ, Rusyn I, Epigenetic alterations induced by genotoxic occupational and environmental human chemical carcinogens: A systematic literature review, Mutat. Res.-Rev. Mutat. Res 768 (2016) 27–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Collotta M, Bertazzi PA, Bollati V, Epigenetics and pesticides, Toxicology 307 (2013) 35–41. [DOI] [PubMed] [Google Scholar]
- [78].Ambatipudi S, Cuenin C, Hernandez-Vargas H, Ghantous A, Le Calvez-Kelm F, Kaaks R, et al. , Tobacco smoking-associated genome-wide DNA methylation changes in the EPIC study, Epigenomics 8(5) (2016) 599–618. [DOI] [PubMed] [Google Scholar]
- [79].Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S, Emerging role of epigenetics in the actions of alcohol, Alcoholism (NY) 32(9) (2008) 1525–1534. [DOI] [PubMed] [Google Scholar]
- [80].Hullar MAJ, Fu BC, Diet, the Gut Microbiome, and Epigenetics, Cancer J 20(3) (2014) 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].McKay JA, Mathers JC, Diet induced epigenetic changes and their implications for health, Acta Physiol 202(2) (2011) 103–118. [DOI] [PubMed] [Google Scholar]
- [82].Katiyar SK, Singh T, Prasad R, Sun Q, Vaid M, Epigenetic Alterations in Ultraviolet Radiation-Induced Skin Carcinogenesis: Interaction of Bioactive Dietary Components on Epigenetic Targets(), Photochemistry and photobiology 88(5) (2012) 1066–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Fernandez AF, Esteller M, Viral epigenomes in human tumorigenesis, Oncogene 29(10) (2010) 1405–1420. [DOI] [PubMed] [Google Scholar]
- [84].Li HP, Leu YW, Chang YS, Epigenetic changes in virus-associated human cancers, Cell Res 15(4) (2005) 262–271. [DOI] [PubMed] [Google Scholar]
- [85].Feinberg AP, Koldobskiy MA, Gondor A, Epigenetic modulators, modifiers and mediators in cancer aetiology and progression, Nat Rev Genet 17(5) (2016) 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P, Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer, Nat Rev Genet 14(11) (2013) 765–780. [DOI] [PubMed] [Google Scholar]
- [87].Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa J-PJ, CpG island methylator phenotype in colorectal cancer, Proceedings of the National Academy of Sciences 96(15) (1999) 8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Hughes LAE, Melotte V, de Schrijver J, de Maat M, Smit VTHBM, Bovée JVMG, et al. , The CpG Island Methylator Phenotype: What’s in a Name?, Cancer Res 73(19) (2013) 5858–5868. [DOI] [PubMed] [Google Scholar]
- [89].Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. , IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype, Nature 483(7390) (2012) 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Langemeijer SMC, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, et al. , Acquired mutations in TET2 are common in myelodysplastic syndromes, Nat Genet 41(7) (2009) 838–842. [DOI] [PubMed] [Google Scholar]
- [91].Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. , Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation, Cancer Cell 18(6) (2010) 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Serra RW, Fang M, Park SM, Hutchinson L, Green MR, A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype, eLife 3 (2014) e02313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Shamma A, Suzuki M, Hayashi N, Kobayashi M, Sasaki N, Nishiuchi T, et al. , ATM Mediates pRB Function To Control DNMT1 Protein Stability and DNA Methylation, Molecular and Cellular Biology 33(16) (2013) 3113–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Teschendorff AE, Yang Z, Wong A, et al. , Correlation of smoking-associated dna methylation changes in buccal cells with dna methylation changes in epithelial cancer, JAMA Oncology 1(4) (2015) 476–485. [DOI] [PubMed] [Google Scholar]
- [95].Mehari BZ, Cédric V, Daniel J, Effect of replication on epigenetic memory and consequences on gene transcription, Physical Biology 12(2) (2015) 026007. [DOI] [PubMed] [Google Scholar]
- [96].Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, et al. , Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population, PLoS Genet 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Teschendorff AE, Liu X, Caren H, Pollard SM, Beck S, Widschwendter M, et al. , The Dynamics of DNA Methylation Covariation Patterns in Carcinogenesis, PLoS Comput Biol 10(7) (2014) e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Teschendorff A, Jones A, Fiegl H, Sargent A, Zhuang J, Kitchener H, et al. , Epigenetic variability in cells of normal cytology is associated with the risk of future morphological transformation, Genome Med 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].De Carvalho DD, Sharma S, You JS, Su S-F, Taberlay PC, Kelly TK, et al. , DNA methylation screening identifies driver epigenetic events of cancer cell survival, Cancer Cell 21(5) (2012) 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Burgess DJ, Epigenetics: Dissecting driving DNA methylations, Nat Rev Cancer 12(7) (2012) 448–449. [DOI] [PubMed] [Google Scholar]
- [101].West J, Widschwendter M, Teschendorff AE, Distinctive topology of age-associated epigenetic drift in the human interactome, Proceedings of the National Academy of Sciences of the United States of America 110(35) (2013) 14138–14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Evidence for an instructive mechanism of de novo methylation in cancer cells, Nat Genet 38 (2006). [DOI] [PubMed] [Google Scholar]
- [103].Aranda S, Mas G, Di Croce L, Regulation of gene transcription by Polycomb proteins, Science Advances 1(11) (2015) e1500737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Saksouk N, Barth Teresa K., Ziegler-Birling C, Olova N, Nowak A, Rey E, et al. , Redundant Mechanisms to Form Silent Chromatin at Pericentromeric Regions Rely on BEND3 and DNA Methylation, Molecular Cell 56(4) (2014) 580–594. [DOI] [PubMed] [Google Scholar]
- [105].Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, et al. , DNA Methylation Alterations Exhibit Intraindividual Stability and Interindividual Heterogeneity in Prostate Cancer Metastases, Science Translational Medicine 5(169) (2013) 169ra10–169ra10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Mazor T, Pankov A, Jun S Song, Joseph F. Costello, Intratumoral Heterogeneity of the Epigenome, Cancer Cell 29(4) (2016) 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Easwaran H, Tsai H-C, Baylin SB, Cancer epigenetics: Tumor Heterogeneity, Plasticity of Stem-like States, and Drug Resistance, Molecular cell 54(5) (2014) 716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Tam WL, Weinberg RA, The epigenetics of epithelial-mesenchymal plasticity in cancer, Nat Med 19(11) (2013) 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Moarii M, Boeva V, Vert J-P, Reyal F, Changes in correlation between promoter methylation and gene expression in cancer, BMC Genomics 16(1) (2015) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, et al. , Aberrant methylation of p16INK4a is an early event in lung cancer and a potential biomarker for early diagnosis, Proceedings of the National Academy of Sciences 95(20) (1998) 11891–11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Suzuki H, Watkins DN, Jair K-W, Schuebel KE, Markowitz SD, Dong Chen W, et al. , Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer, Nat Genet 36(4) (2004) 417–422. [DOI] [PubMed] [Google Scholar]
- [112].Chim C-S, Fung T-K, Cheung W-C, Liang R, Kwong Y-L, SOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway, Blood 103(12) (2004) 4630–4635. [DOI] [PubMed] [Google Scholar]
- [113].van Engeland M, Roemen G, Brink M, Paachen MMM, Weijenberg MP, de Bruine AP, et al. , K-ras mutations and RASSF1A promoter methylation in colorectal cancer, Oncogene 21(23) (2002) 3792–3795. [DOI] [PubMed] [Google Scholar]
- [114].Toyota M, Sasaki Y, Satoh A, Ogi K, Kikuchi T, Suzuki H, et al. , Epigenetic inactivation of CHFR in human tumors, Proceedings of the National Academy of Sciences of the United States of America 100(13) (2003) 7818–7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM, Reprogramming towards pluripotency requires AID-dependent DNA demethylation, Nature 463(7284) (2010) 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Shi JL, Shi W, Ni LC, Xu XD, Su X, Xia L, et al. , OCT4 is epigenetically regulated by DNA hypomethylation of promoter and exon in primary gliomas, Oncol. Rep 30(1) (2013) 201–206. [DOI] [PubMed] [Google Scholar]
- [117].Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, et al. , Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2, Nature 488(7413) (2012) 652–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].You H, Ding W, Rountree CB, Epigenetic regulation of cancer stem cell marker CD133 by transforming growth factor-β, Hepatology 51(5) (2010) 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].McGranahan N, Swanton C, Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution, Cancer Cell 27(1) 15–26. [DOI] [PubMed] [Google Scholar]
- [120].Moelans CB, de Groot JS, Pan X, van der Wall E, van Diest PJ, Clonal intratumor heterogeneity of promoter hypermethylation in breast cancer by MS-MLPA, Mod Pathol 27(6) (2014) 869–874. [DOI] [PubMed] [Google Scholar]
- [121].Siegmund KD, Marjoram P, Woo Y-J, Tavaré S, Shibata D, Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers, Proceedings of the National Academy of Sciences 106(12) (2009) 4828–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Nishino K, Toyoda M, Yamazaki-Inoue M, Fukawatase Y, Chikazawa E, Sakaguchi H, et al. , DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time, PLoS Genet 7(5) (2011) e1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Carmona FJ, Davalos V, Vidal E, Gomez A, Heyn H, Hashimoto Y, et al. , A Comprehensive DNA Methylation Profile of Epithelial-to-Mesenchymal Transition, Cancer Res 74(19) (2014) 5608–5619. [DOI] [PubMed] [Google Scholar]
- [124].Claus R, Gu L, Assenov Y, Hüllein J, Zucknick M, Brocks D, et al. , Heterogeneity and Evolution Of DNA Methylation In Chronic Lymphocytic Leukemia, Blood 122(21) (2013) 1626–1626. [Google Scholar]
- [125].Oakes CC, Claus R, Gu L, Assenov Y, Hüllein J, Zucknick M, et al. , Evolution of DNA Methylation Is Linked to Genetic Aberrations in Chronic Lymphocytic Leukemia, Cancer Discovery 4(3) (2014) 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Frommer M, Mcdonald LE, Millar DS, Collis CM, Watt F, Grigg GW, et al. , A Genomic Sequencing Protocol That Yields a Positive Display of 5-Methylcytosine Residues in Individual DNA Strands, Proceedings of the National Academy of Sciences of the United States of America 89(5) (1992) 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Lewin J, Schmitt AO, Adorjan P, Hildmann T, Piepenbrock C, Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates, Bioinformatics 20(17) (2004) 3005–3012. [DOI] [PubMed] [Google Scholar]
- [128].Colella S, Shen L, Baggerly KA, Issa JP, Krahe R, Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites, Biotechniques 35(1) (2003) 146–50. [DOI] [PubMed] [Google Scholar]
- [129].Taylor KH, Kramer RS, Davis JW, Guo J, Duff DJ, Xu D, et al. , Ultradeep Bisulfite Sequencing Analysis of DNA Methylation Patterns in Multiple Gene Promoters by 454 Sequencing, Cancer Res 67(18) (2007) 8511–8518. [DOI] [PubMed] [Google Scholar]
- [130].Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB, Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands, Proc Natl Acad Sci U S A 93(18) (1996) 9821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Lo YM, Wong IH, Zhang J, Tein MS, Ng MH, Hjelm NM, Quantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reaction, Cancer Res 59(16) (1999) 3899–903. [PubMed] [Google Scholar]
- [132].Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. , MethyLight: a high-throughput assay to measure DNA methylation, Nucleic Acids Res 28(8) (2000) E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Wojdacz TK, Dobrovic A, Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation, Nucleic Acids Res 35(6) (2007) e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Lin TC, Jiang SS, Chou WC, Hou HA, Lin YM, Chang CL, et al. , Rapid Assessment of the Heterogeneous Methylation Status of CEBPA in Patients with Acute Myeloid Leukemia by Using High-Resolution Melting Profile, J Mol Diagn 13(5) (2011) 514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Candiloro ILM, Mikeska T, Hokland P, Dobrovic A, Rapid analysis of heterogeneously methylated DNA using digital methylation-sensitive high resolution melting: application to the CDKN2B (p15) gene, Epigenet Chromatin 1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Pisanic TR, Athamanolap P, Poh W, Chen C, Hulbert A, Brock MV, et al. , DREAMing: a simple and ultrasensitive method for assessing intratumor epigenetic heterogeneity directly from liquid biopsies, Nucleic Acids Research 43(22) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B, Detection and quantification of rare mutations with massively parallel sequencing, Proceedings of the National Academy of Sciences 108(23) (2011) 9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, et al. , Detecting ultralow-frequency mutations by Duplex Sequencing, Nat. Protocols 9(11) (2014) 2586–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. , Human DNA methylomes at base resolution show widespread epigenomic differences, Nature 462(7271) (2009) 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu YP, et al. , Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains, Nature Genetics 44(1) (2012) 40–U62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, et al. , Distinct DNA methylomes of newborns and centenarians, Proceedings of the National Academy of Sciences of the United States of America 109(26) (2012) 10522–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Ziller MJ, Gu HC, Muller F, Donaghey J, Tsai LTY, Kohlbacher O, et al. , Charting a dynamic DNA methylation landscape of the human genome, Nature 500(7463) (2013) 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R, Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis, Nucleic Acids Research 33(18) (2005) 5868–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Guo HS, Zhu P, Wu XL, Li XL, Wen L, Tang FC, Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing, Genome Research 23(12) (2013) 2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, et al. , Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity, Nat Methods 11(8) (2014) 817–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schonegger A, Klughammer J, et al. , Single-Cell DNA Methylome Sequencing and Bioinformatic Inference of Epigenomic Cell-State Dynamics, Cell Rep 10(8) (2015) 1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Weisenberger DJ, Characterizing DNA methylation alterations from The Cancer Genome Atlas, J Clin Invest 124(1) (2014) 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. , Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray, Epigenetics 8(2) (2013) 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Moran S, Arribas C, Esteller M, Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences, Epigenomics 8(3) (2016) 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. , Real-Time DNA Sequencing from Single Polymerase Molecules, Science 323(5910) (2009) 133–138. [DOI] [PubMed] [Google Scholar]
- [151].Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. , Direct detection of DNA methylation during single-molecule, real-time sequencing, Nature Methods 7(6) (2010) 461–U72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, Hegarty R, et al. , Characterizing and measuring bias in sequence data, Genome Biology 14(5) (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Laszlo AH, Derrington IM, Brinkerhoff H, Langford KW, Nova IC, Samson JM, et al. , Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA, Proceedings of the National Academy of Sciences of the United States of America 110(47) (2013) 18904–18909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Shim J, Humphreys GI, Venkatesan BM, Munz JM, Zou XQ, Sathe C, et al. , Detection and Quantification of Methylation in DNA using Solid-State Nanopores, Sci Rep-Uk 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Schreiber J, Wescoe ZL, Abu-Shumays R, Vivian JT, Baatar B, Karplus K, et al. , Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands, Proceedings of the National Academy of Sciences 110(47) (2013) 18910–18915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Laszlo AH, Derrington IM, Brinkerhoff H, Langford KW, Nova IC, Samson JM, et al. , Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA, Proceedings of the National Academy of Sciences 110(47) (2013) 18904–18909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Simpson JT, Workman R, Zuzarte PC, David M, Dursi LJ, Timp W, Detecting DNA Methylation using the Oxford Nanopore Technologies MinION sequencer, bioRxiv (2016). [Google Scholar]