Abstract
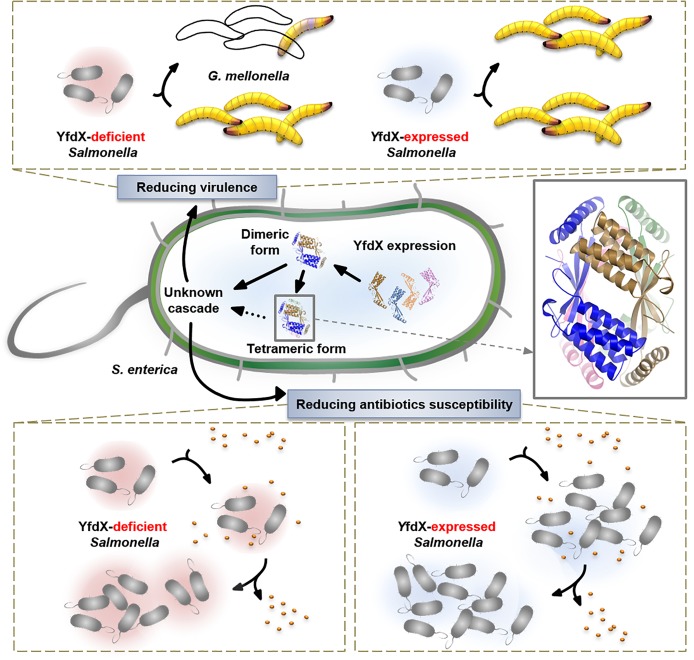
YfdX is a prokaryotic protein encoded by several pathogenic bacteria including Salmonella enterica serovar Typhi, which causes one of the most fatal infectious diseases, typhoid fever. YfdX is a product of the yfdXWUVE operon and is known to be under the control of EvgA, a regulator protein controlling the expression of several proteins involved in response to environmental stress, in Escherichia coli. Nevertheless, unlike other proteins encoded by the same operon, the structural and physiological aspects of YfdX have been poorly characterized. Here, we identified a previously unknown pH-dependent stoichiometric conversion of S. Typhi YfdX between dimeric and tetrameric states; this conversion was further analyzed via determining its structure by X-ray crystallography at high resolution and by small-angle X-ray scattering in a solution state and via structure-based mutant studies. Biologically, YfdX was proven to be critically involved in Salmonella susceptibility to two β-lactam antibiotics, penicillin G and carbenicillin, as bacterial growth significantly impaired by its deficiency upon treatment with each of the two antibiotics was recovered by chromosomal complementation. Furthermore, by using Galleria mellonella larvae as an in vivo model of Salmonella infection, we demonstrated that Salmonella virulence was remarkably enhanced by YfdX deficiency, which was complemented by a transient expression of the wild-type or dimeric mutant but not by that of the monomeric mutant. The present study work provides direct evidence regarding the participation of YfdX in Salmonella antibiotic susceptibility and in the modulation of bacterial virulence, providing a new insight into this pathogen’s strategies for survival and growth.
Keywords: STY3178, YfdX, Salmonella Typhi, antibiotics susceptibility, virulence
Introduction
Salmonella enterica serovar Typhi is a gram-negative bacterium that infects humans only (Hurley et al., 2014), causing systemic typhoid fever. Typhoid is one of the most widespread and hazardous infectious diseases in developing countries, with more than 16 million cases and 200,000 estimated deaths per year (Buckle et al., 2012; Dougan and Baker, 2014; Azmatullah et al., 2015). S. Typhi initially penetrates the small intestinal epithelial cells and then spreads through the bloodstream to other organs such as the spleen, liver, and bone marrow, where this bacterium multiplies and reenters the bloodstream causing symptoms including a high fever (Everest et al., 2001). A variety of antibiotics, such as ampicillin, chloramphenicol, trimethoprim-sulfamethoxazole, and ciprofloxacin, have already been used against S. Typhi (Kalra et al., 2003). Nevertheless, exposure to antibiotics over long periods has allowed S. Typhi to acquire resistance to various antibiotics through genetic changes: a phenomenon called “multiple drug resistance (MDR)” (Rowe et al., 1997; Kalra et al., 2003; Frye and Jackson, 2013). MDR strains exhibit strong resistance to antibiotics, and lead to treatment failure (Rowe et al., 1997; Parkhill et al., 2001; Kariuki et al., 2015; Karkey et al., 2018).
Bacterial resistance to antibiotics is acquired via a combination of a variety of strategies, which include alteration of a target protein, enzymatic deactivation, and restriction of antibiotic accessibility (Nikaido, 2009). How bacteria orchestrate these complicated events accompanying a change in the expression of several proteins has been under intensive investigation. It has been reported that EvgA, one of the response regulator proteins of Escherichia coli, constitutes a two-component system together with the sensor kinase EvgS and controls the expression of a wide range of genes involved in the response to stressors such as antibiotics and pH changes (Nishino and Yamaguchi, 2001, 2002; Masuda and Church, 2002; Nishino et al., 2003). The yfdXWUVE operon is one of the targets upregulated by EvgA (Masuda and Church, 2002; Nishino et al., 2003) and encodes formyl-CoA transferase YfdW (Gruez et al., 2003), oxalyl-CoA decarboxylase YfdU (Werther et al., 2010), acetyl-CoA:oxalate CoA-transferase YfdE (Mullins et al., 2013), putative transporter protein YfdV, and an uncharacterized protein YfdX, whose expression has been reported to be enhanced a 1000-fold by EvgA overproduction (Nishino and Yamaguchi, 2002; Nishino et al., 2003). Homologs of YfdX have been identified in various virulent bacterial species including S. Typhi (Parkhill et al., 2001), S. Typhimurium (McClelland et al., 2001), Hafnia alvei (Lazaro-Diez et al., 2016), Shigella dysenteriae (Kaur et al., 2014), and Klebsiella pneumoniae (Jiang et al., 2010). Furthermore, the crystal structure of K. pneumoniae YfdX in the tetramer-estimated form was determined and deposited at the PDB without accompanying publications (PDB code 3DZA). Recently, it has been reported that a bacterial protein STY3178, an ortholog of YfdX from the S. Typhi representative MDR strain CT18, interacts with three antibiotics ciprofloxacin, ampicillin, and rifampin (Saha et al., 2016b), has a chaperone-like activity (Saha et al., 2016a), and is modeled to interact with the outer-membrane protein STY3179 (Mondal et al., 2017), providing clues for understanding this protein. However, a number of unresolved issues about YfdX remain, with the absence of structural and physiological analyses, including its functional role during bacterial infection as well as its precise stoichiometry. K. pneumoniae YfdX appears to form a homotetramer in the crystal structure, whereas S. Typhi YfdX is proposed to form a trimer in solution according to dynamic light scattering, size exclusion chromatography, and nuclear magnetic resonance experiments (Saha et al., 2016b).
In this study, we attempted to answer such unresolved issues about the YfdX protein through structural, biochemical, and physiological analyses. An unexpected stoichiometric conversion of S. Typhi YfdX between the dimer and tetramer was identified, which was further analyzed via mutational assays based on crystal structure determination. Moreover, we found that Salmonella YfdX plays a significant role in bacterial susceptibility to penicillin G and carbenicillin and is involved in the negative regulation of bacterial virulence in an insect larvae model. These data collectively expand our understanding of this poorly studied protein.
Materials and Methods
Crystallization and Structure Determination of st_YfdX
The DNA fragment coding for residues 10–186 of st_YfdX was cloned into the pET21a plasmid (Novagen). The protein was produced in the E. coli BL21(DE3) RIL strain (Novagen) at 18°C and purified on a Ni-NTA column (QIAGEN) first. The protein was further purified on a HiLoad 26/600 Superdex 75 prep grade gel filtration column (GE Healthcare), equilibrated with a buffer consisting of 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 1 mM DTT. All crystals were obtained via the sitting-drop vapor diffusion method at 18°C by mixing and equilibrating 0.4 μL samples of the protein solution (20 mg/mL) with a precipitant solution as described in Table 1. Before data collection, crystals were immersed briefly in the reservoir solution containing a cryoprotectant reagent as shown in Table 1. Diffraction data were collected on the beamline 5C and 7A at the Pohang Accelerator Laboratory, South Korea, and processed using the HKL 2000 software (Otwinowski and Minor, 1997). The structure was determined by the molecular replacement method with the Phaser software (McCoy et al., 2007) using the structure of kp_YfdX (PDB code 3DZA) as a search model. Programs Coot (Emsley and Cowtan, 2004) and PHENIX (Adams et al., 2010) were used for the model building and refinement, respectively. Crystallographic data statistics are summarized in Table 21.
Table 1.
Crystallization and cryoprotectant reagent conditions.
| Space group | Crystallization condition | Cryoprotectant reagent | |
|---|---|---|---|
| Crystal I | F222 | 0.1 M sodium acetate (pH 4.6), 2.5 M sodium chloride, 12% PEG1500, 1.5% 2-methyl-2,4-pentanediol | 15% glycerol |
| Crystal II | F222 | 0.1 M sodium acetate (pH 4.6), 2.5 M sodium chloride, 12% PEG1500, 1.5% 2-methyl-2,4-pentanediol | 5% glycerol |
| Crystal III | P222 | 4% Tacsimate (pH 5.0), 12% PEG3350 | 20% glycerol |
Table 2.
Data collection and structure refinement statistics.
| PDB code | Crystal I (6A02) | Crystal II (6A07) | Crystal III (6A09) |
|---|---|---|---|
| Space group | F222 | F222 | P222 |
| Unit cell dimensions | |||
| a, b, c (Å) | 88.8, 92.5, 95.5 | 88.8, 92,2, 95.4 | 72.4, 127.9, 170.4 |
| α, β, γ (o) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.9793 | 0.9793 | 0.9793 |
| Resolution (Å) | 50.0-1.4 (1.42-1.40)b | 50.0-1.5 (1.53-1.50)b | 50.0-2.3 (2.34-2.30)b |
| Rsyma | 9.5 (27.1) | 8.1 (28.0) | 8.0 (27.7) |
| I/σ(I) | 41.3 (4.7) | 38.3 (5.6) | 29.5 (4.9) |
| Completeness (%) | 98.8 (96.5) | 99.4 (99.5) | 98.5 (91.2) |
| Redundancy | 9.4 | 6.0 | 5.5 |
| Refinement | |||
| Resolution (Å) | 50.0-1.4 (1.43-1.40) | 50.0-1.5 (1.54-1.50) | 50.0-2.3 (2.35-2.30) |
| Number of reflections | 38,242 | 31,146 | 70,194 |
| Rworkc/Rfree | 18.9/21.3 (28.2/29.3) | 18.2/21.5 (22.2/23.5) | 18.0/23.3 (23.1/27.6) |
| Number of atoms | |||
| Protein/water and ion | 1326/198 | 1337/223 | 10668/587 |
| RMSD | |||
| Bond lengths (Å)/angles (o) | 0.005/0.713 | 0.005/0.720 | 0.008/0.839 |
| Ramachandran plot (%) | |||
| Favored/allowed | 98.2/1.8 | 98.8/1.2 | 98.2/1.8 |
| Average B-values (Å2) | |||
| Protein/water and ion | 17.1/27.2 | 19.1/30.0 | 37.4/38.7 |
| Molprobity score | 1.09 (99th percentile; N= 3363, 1.400 ± 0.25 Å) | 1.05 (99th percentile; N = 4775, 1.501 ± 0.25 Å) | 1.35 (100th percentile; N = 8821, 2.293 ± 0.25 Å) |
aRsym= Σ |Iobs – Iavg|/Iobs, where Iobs is the observed intensity of individual reflection and Iavg is the average over symmetry equivalents. bThe numbers in parentheses are statistics from the shell with the highest resolution. cRwork = Σ ||Fo|– |Fc||/Σ |Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated with 10% of the data.
Preparation of Recombinant Proteins
Each of the DNA fragments encoding mutant st_YfdX proteins containing F42A⋅F45A⋅Y165A or Y100A⋅I137A substitutions and a DNA fragment encoding kp_YfdX was cloned into the pET21a plasmid. The recombinant proteins were produced and purified as was the wild-type st_YfdX.
Size Exclusion Chromatography-Multiangle Light Scattering Experiments
Size exclusion chromatography-multiangle light scattering (SEC-MALS) was performed using Superdex 75 Increase 10/300 GL (GE Healthcare). The differential refractive index spectra were recorded on Optilab T-rEX (Wyatt Technology Corporation), which was combined with high-performance liquid chromatography (Shimadzu) and DAWN HELEOS-II (Wyatt Technology Corporation). The weight-average molar mass was calculated using the ASTRA 6 software (Wyatt Technology Corporation).
Circular Dichroism Spectroscopy
Data were collected on a JASCO model J-815 spectropolarimeter with a 0.1-cm cuvette. The circular dichroism (CD) spectrum was recorded over the range of 200–260 nm in a nitrogen atmosphere with 0.1 mg/mL protein samples dissolved in 40 mM sodium phosphate buffer (pH 6.0 or 7.5). The spectrum comprised the accumulation of three scans at 0.2 nm intervals, which was corrected by subtracting signals from the buffer control. The raw CD signals were converted to mean residue ellipticity [𝜃MRE] (in deg cm2 dmol-1) using the equation [𝜃MRE] = 𝜃obs/Cnl, where 𝜃obs is the observed ellipticity (in millidegrees), C is the protein concentration (in molarity), n is the number of amino acid residues, and l is the pathlength (in millimeters).
Small-Angle X-Ray Scattering Experiments
Small-angle x-ray scattering (SAXS) measurements were carried out using the 4C SAXS II beamline at the Pohang Accelerator Laboratory, South Korea (Kim et al., 2017). Data were collected at 4°C with a sample-to-detector distance of 4,000 and 1,000 mm. Protein samples were prepared at pH 5.5 and 8.0, each of which was diluted to three different concentrations (1, 3, and 5 mg/mL). The SAXS data of each sample were collected in 10 successive frames of 5 s at 0.734 Å wavelength, which were measured in triplicates. Two-dimensional SAXS patterns were averaged and normalized for further analysis. Scattering intensities from the buffer solution were used as the experimental background. The scattering intensity data I(q) as a function of q (q = 4πsin𝜃/λ, where 𝜃 is half of the scattering angle and λ is the wavelength; 0.01 Å-1 < q < 0.7 Å-1) were obtained by radial averaging. CRYSOL (Svergun et al., 1995) and GNOM (Semenyuk and Svergun, 1991) were used to calculate the SAXS curves and distance distribution function P(r), respectively. Molecular envelops were reconstructed using the ab initio shape determination program DAMMIF (Franke and Svergun, 2009). Fifteen independent models were generated, compared, and averaged to obtain the refined models using the programs DAMAVER and DAMSTART. Surface rendering was achieved using the program PyMOL. Structural diagrams were superimposed onto the reconstructed dummy atoms using SUPCOMB (Kozin and Svergun, 2001). Normalized spatial discrepancies were 3.13 for the tetramer and 3.32 for the MolA–Mol B dimer.
Knockout and Complementation of yfdX
Salmonella transformants carrying the pKD46 Red helper plasmid were cultured in the Luria-Bertani (LB) medium containing 100 μg/mL ampicillin and 10 mM L-arabinose at 30°C until turbidity at 600 nm reached 0.35–0.4. The cells were harvested and washed three times with ice-cold 10% glycerol and sterilized water to prepare electro-competent cells. A polymerase chain reaction (PCR) product containing the kanamycin resistance gene (amplified from pKD4) flanked upstream and downstream by sequences of the target gene (yfdX) was obtained using primers yfdX-p1 (5′-TGGCCGCAACAAACATGACTGATAACGTTACTCTGAATAATGACAAGATGTGTAGGCTGGAGCTGCTTC-3′) and yfdX-p2 (5′-GCGCGGCGTCGTGCTGCACGGAGTGCGTGGGTGATTCCTGAACTGACTGAATGGGAATTAGCCATGGTCC-3′). Next, 50 μL of electro-competent cells was mixed with 500 ng of the PCR product containing the 50 bp homologous sequence arms at each end of the kanamycin resistance cassette. Electroporation was performed in a 2-mm cuvette on Gene Pulser 3 (Bio-Rad) set at 2.5 kV, 25 μF, and 200 Ω. Transformants were screened on LB agar containing 50 μg/mL kanamycin. Knockouts were confirmed with primers yfdX-up97 (5′-GTGGGTTACCGGTTCTGAATAG-3′) and yfdX-dn43 (5′-CATAATGCTGCCTGCTGTAATG-3′). The kanamycin cassette was removed by pCP20 transformation (Datsenko and Wanner, 2000). For chromosomal complementation, the yfdX-kanamycin resistance cassette fusion PCR products were prepared using primers yfdX-ATG20 (5′-ATGGTTATCCTGTTTTCAGG-3′) and yfdX-dn_H_P2 (5′-TGTCAGACATAATGCTGCCTGCTGTAATGTATACAGCAGGCTGGGGGATAATGGGAATTAGCCATGGTCC-3′). Homologous recombination was performed as described above. For plasmid complementation, yfdX genes encoding wild-type or each mutant were cloned into the pNM12 vector harboring the ara promoter and ampicillin marker, which were then transformed into the S. Typhimurium UK-1 ΔyfdX strain.
Phenotype Microarrays
BiOLOG phenotypic microarray analyses were conducted following the manufacturer’s instructions (BiOLOG, Inc.). In brief, bacteria were grown at 37°C for 16 h on LB agar media, and then scraped and resuspended in IF-0a. After that, 100 μL of a cell resuspension was inoculated into each well of phenotype microarray plates (#1–20). Respiration signals from 0 to 24 h shown in red were measured by BiOLOG OmniLog and then compared between the two strains. The microarray results were validated following the BiOLOG protocol with small modifications. To determine the minimum inhibitory concentration, serially diluted antibiotics were mixed with cell suspensions (5 × 105 cfu/mL), and then respiration signals were measured. All the experiments were conducted in triplicate.
Insect-Toxicity Assay
Injection assays with Galleria mellonella larvae were performed as described by Flury et al. (2016) with small variations. G. mellonella caterpillars purchased from S-WORM were grown at 30°C until they reached the fourth larval instar. Caterpillars were stored in the dark at 30°C for 3 days for stabilization. For injection, suspensions of overnight cultured bacterial cells were prepared in pH 7.4 PBS consisting of 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.47 mM KH2PO4, and diluted to the desired concentration (105 cfu/μL). Bacterial cells resuspended in 2 μL of PBS were injected into larvae with a 5 μL micro-syringe (MicroliterTM #701, Hamilton). The larvae were kept in Petri dishes at 30°C in the dark. For the evaluation of bacterial virulence, larvae were scored as live or dead for every 6 h up to 72 h. Each experiment consisted of three replicates per treatment with 10 larvae per replicate.
Bacterial RNA Isolation and Real-Time PCR
Salmonella Typhimurium UK1 strains were grown in the LB medium under aerobic conditions at 37°C for 4, 8, and 12 h before RNA isolation. The cells were stabilized using the RNA protect Bacteria Reagent (Qiagen), and total RNA was isolated using the RNeasy Kit (Qiagen). cDNA was synthesized with Superscript III Reverse transcriptase (Invitrogen). The mRNA levels of the coding regions of the yfdX gene were measured by quantification of cDNA using SYBR Green PCR Master Mix (Bio-Rad) with primers Q-ST-yfdX-F (5′-CTAGCAGGCGTCAGTGTTATT-3′) and Q-ST-yfdX-R (5′-ATGCCATCCTGAGCTGATTT-3′) monitored on a CFX connect real-time PCR detection system (Bio-Rad). The mRNA levels of the yfdX gene were normalized to the levels of 16S ribosomal RNA amplified with primers ST-16S-QF (5′-GTTAGCCGGTGCTTCTTCTG-3′) and ST-16S-QR (5′-TAGGCCTTCGGGTTGTAAAGT-3′).
Results
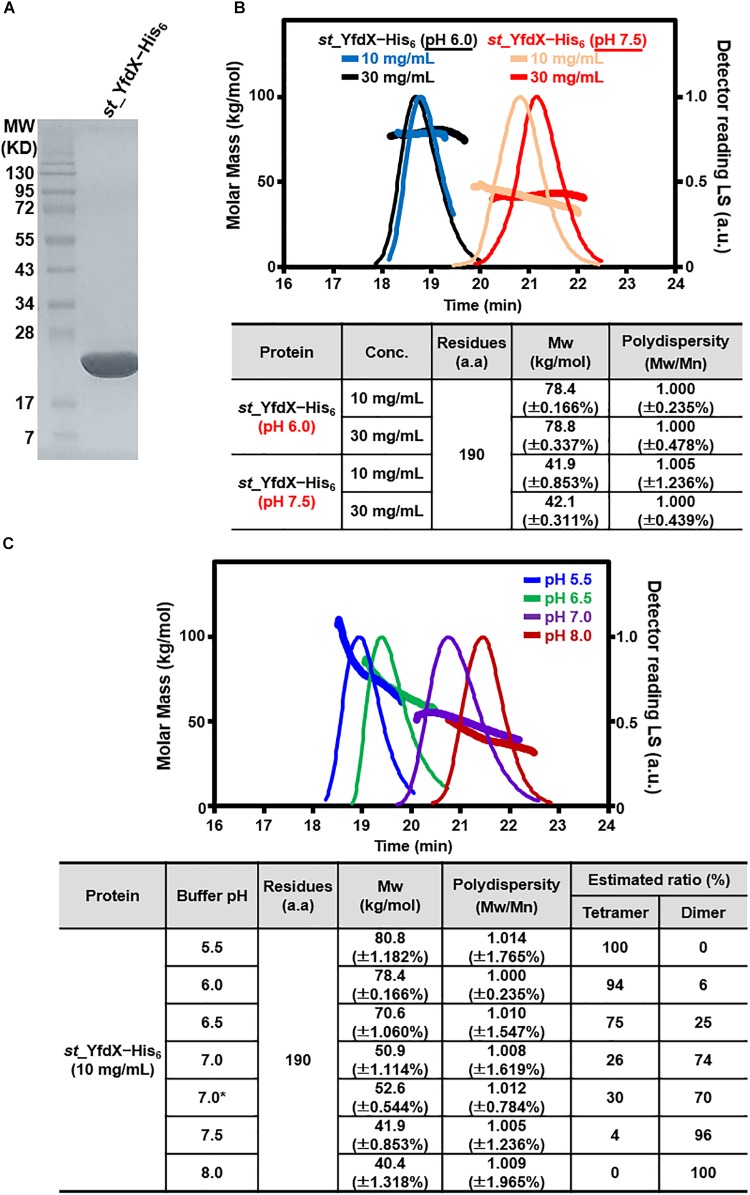
Analysis of Stoichiometry of S. Typhi YfdX by SEC-MALS
The recombinant S. Typhi YfdX protein containing residues 10–186 from its total 199 amino acids, referred to as st_YfdX in this manuscript, was overexpressed in E. coli and purified on a Ni–NTA column and a size-exclusion chromatography column, equilibrated with a final buffer consisting of 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 1 mM DTT (Figure 1A). To verify the stoichiometry of st_YfdX by means of the recombinant protein, we carried out SEC-MALS experiments, which is a useful tool for elucidating the molecular weight and stoichiometry of macromolecules in solution (Sahin and Roberts, 2012). Unexpectedly, the molecular weight of wild-type st_YfdX, which was concentrated to 10 mg/mL, was determined to be ∼42 kDa, suggesting that this protein exists as a dimer in solution, neither a trimer nor a tetramer, under this condition (Figure 1B). Increasing the concentration of the protein sample to 30 mg/mL did not alter its stoichiometry (Figure 1B). Based on the presence of the zinc ion coordination at the center of the K. pneumoniae YfdX (referred to as kp_YfdX) tetrameric structure (not shown), we suspected that a similar coordination might be necessary for the formation of the high-order oligomeric structure of st_YfdX. Therefore, the recombinant st_YfdX protein was dialyzed against a buffer composed of 20 mM Bis-Tris-HCl (pH 6.0), 200 mM NaCl, and 1 mM DTT, with the intention of lowering pH to prevent zinc precipitation (Sayilgan et al., 2010; Park et al., 2015). Prior to zinc treatment, SEC-MALS experiments were conducted with the prepared protein sample, to use the data as a control. Surprisingly, we found that the molecular weights of the st_YfdX samples concentrated to 10 and 30 mg/mL, respectively, are ∼78–79 kDa (Figure 1B), and thus it was shown that st_YfdX forms a tetramer at pH 6.0, even without being treated with zinc chloride. The molecular weight of st_YfdX was then further analyzed by SEC-MALS at various pH conditions ranging from 5.5 to 8.0 at 0.5 intervals. As described in Figure 1C, the molar mass of st_YfdX was shifted from 80.8 to 40.4 kDa, indicating that st_YfdX exists in a state of dynamic equilibrium between tetramer and dimer whose ratio is varied by pH. We also performed SEC-MALS on the recombinant kp_YfdX protein for comparison. As shown in Supplementary Figure S1, kp_YfdX exists as a tetramer at pH 6.0 and 7.5 but as a dimer at pH 10.0, showing that kp_YfdX also undergoes a pH-dependent transition between dimeric and tetrameric forms but the pH range is different from that of st_YfdX. Collectively, these data indicate that the recombinant st_YfdX protein can form a dimer or tetramer in solution, and this process is affected by pH of the solution.
FIGURE 1.
Preparation and SEC-MALS analysis of st_YfdX. (A) Purified st_YfdX–His6 (20 μg) was loaded onto a sodium dodecyl sulfate polyacrylamide gel electrophoresis system and visualized by Coomassie blue staining together with the size markers. (B) SEC-MALS analysis of wild-type st_YfdX–His6. (Top) Molar masses (in kg/mol) are plotted against the elution time (in min) from a size exclusion column. (Bottom) st_YfdX–His6 is in the form of a dimer at pH 7.5 and a tetramer at pH 6.0. Mw, weight-average molar mass; Mn, number-average molar mass. (C) st_YfdX is in pH-dependent dimer–tetramer equilibrium. (Top) SEC-MALS analysis was carried out under various pH conditions. (Bottom) Molecular weight of st_YfdX–His6 measured at the indicated condition are listed. The asterisk indicates the buffer condition (30 mM phosphate buffer pH 7.0 and 150 mM NaCl) used in the previous report (Saha et al., 2016b).
Overall Structural Analysis of st_YfdX
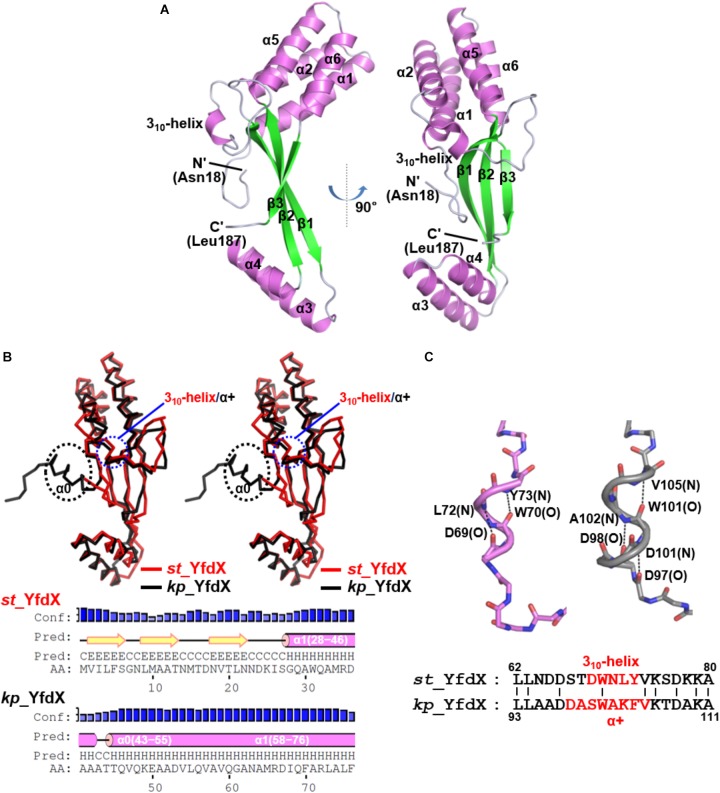
Because structural information about st_YfdX was clearly indispensable for the corroboration of its stoichiometric properties, we attempted to elucidate its crystal structure. The purified recombinant protein was subjected to crystallization trials under 576 crystallization conditions. Rectangular crystals (space group F222) diffracted to the resolution of 1.4 Å were obtained in a crystallization condition with pH 4.6 (Crystal I; Tables 1, 2), implying that this crystal might consist of the st_YfdX tetramer. Its asymmetric unit contains a single st_YfdX molecule that is composed of six α-helices, one 310-helix, and three β-strands (Figure 2A). These units are arranged to form three subdomains: a four-helical bundle (including α1, α2, α5, and α6), an antiparallel β-sheet (including β1–β3), and a two-helical bundle (including α3 and α4; Figure 2A). One 310-helix is located in the middle of the α2–β1 loop (Figure 2A). The st_YfdX monomer overlaps well with the kp_YfdX monomer when superimposed, with a root mean square deviation of 1.27 Å over 154 aligned residues out of a total of 170 residues (Figure 2B, top), which is consistent with high sequence homology between them (35% identity and 52% similarity; Supplementary Figure S2). One noticeable discrepancy is the presence of an additional α-helix (referred to as α0) ahead of α1 of kp_YfdX, which is not shown in st_YfdX (Figure 2B). The PSI-PRED server that accurately predicted the presence of helical portion at that position in kp_YfdX suggested that residues 1–17 of st_YfdX, including residues 1–9 (not contained in the construct used for crystallization) and residues 10–17 (not visible in the crystal structure) might not form the α-helical structure (Figure 2B), supporting that st_YfdX does not contain α0. Another difference at the secondary structural level is that although the short helix of st_YfdX next to α2 is a 310-helix containing i→i+3 hydrogen bonding, the corresponding helix of kp_YfdX is a canonical α-helix (referred to as α+) containing i→i+4 hydrogen bonding (Figure 2C). We noted that minor structural discrepancies between the two proteins are also evident in the 310-helix–β1 and β1–α3 loops (Supplementary Figures 3A,B).
FIGURE 2.
Crystal structure of the st_YfdX monomer. (A) Two views of the st_YfdX monomeric structure are presented as ribbon drawings with the labeled secondary structures. (B) (Top) Stereo views of superimposed st_YfdX (red) and kp_YfdX (black) monomers shown as Cα traces. Remarkable differences at the secondary structural level between the two molecules are indicated. (Bottom) Secondary structure prediction of the amino- terminal region of the two proteins, which was obtained from the PSI-PRED server (http://bioinf.cs.ucl.ac.uk/psipred/). (C) (Top) Structural comparison of the 310-helix of st_YfdX (violet) and α+ of kp_YfdX (gray). Dashed lines highlight helical hydrogen bonding patterns with labels of the involved main chain amide or carbonyl groups. (Bottom) Sequence alignment. The residues constituting 310-helix or α+ helices are highlighted in red.
Structural Analysis of the Oligomeric Form of st_YfdX
To verify the oligomeric state of st_YfdX, the intermolecular interactions and assembly were analyzed among st_YfdX molecules in crystals. Of note, we figured out that a tetramer-shaped oligomeric form of st_YfdX is shown in our crystal lattice (Figure 3A), which can be fairly well-matched to that of kp_YfdX (Supplementary Figure S4). In contrast, we could not find any crystallographic evidence for the formation of a st_YfdX trimer in our structure. We next structurally analyzed the intermolecular binding between st_YfdX monomers in the tetrameric form. The two st_YfdX molecules designated as Mol A and Mol B in Figure 3 interact with each other mainly through very tight hydrophobic contacts. These contacts are mediated by bulky aromatic hydrophobic residues (Phe42, Phe45, and Tyr165) at the tip of a four-helical bundle (especially α1 and α6) of one monomer and a number of hydrophobic residues (Leu21′, Ile26′, Ile94′, Val96′, Ile110′, Ala113′, Met117′, Ile126′, Leu129′, Val134′, and Val136′) in the hydrophobic concave region consisting of a two-helical bundle (α3 and α4) and at the tip of a three-stranded β-sheet of the other monomer (Figure 3B, top). Atomic analysis further revealed 85 intermolecular carbon–carbon contacts (<4.5 Å) in the MolA–MolB binding interface, with more than half being provided by the three aromatic residues Phe42, Phe45, and Tyr165 (51 C–C contacts; 60%). This hydrophobic interaction is well-conserved in the structure of the kp_YfdX tetramer (Supplementary Figure S3C, top) as well, because the contact-involved residues are mostly conserved among YfdX proteins (Supplementary Figure S2; marked with asterisks). It should be noted that residues such as Trp31, Arg35, and Asn91 from the two st_YfdX monomers also reinforce the intermolecular interaction by providing hydrophobic interaction between two tryptophan residues and hydrocarbon portion of two arginine residues and hydrogen bonds between Arg35 and Asn91 (Figure 3B, bottom). Similar but slightly different hydrophobic interactions and hydrogen bonds contribute to the intermolecular interaction between kp_YfdX molecules (Supplementary Figure S3C, bottom).
FIGURE 3.
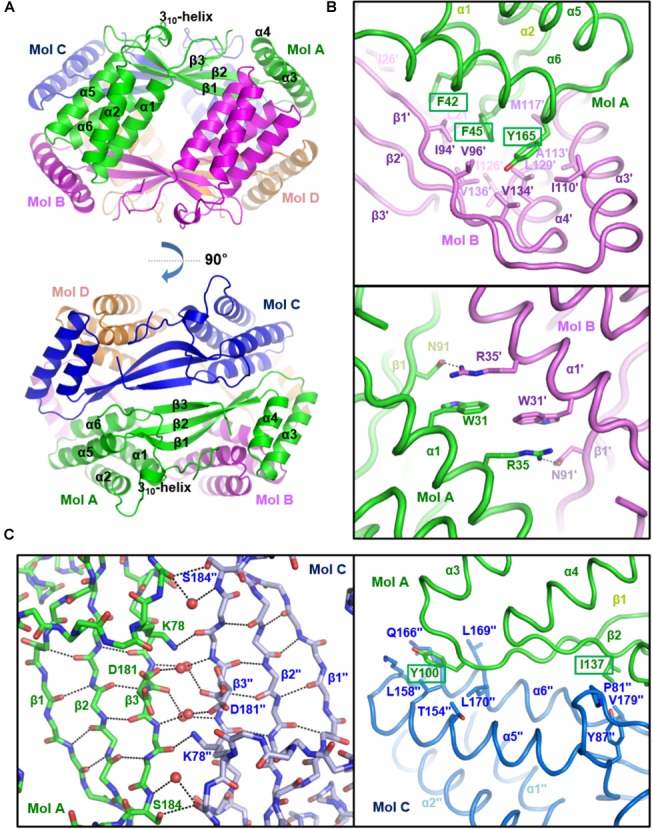
Crystal structure of st_YfdX tetramer. (A) Two views of the st_YfdX tetrameric structure are presented as ribbon models. For clarity, only the secondary structures of Mol A are labeled. (B) Intermolecular interaction between Mol A (green) and Mol B (violet) of st_YfdX. Shown as stick representation and labeled are st_YfdX residues participating in the oligomeric assembly. Three bulky aromatic residues (Phe42, Phe45, and Tyr165) centered at the hydrophobic-interacting interface are marked with rectangles (top). Hydrogen bonds between Arg35 from one molecule and Asn91 form the opposite molecule are presented as dotted lines (bottom). (C) Intermolecular interaction between Mol A (green) and Mol C (blue) of st_YfdX. (Left) Shown as stick representation are the main chain atoms of three-stranded β-sheets from the two molecules and the side chain atoms of Lys78, Asp181, and Ser184. Water molecules mediating intermolecular hydrogen bonds are presented as red circles. Intra- and inter-molecular hydrogen bonds are indicated by dotted lines. (Right) Shown as a stick model and labeled are residues participating in intermolecular hydrophobic interactions. Two hydrophobic residues (Tyr100 and Ile137) that are presumed to play a key role in the Mol A–Mol C binding interaction are marked with rectangles.
In the case of two st_YfdX molecules designated as Mol A and Mol C in Figure 3, at a glance, β-strands from the two monomers appear to form a continuous six-stranded β-sheet. However, we found that no typical hydrogen bond at less than 3.5 Å is present between two facing β-strands (β3 and β3′′) of the two st_YfdX monomers (Figure 3C, left), indicating that the six β-strands are discontinuous. Instead, the intermolecular association between the two three-stranded β-sheets is maintained by several side chains- and water molecules-mediated hydrogen bonds (Figure 3C, left). In the kp_YfdX crystal structure, the corresponding intermolecular interaction between two β-strands was reinforced by zinc ion coordination, in which two oxygen atoms from side chain carboxylates and four oxygen atoms from water molecules are involved (Supplementary Figure S3D, right). However, such a coordination is absent in the st_YfdX structure (Supplementary Figure S3D, left). Hydrophobic interactions provide additional force for the complex formation between Mol A and Mol C, which are mediated by Tyr100 and Ile137 from one molecule and Pro81′′, Tyr87′′, Leu158′′, Leu170′′, Val179′′, and the hydrocarbon part of Thr154′′, Lys163′′, and Gln166′′ from the opposite molecule (Figure 3C, right). Twenty-eight carbon–carbon contacts within 4.5 Å exist in the MolA–MolC binding interface, which is one third of those in the MolA–MolB binding interface. Similar hydrophobic interactions were also observed in the kp_YfdX structure (Supplementary Figure S3E), which are however a bit different from those in the st_YfdX structure along with the structural variation of β1–α3 loops (Supplementary Figure S3B). We found that Tyr100 and Ile137 of st_YfdX are conserved in kp_YfdX as Tyr131 and Ile168 (Supplementary Figure S2; marked with triangles), whereas their associating residues are relatively diverse in sequences between the two proteins. Overall, the interaction between Mol A and Mol B mainly depends on tight hydrophobic interaction and appears to be tight and dense (Figure 3B), whereas that between Mol A and Mol C is maintained by the combination of hydrogen bonds and loose hydrophobic interaction but seems to be relatively weak and detachable (Figure 3C).
SEC-MALS and SAXS Exploration of st_YfdX
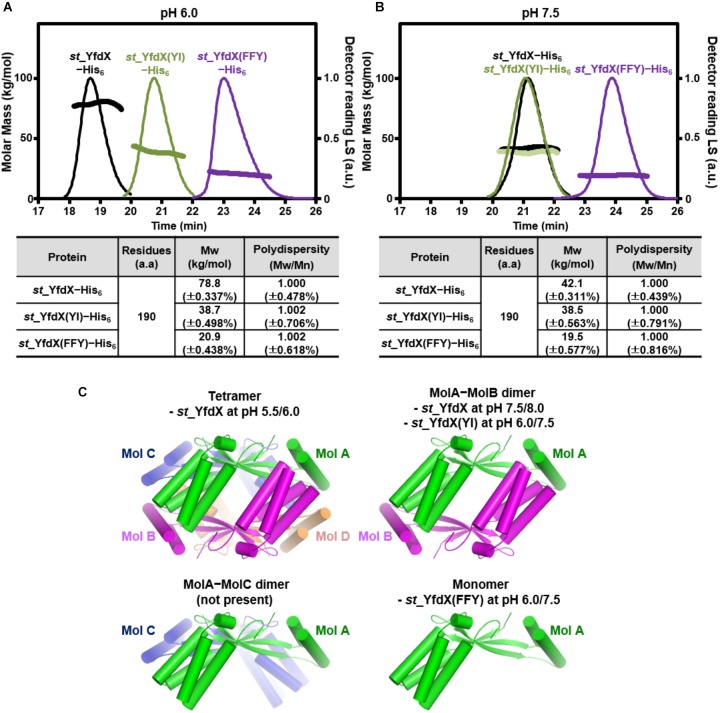
Whether the intermolecular interactions shown in the st_YfdX crystal structure indeed exist in solution was the next issue to be elucidated. Hence, we prepared two mutant st_YfdX proteins: st_YfdX(FFY), in which three core residues (Phe42, Phe45, and Tyr165; see Figure 3B, top) critical for the Mol A–Mol B interaction were substituted with alanine, and st_YfdX(YI), in which two hydrophobic residues (Tyr100 and Ile137; see Figure 3C, right) involved in the Mol A–Mol C interaction were mutated to alanine. These two proteins were expressed, purified, and finally equilibrated in two different solutions buffered with pH 6.0 Bis-Tris-HCl and pH 7.5 Tris-HCl, respectively, as was the wild-type protein. Introduction of such mutations did not affect st_YfdX protein folding, which was confirmed by CD spectroscopic analysis using wild-type and two mutant proteins (Supplementary Figure S5). SEC-MALS analysis was then performed on each protein sample, and the results were compared with one another and with those of wild-type. First, at both pH 6.0 and pH 7.5, the molecular weight of st_YfdX(YI) was ∼39 kDa (Figures 4A,B), indicating that structure-based alanine substitution of Tyr100 and Ile137 indeed abrogates the MolA–MolC interaction and therefore this mutant should exist as the MolA–MolB dimer (Figure 4C, right top). Next, the molecular weight of st_YfdX(FFY) was measured and found to be 20–21 kDa (Figure 4A) at both pH levels, showing that st_YfdX(FFY) exists in a monomeric form (Figure 4C, right bottom) but not as the MolA–MolC dimer (Figure 4C, left bottom). These data strongly indicate that when the very tight hydrophobic interaction-based MolA–MolB binding interface was impaired by the alanine substitution of Phe42, Phe45, and Tyr165, the relatively weak MolA–MolC interaction could not sustain the dimeric interface and thus it is in the monomeric form. As presented in Figure 1B, wild-type st_YfdX forms a tetramer at pH 5.5/6.0 but exists as a dimer at pH 7.5/8.0. We assume that the alteration of proton concentration might affect the formation of hydrogen bonds maintaining the MolA–MolC binding interface. The relatively weak MolA–MolC interaction is presumed not to be sustained at high pH conditions, and thus st_YfdX exists as the MolA–MolB dimer in that condition.
FIGURE 4.
SEC-MALS analysis of mutant st_YfdX. (A,B) Molecular weights of the st_YfdX(FFY) and st_YfdX(YI) mutants were analyzed and compared with each other and with those of the wild-type. All the experiments were conducted on protein samples prepared at 30 mg/mL. (C) Structural diagrams of st_YfdX proteins according to the crystal structure and SEC-MALS experiments.
To further corroborate the pH-dependent conversion of st_YfdX between the two forms, SAXS data were collected using two protein samples equilibrated at pH 5.5 and 8.0, respectively (Table 3). We first confirmed that the calculated scattering intensity curves and distance distribution function P(r) of the tetrameric and the MolA–MolB dimeric structures were nicely matched with the experimental observations (Figures 5A,B). Subsequently, the molecular envelope of the samples was derived from SAXS analysis. The envelope of st_YfdX at pH 5.5 was globular-shaped, in which the tetrameric crystal structure of st_YfdX could be successfully incorporated (Figure 5C, left). In contrast, that of st_YfdX at pH 8.0 was revealed to have a hemisphere-like shape that fitted well the MolA–MolB dimer (Figure 5C, right). Collectively, SEC-MALS and SAXS data demonstrate that st_YfdX switches between two different forms in a pH-dependent manner in the solution, whose conformation could be inferred from the crystal structure.
Table 3.
Structural parameters obtained from the SAXS data of st_YfdX proteins in solution.
| Model/sample | Rg,Gc (Å) | Rg,p(r)d (Å) | Dmaxe (Å) | MMcalculatedf (kDa) | MMSAXg (kDa) | MMMALSh (kDa) |
|---|---|---|---|---|---|---|
| Tetrameric modela | 27.39 ± 0.04 | 26.91 ± 0.04 | 73.4 | 82.4 | – | – |
| MolA-MolB modela | 23.95 ± 0.01 | 23.86 ± 0.14 | 73.5 | 41.2 | – | – |
| st_YfdX-His6 at pH 5.5b | 27.54 ± 0.21 | 26.90 ± 0.09 | 72.5 | – | 80.4 | 80.8 |
| st_YfdX-His6 at pH 8.0b | 23.82 ± 0.21 | 23.84 ± 0.12 | 72.1 | – | 38.2 | 40.4 |
aModels obtained from the st_YfdX structure determined using the crystal I. bSamples subjected to SAXS. cRg,G (radius of gyration) was obtained from the scattering data by the Guinier analysis. dRg,p(r) (radius of gyration) was obtained from the p(r) function by the program GNOM. eDmax (maximum dimension) was obtained from the p(r) function by the program GNOM. fMMcalculated (molecular mass) was obtained from the amino acid sequence of protein. gMMSAXS (molecular mass) was obtained from the SAXS experiments using BSA as a standard. hMMMALS (molecular mass) was obtained from the MALS experiments using BSA as a standard (see Figure 1C).
FIGURE 5.
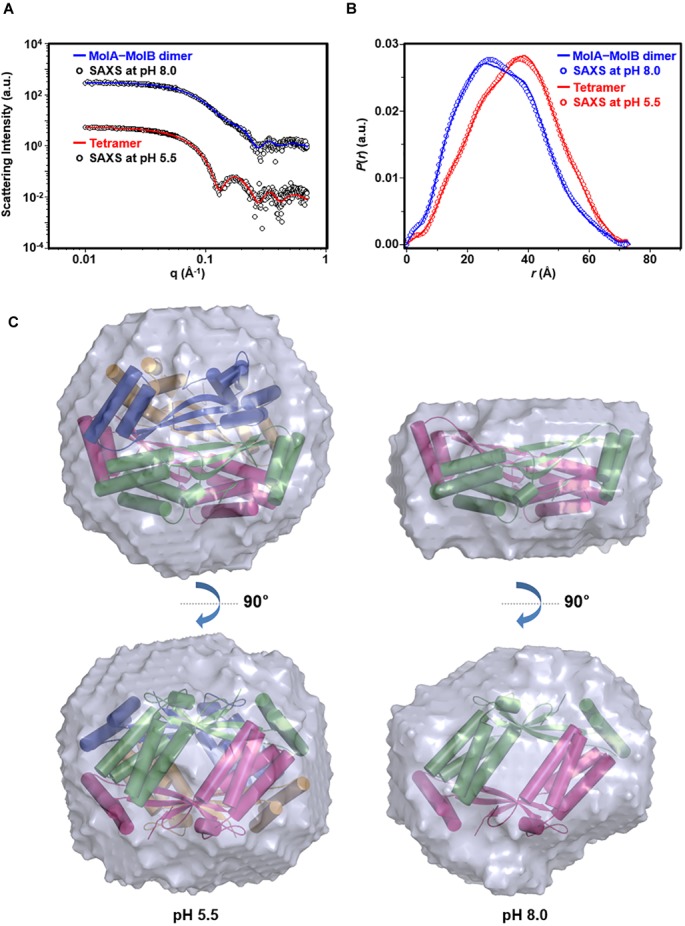
SAXS analysis of st_YfdX. (A) X-ray scattering profiles in solution measured at 4°C. Circles indicate experimental data, whereas solid lines represent the theoretical SAXS curve calculated from the tetrameric (χ2 = 0.583) and dimeric (χ2 = 0.483) models shown in Figure 4C using the program CRYSOL. For clarity, each plot is shifted along the log I(q) axis. (B) Pair distance distribution p(r) functions in aqueous solution that were acquired by analyzing the experimental SAXS data using the program GNOM. Areas under curves were normalized to equal areas for ease of comparison. (C) The tetrameric (left) and dimeric (right) models of st_YfdX were docked into the reconstructed SAXS envelopes that were reconstructed as described in detail in Section “Materials and Methods.”
Crystallization and Structure Determination of ZnCl2-Treated st_YfdX
As mentioned above, the pH-sensitive stoichiometric alteration of st_YfdX was detected during examination of the possibility of zinc coordination-mediated oligomerization. Even though such a zinc coordination was not shown in our crystal structure (Supplementary Figure S3D), we did not exclude such a possibility completely, and thus attempted crystallization of the st_YfdX protein sample equilibrated with the final buffer consisting of 20 mM Bis-Tris-HCl (pH 6.0), 200 mM NaCl, 1 mM DTT, and 1 mM ZnCl2. Two forms of crystals were obtained that were diffracted to high resolution: rectangular crystals diffracted to 1.5 Å (space group F222; crystal II) under the same crystallization condition as that of crystal I, and board-shaped crystals diffracted to 2.3 Å (space group P222; crystal III) under the crystallization condition at pH 5.0 (see Table 1). The asymmetric units of crystals II and III contain one and eight st_YfdX molecules, respectively. We once again determined the structure of st_YfdX using these crystals (Table 2). The st_YfdX structures determined via crystals I–III were confirmed to be almost identical to each other; not only because the st_YfdX monomeric structures from crystals II and III were superimposed on each other and on that from crystal I with root mean square deviations in the range of 0.08–0.77 Å over ∼170 aligned residues, but also because tetramer-shaped oligomeric forms of st_YfdX were detected in the crystal lattices of both crystals II and III, as was the case for crystal I (Supplementary Figure S4). These data support that st_YfdX can form a tetramer. Nevertheless, oligomerization-contributing zinc coordination demonstrated in the kp_YfdX structure (Supplementary Figure S3B, right) was not found in all our crystal structures of st_YfdX (not shown), indicating that the zinc coordination is not necessary for formation of the tetramer of this protein.
Next, to understand the biological function of st_YfdX, a search for homologous structures on the Dali server (Holm and Sander, 1993) was carried out with the monomeric and tetrameric structures of st_YfdX. No homologous protein was identified in this search except for kp_YfdX (Z-score 20.1): a number of proteins containing a helical bundle were listed including Nitrosomonas europaea small metal-binding protein (Z-score 9.9) and E. coli cytochrome b562 (Z-score 8.5), but their structures overlapped with the st_YfdX structure only partially (Supplementary Figure S6), indicating that YfdX adopts a novel protein fold that has not been identified yet.
YfdX Modulates Antibiotic Susceptibility in Salmonella
Previous reports indicate that protein expression of E. coli YfdX is under control of EvgA, the response regulator factor that is intimately associated with the bacterial response to antibiotic stress (Nishino and Yamaguchi, 2001, 2002; Masuda and Church, 2002; Nishino et al., 2003). Even though the homolog of EvgA is absent in the genome of S. Typhi, we assumed that YfdX might also play a role in the response to antibiotics. Therefore, we analyzed the functional and physiological effects of yfdX gene expression in the S. Typhimurium UK-1 strain, whose YfdX (referred to as stm_YfdX in this manuscript) shares 98.0% identity (195 of 199 amino acids) with st_YfdX. First, the cellular expression level of stm_YfdX was examined in a time-dependent manner. As illustrated in Supplementary Figure S7, the transcription level of stm_YfdX was proportionally increased every 4 h and was higher at 12 h than at 4 or 8 h, indicating that cellular stm_YfdX was expressed in the exponential and, particularly, stationary phases. Next, phenotype microarrays were conducted by measuring the bacterial respiration rate to compare physiological changes upon antibiotic treatment between S. Typhimurium UK-1 wild-type and yfdX-deficient (ΔyfdX) strains. We found that the absence of YfdX makes the bacterium more susceptible to two antibiotics that are known to block cell wall synthesis, penicillin G and carbenicillin (Figure 6A). Figure 6B corroborates that the growth of the ΔyfdX strain was remarkably restrained in the presence of each of the two antibiotics at the concentration where the wild-type strain grows well. Moreover, the growth of the ΔyfdX strain was completely (when treated with penicillin G) or considerably (when treated with carbenicillin) recovered by chromosomal complementation of the wild-type S. Typhi yfdX gene but not by that of the yfdX gene containing the FFY mutation (Figure 6B). These data collectively indicate that Salmonella YfdX performs significant functions in basal tolerance of this pathogenic bacterium against β-lactam antibiotics, and that its homooligomerization might be necessary for the functionality of this protein.
FIGURE 6.
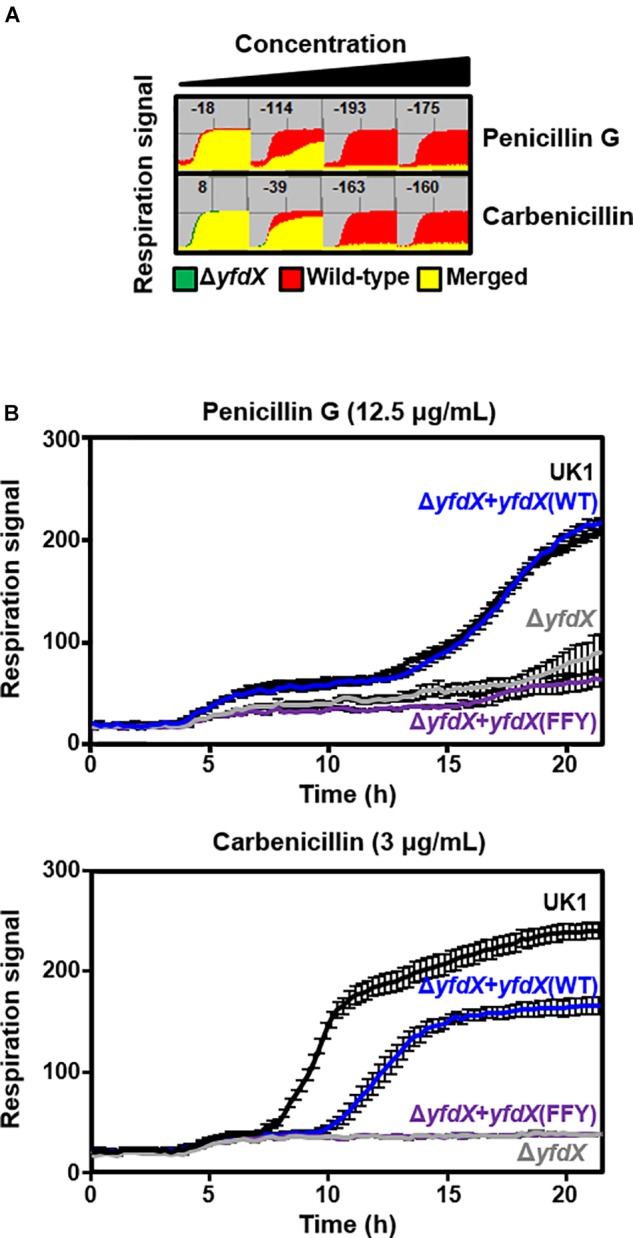
Functionality of YfdX in antibiotic tolerance. (A) Phenotype arrays. Deficiency of YfdX enhanced susceptibility of S. Typhimurium to the indicated antibiotics. (B) Respiration of the S. Typhimurium UK-1 strains from 0 to 25 h was measured in the presence of each antibiotic at the indicated concentration.
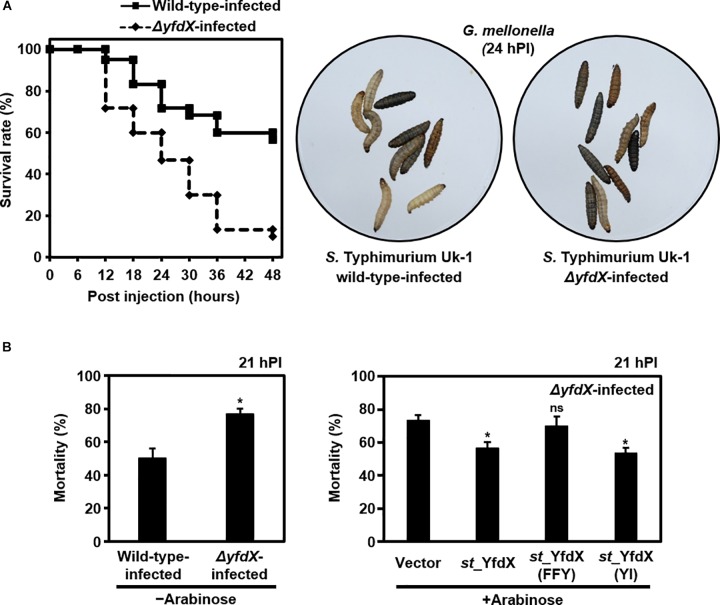
YfdX Deficiency in Salmonella Results in Promoted Bacterial Virulence
Given that various bacterial stress regulators are also known to be involved in bacterial virulence (Fang et al., 2016), we wondered whether Salmonella YfdX is able to modulate microbial virulence as well. A virulence screening system that is based on G. mellonella larvae as the in vivo model for Salmonella infection was employed to determine this issue. After injection of the S. Typhimurium UK-1 wild-type or ΔyfdX strain, the larval survival rate was recorded every 6 h for up to 48 h. Of note, the survival rate of larvae infected by the ΔyfdX strain (∼10% at 48 h) was significantly lower than that of larvae infected by the wild-type strain (∼60% at 48 h), indicating that stm_YfdX plays a role in attenuating bacterial virulence (Figure 7A). To confirm these results, we prepared S. Typhimurium UK-1 ΔyfdX strains in which YfdX-deficiency was complemented by arabinose-induced transient expression of wild-type st_YfdX or crystal structure-based mutant st_YfdX proteins including st_YfdX(FFY) and st_YfdX(YI) (see Figures 3, 4). We first reconfirmed that the ΔyfdX strain-infected larvae show remarkably higher mortality than that of the wild-type strain-infected larvae at 21 h post-injection (Figure 7B, left). Subsequent analysis revealed that the Salmonella virulence was relieved by the transient expression of wild-type st_YfdX or st_YfdX(YI), but not by that of st_YfdX(FFY). Because st_YfdX(YI) and st_YfdX(FFY) exist in a dimeric and monomeric form, respectively (see Figure 4), these results imply that the oligomeric form but not the monomeric form of st_YfdX is functional and necessary to suppress Salmonella virulence. We also confirmed similar effects of st_YfdX on the larval survival rate by means of the S. Typhimurium 14028S strain (Supplementary Figure S8).
FIGURE 7.
YfdX reduces bacterial virulence. (A) yfdX deficiency enhanced Salmonella virulence to Galleria mellonella larvae. (Left) The survival rates of larvae infected by the S. Typhimurium UK-1 wild-type or ΔyfdX strain were measured every 6 h and compared. (Right) Pictures of Salmonella-infected larvae 24 h post-infection. White ones are alive, whereas black ones are dead. (B) Complementation assay. (Left) Mortality rates of wild-type or ΔyfdX Salmonella strain-infected larvae are compared. ∗P < 0.05 (Student’s t-test). (Right) Comparison of mortality rates of ΔyfdX Salmonella-infected larvae with the arabinose-induced transient expression of an empty vector (control), st_YfdX, st_YfdX(YI), or st_YfdX(FFY). ns, not significant; ∗P < 0.05 versus transient expression of empty vector (Student’s t-test).
Discussion
Analysis of the Oligomeric Status of YfdX
In this study, we performed X-ray crystallographic and scattering, biochemical, and physiological analyses of Salmonella-derived YfdX, whose characteristics and biological function remain obscure despite recent research (Saha et al., 2016a,b; Mondal et al., 2017). One of the issues raised by a study on st_YfdX is about its stoichiometry (Saha et al., 2016b): st_YfdX indeed forms a trimer unlike kp_YfdX that appears to form a tetramer despite their high sequence homology. SEC-MALS analysis and structure determination by X-ray crystallographic and scattering analyses were carried out to resolve this issue, which led to the unexpected identification of a pH-related stoichiometric switch of st_YfdX (Figures 1–5). Although both the tetrameric (at pH 7.5/8.0) and dimeric form (at pH 5.5/6.0) were confirmed by SEC-MALS (Figure 1) and SAXS (Figure 5), st_YfdX was found to be in the tetrameric form in all the three crystal structures, presumably because of the increased opportunity of contacts in the crystal lattice (Figure 3 and Supplementary Figure S4). Nevertheless, the dimeric structure of st_YfdX could be inferred from the combination of a structure-based mutational study accompanied by SEC-MALS analysis. Among the two hypothetical dimers illustrated in Figure 4C, the MolA–MolB form, but not the MolA–MolC form, is presumed to exist in solution, for the following reasons. First, the intermolecular interactions between MolA and MolB are clearly tighter and denser than those between MolA and MolC (Figures 3B,C). Second, the MolA–MolB interaction-null st_YfdX(FFY) mutant was revealed to be a monomer, not a dimer (Figure 4B), implying that the MolA–MolC interaction is not strong enough to sustain the complex formation. We speculate that the tetrameric form of st_YfdX observed at pH < 6.0 might involve assembly of two MolA–MolB dimers. SEC-MALS analysis also showed a pH-dependent gradual shift of the calculated molecular mass of st_YfdX from pH 5.5 to 8.0, indicating that st_YfdX is in a state of dynamic equilibrium between tetramer and dimer (Figure 1C). At pH 7.0, the molecular weight of st_YfdX was calculated to be ∼51–53 kDa, which presumably explains the reason that st_YfdX was estimated to form a trimer in the previous report (Saha et al., 2016b). Given the SEC-MALS analysis together with the crystallographic and SAXS structural analysis collectively (Figures 1–5), we consider that st_YfdX undergoes a continuous structural switch between dimer and tetramer rather than exists as a trimer.
Determination of the stoichiometry of st_YfdX was followed by corollary issues concerning the biological importance of such oligomerization. Chromosomal knockout and complementation assay using the S. Typhimurium UK-1 strain demonstrated that the monomer-forming mutant YfdX is non-functional (see Figure 6). The G. mellonella larvae-infection assay showed that the genetic knockout of YfdX could be complemented by the transient expression of the wild-type or the dimer-forming mutant, but not by that of the monomer-promoting mutant (see Figure 7). These data collectively suggest that the oligomerization of st_YfdX might be inevitable for its effectiveness. It is notable that we currently do not rule out the probability that the tetrameric and dimeric forms function differently in other cellular processes in which st_YfdX is involved.
YfdX Acts a Critical Dual Function in Antibiotic Susceptibility and Virulence
Despite previous efforts examining the diverse biochemical properties of st_YfdX (Saha et al., 2016a,b; Mondal et al., 2017), analysis of its functionality in live bacteria has been absent thus far, limiting our understanding of this protein. In this study, we demonstrated that YfdX is indeed involved in Salmonella’s tolerance to antibiotics, because its deficiency caused a remarkable increase in antibiotic susceptibility and impaired bacterial survival upon treatment with penicillin G and carbenicillin, which was compensated by chromosomal complementation with the wild-type yfdX but not with the monomer-promoting mutant yfdX (Figure 6). We also observed that the expression level of YfdX in Salmonella was higher in the stationary phase than that in the exponential phase (Supplementary Figure S7). Because tolerance to a variety of environmental stressors such as antibiotics and osmotic or acidic stress is vital for bacterial species, particularly in the stationary phase where bacterial growth is delayed or arrested, a number of bacterial proteins that are upregulated in this phase were found to be involved in a stress response and/or antibiotic tolerance (Hengge-Aronis, 1993; Battesti et al., 2011), which now should include YfdX. In addition, it was revealed for the first time that YfdX mitigates mortality of the Salmonella infection in our larval model (Figure 7), suggesting that YfdX is a negative regulator of bacterial virulence. Even though antibiotic resistance and bacterial virulence are different in their functionality and applicability, a number of biological processes have been reported to be shared by the two mechanisms, including biofilm formation (Patel, 2005), upregulated expression of an efflux pump (Barbosa and Levy, 2000), regulation of cell permeability (Tsai et al., 2011), and cell wall alteration (Moya et al., 2008). Moreover, in a variety of gram-negative pathogenic bacteria such as Staphylococcus aureus and Streptococcus pneumoniae, bacteria strains resistant to antimicrobial molecules including β-lactams and glycopeptides show attenuated virulence when they infect mice (Rieux et al., 2001; Rudkin et al., 2012) or G. mellonella (Peleg et al., 2009), implying an association between the two mechanisms at the molecular level. Yet, the precise molecular mechanism of YfdX’s action in the control of Salmonella antibiotic tolerance and bacterial virulence remains to be elucidated. One possibility is that YfdX may associate with membrane proteins and affect their function or localization, e.g., proteins involved in cell wall synthesis, as its deficiency led to the increased susceptibility to β-lactam antibiotics such as penicillin G and carbenicillin (Figure 6). Since a recent report shows simulated binding of st_YfdX to the outer-membrane protein STY3179, a Salmonella homolog of an adhesion and invasion locus protein from Yersinia pestis that interacts with the host extracellular matrix protein laminin (Yamashita et al., 2011), another possibility is that YfdX controls bacterial virulence by regulating host–pathogen interaction via binding to STY3179 or other unknown proteins. We suppose that future verification of the binding partner(s) of YfdX will be necessary to resolve this issue.
It has been reported that the EvgS-EvgA system controlling the expression of the yfdXWUVE operon in E. coli regulates the signaling pathways also associated with acid tolerance (Nishino and Yamaguchi, 2001, 2002; Masuda and Church, 2002; Nishino et al., 2003). We, therefore, raise a possibility that st_YfdX might participate not only in antibiotic susceptibility but also in acid tolerance of the bacterium or other processes such as pH homeostasis that is also implicated in Salmonella virulence (Rappl et al., 2003), in which its pH-dependent structural conversion could play some unknown role. However, at least under our assay conditions, the relation of st_YfdX with acid shock was shown to be unclear and inconsistent in the phenotype microarrays and in the minimum inhibitory concentration assays (unpublished data), and this issue remains to be resolved by future research as well.
Conclusion
We characterized various features of the Salmonella-derived YfdX protein, including the crystal structure that contains a novel fold, previously unknown stoichiometric features, proof of contribution to antibiotic response, and participation in the control of bacterial virulence (Figure 8). We believe that our biochemical and physiological findings will provide a rational basis for future research, augmented with the details of molecular mechanisms of the stress response in this pathogenic bacterium, in which much remains to be discovered.
FIGURE 8.
Overall view of the physiological functionality of YfdX.
Author Contributions
BK, C-MR, and SJK conceived and designed the experiments. HSL, SL, J-SK, H-RL, and KSJ performed the experiments. HSL, SL, KSJ, BK, C-MR, and SJK analyzed the data. H-CS, M-SL, and C-HK contributed to reagents, materials, analysis tools. HSL, J-SK, BK, C-MR, and SJK wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study involved the beamlines 4C, 5C, and 7A at the Pohang Accelerator Laboratory in South Korea. We are grateful to Dr. Eunha Hwang (Korea Basic Science Institute, South Korea) for helping with the SEC-MALS experiments and Dr. Myung Hee Kim and Dr. Jungwon Hwang (Korea Research Institute of Bioscience and Biotechnology, South Korea) for helping with the CD experiments.
Abbreviations
- CD
circular dichroism
- DTT
dithiothreitol
- MDR
multiple drug resistance
- PDB
Protein Data Bank
- SAXS
small-angle X-ray scattering
- SEC-MALS
size exclusion chromatography-multiangle light scattering
Funding. This study was supported by the National Research Foundation of Korea (Grant No. NRF_2015M3A9B5030308) and by the BioNano Health-Guard Research Center as the Global Frontier Project (Grant No. H-GUARD_2013M3A6B2078954), which were funded by the Ministry of Science and ICT (MSIT) of South Korea. This study was also supported by the Korea Research Institute of Bioscience and Biotechnology Research Initiative Programs for Creative Research and for Disease Target Structural Research.
The coordinates and the structure factors of the st_YfdX structures determined using crystals I, II, and III have been deposited in the PDB under the accession codes 6A02, 6A07, and 6A09, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03329/full#supplementary-material
References
- Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., et al. (2010). PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66 213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmatullah A., Qamar F. N., Thaver D., Zaidi A. K., Bhutta Z. A. (2015). Systematic review of the global epidemiology, clinical and laboratory profile of enteric fever. J. Glob. Health 5:020407. 10.7189/jogh.05.020407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa T. M., Levy S. B. (2000). Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182 3467–3474. 10.1128/JB.182.12.3467-3474.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A., Majdalani N., Gottesman S. (2011). The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65 189–213. 10.1146/annurev-micro-090110-102946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckle G. C., Walker C. L., Black R. E. (2012). Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010. J. Glob. Health 2:010401. 10.7189/jogh.02.010401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan G., Baker S. (2014). Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu. Rev. Microbiol. 68 317–336. 10.1146/annurev-micro-091313-103739 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Everest P., Wain J., Roberts M., Rook G., Dougan G. (2001). The molecular mechanisms of severe typhoid fever. Trends. Microbiol. 9 316–320. 10.1016/S0966-842X(01)02067-4 [DOI] [PubMed] [Google Scholar]
- Fang F. C., Frawley E. R., Tapscott T., Vazquez-Torres A. (2016). Bacterial stress responses during host infection. Cell Host Microbe 20 133–143. 10.1016/j.chom.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury P., Aellen N., Ruffner B., Pechy-Tarr M., Fataar S., Metla Z., et al. (2016). Insect pathogenicity in plant-beneficial Pseudomonads: phylogenetic distribution and comparative genomics. ISME J. 10 2527–2542. 10.1038/ismej.2016.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D., Svergun D. I. (2009). DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42 342–346. 10.1107/S0021889809000338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye J. G., Jackson C. R. (2013). Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from U.S. food animals. Front. Microbiol. 4:135. 10.3389/fmicb.2013.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruez A., Roig-Zamboni V., Valencia C., Campanacci V., Cambillau C. (2003). The crystal structure of the Escherichia coli YfdW gene product reveals a new fold of two interlaced rings identifying a wide family of CoA transferases. J. Biol. Chem. 278 34582–34586. 10.1074/jbc.C300282200 [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. (1993). Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72 165–168. 10.1016/0092-8674(93)90655-A [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. (1993). Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233 123–138. 10.1006/jmbi.1993.1489 [DOI] [PubMed] [Google Scholar]
- Hurley D., McCusker M. P., Fanning S., Martins M. (2014). Salmonella-host interactions – Modulation of the host innate immune system. Front. Immunol. 5:481. 10.3389/fimmu.2014.00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Yu D., Wei Z., Shen P., Zhou Z., Yu Y. (2010). Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob. Agents Chemother. 54 3967–3969. 10.1128/AAC.00137-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra S. P., Naithani N., Mehta S. R., Swamy A. J. (2003). Current trends in the management of typhoid fever. Med. J. Armed Forces India 59 130–135. 10.1016/S0377-1237(03)80060-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki S., Gordon M. A., Feasey N., Parry C. M. (2015). Antimicrobial resistance and management of invasive Salmonella disease. Vaccine 33(Suppl. 3), C21–C29. 10.1016/j.vaccine.2015.03.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkey A., Thwaites G. E., Baker S. (2018). The evolution of antimicrobial resistance in Salmonella typhi. Curr. Opin. Gastroenterol. 34 25–30. 10.1097/MOG.0000000000000406 [DOI] [PubMed] [Google Scholar]
- Kaur G., Sathyabama S., Arora A., Verma S., Mubin N., Agrewala J. N., et al. (2014). Genome sequencing, annotation and comparative genomic analysis of Shigella dysenteriae strain SD1D. Gut Pathog. 6:28. 10.1186/1757-4749-6-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. W., Kim J., Yun Y. D., Ahn H., Min B., Kim N. H., et al. (2017). Small-angle X-ray scattering beamline BL4C SAXS at pohang light source II. Biodesign 5 24–29. [Google Scholar]
- Kozin M. B., Svergun D. I. (2001). Automated matching of high-and low-resolution structural models. J. Appl. Cryst. 34 33–41. [Google Scholar]
- Lazaro-Diez M., Redondo-Salvo S., Arboleya-Agudo A., Ocejo-Vinyals J. G., Chapartegui-Gonzalez I., Ocampo-Sosa A. A., et al. (2016). Whole-genome sequence of Hafnia alvei HUMV-5920, a human isolate. Genome Announc. 4:e00556. 10.1128/genomeA.00556-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N., Church G. M. (2002). Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 184 6225–6234. 10.1128/JB.184.22.6225-6234.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Sanderson K. E., Spieth J., Clifton S. W., Latreille P., Courtney L., et al. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413 852–856. 10.1038/35101614 [DOI] [PubMed] [Google Scholar]
- McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007). Phaser crystallographic software. J. Appl Crystallogr. 40 658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal M., Chakrabarti J., Ghosh M. (2017). Molecular dynamics simulations on interaction between bacterial proteins: implication on pathogenic activities. Proteins 86 370–378. 10.1002/prot.25446 [DOI] [PubMed] [Google Scholar]
- Moya B., Juan C., Alberti S., Perez J. L., Oliver A. (2008). Benefit of having multiple ampD genes for acquiring beta-lactam resistance without losing fitness and virulence in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52 3694–3700. 10.1128/AAC.00172-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins E. A., Sullivan K. L., Kappock T. J. (2013). Function and X-ray crystal structure of Escherichia coli YfdE. PLoS One 8:e67901. 10.1371/journal.pone.0067901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (2009). Multidrug resistance in bacteria. Annu. Rev. Biochem. 78 119–146. 10.1146/annurev.biochem.78.082907.145923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K., Inazumi Y., Yamaguchi A. (2003). Global analysis of genes regulated by EvgA of the two-component regulatory system in Escherichia coli. J. Bacteriol. 185 2667–2672. 10.1128/JB.185.8.2667-2672.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K., Yamaguchi A. (2001). Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183 1455–1458. 10.1128/JB.183.4.1455-1458.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K., Yamaguchi A. (2002). EvgA of the two-component signal transduction system modulates production of the yhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 184 2319–2323. 10.1128/JB.184.8.2319-2323.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Park S. M., Yoo J. C., Ji S. W., Yang J. S., Baek K. (2015). Selective recovery of dissolved Fe, Al, Cu, and Zn in acid mine drainage based on modeling to predict precipitation pH. Environ. Sci. Pollut. Res. Int. 22 3013–3022. 10.1007/s11356-014-3536-x [DOI] [PubMed] [Google Scholar]
- Parkhill J., Dougan G., James K. D., Thomson N. R., Pickard D., Wain J., et al. (2001). Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413 848–852. 10.1038/35101607 [DOI] [PubMed] [Google Scholar]
- Patel R. (2005). Biofilms and antimicrobial resistance. Clin. Orthop. Relat. Res. 437 41–47. 10.1097/01.blo.0000175714.68624.74 [DOI] [PubMed] [Google Scholar]
- Peleg A. Y., Monga D., Pillai S., Mylonakis E., Moellering R. C., Jr., et al. (2009). Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J. Infect. Dis. 199 532–536. 10.1086/596511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappl C., Deiwick J., Hensel M. (2003). Acidic pH is required for the functional assembly of the type III secretion system encoded by Salmonella pathogenicity island 2. FEMS Microbiol. Lett. 226 363–372. 10.1016/S0378-1097(03)00638-4 [DOI] [PubMed] [Google Scholar]
- Rieux V., Carbon C., Azoulay-Dupuis E. (2001). Complex relationship between acquisition of beta-lactam resistance and loss of virulence in Streptococcus pneumoniae. J. Infect. Dis. 184 66–72. 10.1086/320992 [DOI] [PubMed] [Google Scholar]
- Rowe B., Ward L. R., Threlfall E. J. (1997). Multidrug-resistant Salmonella typhi: a worldwide epidemic. Clin. Infect. Dis. 24(Suppl. 1), S106–S109. [DOI] [PubMed] [Google Scholar]
- Rudkin J. K., Edwards A. M., Bowden M. G., Brown E. L., Pozzi C., Waters E. M., et al. (2012). Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J. Infect. Dis. 205 798–806. 10.1093/infdis/jir845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P., Manna C., Chakrabarti J., Ghosh M. (2016a). Reversible thermal unfolding of a yfdX protein with chaperone-like activity. Sci. Rep. 6:29541. 10.1038/srep29541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P., Manna C., Das S., Ghosh M. (2016b). Antibiotic binding of STY3178, a yfdX protein from Salmonella Typhi. Sci. Rep. 6:21305. 10.1038/srep21305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E., Roberts C. J. (2012). Size-exclusion chromatography with multi-angle light scattering for elucidating protein aggregation mechanisms. Methods Mol. Biol. 899 403–423. 10.1007/978-1-61779-921-1_25 [DOI] [PubMed] [Google Scholar]
- Sayilgan E., Kukrer T., Yigit N. O., Civelekoglu G., Kitis M. (2010). Acidic leaching and precipitation of zinc and manganese from spent battery powders using various reductants. J. Hazard Mater. 173 137–143. 10.1016/j.jhazmat.2009.08.063 [DOI] [PubMed] [Google Scholar]
- Semenyuk A. V., Svergun D. I. (1991). GNOM – A program package for small-angle scattering data processing. J. Appl. Cryst. 24 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D., Barberato C., Koch M. H. J. (1995). CRYSOL – A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28 768–773. [Google Scholar]
- Tsai Y. K., Fung C. P., Lin J. C., Chen J. H., Chang F. Y., Chen T. L., et al. (2011). Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 55 1485–1493. 10.1128/AAC.01275-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werther T., Zimmer A., Wille G., Golbik R., Weiss M. S., Konig S. (2010). New insights into structure-function relationships of oxalyl CoA decarboxylase from Escherichia coli. FEBS J. 277 2628–2640. 10.1111/j.1742-464X.2010.07673.x [DOI] [PubMed] [Google Scholar]
- Yamashita S., Lukacik P., Barnard T. J., Noinaj N., Felek S., Tsang T. M., et al. (2011). Structural insights into ail-mediated adhesion in Yersinia pestis. Structure 19 1672–1682. 10.1016/j.str.2011.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.