Ebola virus and Nipah virus (EBOV and NiV) can cause fatal disease in humans. In contrast, infected fruit bats do not develop symptoms but can transmit the virus to humans. Why fruit bats but not humans control infection is largely unknown. Tetherin is an antiviral host cell protein and is counteracted by the EBOV glycoprotein in human cells. Here, employing model systems, we show that tetherin of fruit bats displays higher antiviral activity than human tetherin and is largely resistant against counteraction by the Ebola virus glycoprotein. Moreover, we demonstrate that induction of tetherin expression is critical for interferon-mediated inhibition of NiV but, for at present unknown reasons, not EBOV spread in fruit bat cells. Collectively, our findings identify tetherin as an antiviral effector of innate immune responses in fruit bats, which might allow these animals to control infection with NiV and potentially other viruses that cause severe disease in humans.
KEYWORDS: Nipah virus, bat, Ebola virus, tetherin
ABSTRACT
Ebola virus (EBOV) and Nipah virus (NiV) infection of humans can cause fatal disease and constitutes a public health threat. In contrast, EBOV and NiV infection of fruit bats, the putative (EBOV) or proven (NiV) natural reservoir, is not associated with disease, and it is currently unknown how these animals control the virus. The human interferon (IFN)-stimulated antiviral effector protein tetherin (CD317, BST-2) blocks release of EBOV- and NiV-like particles from cells and is counteracted by the EBOV glycoprotein (GP). In contrast, it is unknown whether fruit bat tetherin restricts virus infection and is susceptible to GP-driven antagonism. Here, we report the sequence of fruit bat tetherin and show that its expression is IFN stimulated and associated with strong antiviral activity. Moreover, we demonstrate that EBOV-GP antagonizes tetherin orthologues of diverse species but fails to efficiently counteract fruit bat tetherin in virus-like particle (VLP) release assays. However, unexpectedly, tetherin was dispensable for robust IFN-mediated inhibition of EBOV spread in fruit bat cells. Thus, the VLP-based model systems mimicking tetherin-mediated inhibition of EBOV release and its counteraction by GP seem not to adequately reflect all aspects of EBOV release from IFN-stimulated fruit bat cells, potentially due to differences in tetherin expression levels that could not be resolved by the present study. In contrast, tetherin expression was essential for IFN-dependent inhibition of NiV infection, demonstrating that IFN-induced fruit bat tetherin exerts antiviral activity and may critically contribute to control of NiV and potentially other highly virulent viruses in infected animals.
IMPORTANCE Ebola virus and Nipah virus (EBOV and NiV) can cause fatal disease in humans. In contrast, infected fruit bats do not develop symptoms but can transmit the virus to humans. Why fruit bats but not humans control infection is largely unknown. Tetherin is an antiviral host cell protein and is counteracted by the EBOV glycoprotein in human cells. Here, employing model systems, we show that tetherin of fruit bats displays higher antiviral activity than human tetherin and is largely resistant against counteraction by the Ebola virus glycoprotein. Moreover, we demonstrate that induction of tetherin expression is critical for interferon-mediated inhibition of NiV but, for at present unknown reasons, not EBOV spread in fruit bat cells. Collectively, our findings identify tetherin as an antiviral effector of innate immune responses in fruit bats, which might allow these animals to control infection with NiV and potentially other viruses that cause severe disease in humans.
INTRODUCTION
Ebola virus (EBOV), a member of the Filoviridae, is highly virulent in humans and nonhuman primates. The devastating Ebola virus disease epidemic in West Africa claimed more than 11,000 lives (1, 2), and the frequent introduction of the virus into the human population from animal reservoirs during the last decades and its persistence in infected patients (3–5) suggest that similar outbreaks can occur at any time. Moreover, no approved vaccines or therapeutics are available to combat EBOV at present, although testing of certain vaccines in clinical trials yielded encouraging results (6). Thus, EBOV constitutes a serious health threat, and the development of novel countermeasures is called for.
Nipah virus (NiV), a zoonotic paramyxovirus, was first recognized during an outbreak of fatal encephalitis and pneumonia in pig farmers and abattoir workers in Malaysia and Singapore in 1998 (7–9). Since 2001, NiV has caused multiple independent outbreaks in Bangladesh, India, and (potentially) the Philippines (10–15), resulting in more than 600 cases of which more than half had a fatal outcome. Finally, as for EBOV, neither approved vaccines nor therapeutics are available to combat NiV infection, highlighting that NiV is an unmet threat to public health in Southeast Asia.
Asian fruit bats of the genus Pteropus are the natural reservoir of NiV (16–18) and may transmit the virus directly to humans or via pigs, which can serve as intermediate hosts (9, 10, 12, 13, 19). African fruit bats are believed to be the natural reservoir of EBOV; several outbreaks have been associated with the contact of humans with bats (20–23). Analysis of naturally and experimentally infected fruit bats revealed that these animals amplify NiV and EBOV but do not develop disease (20–25). Therefore, understanding how fruit bats control infection by these two viruses might help to define novel targets for antiviral intervention. Recent studies suggest that fruit bats might be equipped with a constitutively active interferon (IFN) system (26), which might constitute a powerful defense against viral spread. IFN can inhibit virus infection by inducing the expression of IFN-stimulated genes (ISGs), many of which encode products with antiviral activity (27). However, it is incompletely understood which ISG-encoded proteins restrict EBOV and NiV infection of human cells. Moreover, EBOV and NiV restricting factors (termed restriction factors) in fruit bat cells have not been identified, although inhibition of an EBOV minireplicon by bat Mx proteins in transfected human cells has been reported (28).
The tetherin protein (CD317, BST-2) is an IFN-induced restriction factor that can block the spread of several enveloped viruses by preventing the release of progeny particles from infected cells (29–31). Tetherin can exert its antiviral activity due to the presence of two membrane anchors, an N-terminal transmembrane domain and a C-terminal glycosylphosphatidylinositol (GPI) anchor. These elements allow tetherin to simultaneously insert into viral and cellular membranes, thereby forming a physical tether between the cell surface and virus particles (32). Human immunodeficiency virus type 1 (HIV-1) and several other viruses encode tetherin antagonizing proteins which interfere with appropriate tetherin expression and/or cellular localization and thus allow viral spread in tetherin-positive cells (30, 31, 33).
The glycoprotein (GP) of EBOV mediates viral entry into target cells and rescues release of VP40-based particles from inhibition by tetherin (34), using a poorly understood mechanism. Inhibition of EBOV release by tetherin has so far only been observed in the context of surrogate systems, and formally it remains to be demonstrated that tetherin inhibits viral release and is counteracted by GP in the context of EBOV-infected cells. Nevertheless, two studies reported that release of EBOV from infected cells is not blocked by human tetherin (35, 36), suggesting that GP-dependent tetherin antagonism might help the virus to evade control by the human IFN system. In contrast, the contribution of tetherin to the innate defenses of fruit bats against EBOV is unknown. Similarly, little information is available regarding the role of tetherin in NiV infection. Two studies reported that release of NiV-like particles, produced by directed expression of the NiV matrix protein, is reduced when tetherin is coexpressed (36, 37). However, it is unknown whether the NiV surface glycoproteins F and G, like EBOV-GP, can antagonize tetherin and whether tetherin is able to restrict spread of authentic NiV.
Here, we report that expression of fruit bat tetherin is stimulated by IFN and show that the protein efficiently restricts release of EBOV-like particles from cells. Furthermore, we reveal that EBOV-GP fails to efficiently antagonize fruit bat tetherin upon directed expression. Finally, we provide evidence that tetherin is essential for efficient IFN-mediated inhibition of fruit bat cell infection by vesicular stomatitis virus (VSV), a prototype RNA virus from the Rhabdoviridae family, and NiV, while, unexpectedly, EBOV spread in fruit bat cells was only moderately affected by tetherin.
RESULTS
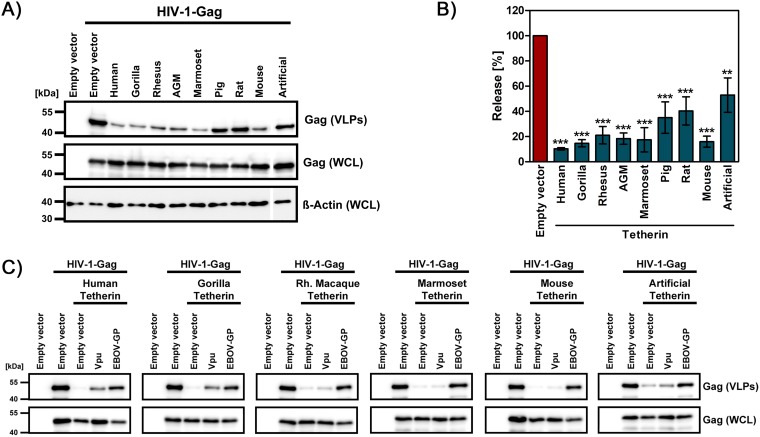
EBOV-GP antagonizes human, nonhuman primate, rodent, and artificial tetherin.
We first investigated whether EBOV-GP can antagonize tetherin orthologues of diverse species, including nonhuman primate and rodent tetherin, as well as artificial tetherin, which has no sequence homology with human tetherin (32). For this, we used a previously described HIV-1 Gag-based virus-like particle (VLP) release assay (35), which is commonly used in the field. All tetherin proteins tested in this assay reduced release of VLPs (Fig. 1A and B). The HIV-1 Vpu protein, a prototypic tetherin antagonist, counteracted human and the closely related gorilla tetherin but was largely inactive against the other tetherin orthologues tested (Fig. 1C), in agreement with previous findings (38, 39). In contrast, EBOV-GP counteracted the antiviral activity of all tetherin proteins tested, including artificial tetherin (Fig. 1C). These findings, which confirm and extend a previous report (40), indicate a broad and potentially sequence independent anti-tetherin activity of EBOV-GP.
FIG 1.
EBOV-GP antagonizes tetherin orthologues from diverse species. (A) HEK293T cells were transfected with expression plasmids for HIV-1 Gag and the indicated tetherin orthologues, artificial tetherin, or empty plasmid as a control. The release of Gag-derived VLPs in culture supernatants and Gag expression in WCLs were analyzed by immunoblotting. Expression of β-actin was determined as a loading control. A representative blot is shown from which irrelevant lanes were excised. (B) The average of four experiments conducted as described for panel A and quantified via the ImageJ program is shown. The release of Gag in the absence of tetherin was set as 100%. Error bars indicate the standard error of the mean (SEM). One-way analysis of variance (ANOVA) with Bonferroni posttest analysis was performed to test statistical significance (**, P ≤ 0.005; ***, P ≤ 0.001). (C) A release assay was conducted as described for panel A, but the tetherin antagonists HIV-1 Vpu and EBOV-GP were included. The results of a representative blot are shown and were confirmed in a separate experiment.
Fruit bat tetherin is a potent antiviral factor.
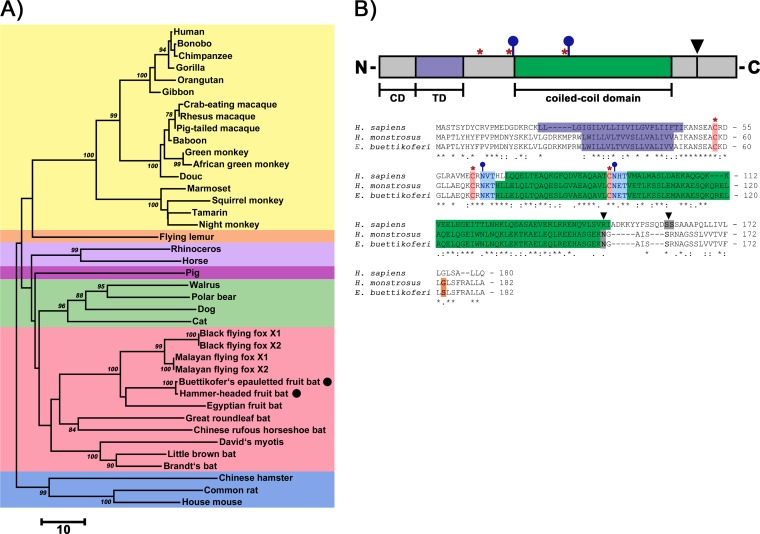
We next investigated whether tetherin orthologues from an assumed EBOV reservoir, Hypsignathus monstrosus (21), and a related fruit bat species, Epomops buettikoferi, can inhibit VLP release and are susceptible to EBOV-GP-mediated antagonism. For this, we PCR amplified and cloned the complete tetherin open reading frames (ORFs) from EpoNi/22.1 (E. buettikoferi, Epo) and HypNi/1.1 (H. monstrosus, Hyp) cells. Sequence analysis showed that both fruit bat tetherins cluster phylogenetically with predicted tetherin proteins of bats and that human and fruit bat tetherin display an identical domain organization, including conserved cysteine residues and N-glycosylation motifs (Fig. 2A and B).
FIG 2.
Fruit bat tetherin and human tetherin exhibit the same domain organization. (A) Phylogenetic relationship of mammalian tetherin proteins (see Table S1 in the supplemental material for additional information). Protein sequences of tetherin proteins from the indicated species were aligned, and a phylogenetic tree was constructed using MEGA6 (v6.06). Small numbers at the nodes reflect bootstrap values, and the scale bar indicates the number of amino acid substitutions per site (for simplicity, only values of >75 are shown). Black circles indicate the fruit bat species whose tetherin proteins were investigated in the present study. The identifiers “X1” and “X2” refer to different isoforms present in the NCBI database. Background colors: yellow, primates; orange, colugos; light purple, odd-toed ungulates; dark purple, even-toed ungulate; green, carnivorans; red, bats and flying foxes; blue, rodents. (B) Domain organization (at the top) and sequence alignment of fruit bat and human tetherin. Conserved cysteine residues (red, asterisks), glycosylation motifs (blue, sticks and circles), GPI anchor (black arrowhead, gray box), and the only amino acid variation between the two fruit bat tetherins studied (position 174, glycine or serine; orange box) are indicated. CD, cytoplasmic domain; TD, transmembrane domain.
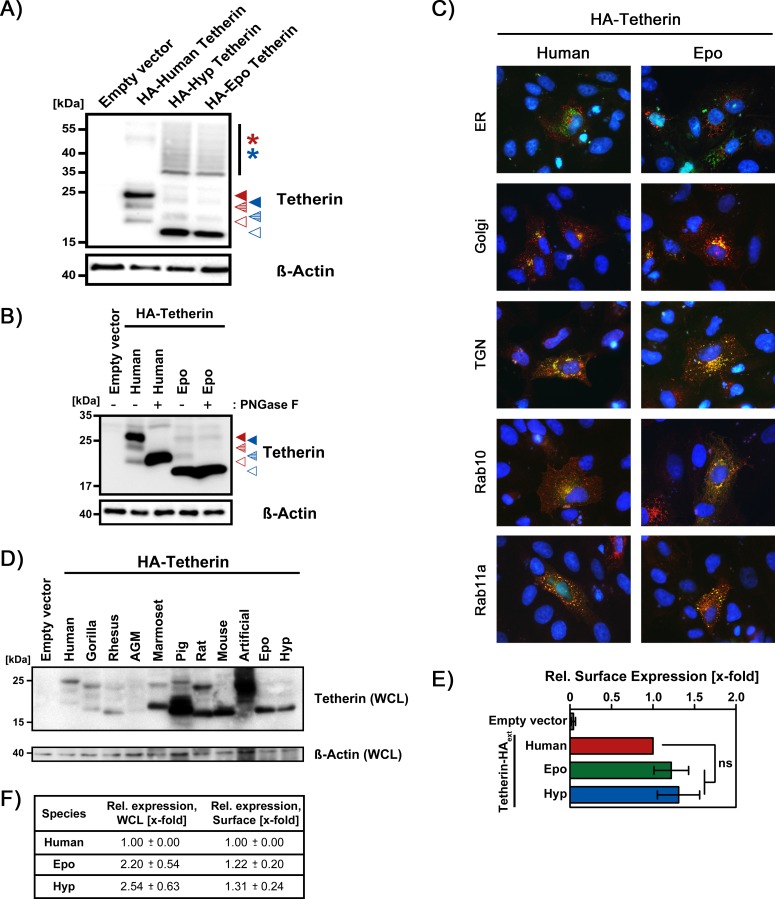
We next sought to determine whether the similarities in sequence (human/Hyp, 46.7%; human/Epo, 46.2%) and domain organization between human and fruit bat tetherins resulted in comparable expression and antiviral activity. Both fruit bat tetherins showed increased formation of higher-order multimers (Fig. 3A), and Epo tetherin was less efficiently N glycosylated (Fig. 3B) compared to human tetherin, but no appreciable differences in the subcellular localization of human and Epo tetherin were observed (Fig. 3C). The total expression (immunoblot) of fruit bat tetherins exceeded that of human tetherin by ∼2-fold (Fig. 3D and F). However, the total expression of fruit bat tetherins was less efficient than that observed for several other tetherin orthologues shown to be susceptible to counteraction by EBOV-GP, including murine (Fig. 1C; the present study) and porcine (41) tetherin (Fig. 3D). Importantly, human and fruit bat tetherin were comparably expressed at the cell surface (flow cytometry, Fig. 3E and F; the surface levels of tetherins examined in Fig. 1 were not determined), indicating that human and fruit bat tetherin are equally well expressed at the place where tetherin unfolds its antiviral activity.
FIG 3.
Fruit bat tetherin displays similar subcellular localization and surface expression as human tetherin. (A) Expression of human and fruit bat tetherin in transfected HEK293T cells was analyzed by immunoblotting (the amounts of the expression plasmids were adjusted to allow for comparable expression levels). An antibody directed against an N-terminal HA antigenic tag added to the tetherin orthologues was used for detection of tetherin expression. The expression of β-actin was determined as a loading control. Similar results were obtained in a separate experiment. Hyp, Hypsignathus monstrosus; Epo, Epomops buettikoferi. Red (human) and blue (fruit bat) arrowheads indicate unglycosylated (open arrowheads), partially glycosylated (dashed arrowheads), and fully glycosylated (filled arrowheads) tetherin, respectively, while asterisks indicate multimeric forms. (B) Cell lysates of HEK293T cells transfected with human or fruit bat (Epo) tetherin were either untreated or treated with PNGase F to enzymatically remove potential N-glycans and subsequently analyzed by immunoblotting (as described for panel A). A representative blot is shown, and similar results were obtained in two separate experiments. (C) Vero 76 cells were cotransfected with identical amounts of expression plasmids coding for human or fruit bat tetherin (containing an N-terminal HA tag) and marker proteins for either the endoplasmic reticulum (ER), Golgi apparatus (Golgi), trans-Golgi network (TGN), or Rab10- or Rab11a-positive recycling endosomes (all fused to eGFP). At 24 h posttransfection, the cells were fixed, permeabilized, and subsequently incubated with an HA-specific primary antibody and Alexa Fluor 594-labeled secondary antibody. Finally, cellular nuclei were stained with DAPI, and samples were processed for fluorescence microscopy. Shown are representative pictures at a 100-fold magnification, with all fluorescence channels merged into one picture. Similar results were obtained in a separate experiment. (D) Identical amounts of expression plasmids for the indicated tetherin orthologues equipped with an N-terminal HA epitope were transfected into HEK293T cells, and tetherin expression in whole-cell lysates (WCLs) was investigated by Western blot analysis with an HA-specific antibody. Cells transfected with empty expression vector served as a control. Similar results were obtained in two separate experiments. (E) The experiment was performed as described for panel C, but human and fruit bat tetherin proteins with an HA tag within the extracellular domain (located upstream of the GPI-anchoring signal) were used, and tetherin expression was analyzed by flow cytometry. The average of eight independent experiments is shown, for which the geometric mean channel fluorescence measured for cells expressing human tetherin was set as 1. Error bars indicate the SEM. Statistical significance was analyzed by one-way ANOVA with Bonferroni posttest analysis (ns, P > 0.05). (F) Normalized data from experiments examining tetherin expression in WCLs (WCL, immunoblot, n = 7) and at the cell surface (Surface, flow cytometry, n = 8) are shown. The expression of human tetherin was set as 1, and the expression of fruit bat tetherin was calculated as x-fold changes ± the SEM.
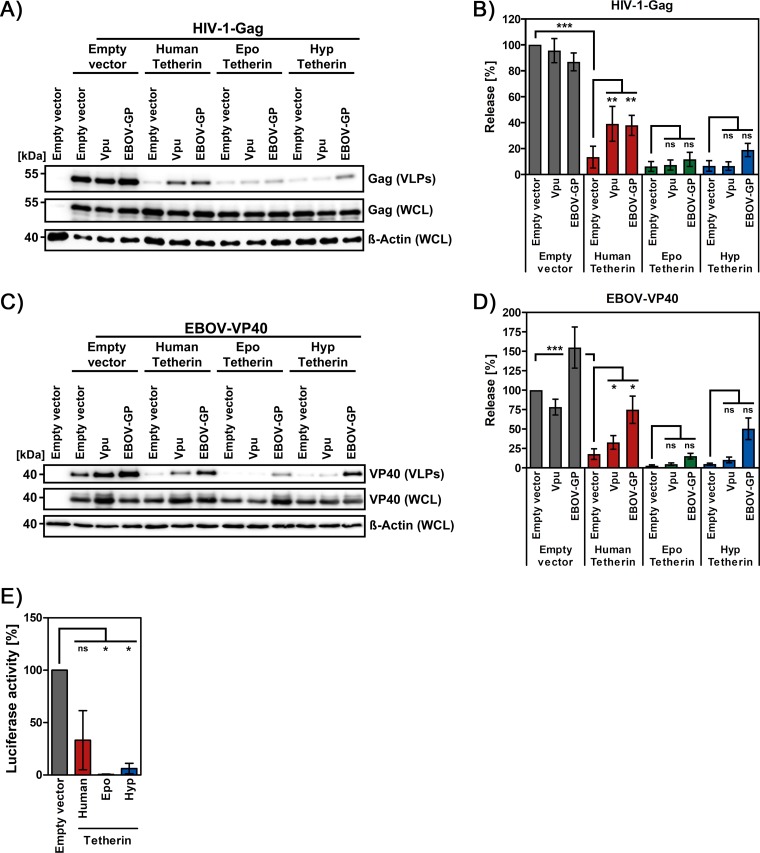
Both human and fruit bat tetherin proteins robustly interfered with release of HIV-1 Gag- and EBOV-VP40-based particles (Fig. 4A to D), and release of the latter is believed to adequately mirror important aspects of release of EBOV from infected cells. Moreover, both fruit bat tetherins exhibited higher resistance against counteraction by HIV-1 Vpu than human tetherin (Fig. 4A to D). Notably, Epo tetherin was also largely resistant against counteraction by EBOV-GP under the conditions chosen, while the resistance of Hyp tetherin was less pronounced. In keeping with these findings, both fruit bat tetherins were more potent than human tetherin in inhibiting the release of replication-competent EBOV-like particles in a system that, unlike the VP40-based particles studied above, faithfully mimics most steps of EBOV infection (42) (Fig. 4E). Finally, the resistance of fruit bat tetherin to counteraction by EBOV-GP was dependent on the amount of tetherin plasmid transfected and thus on tetherin expression levels (not shown). Collectively, these results indicate that fruit bat tetherins, like human tetherin, are potent antiviral factors that can be largely resistant to counteraction by EBOV-GP when expressed at high levels.
FIG 4.
Fruit bat tetherin is largely resistant against counteraction by EBOV-GP. (A) HEK293T cells were cotransfected with expression plasmids for Gag, human tetherin or the indicated tetherin orthologues, and tetherin antagonist (Vpu, EBOV-GP). Cells transfected with empty expression vector instead of tetherin and/or antagonist served as controls. The release of Gag-derived VLPs in culture supernatants and Gag expression in WCLs was analyzed by immunoblotting. The expression of β-actin was determined as a loading control. (B) The average of four experiments conducted as described for panel A and quantified via the ImageJ program is shown. The release of Gag in the absence of tetherin and antagonist was set as 100%. Error bars indicate the SEM. (C) The release assay was conducted as described for panel A, but EBOV-VP40 was used instead of HIV-1-Gag. Detection of VP40 was carried out using an antibody targeting the N-terminal cMYC tag. (D) The average of six experiments conducted as described for panel C and quantified via the ImageJ program is shown. The release of VP40 in the absence of tetherin and antagonist was set as 100%. Error bars indicate the SEM. (E) Transcription and replication-competent VLPs (trVLP system) were produced in HEK293T cells in the absence or presence of the indicated tetherin proteins and used to transduce fresh HEK293T cells expressing DC-SIGN and the EBOV polymerase complex (EBOV-NP, EBOV-VP35, EBOV-VP30, and EBOV-L). The activity of Renilla luciferase, encoded by the viral minigenome, as an indicator for trVLP infection was measured. For normalization, the luciferase activity for trVLP produced in the absence of tetherin was set as 100%. Presented are the combined results from three independent experiments with error bars indicating the SEM. The statistical significance of the data presented in panels B, D, and E was tested by one-way ANOVA with Bonferroni posttest analysis (ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Endogenous expression of fruit bat tetherin is induced by IFN and inhibits VSV infection.
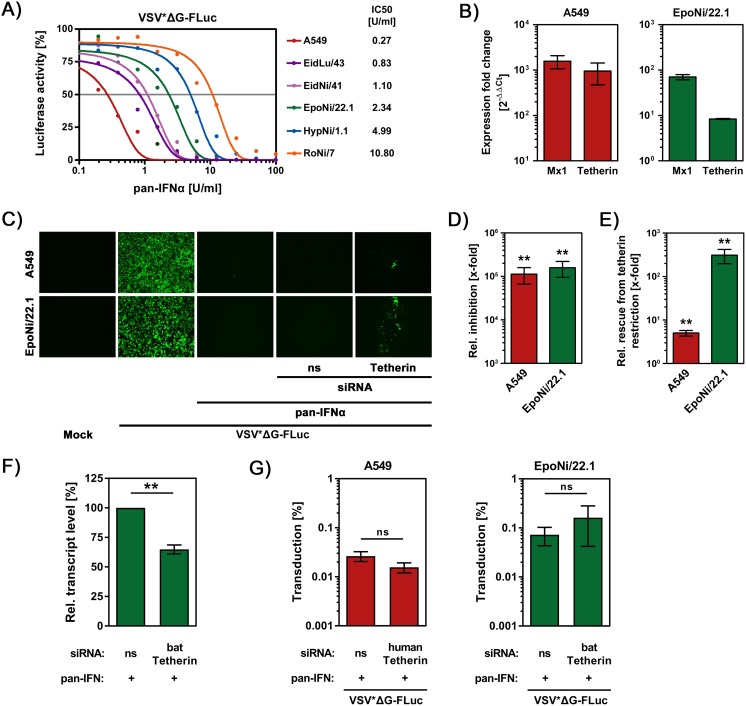
All previously discussed results were obtained upon directed expression of tetherin. Therefore, we sought to determine whether endogenous fruit bat tetherin also exerts antiviral activity. To this end, we first examined whether EpoNi/22.1 (established from kidney of E. buettikoferi) and HypNi/1.1 cells (established from kidney of H. monstrosus) (43, 44), from which tetherin was cloned, as well as other fruit bat cell lines, were responsive to treatment with pan-IFN-α. Human A549 cells were included as a positive control, since these cells are known to be highly IFN sensitive. Treatment of all cell lines with IFN at noncytotoxic concentrations (data not shown) markedly reduced transduction by a single-cycle VSV vector in a concentration-dependent manner (Fig. 5A). Since IFN-mediated reduction of transduction was more prominent for EpoNi/22.1 cells (50% inhibitory concentration [IC50] = 2.34 U/ml) compared to HypNi/1.1 cells (IC50 = 4.99 U/ml), the former cell line was selected for subsequent analyses. Quantitative RT-PCR (qRT-PCR) using myxovirus resistance protein 1 (Mx1) as a positive control for IFN-stimulated gene expression showed that tetherin mRNA expression was highly upregulated in A549 cells upon IFN treatment (Fig. 5B), as expected. Similarly, IFN stimulation upregulated tetherin mRNA levels in EpoNi/22.1 cells, although not to the same extent as Mx1 encoding mRNA (Fig. 5B). Thus, fruit bat cells transit into an antiviral state upon IFN treatment, and tetherin expression is induced by IFN in these cells, allowing us to determine whether tetherin contributes to the antiviral effect of IFN treatment. For this, we utilized a replication-competent VSV variant encoding enhanced green fluorescent protein (eGFP), since this virus is known to be highly IFN sensitive. Indeed, treatment of A549 and EpoNi/22.1 cells with IFN decreased VSV infection (Fig. 5C) and reduced production of progeny virus by more than 100,000-fold (for A549 cells, the mean titers dropped from ∼1.6 × 109 focus-forming units [FFU]/ml to ∼2.9 × 104 FFU/ml upon IFN stimulation, whereas for EpoNi/22.1 cells the titers dropped from ∼7.5 × 108 FFU/ml to ∼5.5 × 103 FFU/ml upon IFN stimulation) (Fig. 5D). Using this system in conjunction with small interfering RNA (siRNA)-mediated gene knockdown, we next sought to determine whether tetherin contributes to the IFN-mediated antiviral state. The IFN-induced block of infection of A549 cells was modestly (∼5-fold) rescued upon pretreatment of cells with siRNA against human tetherin (mean titer, ∼1.3 × 104 FFU/ml) compared to cells treated with control siRNA (mean titer, ∼6.4 × 104 FFU/ml) (Fig. 5C and E), suggesting that expression of other ISGs can facilitate efficient inhibition of VSV in the absence of tetherin in this cell line. More strikingly, siRNAs against fruit bat tetherin but not nonsense control siRNA reduced tetherin mRNA expression (Fig. 5F) and strongly (∼150-fold) rescued the generation of infectious VSV in fruit bat cells (mean titers, ∼1.3 × 103 FFU/ml [control siRNA] and ∼1.9 × 105 FFU/ml [tetherin-specific siRNA]) (Fig. 5E). The rescue most likely occurred at the stage of viral release, the target of tetherin antiviral activity, since the transduction, genome transcription, and translation of virally encoded proteins in IFN-treated cells were not impacted by the siRNA (Fig. 5G). In sum, our results indicate that tetherin expression in fruit bat cells is IFN inducible and is to a significant part responsible for the IFN-mediated blockade of VSV infection.
FIG 5.
Expression of fruit bat tetherin is IFN-inducible and required for robust IFN-mediated inhibition of VSV infection. (A) A549 cells and the indicated fruit bat cell lines were incubated with increasing amounts of pan-IFN-α, inoculated with a single-cycle VSV vector bearing VSV-G and encoding luciferase (VSV*ΔG-FLuc), and the luciferase activity in cell lysates was quantified. The luciferase activity measured for cells treated with medium without pan-IFN-α was set as 100%. The results of a representative experiment performed with triplicate samples are shown and were confirmed in a separate experiment. Error bars indicate the SEM. The 50% inhibitory concentration (IC50) was calculated for each cell line and is indicated. (B) Human (A549) and fruit bat (EpoNi/22.1) cells were pan-IFN-α or mock treated, and cellular RNA was extracted, reverse transcribed into cDNA, and analyzed for transcript levels of β-actin, Mx1 (myxovirus resistance protein), and tetherin by qRT-PCR. Shown are combined data (given as the fold change in expression upon IFN treatment, normalized to β-actin) of three independent experiments. Error bars indicate the SEM. (C) Human (A549) and fruit bat (EpoNi/22.1) cells were transfected with the indicated siRNAs (ns, nonsense = control) or left untransfected and then either treated with pan-IFN-α or mock treated, followed by inoculation with replication-competent VSV encoding eGFP. Finally, infected cells (as determined by the expression of eGFP) were detected via fluorescence microscopy. Similar results were obtained in five separate experiments. (D) Viral titers in the cellular supernatants of the untransfected, IFN-α- or mock-treated cells described in panel C were quantified. The relative (x-fold) differences between viral titers in the supernatants of pan-IFN-α- and mock-treated cells were calculated. The average of six independent experiments performed with triplicate samples is shown. Error bars indicate the SEM. (E) Viral titers in the cellular supernatants of the siRNA-transfected and IFN-treated cells described in panel C were quantified. The relative (x-fold) differences between viral titers of pan-IFN-α-treated cells transfected with control or tetherin-specific siRNA were calculated. The average of six independent experiments performed with triplicate samples is shown. Error bars indicate the SEM. (F) Relative tetherin transcript levels in IFN-treated fruit bat cells (EpoNi/22.1), which were previously transfected with either control (ns) or fruit bat tetherin-specific (batTetherin) siRNA, were compared by qRT-PCR (normalized against β-actin transcript levels). Shown are the combined results of three independent experiments performed with triplicate samples, in which tetherin transcript levels of IFN-treated cells that received control siRNA were set as 100%. Error bars indicate the SEM. (G) Human (A549) and fruit bat (EpoNi/22.1) cells were transfected with control or tetherin-specific siRNA prior to treatment with pan-IFN-α. At 48 h posttransfection, the cells were inoculated with single-cycle VSV vector pseudotyped with VSV-G (VSV*ΔG-FLuc). After incubation for 1 h, the cells were washed and further incubated for 8 h before virus-encoded luciferase activity was quantified in the cell lysates. Shown are normalized data from three independent experiments performed with quadruplicate samples in which transduction of cells without prior IFN treatment was set as 100%. Error bars indicate the SEM. The statistical significance of the data presented in panels D and E was analyzed by using a Mann-Whitney U test, while the data presented in panels F and G were analyzed by using a paired, two-tailed Student t test (ns, P > 0.05; **, P ≤ 0.01).
Fruit bat tetherin is required for efficient inhibition of NiV but not EBOV infection by IFN.
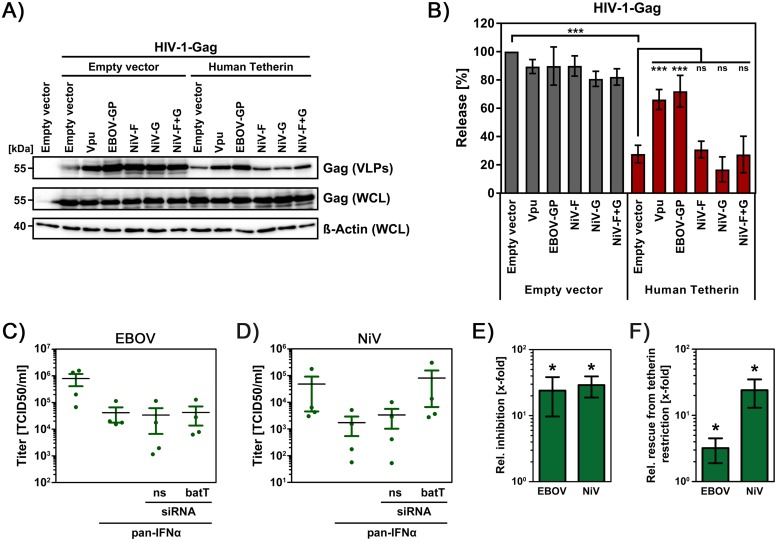
In light of the important contribution of tetherin to control of VSV infection in fruit bat cells, we investigated whether tetherin also contributes to the blockade of EBOV and NiV infection by IFN. NiV was included in the analysis, since the virus uses fruit bats as a natural reservoir (18), is highly pathogenic in humans and may be tetherin sensitive. Thus, tetherin was previously shown to inhibit the release of NiV-like particles (36, 37), and analysis of the viral surface proteins, NiV-F and NiV-G, revealed that they do not antagonize tetherin (Fig. 6A and B). IFN pretreatment reduced EBOV and NiV spread in EpoNi/22.1 cells by roughly 20- and 30-fold, respectively (Fig. 6C to E). For NiV, transfection of tetherin siRNA rescued this blockade almost entirely as it increased titers of free virus in the culture supernatant by ∼20-fold (Fig. 6D and F). In contrast, EBOV spread was only slightly rescued (∼2-fold) (Fig. 6C and F). These findings suggest that tetherin may be a major contributor to control of NiV infection in fruit bats, the natural reservoir, whereas tetherin’s contribution to the inhibition of EBOV spread in the putative natural reservoir might be moderate.
FIG 6.
Fruit bat tetherin is required for robust IFN-mediated inhibition of NiV but not EBOV spread. (A) HEK293T cells were cotransfected with expression plasmids for Gag and human tetherin, and either HIV-1-Vpu, EBOV-GP, NiV-F, NiV-G, or NiV-F+G. Cells transfected with empty expression plasmid instead of tetherin and/or (potential) antagonist served as controls. The release of Gag-derived VLPs in culture supernatants and Gag expression in WCLs was analyzed by immunoblotting. The expression of β-actin was determined as a loading control. (B) The average of three experiments conducted as described for panel A and quantified via the ImageJ program is shown. The release of Gag in the absence of tetherin was set as 100%. Error bars indicate the SEM. (C and D) EpoNi/22.1 cells that were transfected with control (ns) or fruit bat tetherin-specific siRNA (batT) and subsequently pan-IFN-α treated or mock treated were inoculated with EBOV (C) or NiV (D). Subsequently, the cells were washed and incubated, and viral titers in culture supernatants were quantified by measuring the TCID50/ml. Panels C and D show the mean and individual titers for all experimental conditions measured in four independent experiments (performed with triplicate samples). Error bars indicate the SEM. (E) The data from panels C and D were used to calculate the relative (x-fold) reduction of viral spread upon IFN treatment. The combined data from four independent experiments performed with triplicate samples are shown. Error bars indicate the SEM. (F) The data from panels C and D were used to calculate the relative (x-fold) differences between viral titers in the supernatants of pan-IFN-α-treated cells that were transfected with either control or tetherin-specific siRNA. The combined data from four independent experiments performed with triplicate samples are shown. Error bars indicate the SEM. The statistical significance of the data presented in panel B was determined by one-way ANOVA with Bonferroni posttest analysis, while the data presented in panels E and F were analyzed using a Mann-Whitney U test (ns, P > 0.05; *, P ≤ 0.05; ***, P ≤ 0.001).
DISCUSSION
Fruit bats, the suspected natural reservoir of EBOV and the proven reservoir of NiV, control EBOV and NiV infection by poorly understood means, although a contribution of the IFN system has been suspected (26). Here, we show that expression of fruit bat-encoded tetherin is IFN stimulated and associated with robust antiviral activity. Moreover, we demonstrate that fruit bat tetherin critically contributes to IFN-dependent control of VSV and NiV but not EBOV infection of fruit bat cells.
The antiviral activity of tetherin was first reported by Neil et al. in the context of HIV-1 infection (30), but it is now well established that tetherin can also inhibit the spread of several other enveloped viruses (31, 33). Tetherin can exert a broad antiviral activity because it targets a host cell-derived component of virions, the viral envelope. The EBOV-GP was shown to counteract tetherin and to promote release of VP40-based VLPs from transfected cells (34). Moreover, two reports demonstrated that human tetherin expression does not appreciably inhibit spread of authentic EBOV (35, 36), indicating that GP-mediated tetherin antagonism might allow for viral amplification in tetherin-positive cells. However, the contribution of tetherin to viral control in the natural reservoir has not been examined.
Our results show that tetherin from E. buettikoferi and H. monstrosus share ∼46% sequence identity with human tetherin. Moreover, the fruit bat tetherin orthologues (99.5% sequence identity) contain all functional elements previously defined for human tetherin: an N-terminal cytoplasmic domain, a transmembrane domain, an extracellular coiled coil region, a C-terminal GPI anchor, two sequons for attachment of N-glycans, and three cysteines available for formation of disulfide bonds. Moreover, fruit bat tetherin orthologues and human tetherin showed a roughly comparable cellular localization, with both proteins being detectable in the endoplasmic reticulum (ER), Golgi apparatus, trans-Golgi network (TGN), and recycling endosomes and at the cell surface. Strikingly, however, tetherins from E. buettikoferi and, to a lesser degree, H. monstrosus were, in contrast to human tetherin, largely resistant against EBOV-GP-mediated counteraction, at least under conditions of high expression. This finding is noteworthy considering that EBOV-GP antagonizes tetherin orthologues of diverse species, as evidenced by the present study and earlier (35, 40), and is even active against artificial tetherin (40). At present, it is unclear why fruit bat tetherin was resistant to EBOV-GP counteraction under the conditions chosen. However, it is noteworthy that fruit bat tetherin was barely N-glycosylated in HEK293T cells, and the role of tetherin N-glycosylation in resistance deserves further investigation.
The robust antiviral activity of fruit bat tetherin in transfected cells raised the question whether endogenous tetherin contributes to viral control in fruit bat cells. After identification of EpoNi/22.1 cells as being responsive to IFN treatment using a single-cycle VSV vector, quantitative PCR (qRT-PCR) revealed that tetherin mRNA was upregulated upon IFN treatment, indicating that fruit bat tetherin, like its human counterpart, is an ISG. Strikingly, IFN stimulation combined with siRNA knockdown showed that tetherin was essential for robust IFN-mediated inhibition of VSV and NiV but not EBOV infection of fruit bat cells. The observation that NiV spread was sensitive to fruit bat tetherin expression is in keeping with the published finding that release of NiV-like particles is inhibited by human tetherin (36, 37) and the new finding that NiV-F and NiV-G fail to antagonize tetherin, at least in an HIV Gag-based assay. However, the major contribution of tetherin to IFN-mediated control of NiV infection is remarkable, since exposure of human and likely also fruit bat cells to IFN stimulates the expression of several hundred genes, many of which encode proteins which exert antiviral activity (27). An explanation for the moderate contribution of fruit bat tetherin to the IFN-mediated control of EBOV spread might reside in tetherin expression levels. Resistance of fruit bat tetherin to counteraction by EBOV-GP in VLP assays was dependent on the amount of tetherin plasmid transfected, and it is conceivable that tetherin expression levels attained upon IFN stimulation of cells might have been insufficient to provide resistance against EBOV-GP-mediated counteraction. Resolving this question requires reagents that allow comparing fruit bat tetherin levels on the surface of transfected and IFN treated cells, which are not available at present. Furthermore, the possibility, although remote, that authentic EBOV (unlike VP40-based EBOV particles) might be intrinsically tetherin resistant should not be discarded. This scenario can only be investigated upon identification of mutations in GP that selectively interfere with tetherin antagonism, and first steps in this direction have recently been made (45).
Collectively, our findings show that tetherin is central to the IFN-mediated control of NiV in fruit bat cells, while the factor(s) required for the robust control of EBOV in these animals (26, 46) remains to be elucidated.
MATERIALS AND METHODS
Cells and viruses.
HEK293T (human, kidney) and A549 (human, lung) cells were cultivated in Dulbecco modified Eagle medium (DMEM; PAN-Biotech) and DMEM/F-12 (Gibco), respectively, supplemented with 10% fetal bovine serum (FBS; Biochrom) and penicillin-streptomycin (PAN-Biotech) at final concentrations of 100 U/ml (penicillin) and 0.1 µg/ml (streptomycin). Vero E6, Vero 76 (both African green monkey, kidney), and BHK-21 (Syrian hamster, kidney) cells were cultivated in DMEM supplemented with 5% FBS and penicillin-streptomycin. The following fruit bat cell lines, kindly provided by C. Drosten and M. A. Müller, were cultivated in DMEM supplemented with 10% FBS and penicillin-streptomycin: EpoNi/22.1 (Buettikofer’s epauletted fruit bat, Epomops buettikoferi; kidney), HypNi/1.1 (hammer-headed fruit bat, Hypsignathus monstrosus; kidney), RoNi/7 (Egyptian fruit bat, Rousettus aegyptiacus; kidney), and EidNi/41 and EidLu/43 (Straw-colored fruit bat, Eidolon helvum; kidney and lung, respectively). For subcultivation and seeding, cells were washed with phosphate-buffered saline (PBS) and detached by incubation in a trypsin/EDTA solution (PAN-Biotech) or by resuspension in DMEM (HEK293T). Cell numbers were determined under a light microscope using a Neubauer chamber. Cultivation of cells was carried out at 37°C in humidified atmosphere containing 5% CO2.
We employed a recombinantly produced vesicular stomatitis virus (VSV, Indiana strain), which expresses eGFP from an additional transcription unit located between the ORFs for the viral glycoprotein (G) and RNA-dependent RNA polymerase (L). The Ebola virus, Mayinga variant (GenBank accession no. NC_002549), used in this study was propagated in Vero E6 cells, and the virus titer was determined by immunoplaque titration. Furthermore, we utilized a Nipah virus (Malaysia strain) that contains an eGFP transcription unit between the NiV-G and NiV-L ORFs and that has been described elsewhere (47).
Cloning of fruit bat tetherin.
Total cellular RNA was isolated from ∼106 HypNi/1.1 and EpoNi/22.1 cells using the RNeasy minikit (Qiagen) according to the manufacturer’s protocol. Next, 1 µg of RNA was used as a template for cDNA synthesis using the SuperScript III first-strand synthesis system (Thermo Fisher Scientific) according to the manufacturer’s protocol (for random hexamers). A fragment of ∼550 bp was amplified using Phusion polymerase (Thermo Fisher Scientific). Primers were designed based on predicted fruit bat tetherin sequences from the National Center for Biotechnology Information (NCBI) database as a template (primer sequences are available upon request). Next, the DNA fragments were separated by agarose gel electrophoresis, extracted from the gel by commercial kits (Macherey Nagel), and inserted into the pCAGGS expression vector using the EcoRI and XhoI sites. Upon transformation into competent Escherichia coli by heat shock, three individual clones, which contained the insert, were subjected to automated sequence analysis (SeqLab).
Plasmids, mutagenesis, and transfection.
Expression plasmids for HIV-1 p55-Gag (Gag), HIV-1 Vpu (Vpu), EBOV-GP, VSV glycoprotein (VSV-G), Nipah virus fusion (F), and attachment glycoprotein (G), EBOV-VP40 harboring an N-terminal cMYC tag, and DC-SIGN have been described elsewhere (35, 44, 48–50). To generate expression plasmids for human, gorilla, African green monkey, pig, rat, mouse, and artificial tetherin, the respective ORFs were amplified from existing plasmids (32, 51, 52) and inserted into the pCAGGS expression vector. Rhesus macaque and marmoset tetherin were PCR-amplified from reverse-transcribed lung RNA as described for fruit bat tetherin and inserted into the pCAGGS expression vector. In addition, tetherin constructs were equipped with an N-terminal HA (YPYDVPDYA) epitope and identity of all PCR-amplified sequences was verified by automated sequence analysis (SeqLab). For the detection of tetherin at the cell surface via flow cytometry, human and fruit bat tetherin constructs with an extracellular hemagglutinin (HA) epitope (located upstream of their respective GPI anchor motifs) were cloned. Furthermore, eGFP-based expression vectors for localization studies targeting the ER and Golgi apparatus (both kindly provided by F. van Kuppeveld [9]), the TGN (TGN integral membrane protein 2)- and Rab10- or Rab11a-positive recycling endosomes were used. We further used a previously described transcription and replication-competent EBOV-like particle system that included pCAGGS-driven expression plasmids for T7 polymerase, EBOV-NP, EBOV-VP35, EBOV-VP30, and EBOV-L and a plasmid-coded minigenome that contains the genetic information for Renilla luciferase (RLuc) as a reporter gene (42). Plasmid transfection of HEK293T cells was carried out using calcium phosphate precipitation, while Vero 76 cells were transfected using ICAFectin441 (In-Cell-Art).
Analysis of tetherin expression and VLP release assays.
Expression of tetherin was analyzed by transfection of HEK293T cells grown in 12-well plates with expression plasmids for different tetherin constructs (2 µg). The impact of tetherin and EBOV-GP on the release of Gag- or VP40-based VLPs was studied essentially as described elsewhere (35, 49). HEK293T cells grown in 12-well plates were cotransfected with combinations of expression plasmids for Gag or VP40 (2 µg), tetherin (0.5 µg), and potential antagonist (2 µg). As controls and for the equilibration of the total DNA amounts, empty pCAGGS expression vector or an eGFP expression plasmid were used. At 16 h posttransfection, the transfection medium was replaced by fresh culture medium, and cells were incubated for an additional 32 h. The supernatants were then collected and cleared from cellular debris by centrifugation, and VLPs were pelleted from cleared supernatants by high-speed centrifugation through a 20% sucrose cushion. Next, 50 µl of 2× sodium dodecyl sulfate (SDS) loading buffer was added to concentrated VLPs, and samples were incubated at 95°C for 30 min before being directly used for analysis or stored at −20°C until further use. In parallel, whole-cell lysates (WCLs) were prepared by first washing cells with PBS and then lysing them with 100 µl of 2× SDS-containing lysis buffer (30 mM Tris [pH 6.8], 10% glycerol, 2% SDS, 5% β-mercaptoethanol, 0.1% bromophenol blue, 1 mM EDTA). After 15 min of incubation at room temperature, the samples were incubated at 95°C for 30 min and either directly used for analysis or stored at –20°C until further use.
Quantification of VLP release.
Quantification of Gag and VP40 release was carried out using the program ImageJ (Fiji Distribution) (53). For this, Gag or VP40 signals detected in the supernatants (corresponding to VLPs) were normalized against the respective signals obtained in WCLs. To compare multiple samples (e.g., different tetherin orthologues or tetherin antagonists), one sample was set at 100% and designated as a reference (i.e., Gag or VP40 release in the absence of tetherin and antagonist) for all other samples in that experiment. At least three independent immunoblots were used for quantification.
SDS-PAGE and immunoblot analysis.
VLP and WCL samples were separated via SDS-PAGE using gels containing 12.5% polyacrylamide and transferred onto a nitrocellulose membrane (GE Life Sciences, 0.2-μm pore size) using a tank blot system (Bio-Rad). After blotting, the membranes were blocked in 5% milk powder in PBS containing 0.1% Tween 20 (PBS-T) for 1 h at room temperature before antibody incubation was performed as follows: HIV-1 Gag was detected using supernatant of hybridoma cells secreting a mouse anti-Gag antibody (183-H12-5C) at a dilution of 1:100, while EBOV-VP40 containing an N-terminal cMYC tag was detected using supernatant of hybridoma cells secreting a mouse anti-cMYC antibody (9E10) at a dilution of 1:3. To detect tetherin containing an HA epitope, an HA-specific mouse antibody (Sigma-Aldrich) was used at a dilution of 1:1,000. Primary antibody binding was further detected through incubation with an anti-mouse, horseradish peroxidase (HRP)-coupled secondary antibody (Dianova) at a dilution of 1:10,000. Visualization of bound secondary antibodies was achieved using a self-made enhanced chemiluminescence (ECL) solution (0.1 M Tris-HCl [pH 8.6], 250 µg/ml luminol, 1 mg/ml para-hydroxycoumaric acid, 0.3% H2O2) in combination with the ChemoCam imaging system and ChemoStarProfessional software (Intas). After imaging of the Gag, VP40, and tetherin signals in WCLs, the membranes were stripped of bound antibodies by incubation with stripping buffer (0.0625 M Tris-HCl [pH 6.8], 2% SDS, 0.8% β-mercaptoethanol) for 30 min at 50°C, rinsed with water for 1 h, washed with PBS-T, and reblocked. Subsequently, membranes were probed with β-actin-specific rabbit antibody (1:1,000; Sigma-Aldrich) and rabbit-specific, HRP-conjugated secondary antibody (1:10,000; Dianova) for the detection of β-actin levels as a loading control.
Production of transcription and replication-competent EBOV-like particles and transduction of target cells.
To assess the impact of tetherin expression on EBOV-like particles, we used a tetracistronic transcription and replication-competent virus-like particle (trVLP) system that resembles many aspects of the EBOV life cycle (42). Briefly, HEK293T cells grown in 6-well plates were cotransfected with expression plasmids for EBOV-NP (125 ng), EBOV-VP35 (125 ng), EBOV-VP30 (75 ng), EBOV-L (1 µg), p4cis-vRNA-RLuc (250 ng), T7 polymerase (250 ng), and tetherin or empty expression vector as a control (500 ng) (producer cells). At 16 h posttransfection, the transfection medium was replaced by fresh culture medium, and cells were incubated for an additional 56 h. The supernatants were then collected and cleared from cellular debris by centrifugation, and trVLPs were inoculated onto HEK293T cells grown in 96-well plates that were previously (24 h) cotransfected with EBOV-NP (25 ng), EBOV-VP35 (25 ng), EBOV-VP30 (15 ng), EBOV-L (200 ng), and DC-SIGN (250 ng) (target cells). Target cells were further incubated for 72 h. In order to investigate the ability of tetherin to restrict spread of trVLPs, the target cells were lysed in cell culture lysis reagent (Promega) at 50 µl/well. Next, the lysates were transferred to white, opaque-walled 96-well plates and incubated with an in-house-made RLuc substrate, and the RLuc activity was measured in a microplate reader (Hidex).
Flow cytometry.
Analysis of tetherin expression at the cell surface was performed as follows: HEK293T cells were transfected with human or fruit bat tetherin harboring an extracellular HA epitope. Cells transfected with an eGFP expression vector or empty plasmid served as negative controls. At 48 h posttransfection, the cells were washed and resuspended in PBS supplemented with 0.1% BSA (Roth, PBS/BSA). Next, the samples were split into two reaction tubes and probed either with an HA epitope-specific mouse antibody (Sigma-Aldrich) or with an isotype control antibody (Sigma-Aldrich, 1:100) for 1 h at 4°C. Subsequently, the cells were pelleted, washed with PBS/BSA, and incubated with an Alexa Fluor 647-coupled anti-mouse antibody (Thermo Fisher Scientific, 1:100) again for 1 h at 4°C. Afterward, the cells were washed twice with PBS/BSA and fixed with 2% paraformaldehyde, and staining was analyzed by using an LSR II flow cytometer combined with the FACSDiva software (both from BD Biosciences). Further data analysis was performed using the FCS Express 4 Flow research software (De Novo Software).
Phylogenetic and sequence analyses.
Phylogenetic analysis was performed utilizing the MEGA6 (v6.06) software package (54). For this, sequences were aligned (MUSCLE algorithm), and a phylogenetic tree was constructed based on the neighbor-joining method with 1,000 bootstrap iterations. Amino acid sequences of diverse mammalian tetherin orthologues were obtained from the NCBI database and are summarized in Table S1 in the supplemental material. For motif and domain predictions, the following online tools were used: HMMTOP (http://www.enzim.hu/hmmtop/, transmembrane domains), NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/, N-glycosylation motifs), Coiled-Coil Prediction (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_lupas.html, coiled-coil domains), and big-PI Predictor (http://mendel.imp.ac.at/gpi/gpi_server.html, GPI anchor addition sites).
Transduction with single-cycle VSV vectors.
We used a previously described, replication-deficient VSV that lacks the genetic information for VSV-G but instead harbors separate transcription units encoding eGFP and firefly luciferase (FLuc), VSV*ΔG-FLuc (kindly provided by G. Zimmer) (12). Propagation of VSV*ΔG-FLuc and transcomplementation with VSV-G was achieved on a helper cell line that expresses VSV-G in an inducible fashion, BHK-21(G43) (55). To quantify the FLuc activity upon the inoculation of cells with VSV*ΔG-FLuc, cell culture supernatants were removed, and cells washed with PBS, followed by cell lysis using a cell culture lysis reagent (Promega) for 30 min. Subsequently, lysates were transferred into white, opaque-walled 96-well plates. Finally, FLuc substrate (PJK) was added, and luminescence signals were detected using a plate luminometer (Hidex).
Knockdown of endogenous tetherin expression by siRNA.
Human tetherin-specific siRNA and the corresponding control siRNA-A were purchased from Santa Cruz, while custom-designed stealth siRNA specific for fruit bat tetherin and the corresponding medium G+C content control siRNA were obtained from Thermo Fisher Scientific. Delivery of siRNA into A549, HypNi/1.1, and EpoNi/22.1 cells (25 pmol/well, 12-well format) was achieved using RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Quantification of tetherin transcripts by qPCR.
In order to measure the mRNA transcript levels for tetherin, Mx1 (ISG control), and β-actin (housekeeping gene control), qPCR analysis of reverse-transcribed total cellular RNA, which was obtained from cells that were transfected with siRNA and/or stimulated with pan-IFN-α, was performed. First, RNA was extracted from cells using an RNeasy minikit (Qiagen) according to the manufacturer’s protocol. Next, 1 µg of RNA was treated with DNase I (New England Biolabs) to eliminate coisolated genomic DNA and directly used as a template for cDNA synthesis by using the SuperScript III first-strand synthesis system (Thermo Fisher Scientific) according to the manufacturer’s protocol (for random hexamers). Then, 1 µl of cDNA mix was subjected to qPCR on a Rotorgene Q platform (Qiagen) using a QuantiTect SYBR Green PCR kit (Qiagen) with primers targeting either human or fruit bat tetherin (designed using GeneScript [https://www.genscript.com/tools/real-time-pcr-tagman-primer-design-tool]), Mx1, or β-actin (14). Induction of tetherin and Mx1 gene expression after stimulation with pan-IFN-α (displayed as the expression fold change) was analyzed by the 2−ΔΔCT method (15) with β-actin as the housekeeping gene. To assess the efficiency of siRNA-mediated knockdown by qPCR, the relative tetherin transcript levels for control siRNA-treated, pan-IFN-α-stimulated cells were set as 100% and compared to the value for cells transfected with siRNA targeting tetherin and stimulated with pan-IFN-α.
Infection of cells with replication-competent VSV, EBOV, or NiV.
All experiments with live EBOV and NiV were performed under biosafety level 4 conditions at the Institute of Virology, Philipps University of Marburg, by trained personnel and in accordance with national regulations. After removal of the cell culture supernatant, the cells were washed once and subsequently inoculated with VSV (MOI = 0.005) or EBOV or NiV (both, MOI = 0.1). All experiments were performed in 12-well format in triplicates, and mock-infected cells served as controls. At 1 h postinfection (p.i.), the cells were washed and further incubated with fresh culture medium. Viral titers in the supernatant were quantified at 1 h p.i. (all viruses, washing control) and at 24 h (VSV) or 48 h p.i. (EBOV and NiV).
Quantification of viral titers.
VSV titers were quantified on confluently grown BHK-21 cells (96-well format) that were inoculated with 10-fold serial dilutions of the supernatants to be analyzed. At 1 h p.i., the cells were overlaid with culture medium containing 1% methylcellulose (Sigma-Aldrich) to only allow viral spread between neighboring cells, resulting in focus formation. At 18 h p.i., eGFP-positive foci were counted (FFU/ml) under a fluorescence microscope. To quantify the relative inhibition of virus replication by stimulation with pan-IFN-α, the x-fold difference between VSV titers in supernatants of mock- versus pan-IFN-α-treated cells was calculated. In addition, the relative rescue of VSV from tetherin restriction by siRNA-mediated tetherin knockdown in pan-IFN-α-treated cells was quantified by calculating the x-fold difference between VSV titers in the supernatants of control versus tetherin-specific siRNA-transfected cells. EBOV and NiV titers were analyzed on Vero E6 and Vero 76 cells, respectively, inoculated with 10-fold serial dilutions of the supernatants. At 5 days p.i. (NiV) or 14 days p.i. (EBOV), the 50% tissue culture infective dose (TCID50)/ml was calculated based on the formation of cytopathic effects and by using Spearman-Karber analysis (47, 56).
Fluorescence microscopy.
To analyze intracellular localization of human and fruit bat tetherin, Vero 76 cells grown on coverslips were cotransfected with expression plasmids for tetherin constructs harboring an N-terminal HA epitope and the respective marker for the ER, Golgi, TGN, or Rab10- or Rab11a-positive recycling endosomes, all linked to eGFP, using ICAFectin441 (In-Cell-Art) as the transfection reagent according to the manufacturer’s protocol. At 24 h posttransfection, the cells were fixed, permeabilized with 0.1% Triton X-100 in PBS, and subsequently incubated with anti-HA (Sigma-Aldrich, 1:500) and anti-mouse Alexa Fluor 594 (Thermo Fisher Scientific, 1:500) antibodies. Finally, the cells were incubated with DAPI (Roth) to stain the cellular nuclei, and coverslips were mounted on glass slides using Mowiol containing DABCO (Roth) as an antibleaching reagent. Representative pictures were taken at a magnification of ×100 using a Nikon Eclipse Ti fluorescence microscope in combination with NIS elements AR software (both Nikon). To investigate VSV spread in cells transfected with control or tetherin-specific siRNA and subsequently treated with or without pan-IFN-α, cells were fixed at 24 h p.i., and pictures were taken at a magnification of ×10.
Data availability.
Sequences of Epo and Hyp tetherin are available in GenBank under accession numbers MG792836.1 and MG792837.1, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by Deutsche Forschungsgemeinschaft grants PO 716/8-1 (to S.P.), SCHI1073/4-1 (to M.S.), SFB 1021, B04 (to A.M.), and SFB 1021, A02 (to S.B.). In addition, this work was funded by the German Federal Ministry of Education and Research, EBOKON consortium (S.P. and S.B.). The funders had no role in the design of the research and the interpretation of the research results.
We thank F. van Kuppeveld, G. Zimmer, and C. Drosten and M. A. Müller for providing the eGFP-tagged markers for the ER and Golgi apparatus, the replication-deficient VSV vector, and the fruit bat cell lines, respectively.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01821-18.
REFERENCES
- 1.World Health Organization. 2016. Ebola situation report. World Health Organization, Geneva, Switzerland: http://apps.who.int/ebola/current-situation/ebola-situation-report-17-february-2016. [Google Scholar]
- 2.WHO Ebola Response Team. 2016. Ebola virus disease among male and female persons in West Africa. N Engl J Med 374:96–98. doi: 10.1056/NEJMc1510305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biava M, Caglioti C, Bordi L, Castilletti C, Colavita F, Quartu S, Nicastri E, Lauria FN, Petrosillo N, Lanini S, Hoenen T, Kobinger G, Zumla A, Di CA, Ippolito G, Capobianchi MR, Lalle E. 2017. Detection of viral RNA in tissues following plasma clearance from an Ebola virus infected patient. PLoS Pathog 13:e1006065. doi: 10.1371/journal.ppat.1006065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brainard J, Pond K, Hooper L, Edmunds K, Hunter P. 2016. Presence and persistence of Ebola or Marburg virus in patients and survivors: a rapid systematic review. PLoS Negl Trop Dis 10:e0004475. doi: 10.1371/journal.pntd.0004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deen GF, Knust B, Broutet N, Sesay FR, Formenty P, Ross C, Thorson AE, Massaquoi TA, Marrinan JE, Ervin E, Jambai A, McDonald SL, Bernstein K, Wurie AH, Dumbuya MS, Abad N, Idriss B, Wi T, Bennett SD, Davies T, Ebrahim FK, Meites E, Naidoo D, Smith S, Banerjee A, Erickson BR, Brault A, Durski KN, Winter J, Sealy T, Nichol ST, Lamunu M, Stroher U, Morgan O, Sahr F. 2015. Ebola RNA persistence in semen of Ebola virus disease survivors: preliminary report. N Engl J Med. [DOI] [PMC free article] [PubMed]
- 6.Martins KA, Jahrling PB, Bavari S, Kuhn JH. 2016. Ebola virus disease candidate vaccines under evaluation in clinical trials. Expert Rev Vaccines 15:1101–1112. doi: 10.1080/14760584.2016.1187566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control. 1999. Outbreak of Hendra-like virus: Malaysia and Singapore, 1998-1999. JAMA 281:1787–1788. [PubMed] [Google Scholar]
- 8.Paton NI, Leo YS, Zaki SR, Auchus AP, Lee KE, Ling AE, Chew SK, Ang B, Rollin PE, Umapathi T, Sng I, Lee CC, Lim E, Ksiazek TG. 1999. Outbreak of Nipah virus infection among abattoir workers in Singapore. Lancet 354:1253–1256. doi: 10.1016/S0140-6736(99)04379-2. [DOI] [PubMed] [Google Scholar]
- 9.Chua KB, Goh KJ, Wong KT, Kamarulzaman A, Tan PS, Ksiazek TG, Zaki SR, Paul G, Lam SK, Tan CT. 1999. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354:1257–1259. doi: 10.1016/S0140-6736(99)04299-3. [DOI] [PubMed] [Google Scholar]
- 10.Chadha MS, Comer JA, Lowe L, Rota PA, Rollin PE, Bellini WJ, Ksiazek TG, Mishra A. 2006. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis 12:235–240. doi: 10.3201/eid1202.051247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ching PK, de los Reyes VC, Sucaldito MN, Tayag E, Columna-Vingno AB, Malbas FF Jr, Bolo GC Jr, Sejvar JJ, Eagles D, Playford G, Dueger E, Kaku Y, Morikawa S, Kuroda M, Marsh GA, McCullough S, Foxwell AR. 2015. Outbreak of henipavirus infection, Philippines, 2014. Emerg Infect Dis 21:328–331. doi: 10.3201/eid2102.141433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurley ES, Montgomery JM, Hossain MJ, Bell M, Azad AK, Islam MR, Molla MA, Carroll DS, Ksiazek TG, Rota PA, Lowe L, Comer JA, Rollin P, Czub M, Grolla A, Feldmann H, Luby SP, Woodward JL, Breiman RF. 2007. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis 13:1031–1037. doi: 10.3201/eid1307.061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Islam MS, Sazzad HM, Satter SM, Sultana S, Hossain MJ, Hasan M, Rahman M, Campbell S, Cannon DL, Stroher U, Daszak P, Luby SP, Gurley ES. 2016. Nipah virus transmission from bats to humans associated with drinking traditional liquor made from date palm sap, Bangladesh, 2011-2014. Emerg Infect Dis 22:664–670. doi: 10.3201/eid2204.151747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo MK, Lowe L, Hummel KB, Sazzad HM, Gurley ES, Hossain MJ, Luby SP, Miller DM, Comer JA, Rollin PE, Bellini WJ, Rota PA. 2012. Characterization of Nipah virus from outbreaks in Bangladesh, 2008-2010. Emerg Infect Dis 18:248–255. doi: 10.3201/eid1802.111492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luby SP, Hossain MJ, Gurley ES, Ahmed BN, Banu S, Khan SU, Homaira N, Rota PA, Rollin PE, Comer JA, Kenah E, Ksiazek TG, Rahman M. 2009. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001-2007. Emerg Infect Dis 15:1229–1235. doi: 10.3201/eid1508.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chua KB, Koh CL, Hooi PS, Wee KF, Khong JH, Chua BH, Chan YP, Lim ME, Lam SK. 2002. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect 4:145–151. doi: 10.1016/S1286-4579(01)01522-2. [DOI] [PubMed] [Google Scholar]
- 17.Rahman SA, Hassan SS, Olival KJ, Mohamed M, Chang LY, Hassan L, Saad NM, Shohaimi SA, Mamat ZC, Naim MS, Epstein JH, Suri AS, Field HE, Daszak P. 2010. Characterization of Nipah virus from naturally infected Pteropus vampyrus bats, Malaysia. Emerg Infect Dis 16:1990–1993. doi: 10.3201/eid1612.091790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yob JM, Field H, Rashdi AM, Morrissy C, van der Heide B, Rota P, bin AA, White J, Daniels P, Jamaluddin A, Ksiazek T. 2001. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg Infect Dis 7:439–441. doi: 10.3201/eid0703.017312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahman MA, Hossain MJ, Sultana S, Homaira N, Khan SU, Rahman M, Gurley ES, Rollin PE, Lo MK, Comer JA, Lowe L, Rota PA, Ksiazek TG, Kenah E, Sharker Y, Luby SP. 2012. Date palm sap linked to Nipah virus outbreak in Bangladesh, 2008. Vector-Borne Zoonotic Dis 12:65–72. doi: 10.1089/vbz.2011.0656. [DOI] [PubMed] [Google Scholar]
- 20.Hayman DT, Emmerich P, Yu M, Wang LF, Suu-Ire R, Fooks AR, Cunningham AA, Wood JL. 2010. Long-term survival of an urban fruit bat seropositive for Ebola and Lagos bat viruses. PLoS One 5:e11978. doi: 10.1371/journal.pone.0011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Delicat A, Paweska JT, Gonzalez JP, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 22.Leroy EM, Epelboin A, Mondonge V, Pourrut X, Gonzalez JP, Muyembe-Tamfum JJ, Formenty P. 2009. Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, Democratic Republic of Congo, 2007. Vector-Borne Zoonotic Dis 9:723–728. doi: 10.1089/vbz.2008.0167. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa H, Miyamoto H, Nakayama E, Yoshida R, Nakamura I, Sawa H, Ishii A, Thomas Y, Nakagawa E, Matsuno K, Kajihara M, Maruyama J, Nao N, Muramatsu M, Kuroda M, Simulundu E, Changula K, Hang’ombe B, Namangala B, Nambota A, Katampi J, Igarashi M, Ito K, Feldmann H, Sugimoto C, Moonga L, Mweene A, Takada A. 2015. Seroepidemiological prevalence of multiple species of filoviruses in fruit bats (Eidolon helvum) migrating in Africa. J Infect Dis 212:S101–S108. doi: 10.1093/infdis/jiv063. [DOI] [PubMed] [Google Scholar]
- 24.Paweska JT, Storm AA, Grobbelaar W, Markotter A, Kemp JV. 2016. Experimental inoculation of egyptian fruit bats (Rousettus aegyptiacus) with Ebola virus. Viruses 8:29. doi: 10.3390/v8020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Middleton DJ, Morrissy CJ, van der Heide BM, Russell GM, Braun MA, Westbury HA, Halpin K, Daniels PW. 2007. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus). J Comp Pathol 136:266–272. doi: 10.1016/j.jcpa.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Zhou P, Tachedjian M, Wynne JW, Boyd V, Cui J, Smith I, Cowled C, Ng JH, Mok L, Michalski WP, Mendenhall IH, Tachedjian G, Wang LF, Baker ML. 2016. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. Proc Natl Acad Sci U S A 113:2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchs J, Hölzer M, Schilling M, Patzina C, Schoen A, Hoenen T, Zimmer G, Marz M, Weber F, Müller MA, Kochs G. 2017. Evolution and antiviral specificities of interferon-induced Mx proteins of bats against Ebola, influenza, and other RNA viruses. J Virol 91:e00361-17. doi: 10.1128/JVI.00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. 2007. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 31.Neil SJ. 2013. The antiviral activities of tetherin. Curr Top Microbiol Immunol 371:67–104. doi: 10.1007/978-3-642-37765-5_3. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauter D. 2014. Counteraction of the multifunctional restriction factor tetherin. Front Microbiol 5:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. 2009. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci U S A 106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kühl A, Banning C, Marzi A, Votteler J, Steffen I, Bertram S, Glowacka I, Konrad A, Sturzl M, Guo JT, Schubert U, Feldmann H, Behrens G, Schindler M, Pöhlmann S. 2011. The Ebola virus glycoprotein and HIV-1 Vpu employ different strategies to counteract the antiviral factor tetherin. J Infect Dis 204:S850–S860. doi: 10.1093/infdis/jir378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radoshitzky SR, Dong L, Chi X, Clester JC, Retterer C, Spurgers K, Kuhn JH, Sandwick S, Ruthel G, Kota K, Boltz D, Warren T, Kranzusch PJ, Whelan SP, Bavari S. 2010. Infectious Lassa virus, but not filoviruses, is restricted by BST-2/tetherin. J Virol 84:10569–10580. doi: 10.1128/JVI.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong WS, Irie T, Yoshida A, Kawabata R, Kadoi T, Sakaguchi T. 2012. Inhibition of virus-like particle release of Sendai virus and Nipah virus, but not that of mumps virus, by tetherin/CD317/BST-2. Hiroshima J Med Sci 61:59–67. [PubMed] [Google Scholar]
- 38.Kobayashi T, Ode H, Yoshida T, Sato K, Gee P, Yamamoto SP, Ebina H, Strebel K, Sato H, Koyanagi Y. 2011. Identification of amino acids in the human tetherin transmembrane domain responsible for HIV-1 Vpu interaction and susceptibility. J Virol 85:932–945. doi: 10.1128/JVI.01668-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog 5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez LA, Yang SJ, Hauser H, Exline CM, Haworth KG, Oldenburg J, Cannon PM. 2010. Ebola virus glycoprotein counteracts BST-2/Tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J Virol 84:7243–7255. doi: 10.1128/JVI.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brinkmann C, Hoffmann M, Lubke A, Nehlmeier I, Kramer-Kuhl A, Winkler M, Pöhlmann S. 2017. The glycoprotein of vesicular stomatitis virus promotes release of virus-like particles from tetherin-positive cells. PLoS One 12:e0189073. doi: 10.1371/journal.pone.0189073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watt A, Moukambi F, Banadyga L, Groseth A, Callison J, Herwig A, Ebihara H, Feldmann H, Hoenen T. 2014. A novel life cycle modeling system for Ebola virus shows a genome length-dependent role of VP24 in virus infectivity. J Virol 88:10511–10524. doi: 10.1128/JVI.01272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann M, Müller MA, Drexler JF, Glende J, Erdt M, Gützkow T, Losemann C, Binger T, Deng H, Schwegmann-Weßels C, Esser K-H, Drosten C, Herrler G. 2013. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS One 8:e72942. doi: 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann M, Gonzalez HM, Berger E, Marzi A, Pöhlmann S. 2016. The glycoproteins of all filovirus species use the same host factors for entry into bat and human cells but entry efficiency is species dependent. PLoS One 11:e0149651. doi: 10.1371/journal.pone.0149651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez-Hernandez M, Hoffmann M, Brinkmann C, Nehls J, Winkler M, Schindler M, Pöhlmann S. 2018. A GXXXA motif in the transmembrane domain of the Ebola virus glycoprotein is required for tetherin antagonism. J Virol 92:e00403-18. doi: 10.1128/JVI.00403-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuzmin IV, Schwarz TM, Ilinykh PA, Jordan I, Ksiazek TG, Sachidanandam R, Basler CF, Bukreyev A. 2017. Innate immune responses of bat and human cells to filoviruses: commonalities and distinctions. J Virol 91:e02471-16. doi: 10.1128/JVI.02471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dietzel EL, Kolesnikova B, Sawatsky A, Heiner M, Weis GP, Kobinger S, Becker M, von V, Maisner A. 2016. Nipah virus matrix protein influences fusogenicity and is essential for particle infectivity and stability. J Virol 90:2514–2522. doi: 10.1128/JVI.02920-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brinkmann C, Nehlmeier I, Walendy-Gnirß K, Nehls J, González Hernández M, Hoffmann M, Qiu X, Takada A, Schindler M, Pöhlmann S. 2016. The tetherin antagonism of the Ebola virus glycoprotein requires an intact receptor-binding domain and can be blocked by GP1-specific antibodies. J Virol 90:11075–11086. doi: 10.1128/JVI.01563-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gnirß K, Fiedler M, Krämer-Kühl A, Bolduan S, Mittler E, Becker S, Schindler M, Pöhlmann S. 2014. Analysis of determinants in filovirus glycoproteins required for tetherin antagonism. Viruses 6:1654–1671. doi: 10.3390/v6041654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamp B, Dietzel E, Kolesnikova L, Sauerhering L, Erbar S, Weingartl H, Maisner A. 2013. Nipah virus entry and egress from polarized epithelial cells. J Virol 87:3143–3154. doi: 10.1128/JVI.02696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goffinet C, Allespach I, Homann S, Tervo HM, Habermann A, Rupp D, Oberbremer L, Kern C, Tibroni N, Welsch S, Krijnse-Locker J, Banting G, Krausslich HG, Fackler OT, Keppler OT. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5:285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis, v6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanika A, Larisch B, Steinmann E, Schwegmann-Wessels C, Herrler G, Zimmer G. 2005. Use of influenza C virus glycoprotein HEF for generation of vesicular stomatitis virus pseudotypes. J Gen Virol 86:1455–1465. doi: 10.1099/vir.0.80788-0. [DOI] [PubMed] [Google Scholar]
- 56.Krähling V, Becker D, Rohde C, Eickmann M, Eroğlu Y, Herwig A, Kerber R, Kowalski K, Vergara-Alert J, Becker S. 2016. Development of an antibody capture ELISA using inactivated Ebola Zaire Makona virus. Med Microbiol Immunol 205:173–183. doi: 10.1007/s00430-015-0438-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences of Epo and Hyp tetherin are available in GenBank under accession numbers MG792836.1 and MG792837.1, respectively.