After exposure to influenza virus, the host generates neutralizing anti-hemagglutinin (anti-HA) antibodies against that specific infecting influenza strain. These antibodies can also neutralize some, but not all, cocirculating strains. The goal of next-generation influenza vaccines, such as HA head-based COBRA, is to stimulate broadly protective neutralizing antibodies against all strains circulating within a subtype, in particular those that persist over multiple influenza seasons, without requiring an update to the vaccine. To mimic the human condition, COBRA HA virus-like particle vaccines were tested in ferrets that were previously exposed to historical H3N2 influenza viruses. In this model, these vaccines elicited broadly protective antibodies that neutralized cocirculating H3N2 influenza viruses isolated over a 20-year period. This is the first study to show the effectiveness of H3N3 COBRA HA vaccines in a host with preexisting immunity to influenza.
KEYWORDS: COBRA, H3N2, broadly protective vaccine, ferrets, hemagglutination inhibition, influenza, neutralization
ABSTRACT
The vast majority of people already have preexisting immune responses to influenza viruses from one or more subtypes. However, almost all preclinical studies evaluate new influenza vaccine candidates in immunologically naive animals. Recently, our group demonstrated that priming naive ferrets with broadly reactive H1 COBRA HA-based vaccines boosted preexisting antibodies induced by wild-type H1N1 virus infections. These H1 COBRA hemagglutinin (HA) antigens induced antibodies with HAI activity against multiple antigenically different H1N1 viral variants. In this study, ferrets, preimmune to historical H3N2 viruses, were vaccinated with virus-like particle (VLP) vaccines expressing either an HA from a wild-type H3 influenza virus or a COBRA H3 HA antigen (T6, T7, T10, or T11). The elicited antisera had the ability to neutralize virus infection against either a panel of viruses representing vaccine strains selected by the World Health Organization or a set of viral variants that cocirculated during the same time period. Preimmune animals vaccinated with H3 COBRA T10 HA antigen elicited sera with higher hemagglutination inhibition (HAI) antibody titers than antisera elicited by VLP vaccines with wild-type HA VLPs in preimmune ferrets. However, while the T11 COBRA vaccine did not elicit HAI activity, the elicited antibodies did neutralize antigenically distinct H3N2 influenza viruses. Overall, H3 COBRA-based HA vaccines were able to neutralize both historical H3 and contemporary, as well as future, H3N2 viruses with higher titers than vaccines with wild-type H3 HA antigens. This is the first report demonstrating the effectiveness of a broadly reactive H3N3 vaccine in a preimmune ferret model.
IMPORTANCE After exposure to influenza virus, the host generates neutralizing anti-hemagglutinin (anti-HA) antibodies against that specific infecting influenza strain. These antibodies can also neutralize some, but not all, cocirculating strains. The goal of next-generation influenza vaccines, such as HA head-based COBRA, is to stimulate broadly protective neutralizing antibodies against all strains circulating within a subtype, in particular those that persist over multiple influenza seasons, without requiring an update to the vaccine. To mimic the human condition, COBRA HA virus-like particle vaccines were tested in ferrets that were previously exposed to historical H3N2 influenza viruses. In this model, these vaccines elicited broadly protective antibodies that neutralized cocirculating H3N2 influenza viruses isolated over a 20-year period. This is the first study to show the effectiveness of H3N3 COBRA HA vaccines in a host with preexisting immunity to influenza.
INTRODUCTION
Type A and B influenza viruses infect people and cause morbidity and mortality each viral season (1). For influenza type A, the subtypes H3N2 and H1N1 are currently circulating in the human population. In general, influenza seasons in which H3N2 viruses are predominant tend to be more severe, with a greater number of hospitalizations and deaths. This is reflected in the number of elderly people that are infected with H3N2 viruses and lead to enhanced hospitalizations, whereas in influenza seasons with more H1N1 influenza virus cases, more children and younger adults are typically affected with fewer hospitalizations and fewer deaths. In 2017, countries in the Southern Hemisphere, including Australia and South Africa, had a relatively severe influenza season due to strains of the H3N2 influenza subtype. The severity of influenza season in one hemisphere is often a prelude for the severity for the next influenza season for the alternating hemisphere and during the 2017 to 2018 influenza season it was more severe than most prior Northern Hemisphere seasons (2).
There has been much speculation regarding the reasons for this enhanced influenza pathology associated with this season’s H3N2 strains, including sequence changes in the hemagglutinin (HA) due to egg adaptation (3). However, phylogenetic analysis of the HA genes from 256 A(H3N2) viruses from 2017 revealed extensive genetic diversity with multiple cocirculating clades/subclades. The HA genes of circulating viruses belonged predominately to clade 3C.2a and subclade 3C.2a1. Approximately 93 to 96% of these H3N2 influenza viruses were antigenically similar to clade (3C.2a1) viruses, the vaccine component in the quadrivalent inactivated split vaccine used in the Northern Hemisphere in 2017 to 2018 (2, 4). Influenza vaccine effectiveness is undermined by antigenic variation in the circulating viral strains, particularly in the HA and neuraminidase (NA) proteins. However, each season, the HA and NA components of the annual human influenza vaccine may be updated to ensure that they antigenically match circulating influenza strains (5, 6). Annually, in the United States, the Advisory Committee on Immunization Practices (ACIP) recommends the inclusion of two influenza A (H1N1 and H3N2) and two influenza B (one each from the Yamagata and Victoria lineages). Developing an influenza vaccine that provides broad and long-lasting protective antibody responses remains the central challenge for influenza vaccine research. Previously, we reported the development of broadly reactive influenza hemagglutinin (HA) vaccines for H5, H1, and H3 subtypes using an algorithm called COBRA (computationally optimized broadly reactive antigen) (7–13).
These vaccines elicit antibodies that block HA-specific receptor binding and inhibit infection and virus-induced pathogenesis in immunologically naive mice and ferrets (14–16). The vaccine-elicited antibodies have hemagglutination inhibition (HAI) activity against of a range of seasonal and pandemic strains. For the H3N2 subtype, the most effective COBRA HA vaccine regimens elicited antibodies with broader HAI activity against a panel of H3N2 viruses compared to wild-type H3 HA vaccines, including against all cocirculating variants from 2004 to 2007 (13).
Despite the effectiveness of these vaccine candidates, influenza immunologically naive hosts do not represent the immune state of most humans. Almost all adults and older children have preexisting immune responses to influenza viral antigens due to previous imprinting (17). Recently, our group demonstrated that priming naive ferrets with historical influenza H1 strains, and then vaccinating with COBRA HA-based vaccines boosted preexisting antibodies induced by wild-type H1N1 viruses. COBRA HA antigens elicited sera with the broadest HAI reactivity against multiple antigenic H1N1 viral variants compared to wild-type HA-based vaccines (7). In this study, we primed ferrets with historical H3N2 viruses from different eras, and then vaccinated these preimmune ferrets with COBRA HA or wild-type HA virus-like particle (VLP) vaccines. The effectiveness of each vaccine to induce broadly reactive antibodies with HAI activity against not only historical vaccine strains but also contemporary and future cocirculating H3N2 isolates from 2010 to 2016 was determined in a preimmune ferret model.
RESULTS
COBRA VLP vaccination of ferrets that were preimmune to multiple influenza viruses.
Previous studies from our laboratory used the COBRA methodology to design H3 HA sequences to account for the evolution of H3N2 influenza viruses isolated in humans over the past 45 years (13). Multilayered consensus building approach was applied to 6,340 human H3N2 HA amino acid sequences to generate unique COBRA HA sequences that resulted in four lead COBRA vaccine candidates (T6, T7, T10, and T11) that elicited antibodies with a breadth of HAI activity against both historical and drifted cocirculating strains (13). COBRA T1-T5, T8, and T9 VLP vaccines were not included in this study since these vaccine candidates displayed limited HAI breadth in naive vaccinated mice (13). The lead vaccines candidates represented various input sequences from viruses isolated between 1998 to 2013 (T6, T7, and T10) and isolates collected between only 2011 to 2013 (T11). These H3 HA COBRA vaccine candidates were tested in a host with preexisting anti-influenza immune responses (Table 1) to determine the ability of elicited antibodies to neutralize a panel of H3N2 influenza viruses representing historical, contemporary, and future cocirculating viral variants.
TABLE 1.
Vaccination and infection schema of naive and preimmune ferretsa
| Virus prime D0 | Vaccinate D84 | Vaccinate D168 |
|---|---|---|
| Virus infection group | ||
| A/Panama/2007/1999 | Mock | Mock |
| A/Wisconsin/67/2005 | Mock | Mock |
| A/Texas/50/2012 | Mock | Mock |
| VLP vaccination group | ||
| Mock 1 | T-6 | T-6 |
| Mock 2 | T-7 | T-7 |
| Mock 3 | T-10 | T-10 |
| Mock 4 | T-11 | T-11 |
| Mock 5 | Wisc/05 | Wisc/05 |
| Mock 6 | TX/12 | TX/12 |
| Preimmune group | ||
| A/Panama/2007/1999 | T-6 | T-6 |
| A/Panama/2007/1999 | T-7 | T-7 |
| A/Panama/2007/1999 | T-10 | T-10 |
| A/Panama/2007/1999 | T-11 | T-11 |
| A/Panama/2007/1999 | Wisc/05 | Wisc/05 |
| A/Panama/2007/1999 | TX/12 | TX/12 |
Three groups (n = 4) of immunologically naive ferrets were infected intranasally (day 0) with 106 PFU of Pan/99, Wisc/05, or TX/12 H3N2 influenza viruses (virus infection group). Six groups of immunologically naive ferrets (n = 4) were mock infected (day 0) with PBS and vaccinated intramuscularly twice (days 84 and 168) with one of the four COBRA H3N3 VLP vaccines (T6, T7, T10, or T11) or H3N3 VLP vaccines expressing wild-type HA proteins from Wisc/05 or TX/12 (VLP vaccination group). Six groups of immunologically naïve ferrets (n = 4) were infected intranasally with 1e7PFU of Pan/99 (day 0) and vaccinated intramuscularly twice (days 84 and 168) with one of the four COBRA H3N3 VLP vaccines (T6, T7, T10, or T11) or H3N3 VLP vaccines expressing wild-type HA proteins from Wisc/05 or TX/12 (preimmune group). Blood was collected from all animals at days 0, 14, 84, 96, 168, and 182 postinfection.
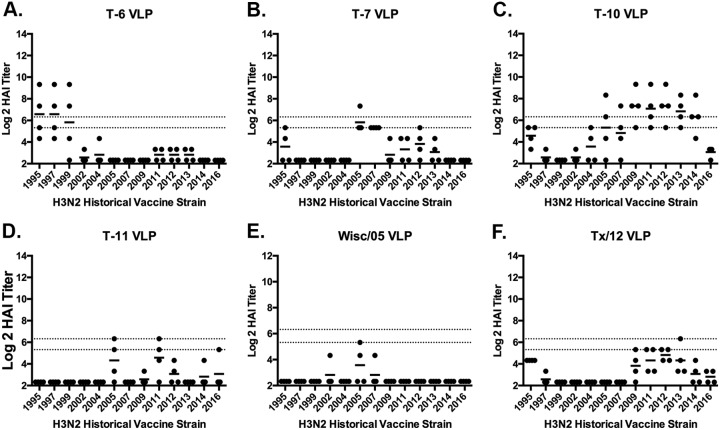
Based upon previous vaccine studies in mice (13), fitch ferrets (n = 4/group) were vaccinated with one of the four leading H3 COBRA HA VLP candidates (T6, T7, T10, or T11). Antisera collected from ferrets following two vaccinations was tested for HAI activity against a panel of 13 H3N2 vaccine virus strains isolated between 1995 and 2016. Ferrets vaccinated with the T6 COBRA VLP vaccine had antisera with HAI activity against the three oldest viruses in the panel isolated in 1995, 1997, and 1999 (Fig. 1A). Ferrets vaccinated with T7 had antibodies with low levels of HAI activity against the 2005 and 2007 viruses (Fig. 1B). Whereas ferrets vaccinated with T10 had antisera with HAI activity against the seven vaccine viruses isolated between 2005 to 2014 (Fig. 1C) and T11 elicited antisera had HAI activity against only two viruses (Fig. 1D), which was similar to the breadth pattern in sera collected from Wisc/05 or TX/12 VLP-vaccinated ferrets (Fig. 1E and F).
FIG 1.
HAI serum antibody titers induced by vaccination of ferrets with wild-type H3N3 VLP vaccines. HAI titers were determined for each group of immunologically naive ferrets (n = 4) vaccinated two times (days 84 and 168) with one of the four COBRA H3N3 VLP vaccines (T6, T7, T10, or T11) or H3N3 VLP vaccines expressing wild-type HA proteins from Wisc/05 or TX/12 against a panel of 13 H3N2 influenza viruses. Values are the log2 HAI titers of each individual animal from antisera collected on day 182. The two dotted lines indicate the 1:40 to 1:80 HAI titer range. (A) T6 VLP; (B) T7 VLP; (C) T10 VLP; (D) T11 VLP; (E) Wisc/05 VLP; (F) TX/12 VLP.
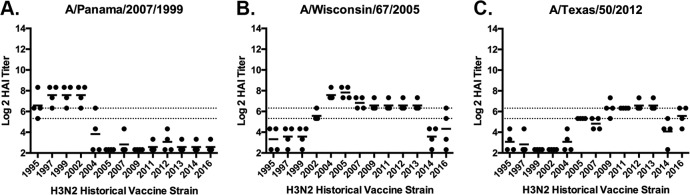
Another set of ferrets (n = 4/group) were infected with one of three different human H3N2 virus isolated in 1999 (Pan/99), 2005 (Wisc/05), or 2012 (TX/12) (Table 1). Sera collected from ferrets infected with Wisc/05 at day 84 postinfection had antibodies with HAI activity against the all the H3N2 viruses, representing vaccine strains from 2004 to 2013 (Fig. 2). The HAI titers were lower against H3N2 viruses isolated earlier than 2002, as well as the HK/14 virus (Fig. 2B). Similar, albeit lower, HAI titers were observed in sera collected from ferrets infected with TX/12 virus (Fig. 2C). However, ferrets infected with Pan/99 virus had antisera with high HAI titers against the four vaccine strains isolated between 1995 and 2002 (Fig. 2A). The HAI titers against the eight vaccine strains isolated between 2004 and 2014 were low to undetectable from antisera collected from ferrets infected with Pan/99.
FIG 2.
HAI serum antibody titers induced by H3N2 viral infections. HAI serum antibody titers induced by H3N2 influenza virus infection of immunologically naive ferrets were determined. Ferrets were infected with Pan/99 (A), Wisc/05 (B), or TX/12 (C) H3N2 influenza viruses. At day 84, serum samples were collected, and HAI titers were determined for each group of ferrets against a panel of 13 H3N2 influenza viruses. Values are the log2 HAI titers of each individual animal. Dotted lines indicate the 1:40 to 1:80 HAI titer range.
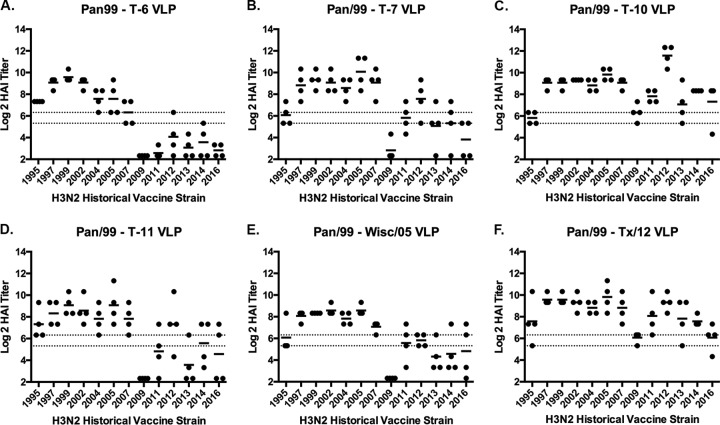
A second set of ferrets (n = 24) was infected with Pan/99 virus and, at day 84 postinfection, these Pan/99 preimmune ferrets were divided into six groups (n = 4) and vaccinated with one of six VLP vaccines expressing either COBRA T6, T7, T10, or T11 or the wild-type Wisc/05 or TX/12 HA proteins. Each ferret was boosted with the same vaccine at day 168, and sera were collected 2 weeks postboost at day 182 and tested for HAI activity (Fig. 3). All ferrets previously infected with the Pan/99 virus and vaccinated with any of the six VLP vaccines had antibodies with HAI activity against the seven H3N2 viruses isolated from 1995 to 2007 (Fig. 3). However, Pan/99 preimmune ferrets vaccinated with T6 COBRA VLP vaccine did not expand the HAI activity of the elicited antisera to the six strains isolated from 2009 to 2016 (Fig. 3A), whereas Pan/99 preimmune ferrets vaccinated with T10 COBRA VLP or TX/12 VLP vaccines had antisera with high HAI activity against most of the 13 H3N2 strains in the panel (Fig. 3C and F). Pan/99 preimmune ferrets vaccinated with the T7, T11, or Wisc/05 VLP vaccines had antisera with similar HAI reactivity profiles. Some, but not all, of the ferrets in these groups had HAI activity against the H3N2 vaccine virus strains isolated in 2011, 2012, 2013, and 2014 (Fig. 3B, D, and E).
FIG 3.
HAI serum antibody titers induced by COBRA HA and wild-type HA H3N3 VLP vaccination in Pan/99 preimmune ferrets. Immunologically naive ferrets were infected with the Pan/99 influenza virus. At days 84 and 168 postinfection these preimmune ferrets were vaccinated with H3N3 VLP vaccines expressing different HA proteins. (A) T6 VLP; (B) T7 VLP; (C) T10 VLP; (D) T11 VLP; (E) Wisc/05 VLP; (F) TX/12 VLP. HAI titers were determined for each group of ferrets at day 182 against a panel of 13 H3N2 influenza viruses isolated between 1995 and 2016. Values are the log2 HAI titers of each individual animal from antisera collected on day 182. Dotted lines indicate the 1:40 to 1:80 HAI titer range.
Neutralization titers.
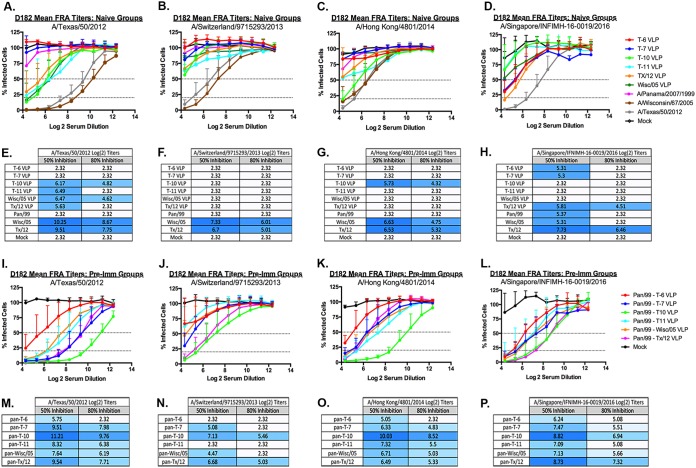
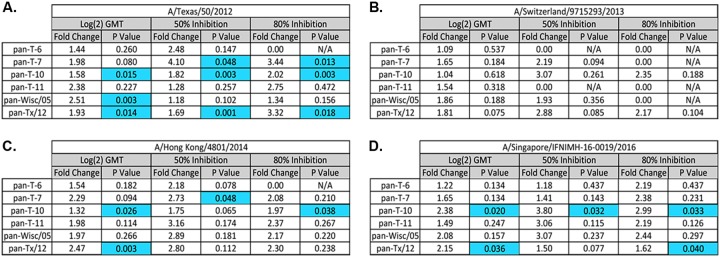
To evaluate the ability of the elicited antibodies to block live virus infection, serum was tested in a focal reduction assay (FRA). Naive ferrets vaccinated with any of the wild-type or COBRA VLP vaccines had low to undetectable neutralizing antibody titers against the TX/12, Switz/13, or HK/14 (Fig. 4A to C). Sera collected from ferrets vaccinated with T10, T11, Wisc/05 or TX/12 VLPs had a log2 serum dilution of 5.6 to 6.5 (50% inhibition) against TX/12, whereas sera from ferrets vaccinated with T6 or T7 VLP vaccines had no detectable titer, which was similar to ferrets infected with Pan/99 virus (Fig. 4A). Pan/99 preimmune ferrets vaccinated with T7, T10, or TX/12 VLPs had a significantly higher fold change between pre- and postneutralization titers against TX/12 compared to nonpreimmune ferrets vaccinated with these same vaccines (Fig. 5A). Sera collected from ferrets vaccinated with any of the five VLP vaccines had minimal FRA activity against Switz/13 (Fig. 4B), but the T10 VLP-vaccinated ferrets had detectable neutralization titers against the HK/14 virus similar to those observed in wild-type-infected ferrets (Fig. 4C). Ferrets infected with Wisc/05 or TX/12 viruses had higher log2 titers against TX/12 (10.25 to 9.51, respectively), as well as Switz/13 (7.33. to 6.7, respectively) and HK/14 virus (6.63 to 6.53, respectively) (Fig. 4E to G). In general, Pan/99 preimmune ferrets vaccinated with T7 or T10 VLPs had a significantly higher fold change between pre- and postneutralization titers against HK/14 compared to nonpreimmune ferrets vaccinated with these same vaccines, and T10 and TX/12 VLPs had a significantly higher fold change against Sing/16 in the preimmune setting (Fig. 5C and D). There was no significant fold change against Switz/13 between preimmune and naive vaccinated animals (Fig. 5B).
FIG 4.
FRA. FRA neutralizing titers were determined for each group of immunologically naive ferrets (n = 4) vaccinated two times (days 84 and 168) with one of the four COBRA H3N3 VLP vaccines (T6, T7, T10, or T11) or H3N3 VLP vaccines expressing wild-type HA proteins from Wisc/05 or TX/12. At day 182, sera were collected and tested in an FRA against four H3N2 viruses isolated between 2012 and 2016 representing clades 3c.1 (TX/12) (A and I), 3c.3a (Switz/13) (B and J), 3c.2a (HK/14) (C and K), and 3c.2a1 (Sing/16) (D and L). Alternatively, ferrets were infected with wild-type H3N2 influenza viruses (Pan/99, Wisc/05, or TX/12), and sera collected at day 182 postinfection were also tested in an FRA assay against the four H3N2 viruses (A to D). Ferrets infected with Pan/99 were also subsequently vaccinated with H3N3 VLPs expressing one of four COBRA H3 HA antigens or the Wisc/05 or TX/12 wild-type HA proteins. Collected sera was assayed against the four H3N2 viruses (I to L). For each virus, the virus concentration was standardized to 1.2 × 104 FFU/ml. The dotted lines represent the 50% inhibition and the 80% inhibition of viral infection by antisera compared to virus-only control wells (A to D and I to L). A heat map of the log2 serum dilution titer for the 50 and 80% inhibition per virus for each group of ferrets (E to H and M to P) was also generated. Colors range from white (lowest level of inhibition) to dark blue (highest level of inhibition).
FIG 5.
Comparison of log2-fold change in HAI GMT, and 50 and 80% FRA neutralization titers between VLP-vaccinated and preimmune vaccinated ferrets. Fold changes in HAI and FRA at day 182 were compared between immunologically naive ferrets (n = 4) vaccinated two times (days 84 and 168) with one of the four COBRA H3N3 VLP vaccines (T6, T7, T10, or T11) or H3N3 VLP vaccines expressing wild-type HA proteins from Wisc/05 or TX/12 and preimmune ferrets that were vaccinated two times (days 84 and 168) with H3N3 VLP vaccines expressing the same HA proteins used to immunize the naive ferrets. Fold change differences were compared against four H3N2 viruses isolated between 2012 and 2015 representing clades 3c.1 (TX/12) (A), 3c.3a (Sz/13) (B), 3c.2a (HK/14) (C), and 3c.2a1 (Sing/16) (D). Statistical analyses were performed using unpaired parametric t tests to determine significance of vaccination in the preimmune setting compared to naive ferrets. P values of <0.05 are considered significant and are colored blue.
Ferrets preimmune to Pan/99 and then vaccinated with one of the wild-type or COBRA VLP vaccines had considerably higher neutralization titers compared to naive ferrets (Fig. 4). Pan/99 preimmune ferrets that were vaccinated with T10 VLPs had antisera with the highest 50 and 80% neutralizing titers against TX/12, Switz/13, and HK/14. This group also had the highest 50% neutralization titer against Sing/16 comparable to that of the Pan-TX/12-vaccinated group (Fig. 4P). Sera from Pan/99 preimmune/T10 VLP-vaccinated ferrets had had log2 serum dilution neutralization titer of 11.2 (50% inhibition) against TX/12 and log2 serum dilution 10.0 against HK/14 (Fig. 4M and O). These titers were statistically higher (P < 0.05) than sera collected from Pan/99 preimmune TX/12 VLP-vaccinated ferrets. Sera collected from these ferrets had titers of 9.54 against TX/12, 6.68 against Switz/13, 6.49 against HK/14 virus, and 8.73 against the Sing/16 virus (Fig. 4M to P). Interestingly, even though T7 VLP-vaccinated ferrets had no HAI (Fig. 1B) or 50% FRA inhibition activity (Fig. 4A to C) against TX/12, Switz/13, or HK/14 viruses, T7 VLP-vaccinated Pan/99 preimmune ferrets had FRA activity similar to Pan/99 preimmune ferrets vaccinated with TX/12 VLPs against TX/12 virus (Fig. 4I) or T11, Wisc/05, or TX/12 VLP vaccines against HK/14 (Fig. 4K).
Panel of deep dive viruses and HAI activity.
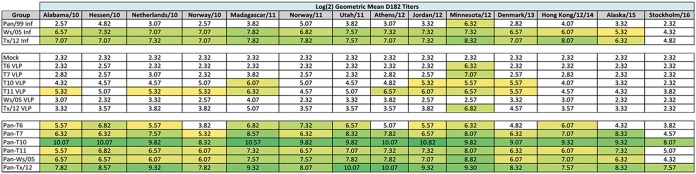
In order to demonstrate the breadth of antibody activity against cocirculating H3N2 influenza strains between 2010 and 2016, HAI activity from the collected antisera from the preimmune vaccinated ferrets was assessed. HA sequences were identified from the GISAID database and reports from Francis Crick Institute to determine the frequency of related strains per season, as previously described (13). Representative strains from each cluster of viruses (Fig. 6) were used to generate a comprehensive list of representative sequences that were assessed for genetic relatedness to reference antigens (Table 2), and these were complied with vaccine strains and COBRA vaccine HA sequences into a phylogenetic tree (Fig. 7). Fourteen drift viruses were identified, including two viruses representing future strains, AK/15 and Stock/16. Ferrets infected with wild-type H3N2 viruses Wisc/05 or TX/12 had antisera with HAI activity against 12 of the 13 viruses in the panel (Table 3) achieving a log2 titer above 5.32. Ferrets vaccinated with VLPs containing the wild-type HA from Wisc/05 or TX/12 had antisera with little or no HAI activity against the same variant virus panel. TX/12 VLP-vaccinated ferrets were able to elicit a titer of 6.82 against MN/12; however, all experimental groups except Wisc/05 VLP- and mock-vaccinated animals were able to achieve a HAI titer greater than 5.32 against this virus. Ferrets vaccinated with VLPs containing the COBRA HA sequence from T6 or T7 also had antisera with little HAI activity against the panel of viruses. However, ferrets vaccinated with T10 VLPs had HAI activity against 4 of the viruses, and ferrets vaccinated with T11 VLPs had antibodies against 8 of 14 viruses in the panel, albeit at low titers. Ferrets that were preimmune to the Pan/99 virus and vaccinated with VLPs expressing either Wisc/05, TX/12, or one of the COBRA HA antigens had high HAI titers against almost all the H3N2 viruses in the panel. Only Pan/99 preimmune ferrets that were vaccinated with T10 or TX/12 had antibodies against the 2016 strain. Interestingly, preimmune ferrets vaccinated with T10 had HAI titers that were consistently higher (2- to 4-fold) than the HAI titers from preimmune ferrets vaccinated with TX/12 VLPs.
FIG 6.
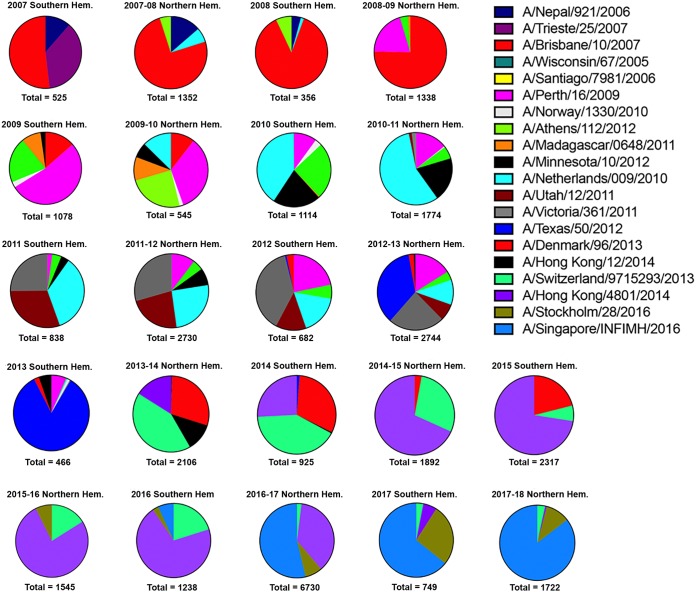
Frequencies of influenza HA clusters in consecutive seasons between 2007 and 2016. Influenza HA sequences posted in GISAID databases were aligned and clustered into families per Northern and Southern Hemisphere seasons. The clusters in each pie chart that differ by 5% in amino acids in the HA sequence are depicted by a different color and slice of the pie chart. Each influenza season for the Northern and Southern Hemispheres from 2007 to 2017 to 2018 is depicted. Representative influenza viruses from each cluster in the pie charts are listed and match the color of the pie charts.
TABLE 2.
H3 cocirculating strain panel (2010 to 2016)a
| H2N2 virus strain | Clade |
|---|---|
| A/Alabama/5/2010 | Perth/16 |
| A/Hessen/5/2010 | Perth/16 |
| A/Netherlands/009/2010 | Vic/208 |
| A/Norway/1330/2010 | Perth/16 |
| A/Madagascar/0648/2011 | Vic/208 |
| A/Norway/1186/2011 | Perth/16 |
| A/Utah/12/2011 | Perth/16 |
| A/Athens/112/2012 | 3B |
| A/Jordan/30502/2012 | 3c.1 |
| A/Minnesota/10/2012 | Clade 7 |
| A/Denmark/96/2013 | 3c.3 |
| A/Hong Kong/12/2014 | 3c.2 |
| A/Alaska/232/2015 | 3c.2a1 |
| A/Stockholm/28/2016 | 3c.2a1a |
The first column lists the 14 H3N2 viruses in the panel. The second column indicates the clade with which each virus is associated.
FIG 7.
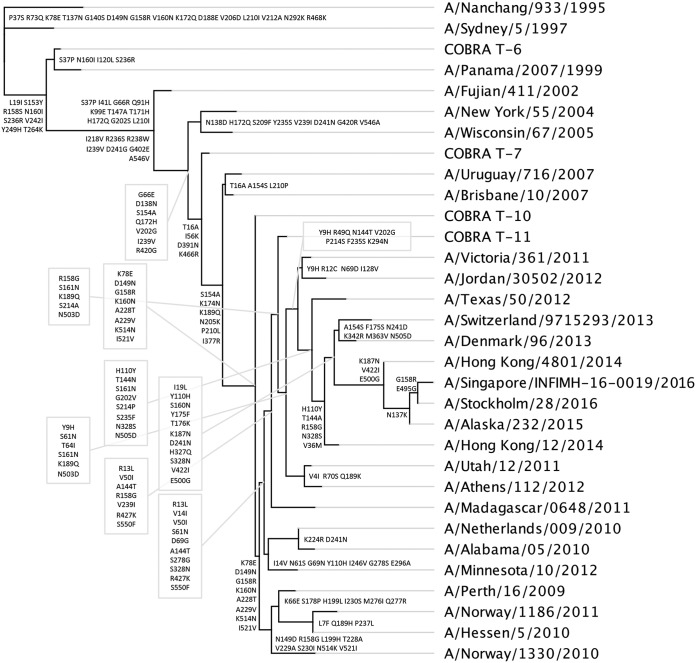
Phylogenetic tree of H3 HA sequences. The rooted (A/Nangchang/933/1995) phylogenetic tree was inferred from COBRA HA and wild-type HA amino acid sequences derived from the representative H3N2 variant viruses from 1995 to 2015 using the maximum-likelihood method. Sequences were aligned with MUSCLE 3.7 software, and the alignment was refined by Gblocks 0.91b software. Phylogeny was determined using the maximum-likelihood method with PhyML software. Trees were rendered using TreeDyn 198.3 software (45).
TABLE 3.
Heat map of day 182 log2 GMTs induced by vaccination in naive and preimmune ferrets with H3N3 VLP vaccines against a panel of cocirculating H3N2 viral strains isolated from 2010 to 2016a
HAI assays were performed against a panel of cocirculating viruses isolated from 2010 to 2016 with day 182 serum samples from all ferrets in the study. The average log2 geometric mean HAI serum antibody titers (GMTs) were determined for each group of ferrets (n = 4) and are presented in heat map form. Colored cells represent groups that achieved a GMT of ≥5.32, which correlates to an average antibody titer of ≥1:40. Values closest to 5.32 are yellow and become more green as the values increase. Cells with no color represent groups that did not achieve a GMT of ≥5.32.
DISCUSSION
Influenza viruses of the H3N2 subtype have been responsible for the majority of seasonal influenza epidemics since 1968 (18). Prior to its introduction into the human population, there was no documentation of H3N2 influenza viruses circulating in humans. Over the past 40 years, the World Health Organization (WHO) has recommended 28 H3N2 vaccine strain changes as a result of the extensive genetic and antigenic evolution of these viruses (19). In the most recent 2017/2018 Northern Hemisphere influenza season, the interim estimate of 25% vaccine efficacy (VE) against H3N2 influenza A viruses in the United States (2) was similar to interim estimates from Canada (17%) and Australia (10%) (4, 20). This VE was similar to final (32%) VE estimates. Current licensed influenza seasonal vaccines are available as inactivated, live-attenuated, and recombinant-HA vaccines. Inactivated influenza vaccine includes whole inactivated virus, split virion, or subunit formulations. These vaccines induce mainly strain-specific serum antibodies against H3N2 influenza viruses isolated during the 2016/2017 season (2). However, among children (6 months to 8 years of age) during the 2017/2018 season, the VE against H3N2 influenza viruses and associated medically attended influenza illness was reduced by more than half (59%) among vaccinated children.
The rapid evolution of influenza viruses creates difficulties to recognize and predict current and future epidemiological threats (21). Most of the current split-inactivated, recombinant, and live-attenuated influenza vaccines target the surface viral glycoproteins HA and NA (22). The goal of a broadly reactive COBRA HA-based vaccine(s) is to stimulate immune responses that react against most, if not all, circulating influenza strains, over a long period of time and in all populations of people. Humans are rarely immunologically naive to influenza viruses; however, almost all preclinical studies test new influenza vaccine formulations in animals without preexisting immune responses to influenza viruses (23). Recently, our group demonstrated that priming naive ferrets with broadly reactive H1 COBRA HA-based vaccines boosted preexisting antibodies induced by wild-type H1N1 viruses (7). These H1 COBRA HA antigens induced antibodies with the broadest HAI reactivity against multiple antigenically different H1N1 viral variants (7). In this study, ferrets, preimmune to historical H3N2 viruses were vaccinated with VLP vaccines expressing either an HA from a wild-type H3 influenza virus or a COBRA H3 HA antigen. The elicited antisera were tested for HAI activity and the ability to neutralize virus infection against a panel of viruses representing 13 vaccine strains selected by the WHO or against a set of 14 viral variants that cocirculated during the same time period.
Preimmunity was established in naive ferrets by infecting them with one of three historical H3N2 influenza viruses isolated in 1999, 2005, or 2012. These animals were housed for 3 months and tested for HAI activity against a panel of 13 WHO H3N2 vaccine viruses representing the years 1995 to 2016. Ferrets infected with the 2005 or 2012 viruses elicited a wide breadth in HAI activity against H3N2 viruses from the 2000s and 2010s (Fig. 2). This HAI activity is most likely a reflection of the high level of antigenic relatedness of the selected vaccine strains from this era. Interestingly, ferrets infected with either of these viruses did not efficiently neutralize the 2014 isolate, HK/14. In contrast, the Pan/99-infected ferrets had HAI activity against viruses from the 1990s era but elicited few or no antibodies with HAI activity against the viruses in later decades. Therefore, we chose to use the Pan/99 virus as the primary isolate to establish preimmunity for the experiments in this study and to determine whether the vaccines would stimulate HAI activity against “future” H3N2 vaccine viruses isolated in the 2000s and 2010s. This option was not possible by establishing preimmunity using the 2005 or 2012 viruses due to the high-titer cross-reactive HAI antibodies against most of the vaccine strains during this era.
After infecting ferrets with the 1999 virus, the T7, T10, and T11 COBRA HA VLP vaccines elicited antibodies with HAI activity against almost all the vaccine viruses in the panel, which was similar to the breadth of HAI activity following vaccination with Wisc/05 and TX/12 HA-based VLP vaccines (Fig. 3). In contrast, immunologically naive ferrets vaccinated with the same vaccines had polyclonal sera with a narrow recognition of H3N2 viruses. T10 COBRA HA VLPs were the most effective at eliciting HAI activity in naive ferrets with their antisera having HAI activity against 6 of the 13 viruses in the panel (Fig. 1C). No ferret group vaccinated with VLPs expressing a wild-type HA antigen had an average HAI titer above 1:40 against any of the viruses in the panel.
HAI-specific antibodies are an indication of the ability of the vaccine to elicit antibodies against the receptor binding site on the head of the HA and prevent the HA from binding to its sialic acid receptor (24). In order to determine whether COBRA HA-based vaccines elicited antibodies against other parts of the HA molecule, an FRA was performed to demonstrate the ability to prevent the infection of susceptible cells (13). Neutralizing antibodies against the neuraminidase could be ruled out as a factor, since the VLP vaccines used an N3 NA from an avian influenza virus, and the viruses used in the FRA were wild-type H3 viruses with an N2 NA protein. Vaccines that were effective in eliciting antibodies with HAI activity were also effective at eliciting antibodies that neutralized viral infection in the FRA. The T11 COBRA vaccine did not elicit antibodies with HAI activity, but it was particularly effective neutralizing antigenically distinct influenza viruses (Fig. 1D and E). The T11 COBRA HA could elicit antibodies directed against the fusion domain, HA stem, or other epitopes on wild-type H3 HA molecules. These induced antibodies may mediate viral neutralization through non-HAI mechanisms such as antibody-dependent cellular cytotoxicity or other antibody Fc effector functions (25, 26).
In naive ferrets, viral infections with wild-type HA antigens from 2005 or 2012 isolates elicited the greatest breadth of neutralizing activity compared to VLP vaccinations with the same HA. In preimmune ferrets, VLP vaccines, particularly T10 and T11 COBRA HA vaccines, elicited higher-titer, broadly neutralizing antibodies in preimmune ferrets compared to vaccination in naive ferrets (Fig. 4E to H). The T10 and T11 COBRA HA proteins have epitopes in the five antigenic sites that are similar to the four viruses in the FRA panel with a dominant p-epitope values (range, 0.148 to 0.185) indicative of predictive vaccine effectiveness against these four strains of 0.077 (27). Vaccine efficacy has a linear correlation with the antigenic distance between the vaccine strain and the circulating virus strain (28). The dominant p-epitope values correlate well with influenza vaccine efficacy in humans. For example, when the p-epitope is larger than 0.19, the vaccine no longer offers efficient protection (27, 29–31). In contrast to COBRA T10 HA, TX12 HA, which elicits a pattern of HAI activity similar to that expressed by T10 in preimmune ferrets, had p-epitope values that would not predict efficient vaccine effectiveness, and the elicited antibodies were not as efficient at neutralizing viral infection. Direct challenge of vaccinated ferrets would be another measure of the efficacy of these vaccines. However, ferrets infected with most modern wild-type H3N2 influenza viruses, such as HK/14, do not show signs of morbidity, and virus recovery from nasal washes is low to undetectable (data not shown). Therefore, the FRA is a suitable substitute assay for assessing viral infection of mammalian cells and the ability of these vaccines to elicit antibodies that block viral infection.
Viral variants within each subtype of influenza A cocirculate with the dominant cluster of strains due to antigenic drift (32–34). Each influenza season, wild-type influenza viruses and, subsequently, the HA associated with each virus are used by vaccine manufacturers to generate the seasonal influenza vaccine. However, the selected strain in the seasonal vaccine is similar to only a percentage of viral variants in any given season. Using a retrospective sequence analysis of HA sequences, the frequency of related strains per season was used to determine a set of variant clusters, and a representative influenza virus was selected from each cluster (13).
This technique allows for the selection of a set of viruses that can be used to assess the effectiveness of vaccines to elicit antibodies that will not only neutralize viruses in the dominant cluster but also the viruses representing minor cluster variants. Antibodies elicited by COBRA HA in preimmune ferrets had high HAI activity not only against viruses from the predominant cluster of viruses represented by the HA in the selected vaccine candidate but also the cocirculating viral variants. Not only did the antibodies elicited by these vaccines have HAI activity against a broad number of viral variants, but the overall magnitude of the response was high (Fig. 5 and Table 3). This is a significant observation because the breadth of the HAI activity against different strains may be dependent on the height of the antibody response. In preimmune animals, the COBRA vaccines that were most effective had 2- to 4-fold-higher titers in preimmune ferrets than in naive ferrets (Fig. 5). These viral variants represented a period of circulation between 2010 and 2016 but were distinct from the cluster of viruses that the vaccine strains were selected for each of the 14 combined Northern and Southern Hemisphere influenza seasons during this time period. The H3N3 COBRA HA constructs used in this study were highly effective when used to vaccinate naive mice (13) but elicited low HAI titers in naive ferrets (Fig. 1). In contrast, these same COBRA HA vaccines were highly effective at eliciting antibodies in preimmune ferrets against all viruses in the panel (Fig. 3). Most likely, the COBRA HA-based vaccines are more effective at recalling memory B cells that were elicited from previous H3N2 infection compared to vaccines containing wild-type HA antigens. Often, the first influenza virus to which a host is exposed educates or imprints onto the immune system and leads to the induction of memory cells. Vaccination recalls these memory B cells with broad or narrow antibody specificities. Infection leaves an immunological imprint that can be subsequently boosted by COBRA HA vaccines, resulting in the elicitation of antibody responses against variant viruses. Priming by infection, but not immunization, can confer superior immune responses that are readily recruited following subsequent vaccination or infection (35).
We demonstrated here that the H3 COBRA HA antigens are superior at eliciting higher HAI titers and more neutralizing antibodies than vaccines with wild-type HA protein in ferrets preimmune to a historical H3N2 influenza virus. COBRA VLP vaccination in the preimmune setting also elicited significantly higher and more broadly reactive antibody titers than those tested in the naive animal model. In particular, COBRA T10 VLPs were superior to the other comparators at eliciting high HAI and neutralizing antibody titers against viruses from 2012 to 2016 (Fig. 3C and Fig. 4M to P). Notably, in this preimmune setting, the TX/12, T7, and T11 VLP vaccines were also able to generate higher HAI and neutralizing antibody titers against viruses from this same time period. Testing broadly reactive or universal influenza vaccine candidates in immunologically naive ferrets may not reflect the performance of these vaccines in humans. While a single infection with a historical H3N2 influenza virus cannot recapitulate the complex influenza memory B and T cell responses from past infections and vaccinations in a human, this model does allow for the assessment of broadly reactive/universal candidates for effectiveness before using in human trials. The performance of H3 COBRA HA proteins were tested in this H3N2 preimmune model and elicited higher, long-lasting HAI titers and more neutralizing antibody titers against a panel of vaccine and cocirculating strains compared to the Wisc/05 standard-of-care vaccine. HAI assays were performed with sera collected from the mock-vaccinated animals at 14 days after their initial infection. The HAI titers were not significantly different from the titers collected on D182 against the panel of H3 viruses. Similar results were observed this same phenomenon using H1 viruses and vaccines as previously reported (7, 14). An improved and more broadly protective influenza vaccine against H3 isolates that protects close to 100% of the cocirculating variants in the human population over multiple influenza seasons will be highly useful and improve our annual vaccine efficacy.
MATERIALS AND METHODS
Vaccine preparation.
Mammalian 293T cells were transfected with each of three plasmids expressing the influenza neuraminidase (A/mallard/Alberta/24/01, H7N3), the HIV p55 Gag sequences and one of the various H3 wild-type or COBRA HA (U.S. patent application 15/762,050) expressing plasmids on previously described mammalian expression vectors (36). After 72 h of incubation at 37°C, supernatants from transiently transfected cells were collected, centrifuged to remove cellular debris, and filtered through a 0.22-μm-pore-size membrane. Mammalian VLPs were purified and sedimented by ultracentrifugation on a 20% glycerol cushion at 135,000 × g for 4 h at 4°C. VLPs were resuspended in phosphate-buffered saline (PBS), and the total protein concentration was assessed by a conventional bicinchoninic acid assay. The hemagglutination activity of each preparation of VLPs was determined by adding equal volume turkey red blood cells (RBCs) to a V-bottom 96-well plate, followed by incubation with serially diluted volumes of VLPs for 30 min at room temperature. The highest dilution of VLP with full agglutination of RBCs was considered the endpoint HA titer.
Determination of HA content.
A high-affinity, 96-well flat-bottom enzyme-linked immunosorbent assay (ELISA) plate was coated with 5 to 10 μg of total protein of VLP and serial dilutions of a recombinant H3 antigen (3006_H3_Vc; Protein Sciences, Meriden, CT) in ELISA carbonate buffer (50 mM carbonate buffer [pH 9.5]), and the plate was incubated overnight at 4°C on a rocker. The next morning, the plates were washed in PBS with 0.05% Tween 20 (PBST), and nonspecific epitopes were blocked with 1% bovine serum albumin (BSA) in PBST solution for 1 h at room temperature. Buffer was removed, and then stalk-specific group 2 antibody CR8020 (37) was added to the plate, followed by incubation for 1 h at 37°C. The plates were washed, and probed with goat anti-human IgG horseradish peroxidase-conjugated secondary antibody (2040-05; Southern Biotech, Birmingham, AL) for 1 h at 37°C. The plates were washed, and freshly prepared o-phenylenediamine dihydrochloride (P8287; Sigma, St. Louis, MO) substrate in citrate buffer (P4922; Sigma) was added to the wells, followed by 1 N H2SO4 stopping reagent. Next, the plates were read at 492 nm absorbance using a microplate reader (Powerwave XS; BioTek, Winooski, VT), and the background was subtracted from negative wells. Linear regression standard curve analysis was performed using known concentrations of recombinant standard antigen to estimate the HA content in the VLP lots.
Viruses and HA antigens.
H3N2 viruses were obtained through the Influenza Reagents Resource (IRR), BEI Resources, the Centers for Disease Control (CDC), or Sanofi-Pasteur. Viruses were passaged once, under the same growth conditions as they were received, in semiconfluent Madin-Darby canine kidney (MDCK)-SAIT cell culture according to the instructions provided the WHO (38). Titers of virus lots were determined with both guinea pig and turkey erythrocytes and divided into aliquots for single-use applications.
H3N2 viruses were obtained through the IRR, BEI Resources, the CDC, or Sanofi-Pasteur. Viruses were passaged once, under the same growth conditions as they were received, in semiconfluent MDCK-SAIT cell culture according to the instructions provided the WHO (38). Titers of virus lots were determined with both guinea pig and turkey erythrocytes and divided into aliquots for single-use applications. The H3N2 vaccine panel included the following strains: A/Nanchang/933/1995 (Nan/95; EPI_ISL_111174) passage 2 (P2), A/Sydney/05/1997 (Syd/97; EPI_ISL_111337) P1, A/Panama/2007/1999 (Pan/99; EPI_ISL_111375) P4, A/Fujian/411/2002 (Fuj/02; EPI_ISL_111384) P2, A/New York/55/2004 (NY/04; EPI_ISL_123227) P6, A/Wisconsin/67/2005 (Wis/05; EPI_ISL_115646) P4, A/Brisbane/10/2007 (Bris/07; EPI_ISL_176548) P3, A/Perth/16/2009 (Per/09; EPI_ISL_167306) P4, A/Victoria/361/2011 (Vic/11; EPI_ISL_118588) P4, A/Texas/50/2012 (TX/12; EPI_ISL_129858) P4, A/Switzerland/9715293/2013 (Switz/13; EPI_ISL_162149) P4, A/Hong Kong/4801/2014 (HK/14; EPI_ISL_153117) P11, and A/Singapore/INFIMH-16-0019/2016 (Sing/16; EPI_ISL_285898) P3.
A panel of 14 cocirculating H3N2 variants from the period of 2010 to 2016 included: A/Alabama/05/2010 (AL/10; EPI_ISL_81377) P2, A/Netherlands/009/2010 (NL/10; EPI_ISL_115774) P2, A/Hessen/5/2010 (Hes/10; EPI_ISL_85730) P3, A/Norway/1330/2010 (Nor/10; EPI_ISL_86071) P3, A/Madagascar/0648/2011 (Mad/11; EPI_ISL_90628) P2, A/Utah/12/2011 (Utah/11; EPI_ISL_100450) P2, A/Norway/1186/2011 (Nor/11; EPI_ISL_93711) P2, A/Athens/112/2012 (Ath/12; EPI_ISL_107823) P2, A/Jordan/30502/2012 (Jor/12; EPI_ISL_138476) P2, A/Minnesota/10/2012 (MN/12; EPI_ISL_121523) P2, A/Denmark/96/2013 (Den/13; EPI_ISL_153117) P2, A/Hong Kong/12/2014 (HK/12/14; EPI_ISL_154026) P2, A/Alaska/232/2015 (AK/15; EPI_ISL_239800) P2, and A/Stockholm/28/2016 (Stock/16; EPI_ISL_226095) P2. COBRA HA sequences have been previously published in Wong et al. (13). Briefly, the years that input sequences were used to design each COBRA HA sequence were as follows: T6, 1998 to 2001; T7, 2002 to 2010; T10, 2002 to 2013; and T11, 2011 to 2013 (13).
Viral infection and COBRA VLP vaccination of ferrets.
Fitch ferrets (Mustela putorius furo, spayed, female, 6 to 12 months of age), negative for antibodies to circulating influenza A (H1N1, H3N2) and influenza B viruses, were de-scented and purchased from Triple F Farms (Sayre, PA). Ferrets were pair housed in stainless steel cages (Shor-Line, Kansas City, KS) containing Sani-Chips laboratory animal bedding (P. J. Murphy Forest Products, Montville, NJ). Ferrets were provided with Teklad Global Ferret Diet (Harlan Teklad, Madison, WI) and fresh water ad libitum. The University of Georgia Institutional Animal Care and Use Committee approved all experiments, which were conducted in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals, The Animal Welfare Act, and the CDC/NIH’s Biosafety in Microbiological and Biomedical Laboratories guide. Ferrets (n = 4) were preinfected with one of three seasonal H3N2 influenza viruses (Pan/99, Wisc/05, or TX/12 at 107 PFU) intranasally. Preimmune groups were infected with seasonal isolate A/Panama/2007/1999. Animals were monitored daily during the infection for adverse events, including weight loss, loss of activity, nasal discharge, sneezing, and diarrhea, and allowed to recover for 84 days. Ferrets were then vaccinated with one of four H3N3 COBRA VLP vaccines (T6, T7, T10, and T11) (13), one of two wild-type H3N3 VLP vaccines (Wisc/05, TX/12), or PBS alone as a mock vaccination. Vaccines (15-μg doses based upon HA content) were formulated with an emulsified squalene-in-water AF03 adjuvant (Sanofi Pasteur, Lyon, France) in a final 1:1 mixture with VLPs. Ferrets were boosted 84 days after initial vaccination (day 168). Blood was harvested from all anesthetized ferrets via the anterior vena cava at days 0, 14, 84, 96, 168, and 182. Serum was transferred to a centrifuge tube and centrifuged at 6,000 rpm. Clarified serum was removed and frozen at −20 ± 5°C.
HAI assay.
A hemagglutination inhibition (HAI) assay was used to assess functional antibodies to the HA able to inhibit agglutination of guinea pig erythrocytes. The protocols were adapted from the WHO laboratory influenza surveillance manual (38) and use the host species that is frequently used to characterize contemporary H3N2 strains that have preferential binding to α-(2,6)-linked sialic acid receptors (39, 40). To inactivate nonspecific inhibitors, day 84 and day 182 serum samples were treated with receptor-destroying enzyme (RDE; Denka Seiken, Co., Japan) prior to testing. Briefly, three parts of RDE were added to one part of serum, followed by incubation overnight at 37°C. RDE was inactivated by incubation at 56°C for 30 min. RDE-treated sera were diluted in a series of 2-fold serial dilutions in V-bottom microtiter plates. An equal volume of each H3N2 virus, adjusted to approximately 8 hemagglutination units (HAU)/50 μl in the presence of 20 nM oseltamivir, was added to each well. The plates were covered and incubated at room temperature for 30 min, and then 0.75% guinea pig erythrocytes (Lampire Biologicals, Pipersville, PA) in PBS were added. The RBCs were washed with PBS, stored at 4°C, and used within 24 h of preparation. The plates were mixed by agitation and covered, and the RBCs were allowed to settle for 1 h at room temperature. The HAI titer was determined by the reciprocal dilution of the last well that contained nonagglutinated RBCs. Positive and negative serum controls were included for each plate. All ferrets were negative (HAI ≤ 1:10) for preexisting antibodies to currently circulating human influenza viruses prior to vaccination, and seroprotection was defined as an HAI titer of >1:40 and seroconversion as a 4-fold increase in titer compared to baseline, in according with WHO and European Committee for Medicinal Products guidelines for evaluating influenza vaccines (41); however, we often examined a more stringent threshold of >1:80. Since the ferrets were naive and seronegative at the time of vaccination, the seroconversion and seroprotection rates are interchangeable in this study.
FRA.
The focus reduction assay (FRA) used in this study was initially developed by the WHO Collaborating Centre in London (42, 43) and modified by the CDC (Thomas Rowe, unpublished data). MDCK-SIAT1 cells were cells plated at 2.5 × 105 to 3 × 105 cells/ml (100 μl/well in a 96-well plate) on the day before the assay was run. Cells need to be 95 to 100% confluence at the time of the assay in 96-well plates overnight to form a confluent monolayer in Dulbecco modified Eagle medium (DMEM) containing 5% heat-inactivated fetal bovine serum and antibiotics. The following day, the cell monolayers were rinsed with 0.01 M PBS (pH 7.2; Gibco), followed by the addition of 2-fold serially diluted RDE-treated serum at 50 μl per well, starting with a 1:20 dilution in virus growth medium containing 1 μg/ml TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin and VGM-T (DMEM containing 0.1% BSA, penicillin-streptomycin, and 1 µg/ml TPCK-treated trypsin [Sigma]). Afterward, 50 μl of virus standardized to 1.2 × 104 focus-forming units (FFU)/ml (corresponding to 600 FFU/50 μl) in VGM-T was added to each plate or VGM-T was added to cell control wells. The virus stocks were standardized by previous titration in the FRA. After 2 h of incubation at 37°C with 5% CO2, the cells in each well were then overlaid with 100-μl equal volumes of 1.2% Avicel RC/CL (type RC581 NF; FMC Health and Nutrition, Philadelphia, PA) (42) in 2× modified Eagle medium containing 1 µg/ml TPCK-treated trypsin, 0.1% BSA, and antibiotics. Plates were incubated for 18 to 22 h at 37°C and 5% CO2. The overlays were then removed from each well, and the monolayer was washed once with PBS to remove any residual Avicel. The plates were fixed with ice-cold 4% formalin in PBS for 30 min at 4°C, followed by a PBS wash and permeabilization using 0.5% Triton X-100 in PBS/glycine at room temperature for 25 min. The plates were washed three times with wash buffer (PBS, 0.1% Tween 20 [PBST]) and incubated for 1 h with a monoclonal antibody against influenza A nucleoprotein (IRR, FR-1217) (44) in ELISA buffer (PBS, 10% horse serum, 0.1% Tween 80). After three washes with PBST, the cells were incubated with goat anti-mouse peroxidase-labeled IgG (474-1802; SeraCare, Inc., Milford, MA) in ELISA buffer for 1 h at room temperature. The plates were then washed three times with PBST, and infectious foci (spots) were visualized using TrueBlue substrate (SeraCare) containing 0.03% H2O2 and incubated at room temperature for 10 min. The reaction was stopped by five washes with distilled water. Plates were dried, and foci were enumerated using a BioSpot analyzer with ImmunoCapture 6.4.87 software (CTL, Shaker Heights, OH). The FRA titer was reported as the reciprocal of the highest dilution of serum corresponding to 50% focus reduction compared to the virus control minus the cell control.
For a plate to pass quality control, both the average of the octuplet virus control wells (VC) and the average of the octuplet cell control wells (CC) must pass. The virus controls initially were between 150 to 650 foci (spots), and the cell controls must be less than 21 foci. The virus control wells were subsequently expanded to between 200 and 1,600 spots. In addition, the reference vaccine strain virus was assessed in triplicate plates in each individual assay; at least two of three plates must pass VC and CC criteria, and homologous ferret antisera must have the same titer. Each assay plate (one virus per plate) contained a panel of reference antisera, as well as human vaccine serum control, to assess the overall assay consistency.
Phylogenetic comparison of cocirculating variants within an influenza season (1999 to 2017).
Multiple sequence alignments were performed on extracted HA sequences that retained the majority of the HA1 domain; sequences were organized based on collection during the Northern Hemisphere or the Southern Hemisphere influenza seasons. Neighbor-joining Jukes-Cantor phylogenetic tree models were assembled using Geneious, and branches were compared to their sequence similarity to a panel of 14 drift viruses described above. The number of sequences that branched within 98.9% HA1 sequence identity of one of the 14 antigens was counted, and frequencies were calculated based on the total number of sequences available per season.
ACKNOWLEDGMENTS
We thank Amanda Skarlupka for technical assistance. Some of the H3N2 influenza A viruses were obtained through the Influenza Reagent Resource, Influenza Division, WHO Collaborating Center for Surveillance, Epidemiology, and Control of Influenza, Centers for Disease Control and Prevention, Atlanta, GA. We thank the University of Georgia Animal Resource staff, technicians, and veterinarians for the excellent animal care.
This study was funded, in part, by the University of Georgia (UGA; MRA-001) and by Sanofi Pasteur (CRA UGA 001). In addition, T.M.R. is supported by the Georgia Research Alliance as an Eminent Scholar.
REFERENCES
- 1.Nicholson KG, Wood JM, Zambon M. 2003. Influenza. Lancet 362:1733–1745. doi: 10.1016/S0140-6736(03)14854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flannery B, Chung JR, Belongia EA, McLean HQ, Gaglani M, Murthy K, Zimmerman RK, Nowalk MP, Jackson ML, Jackson LA, Monto AS, Martin ET, Foust A, Sessions W, Berman S, Barnes JR, Spencer S, Fry AM. 2018. Interim estimates of 2017–18 seasonal influenza vaccine effectiveness—United States, February 2018. MMWR Morb Mortal Wkly Rep 67:180–185. doi: 10.15585/mmwr.mm6706a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen JD, Ross TM. 2018. H3N2 influenza viruses in humans: viral mechanisms, evolution, and evaluation. Hum Vaccin Immunother 14(8):1840–1847. doi: 10.1080/21645515.2018.1462639:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skowronski DM, Chambers C, De Serres G, Dickinson JA, Winter AL, Hickman R, Chan T, Jassem AN, Drews SJ, Charest H, Gubbay JB, Bastien N, Li Y, Krajden M. 2018. Early season cocirculation of influenza A(H3N2) and B(Yamagata): interim estimates of 2017/18 vaccine effectiveness, Canada. Euro Surveill 23. doi: 10.2807/1560-7917.ES.2018.23.5.18-00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monto AS. 2010. Seasonal influenza and vaccination coverage. Vaccine 28(Suppl 4):D33–D44. doi: 10.1016/j.vaccine.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Ellebedy AH, Webby RJ. 2009. Influenza vaccines. Vaccine 27(Suppl 4):D65–D68. doi: 10.1016/j.vaccine.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter DM, Darby CA, Johnson SK, Carlock MA, Kirchenbaum GA, Allen JD, Vogel TU, Delagrave S, DiNapoli J, Kleanthous H, Ross TM. 2017. Elicitation of protective antibodies against a broad panel of H1N1 viruses in ferrets preimmune to historical H1N1 influenza viruses. J Virol 91. doi: 10.1128/JVI.01283-17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Carter DM, Darby CA, Lefoley BC, Crevar CJ, Alefantis T, Oomen R, Anderson SF, Strugnell T, Cortes–Garcia G, Vogel TU, Parrington M, Kleanthous H, Ross TM. 2016. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J Virol 90:4720–4734. doi: 10.1128/JVI.03152-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crevar CJ, Carter DM, Lee KY, Ross TM. 2015. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum Vaccin Immunother 11:572–583. doi: 10.1080/21645515.2015.1012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giles BM, Bissel SJ, Dealmeida DR, Wiley CA, Ross TM. 2012. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin Vaccine Immunol 19:128–139. doi: 10.1128/CVI.05533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, Wiley CA, Ross TM. 2012. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis 205:1562–1570. doi: 10.1093/infdis/jis232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giles BM, Ross TM. 2011. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29:3043–3054. doi: 10.1016/j.vaccine.2011.01.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong TM, Allen JD, Bebin-Blackwell AG, Carter DM, Alefantis T, DiNapoli J, Kleanthous H, Ross TM. 2017. Computationally optimized broadly reactive hemagglutinin elicits hemagglutination inhibition antibodies against a panel of H3N2 influenza virus cocirculating variants. J Virol 91:e01581-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter DM, Bloom CE, Nascimento EJ, Marques ET, Craigo JK, Cherry JL, Lipman DJ, Ross TM. 2013. Sequential seasonal H1N1 influenza virus infections protect ferrets against novel 2009 H1N1 influenza virus. J Virol 87:1400–1410. doi: 10.1128/JVI.02257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirchenbaum GA, Carter DM, Ross TM. 2016. Sequential infection in ferrets with antigenically distinct seasonal H1N1 Influenza viruses boosts hemagglutinin stalk-specific antibodies. J Virol 90:1116–1128. doi: 10.1128/JVI.02372-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirchenbaum GA, Ross TM. 2014. Eliciting broadly protective antibody responses against influenza. Curr Opin Immunol 28:71–76. doi: 10.1016/j.coi.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Wilson IA, Cox NJ. 1990. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol 8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 18.Alymova IV, York IA, Air GM, Cipollo JF, Gulati S, Baranovich T, Kumar A, Zeng H, Gansebom S, McCullers JA. 2016. Glycosylation changes in the globular head of H3N2 influenza hemagglutinin modulate receptor binding without affecting virus virulence. Sci Rep 6:36216. doi: 10.1038/srep36216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y, Wharton SA, Whittaker L, Dai M, Ermetal B, Lo J, Pontoriero A, Baumeister E, Daniels RS, McCauley JW. 2017. The characteristics and antigenic properties of recently emerged subclade 3C.3a and 3C.2a human influenza A(H3N2) viruses passaged in MDCK cells. Influenza Other Respir Viruses 11:263–274. doi: 10.1111/irv.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan SGCM, Carville KS, Deng YM, Grant KA, Higgins G, Komadina N, Leung VK, Minney-Smith CA, Teng D, Tran T, Stocks N, Fielding JE. 2018. Low interim influenza vaccine effectiveness, Australia. Euro Surveill 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blackburne BP, Hay AJ, Goldstein RA. 2008. Changing selective pressure during antigenic changes in human influenza H3. PLoS Pathog 4:e1000058. doi: 10.1371/journal.ppat.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou F, Trieu MC, Davies R, Cox RJ. 2018. Improving influenza vaccines: challenges to effective implementation. Curr Opin Immunol 53:88–95. doi: 10.1016/j.coi.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Tripp RA, Tompkins SM. 2009. Animal models for evaluation of influenza vaccines. Curr Top Microbiol Immunol 333:397–412. doi: 10.1007/978-3-540-92165-3_19. [DOI] [PubMed] [Google Scholar]
- 24.Byrd-Leotis L, Cummings RD, Steinhauer DA. 2017. The interplay between the host receptor and influenza virus hemagglutinin and neuraminidase. Int J Mol Sci 18. doi: 10.3390/ijms18071541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Vries RD, Nieuwkoop NJ, van der Klis FRM, Koopmans MPG, Krammer F, Rimmelzwaan GF. 2017. Primary human influenza B virus infection induces cross-lineage hemagglutinin stalk-specific antibodies mediating antibody-dependent cellular cytotoxicity. J Infect Dis 217:3–11. doi: 10.1093/infdis/jix546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Vries RD, Nieuwkoop NJ, Pronk M, de Bruin E, Leroux-Roels G, Huijskens EGW, van Binnendijk RS, Krammer F, Koopmans MPG, Rimmelzwaan GF. 2017. Influenza virus-specific antibody dependent cellular cytoxicity induced by vaccination or natural infection. Vaccine 35:238–247. doi: 10.1016/j.vaccine.2016.11.082. [DOI] [PubMed] [Google Scholar]
- 27.Bonomo ME, Deem MW. 2018. Predicting influenza H3N2 vaccine efficacy from evolution of the dominant epitope. Clin Infect Dis doi: 10.1093/cid/ciy323. [DOI] [PubMed] [Google Scholar]
- 28.Gupta V, Earl DJ, Deem MW. 2006. Quantifying influenza vaccine efficacy and antigenic distance. Vaccine 24:3881–3888. doi: 10.1016/j.vaccine.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz ET, Deem MW. 2005. Epitope analysis for influenza vaccine design. Vaccine 23:1144–1148. doi: 10.1016/j.vaccine.2004.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Deem MW. 2016. Influenza evolution and H3N2 vaccine effectiveness, with application to the 2014/2015 season. Protein Eng Des Sel 29:309–315. doi: 10.1093/protein/gzw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deem MW, Pan K. 2009. The epitope regions of H1-subtype influenza A, with application to vaccine efficacy. Protein Eng Des Sel 22:543–546. doi: 10.1093/protein/gzp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M, Boulton R, Haddy L, Biggerstaff M, Brammer L, Trock S, Burns E, Gomez T, Wong KK, Katz J, Lindstrom S, Klimov A, Bresee JS, Jernigan DB, Cox N, Finelli L. 2013. Human infections with influenza A(H3N2) variant virus in the United States, 2011-2012. Clin Infect Dis 57(Suppl 1):S4–S11. doi: 10.1093/cid/cit272. [DOI] [PubMed] [Google Scholar]
- 33.Huang JW, Yang JM. 2011. Changed epitopes drive the antigenic drift for influenza A (H3N2) viruses. BMC Bioinformatics 12(Suppl 1):S31. doi: 10.1186/1471-2105-12-S1-S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao Y, Moghadas SM. 2013. Impact of viral drift on vaccination dynamics and patterns of seasonal influenza. BMC Infect Dis 13:589. doi: 10.1186/1471-2334-13-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JH, Liepkalns J, Reber AJ, Lu X, Music N, Jacob J, Sambhara S. 2016. Prior infection with influenza virus but not vaccination leaves a long-term immunological imprint that intensifies the protective efficacy of antigenically drifted vaccine strains. Vaccine 34:495–502. doi: 10.1016/j.vaccine.2015.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green TD, Montefiori DC, Ross TM. 2003. Enhancement of antibodies to the human immunodeficiency virus type 1 envelope by using the molecular adjuvant C3d. J Virol 77:2046–2055. doi: 10.1128/JVI.77.3.2046-2055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan GS, Lee PS, Hoffman RM, Mazel-Sanchez B, Krammer F, Leon PE, Ward AB, Wilson IA, Palese P. 2014. Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. J Virol 88:13580–13592. doi: 10.1128/JVI.02289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization. 2011. Manual for the laboratory diagnosis and virological surveillance of influenza. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 39.Katz JM, Hancock K, Xu X. 2011. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert Rev anti Infect Ther 9:669–683. doi: 10.1586/eri.11.51. [DOI] [PubMed] [Google Scholar]
- 40.Oh DY, Barr IG, Mosse JA, Laurie KL. 2008. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J Clin Microbiol 46:2189–2194. doi: 10.1128/JCM.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.European Medicines Agency. 2014. Guideline on influenza vaccines: nonclinical and clinical module [draft]. Publisher, London, United Kingdom. [Google Scholar]
- 42.Matrosovich M, Matrosovich T, Garten W, Klenk HD. 2006. New low-viscosity overlay medium for viral plaque assays. Virol J 3:63. doi: 10.1186/1743-422X-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan K, Kloess J, Qian C, Bell D, Hay A, Lin YP, Gu Y. 2012. High-throughput virus plaque quantitation using a flatbed scanner. J Virol Methods 179:81–89. doi: 10.1016/j.jviromet.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Walls HH, Harmon MW, Slagle JJ, Stocksdale C, Kendal AP. 1986. Characterization and evaluation of monoclonal antibodies developed for typing influenza A and influenza B viruses. J Clin Microbiol 23:240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]