The two human gammaherpesviruses, Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV), can cause aggressive forms of cancer. These herpesviruses are strictly host specific, and therefore the homolog murine gammaherpesvirus 68 (MHV68) is a widely used model to obtain in vivo insights into the interaction between these two gammaherpesviruses and their host. Like EBV and KSHV, MHV68 establishes lifelong latency in B cells. The innate immune system serves as one of the first lines of host defense, with pattern recognition receptors such as the Toll-like receptors playing a crucial role in mounting a potent antiviral immune response to various pathogens. Here, we shed light on a yet unanticipated role of Toll-like receptor 7 in the recognition of MHV68 in a subset of immune cells called plasmacytoid dendritic cells, as well as on the control of this virus in its host.
KEYWORDS: herpesviruses, MHV68, Toll-like receptor, dendritic cells, murine gammaherpesvirus 68, pattern recognition receptor, type I interferon
ABSTRACT
Murine gammaherpesvirus 68 (MHV68) is a small-animal model suitable for study of the human pathogens Epstein-Barr virus and Kaposi’s sarcoma-associated herpesvirus. Here, we have characterized the roles of the endosomal Toll-like receptor (TLR) escort protein UNC93B, endosomal TLR7, -9, and -13, and cell surface TLR2 in MHV68 detection. We found that the alpha interferon (IFN-α) response of plasmacytoid dendritic cells (pDC) to MHV68 was reduced in Tlr9−/− cells compared to levels in wild type (WT) cells but not completely lost. Tlr7−/− pDC responded similarly to WT. However, we found that in Unc93b−/− pDC, as well as in Tlr7−/− Tlr9−/− double-knockout pDC, the IFN-α response to MHV68 was completely abolished. Thus, the only pattern recognition receptors contributing to the IFN-α response to MHV68 in pDC are TLR7 and TLR9, but the contribution of TLR7 is masked by the presence of TLR9. To address the role of UNC93B and TLR for MHV68 infection in vivo, we infected mice with MHV68. Lytic replication of MHV68 after intravenous infection was enhanced in the lungs, spleen, and liver of UNC93B-deficient mice, in the spleen of TLR9-deficient mice, and in the liver and spleen of Tlr7−/− Tlr9−/− mice. The absence of TLR2 or TLR13 did not affect lytic viral titers. We then compared reactivation of MHV68 from latently infected WT, Unc93b−/−, Tlr7−/− Tlr9−/−, Tlr7−/−, and Tlr9−/− splenocytes. We observed enhanced reactivation and latent viral loads, particularly from Tlr7−/− Tlr9−/− splenocytes compared to levels in the WT. Our data show that UNC93B-dependent TLR7 and TLR9 cooperate in and contribute to detection and control of MHV68 infection.
IMPORTANCE The two human gammaherpesviruses, Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV), can cause aggressive forms of cancer. These herpesviruses are strictly host specific, and therefore the homolog murine gammaherpesvirus 68 (MHV68) is a widely used model to obtain in vivo insights into the interaction between these two gammaherpesviruses and their host. Like EBV and KSHV, MHV68 establishes lifelong latency in B cells. The innate immune system serves as one of the first lines of host defense, with pattern recognition receptors such as the Toll-like receptors playing a crucial role in mounting a potent antiviral immune response to various pathogens. Here, we shed light on a yet unanticipated role of Toll-like receptor 7 in the recognition of MHV68 in a subset of immune cells called plasmacytoid dendritic cells, as well as on the control of this virus in its host.
INTRODUCTION
The double-stranded DNA virus murid gammaherpesvirus 4, also known as murine gammaherpesvirus 68 (MHV68), was isolated in 1980 (1). Due to strict host species specificity of the gammaherpesviruses, MHV68 is used as a small-animal model for the human gammaherpesviruses Kaposi’s sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV) (2). The MHV68 life cycle is typical for herpesviruses and includes both lytic and latent phases.
The pattern recognition receptors (PRR) are a crucial first line of the host defense against pathogens, including viruses. One class of PRR is comprised of the Toll-like receptors (TLR) that are located on the cell surface and intracellularly in endosomes and detect a wide range of nonself patterns. Cell surface TLR typically recognize exposed surface molecules on bacteria or viruses. TLR2 recognizes bacterial lipoproteins and lipoteichoic acid, as well as viral structural proteins (3). Recently, TLR2 was found to recognize repeating protein subunit patterns, including those present in viral capsids (4).
The endosomal TLR sense nucleic acids. TLR3, TLR7, and TLR9 recognize double-stranded RNA (dsRNA), single-stranded RNA (ssRNA), and CpG-rich dsDNA, respectively (5). TLR13 detects a specific sequence of bacterial 23S rRNA and also responds to vesicular stomatitis virus (VSV) (6, 7). Proper trafficking and signaling of endosomal TLR are dependent on the protein UNC93B; mutation or deletion of UNC93B eliminates endosomal TLR signaling (8–12).
To date, 12 murine TLR have been described: TLR1 to TLR9 and TLR11 to TLR13. However, very few have been characterized during MHV68 infection. So far, surface TLR2 and endosomal TLR9 have been found to be important for detection of MHV68. TLR2 contributed to interleukin-6 (IL-6) and alpha interferon (IFN-α) production in murine embryonic fibroblasts, and elevated titers were observed in the lungs of Tlr2−/− mice after intranasal (i.n.) infection (13). TLR9 was important for control of viral replication after intraperitoneal infection, as titers were elevated in the spleens of Tlr9−/− mice compared to those in the wild-type (WT) controls (14). Reactivation and latent viral load were correspondingly increased in Tlr9−/− splenocytes (14). TLR9 also partially contributes to the IFN-α response to MHV68 in primary FMS-related tyrosine kinase 3 ligand (Flt-3L)-induced dendritic cells (FLDC) (14). The role of TLR in detection of human gammaherpesviruses has been analyzed in more detail. In humans, 10 TLRs, TLR1 to TLR10, have been described. To date, TLR2, TLR3, TLR7, and TLR9 have been shown to recognize at least one human gammaherpesvirus (15). Whether TLR13 can detect herpesviral infection is not yet known.
With the exception of TLR3, TLR recruit the adapter protein MyD88 to activate a signaling cascade, resulting in production of type I interferon (IFN) and proinflammatory cytokines (16). The role of MyD88 during MHV68 infection is somewhat unclear; one study found that viral titers were elevated in the lungs of Myd88−/− mice at 3 and 5 days postinfection (13), while another study found that on days 4 and 9 postinfection, replication in the lungs of Myd88−/− mice was unaffected (17). The latent viral load was markedly decreased in Myd88−/− spleens after intranasal infection, with a concomitant decrease in reactivation. However, upon intraperitoneal infection, the latent viral load in peritoneal exudate cells (PEC) and splenocytes was similar in Myd88−/− and WT mice, and reactivation was increased from PEC in the absence of MyD88 but decreased from Myd88−/− splenocytes (17).
To clarify the role of additional individual and multiple TLR in detection of MHV68 in vitro, we analyzed the type I IFN response to MHV68 of primary plasmacytoid dendritic cells (pDC) derived from the bone marrow of WT, Unc93b−/−, Tlr2−/−, Tlr7−/−, Tlr9−/−, Tlr13−/−, and Tlr7−/− Tlr9−/− mice and identified a novel role for TLR7. To understand the role of TLR during the acute phase of infection, we analyzed lytic viral titers in the WT and all six knockout (KO) genotypes mentioned. We observed that lytic replication was enhanced in the lung, spleen, and liver of Unc93b−/− mice, in the spleen of Tlr9−/− mice, and in the liver and spleen of Tlr7−/− Tlr9−/− mice. Last, we analyzed latent genome copy numbers and ex vivo reactivation of MHV68 in WT and Unc93b−/−, Tlr7−/−, Tlr9−/−, and Tlr7−/− Tlr9−/− mice and found that they were increased, particularly in the absence of both TLR7 and TLR9. Our data have identified a novel role for TLR7 in detection of MHV68 in vitro and clarify the cooperative contribution of TLR7 and TLR9 to detection and control of MHV68 replication in vivo.
RESULTS
MHV68 is detected by both TLR7 and TLR9 in pDC.
A previous study has shown that the production of type I interferon in response to MHV68 stimulation is partially dependent on TLR9; in primary murine FLDC derived from Tlr9−/− mice, the IFN-α response to MHV68 is abrogated but not completely lost (14). When murine bone marrow cells are cultured with FMS-like tyrosine kinase 3 ligand (Flt-3L), cells differentiate into conventional and plasmacytoid dendritic cells (cDC and pDC, respectively). Of these, the pDC are the major type I IFN-producing cells (18), and detection of infection in pDC is almost exclusively dependent on TLR, suggesting that another TLR besides TLR9 may be responsible for the remaining IFN-α response to MHV68 in Tlr9−/− FLDC. To identify the TLR that also contribute(s) to detection of MHV68 in pDC, we analyzed the response to MHV68 infection of pDC purified from primary FLDC cultures derived from WT and TLR-deficient mice.
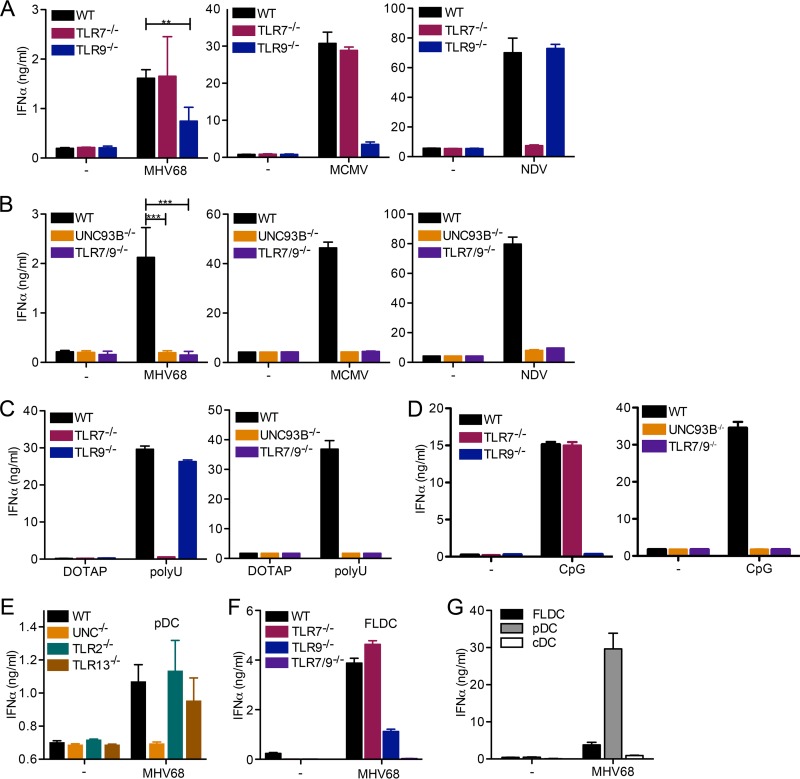
First, we infected WT, Tlr7−/−, or Tlr9−/− pDC with MHV68 and measured IFN-α secretion by enzyme-linked immunosorbent assay (ELISA). We found that Tlr9−/− pDC produced significantly less IFN-α than WT pDC in response to MHV68 treatment (Fig. 1A) as previously shown for FLDC (14). In contrast, Tlr7−/− pDC responded to MHV68 similarly to WT cells (Fig. 1A). A similar pattern was observed in response to murine cytomegalovirus (MCMV) infection (Fig. 1A). The response to Newcastle disease virus (NDV), an RNA virus, was almost entirely dependent on the presence of the RNA sensor TLR7, as expected (Fig. 1A). We next analyzed the IFN-α response to MHV68 in pDC deficient in multiple endosomal TLR. We used either Unc93b−/− pDC, deficient in all endosomal TLR signaling, or Tlr7−/− Tlr9−/− pDC, lacking only TLR7 and TLR9. Interestingly, Unc93b−/− and Tlr7−/− Tlr9−/− pDC did not mount an IFN-α response upon infection with MHV68. The same result was observed upon MCMV and NDV infection (Fig. 1B). We successfully verified the cell phenotypes using the synthetic TLR7 ligand poly(U) and the TLR9 ligand CpG DNA (Fig. 1C and D). Finally, we analyzed the IFN-α response to MHV68 in pDC lacking TLR2 or TLR13, with WT and Unc93b−/− pDC as controls (Fig. 1E). We found that Tlr2−/− pDC and Tlr13−/− pDC were not impaired in their IFN-α response compared to that of WT pDC (Fig. 1E).
FIG 1.
Both TLR7 and TLR9 are required for the IFN-α response to MHV68 in pDC. (A to D) Bone marrow cells were cultured in the presence of Flt-3 ligand (Flt-3L) for 8 days to produce FLDC, and pDC were MACS purified. WT, Tlr7−/−, and TLR9−/− (A) or WT, Unc93b−/−, or Tlr7−/− Tlr9−/− (B) pDC were mock infected or infected with MHV68, MCMV, or NDV as indicated. (C and D) pDC of the indicated genotype were treated with DOTAP (control) or the TLR7 agonist poly(U) complexed with DOTAP (C) and medium (−) or the TLR9 agonist CpG DNA (D), as indicated. (E) pDC from WT, Unc93b−/−, Tlr2−/−, and Tlr13−/− FLDC were prepared by MACS negative selection. (F) FLDC from WT, Tlr7−/−, Tlr9−/−, and Tlr7−/− Tlr9−/− mice were prepared. (G) WT FLDC were reserved or stained with APC-coupled anti-mouse/human CD45R/B220 and PE-coupled anti-mouse CD11c and then separated into CD45R/B220+ CD11cmid plasmacytoid dendritic cells (pDC) and CD45R/B220− CD11chigh conventional dendritic cells (cDC) by FACS. For panels E to G, equal cell numbers were plated and mock infected or infected with MHV68. For all panels, the IFN-α response was measured by ELISA. In panels A and B, combined duplicates from two independent experiments are shown for MHV68, and log-transformed data for MHV68 were analyzed by a two-tailed unpaired t test (**, P < 0.01; ***, P < 0.001). For all other graphs (MCMV and NDV in panels A and B and C to G), duplicates from one representative experiment are shown.
FLDC cultures include pDC as well as conventional dendritic cells (cDC). To clarify whether pDC alone are responsible for the observed phenotype or understand if cDC also contribute, we next compared the response to MHV68 in total FLDC cultures derived from WT, Tlr7−/−, Tlr9−/−, and Tlr7−/− Tlr9−/− bone marrow. The results were similar to those observed for purified pDC; the response was reduced in Tlr9−/− FLDC and completely lost in Tlr7−/− Tlr9−/− FLDC (Fig. 1F). When we compared the IFN-α response from WT FLDC to fluorescence-activated cell sorter (FACS)-purified CD11cmid B220+ pDC (expressing intermediate levels of CD11c) and CD11c+ B220− cDC, we found that the pDC were primarily responsible for the IFN-α response upon MHV68 infection (Fig. 1G). These data show that both TLR7 and TLR9, but not TLR2 or TLR13, are required for IFN-α production after MHV68 infection of pDC. Interestingly, the contribution of TLR7 was observed only in the absence of TLR9.
The absence of endosomal TLR does not affect weight loss or lytic replication in organs after intranasal infection with MHV68.
Next, we wanted to determine the role of TLR during in vivo infection with MHV68. The natural route of infection in wild rodents is still unclear. Many routes of infection, including intranasal and intravenous (i.v.), are used in the literature (19). Intranasal infection is often described as a more natural route than intravenous infection, and it is anticipated that murine noses play an important role in animal interactions (19, 20). However, horizontal transmission among female laboratory mice has not been observed (21, 22). In contrast, sexual transmission has been reported (22). MHV68 DNA has been identified in wild ticks (23), and there is experimental evidence for tick-borne transmission of MHV68 (24). Both sexual and tick-borne transmission are more similar to intravenous or blood-borne infection than they are to intranasal infection. In humans, transmission of EBV and KSHV can be salivary or sexual (reviewed in reference 25). To comprehensively address the role of TLR during MHV68 infection, we utilized both intranasal and intravenous infection.
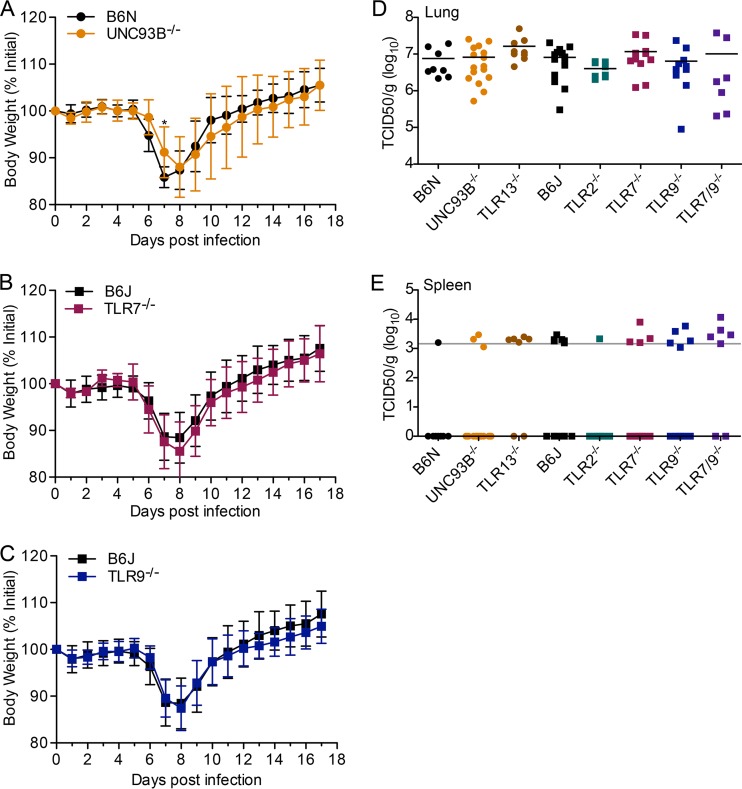
First, we infected WT, Unc93b−/−, Tlr7−/−, and Tlr9−/− mice intranasally and monitored body weight. We observed minimal differences between Unc93b−/− mice and their respective B6N controls (Fig. 2A). There were no differences observed in body weight change of Tlr7−/− or Tlr9−/− mice compared to weights of their respective B6J controls (Fig. 2B and C). Mice of all genotypes lost weight but recovered.
FIG 2.
Early acute replication of MHV68 after intranasal infection is TLR independent and limited to the lungs. Mice of the indicated genotype were infected intranasally with 2 × 106 PFU of MHV68. (A to C) Body weight was measured daily. Data are combined from at least two independent experiments, with 8 or more mice total per genotype. Data were analyzed by a two-tailed Mann-Whitney test (*, P < 0.05). (D and E) Lungs and spleen were harvested at 3 days postinfection, and viral titers were determined using the median tissue culture infectious dose (TCID50) on M2-10B4 cells. Splenic titers were almost exclusively around the average detection limit (approximate line in gray) or negative (indicated by log10 TCID50/g = 0). Each data point represents one mouse.
Next, we determined acute lytic viral titers in the lungs and spleens 3 days after intranasal infection. We included endosomal TLR-deficient Unc93b−/− mice, Tlr13−/−, Tlr2−/−, Tlr7−/−, Tlr9−/−, and Tlr7−/− Tlr9−/− mice, as well as B6N and B6J WT mice as controls. There were no significant differences in viral titers in the lungs among the genotypes examined (Fig. 2D). Very little lytic virus was detected in spleen samples, as expected based on the kinetics of viral spread; many samples were around the approximate detection limit (Fig. 2E, horizontal gray line). Many were negative, and those samples are displayed as points on the x axis, where the log10 value is 0 (Fig. 2E).
To summarize, we did not observe a role of TLR in control of lytic replication upon intranasal infection, but this finding was not surprising since endosomal TLR are not the predominant pattern recognition receptors expressed in the lung. Previously, TLR9 was not found to play a role during intranasal infection, but it was important in control of replication upon intraperitoneal infection (14). Like intraperitoneal infection, intravenous infection allows direct infection of the spleen without initial viral replication in the lung.
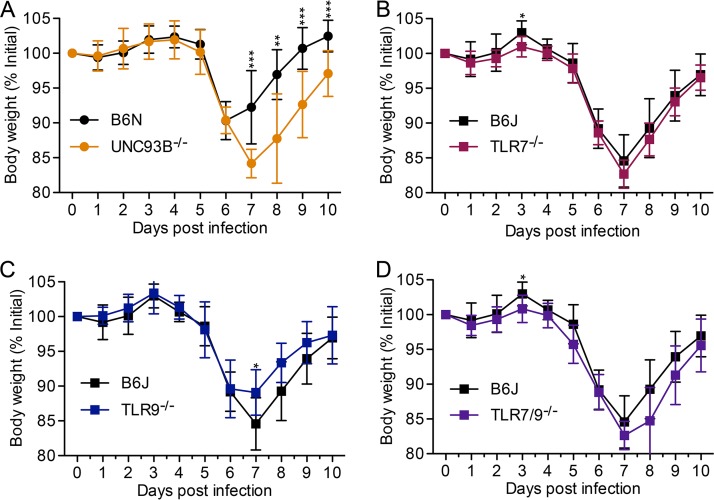
We thus utilized intravenous infection for further experiments. We found that Unc93b−/− mice lost significantly more weight and recovered more slowly from weight loss than their WT B6N counterparts upon MHV68 infection (Fig. 3A). In general, Tlr7−/−, Tlr9−/−, and Tlr7−/− Tlr9−/− mice lost weight at levels similar to those of WT B6J mice (Fig. 3B to D).
FIG 3.
The absence of endosomal TLR minimally affects weight loss after intravenous infection with MHV68. Mice of the indicated genotype were infected intravenously with 2 × 106 PFU of MHV68. Body weight was measured daily. Data are combined from at least two independent experiments, with 8 or more mice total per genotype. Statistical significance was determined by a two-tailed Mann-Whitney test for days 3, 7, 8, 9, and 10 (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
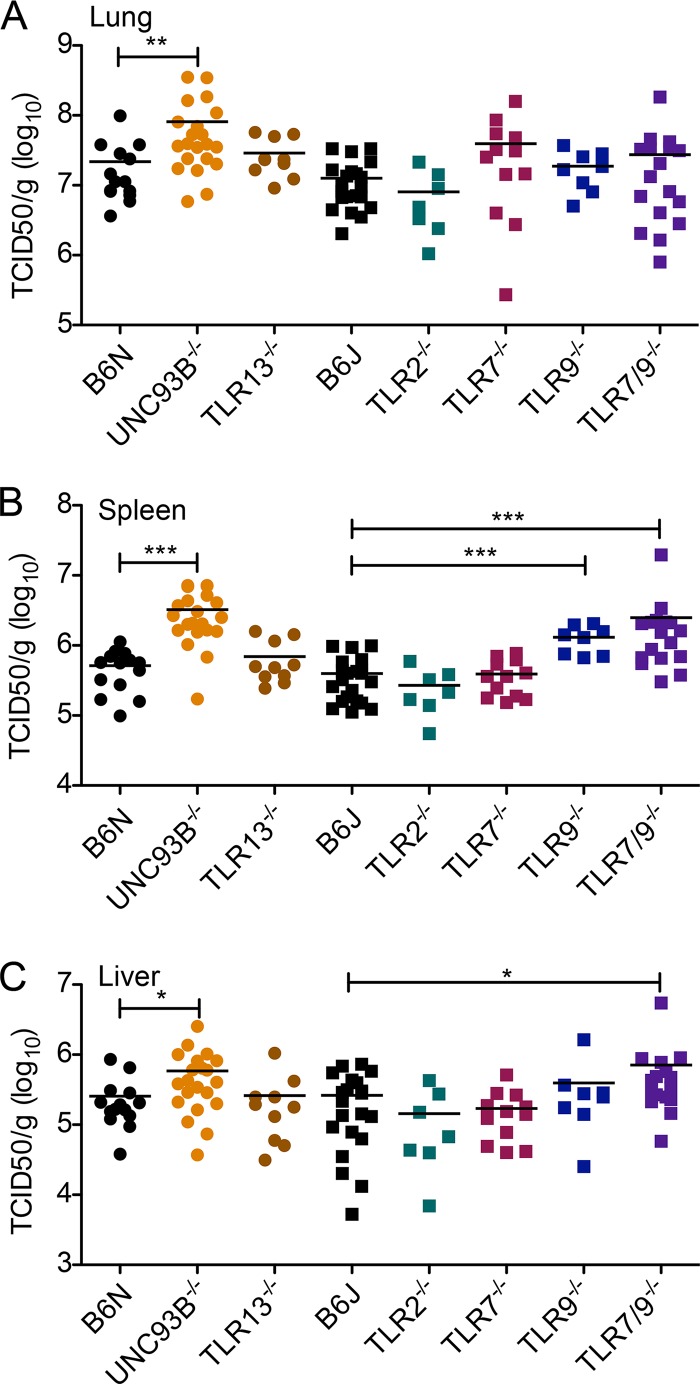
We next examined replication of MHV68 in organs at day 3 post-intravenous infection. We compared lytic titers in the lungs (Fig. 4A), spleen (Fig. 4B), and liver (Fig. 4C) of WT, Unc93b−/−, Tlr2−/−, Tlr7−/−, Tlr9−/−, Tlr13−/−, and Tlr7−/− Tlr9−/− mice. Titers in the lungs of Unc93b−/− mice were increased 4-fold on average, a statistically significant increase compared to levels in their B6N controls (Fig. 4A). Average titers from Tlr7−/−, Tlr9−/−, Tlr7−/− Tlr9−/−, and Tlr13−/− lungs were increased 3-, 1.5-, 2.2-, and 1.3-fold, respectively, compared to titers of B6J WT mice, but these increases were not statistically significant (Fig. 4A). As previously reported for TLR9 upon intraperitoneal infection, we found that titers in the spleens from Tlr9−/− mice were significantly increased upon intravenous infection; we observed an approximately 3.3-fold increase in titers (Fig. 4B). Additionally, titers in spleens derived from Unc93b−/− and Tlr7−/− Tlr9−/− mice were significantly elevated compared to levels in their B6N and B6J controls, with approximately a 6.3-fold increase in both cases (Fig. 4B). Finally, we found that lytic titers were significantly elevated in the livers of Unc93b−/−and Tlr7−/− Tlr9−/− mice, by approximately 2.3- and 2.7-fold, respectively, but not in Tlr9−/− livers (Fig. 4C). In summary, after intravenous infection, viral titers were elevated in the lungs of Unc93b−/− mice. TLR9 is important for control of MHV68 replication in the spleen, and both TLR7 and TLR9 are important for control of MHV68 replication in the liver.
FIG 4.
Early acute replication of MHV68 after intravenous infection is enhanced in the absence of endosomal TLR, especially TLR9. Mice of the indicated genotypes were infected intravenously with 2 × 106 PFU MHV68. At 3 days postinfection, lungs, spleen, or liver was harvested, as indicated, and viral titers were determined by TCID50 on M2-10B4 cells. Data were analyzed by a two-tailed Mann-Whitney test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
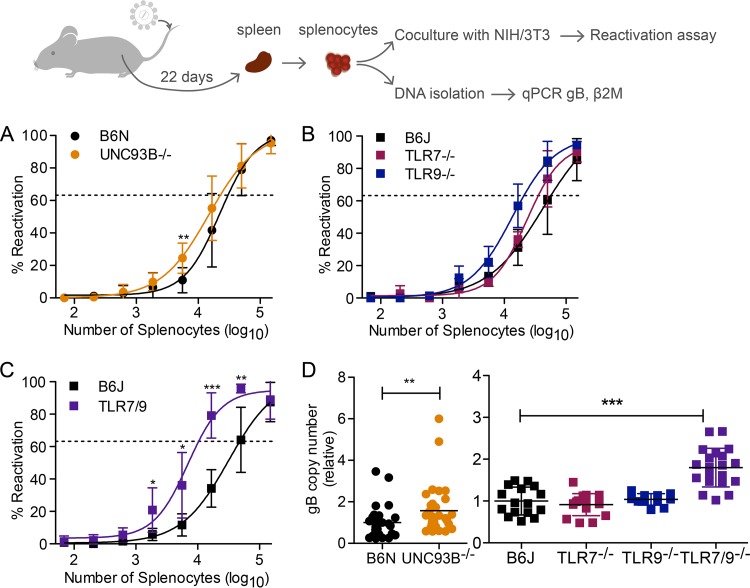
Like all herpesviruses, after a period of lytic replication, MHV68 establishes latency in vivo. During MHV68 infection, independently of the route of infection, latency is established in the spleen, predominantly in activated germinal center B220+ PNAhigh B cells (expressing high levels of peanut agglutinin [PNA]) (26). The coculture of latently infected B cells or total splenocytes with NIH 3T3 fibroblasts leads to reactivation and spread of lytic virus to fibroblasts, which ae then scored for cytopathic effect (CPE) (27, 28). By comparing serial dilutions of splenocytes from latently infected mice, the reactivation frequency can be determined.
We next wanted to analyze the role of TLR signaling in the reactivation of MHV68. We infected mice intravenously with MHV68 and then waited 22 days for the establishment of latency. For an ex vivo reactivation assay, total splenocytes were isolated as a single cell suspension, diluted in 3-fold serial dilutions, and 24 replicates were plated per dilution. The frequency of reactivating wells at each dilution was then determined by analyzing CPE. To compare reactivation frequencies, the regression line intersect was set to 63.2% based on the Poisson distribution. Slightly more reactivation was observed for Unc93b−/− splenocytes than for B6N controls. Approximately 1 in 22,719 Unc93b−/− splenocytes reactivated whereas 1 in 30,170 B6N splenocytes reactivated, for a slight 1.33-fold increase in reactivation of Unc93b−/− compared to levels of B6N splenocytes (Fig. 5A). For B6J, Tlr7−/−, and Tlr9−/− splenocytes, we found that approximately 1 in 47,982 B6J, 1 in 33,859 Tlr7−/−, and 1 in 22,145 Tlr9−/− splenocytes reactivated, for a 1.42-fold increase in reactivation of Tlr7−/− splenocytes over the B6J level and a 2.17-fold increase in reactivation of Tlr9−/− splenocytes compared to levels in B6J (Fig. 5B). Next, we examined reactivation of Tlr7−/− Tlr9−/− splenocytes. At multiple dilutions, reactivation was significantly more frequent than that of WT splenocytes. The frequency of reactivation was approximately 1 in 10,350, for a 4.64-fold increase in reactivation of Tlr7−/− Tlr9−/− splenocytes compared to the WT level (Fig. 5C).
FIG 5.
Reactivation and latent viral load are increased in the absence of endosomal TLR. Mice of the indicated genotypes were infected with 2 × 106 PFU of MHV68 intravenously. At 21 days postinfection, spleens were harvested, and single-cell splenocyte suspensions pooled from multiple mice were prepared. (A to C) Three-fold serial dilutions of splenocytes were cocultured with NIH 3T3 fibroblasts for 2 weeks, and reactivation was determined based on the presence of cytopathic effect in the NIH 3T3 monolayer. Curve fit lines were derived from nonlinear regression analysis based on the variable slope sigmoidal dose-response equation. The regression line intersect was set to 63.2% based on the Poisson distribution. (D) DNA was isolated from pooled splenocytes, and the gB copy number was determined relative to that of β2M and scaled to the average of the B6N or B6J WT control as appropriate. For each set of pooled splenocytes, copy numbers were determined for 3 to 4 replicates. Data points represent individual replicates. Log-transformed data (data points for >103 splenocytes in panels A to C and all samples in panel D) were analyzed by two-tailed unpaired t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Enhanced reactivation could be due to an increased ability of individual cells to reactivate or to an increase in the number of latent genome copies per spleen or cell. To differentiate between an improved ability to reactivate and an increase in the number of latent genome copies, we performed quantitative PCR (qPCR) for the MHV68 glycoprotein B (gB) gene to determine the latent viral load in splenocytes. We normalized levels to the value of the murine β-2-microglobulin (β2M) gene to control for the number of cells. For ease of comparison, we then scaled the gB values to the average of the appropriate B6 control. We found that for an equal number of splenocytes, Unc93b−/− splenocytes contained about 1.57-fold more gB copies than B6N WT splenocytes (Fig. 5D). In contrast, the number of gB-positive Tlr7−/− and Tlr9−/− splenocytes did not significantly differ from the number of gB-positive B6J splenocytes (0.91-fold and 1.04-fold, respectively) (Fig. 5D). However, when both TLR7 and TLR9 were absent, the number of gB-positive splenocytes was significantly increased compared to levels of B6J splenocytes, with Tlr7−/− Tlr9−/− splenocytes containing about 1.8-fold more gB copies than B6J splenocytes (Fig. 5D). Collectively, both the latent viral load and reactivation frequency of Unc93b−/− and Tlr7−/− Tlr9−/− splenocytes were elevated compared to levels in the WT controls. In the case of Unc93b−/− splenocytes, the increase in reactivation and increase in latent viral load were similar, suggesting a direct relationship between viral load and reactivation in B6N-background mice. However, in the case of Tlr7−/− Tlr9−/− splenocytes, genome copy numbers were increased 1.8-fold, while reactivation was increased 4.6-fold, suggesting that increased reactivation is only partially due to increased latent viral load on the B6J background.
TLR7 is an important pattern recognition receptor that senses multiple viruses. So far, our data indicate that TLR7 is important for detection and control of MHV68, but the ligand is unknown. Furthermore, the effect of TLR7 appears to be masked by the presence of TLR9, particularly in primary dendritic cell cultures. Thus, to analyze various ligands for their contribution to TLR7 signaling during MHV68 infection, we utilized stimulation of primary Tlr9−/− FLDC with various MHV68 mutants.
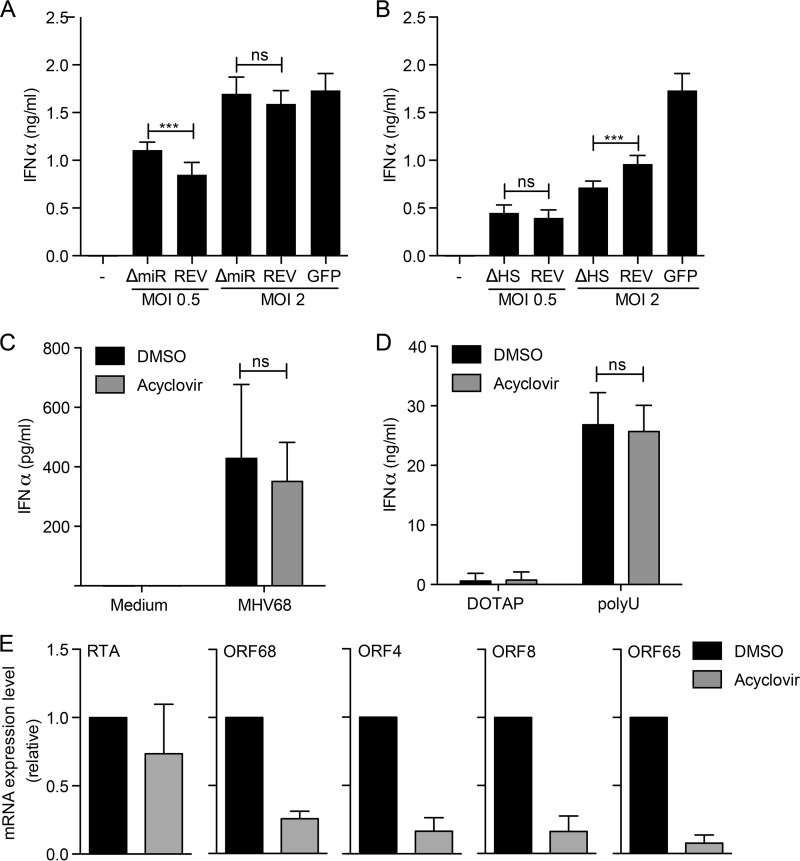
One potential TLR7 ligand during MHV68 infection is viral microRNAs (miRNA). In MHV68, pre-miRNA stem-loops give rise to 28 mature miRNAs (29–31). The stem-loops, however, also contain viral genome-encoded 5′ tRNAs (vtRNAs) (32). Due to the tight linkage of vtRNAs and miRNAs, an MHV68 virus mutant lacking small noncoding RNAs (sncRNAs; ΔmiR strain) lacks both miRNAs and vtRNAs (33). We stimulated Tlr9−/− FLDC with MHV68 ΔmiR, the corresponding revertant virus (REV), or MHV68 expressing green fluorescent protein (MHV68-GFP) as a control and then determined the IFN-α response (Fig. 6A). All three viruses induced an IFN-α response in Tlr9−/− FLDC, strongly indicating that miRNAs are not the sole TLR7 ligand during MHV68 infection (Fig. 6A).
FIG 6.
MHV68 miRNA, MHV68-mediated host shutoff, and MHV68 DNA polymerase activity minimally affect TLR7 signaling. Tlr9−/− bone marrow cells were cultured in the presence of Flt-3L for 8 days to produce FLDC. (A and B) Cells were mock infected or stimulated with miRNA-deficient MHV68 (ΔmiR), its revertant (REV), or MHV68-GFP (GFP) or with host shutoff (HS)-deficient MHV68 (ΔHS), its revertant (REV), or MHV68-GFP (GFP) at the indicated MOI. The IFN-α response was measured by ELISA. Combined quadruplicates from three independent experiments are shown. (C to E) Tlr9−/− FLDC were sorted into B220+CD11cmid pDC and pretreated with DMSO or acyclovir for three hours. Cells were then infected with MHV68 (C and E) or stimulated with the TLR7 ligand poly(U) (D) for 24 h. The IFN-α response was measured by ELISA, as indicated. Combined duplicates from three independent experiments are shown in panels C and D. (E) Viral mRNA expression of RTA, ORF68, ORF4, ORF8, and ORF65 was measured by qRT-PCR, normalized to the level of 18S rRNA, and compared using the 2−ΔΔCT method. Combined duplicates from two independent experiments are shown. In panels A to D log-transformed data were analyzed by two-tailed unpaired t test (ns, not significant; ***, P < 0.001).
Another potential TLR7 ligand during MHV68 infection is cleaved cellular mRNA. During lytic replication, the MHV68 protein muSOX induces host shutoff via global mRNA degradation. A host shutoff-deficient muSOX mutant (ΔHS) lacks the ability to degrade mRNA due to a single amino acid change but is well-expressed and retains DNase activity (34). We stimulated Tlr9−/− FLDC with MHV68 ΔHS, the corresponding revertant virus (REV), or MHV68-GFP for comparison (Fig. 6B). At a multiplicity of infection (MOI) of 0.5, ΔHS and REV induced a similar IFN-α responses. At an MOI of 2, the response was slightly reduced upon stimulation with ΔHS compared to that with REV, but a response was still present (Fig. 6B).
It is possible that the TLR7 ligand is packaged into MHV68 virions or transcribed before the onset of viral replication or expressed later during infection in pDC. To help clarify this question, we treated FACS-sorted Tlr9−/− pDC with the viral DNA polymerase inhibitor acyclovir or with dimethyl sulfoxide (DMSO) as a control and then infected them with MHV68 (Fig. 6C). The IFN-α responses to MHV68 were similar in both the presence and absence of acyclovir (Fig. 6C). To confirm that acyclovir treatment did not affect the IFN-α response in general, we also determined the response to the TLR7 agonist poly(U) in the presence of DMSO and acyclovir. No differences were observed (Fig. 6D). To verify that acyclovir treatment inhibited viral replication, we measured mRNA levels for immediate early RTA, early ORF68, early-late ORF4, and late ORF8 and ORF65/M9 (Fig. 6E). In the presence of acyclovir, the levels of early, early-late, and late mRNAs were greatly reduced.
In summary, as both the ΔmiR and ΔHS viruses induced an IFN-α response in Tlr9−/− FLDC, our results suggest that neither muSOX nor cleaved cellular mRNAs are exclusively responsible for the induction of IFN-α in Tlr9−/− FLDC. Furthermore, experiments with acyclovir suggest that the TLR7 ligand during MHV68 infection is packaged into virions, expressed before the onset of viral DNA replication, or is not of viral origin. In summary, we have identified TLR7 as an important TLR for detection of MHV68 in vitro and control of MHV68 infection in vivo, and it works cooperatively with TLR9 in detection of MHV68.
DISCUSSION
Our data identify the RNA sensor TLR7 as an important pattern recognition receptor during infection with the DNA virus MHV68. Interestingly, we have found that the role of TLR7 is frequently masked by the presence of TLR9. In Tlr9−/− pDC, however, we observed IFN-α production that is entirely dependent on TLR7 as the response was completely lost in Tlr7−/− Tlr9−/− pDC. pDC have long been known to be the major IFN-α-producing cells in response to viral infection. Additionally, pDC detection of infection is dependent on their expression of TLR7 and TLR9 (reviewed in reference 35). pDC have been shown to detect both MCMV (36) and MHV68 (37). However, until now, the two pattern recognition receptors required for MHV68 recognition in pDC were not fully known.
While the role of TLR9 has been described previously (14), TLR7 had not been studied in conjunction with TLR9 until now. Similar to our in vitro results, in vivo we also see that both TLR7 and TLR9 contribute to MHV68 recognition, but the strongest phenotype is present when both TLR7 and TLR9 are absent. Reactivation has been previously studied in splenocytes derived from Tlr9−/− (14, 38) and Tlr7−/− mice (38). While the authors of these previous studies observed that reactivation was enhanced in the absence of TLR9, as we also show, no effect was seen when TLR7 was absent. This is likely due to the fact that the presence of TLR9 masks the contribution of TLR7, which we now see in this report. A similar overlapping role for TLR7 and TLR9 in detection of MCMV has also been described (39). In Tlr7−/− Tlr9−/− pDC, there is a markedly reduced response to MCMV, as we also show, and in vivo, Tlr7−/− Tlr9−/− mice are more susceptible to MCMV infection (39). The interferon response we observed in pDC infected with MHV68 was clearly lower than the interferon response we detected upon MCMV or NDV infection. This has previously been shown for MCMV and MHV68 in wild-type FLDC due to deamination and, thus, suppression of stimulatory CpG motifs in the MHV68 genome but not in that of MCMV (37).
Here, we have identified a role for TLR7 and TLR9 in detection of MHV68. Endosomal TLR have been studied during KSHV and EBV infection and are important for detection and control of reactivation. For example, primary human pDC detect KSHV in a primarily TLR9-dependent manner; the use of a TLR9 inhibitor reduced or eliminated the IFN-α response with donor-to-donor variability (40). Although the role of TLR7 could not be tested, in the donors where a response was still present despite TLR9 inhibition, the remaining response could be due to TLR7 signaling (40), as we have observed in the case of MHV68 infection of pDC. A role for TLR7 has been shown during reactivation of KSHV; TLR7 ligands or VSV infection induced lytic gene transcription and replication (41). Similarly, in the case of EBV infection, both TLR7 and TLR9 contribute to pDC production of IFN-α (42, 43). TLR9 also recognizes EBV in monocytes (43), but whether TLR7 plays a role in these cells has not been studied.
One question raised by this study is the source of the TLR7 ligand in MHV68. Infection induces transcription of many different cellular and viral RNAs, including mRNA, microRNAs (miRNAs), and long noncoding RNAs, among others. First, we considered miRNAs, which are RNA molecules about 22 nucleotides long and are important for posttranscriptional regulation of mRNA. miRNAs control many cellular processes, including innate immunity (44), so it is not surprising that many viruses, like their hosts, encode miRNAs (45). MHV68 is no exception; it encodes 14 pre-miRNA stem-loops that form 28 mature miRNAs (29, 31, 32). Viral miRNAs were, thus, a potential source of TLR7 activation during infection. In Tlr9−/− FLDC, however, the response to infection with MHV68 ΔmiR or its revertant led to similar IFN-α responses, suggesting that miRNAs are not responsible for the TLR7 response to MHV68.
Next, we considered the MHV68 muSOX protein and its function as a possible source of TLR7 ligands during infection. The human gammaherpesviruses KSHV and EBV encode a conserved alkaline exonuclease gene that has an additional function in repression of host gene expression or host shutoff. This function is also conserved in the MHV68 homolog muSOX. We hypothesized that cleaved cellular mRNAs could be a TLR7 ligand during MHV68 infection (46, 47). While our data do not exclude that TLR7 activation during MHV68 infection is partially due to muSOX-dependent cleavage of host cell mRNA, our data verify that cleaved host cell mRNA is not the sole TLR7 ligand during MHV68 infection. If cleaved cellular mRNA were the only TLR7 ligands present during MHV68 infection, no IFN-α response would be anticipated in Tlr9−/− FLDC. In contrast, an IFN-α response was still observed in Tlr9−/− FLDC infected with MHV68 ΔHS.
In general, it is inadvisable for a cell to detect its own nucleic acid sequences as that could potentially cause unnecessary inflammation. However, cleaved cellular mRNAs could be an exception to this standard as they are not normally present in a healthy cell. One possible explanation is that to prevent autoimmune responses, TLR7 cannot detect these cleaved host-derived mRNAs, in which case, other potential ligands must be considered. A second possibility is that these cleaved mRNAs are simply not produced in pDC. However, this would not change the interpretation of our data. If cleaved mRNAs are not produced, they cannot be the ligand. Whether muSOX functions in primary pDC is not currently known. Studies comparing the functions of ΔHS and revertant viruses primarily used murine embryonic fibroblasts, NIH 3T3 cells, and the dendritic cell line DC2.4 (48) but did not compare them in primary pDC.
Another open question is whether the TLR7 ligand is MHV68 RNA that is packaged into virions, viral RNA that is transcribed prior to viral DNA replication, or viral RNA that is transcribed later during the life cycle. To help address this question, we treated Tlr9−/− pDC with the viral DNA polymerase inhibitor acyclovir and measured the IFN-α response to MHV68. We found that acyclovir treatment did not affect the IFN-α response. We verified DNA polymerase inhibition by quantitative reverse transcription PCR (qRT-PCR) and found that expression of multiple viral genes was inhibited by acyclovir. The observed effect of acyclovir on gene expression was in agreement with a study that used the DNA polymerase inhibitor phosphonoacetic acid (PAA) (49).
It is possible that even a limited amount of immediate early gene expression is sufficient to activate the TLR7-dependent IFN-α response in pDC. It is also plausible that packaged RNA induces the response; we cannot distinguish between these possibilities. Unfortunately, well-characterized immediate early inhibitors of MHV68 replication are lacking. As MHV68 uses host RNA polymerase II for mRNA synthesis, we cannot inhibit all viral mRNA synthesis; since we would also inhibit cellular mRNA synthesis, we would not be able to measure host IFN-α. Further studies will need to use a global approach to conclusively identify the TLR7 ligand during MHV68 infection, but this is beyond the scope of this study.
In vitro, we found that Unc93b−/− and Tlr7−/− Tlr9−/− pDC responded identically to MHV68 infection. In vivo, however, we observed differences between the two strains. One obvious explanation is that the Unc93b−/− and Tlr7−/− Tlr9−/− mice are on different genetic backgrounds. The Unc93b−/− mice were created as part of the Knockout Mouse Project (KOMP), and like all International Knockout Mouse Consortium mutant clones, they were generated in C57BL/6N embryonic stem (ES) cells (50). Chimaeras were then bred to C57BL/6N mice. In contrast, the Tlr7−/− and Tlr9−/− mice were made using genomic DNA from 129/Sv mice and most likely 129/Ola ES cells that were then injected into C57BL/6 blastocysts (51, 52). The Tlr7−/− and Tlr9−/− mice were then backcrossed for a number of generations onto C57BL/6J mice.
While C57BL/6N and C57BL/6J are closely related, the two strains diverged around 220 generations ago (53). There are a number of single-nucleotide polymorphisms, including one in the NOD-like receptor (NLR) family pyrin domain-containing 12 (Nlrp12) gene that leads to an amino acid change in NLRP12, as well as small indels and structural variants (53). C57BL/6J mice exhibit impaired glucose tolerance and have increased lean mass compared with C57BL/6N mice, and control of Listeria monocytogenes infection varied by strain. Furthermore, the activity of splenic natural killer (NK) cells from C57BL/6N mice was reduced compared to that of C57BL/6J cells (53). Taken together, these differences in NLRP12, metabolism, and NK cell activity may help clarify differences we observed between Unc93b−/− and Tlr7−/− Tlr9−/− mice.
We observed route-dependent differences in viral replication in our studies that can be explained by the distribution of TLR and UNC93B expression in the mouse. Levels of full-length TLR9 and UNC93B are greatly reduced in the lung compared to those in the spleen, and cleaved TLR9, required for signaling upon detection of viral DNA, is not detectable in the lung (11, 54). Likewise, TLR7 is strongly expressed in the spleen but not the lung (51). In contrast, mRNA for MAb-21 domain-containing 1, which encodes the PRR cyclic GMP-AMP synthase (cGAS), is expressed similarly in the lung and spleen (55). MHV68 infection of cGAS-deficient mice leads to a 2-fold increase of viral titers in the spleen and a 3.5-fold increase of viral titers in the lung compared to titers in WT mice (56). During primary infection of the lung after intranasal infection, MHV68 replication is thus controlled primarily in a nonendosomal TLR-dependent manner; cGAS is the most likely PRR involved in detection although RIG-I and other non-TLR PRR may also be involved. As all of the mice analyzed in our experiments were TLR deficient, we would not expect to see a difference in levels of primary viral replication in these mice upon intranasal infection. In contrast, upon intravenous infection, the spleen is one of the first sites of viral replication (19). In the spleen, UNC93B and endosomal TLR expression levels are high, leading to an effect of TLR on early viral replication.
At least three publications have examined TLR ligand stimulation of latently infected cell lines and mice (38, 57, 58). In one of these studies, treatment of infected A20 B cell lines with TLR3, -4, -5, or -9 ligands enhanced MHV68 reactivation (57). In a separate study, TLR7 and TLR9 ligand stimulation inhibited reactivation of S11 cells, which are derived from naturally occurring MHV68-induced lymphoproliferation (38). These results likely differ due to the different cell types or establishment of infection in them. In vivo, however, repeated stimulation of MHV68-infected WT mice with the TLR7 ligand R848 increased the number of latently infected splenocytes (38). Likewise, injection of latently infected mice with the TLR9 ligand CpG DNA resulted in increased MHV68 genome copy numbers and enhanced reactivation of MHV68 in an ex vivo reactivation assay (58).
These results regarding TLR stimulation and latency may seem at odds with the increased frequency of latent genome copies we have observed in Tlr7−/− Tlr9−/− mice. However, these are two separate mechanisms. When multiple endosomal TLR are nonfunctional, for example, in Unc93b−/− mice, viral replication after intravenous infection is already increased compared to that in WT mice. The presence of more virus increases latent genome copy numbers of MHV68, which we observed. In WT mice, however, viral replication is controlled at the beginning. Establishment of latency occurs but at a reduced frequency compared to that in Tlr−/− mice. However, when TLR signaling is activated in WT cells with synthetic ligands, latently infected cells are reactivated, allowing increased viral spread, leading to establishment of latency in additional cells.
TLR signaling is initiated via the adaptor protein MyD88, but, as mentioned, whether MyD88 is important for MHV68 detection or control is unclear. In a study that reported no effect on early replication, the authors observed a decreased anti-MHV68 B cell response in Myd88−/− mice and a decreased ability of MHV68 to establish latency in the spleen of Myd88−/− mice after intranasal infection (17). The authors found that this effect was directly related to the absence of MyD88 in B cells and was independent of IL-1R signaling (17). However, after intraperitoneal infection, latent viral loads were not affected. Interestingly, though, reactivation was actually enhanced in Myd88−/− peritoneal exudate cells (PEC), and the authors suggested that MyD88 may be important for suppressing reactivation from latently infected PEC (17). Likewise, we found that two MyD88-dependent TLR, TLR7 and TLR9, may be important for suppressing reactivation but in a different cell type. Further study to understand this apparent discrepancy is required, but differences in infection route and infectious dose are possible explanations.
In summary, two TLR, TLR7 and TLR9, cooperate in recognition of MHV68. This is the first report of the contribution of TLR7 to MHV68 detection and demonstrates the complexity of overlapping and redundant pattern recognition pathways. Further studies on TLR and PRR will likely benefit from analyzing combinations of PRR rather than individual PRR alone.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were performed in compliance with the German animal protection law (TierSchG BGBI S. 1105; 25.05.1998). The mice were handled in accordance with good animal practice as defined by Federation for Laboratory Animal Science Associations (FELASA) and Die Gesellschaft für Versuchstierkunde/Society of Laboratory Animal Science (GV-SOLAS). All animal experiments were approved by the responsible state office (Lower Saxony State Office of Consumer Protection and Food Safety) under permit numbers 33.9-42502-04-12/0930 and 33.19-42502-04-17/2657.
Mice.
WT and KO mice were bred and maintained under specific-pathogen-free conditions at the animal facility of the Helmholtz Centre for Infection Research in Braunschweig, Germany. WT C57BL/6J and C57BL/6NCrl were originally obtained from Charles River Laboratories. For most experiments, WT controls were bred in-house. For some experiments, C57BL/6J Rj mice were purchased from Janvier Labs, and C57BL/6NCrl were purchased from Charles River Laboratories.
Tlr13−/− [C57BL/6N-Tlr13tm1(KOMP)Vlcg] and Unc93b−/− [B6.Unc93b1tm1(KOMP)Vlcg] mice are on the C57BL/6NTac background and were originally obtained as Tlr13−/− ES cells, clone Tlr13_BC7, or Unc93b−/− sperm from the mouse strain derived from ES cell clone 10049A-G9 from the NCRR-NIH-supported mouse strain KOMP Repository (https://www.komp.org) (grant U42-RR024244). VelociGene targeted alleles (50) were generated by Regeneron Pharmaceuticals, Inc., for KOMP. Germline-positive Tlr13−/− mice were backcrossed for two generations onto C57BL/6NTac mice and then interbred and maintained as a homozygous colony. For Unc93b−/− mice, oocytes of C57BL/6NCrl mice were in vitro fertilized. Heterozygous offspring were interbred and maintained as a homozygous colony.
Tlr2−/− (B6.129-Tlr2tm1Kir/J) mice (59) were backcrossed to C57BL/6J mice for 9 generations and were originally obtained from the Jackson Laboratory (004650) (60). Tlr7−/− (B6.129P2-Tlr7tm1Aki) mice (51) were kindly provided by Stefan Bauer (Marburg, Germany) and were backcrossed to C57BL/6J for 10 generations (61). Tlr9−/− (B6.129P2-Tlr9tm1Aki) mice (52), backcrossed to C57BL/6J mice for 12 generations, were kindly provided by Stefan Bauer (Marburg, Germany). Tlr7−/− Tlr9−/− double-knockout mice were bred for this study by crossing Tlr7−/− and Tlr9−/− mice.
Cell lines and cell culture.
Murine bone marrow stromal M210-B4 (ATCC CRL-1972) and murine embryonic fibroblast NIH 3T3 (ATCC CRL-1658) cells were obtained from the American Type Culture Collection (ATCC) and cultured in basal medium of high-glucose Dulbecco’s modified Eagle’s medium (DMEM) containing glutamine, supplemented with 8% fetal calf serum (FCS), 1% penicillin-streptomycin, and, in the case of NIH 3T3 cells, 1 mM sodium pyruvate and 1% nonessential amino acids. B16 Flt-3L cells, expressing FMS-like tyrosine kinase 3 ligand (Flt-3L), were made by H. Chapman (62), and as for primary Flt-3L-induced dendritic cells (FLDC), were cultured in RPMI medium containing glutamine and supplemented with 10% FCS, 1% penicillin-streptomycin, and 50 µM β-mercaptoethanol. All cells were cultured at 37°C in a humidified 7.5% CO2 incubator.
Virus stock preparation.
The MHV68 WUMS strain was from the ATCC (VR-1465). Recombinant viruses MHV68-GFP (63), the MHV68 sncRNA KO lacking all known sncRNAs (ΔmiR) and its revertant (REV) (33), the MHV68 muSOX mutant deficient in host shutoff activity, ΔHS, and its revertant (REV) (34), and MCMV-GFP (64) have all been previously described. Virus stocks were amplified in M210-B4 cells, concentrated, purified over a 10% Nycodenz cushion in virus standard buffer (VSB) (50 mM Tris-HCl, 12 mM KCl, 5 mM Na-EDTA, pH 7.8), resuspended in VSB, and flash frozen as previously described (65–68). Virus stock titers were determined on M210-B4 cells by using the median (50%) tissue culture infective dose (TCID50) for MHV68 strains and by plaque assay for MCMV. Newcastle disease virus LaSota strain (NDV) was kindly provided by Andrea Kröger (Helmholtz Centre for Infection Research, Braunschweig, Germany).
Preparation and sorting of primary FLDC.
To obtain sufficient pDC numbers after magnetically activated cell sorting (MACS) or FACS, bone marrow was combined from multiple mice of the same genotype for all experiments. Briefly, the femur and tibia of mice of the genotypes indicated in the text and figures were cleaned of tissue, and bone marrow was either extruded using a medium-filled syringe equipped with a 25-gauge needle or by centrifugation. After resuspension in RPMI medium, red blood cells were lysed. Cells were cultured for 8 days at 1.5 × 106 cells/ml in complete RPMI medium supplemented with 2.5% conditioned medium from B16 Flt-3L cells. The resulting FLDC were then either seeded for experiments directly or sorted using MACS or FACS into plasmacytoid or conventional dendritic cells (pDC or cDC, respectively). For MACS sorting, pDC were enriched from FLDC using a discontinued negative-selection plasmacytoid dendritic cell isolation kit II (130-092-786; Miltenyi Biotec) according to the manufacturer’s instructions. For FACS sorting, FLDC were labeled with allophycocyanin (APC)-coupled anti-mouse/human CD45R/B220, clone RA3-6B2 (103212; Biolegend), and phycoerythrin (PE)-coupled anti-mouse CD11c, clone N418 (117308; Biolegend), and then sorted on a FACSAria into CD45R/B220+ CD11cmid pDC and CD45R/B220−CD11chigh cDC (expressing high levels of CD11c).
Stimulation of primary DC.
Equal cell numbers were seeded in a 100-µl volume in 96-well plates in each experiment, and then mock treated, infected at an MOI of 0.5 or 2, or stimulated with TLR agonists by addition of 100 µl of the appropriate control or stimulus diluted in FLDC medium as indicated in the text and figures. For TLR9 activation, cells were treated with a 1 µM final concentration CpG DNA ODN 2336. For stimulation of TLR7 with poly(U) (tlrl-sspu; Invivogen), poly(U) was first complexed with dioleoyl trimethylammonium propane (DOTAP) (11811177001; Roche). Two micrograms of poly(U) was diluted to a final volume of 20 µl with HBS. Separately, 12 µl of DOTAP was diluted to 40 µl with HEPES-buffered saline (HBS). Diluted DOTAP and poly(U) were combined, incubated for 15 min, and diluted to a 1-ml total volume with FLDC medium, after which 100 µl was added per well, for a final concentration of 1 µg/ml. Cells were stimulated with TLR ligands for 18 h or with viruses for 24 h, and then supernatants were harvested, clarified, and stored at −20°C for ELISA.
Antiviral treatment of pDC.
FACS-sorted Tlr9−/− pDC were seeded into 96-well plates for ELISA (3 × 105 cells in 150 µl/well) or six-well plates for qRT-PCR and ELISA (3 × 106 cells in 1.5 ml/well). Cells were then pretreated for approximately 3 h with 20 µM acyclovir in DMSO in a 0.1% final DMSO concentration or with 0.1% DMSO as a vehicle control. Acyclovir at 20 µM is approximately 3× the 50% effective concentration (EC50) for MHV68 and about 10× less than the 50% inhibitory concentration (IC50) (69). Cells were then infected with MHV68 or stimulated with DOTAP or poly(U) with the final concentrations described the paragraph “Stimulation of primary DC.”
Cells were incubated for 24 h postinfection or stimulated with a final concentration of 15 µM acyclovir in 0.75% DMSO or with 0.75% DMSO as a vehicle control. Supernatants were harvested, clarified, and stored at −20°C for ELISA. Cells from six-well plates were collected for RNA extraction and qRT-PCR.
IFN-α ELISA
IFN-α levels were measured by ELISA. For most experiments, a rat anti-mouse IFN-α capture antibody (22100-1; PBL), a rabbit anti-mouse IFN-α detection antibody (32100-1; PBL), and a horseradish peroxidase (HRP)-coupled donkey anti-rabbit IgG antibody (711-036-152; Jackson ImmunoResearch) were used with mouse alpha interferon standard (12100-1; PBL) at concentrations between 78 and 5,000 pg/ml. Similar results were obtained with the more sensitive PBL Verikine mouse IFN-α ELISA kit with IFN-α standard concentrations between 12.5 and 400 pg/ml.
Quantitative reverse transcription PCR analysis of viral gene expression.
RNA was isolated using an innuPREP RNA minikit (Analytik Jena). The kit includes a genomic DNA removal column step. RNAs were diluted to equal concentrations, and cDNA was synthesized using an iScript genomic DNA (gDNA) clear cDNA synthesis kit (Bio-Rad). Transcript levels were determined using GoTaq qPCR master mix (Promega) and oligonucleotide pairs specific for ORF50/RTA, ORF68, ORF4, ORF8, ORF65/M9, and 18S rRNA (48, 65). The viral mRNA expression levels were normalized to the level of the 18S rRNA and compared using the 2−ΔΔCT (where CT is threshold cycle) method (70).
In vivo infections and analysis of lytic titers in organs.
Mice were anesthetized with ketamine-xylazine and infected intranasally (i.n.) with 2 × 106 PFU of MHV68 WUMS in 20 µl of phosphate-buffered saline (PBS)/mouse. Alternatively, mice were humanely restrained and infected intravenously (i.v.) with 2 × 106 PFU of MHV68 WUMS in 100 µl of PBS/mouse. At 3 days postinfection, mice were euthanized with CO2. Organs were harvested, weighed, homogenized using FastPrep 24 (MP Biomedicals), and frozen at −70°C. To determine lytic titers, homogenates were thawed and mixed, and titers of clarified supernatants were determined by TCID50 on M210-B4 cells. The limit of detection was 102.10 TCID50s/ml for spleen and lungs and 103.10 TCID50s/ml for liver, based on organ toxicity and the minimum number of positive wells. Data are combined from multiple independent experiments; results for individual mice are shown.
Reactivation assay and determination of latent viral load.
Intravenously infected mice were maintained for 22 days to allow establishment of MHV68 latency, and reactivation assays were performed essentially as described previously (14). On day 21, 10,000 NIH 3T3 cells were plated per well in 96-well plates in 100 µl/well. On day 22, two to four spleens per genotype were harvested, disrupted over 70-µm-pore-size mesh filters, and pooled to produce single-cell suspensions. After red blood cell lysis, splenocytes were serially diluted 3-fold from 1.5 × 106 cells/ml to 690 cells/ml. Twenty-four 100-µl replicates of each dilution were seeded onto NIH 3T3 cells and cocultured for 14 days. Wells were scored for CPE, and the percentage of positive wells was determined. To control for preformed infectious virus, splenocyte dilutions were twice frozen and thawed and plated similarly onto NIH 3T3 cells; values for positive wells were subtracted from the value of the relevant live splenocyte dilution. Reactivation assays were performed at least three independent times. To determine the frequency of reactivation, a variable slope sigmoidal dose-response equation was used with the regression line intersect set to 63.2% based on the Poisson distribution. To determine the latent viral load, DNA was isolated in duplicate from 2 × 107 splenocytes using a modified version of a published protocol (71). First, splenocytes were digested overnight at 56°C with proteinase K in tail lysis buffer (100 mM Tris-HCl, pH 8.5, 200 mM NaCl, 5 mM EDTA, 0.2% SDS, supplemented with 5 µl 20 mg/ml proteinase K per ml buffer), followed by isopropanol precipitation. DNA was then wound around a pipette tip, transferred to a fresh tube, dried, and resuspended in 500 µl of buffer (10 mM Tris-HCl, 10 mM EDTA, pH 8.0). Fifty nanograms of DNA was then used to determine copy numbers, usually in duplicate, for MHV68 gB and murine β2M by qPCR using the Roche Universal Probe Library and specific primers (MHV68 gB, CGTATGTCAGCCAACCACTCT and AGTCTCTGTTGGAGCGTCCT and probe 46; β2M, GTGGTGCCAGCAGAGACTTA and GGACAGTGGGTAGGGAACTG and probe 40). gB copy numbers were normalized to those of β2M for each sample and then scaled to the average of the appropriate control. Per pooled splenocyte sample analyzed for reactivation, three to four qPCRs were analyzed. Each data point represents one qPCR.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism. Significance for ELISA data and MHV68 genome copy numbers was determined by a two-tailed unpaired t test of log-transformed data. For weight loss and viral titers in organs, significance was determined by a two-tailed Mann-Whitney test.
ACKNOWLEDGMENTS
We thank the HZI Central Animal Facility and all animal caretakers, as well as Lothar Gröbe and the HZI Flow Cytometry and Cell Sorting Platform. We thank Christine Standfuß-Gabisch and Georg Wolf for excellent technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft grants CRC900 and BR 3432/3-1 and Helmholtz Association grant VH-NG-637 (M.M.B.), by intramural grants from the Helmholtz Association (Program Infection and Immunity) and the grant Infection Challenge in the German Mouse Clinic from the German Ministry of Education and Research (K.S.), and by NIH grant R01CA136367 (B.G.). This work has been carried out within the framework of the SMART BIOTECS alliance between the Technische Universität Braunschweig and the Leibniz Universität Hannover, an initiative that is supported by the Ministry of Science and Culture (MWK) of Lower Saxony, Germany.
REFERENCES
- 1.Blaskovic DS M, Svobodova J, Kresakova J. 1980. Isolation of five strains of herpesvirus from two species of free living small rodents. Acta Virologica 28:225–231. [PubMed] [Google Scholar]
- 2.Barton E, Mandal P, Speck SH. 2011. Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annu Rev Immunol 29:351–397. doi: 10.1146/annurev-immunol-072710-081639. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Shepardson KM, Schwarz B, Larson K, Morton RV, Avera J, McCoy K, Caffrey A, Harmsen A, Douglas T, Rynda-Apple A. 2017. Induction of antiviral immune response through recognition of the repeating subunit pattern of viral capsids is Toll-like receptor 2 dependent. mBio 8:e01356-17. doi: 10.1128/mBio.01356-17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 6.Li XD, Chen ZJ. 2012. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. Elife 1:e00102. doi: 10.7554/eLife.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Z, Cai Z, Sanchez A, Zhang T, Wen S, Wang J, Yang J, Fu S, Zhang D. 2011. A novel Toll-like receptor that recognizes vesicular stomatitis virus. J Biol Chem 286:4517–4524. doi: 10.1074/jbc.M110.159590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, Shamel L, Herskovits AA, Portnoy DA, Cooke M, Tarantino LM, Wiltshire T, Steinberg BE, Grinstein S, Beutler B. 2006. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol 7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 9.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. 2007. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Physiol 177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. 2008. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 11.Avalos AM, Kirak O, Oelkers JM, Pils MC, Kim YM, Ottinger M, Jaenisch R, Ploegh HL, Brinkmann MM. 2013. Cell-specific TLR9 trafficking in primary APCs of transgenic TLR9-GFP mice. J Immunol 190:695–702. doi: 10.4049/jimmunol.1202342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelka K, Bertheloot D, Reimer E, Phulphagar K, Schmidt SV, Christ A, Stahl R, Watson N, Miyake K, Hacohen N, Haas A, Brinkmann MM, Marshak-Rothstein A, Meissner F, Latz E. 2018. The chaperone UNC93B1 regulates Toll-like receptor stability independently of endosomal TLR transport. Immunity 48:911–922 e7. doi: 10.1016/j.immuni.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michaud F, Coulombe F, Gaudreault E, Kriz J, Gosselin J. 2010. Involvement of TLR2 in recognition of acute gammaherpesvirus-68 infection. PLoS One 5:e13742. doi: 10.1371/journal.pone.0013742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guggemoos S, Hangel D, Hamm S, Heit A, Bauer S, Adler H. 2008. TLR9 contributes to antiviral immunity during gammaherpesvirus infection. J Immunol 180:438–443. doi: 10.4049/jimmunol.180.1.438. [DOI] [PubMed] [Google Scholar]
- 15.West JA, Gregory SM, Damania B. 2012. Toll-like receptor sensing of human herpesvirus infection. Front Cell Infect Microbiol 2:122. doi: 10.3389/fcimb.2012.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway CA. Jr, 1998. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell 2:253–258. doi: 10.1016/S1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 17.Gargano LM, Moser JM, Speck SH. 2008. Role for MyD88 signaling in murine gammaherpesvirus 68 latency. J Virol 82:3853–3863. doi: 10.1128/JVI.02577-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naik SH, Corcoran LM, Wu L. 2005. Development of murine plasmacytoid dendritic cell subsets. Immunol Cell Biol 83:563–570. doi: 10.1111/j.1440-1711.2005.01390.x. [DOI] [PubMed] [Google Scholar]
- 19.Sunil-Chandra NP, Efstathiou S, Arno J, Nash AA. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J Gen Virol 73:2347–2356. doi: 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- 20.Milho R, Smith CM, Marques S, Alenquer M, May JS, Gillet L, Gaspar M, Efstathiou S, Simas JP, Stevenson PG. 2009. In vivo imaging of murid herpesvirus-4 infection. J Gen Virol 90:21–32. doi: 10.1099/vir.0.006569-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aligo J, Brosnan K, Walker M, Emmell E, Mikkelsen SR, Burleson GR, Burleson FG, Volk A, Weinstock D. 2015. Murine gammaherpesvirus-68 (MHV-68) is not horizontally transmitted amongst laboratory mice by cage contact. J Immunotoxicol 12:330–341. doi: 10.3109/1547691X.2014.980020. [DOI] [PubMed] [Google Scholar]
- 22.Francois S, Vidick S, Sarlet M, Desmecht D, Drion P, Stevenson PG, Vanderplasschen A, Gillet L. 2013. Illumination of murine gammaherpesvirus-68 cycle reveals a sexual transmission route from females to males in laboratory mice. PLoS Pathog 9:e1003292. doi: 10.1371/journal.ppat.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vrbova M, Belvoncikova P, Kovalova A, Matuskova R, Slovak M, Kudelova M. 2016. Molecular detection of murine gammaherpesvirus 68 (MHV-68) in Haemaphysalis concinna ticks collected in Slovakia. Acta Virol 60:426–428. doi: 10.4149/av_2016_04_426. [DOI] [PubMed] [Google Scholar]
- 24.Hajnicka V, Kudelova M, Stibraniova I, Slovak M, Bartikova P, Halasova Z, Pancik P, Belvoncikova P, Vrbova M, Holikova V, Hails RS, Nuttall PA. 2017. Tick-borne transmission of murine gammaherpesvirus 68. Front Cell Infect Microbiol 7:458. doi: 10.3389/fcimb.2017.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cieniewicz B, Santana AL, Minkah N, Krug LT. 2016. Interplay of murine gammaherpesvirus 68 with NF-κB signaling of the host. Front Microbiol 7:1202. doi: 10.3389/fmicb.2016.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flano E, Husain SM, Sample JT, Woodland DL, Blackman MA. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J Immunol 165:1074–1081. doi: 10.4049/jimmunol.165.2.1074. [DOI] [PubMed] [Google Scholar]
- 27.Flano E, Woodland DL, Blackman MA. 1999. Requirement for CD4+ T cells in Vβ4+ CD8+ T cell activation associated with latent murine gammaherpesvirus infection. J Immunol 163:3403–3408. [PubMed] [Google Scholar]
- 28.Weck KE, Kim SS, Virgin HI, Speck SH. 1999. B cells regulate murine gammaherpesvirus 68 latency. J Virol 73:4651–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. 2005. Identification of microRNAs of the herpesvirus family. Nat Methods 2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 30.Reese TA, Xia J, Johnson LS, Zhou X, Zhang W, Virgin HW. 2010. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J Virol 84:10344–10353. doi: 10.1128/JVI.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu JY, Strehle M, Frohn A, Kremmer E, Hofig KP, Meister G, Adler H. 2010. Identification and analysis of expression of novel microRNAs of murine gammaherpesvirus 68. J Virol 84:10266–10275. doi: 10.1128/JVI.01119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. 2010. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell 37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steer B, Strehle M, Sattler C, Bund D, Flach B, Stoeger T, Haas JG, Adler H. 2016. The small noncoding RNAs (sncRNAs) of murine gammaherpesvirus 68 (MHV-68) are involved in regulating the latent-to-lytic switch in vivo. Sci Rep 6:32128. doi: 10.1038/srep32128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richner JM, Clyde K, Pezda AC, Cheng BY, Wang T, Kumar GR, Covarrubias S, Coscoy L, Glaunsinger B. 2011. Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency. PLoS Pathog 7:e1002150. doi: 10.1371/journal.ppat.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilliet M, Cao W, Liu YJ. 2008. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol 8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 36.Dalod M, Hamilton T, Salomon R, Salazar-Mather TP, Henry SC, Hamilton JD, Biron CA. 2003. Dendritic cell responses to early murine cytomegalovirus infection: subset functional specialization and differential regulation by interferon alpha/beta. J Exp Med 197:885–898. doi: 10.1084/jem.20021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pezda AC, Penn A, Barton GM, Coscoy L. 2011. Suppression of TLR9 immunostimulatory motifs in the genome of a gammaherpesvirus. J Immunol 187:887–896. doi: 10.4049/jimmunol.1003737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas F, Yamauchi K, Murat M, Bernasconi M, Yamanaka N, Speck RF, Nadal D. 2014. Activation of NF-kappaB via endosomal Toll-like receptor 7 (TLR7) or TLR9 suppresses murine herpesvirus 68 reactivation. J Virol 88:10002–10012. doi: 10.1128/JVI.01486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zucchini N, Bessou G, Traub S, Robbins SH, Uematsu S, Akira S, Alexopoulou L, Dalod M. 2008. Cutting edge: Overlapping functions of TLR7 and TLR9 for innate defense against a herpesvirus infection. J Immunol 180:5799–5803. doi: 10.4049/jimmunol.180.9.5799. [DOI] [PubMed] [Google Scholar]
- 40.West JA, Gregory SM, Sivaraman V, Su L, Damania B. 2011. Activation of plasmacytoid dendritic cells by Kaposi’s sarcoma-associated herpesvirus. J Virol 85:895–904. doi: 10.1128/JVI.01007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory SM, West JA, Dillon PJ, Hilscher C, Dittmer DP, Damania B. 2009. Toll-like receptor signaling controls reactivation of KSHV from latency. Proc Natl Acad Sci U S A 106:11725–11730. doi: 10.1073/pnas.0905316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quan TE, Roman RM, Rudenga BJ, Holers VM, Craft JE. 2010. Epstein-Barr virus promotes interferon-alpha production by plasmacytoid dendritic cells. Arthritis Rheum 62:1693–1701. doi: 10.1002/art.27408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiola S, Gosselin D, Takada K, Gosselin J. 2010. TLR9 contributes to the recognition of EBV by primary monocytes and plasmacytoid dendritic cells. J Immunol 185:3620–3631. doi: 10.4049/jimmunol.0903736. [DOI] [PubMed] [Google Scholar]
- 44.Nejad C, Stunden HJ, Gantier MP. 2018. A guide to miRNAs in inflammation and innate immune responses. FEBS J doi: 10.1111/febs.14482. [DOI] [PubMed] [Google Scholar]
- 45.Kincaid RP, Sullivan CS. 2012. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog 8:e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glaunsinger B, Chavez L, Ganem D. 2005. The exonuclease and host shutoff functions of the SOX protein of Kaposi’s sarcoma-associated herpesvirus are genetically separable. J Virol 79:7396–7401. doi: 10.1128/JVI.79.12.7396-7401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rowe M, Glaunsinger B, van Leeuwen D, Zuo J, Sweetman D, Ganem D, Middeldorp J, Wiertz EJ, Ressing ME. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc Natl Acad Sci U S A 104:3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abernathy E, Clyde K, Yeasmin R, Krug LT, Burlingame A, Coscoy L, Glaunsinger B. 2014. Gammaherpesviral gene expression and virion composition are broadly controlled by accelerated mRNA degradation. PLoS Pathog 10:e1003882. doi: 10.1371/journal.ppat.1003882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez-Guzman D, Rickabaugh T, Wu TT, Brown H, Cole S, Song MJ, Tong L, Sun R. 2003. Transcription program of murine gammaherpesvirus 68. J Virol 77:10488–10503. doi: 10.1128/JVI.77.19.10488-10503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, Chernomorsky R, Boucher M, Elsasser AL, Esau L, Zheng J, Griffiths JA, Wang X, Su H, Xue Y, Dominguez MG, Noguera I, Torres R, Macdonald LE, Stewart AF, DeChiara TM, Yancopoulos GD. 2003. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol 21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 51.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. 2002. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 52.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 53.Simon MM, Greenaway S, White JK, Fuchs H, Gailus-Durner V, Wells S, Sorg T, Wong K, Bedu E, Cartwright EJ, Dacquin R, Djebali S, Estabel J, Graw J, Ingham NJ, Jackson IJ, Lengeling A, Mandillo S, Marvel J, Meziane H, Preitner F, Puk O, Roux M, Adams DJ, Atkins S, Ayadi A, Becker L, Blake A, Brooker D, Cater H, Champy M-F, Combe R, Danecek P, di Fenza A, Gates H, Gerdin A-K, Golini E, Hancock JM, Hans W, Hölter SM, Hough T, Jurdic P, Keane TM, Morgan H, Müller W, Neff F, Nicholson G, Pasche B, Roberson L-A, Rozman J, Sanderson M, et al. 2013. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol 14:R82. doi: 10.1186/gb-2013-14-7-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, Arora M, Yarlagadda M, Oriss TB, Krishnamoorthy N, Ray A, Ray P. 2006. Distinct responses of lung and spleen dendritic cells to the TLR9 agonist CpG oligodeoxynucleotide. J Immunol 177:2373–2383. doi: 10.4049/jimmunol.177.4.2373. [DOI] [PubMed] [Google Scholar]
- 55.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams BA, Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See L-H, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, Lagarde J, Bussotti G, Tanzer A, Denas O, Li K, Bender MA, Zhang M, Byron R, Groudine MT, McCleary D, Pham L, Ye Z, Kuan S, Edsall L, Wu Y-C, Rasmussen MD, Bansal MS, Kellis M, Keller CA, Morrissey CS, Mishra T, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gargano LM, Forrest JC, Speck SH. 2009. Signaling through Toll-like receptors induces murine gammaherpesvirus 68 reactivation in vivo. J Virol 83:1474–1482. doi: 10.1128/JVI.01717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ptaschinski C, Wilmore J, Fiore N, Rochford R. 2010. In vivo activation of toll-like receptor-9 induces an age-dependent abortive lytic cycle reactivation of murine gammaherpesvirus-68. Viral Immunol 23:547–555. doi: 10.1089/vim.2010.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Werts C, Tapping RI, Mathison JC, Chuang TH, Kravchenko V, Saint Girons I, Haake DA, Godowski PJ, Hayashi F, Ozinsky A, Underhill DM, Kirschning CJ, Wagner H, Aderem A, Tobias PS, Ulevitch RJ. 2001. Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat Immunol 2:346–352. doi: 10.1038/86354. [DOI] [PubMed] [Google Scholar]
- 60.Loof TG, Goldmann O, Medina E. 2008. Immune recognition of Streptococcus pyogenes by dendritic cells. Infect Immun 76:2785–2792. doi: 10.1128/IAI.01680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stegemann-Koniszewski S, Behrens S, Boehme JD, Hochnadel I, Riese P, Guzman CA, Kroger A, Schreiber J, Gunzer M, Bruder D. 2018. Respiratory influenza A virus infection triggers local and systemic natural killer cell activation via Toll-Like receptor 7. Front Immunol 9:245. doi: 10.3389/fimmu.2018.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi GP, Villadangos JA, Dranoff G, Small C, Gu L, Haley KJ, Riese R, Ploegh HL, Chapman HA. 1999. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity 10:197–206. doi: 10.1016/S1074-7613(00)80020-5. [DOI] [PubMed] [Google Scholar]
- 63.Adler H, Messerle M, Wagner M, Koszinowski UH. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol 74:6964–6974. doi: 10.1128/JVI.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mathys S, Schroeder T, Ellwart J, Koszinowski UH, Messerle M, Just U. 2003. Dendritic cells under influence of mouse cytomegalovirus have a physiologic dual role: to initiate and to restrict T cell activation. J Infect Dis 187:988–999. doi: 10.1086/368094. [DOI] [PubMed] [Google Scholar]
- 65.Bussey KA, Lau U, Schumann S, Gallo A, Osbelt L, Stempel M, Arnold C, Wissing J, Gad HH, Hartmann R, Brune W, Jansch L, Whitehouse A, Brinkmann MM. 2018. The interferon-stimulated gene product oligoadenylate synthetase-like protein enhances replication of Kaposi’s sarcoma-associated herpesvirus (KSHV) and interacts with the KSHV ORF20 protein. PLoS Pathog 14:e1006937. doi: 10.1371/journal.ppat.1006937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan B, Gonçalves Magalhães V, Lemmermann NAW, Juranić Lisnić V, Stempel M, Bussey KA, Reimer E, Podlech J, Lienenklaus S, Reddehase MJ, Jonjić S, Brinkmann MM. 2017. The murine cytomegalovirus M35 protein antagonizes type I IFN induction downstream of pattern recognition receptors by targeting NF-kappaB mediated transcription. PLoS Pathog 13:e1006382. doi: 10.1371/journal.ppat.1006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bussey KA, Reimer E, Todt H, Denker B, Gallo A, Konrad A, Ottinger M, Adler H, Sturzl M, Brune W, Brinkmann MM. 2014. The gammaherpesviruses Kaposi’s sarcoma-associated herpesvirus and murine gammaherpesvirus 68 modulate the Toll-like receptor-induced proinflammatory cytokine response. J Virol 88:9245–9259. doi: 10.1128/JVI.00841-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scheibe E, Lienenklaus S, May T, Magalhaes VG, Weiss S, Brinkmann MM. 2013. Measurement of mouse cytomegalovirus-induced interferon-beta with immortalized luciferase reporter cells. Methods Mol Biol 1064:355–366. doi: 10.1007/978-1-62703-601-6_25. [DOI] [PubMed] [Google Scholar]
- 69.Neyts J, De Clercq E. 1998. In vitro and in vivo inhibition of murine gamma herpesvirus 68 replication by selected antiviral agents. Antimicrob Agents Chemother 42:170–172. doi: 10.1128/AAC.42.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 71.Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res 19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]