Coevolution of multicellular organisms and their natural viruses may lead to an intricate relationship in which host survival requires effective immunity and virus survival depends on evasion of such responses. Insect antiviral immunity and reciprocal virus immunosuppression tactics have been well studied in Drosophila melanogaster, primarily during RNA, but not DNA, virus infection. Therefore, we describe interactions between a recently isolated Drosophila DNA virus (Kallithea virus [KV]) and immune processes known to control RNA viruses, such as RNA interference (RNAi) and Imd pathways. We found that KV suppresses the Toll pathway and identified gp83 as a KV-encoded protein that underlies this suppression. This immunosuppressive ability is conserved in another nudivirus, suggesting that the Toll pathway has conserved antiviral activity against DNA nudiviruses, which have evolved suppressors in response. Together, these results indicate that DNA viruses induce and suppress NF-κB responses, and they advance the application of KV as a model to study insect immunity.
KEYWORDS: innate immunity, Drosophila melanogaster, immune suppression, NF-κB, RNA interference
ABSTRACT
Interactions between the insect immune system and RNA viruses have been extensively studied in Drosophila, in which RNA interference, NF-κB, and JAK-STAT pathways underlie antiviral immunity. In response to RNA interference, insect viruses have convergently evolved suppressors of this pathway that act by diverse mechanisms to permit viral replication. However, interactions between the insect immune system and DNA viruses have received less attention, primarily because few Drosophila-infecting DNA virus isolates are available. In this study, we used a recently isolated DNA virus of Drosophila melanogaster, Kallithea virus (KV; family Nudiviridae), to probe known antiviral immune responses and virus evasion tactics in the context of DNA virus infection. We found that fly mutants for RNA interference and immune deficiency (Imd), but not Toll, pathways are more susceptible to Kallithea virus infection. We identified the Kallithea virus-encoded protein gp83 as a potent inhibitor of Toll signalling, suggesting that Toll mediates antiviral defense against Kallithea virus infection but that it is suppressed by the virus. We found that Kallithea virus gp83 inhibits Toll signalling through the regulation of NF-κB transcription factors. Furthermore, we found that gp83 of the closely related Drosophila innubila nudivirus (DiNV) suppresses D. melanogaster Toll signalling, suggesting an evolutionarily conserved function of Toll in defense against DNA viruses. Together, these results provide a broad description of known antiviral pathways in the context of DNA virus infection and identify the first Toll pathway inhibitor in a Drosophila virus, extending the known diversity of insect virus-encoded immune inhibitors.
IMPORTANCE Coevolution of multicellular organisms and their natural viruses may lead to an intricate relationship in which host survival requires effective immunity and virus survival depends on evasion of such responses. Insect antiviral immunity and reciprocal virus immunosuppression tactics have been well studied in Drosophila melanogaster, primarily during RNA, but not DNA, virus infection. Therefore, we describe interactions between a recently isolated Drosophila DNA virus (Kallithea virus [KV]) and immune processes known to control RNA viruses, such as RNA interference (RNAi) and Imd pathways. We found that KV suppresses the Toll pathway and identified gp83 as a KV-encoded protein that underlies this suppression. This immunosuppressive ability is conserved in another nudivirus, suggesting that the Toll pathway has conserved antiviral activity against DNA nudiviruses, which have evolved suppressors in response. Together, these results indicate that DNA viruses induce and suppress NF-κB responses, and they advance the application of KV as a model to study insect immunity.
INTRODUCTION
Innate antiviral immunity in insects has been best studied in response to RNA virus infections of Drosophila melanogaster. Antiviral immune mechanisms that target RNA viruses include RNA-mediated defenses such as RNA interference (RNAi) and RNA decay pathways, cellular defenses such as apoptosis, phagocytosis, and autophagy, and other effectors of resistance and tolerance that are transcriptionally induced following infection. The latter are primarily mediated by Janus kinase/signal transducers and activators of transcription (JAK-STAT) and nuclear factor κB (NF-κB) pathways (reviewed in references 1–5).
The insect response to DNA viruses is less well studied, but RNAi and apoptosis have demonstrated antiviral activity (6–8) and the JAK-STAT pathway is active during infection, possibly mediating a tolerance response (9). Baculovirus, nudivirus, and iridovirus infections of Drosophila all give rise to virus-derived small interfering RNA (vsiRNAs), which regulate DNA virus gene expression (7, 8, 10, 11), and mutants for RNAi effectors Dicer-2 (Dcr-2) and Argonaute-2 (AGO2) are hypersensitive to invertebrate iridescent virus 6 (IIV6; an iridovirus) infection. This suggests that RNAi is also an important defense against DNA viruses, and IIV6 correspondingly encodes a suppressor of RNAi (7, 12). Virus-encoded suppressors of apoptosis are also widespread in DNA viruses, acting through binding and inhibition of cellular caspases (e.g., p35) or stabilization of cellular inhibitors of apoptosis (e.g., the IAP gene family [13–15]). In contrast, the contribution of transcriptional responses, such as the NF-κB pathways, to DNA viruses has not yet been elucidated.
There are two NF-κB pathways in Drosophila: Toll and Imd, which primarily function in antibacterial (Toll, Gram positive, and Imd, Gram negative) and antifungal (Toll) defense, although both provide protection against some RNA viruses (reviewed in references 1, 4, 5, 16, and 17). The Toll and Imd pathways are activated following recognition of a pathogen-associated molecular patterns (PAMP; e.g., bacterial peptidoglycan), leading to the phosphorylation and degradation of the inhibitor of kappa B (IκB; encoded by cactus for Toll signalling and by the relish C terminus in Imd signalling) (reviewed in references 16 and 17). Under nonsignalling conditions, IκB sequesters NF-κB transcription factors in the cytoplasm. These transcription factors are encoded by dorsal (dl) and Dorsal immune-related factor (Dif) in Toll signalling and Relish (Rel) in Imd signalling, and all translocate to the nucleus to induce gene expression following IκB degradation (reviewed in references 16 and 17). Although the mechanism by which Toll and Imd recognize RNA viruses is unclear, both are active and provide immunity against some viral infections in insects, most likely through induction of antiviral effector responses. For example, Toll is broadly antiviral against RNA viruses such as Drosophila C virus, Nora virus, and Flock House virus in Drosophila during orally acquired, but not systemic, infections and in Aedes mosquitoes against dengue virus (18–21). Additionally, Imd is antiviral against a subset of viruses in Drosophila, such as cricket paralysis virus, Drosophila C virus, and Sindbis virus and in Aedes cell culture against the alphaviruses Semliki Forest virus and O’nyong’nyong virus (22–26).
Although the effect of NF-κB signalling on DNA virus infection in insects has not been directly tested, polydnaviruses, ascoviruses, baculoviruses, and entomopoxviruses have acquired suppressors of NF-κB signalling by horizontal gene transfer, providing indirect evidence for anti-DNA virus activity of NF-κB pathways (27, 28). First, a polydnavirus encoded in the genome of the Braconid parasitoid wasp Microplitis demolitor has acquired homologs of IκB, some of which inhibit Dif and Rel by direct binding (27). However, this is a domesticated endogenous viral element that forms viral particles injected into the parasitoid’s host, and as these IκB homologs are not found in related nudiviruses, baculoviruses, or hytrosaviruses, it seems likely that they were acquired to inhibit antiparasitoid immune responses in the host of the parasitoid wasp, rather than the antiviral immune response of the wasp itself (29, 30). Second, homologs of diedel, which encode a cytokine that inhibits apoptosis and the Imd pathway in Drosophila, are similarly found in ascoviruses, baculoviruses, and entomopoxviruses, likely through independent horizontal transfer from arthropod hosts (28). Virus-encoded diedel phenocopies fly-encoded diedel, suggesting that viral diedel has retained an Imd-suppressive function and that the Imd pathway likely interacts with these DNA viruses (28, 31). However, it is still unclear whether antiviral Toll signalling is targeted by insect virus-encoded immunosuppressors and whether these hijacked host pathway inhibitors represent a subset of a greater diversity of NF-κB immune inhibitors or reflect evasion of virus-specific immune mechanisms.
The recent isolation of Kallithea virus (KV) (11, 32), a nudivirus that naturally infects Drosophila melanogaster at high prevalence in the wild, provides a tractable system to study host-DNA virus interactions and to identify immune evasion strategies in DNA viruses. Nudiviruses are large double-stranded DNA (dsDNA) viruses (100 to 200 kb, including roughly 100 to 150 genes) that most often infect the arthropod midgut and fat body and are transmitted fecal-orally (33–39). Because some virus-encoded immunosuppressors have been found to be highly host specific, the use of native host-virus pairs is vital to our understanding of viral immune evasion (for examples, see references 40–45). In this study, we used this system to analyze the interaction between antiviral immune pathways and a DNA virus in Drosophila. Using mutant fly lines, we found that the RNAi and Imd pathways mediate antiviral protection against KV in vivo but that abrogation of Toll signalling has no effect on virus replication. Through reanalysis of previous RNA sequencing data, we observed a broad downregulation of NF-κB-responsive antimicrobial peptides following KV infection and performed a small-scale screen for KV-encoded immune inhibitors. We identified viral protein gp83 as having a complex interaction with NF-κB signalling, leading to induction of Imd signalling but potent suppression of Toll signalling. This suppression acts directly through, or downstream of, NF-κB transcription factors. Finally, through analysis of the related Drosophila innubila nudivirus (DiNV) gp83 ortholog, we showed that the immunosuppressive activity of gp83 against D. melanogaster NF-κB signalling is conserved.
(This article was submitted to an online preprint archive [46].)
RESULTS AND DISCUSSION
The RNAi and Imd pathways are antiviral against KV in vivo.
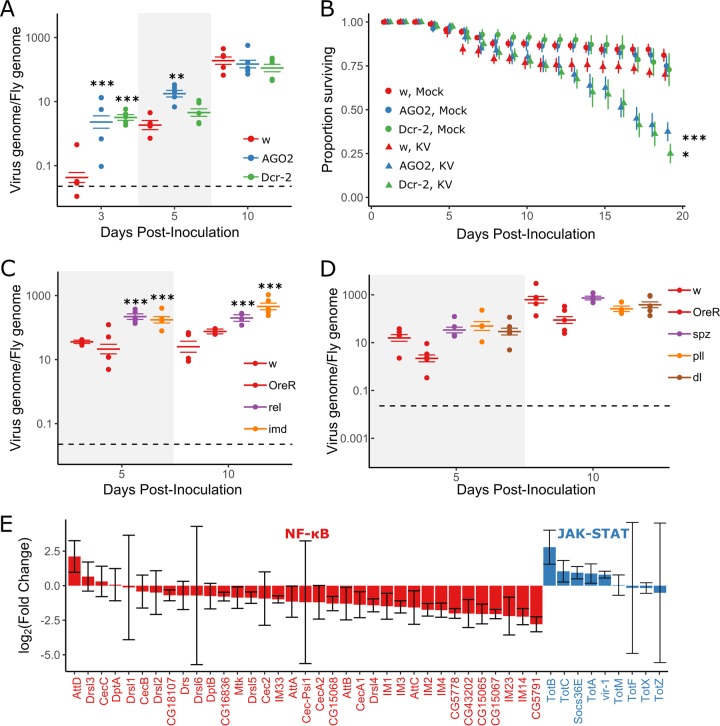
The RNAi pathway provides antiviral activity against the DNA virus IIV6, and KV-derived vsiRNAs are produced upon infection of adult naturally infected Drosophila (7, 11, 12). However, the contributions of the Imd and Toll pathways to anti-DNA virus immunity have not been described. We used fly lines mutant for RNAi, Imd, and Toll pathway components to assess whether these pathways fulfill an antiviral function during KV infection. First, we infected mutants for RNAi genes Dcr-2 and AGO2 with KV and measured viral titer and mortality following infection. Following KV infection, both Dcr-2 and AGO2 mutants exhibited significantly greater KV titers at 3 days postinfection (dpi), with KV titers 78-fold greater in Dcr-2 mutants (95% highest posterior density [HPD] intervals, 18- to 281-fold; P value as determined by MCMCglmm [MCMCp] < 0.001) and 55-fold greater in AGO2 mutants (13- to 237-fold, MCMCp < 0.001 [Fig. 1A]). However, the increased KV replication in RNAi mutants was not sustained at later infection time points. At 5 dpi, Dcr-2 mutants did not have significantly different KV titers from the controls (MCMCp = 0.22), but titers were still increased in AGO2 mutants, albeit to a lesser extent that at 3 dpi (12-fold increase; 2.5- to 43-fold; MCMCp < 0.001 [Fig. 1A]). By 10 dpi, there was no significant difference between viral titer in control flies and either Dcr-2 mutants (MCMCp = 0.43) or AGO2 mutants (MCMCp = 0.7). Therefore, either the antiviral effect of RNAi is short-lived (for example, a viral suppressor of RNAi may eventually be expressed in vivo), other immune pathways take over as the dominant antiviral force, or KV negatively regulates its own replication or depletes a resource. Nevertheless, despite the similar titers during late infection, there was still a significant increase in KV-induced mortality in Dcr-2 and AGO2 mutants, where 70% of control flies were alive at 19 dpi, compared to 25% in Dcr-2 mutants (MCMCp = 0.014) and 38% in AGO2 mutants (MCMCp = 0.004) (Fig. 1B). Increased late life mortality in RNAi mutants could be due to early host damage or to increased expression of virulence factors throughout infection, expression of which could be regulated by RNAi, independent of KV titer (for an example, see reference 10). These results extend the antiviral role of the RNAi pathway to KV infection.
FIG 1.
RNAi and Imd pathways provide antiviral defense against Kallithea virus. Mutants for RNAi (A and B) and NF-κB (C and D) pathways were assayed for viral titer (A, C, and D) and mortality (B) following KV infection. Oregon R (OreR) and w1118 flies were used as wild-type controls. Viral titer was measured by qPCR, relative to Rpl32 DNA, where each data point represents a vial of 5 flies, and colored horizontal lines correspond to the mean titer and associated SE (A, C, and D). Horizontal dotted lines (A, C, and D) represent the amount of virus injected. (B) RNAi mutants (AGO2 and Dcr2) and w1118 controls were injected with chloroform-treated KV (mock) or KV, and survival was monitored each day. Each point is the mean number of surviving flies across 10 vials of 10 flies, with associated standard errors. (E) Log-transformed fold changes of presumed NF-κB-responsive genes (colored red; Cecropin, Diptericin, Attacin, Metchnikowin, Drosomycin and Drosomycin-like genes, Bomamins [i.e., IM1, CG18107, IM2, IM3, CG15065, CG15068, CG43202, CG16836, CG5778, IM23, CG15067, and CG5791)], and other IM genes) and JAK-STAT-responsive genes (colored blue; Socs36E, vir-1, and Turandot [Tot] family) following KV infection of OreR flies at 3 dpi, relative to uninfected controls (ERP023609; n = 5 libraries per treatment, with n = 10 flies per library [32]). Error bars show SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (statistical tests were performed in MCMCglmm).
We next infected Imd and Toll pathway mutants with KV and assessed KV DNA levels by quantitative PCR (qPCR) at 5 and 10 dpi. We found that Imd pathway mutants had significantly greater viral titers than two control lines, with imd mutants having 6-fold-greater KV titers at 5 and 10 dpi (2.7- to 13.7-fold; MCMCp < 0.001) and Rel mutants having 8-fold-greater viral titers at 5 and 10 dpi (3.1- to 15.9-fold; MCMCp < 0.001 [Fig. 1C]). Because the Imd effect spans 5 and 10 dpi, and we have previously measured KV titers in 125 inbred lines of the Drosophila Genetic Reference Panel at 8 dpi (32, 47), we attempted to account for genetic background by comparing the average effects of Imd mutants to the distribution of effects consistent with natural variation in the genetic background. This analysis indicated that the increased titer observed in Imd mutants is unlikely to be due to genetic background (P = 0.01). We also infected flies mutant for the Toll pathway components spz, pll, and dl. Viral titer was unchanged in Toll pathway mutants compared to controls, and the pathway-level effect of Toll mutants was within the expected distribution of effects caused by differences in genetic background (P = 0.28). We conclude that the Imd pathway is antiviral against KV but that abrogation of Toll function has no effect on KV growth. This could indicate that Toll is not antiviral against this DNA virus or that the pathway is efficiently suppressed by virus infection. The latter is consistent with our observation that genes encoding antimicrobial peptides are generally downregulated in KV-infected flies compared to uninfected controls (Fig. 1E), and we therefore explored the capability of KV to suppress innate immune pathways using a cell culture model of immunosuppression.
KV replicates efficiently in some Drosophila cell lines.
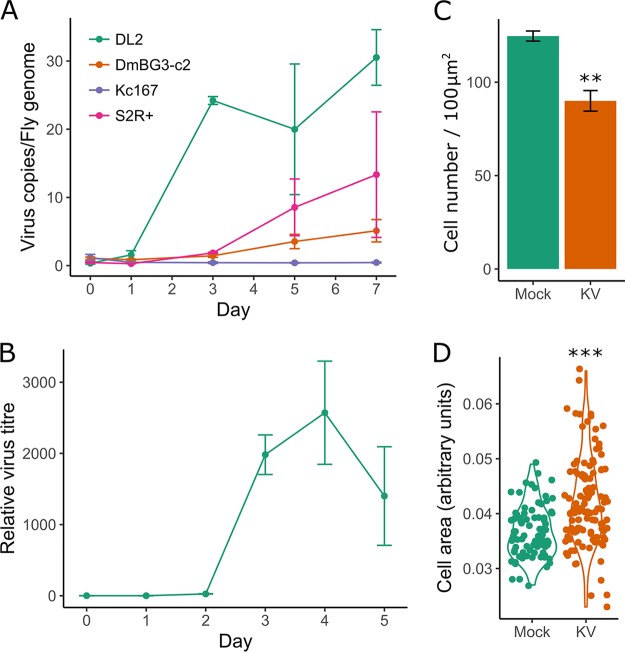
To establish a cell culture model for KV infection, we analyzed viral replication in five commonly used D. melanogaster cell lines. We found variation in the ability of KV to infect these cells, with efficient replication in several Drosophila S2 cell clones, including S2 (data not shown), S2R+, and DL2 cells, but no or inefficient replication in Kc167 and Dm-BG3-c2 cells (Fig. 2A). In S2 cells, which we used for further analyses, KV was released into the medium at substantial levels starting from 3 dpi (Fig. 2B). Therefore, in all subsequent experiments, we assayed cells at 4 dpi, assuming that a high proportion of cells would be infected at this time point. We did not observe any overt cytopathic effects of KV-infected cells within 14 days of infection. However, when KV-infected cells were passaged at 7 dpi, we observed larger (MCMCp < 0.001) and fewer (MCMCp < 0.001) cells, likely due to a decrease in cell proliferation (Fig. 2C and D).
FIG 2.
(A) KV replicates in cell culture. KV growth was assessed in various D. melanogaster cell lines by qPCR against the KV genome, relative to the fly gene Rpl32 (n = 3 for each time point). (B) KV release from S2 cells into the culture medium was assessed by DNA extraction of 50 μl of culture medium and qPCR against the KV genome, plotted relative to the amount of KV in the medium directly following infection (i.e., zero time point is equal to 1). (C) Cell density (number of cells per approximately 100 μm2 in KV versus mock-treated cells) at 10 dpi (n = 3). (D) Size of mock- or KV-infected cells at 10 dpi. Each dot represents a single cell, and the data distribution is presented as a violin plot. Error bars show SEMs.
KV leads to downregulation of JAK-STAT and Toll and induction of Imd signalling in cell culture.
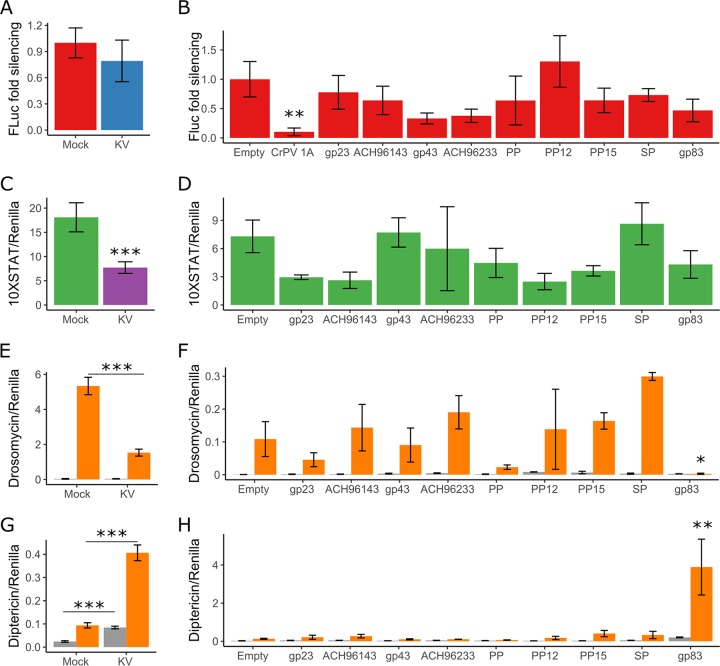
We used previously established luciferase reporter-based assays to describe the effect of KV infection on the RNAi, JAK-STAT, Toll, and Imd pathways in cell culture. To determine if KV suppresses RNAi, we measured the RNAi silencing efficiency of cells inoculated with KV or chloroform-inactivated KV (here referred to as mock treated) by cotransfecting an expression plasmid encoding firefly luciferase (FLuc) with either green fluorescent protein (GFP) dsRNA or FLuc dsRNA. In both mock- and KV-treated cells, FLuc dsRNA caused a 95% reduction in FLuc activity compared with that in GFP dsRNA-treated cells, indicating that KV infection does not inhibit RNAi in cell culture (MCMCp = 0.9) (Fig. 3A). Many viruses studied in Drosophila encode a suppressor of RNAi (for examples, see references 12, 44, and 48–52), and therefore, the absence of KV-induced RNAi suppression is somewhat surprising. It is possible that KV-RNAi interactions are different in the cell types that are naturally infected by KV and that our inability to observe RNAi suppressive activity is a limitation of the cell culture model. Alternatively, if KV transmission does not occur until later stages of infection, there may be limited selective pressure to evade RNAi, as RNAi mutants and control flies have similar titers during late infection.
FIG 3.
Kallithea virus gp83 suppresses Toll and induces Imd signalling. The ability of KV (4 dpi) and 9 highly expressed KV genes to inhibit the RNAi (A and B), JAK-STAT (C and D), Toll (E and F), and Imd (G and H) pathways was assessed. For RNAi suppression assays (A and B), RNAi efficiency was assessed by transfecting S2 cells with plasmids expressing FLuc and, as a normalization control, Renilla luciferase (RLuc), along with dsRNA targeting either FLuc or GFP. Data are expressed as fold silencing in cells treated with GFP dsRNA relative to those treated with FLuc dsRNA, normalized to 1 in mock-infected cells. The CrPV suppressor of RNAi, protein 1A, was used as a positive control (data combined from 2 experiments). For JAK-STAT suppression assays (C and D), S2 cells were transfected with a plasmid encoding FLuc under the control of 10 STAT binding sites (10×STAT-FLuc). In contrast to the JAK-STAT pathway, the Toll and Imd pathways are not endogenously active in S2 cells (gray bars in E, F, G, and H) but can be activated by expression of TollLRR (orange bars in E and F) or PGRP-LC (orange bars in G and H). For Toll suppression assays (E and F), S2 cells were transfected with the Drs-FLuc reporter, encoding FLuc under the control of a Drosomycin promoter, with either pAc5.1-TollLRR or an empty control plasmid (gray bars). For Imd suppression assays (G and H), S2 cells were transfected with the Dpt-FLuc reporter, encoding FLuc under the control of a Diptericin promoter, with either pMT (empty) or pMT-PGRP-LC. All FLuc luciferase values were normalized to RLuc values, driven by a constitutively active Actin promoter from a cotransfected plasmid. PP, putative protein; SP, serine protease. Error bars show SEMs, calculated from 5 biological replicates for panels A, C, E, and G and at least 3 biological replicates for panels B, D, F, and H.
The JAK-STAT pathway has an antiviral role during Drosophila C virus infection (53) and mediates tolerance to the DNA virus IIV6, evidenced by upregulation of the vir-1 and Turandot (Tot) genes (9). However, previous in vivo transcriptional profiling did not identify strong differential expression of STAT-responsive genes following infection with KV (Fig. 1E) (32). We assessed JAK-STAT activity in mock- and KV-treated cells with a FLuc reporter driven by a promoter containing 10 STAT binding sites (54). This reporter is endogenously active in S2 cells (54), but KV infection led to a 58% reduction in STAT-mediated FLuc activity (37 to 74%; MCMCp < 0.001 [Fig. 3C]), indicating that JAK-STAT is downregulated or inhibited following KV infection. However, in addition to mediating a transcriptional immune response, the JAK-STAT pathway is involved in cell proliferation (55), which also decreases following KV infection in cell culture (Fig. 2), making cause and effect difficult to distinguish.
We next assayed the effect of KV on Toll and Imd signalling. However, these pathways are not constitutively active in S2 cells. To measure KV suppression of these pathways, we therefore cotransfected TollLRR (a Toll receptor lacking the leucine-rich repeats in the extracellular domain) or PGRP-LC (an Imd pathway receptor) with Drosomycin (Drs) or Diptericin (Dpt) luciferase reporters to artificially induce signalling of the Toll and Imd pathways, respectively. Transfection of TollLRR increased Drs-Fluc 243-fold (MCMCp < 0.001), consistent with previous reports (56). However, KV infection reduced the maximum level of TollLRR-mediated Drs activity by 81% (38% to 93%; MCMCp < 0.001 [Fig. 3E]), indicating that KV can inhibit Toll signalling. Overexpression of PGRP-LC led to a 4-fold increase in Dpt-FLuc (3- to 5-fold; MCMCp < 0.001). In contrast to the effect on Toll signalling, KV infection led to a 3.6-fold increase (2.6- to 4.8-fold; MCMCp < 0.001) in Dpt-FLuc, which additively increased when PGRP-LC-overexpressing cells were infected with KV (17-fold increase compared to the value for Imd-inactive, mock-treated cells; 12- to 23-fold [Fig. 3G]). These results suggest that KV infection in S2 cell culture leads to downregulation or suppression of Toll signalling but induction of Imd signalling.
KV-encoded gp83 modifies NF-κB signalling during infection.
The immunosuppressive function of nudivirus genes has not previously been explored. Because we observed KV-mediated downregulation of NF-κB-regulated antimicrobial peptides (AMPs) in vivo and downregulation of JAK-STAT and Toll reporters in vitro, we wished to identify potential KV-encoded suppressors of canonical immune pathways. Therefore, we cloned 9 uncharacterized KV genes that are highly expressed at 3 dpi in adult flies (32) and performed immunosuppression assays for the RNAi, JAK-STAT, Toll, and Imd pathways. We were unable to identify KV-encoded suppressors of RNAi or JAK-STAT among these 9 genes, although we confirmed that cricket paralysis virus protein 1A potently suppressed RNAi in these assays, as expected (51) (MCMCp = 0.006) (Fig. 3B and D). However, we found that gp83—a KV gene encoding no recognizable protein domains, named for its homology to the Gryllus bimaculatus nudivirus (GbNV) gp83 locus (57)—significantly reduced TollLRR-dependent Drs-FLuc expression (Fig. 3F). In this experiment, TollLRR expression induced Drs-FLuc 24-fold (8- to 66-fold), but only 1.9-fold (0.3- to 8-fold; MCMCp = 0.02) when gp83 was coexpressed. We further found that expression of gp83 caused a 5-fold (1.5- to 18-fold) increase in Imd-mediated Dpt-FLuc expression, with or without PGRP-LC overexpression (MCMCp = 0.008) (Fig. 3H).
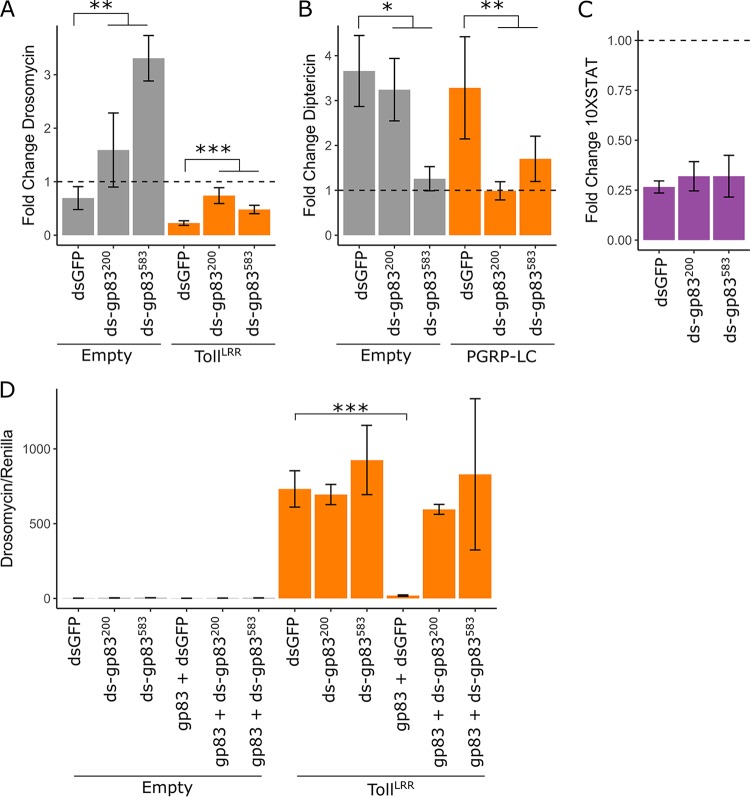
We next aimed to confirm that the interactions between the transfected KV gene gp83 and NF-κB pathways are representative of the function of gp83 during KV infection. Therefore, we silenced gp83 during KV infection using dsRNA and measured associated changes in Toll, Imd, and JAK-STAT signalling. Cotransfection of gp83 plasmid with independent dsRNAs targeting gp83 completely reversed inhibition of Drs-FLuc compared with transfection of GFP dsRNA, indicating that these dsRNAs effectively silence gp83 (MCMCp < 0.001 for both dsRNAs [Fig. 4D]). As reported above (Fig. 3E), KV infection had no effect on Drs-FLuc in the absence of TollLRR (MCMCp = 0.26) but inhibited TollLRR-induced signalling (MCMCp < 0.001). Knockdown of gp83 during KV infection of TollLRR-expressing cells led to increased Drs-FLuc (MCMCp < 0.001; orange bars in Fig. 4A). Surprisingly, Drs-FLuc was also slightly increased in Toll-inactive cells upon KV infection and gp83 knockdown (MCMCp = 0.004; gray bars in Fig. 4A). Likewise, knockdown of gp83 in KV-infected cells expressing PGRP-LC caused a decrease in Dpt-FLuc expression (MCMCp = 0.006; orange bars in Fig. 4B), and this effect was also noticeable in controls that do not express PGRP-LC (MCMCp = 0.03; gray bars in Fig. 4B). Consistent with a specific interaction with NF-κB signalling, gp83 knockdown had no effect on the ability of KV to downregulate JAK-STAT signalling in S2 cells (MCMCp = 0.63) (Fig. 4C). Together, these observations indicate that gp83 is responsible for Toll suppression and Imd activation during KV infection.
FIG 4.
KV induction and suppression of NF-ΚB pathways are mediated by gp83. The ability of KV to inhibit Toll (A), induce Imd (B), and inhibit JAK-STAT (C) was assessed during gp83 knockdown, using two independent dsRNAs against gp83 (labeled ds-gp83200 and ds-gp83583). Drosomycin, diptericin, and 10×STAT activities were measured as Drs-FLuc, Dpt-FLuc, and 10×STAT-FLuc expression, relative to RLuc expression as described in the legend to Fig. 3. For each, data are presented as fold change in signalling following KV infection relative to mock infection (chloroform-treated KV) (4 dpi), where 1 (horizontal dotted line) represents no induction or suppression of the pathway by KV infection. (A) Fold change in Drs-FLuc expression following KV infection of S2 cells with (orange bars) or without (gray bars) activation of the pathway by TollLRR expression. (B) Fold change in Dpt-FLuc expression following KV infection of S2 cells with (orange bars) or without (gray bars) pathway activation by PGRP-LC expression. (C) Fold change in 10×STAT FLuc expression following KV infection of S2 cells. (D) Efficiency of gp83 knockdown was assessed by cotransfection of an expression plasmid encoding gp83 with two independent dsRNAs against gp83 and Drs-FLuc reporter plasmids. Error bars show SEMs (n = 5 biological replicates for panels A to C and n = 3 biological replicates for panel D).
The immunosuppressive function of gp83 on Toll signalling in vitro is consistent with the observed downregulation of AMPs following KV infection in vivo and substantiates the hypothesis that Toll is antiviral and suppressed during infection. However, the induction of antiviral Imd signalling by a single viral protein is unexpected, and it is unclear why KV has not evolved to avoid or suppress Imd activation as seen for other insect-infecting DNA viruses (28). Assuming that Imd activation is detrimental to virus transmission, this could indicate a trade-off between suppression of Toll and activation of Imd or that gp83 is directly recognized by the fly immune system. Additionally, gp83-mediated Imd activation in vitro is at odds with the observed broad downregulation of AMPs in vivo, which are controlled, in part, by Imd signalling. This could be explained by differences in the intracellular versus systemic effects of KV on Imd signalling, or tissue-specific responses to KV, either of which could mask an excitatory effect of gp83 on Imd in vivo. Because of these inconsistencies, we chose to focus specifically on the Toll immunosuppressive effect of gp83, because the in vitro data are consistent with observed AMP expression patterns in vivo. We conclude that KV-encoded gp83 is involved in mediating complex interactions with NF-κB signalling in vitro, including suppression of Toll signalling and induction of Imd signalling.
Immunosuppression by gp83 occurs downstream of Toll transcription factors.
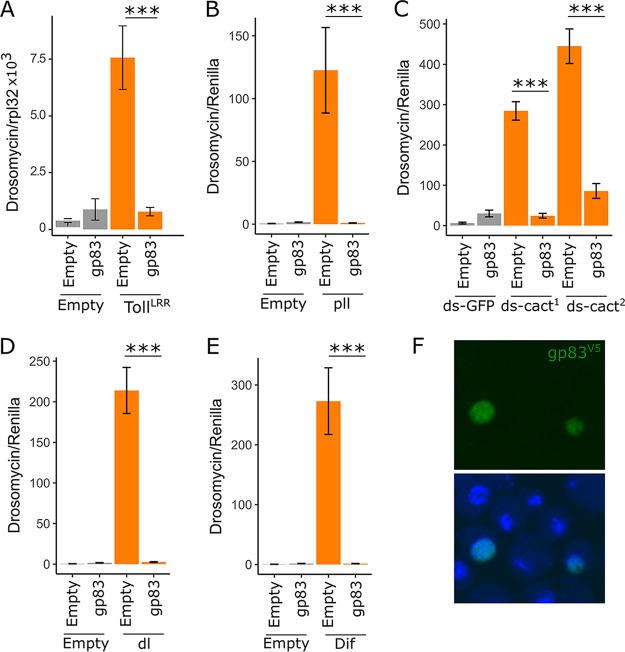
Previously described polydnavirus-encoded Toll pathway inhibitors imitate IκB, blocking the nuclear entry of NF-κB transcription factors (27). Although the precise mechanism of interaction between gp83 and Toll signalling is unknown, suppression of TollLRR-induced signalling indicates that gp83 functions downstream of Toll and interferes with intracellular Toll signalling. We therefore performed genetic interaction experiments between gp83 and downstream Toll components to narrow down the point in the Toll signalling pathway at which gp83 acts. As observed before with reporter assays, gp83 inhibited TollLRR-mediated signalling; in this experiment, we assessed this by qRT-PCR of endogenous Drs expression (MCMCp < 0.001 [Fig. 5A]). Additionally, Drs-FLuc was greatly increased by overexpressing pll (240-fold [131- to 414-fold] induction of Drs-FLuc), silencing cact (75-fold [33- to 161-fold] induction of Drs-FLuc), overexpressing Dif (563-fold [317- to 1,002-fold] induction of Drs-FLuc), and overexpressing dl (459-fold [257- to 778-fold] induction of Drs-FLuc). Coexpression of gp83 potently reduced Drs-FLuc in each of these scenarios (MCMCp < 0.001 for each): pll/gp83 coexpression led to a 0.55-fold change in Drs-FLuc (0.31- to 0.99-fold), cactdsRNA/gp83 led to a 1.73-fold change in Drs-FLuc (0.75- to 3.5-fold), Dif/gp83 led to a 0.86-fold change in Drs-FLuc (0.5- to 1.5-fold), and dl/gp83 led to a 1.5-fold change in Drs-FLuc (0.9- to 2.5-fold) relative to baseline Drs-FLuc expression (Fig. 5B to E). Additionally, staining of V5 epitope-tagged gp83 revealed that it is a nuclear protein (Fig. 5F). Together, these results indicate that gp83 either inhibits NF-κB transcription factors or acts downstream of them to suppress Toll signalling in vitro.
FIG 5.
gp83 inhibits Toll signalling downstream of Dif and dorsal. (A) The ability of gp83 to inhibit endogenous Drosomycin expression was assessed by transfection of S2 cells with pAc-gp83 or empty control plasmid, and the Toll pathway was activated by cotransfection of pAc-TollLRR or control plasmid. Drosomycin expression levels were measured relative to Rpl32 expression by qRT-PCR. (B to E) The Toll pathway was activated downstream of the Toll receptor by transfection of a plasmid encoding pll (B), knockdown of cactus with two independent, nonoverlapping dsRNAs (labeled ds-cact1 and ds-cact2) (C), and transfection of plasmids encoding the transcription factors dl and Dif (D and E). Activation of the pathway was assessed using the Drs-FLuc reporter, relative to RLuc expression (orange bars in panels B to E; gray bars represent controls in which empty plasmids [B, D, and E] or dsRNA targeting GFP [C] were transfected). Suppression of the Toll pathway at different stages by gp83 was assessed by cotransfection of pAc-gp83 or an empty control plasmid (B to E). (F) Representative confocal image of S2 cells expressing V5 epitope-tagged gp83 stained with a V5 antibody (top) and a merged image in which nuclei are stained with Hoechst (bottom). Error bars show SEMs (n = 5 biological replicates).
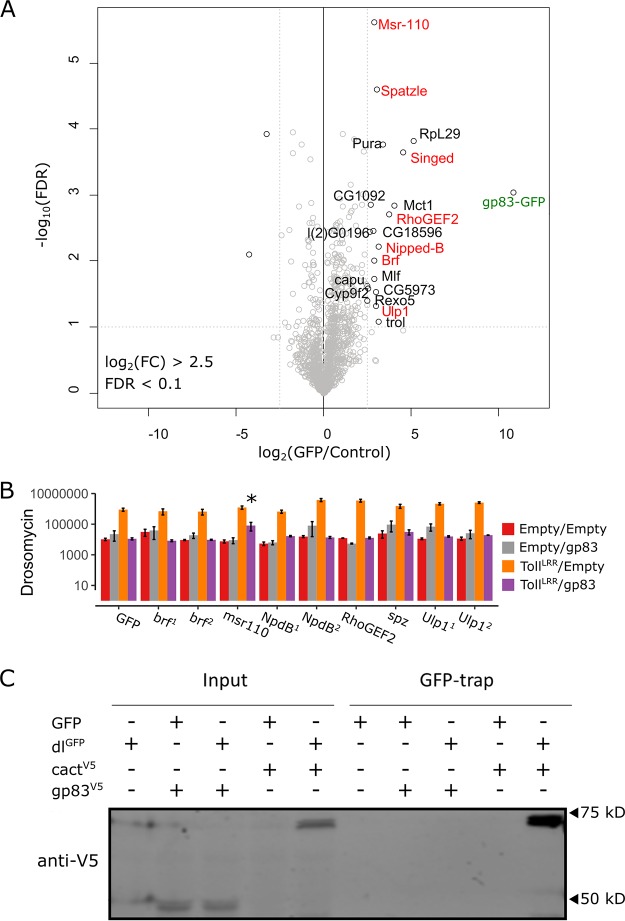
Virus-encoded inhibitors of NF-κB in mammals have been reported to operate by promoting degradation of NF-κB transcription factors, blocking NF-κB access to the nucleus, or interfering with transcriptional coactivators to evade the interferon response (reviewed in reference 58). In order to better define the mechanism of the immunosuppressive action of gp83, we searched for direct host interactions that may mediate Toll suppression. Because our genetic interaction experiments indicate that gp83 acts on or downstream of dl, we first tested for a physical interaction between dl and gp83 using coimmunoprecipitation and subsequent Western blotting. Following immunoprecipitation of GFP-tagged dl, we were able to detect cact as an interacting positive control, but we did not detect gp83 in GFP-dl immunoprecipitation (Fig. 6C). Thus, to identify host-interacting proteins of gp83 in an unbiased manner, we created an S2 cell line stably expressing GFP-tagged gp83, immunoprecipitated gp83GFP, and performed quantitative mass spectrometry on interacting partners. We identified 19 D. melanogaster proteins, including 4 nuclear proteins (Nipped-B, Brf, Mlf, and Ulp1), that were enriched in the gp83 immunoprecipitate (log2 fold enrichment > 2.5; false-discovery rate [FDR] < 0.1 [Fig. 6A]). While we did not identify known downstream NF-κB pathway factors, the extracellular Toll ligand spz was enriched, despite the nuclear localization of gp83. However, peptide coverage of spz was poor and dsRNA knockdown of spz did not rescue the immunosuppressive effect of gp83, indicating that this interaction may not occur in live cells or that it is not required for gp83 to inhibit Toll signalling (Fig. 6B). Further, dsRNA-mediated knockdown of a subset of the enriched genes, including 3 of the 4 identified nuclear proteins, was unable to rescue the gp83 immunosuppressive effect (Fig. 6B), suggesting that gp83 may not form stable complexes with host proteins to interfere with NF-κB signalling.
FIG 6.
Identification of host interactors of gp83. (A) Identification of gp83 interacting proteins in S2 cell lysates by label-free quantitative (LFQ) mass spectrometry. Permutation-based FDR-corrected t tests were used to determine proteins that are statistically enriched in gp83-GFP immunoprecipitation (IP). The log2 LFQ intensity of gp83-GFP IP over control IP (cells that do not express gp83-GFP) is plotted against the −log10 FDR. The gp83-GFP bait (labeled in green) and interactors with an enrichment of fold change of >2.5; −log10 FDR values of >1 are indicated. (B) Drs-FLuc expression was measured following cotransfection of pAc-gp83, pAc-TollLRR, or empty control plasmids, along with dsRNA targeting brf, msr-110, Nipped-B, RhoGEF2, spätzle, and Ulp1 (labeled red in panel A), with dsRNA targeting GFP as a control. Genes are superscripted with “1” or “2” when two independent dsRNAs were used to knock down the gene. Although msr-110 knockdown appears to partially rescue gp83 immunosuppression, subsequent experiments did not reproduce this effect. Error bars represent SEMs (n = 3). Statistical tests were performed in MCMCglmm. (C) V5-tagged gp83 or V5-tagged cact (an IκB protein known to interact with dl) were expressed alongside GFP-tagged dl or GFP and GFP-associated complexes were immunoprecipitated with GFP-trap magnetic beads and analyzed by Western blotting using V5 antibodies. Note that cact appears to be stabilized when coexpressed with dl compared to when it is expressed alone.
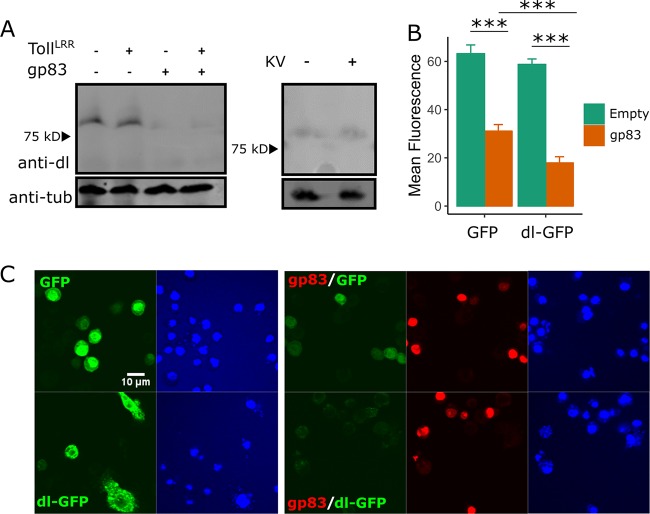
Although we did not detect a direct association between dl and gp83, we observed a reduction in dl protein levels upon gp83 overexpression that is not dependent on Toll signalling (Fig. 7A). We quantified this effect by transfecting either GFP or GFP-tagged dl, in the absence or presence of gp83, and measuring fluorescence by confocal microscopy. We found that while gp83 caused a 53% reduction in GFP levels (42% to 62%; MCMCp < 0.001), possibly due to a dl binding site in the actin 5 C promoter of this construct (59), gp83 caused a significantly greater reduction in dlGFP (73% reduction; 66% to 78%; MCMCp < 0.001 [Fig. 7B and C]). However, KV infection did not decrease dl protein levels, indicating that this may not be the primary mechanism by which KV inhibits Toll signalling (Fig. 7A). Instead, we hypothesize that gp83 interferes with the access of dl either to the nucleus or to NF-κB binding sites, which indirectly affects dl localization and results in increased turnover. We prefer the latter explanation, that gp83 directly interferes with the Toll pathway transcriptional response, because overexpression of gp83 simultaneously induced the Dpt reporter (Fig. 2H) and reduced dl-responsive promoters (Drs-FLuc and Act5C-GFP) (Fig. 3F and Fig. 7B and C). These observations implicate gp83 in regulating transcription at diverse loci responsive to both dl and Rel and suggest an interaction between gp83 and NF-κB-responsive genes, possibly by directly interacting with DNA.
FIG 7.
Overexpression of gp83 may reduce dorsal levels. (A) Western blots show endogenous dl protein levels in S2 cells transfected with a plasmid encoding gp83 or empty control plasmid (left) and in S2 cells infected with KV (4 dpi) (right). The Toll pathway was activated by expression of pAc-TollLRR, as indicated. Western blot analysis using anti-tubulin antibody was used to verify equal loading. (B and C) The effect of gp83 was analyzed by confocal microscopy of S2 cells transfected with plasmid encoding gp83 or control plasmid and plasmids encoding either GFP or dl-GFP. ImageJ-based quantification of mean GFP fluorescence for individually outlined cells is shown (n ≥ 20 cells for each condition; error bars show SEMs). (C) A representative image from panel B, showing GFP (top) and dl-GFP (bottom) expression with or without gp83. Nuclei were visualized using Hoechst.
Immunosuppressive function of gp83 depends on conserved residues and is conserved in other nudiviruses.
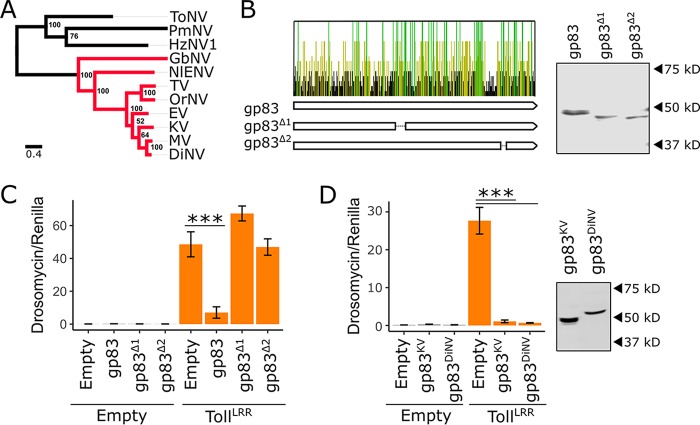
Conflict between the host immune system and virus-encoded immune inhibitors may result in an evolutionary arms race, leading to recurrent positive selection and eventual host specialization (for examples, see references 60–62). Consistent with this, some immune inhibitors are effective only against their native host species, thereby defining the viral host range (for examples, see references 40–45). We tested whether the immunosuppressive function of gp83 is conserved and whether gp83 acts in a species-specific manner. The gp83 locus is absent from nudiviruses distantly related to KV, such as Heliothis zea nudivirus 1 (HzNV1), Tipula oleracea nudivirus (ToNV), and Peneaus monodon nudivirus (PmNV), but gp83 homologs are found in the more closely related GbNV, Nilaparvata lugens endogenous nudivirus (NlENV), Oryctes rhinoceros nudivirus (OrNV), Drosophila innubila nudivirus (DiNV), Tomelloso virus (TV), Mauternbach virus (MV), and Esparto virus (EV) (Fig. 8A). Although gp83 lacks recognizable protein domains, several regions are strongly conserved among these nudiviruses, suggesting functional conservation (Fig. 8B). To test whether gp83 function depends on these conserved domains, we made two gp83 deletion constructs (gp83Δ1 and gp83Δ2) that remove conserved regions of, respectively, 18 and 8 amino acids without substantially altering protein stability, and transfected these alongside TollLRR with the Drs-FLuc reporter. Although detectable by Western blotting (Fig. 8B), gp83Δ1 (MCMCp = 0.67) and gp83Δ2 (MCMCp = 0.79) were unable to inhibit Toll signalling, indicating that these conserved residues are important for the immunosuppressive function of gp83 (Fig. 8C).
FIG 8.
The immunosuppressive function of gp83 is evolutionarily conserved. (A) Maximum likelihood phylogeny inferred from a protein alignment of nudivirus-encoded DNA polymerase B using PhyML (83), with an LG substitution model and gamma-distributed rate parameter. Support for each node was assessed by bootstrapping, and the scale bar represents substitutions per site. Nudivirus species that encode gp83 homologs are colored in red. (B) Conservation of the gp83 amino acid sequence across 7 species of nudivirus (all red-labeled viruses in panel A, except the endogenized virus NlENV). Each bar represents an amino acid, and bars are colored yellow if the residue is conserved in ≥50% of the species, green if conserved in 100% of the species, and black if conserved in <50% of the species. Two V5-tagged gp83 constructs were created with deletions that span regions with an excess of conserved residues: gp83Δ1 and gp83Δ2. Western blotting and subsequent V5 antibody staining show that both deletion constructs accumulate to levels similar to those of full-length gp83 following transfection of S2 cells. (C) Full-length gp83, gp83Δ1, or gp83Δ2 was coexpressed with TollLRR, and Drs-FLuc expression was measured relative to pAct-FLuc expression. (D) V5-tagged gp83 from KV and DiNV were coexpressed with TollLRR to assess suppression of Drs-FLuc expression (relative to pAct-FLuc expression) in D. melanogaster S2 cells. Western blot analysis using V5 antibody was used to confirm gp83 expression. Error bars show SEMs (n = 5 biological replicates).
To test whether gp83 function is conserved among viruses, we cloned gp83 from DiNV, which has not been found to be associated with D. melanogaster (11), and performed Toll immunosuppression assays. The gp83 homolog from DiNV was able to completely inhibit D. melanogaster Toll signalling in S2 cells (MCMCp < 0.001), despite only 57% amino acid identity with KV gp83, demonstrating that the immunosuppressive function of gp83 is conserved in other nudiviruses and that it is not highly host specific (Fig. 8D). This observation suggests that the Toll-gp83 interaction may not be a hot spot of antagonistic “arms race” coevolution and has not led to specialization of DiNV gp83 to the D. innubila immune system at the expense of its ability to function in D. melanogaster. This could be because gp83 has relatively few direct interactions with host proteins (Fig. 6A) and may instead interact directly with transcription factor binding sites which are under high constraint and therefore unable to evolve resistance to the immunosuppressive effect of gp83 (63).
Conclusions.
In this study, we investigated the role of known antiviral immune pathways in the context of DNA virus infection, including the RNAi, JAK-STAT, Imd, and Toll pathways. Our data support an antiviral role for RNAi and Imd against KV, consistent with previously described antiviral RNAi against IIV6 and DNA virus-encoded suppressors of Imd (7, 8, 28). Furthermore, we identified gp83 as a KV-encoded Toll suppressor that acts downstream of NF-κB transcription factor release of IκB in cell culture, suggesting that Toll signalling can be antiviral during DNA virus infection in insects. The immunosuppressive effect of gp83 is conserved in other nudiviruses, and has not evolved host specificity in DiNV, indicating that the Toll-gp83 interaction is unlikely to be a hot spot of reciprocal host-virus adaptation and that other KV genes may be more important in determining host range.
MATERIALS AND METHODS
Fly strains, virus growth, and mortality experiments.
All fly lines were maintained and crossed on standard cornmeal medium at 25°C. Viral titer and mortality were measured following KV infection in two control lines (w1118 and Oregon R) and in mutant lines compromised in the following immune signalling pathways: RNAi (Dcr-2L811fsX [64] and AGO2414 [65]), Toll (spz4 [66], dl1 [67], and pll2/pll21 trans-heterozygotes [68, 69]), and Imd (rele20 [70] and imd10191 [71]).
For mortality assays, 100 female flies of each genotype were injected with 50 nl of either KV suspension (105 50% infectious doses [ID50], as described in reference 32) or chloroform-treated KV suspension (which inactivates KV through the destruction of the membrane [32]). For chloroform treatment, the KV suspension was mixed with an equal volume of chloroform, vortexed for 30 s, and centrifuged for 5 min at 6,000 × g, and the aqueous phase was taken for downstream experiments. Injected flies were transferred to sucrose agar vials in groups of 10, and the number of surviving flies was recorded daily. While maintenance of flies on a protein-free diet likely affects some aspects of the immune response, we have assumed that this is similarly tolerated across the fly lines used. Each group of flies was transferred to fresh food each week. Per-day mortality was analyzed as a binomial response variable with the Bayesian generalized linear mixed modeling R package, MCMCglmm (72), with days postinoculation (dpi), dpi2 (to allow for nonlinear changes in mortality), and genotype as fixed effects, and vial as a random effect, as described previously (32). All confidence intervals are reported as 95% highest posterior density (HPD) intervals.
Viral titer was measured in each line after intra-abdominal injection of 50 nl of KV suspension. Infected female flies of each line (n = 50) were transferred to 10 sucrose agar vials in groups of 5, and 5 vials of each genotype were homogenized in TRIzol (Invitrogen) at 5 and 10 dpi. For RNAi mutants, flies were also assayed at 3 dpi. DNA was extracted by phenol-chloroform precipitation and viral titer estimated by quantitative PCR relative to host genomic DNA, using previously described primers (rpl32 [32]). Log-transformed viral titer was analyzed as a Gaussian response variable using MCMCglmm (72), with genotype, dpi, and genotype-by-dpi interactions as fixed effects. Titer in RNAi and NF-κB mutants were assayed in separate experiments and therefore analyzed independently. A statistical approach was used to account for the impact of differing genetic backgrounds between mutant lines, using the range of KV titers seen previously across 120 different natural genetic backgrounds from the Drosophila Genetic Reference Panel (DGRP) (32). Specifically, considering w1118 and Oregon R as controls and mutants of each pathway as the “experimental” group, a null distribution of effect sizes expected only from differences in genetic background was created by randomly choosing two DGRP lines to serve as controls and additional DGRP lines reflecting the mutant lines used in each pathway. For each null draw, the same model was fitted as described above, the absolute value of the effect size was recorded, and this was repeated 1,000 times to obtain a distribution. If the average effect size associated with mutants in a pathway was greater than the highest 5% of effect sizes, we concluded that the observed differences in KV titer were due to mutations in the tested pathway.
Cell culture and virus propagation.
S2 cells (Invitrogen) were cultured at 25°C in Schneider’s Drosophila medium with 10% heat-inactivated fetal bovine serum and 50 U/ml of penicillin and 50 μg/ml of streptomycin (Life Technologies). KV was purified from flies 10 days after initial infection as previously described (32). Briefly, KV was injected into 2,000 Oregon R adult flies, which were incubated at 25°C for 10 days, homogenized in 5 ml of 10 mM Tris-HCl, filtered through cheesecloth, centrifuged twice for 10 min at 6,000 × g, filtered through a 0.22-µm polyvinylidene fluoride syringe filter, and subjected to gradient centrifugation in an iodixanol (Optiprep) gradient (32). KV-positive fractions of the gradient, as assessed by qPCR, were kept as the KV isolate. To measure the effects of KV on cell size and number, 5 × 104 S2 cells were seeded in 96-well plates, followed by the immediate addition of 5 μl of either KV suspension (103 ID50) or chloroform-treated KV. Cells were split once 7 dpi, and cell size and number were measured using FIJI 10 dpi (73).
Cloning.
We selected 9 KV genes identified as highly expressed at 3 dpi (32) to screen for KV-encoded immunosuppressors. These were gp23, gp43, gp83, ACH96233.1-like, ACH96143.1-like, putative protein 1, putative protein 12, putative protein 15, and putative serine protease (corresponding to GenBank accession numbers AKH40365.1, AKH40394.1, AKH40369.1, AKH40392.1, AKH40340.1, AQN78560.1, AKH40392.1, AKH40404.1, and AQN78556.1). Each KV gene was amplified using the Qiagen long-range PCR kit as per the manufacturer’s instructions, with primers that introduced restriction sites and the Drosophila Kozak sequence (restriction enzymes and primers used are listed in Table 1), and cloned into a pAc5.1 vector (Invitrogen) with a C-terminal V5-His tag. The KV gene gp83 was also cloned into pAc5.1 vector with GFP instead of V5-His to introduce a C-terminal GFP tag. Deletion constructs for gp83 were created by separately amplifying 2 segments of gp83 with primers that span the desired deletion and performing a second PCR with these segments as a template and the forward and reverse primers from the 5′ and 3′ segments, respectively (Table 1) (gp83Δ1, CGLIECSELLRDRLCSKL deletion; gp83Δ2, WSDRLNLI deletion). The resulting amplicons with deletions were cloned as described above. The gp83 gene from DiNV (35, 74) was also cloned as described above (Table 1).
TABLE 1.
Primers for cloning and dsRNA synthesisa
| Use and source | Amplicon or targeted gene | Primer F name | Primer F sequence | Primer R name | Primer R sequence |
|---|---|---|---|---|---|
| Cloning | |||||
| KV | Putative protein 12 | PutPro12_Acc651_0F | actgGGTACCaacATGATCAACCACCAAGGTATCG | PutPro12_XbaI_273R | tgacTCTAGAATATTCCAGTTTTATCATCAGATGTTTG |
| KV | Putative protein 15 | PutPro15_Acc651_0F | actgGGTACCaacATGTCTTGCAATAAAGTCGAATC | PutPro15_XbaI_819R | tgacTCTAGAGATTTTAGATGGTTTGCGGTTA |
| KV | Putative protein | PutPro_Acc651_0F | actgGGTACCaacATGTTCAAGTATTCAAACTCAAAATA | PutPro_XhoI_378R | tgacCTCGAGATTTGATGAATATCCTTGATCTAAAAG |
| KV | Putative serine protease | PutSerProt_Acc651_0F | actgGGTACCaacATGTTGCCAATTATAAGTTCG | PutSerProt_XbaI_960R | tgacTCTAGAAACATTTGATTGGCAAATG |
| KV | gp23-like | gp23-like_Acc651_0F | actgGGTACCaacATGGCGGACAAAAAAATATTC | gp23-like_XbaI_1878R | tgacTCTAGATTCCATTTTTAAACGTTTACTCTG |
| KV | gp83-like | gp83-like_Acc651_0F | actgGGTACCaacATGTCAGAATCAAAGCTGCAAC | gp83-like_XbaI_1278R | tgacTCTAGATTTCTTATTATCGGATTTTTTCAATG |
| KV | ach96233-like | ach96233_Acc651_0F | actgGGTACCaacATGCGTGTGTATAGACCTACAATGAAG | ach96233_XbaI_3009R | tgacTCTAGAGGGAAAAATGCTTACAACTGTCG |
| KV | ach96143-like | ach96143_Acc651_0F | actgGGTACCaacATGACTTCGACAACCGTTAAC | ach96143_XbaI_585R | tgacTCTAGAGATTTTATCGTATATCTTCTTAATTACTTTTTC |
| KV | gp72-like | gp72-like_BsiWI_0F | actgCGTACGaacATGAACGTTTCAATTGGAAATATTAGC | gp72-like-XhoI_662R | tgacCTCGAGAGCTACGGCCACTGGTCTTG |
| KV | gp43-like | gp43-like_BsiWI_0F | actgCGTACGaacATGAGTACATTGTTGAATTTGTGCG | gp43-like_XbaI_1296R | tgacTCTAGACAAAAGTATATTATCAAATACATCATCATTGTTG |
| KV | gp83-delta1 fragment 1 | gp83-like_Acc651_0F | actgGGTACCaacATGTCAGAATCAAAGCTGCAAC | gp83_del1_R | ACTAACCGATATTGCAATTAATTGCTTTCG |
| KV | gp83-delta1 fragment 2 | gp83_del1_F | CGAAAGCAATTAATTGCAATATCGGTTAGT | gp83-like_XbaI_1278R | tgacTCTAGATTTCTTATTATCGGATTTTTTCAATG |
| KV | gp83-delta2 fragment 1 | gp83-like_Acc651_0F | actgGGTACCaacATGTCAGAATCAAAGCTGCAAC | gp83_del2_R | TTTTGTACGGAATCGAGACATGCGATCGTA |
| KV | gp83-delta2 fragment 2 | gp83_del2_F | TACGATCGCATGTCTCGATTCCGTACAAAA | gp83-like_XbaI_1278R | tgacTCTAGATTTCTTATTATCGGATTTTTTCAATG |
| DiNV | DiNV gp83 | DiNV_gp83_Acc651_0F | actgGGTACCaacATGGATAGCAAAACAGAAACAAC | DiNV_gp83_XbaI_1350R | tgacTCTAGACAATCCTGCTTTATTATCGG |
| Dmel | cact | cact_Acc651_0F | actgGGTACCaacATGCCGAGCCCAACAAAAG | cact_XhoI_1503R | tgacCTCGAGGGCAACTGTCATGGGATTG |
| Dmel | Dif | Dif_Acc651_0F | actgGGTACCaacATGTTTGAGGAGGCTTTCG | Dif_XbaI_2001R | tgacTCTAGATTTGAATGGCTGAATTCCC |
| Dmel | dorsal | dl_Acc651_0F | actgGGTACCaacATGTTTCCGAACCAGAAC | dl_XbaI_2031R | tgacTCTAGACGTGGATATGGACAGGTTC |
| Dmel | pelle | pll_Acc651_0F | actgGGTACCaacATGAGTGGCGTCCAGAC | pll_XbaI_1503R | tgacTCTAGAGTCGGTAACAAACGGTTC |
| Dmel | tube | tub_Acc651_0F | actgGGTACCaacATGGCGTATGGCTGGAAC | tub_XbaI_1386R | tgacTCTAGATTGCTGCAGCTCACTCAAG |
| dsRNA synthesis | |||||
| Dmel | msr-110 | msr-110_T7_210F | taatacgactcactatagggATCCTTCATCCTGGCCTCCT | msr-110_T7_627R | taatacgactcactatagggCGAGTCGACAATCTCCTCGG |
| Dmel | msr-110 | msr-110_T7_608F | taatacgactcactatagggCGAGTCGACAATCTCCTCGG | msr-110_T7_1127R | taatacgactcactatagggTGGATCTCACGGAAGGGGAT |
| Dmel | brf | brf_T7_921F | taatacgactcactatagggAGATATGGGCGAGCATGAGC | brf_T7_1248R | taatacgactcactatagggGGTCACCCTGCAAATAGCCT |
| Dmel | brf | brf_T7_1229F | taatacgactcactatagggAGGCTATTTGCAGGGTGACC | brf_T7_1734R | taatacgactcactatagggTGCCCTATTGCCCCTACTCT |
| Dmel | Nipped-B | NippedB_T7_1390F | taatacgactcactatagggGTGAAGGAGGAAGTCACCCG | NippedB_T7_1876R | taatacgactcactatagggCCGTGTAAACCGGGTCATGA |
| Dmel | Nipped-B | NippedB_T7_5103F | taatacgactcactatagggATTCGGGCTACAGCTTTGCT | NippedB_T7_5555R | taatacgactcactatagggTCCGTCAAATCTTCGGGCAA |
| Dmel | RhoGEF2 | RhoGEF2_T7_557F | taatacgactcactatagggACAATGCGAGCCACAACAAC | RhoGEF2_T7_1036R | taatacgactcactatagggCTGATACCGAAGAGGGTCGC |
| Dmel | RhoGEF2 | RhoGEF2_T7_4521F | taatacgactcactatagggGGAGAGCGAGGATGAAGACG | RhoGEF2_T7_4906R | taatacgactcactatagggCCGCAAACTCGCAAAGAACA |
| Dmel | singed | singed_T7_88F | taatacgactcactatagggATCAACGGCCAGCACAAGTA | singed_T7_511R | taatacgactcactatagggACTCCGACAAATGGGCGAAT |
| Dmel | singed | singed_T7_1133F | taatacgactcactatagggATCTGTTTGCCACCTCGGAG | singed_T7_1444R | taatacgactcactatagggTTATGCAGATCCTGGTCGGC |
| Dmel | spz | spz_T7_553F | taatacgactcactatagggTTCTGCACGAATGTGGACGA | spz_T7_894R | taatacgactcactatagggGGTCTGCTGTGTGTAGTGCT |
| Dmel | spz | spz_T7_389F | taatacgactcactatagggAATCCGAACAGCCGATACCC | spz_T7_718R | taatacgactcactatagggACACCAGCTTCCTGATGCTC |
| Dmel | Ulp1 | Ulp1_T7_186F | taatacgactcactatagggCAGCACGTTTCCAAGTTGGG | Ulp1_T7_617R | taatacgactcactatagggGCGACCTGGTTGTTTTGGTC |
| Dmel | Ulp1 | Ulp1_T7_2991F | taatacgactcactatagggTAATGGCATCTCCGAGTCGC | Ulp1_T7_3512R | taatacgactcactatagggTCGTTGGCTTCCTTGACCTC |
| Dmel | cact | cact_T7_1F | taatacgactcactatagggGAGAACGCTGTGTGCATTTG | cact_T7_1R | taatacgactcactatagggGCTTCTCCAGGATGTTCTGC |
| Dmel | cact | cact_T7_318F | taatacgactcactatagggCCAAAAGGAACAGCCCGTTG | cact_T7_902R | taatacgactcactatagggGTGTTTCCATGACGATCGCG |
| KV | gp83 | gp83_T7_200F | taatacgactcactatagggCCTCGAATCGTGCCAAATCG | gp83_T7_602R | taatacgactcactatagggTTGGAACAGAGACGGTCACG |
| KV | gp83 | gp83_T7_583F | taatacgactcactatagggCGTGACCGTCTCTGTTCCAA | gp83_T7_882R | taatacgactcactatagggTTGGTCGGGTTTGAAAACGC |
| Plasmid | Firefly Luc | T7-Luc-F | taatacgactcactatagggAGATATGAAGAGATACGCCCTGGTT | T7-Luc-R | taatacgactcactatagggAGATAAAACCGGGAGGTAGATGAGA |
| Plasmid | GFP | T7-GFP-F | taatacgactcactatagggAGAAGCTGACCCTGAAGTTCATCTG | T7-GFP-R | taatacgactcactatagggAGAGGTGTTCTGCTGGTAGTGGTC |
Templates and primer names and sequences for cloning and dsRNA synthesis. Primer names include target amplicon, base coordinates of the target sequence that the 3′ end of the primer anneals to, and restriction enzyme or T7 binding site additions to the 5′ primer ends. For cloning, amplicons are shown, and for dsRNA synthesis, gene targets are shown. F and R, forward and reverse primers, respectively. Lowercase sequences are primer tails that include restriction enzyme cut-site, T7 polymerase promoter, and Kozak sequences; uppercase primer sequences are those that match the gene of interest.
Additionally, Toll pathway components pll, tube, cact, Dif, and dl were cloned into the pAc5.1 vector, as described above (Table 1). Other Toll and Imd pathway constructs have been described before: pAc5.1-TollLRR (56), pAc5.1-dl-GFP (75), pMT-PGRP-LCx (76), pAc5.1-rel-GFP (77), and the firefly luciferase (FLuc) reporter plasmids with promoter sequences from Drosomycin (Drs), Diptericin (Dpt), and Attacin-A (Att-A) (56) or with 10× STAT binding sites (54).
Transfection and RNAi knockdown in S2 cells.
S2 cells were transfected using Effectene transfection reagent, as per the manufacturer’s instructions. Double-stranded RNA (dsRNA) was synthesized against cactus, gp83, FLuc, renilla luciferase (RLuc), and GFP for RNAi-mediated knockdown. Primers with flanking T7 sequences were used to amplify regions of each gene (Table 1) and dsRNA was synthesized from the resulting PCR products with T7 RNA polymerase and purified using GenElute total RNA minikit (Qiagen) (78).
Immunosuppression assays.
The 9 cloned KV genes were tested for their ability to suppress RNAi, JAK-STAT, Toll, or Imd activity. RNAi suppression assays were performed as described previously (78). Briefly, 5 × 104 S2 cells were seeded in a 96-well plate and 24 h later transfected with 33 ng of pMT-FLuc, 33 ng of pMT-Rluc, and 33 ng of either pAc5.1 empty vector or the pAc5.1 expression plasmid encoding a KV gene. Two days later, 400 ng of either GFP or FLuc dsRNA was added to each well, and CuSO4 was added 8 h later to a final concentration of 500 µM to induce expression of the luciferase reporters. RLuc and FLuc luciferase activities were measured using a dual-luciferase assay kit (Promega).
For JAK-STAT immunosuppression assays, 5 × 104 S2 cells were seeded in a 96-well plate and transfected 24 h later with 30 ng of 10×STAT-FLuc, 20 ng of pAc5.1-Rluc, and 50 ng of either pAc5.1 empty vector or the pAc5.1 expression plasmid carrying a KV gene. Luciferase activity was measured at 48 h following transfection.
For NF-κB immunosuppression assays, a plasmid encoding the Imd receptor PGRP-LC (isoform x; pMT-PGRP-LCx) (76, 79) or a constitutively active Toll construct lacking the extracellular leucine-rich repeat domain, pAc5.1-TollLRR (56), was transfected alongside each KV gene and an NF-κB-responsive FLuc reporter containing either the Dpt (Imd) or Drs (Toll) promoter sequence (56). For Toll immunosuppression assays, 5 × 104 S2 cells were seeded in 96-well plates and 24 h later transfected with 50 ng of either empty pAc5.1 vector or a pAc5.1 KV gene expression construct, 20 ng of either pAc5.1 or pAc5.1-TollLRR, 10 ng of Drs-FLuc, and 10 ng of pAc5.1-Rluc. Imd immunosuppression assays were performed in the same manner, except that pMT, pMT-PGRP-LCx, and Dpt-FLuc were substituted for pAc5.1, pAc5.1-TollLRR, and Drs-FLuc, respectively, and CuSO4 was added immediately following transfection. Analogous experiments were performed using pAc5.1-dl, pAc5.1-Dif, and pAc5.1-pll instead of pAc5.1-TollLRR or by transfecting 5 ng of cact dsRNA. In the latter case, 70 ng of KV gene expression construct was transfected instead of 50 ng. RLuc and FLuc activities were assayed 48 h after transfection.
Immunosuppression assays were also performed using KV-infected cells. A total of 5 × 104 cells were seeded in 96-well plates, followed by the immediate addition of 5 μl of either KV suspension (103 ID50) or chloroform-treated KV, and transfected the next day. For RNAi suppression assays with KV, 50 ng of pMT-RLuc, 50 ng of pMT-FLuc (78), and 5 ng of either GFP or GL3 dsRNA were transfected 2 dpi and CuSO4 was added 8 h later. To measure JAK-STAT activity following KV infection, 70 ng of 10×STAT-FLuc and 30 ng of pAc5.1-Rluc (48) were transfected. For Toll suppression assays, 70 ng of either pAc5.1 or pAc5.1-TollLRR, 20 ng of Drs-FLuc, and 10 ng of pAc-RLuc were transfected. Finally, to measure Imd activity following KV infection, 70 ng of either pMT or pMT-PGRP-LCx, 20 ng of Dpt-FLuc, and 10 ng of pAc-RLuc were transfected, and CuSO4 was added immediately following transfection. Luciferase activity was measured at 4 dpi.
The R package MCMglmm was used to determine significance in immunosuppression assays, with the RLuc-normalized FLuc values as a Gaussian response variable. In the original screen for immunosuppressors, any experimental induction of an immune pathway was treated as a fixed effect (e.g., addition of dsRNA against FLuc in the RNAi suppression assay, PGRP-LC overexpression in the Imd suppression assay, and TollLRR transgene expression in the Toll suppression assay), each KV gene was treated as a random effect, and the interaction between KV gene and the induced experimental change to signalling output was treated as a random effect. In subsequent NF-κB suppression experiments, where the only tested KV gene was the gp83 gene, gp83 and the interaction between gp83 and overexpression of NF-κB receptors were treated as fixed effects. Likewise, when immunosuppression experiments were carried out with KV-infected cells instead of cells expressing individual KV transgenes, KV infection status, the induction of an immune pathway, and the interaction between these were treated as fixed effects.
Immunoprecipitation and Western blotting.
To test whether gp83 directly interacted with dl, 2 × 106 S2 cells were seeded in 6-well plates and transfected with 150 ng of either pAc5.1 empty vector, pAc5.1 encoding V5-tagged gp83, or V5-tagged cact alongside 150 ng of the expression plasmid (pAc5.1) encoding GFP or GFP-tagged dl. Two days posttransfection, two wells per treatment were resuspended in lysis buffer (0.1% NP-40, 30 mM HEPES-KOH, 150 mM NaCl, 2 mM MgOAc) supplemented with cOmplete protease inhibitor cocktail (Roche) and 5 mM dithiothreitol (DTT), and disrupted 30 times through a 25-gauge needle. After 10 min of incubation on ice, cell debris was pelleted by centrifugation at 16,000 × g for 30 min and the supernatant was either stored as an input control or collected and incubated for 5 h at 4°C with magnetic control beads. Binding control beads were removed and the resulting supernatant was incubated with GFP-trap magnetic beads (Chromotek) overnight at 4°C. Beads were washed 3 times in lysis buffer and 3 times in 25 mM Tris-HCl–150 mM NaCl solution, and protein complexes were eluted by boiling for 10 min at 95°C in Laemmli buffer.
Whole cellular protein extracts were prepared by heating S2 cells for 10 min at 95°C in Laemmli buffer. Whole cellular extracts or immunoprecipitated proteins were separated on a 12% SDS-PAGE gel and transferred to a nitrocellulose membrane. Nonspecific binding was blocked with blocking solution (phosphate-buffered saline [PBS] with 0.1% Triton-X [PBT] and 5% dry milk). Proteins of interest were probed with primary antibody diluted in blocking solution overnight at 4°C and visualized with a 1-h incubation of secondary antibody in blocking solution. Membranes were washed 3 times in PBT before and after each step. The following antibodies were used: mouse anti-dl (1:100 dilution; Developmental Studies Hybridoma Bank), mouse anti-V5 (1:1,000 dilution; Invitrogen), rat anti-tubulin α (1:1,000 dilution; SanBio), and rabbit anti-GFP (1:1,500 dilution; Abcam; ab6556) as primary antibodies and goat anti-mouse IR-Dye 680 (1:15,000 dilution; LI-COR), goat anti-rat IR-Dye 800 (1:15,000 dilution; LI-COR), and goat anti-rabbit IR-Dye 800 (1:15,000; LI-COR) as secondary antibodies. An Odyssey infrared imager (LI-COR) was used to image blots.
Mass spectrometry.
A total of 106 S2 cells were cotransfected with pCoBLAST and pAc5.1-gp83GFP plasmid at a 1:19 ratio (125 ng and 2.38 μg, respectively). Medium was replaced 3 h posttransfection and again at 48 h posttransfection with medium supplemented with blasticidin (20 μg/ml). Another 48 h later, cells were refreshed with medium containing 4 μg/ml of blasticidin, which was thereafter replaced every 3 to 4 days with medium containing 4 μg/ml of blasticidin, resulting in a polyclonal cell line.
For mass spectrometry, wild-type S2 cells or S2 cells stably expressing GP83GFP were lysed in 50 mM Tris-HCl (pH 7.8), 150 mM NaCl, 1% NP-40, 0.5 mM DTT, 10% glycerol, and protease inhibitor cocktail (Roche). Approximately 4 mg of protein lysate was subjected to GFP affinity purification using 7.5 μl of GFP-trap beads (Chromotek) for approximately 1.5 h at 4°C. Beads were washed twice in lysis buffer, twice in PBS containing 1% NP-40, and three times in PBS, followed by on-bead trypsin digestion as described previously (80). Afterwards, tryptic peptides were acidified and desalted using Stagetips, eluted, and brought onto an EASY-nLC 1000 liquid chromatograph (Thermo Scientific). Mass spectra were recorded on a QExactive mass spectrometer (Thermo Scientific), and mass spectrometry (MS) and MS2 data were recorded using TOP10 data-dependent acquisition. Maxquant (v1.5.1.0) was used to analyze raw data, using recommended settings (81). LFQ, IBAQ, and match between runs were enabled. The peptides were mapped to D. melanogaster proteins (UniProt; June 2017), and contaminants and reverse hits were filtered with Perseus (v1.3.0.4) (82). Missing values were imputed, assuming a normal distribution, and significance was determined by a t test on log-transformed LFQ values between wild-type and gp83-expressing S2 cells.
Immunofluorescence microscopy.
A total of 5 × 105 S2 cells were seeded in 12-well plates with glass coverslips in each well. Cells were transfected with 100 ng of pAc5.1 or pAc5.1-gp83-V5 and 100 ng of pAc5.1-dl-GFP. Two days after transfection, cells were fixed with 4% paraformaldehyde, washed twice in PBS and once with PBT, and blocked with PBT with 10% goat serum. Cells were stained by incubation with mouse anti-V5 (1:400; Invitrogen) for 1 h, followed by fluorophore-containing goat anti-mouse secondary antibody (1:400, Alexa Fluor) with 10 μg/ml Hoechst for 1 h. Finally, cells were washed twice in PBT and twice in PBS, mounted on slides with Fluoromount-G (eBiosciences), and imaged with an Olympus FluoView FV1000. Fluorescence was measured in whole cells or separately in the cytoplasm and nuclei by outlining the region of interest in Fiji (73) to calculate the mean fluorescence.
Data availability.
All data presented in this article, and associated code to fit statistical models, are provided via Figshare (https://doi.org/10.6084/m9.figshare.c.4151009).
ACKNOWLEDGMENTS
We thank Maria-Carla Saleh, Marc Dionne, François Leulier, David Finnegan, and Bruno Lemaitre for kindly sharing RNAi, Toll, and Imd pathway mutant fly lines. We thank Pascale Dijkers, Jean-Luc Imler, Neal Silverman, Edan Foley, and Norbert Perrimon (Addgene plasmid 37393) for kindly sharing Toll, Imd, and JAK-STAT constructs. We thank Rob Unckless for sharing a DiNV DNA sample with us. We thank the Ruth Steward and the Developmental Studies Hybridoma Bank for making the dorsal antibody available.
W.H.P. is supported by the Darwin Trust of Edinburgh and by an EMBO Short-Term Fellowship in R.P.V.R.’s laboratory (Grant Number 7095). Work in R.P.V.R.’s laboratory is supported by a European Research Council Consolidator Grant under the European Union’s Seventh Framework Program (grant number ERC CoG 615680) and a VICI grant from the Netherlands Organization for Scientific Research (grant number 016.VICI.170.090).
REFERENCES
- 1.Merkling SH, van Rij RP. 2013. Beyond RNAi: antiviral defense strategies in Drosophila and mosquito. J Insect Physiol 59:159–170. doi: 10.1016/j.jinsphys.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Cherry S. 2014. Viruses and antiviral immunity in Drosophila. Dev Comp Immunol 42:67–84. doi: 10.1016/j.dci.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bronkhorst AW, van Rij RP. 2014. The long and short of antiviral defense: small RNA-based immunity in insects. Curr Opin Virol 7:19–28. doi: 10.1016/j.coviro.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Lamiable O, Imler JL. 2014. Induced antiviral innate immunity in Drosophila. Curr Opin Microbiol 20:62–68. doi: 10.1016/j.mib.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer WH, Varghese FS, van Rij RP. 2018. Natural variation in resistance to virus infection in dipteran insects. Viruses 10:E118. doi: 10.3390/v10030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clem RJ. 2001. Baculoviruses and apoptosis: the good, the bad, and the ugly. Cell Death Differ 8:137–143. doi: 10.1038/sj.cdd.4400821. [DOI] [PubMed] [Google Scholar]
- 7.Bronkhorst AW, van Cleef KWR, Vodovar N, Ince IA, Blanc H, Vlak JM, Saleh M-C, van Rij RP. 2012. The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc Natl Acad Sci U S A 109:E3604–E3613. doi: 10.1073/pnas.1207213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, Dostert C, Troxler L, Hetru C, Meignin C, Pfeffer S, Hoffmann JA, Imler J-L. 2013. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol 190:650–658. doi: 10.4049/jimmunol.1102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West C, Silverman N. 2018. p38b and JAK-STAT signaling protect against Invertebrate iridescent virus 6 infection in Drosophila. PLoS Pathog 14:e1007020. doi: 10.1371/journal.ppat.1007020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayachandran B, Hussain M, Asgari S. 2012. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J Virol 86:13729–13734. doi: 10.1128/JVI.02041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webster CL, Waldron FM, Robertson S, Crowson D, Ferrari G, Quintana JF, Brouqui J-M, Bayne EH, Longdon B, Buck AH, Lazzaro BP, Akorli J, Haddrill PR, Obbard DJ. 2015. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol 13:e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bronkhorst AW, van Cleef KWR, Venselaar H, van Rij RP. 2014. A dsRNA-binding protein of a complex invertebrate DNA virus suppresses the Drosophila RNAi response. Nucleic Acids Res 42:12237–12248. doi: 10.1093/nar/gku910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P. 1995. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science 269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 14.Xue D, Horvitz HR. 1995. Inhibition of the Caenorhabditis elegans cell-death protease CED-3 by a CED-3 cleavage site in baculovirus p35 protein. Nature 377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 15.Byers NM, Vandergaast RL, Friesen PD. 2016. Baculovirus inhibitor-of-apoptosis Op-IAP3 blocks apoptosis by interaction with and stabilization of a host insect cellular IAP. J Virol 90:533–544. doi: 10.1128/JVI.02320-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valanne S, Wang J-H, Rämet M. 2011. The Drosophila Toll signaling pathway. J Immunol 186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- 17.Myllymäki H, Valanne S, Rämet M. 2014. The Drosophila Imd signaling pathway. J Immunol 192:3455–3462. doi: 10.4049/jimmunol.1303309. [DOI] [PubMed] [Google Scholar]
- 18.Zambon RA, Nandakumar M, Vakharia VN, Wu LP. 2005. The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci U S A 102:7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xi Z, Ramirez JL, Dimopoulos G. 2008. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog 4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramirez JL, Dimopoulos G. 2010. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev Comp Immunol 34:625–629. doi: 10.1016/j.dci.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira ÁG, Naylor H, Esteves SS, Pais IS, Martins NE, Teixeira L. 2014. The Toll-Dorsal pathway is required for resistance to viral oral infection in Drosophila. PLoS Pathog 10:e1004507. doi: 10.1371/journal.ppat.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fragkoudis R, Chi Y, Siu RWC, Barry G, Attarzadeh-Yazdi G, Merits A, Nash AA, Fazakerley JK, Kohl A. 2008. Semliki Forest virus strongly reduces mosquito host defence signaling. Insect Mol Biol 17:647–656. doi: 10.1111/j.1365-2583.2008.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costa A, Jan E, Sarnow P, Schneider D. 2009. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One 4:e7436. doi: 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. 2009. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog 5:e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waldock J, Olson KE, Christophides GK. 2012. Anopheles gambiae antiviral immune response to systemic O’nyong-nyong infection. PLoS Negl Trop Dis 6:e1565. doi: 10.1371/journal.pntd.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sansone CL, Cohen J, Yasunaga A, Xu J, Osborn G, Subramanian H, Gold B, Buchon N, Cherry S. 2015. Microbiota-dependent priming of antiviral intestinal immunity in Drosophila. Cell Host Microbe 18:571–581. doi: 10.1016/j.chom.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thoetkiattikul H, Beck MH, Strand MR. 2005. Inhibitor B-like proteins from a polydnavirus inhibit NF-κB activation and suppress the insect immune response. Proc Natl Acad Sci U S A 102:11426–11431. doi: 10.1073/pnas.0505240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamiable O, Kellenberger C, Kemp C, Troxler L, Pelte N, Boutros M, Marques JT, Daeffler L, Hoffmann JA, Roussel A, Imler J-L. 2016. Cytokine Diedel and a viral homologue suppress the IMD pathway in Drosophila. Proc Natl Acad Sci U S A 113:698–703. doi: 10.1073/pnas.1516122113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bitra K, Suderman RJ, Strand MR. 2012. Polydnavirus Ank proteins bind NF-κB homodimers and inhibit processing of Relish. PLoS Pathog 8:e1002722. doi: 10.1371/journal.ppat.1002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herniou EA, Huguet E, Thézé J, Bézier A, Periquet G, Drezen J-M. 2013. When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Philos Trans R Soc Lond B Biol Sci 368:20130051. doi: 10.1098/rstb.2013.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mlih M, Khericha M, Birdwell C, West AP, Karpac J. 2018. A virus-acquired host cytokine controls systemic aging by antagonizing apoptosis. PLoS Biol 16:e2005796. doi: 10.1371/journal.pbio.2005796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer WH, Medd NC, Beard PM, Obbard DJ. 2018. Isolation of a natural DNA virus of Drosophila melanogaster, and characterisation of host resistance and immune responses. PLoS Pathog 14:e1007050. doi: 10.1371/journal.ppat.1007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huger AM. 2005. The Oryctes virus: its detection, identification, and implementation in biological control of the coconut palm rhinoceros beetle, Oryctes rhinoceros (Coleoptera: Scarabaeidae). J Invertebr Pathol 89:78–84. doi: 10.1016/j.jip.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Jehle JA. 2009. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J Invertebr Pathol 101:187–193. doi: 10.1016/j.jip.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Unckless RL. 2011. A DNA virus of Drosophila. PLoS One 6:e26564. doi: 10.1371/journal.pone.0026564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huger AM. 1985. A new virus disease of crickets (Orthoptera: Gryllidae) causing macronucleosis of fatbody. J Invertebr Pathol 45:108–111. doi: 10.1016/0022-2011(85)90055-2. [DOI] [Google Scholar]
- 37.Zelazny B. 1973. Studies on Rhabdionvirus oryctes: II. Effect on adults of Oryctes rhinoceros. J Invertebr Pathol 22:122–126. doi: 10.1016/0022-2011(73)90020-7. [DOI] [Google Scholar]
- 38.Zelazny B. 1972. Studies on Rhabdionvirus oryctes: I. Effect on larvae of Oryctes rhinoceros and inactivation of the virus. J Invertebr Pathol 20:235–241. doi: 10.1016/0022-2011(72)90150-4. [DOI] [Google Scholar]
- 39.Huger AM. 1966. A virus disease of the Indian rhinoceros beetle, Oryctes rhinoceros (Linnaeus), caused by a new type of insect virus, Rhabdionvirus oryctes gen. n., sp. n. J Invertebr Pathol 8:38–51. doi: 10.1016/0022-2011(66)90101-7. [DOI] [PubMed] [Google Scholar]
- 40.Parisien J-P, Lau JF, Horvath CM. 2002. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J Virol 76:6435–6441. doi: 10.1128/JVI.76.13.6435-6441.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, Münk C, Nymark-McMahon H, Landau NR. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31. doi: 10.1016/S0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 42.Goffinet C, Allespach I, Homann S, Tervo H-M, Habermann A, Rupp D, Oberbremer L, Kern C, Tibroni N, Welsch S, Krijnse-Locker J, Banting G, Kräusslich H-G, Fackler OT, Keppler OT. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5:285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villán E, García-Sastre A, Gack MU. 2012. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog 8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Mierlo JT, Overheul GJ, Obadia B, van Cleef KWR, Webster CL, Saleh MC, Obbard DJ, van Rij RP. 2014. Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS Pathog 10:e1004256. doi: 10.1371/journal.ppat.1004256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stabell AC, Meyerson NR, Gullberg RC, Gilchrist AR, Webb KJ, Old WM, Perera R, Sawyer SL. 2018. Dengue viruses cleave STING in humans but not in nonhuman primates, their presumed natural reservoir. Elife 7:e31919. doi: 10.7554/eLife.31919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer WH, Joosten J, Overheul GJ, Jansen PW, Vermeulen M, Obbard DJ, Van Rij RP. 2018. Induction and suppression of NF-κB signalling by a DNA virus of Drosophila. bioRxiv 10.1101/358176. [DOI] [PMC free article] [PubMed]
- 47.Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, Richardson MF, Anholt RRH, Barrón M, Bess C, Blankenburg KP, Carbone MA, Castellano D, Chaboub L, Duncan L, Harris Z, Javaid M, Jayaseelan JC, Jhangiani SN, Jordan KW, Lara F, Lawrence F, Lee SL, Librado P, Linheiro RS, Lyman RF, Mackey AJ, Munidasa M, Muzny DM, Nazareth L, Newsham I, Perales L, Pu L-L, Qu C, Ràmia M, Reid JG, Rollmann SM, Rozas J, Saada N, Turlapati L, Worley KC, Wu Y-Q, Yamamoto A, Zhu Y, Bergman CM, Thornton KR, Mittelman D, Gibbs RA. 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Cleef KWR, Van Mierlo JT, Miesen P, Overheul GJ, Fros JJ, Schuster S, Marklewitz M, Pijlman GP, Junglen S, Van Rij RP. 2014. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res 42:8732–8744. doi: 10.1093/nar/gku528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Li WX, Ding SW. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 50.Van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. 2006. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nayak A, Berry B, Tassetto M, Kunitomi M, Acevedo A, Deng C, Krutchinsky A, Gross J, Antoniewski C, Andino R. 2010. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol 17:547–554. doi: 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Mierlo JT, Bronkhorst AW, Overheul GJ, Sadanandan SA, Ekström J-O, Heestermans M, Hultmark D, Antoniewski C, van Rij RP. 2012. Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathog 8:e1002872. doi: 10.1371/journal.ppat.1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL. 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol 6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 54.Baeg G-H, Zhou R, Perrimon N. 2005. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev 19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arbouzova NI, Zeidler MP. 2006. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development 133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- 56.Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. 2000. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci U S A 97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Kleespies RG, Huger AM, Jehle JA. 2007. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J Virol 81:5395–5406. doi: 10.1128/JVI.02781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao J, He S, Minassian A, Li J, Feng P. 2015. Recent advances on viral manipulation of NF-κB signaling pathway. Curr Opin Virol 15:103–111. doi: 10.1016/j.coviro.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zehavi Y, Sloutskin A, Kuznetsov O, Juven-Gershon T. 2014. The core promoter composition establishes a new dimension in developmental gene networks. Nucleus 5:298–303. doi: 10.4161/nucl.29838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM. 2009. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci 364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sawyer SL, Elde NC. 2012. A cross-species view on viruses. Curr Opin Virol 2:561–568. doi: 10.1016/j.coviro.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brockhurst MA, Chapman T, King KC, Mank JE, Paterson S, Hurst GDD. 2014. Running with the Red Queen: the role of biotic conflicts in evolution. Proc Biol Sci 281:20141382. doi: 10.1098/rspb.2014.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nitta KR, Jolma A, Yin Y, Morgunova E, Kivioja T, Akhtar J, Hens K, Toivonen J, Deplancke B, Furlong EEM, Taipale J. 2015. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. Elife 4:e04837. doi: 10.7554/eLife.04837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117:69–81. doi: 10.1016/S0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 65.Okamura K, Ishizuka A, Siomi H, Siomi MC. 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weber ANR, Gangloff M, Moncrieffe MC, Hyvert Y, Imler J-L, Gay NJ. 2007. Role of the Spätzle Pro-domain in the generation of an active Toll receptor ligand. J Biol Chem 282:13522–13531. doi: 10.1074/jbc.M700068200. [DOI] [PubMed] [Google Scholar]
- 67.Nusslein-Volhard C. 1979. Maternal effect mutations that alter the spatial coordinates of the embryo of Drosophila melanogaster, p 185–211. In Subtelny S, Konigsberg IR (ed), Determinants of spatial organization. Academic Press, New York, NY. [Google Scholar]
- 68.Anderson KV, Nüsslein-Volhard C. 1984. Information for the dorsal-ventral pattern of the Drosophila embryo is stored as maternal mRNA. Nature 311:223–227. doi: 10.1038/311223a0. [DOI] [PubMed] [Google Scholar]
- 69.Hecht PM, Anderson KV. 1993. Genetic characterization of tube and pelle, genes required for signaling between Toll and dorsal in the specification of the dorsal-ventral pattern of the Drosophila embryo. Genetics 135:405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, Hultmark D. 1999. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell 4:827–837. doi: 10.1016/S1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 71.Pham LN, Dionne MS, Shirasu-Hiza M, Schneider DS. 2007. A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog 3:e26. doi: 10.1371/journal.ppat.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw 33:1–22.20808728 [Google Scholar]
- 73.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hill T, Unckless RL. 2018. The dynamic evolution of Drosophila innubila nudivirus. Infect Genet Evol 57:151–157. doi: 10.1016/j.meegid.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y-X, Dijkers PF. 2015. Specific calcineurin isoforms are involved in Drosophila Toll immune signaling. J Immunol 194:168–176. doi: 10.4049/jimmunol.1401080. [DOI] [PubMed] [Google Scholar]
- 76.Kaneko T, Yano T, Aggarwal K, Lim J-H, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh B-H, Kurata S, Silverman N. 2006. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol 7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 77.Foley E, O’Farrell PH. 2004. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol 2:e203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Cleef KWR, van Mierlo JT, van den Beek M, van Rij RP. 2011. Identification of viral suppressors of RNAi by a reporter assay in Drosophila S2 cell culture. Methods Mol Biol 721:201–213. doi: 10.1007/978-1-61779-037-9_12. [DOI] [PubMed] [Google Scholar]
- 79.Werner T, Borge-Renberg K, Mellroth P, Steiner H, Hultmark D. 2003. Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J Biol Chem 278:26319–26322. doi: 10.1074/jbc.C300184200. [DOI] [PubMed] [Google Scholar]
- 80.Smits AH, Jansen PWTC, Poser I, Hyman AA, Vermeulen M. 2013. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res 41:e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 82.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 83.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data presented in this article, and associated code to fit statistical models, are provided via Figshare (https://doi.org/10.6084/m9.figshare.c.4151009).