Abstract
Background & Aims:
Diverticulitis is the chronic inflammation of diverticula. Whether the pathophysiology of earlier-onset patients differs from later-onset patients is unknown. We profiled the colonic transcriptomes of these two patient populations to gain insight into the molecular underpinnings of diverticulitis.
Methods:
We conducted deep RNA sequencing (RNA-seq) on colonic segments surgically resected from earlier-onset (<42 years old, n=13) and later-onset (>65 years old, n=13) diverticulitis patients. We used bioinformatic approaches to cluster the patients based on the relationship of differentially expressed genes and to inform on the molecular pathways that segregated the clusters.
Results:
Principal component analysis identified three patient clusters; diverticulitis later-onset (DVT-LO), diverticulitis mixed-onset (DVT-MO), and diverticulitis earlier-onset (DVT-EO). The patients comprising DVT-EO, which was the majority of earlier-onset patients, displayed increased expression of anti-viral response genes. This finding was confirmed using an independent weighted co-expression network analysis (WGCNA) of differentially expressed genes.
Conclusions:
We found that the majority of patients with earlier-onset disease contained elevated expression of host genes involved in the anti-viral response. Thus, susceptibility to a viral pathogen may offer one explanation why some individuals develop diverticulitis at an earlier age.
Keywords: diverticulitis, earlier-onset, RNA-seq, viral response
INTRODUCTION
Diverticulosis is an asymptomatic condition defined by the development of diverticula or outpouchings of the colonic mucosa and submucosa through the muscularis [1]. It is estimated that about 4% of individuals with diverticulosis develop diverticular inflammation, known as diverticulitis [2]. Diverticular disease, encompassing both diverticulosis and diverticulitis, is a significant healthcare burden in the United States, contributing to more inpatient and outpatient healthcare visits than inflammatory bowel disease (IBD) [3]. Diverticulosis is associated with the aging population, affecting approximately 50% of individuals aged 60 [4]. However, adults can be afflicted with diverticulitis at an earlier age and in this population, there is a higher risk of disease recurrence [5]. Whether the pathophysiology of earlier-onset disease is distinct from later-onset disease is unknown.
Although diverticular disease is highly prevalent in the adult population, the disease etiology is poorly understood. As antibiotics are used for the medical management of acute, uncomplicated diverticulitis, the disease is thought to be caused by an infectious process [6]. Thus, studies evaluating the role of the intestinal microbiome have recently become of interest and potential microbial populations contributing to diverticulitis have been identified [7, 8]. As randomized clinical trials found no differences in outcomes for acute diverticulitis patients treated with or without antibiotics, the precise role of bacteria in disease pathogenesis is uncertain [9, 10]. It is currently unknown whether viruses contribute to the pathogenesis of diverticulitis.
Due to the lack of a reliable animal model [11, 12], studying diverticulitis requires access to human colonic tissues. Obtaining inflamed diverticulum is a challenge as colonoscopy and biopsies are contraindicated during diverticulitis and recruiting patients for surgical tissue during an emergent surgery is often not feasible. The availability of resected tissue samples obtained at the time of surgery during disease quiescence is a valuable resource to gain insight into novel mechanisms contributing disease. In a recent study, we used RNA deep sequencing (RNA-seq) to define the colonic transcriptomes in diverticulitis patients versus non-diverticulosis controls [13]. Although the tissue analyzed lacked gross evidence of inflammation, molecular pathways associated with the immune response were enriched in diverticulitis patients [13]. The aim of this study was to use RNA-seq to investigate molecular signatures that distinguish patients who develop diverticulitis at an earlier versus later age.
METHODS
Specimen collection
The Pennsylvania State University College of Medicine Institutional Review Board approved this study. Patients that underwent elective sigmoid resection for multiple previous episodes of diverticulitis were identified and consented to have their tissue collected into the Penn State Hershey Colorectal Diseases Biobank following sigmoid resection. Previous episodes of diverticulitis were confirmed by computed tomography scans and surgical pathology. Surgical specimens were immediately transferred from the operating room to the surgical pathology laboratory. Full thickness sections of sigmoid colon were obtained away from any evident sites of active disease and stored in either RNAlater (Invitrogen, Carlsbad, CA) or formalin-fixed and paraffin-embedded. Diverticulitis patients were excluded if they had a history of IBD, cancer, or dysplasia.
RNA-sequencing
We subjected resected colonic tissues from an independent cohort of diverticulitis patients (n=26) to RNA-seq using protocols previously described, with the exception that NEXTflex qRNA-Seq unique molecular indices (Bioo Scientific, Austin, TX) were incorporated into the libraries [13]. Quality of the sequenced reads was assessed using FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects.fastqc). FASTQ files were uploaded to the Galaxy web platform at usegalaxy.org and the public server was used for pre-processing steps [14]. Trimmomatic (Galaxy v0.36.3) was used to remove the molecular indices from the sequencing reads, resulting in a final read length of 51 bp [15]. Filtered reads maintained a mean per base quality score >37. Reads were aligned to GRCh37.p13 (GENCODE release 19) using RNA STAR (Galaxy v2.5.2b-0) set to default parameters [16, 17]. Reads were counted using HTSeq-Count (Galaxy v0.6.1galaxy3) set to union mode with a minimum alignment quality of 10 [18]. The datasets generated and analyzed are available in the Gene Expression Omnibus (GEO) repository under the following accession number GSE111819 (https://www.ncbi.nlm.nih.gov/geo/).
Differential gene expression analysis
As controls, gene read counts (v7) generated from the RNA-seq analysis of sigmoid colon tissue from healthy individuals were downloaded from the Genotype-Tissue Expression project (GTEx) portal [19, 20]. In total, RNA-seq data was available from the sigmoid colon tissue of 233 GTEx subjects. GTEx subjects (n=52) were age- and sex-matched 2:1 to our diverticulitis cohort. Reads were evaluated for differential expression using DESeq2 in R v3.4.3 (R Software for Statistical Computing, Vienna, Austria) [21]. Genes were excluded if expression was not detected in at least 10% of the entire study cohort. Differentially expressed genes were visualized by volcano plot using the R package plotly [22]. Gene set enrichment analysis of Gene Ontology Biological Processes was performed using Enrichr from a candidate gene list comprised of differentially expressed genes (∣log2 fold change∣ >0.60 and q-value <0.05) and top ontologies reported by q-value [23, 24].
Hierarchical clustering on principal components
Mean absolute deviation (MAD) was calculated from normalized expression values using the variance stabilizing transformation (VST) method in DESeq2 and the genes with the highest variability were identified or those falling below the mean three times were removed, accounting for about 4% of the total genes analyzed. Hierarchical Clustering on Principal Components (HCPC) was performed on the genes with the highest MAD using the R packages FactoMineR and factoextra [25, 26]. Three-dimensional principal component analysis (PCA) plots were constructed using the R package pca3d [27]. Gene expression was compared between HCPC clusters using one-way analysis of variance (ANOVA) followed by Tukey’s honest significance difference (HSD) multiple comparison test. Results are presented by boxplot using the R package ggplot2 [28].
Weighted Gene Co-Expression Network Analysis
Following VST, normalized read counts were used from 12,488 protein coding genes for signed network analysis using the weighted gene co-expression network analysis (WGCNA) package in R [29]. Using a soft thresholding power of 7, clustering was performed as previously described using topological overlap matrix (TOM)-based dissimilarity followed by a clustering of consensus module eigengenes (MEs) [13]. Pearson correlation was used to evaluate the relationship between MEs and clinical traits, including age at diagnosis, age at surgery, sex, smoking status, history of fistula, and history of perforation/abscess.
RESULTS
A unique gene signature distinguishes earlier-onset from later-onset diverticulitis patients
RNA-seq profiles the transcriptome at a genome-wide level and captures the effect of both host genetics and the environment on gene expression [30]. We deep-sequenced colonic mRNAs from an independent cohort of patients who underwent sigmoid colon resection for diverticulitis (n=26) (Table I). Most patients (57.5%) required a surgical resection for complicated (perforation or abscess) diverticulitis, but were done electively subsequent to acute presentation. However, two individuals received an urgent or semi-elective procedure. Patients were selected to represent two distinct age classes of diverticulitis, those diagnosed with earlier-onset disease (<42 years old, n=13) and those diagnosed with later-onset disease (>65 years old, n=13). Groups were matched for sex, smoking status, history of familial diverticular disease, and history of complicated diverticulitis (Table I). Of the 12,488 protein coding genes evaluated, a total of 2,627 (21.0%) were differentially expressed with a ∣log2 fold change∣ >0.60 and q <0.05, of which 1,326 and 1,301 were up- and down-regulated, respectively (Fig. 1A). We performed a gene set enrichment analysis to identify molecular pathways associated with the differentially expressed genes. In earlier-onset patients, we found an enrichment of gene ontologies associated with viral response (Suppl. Table I) and a down-regulation of those involved in the nervous system (Suppl. Table II).
Table I.
Patient demographics
| Parameter | Early onset diverticulitis |
Later onset diverticulitis |
P-value |
|---|---|---|---|
| Patients (n) | 13 | 13 | n.a. |
| Sex, male, n (%), female, n (%) | 8 (61.5), 5 (38.5) | 4 (30.8), 9 (69.2) | 0.238 |
| *Smoking status, positive, n (%), negative, n (%) | 8 (61.5), 5 (38.5) | 7 (53.8), 6 (46.2) | 1 |
| Age of first diverticulitis episode, yrs, mean, SD | 37.1 ± 4.7 | 73.2 ± 4.7 | 3.14 × 10−16 |
| Type of surgery, elective, n (%), urgent/emergent, n (%) | 11 (84.6), 2 (15.4) | 15 (100), 0 (0) | 0.206 |
| Age of sigmoid resection, years, mean, SD | 42.6 ± 7.3 | 74.6 ± 5.0 | 1.29 × 10−11 |
| Body mass index, kg/m2, mean, SD | 30.5 ± 8.0 | 31.1 ± 4.9 | 0.817 |
| Familial diverticular disease, yes, n (%), no, n (%) | 4 (30.8), 9 (69.2) | 4 (30.8), 9 (69.2) | 1 |
| History of perforation/abscess, yes, n (%), no, n (%) | 6 (46.2), 7 (53.8) | 8 (61.5), 5 (35.8) | 0.695 |
| History of fistula, yes, n (%), no, n (%) | 3 (23.1), 10 (76.9) | 6 (46.2), 7 (53.8) | 0.411 |
Positive smoking status includes both current and former smokers at the time of surgery.
Continuous variables analyzed by two-tailed t-test, categorical variables analyzed by two-tailed Fisher’s Exact Test.
SD: standard deviation.
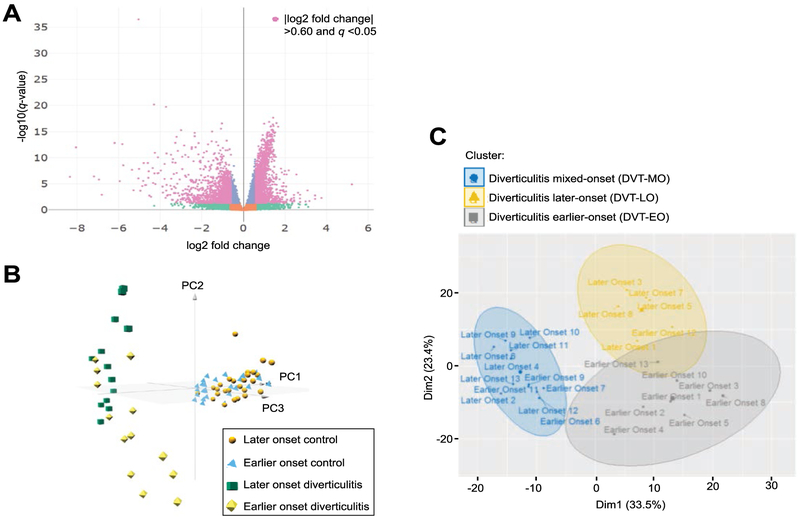
Fig. 1.
A unique gene signature distinguishes earlier-onset from later-onset diverticulitis patients. (A) Volcano plot of differentially expressed genes in earlier-onset (<42 years old at diagnosis, n=13) versus later-onset diverticulitis patients (>65 years old at diagnosis, n=13). Each gene is represented by a point with colors indicating significance of differential expression, where green are genes with a ∣log2 fold change∣ >0.60, purple is significant at a q <0.05, and pink are significant with a ∣log2 fold change∣ >0.60 and q <0.05. Genes shown in orange are non-significant and do not meet any of the aforementioned criteria. (B) Three-dimensional principal component analysis plot of diverticulitis patients (n=26) and age- and sex-matched controls (n=52). Diverticulitis patients are delineated into earlier-onset (green) and later-onset (yellow). GTEx controls were age-matched to diverticulitis patients: earlier-onset (blue) and later-onset (orange). (C) Hierarchical Clustering on Principal Components of diverticulitis patients (n=26) using the genes with the highest mean absolute deviation (n=374). Three clusters were identified and designated DVT-MO (blue), DVT-LO (yellow), DVT-EO (gray).
To determine whether the differences between earlier-onset and later-onset diverticulitis patients were related to aging, we identified a cohort of age- and sex-matched healthy sigmoid colon tissues that was generated by the GTEx project [19, 20]. RNA-seq datasets generated from GTEx controls and our diverticulitis patients were found to occupy distinct spaces in three-dimensional principal component analysis (PCA) (Fig. 1B). Although the GTEx controls showed tight clustering of all individuals, a wide spread was seen amongst our diverticulitis cohort that segregated by age. Thus, distinct disease mechanisms rather than the aging process likely drove gene expression differences in earlier-onset and later-onset diverticulitis patients.
As PCA identified an age-associated distribution of diverticulitis patients, we used Hierarchical Clustering on Principal Components (HCPC) to identify distinct disease phenotypes. Three clusters or phenotypes were distinguished within our diverticulitis cohort (Fig. 1C). The first cluster, DVT-LO, contained primarily patients that developed disease at a later age. The second cluster, DVT-MO, contained both earlier-onset and later-onset patients. The third cluster, designated DVT-EO, contained only those patients with earlier-onset diverticulitis. Importantly, the DVT-EO cluster contained the majority of earlier-onset diverticulitis patients (8 out of 12) analyzed in our study.
A viral response signature distinguishes DCT-EO from DVT-MO and DVT-LO
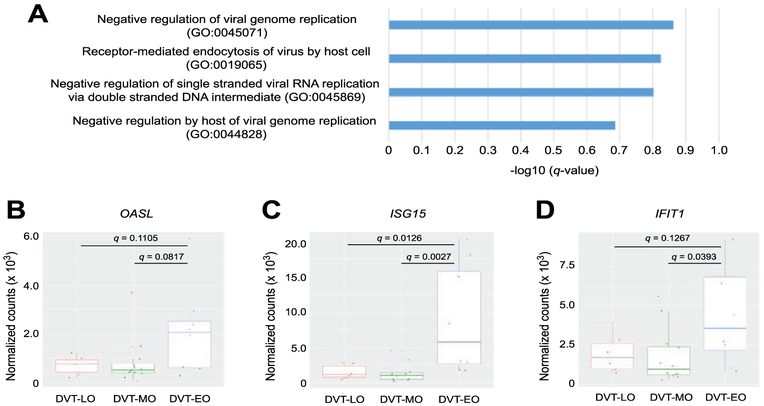
Based upon gene expression profiles resulting in patient clustering, the main physiological feature segregating DVT-MO and DVT-LO were genes and pathways associated with muscle contraction (Suppl. Table III). In particular, down-regulation of muscle contraction pathways, including smooth muscle, was found in DVT-MO whereas up-regulation of these pathways was found in DVT-LO. Notably, gene set enrichment analysis of those transcripts defining the DVT-EO cluster identified viral response pathways (Fig. 2A). We found enrichment of the OAS family member 2’-5’-oligoadenylate synthetase-like (OASL) in DVT-EO (Fig. 2B). Furthermore, genes induced by interferons were also up-regulated in DVT-EO, including interferon-stimulated gene 15 (ISG15) and IFIT1 (Fig. 2 C and D). Aside from age of diagnosis and surgery, no overt clinical differences were noted amongst the three clusters (Suppl. Table IV).
Fig. 2.
A viral response signature distinguishes DVT-EO from DVT-MO and DVT-LO. (A) Gene set enrichment analysis of enriched genes contributing to DVT-EO (n=151). Only those associated with the host viral response and are significant at a discovery threshold q <0.25 are presented. (B) OASL read counts from RNA-seq (n=26) were normalized by variance stabilizing transformation and delineated according to patient clustering profiles; DVT-LO, DVT-MO, and DVT-EO. Data is presented as a boxplot and displays the median values for each cluster. Groups were compared using one-way ANOVA followed by Tukey HSD multiple comparison test. (C) and (D) Analysis of ISG15 and IFIT1 gene expression levels, respectively.
Earlier-onset diverticulitis is associated with a host response to virus
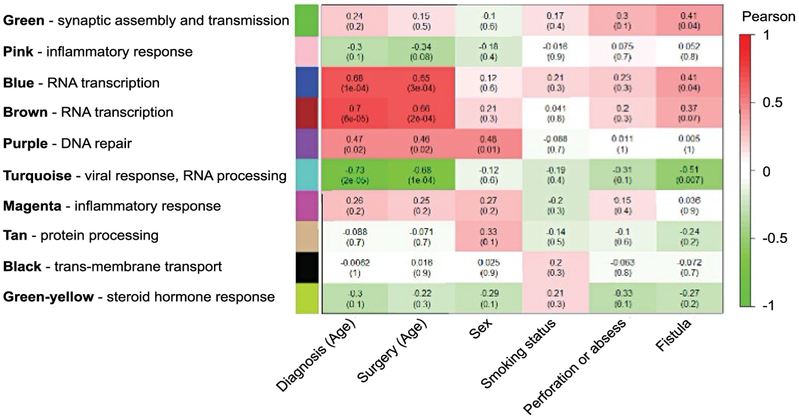
To assess whether differences in the transcriptome correlate with clinical traits, we performed a signed WGCNA [29]. WGCNA was conducted on 12,488 protein coding genes that were differentially-expressed between earlier-onset and later-onset patients and the following patient clinical metadata: age at diagnosis, age at surgery, sex, smoking status, history of fistula, or history of perforation/abscess. Ten modules were established and the genes assigned to each module were subjected to gene set enrichment analysis to identify the biological mechanisms contributing to these correlations (Fig. 3). We found that genes associated with the host response to the virus comprised the turquoise module corresponding to pathways such as viral transcription (GO:0019083, q=1.77×10−9) and viral entry into the host cell (GO:0046718, q=0.0004). Importantly, we found a negative correlation between the turquoise ME and age of surgery (r=−0.68, P=1×10−4) and age of diagnosis (r=−0.73, P=2×10−5), confirming our previous finding of an increased viral response in the earlier-onset diverticulitis cohort (Fig. 2). These findings further substantiate that the host defense response to virus is associated with earlier-onset diverticulitis.
Fig. 3.
Earlier-onset diverticulitis is associated with a host response to virus. Signed weighted gene co-expression network analysis of RNA-seq transcripts from diverticulitis patients (n=26). Genes were segregated into ten modules designated by color. Gene set enrichment analysis was performed on each module and the representative molecular pathways are presented. The heat map represents the Pearson correlation coefficient where red indicates a positive correlation and green indicates a negative correlation. P-values associated with the correlation analysis are presented in parenthesis with the Pearson correlation coefficient.
DISCUSSION
In our analysis of transcripts derived from intestinal tissues of a distinct cohort of earlier-onset and later-onset diverticulitis patients, we evaluated gene expression profiles and did not see a distinct clustering of patients solely by age. Rather, diverticulitis patients segregated in a separate threedimensional space from GTEx control colon tissues and were delineated into three phenotypes. Unexpectedly, the cluster comprised solely of DVT-EO patients was segregated, in part, due to genes involved in host viral response, suggesting that a different pathological mechanism may contribute to disease in these individuals. This association of the host response to virus was confirmed through WGCNA. Given that earlier-onset individuals are more likely to develop recurrent diverticulitis, a model for intestinal viral infection in these individuals is reasonable [5]. While some viruses are highly infectious and depend on rapid transmission of virions for primary infections, others establish persistence due to life-long latency in the host and undergo periodic reactivation [31].
A prior study in Sweden uncovered a high prevalence of human cytomegalovirus (HCMV) early proteins in resected tissues from diverticulitis patients [32]. Moreover, anti-HCMV antibodies were detected in serum from all patients tested [32]. The authors speculated that the inflammatory microenvironment could contribute to reactivation of latent HCMV in diverticulitis and that an active virus could potentiate expression of pro-inflammatory cytokines. While our efforts to identify HCMV were unsuccessful, it is possible that the differences in patient populations between the aforementioned study and ours could account for this discrepancy. Perhaps a more thorough and longitudinal study of biopsies collected from diverticulitis patients during periods of acute and quiescent disease would shed light on the role of HCMV, or other viruses, in the pathogenesis of diverticulitis.
Using hierarchical clustering, we found that viral response genes were one component that segregated the DVT-EO cluster from DVT-MO and DVT-LO. However, differences in expression of genes involved in smooth muscle contraction were also a contributor to the clustering patterns. Both DVT-EO and DVT-LO demonstrated an up-regulation of smooth muscle contraction pathways compared to DVT-MO. That deregulated muscle contraction is associated with diverticulitis is not surprising as prior evidence suggested hypermotility in the pathogenesis of disease [33]. However, as these pathways are distinguishing DVT-EO and DVT-LO from DVT-MO, perhaps this is not a universal effect on the colon and more work is required to appreciate how muscle contraction processes contribute to diverticular disease.
In patients with Crohn’s disease, the formation of a fistula stems from impairment of wound healing [34]. In our analysis of gene expression and clinical metadata, we identified three MEs that correlated with a history of fistula, which comprised genes involved in RNA processing and transcription as well as synaptic assembly and transcription (Fig. 3). Additional work is required to dissect the contribution of genes comprising these modules to the formation of fistulas. Conversely, the similarity between the fistula and age-related modules is intriguing as the prevalence of fistulas in our younger diverticulitis group is lower (23.1%) than those in the older group (46.2%) (Table I). However, as this correlation did not reach statistical significance, we cannot exclude the possibility that this finding reflects a difference in age rather than a disease process. We also failed to identify gene expression patterns that correlated with a history of perforation or abscess. The tissue utilized in this study was not acutely inflamed which may offer one explanation why we did not identify a gene signature that correlated with these phenotypes.
We found that expression of the type I interferon-stimulated genes IFIT1 and ISG15 were up-regulated in early-onset diverticulitis patients [35]. Expression of OASL, that potentiates a type I response, was also elevated in this patient population [36]. As type I interferon signaling is a well established anti-viral host defense, it is likely that the colonic mucosa is responding to a viral pathogen. However, as bacteria and fungi can induce interferon expression, potential involvement of these microbial pathogens must be considered [37]. It is possible that viruses initiate disease in complicated diverticulitis cases requiring surgery and that colonic bacteria superimpose a second layer of insult following perforation of the bowel wall. As recent clinical trial data demonstrated similar outcomes for acute, uncomplicated diverticulitis patients treated with and without antibiotic management, their use should be reserved for complicated cases [9, 10].
CONCLUSION
Using RNA-seq analysis to define the colonic transcriptomes of diverticulitis patients, we found that the majority of those with early-onset disease displayed an enhanced antiviral response. Although the intestinal virome is poorly characterized in humans, studies are required that evaluate the viral contribution to the human intestinal microbiome and how this changes during the inflammatory processes associated with diverticulitis. As earlier-onset patients are typically afflicted with recurring disease [5], identifying a causal viral agent may lead to better therapeutic management which could delay or prevent the need for surgical intervention.
Supplementary Material
Acknowledgements:
We would like to thank Drs. John Wills and Jianming Hu for their thoughtful insight throughout the project and Yuka Imamura and Anna Salzberg for performing the RNA-seq. This project was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1 TR002014 and, in part, by the Carlino Fund for IBD Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations:
- DVT-EO
diverticulitis earlier-onset
- DVT-MO
diverticulitis mixed-onset
- DVT-LO
diverticulitis later-onset
- GO
gene ontology
- GTEx
Genotype-Tissue Expression
- IBD
inflammatory bowel disease
- IFIT
interferon-induced protein with tetratricopeptide
- ISG15
interferon-stimulated gene fifteen
- ME
module eigengene
- OASL
2’-5’ oligoadenylate synthetase-like
- PCA
principal component analysis
- RNA-seq
RNA deep sequencing
- WGCNA
weighted gene co-expression network analysis
Footnotes
Conflicts of interest: No conflicts to declare.
Authors’ contributions: K.M.S., W.A.K., G.S.Y.: study concept and design; K.M.S., B.P.K., L.R.H., S.D.: acquisition of data; K.M.S., B.P.K., W.A.K., G.S.Y.: analysis and interpretation of data; K.M.S., G.S.Y.: drafting of the manuscript; K.M.S., B.P.K., W.A.K., G.S.Y.: critical revision of the manuscript for important intellectual content; K.M.S.: statistical analysis; W.A.K., G.S.Y.: administrative, technical, or material support; W.A.K., G.S.Y.: study supervision. All authors approved the final version of the manuscript for publication.
Supplementary material: To access the supplementary material visit the online version of the J Gastrointestin Liver Dis at http://www.jgld.ro/wp/archive/y2018/n3/a9 and http://dx.doi.org/10.15403/jgld.2014.1121.273.sch
REFERENCES
- 1.Brian West A. The pathology of diverticulosis: classical concepts and mucosal changes in diverticula. J Clin Gastroenterol 2006;40 Suppl 3:S126–S131. doi: 10.1097/01.mcg.0000225508.90417.07 [DOI] [PubMed] [Google Scholar]
- 2.Strate LL, Modi R, Cohen E, Spiegel BM. Diverticular disease as a chronic illness: evolving epidemiologic and clinical insights. Am J Gastroenterol 2012;107:1486–1493. doi: 10.1038/ajg.2012.194 [DOI] [PubMed] [Google Scholar]
- 3.Peery AF, Dellon ES, Lund J, Crockett SD, et al. Burden of gastrointestinal disease in the United States: 2012 update. Gastroenterology 2012;143:1179–1187.e3. doi: 10.1053/j.gastro.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Everhart JE, Ruhl CE. Burden of digestive diseases in the United States part II: lower gastrointestinal diseases. Gastroenterology 2009;136:741–754. doi: 10.1053/j.gastro.2009.01.015 [DOI] [PubMed] [Google Scholar]
- 5.Katz LH, Guy DD, Lahat A, Gafter-Gvili A, Bar-Meir S. Diverticulitis in the young is not more aggressive than in the elderly, but it tends to recur more often: systematic review and meta-analysis. J Gastroenterol Hepatol 2013;28:1274–1281. doi: 10.1111/jgh.12274 [DOI] [PubMed] [Google Scholar]
- 6.Boynton W, Floch M. New strategies for the management of diverticular disease: insights for the clinician. Therap Adv Gastroenterol 2013;6:205–213. doi: 10.1177/1756283X13478679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniels L, Budding AE, de Korte N, et al. Fecal microbiome analysis as a diagnostic test for diverticulitis. Eur J Clin Microbiol Infect Dis 2014;33:1927–1936. doi: 10.1007/s10096-014-2162-3 [DOI] [PubMed] [Google Scholar]
- 8.Schieffer KM, Sabey K, Wright JR, et al. The Microbial Ecosystem Distinguishes Chronically Diseased Tissue from Adjacent Tissue in the Sigmoid Colon of Chronic, Recurrent Diverticulitis Patients. Sci Rep 2017;7:8467. doi: 10.1038/s41598-017-06787-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels L, Unlu C, de Korte N, et al. Randomized clinical trial of observational versus antibiotic treatment for a first episode of CT-proven uncomplicated acute diverticulitis. Br J Surg 2017;104:52–61. doi: 10.1002/bjs.103090 [DOI] [PubMed] [Google Scholar]
- 10.Shabanzadeh DM, Wille-Jorgensen P. Antibiotics for uncomplicated diverticulitis. Cochrane Database Syst Rev 2012;11:CD009092. doi: 10.1002/14651858.CD009092.pub2 [DOI] [PubMed] [Google Scholar]
- 11.Fisher N, Berry CS, Fearn T, Gregory JA, Hardy J. Cereal dietary fiber consumption and diverticular disease: a lifespan study in rats. Am J Clin Nutr 1985;42:788–804. doi: 10.1093/ajcn/42.5.788 [DOI] [PubMed] [Google Scholar]
- 12.Wess L, Eastwood M, Busuttil A, Edwards C, Miller A. An association between maternal diet and colonic diverticulosis in an animal model. Gut 1996;39:423–427. doi: 10.1136/gut.39.3.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schieffer KM, Choi CS, Emrich S, et al. RNA-seq implicates deregulation of the immune system in the pathogenesis of diverticulitis. Am J Physiol Gastrointest Liver Physiol 2017;313:G277–G284. doi: 10.1152/ajpgi.00136.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afgan E, Baker D, van den Beek M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 2016;44(W1):W3–W10. doi: 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012;22:1760–1774. doi: 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium GTEx. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; et al. Genetic effects on gene expression across human tissues. Nature 2017;550:204–213. doi: 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plotly Technologies Inc; Collaborative data science. Montreal, QC, 2015. https://plot.ly [Google Scholar]
- 23.Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128. doi: 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kassambara A, Mundt F. Factoextra: Extract and Visualize the Results of Multivariate Data Analysis. 2017. Available at: https://CRAN.R-project.org/package=factoextra [Google Scholar]
- 26.Le S, Josse J, Husson F. FactoMineR : An R Package for Multivariate Analysis. J Stat Softw 2008;25:1–18. doi: 10.18637/jss.v025.i01 [DOI] [Google Scholar]
- 27.Weiner J. Package pca3d: Three Dimensional PCA Plots. 2017. URL: http://logfc.wordpress.com [Google Scholar]
- 28.Wickham H. ggplot 2 - Elegant Graphics for Data Analysis. Springer-Verlag, New York: 2009. doi: 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- 29.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63. doi: 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villarreal LP, Defilippis VR, Gottlieb KA. Acute and persistent viral life strategies and their relationship to emerging diseases. Virology 2000;272:1–6. doi: 10.1006/viro.2000.0381 [DOI] [PubMed] [Google Scholar]
- 32.Hollink N, Dzabic M, Wolmer N, Bostrom L, Rahbar A. High prevalence of an active human cytomegalovirus infection in patients with colonic diverticulitis. J Clin Virol 2007;40:116–119. doi: 10.1016/j.jcv.2007.07.008 [DOI] [PubMed] [Google Scholar]
- 33.Matrana MR, Margolin DA. Epidemiology and pathophysiology of diverticular disease. Clin Colon Rectal Surg 2009;22:141–146. doi: 10.1055/s-0029-1236157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scharl M, Rogler G. Pathophysiology of fistula formation in Crohn’s disease. World J Gastrointest Pathophysiol 2014;5:205–212. doi: 10.4291/wjgp.v5.i3.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leisching G, Wiid I, Baker B. The Association of OASL and Type I Interferons in the Pathogenesis and Survival of Intracellular Replicating Bacterial Species. Front Cell Infect Microbiol 2017;7:196. doi: 10.3389/fcimb.2017.00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovarik P, Castiglia V, Ivin M, Ebner F. Type I Interferons in Bacterial Infections: A Balancing Act. Front Immunol 2016;7:652. doi: 10.3389/fimmu.2016.00652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.