Abstract
Metabolic adaptation to the host environment has been recognized as an essential mechanism of pathogenicity and the growth of Mycobacterium tuberculosis (Mtb) in the lungs for decades. The Mtb uses CO2 as a source of carbon during the dormant or non-replicative state. However, there is a lack of biochemical knowledge of its metabolic networks. In this study, we investigated the CO2 fixation pathways (such as ko00710 and ko00720) most likely involved in the energy production and conversion of CO2 in Mtb. Extensive pathway evaluation of 23 completely sequenced strains of Mtb confirmed the existence of a complete list of genes encoding the relevant enzymes of the reductive tricarboxylic acid (rTCA) cycle. This provides the evidence that an rTCA cycle may function to fix CO2 in this bacterium. We also proposed that as CO2 is plentiful in the lungs, inhibition of CO2 fixation pathways (by targeting the relevant CO2 fixation enzymes) could be used in the expansion of new drugs against the dormant Mtb. In support of the suggested hypothesis, the CO2 fixation enzymes were confirmed as a potential drug target by analyzing a number of attributes necessary to be a good bacterial target.
Keywords: Tuberculosis, CO2 fixation enzyme, rTCA cycle, dormant phase, drug discovery
1. Introduction
Tuberculosis (TB), caused by the bacterium Mycobacterium tuberculosis (Mtb) is one of the most feared diseases of present times, and is one of the top 10 causes of death worldwide. As per World Health Organization (WHO) records in 2015, the estimated number of TB cases was 10.4 million, followed by 1.8 million TB related deaths annually (http://www.who.int/gho/tb/en/). Though the rate of new TB cases has shrunk, the synergistic relationship between drug resistance TB [multi drug-resistant (MDR); extensively drug-resistant (XDR)] and HIV (TB/HIV coinfection) has transformed TB into a serious public health threat [1], [2], [3]. As an intracellular pathogen, Mtb is exposed to a strong host response, including low pH (H+), hydrogen peroxide (H2O2), hypochlorous acid (HOCl) and toxic gasses such as nitric oxide (NO), carbon monoxide (CO), and superoxide during the infection [4]. In response to host attack, Mtb has evolved a variety of defense mechanisms to counterattack the toxic environment. For example, Mtb could utilize superoxide dismutase (SOD, EC 1.15.1.1) and catalase (hydroxyperoxidases, EC 1.11.1.6) enzymes to transform the toxic reactive oxygen intermediates (such as superoxide and H2O2) into water and oxygen [5], [6], [7], [8], [9]. Dormancy of Mtb is an alternative counter mechanism which allows Mtb to survive in the host as well as in the unhostile environment for decades [10], [11], [12], [13], [14], [15]. To adjust to the low oxygen in the dormancy phase, Mtb triggers a metabolic switch from the aerobic to anaerobic respiration [16], [17] allowing the survival, growth and persistence of the pathogen. Mtb has evolved genes for NO and CO resistance to counterattack NO [5], [6], [7], [8], [9] and CO toxicity [4], [18], [19], vital for long-term survival of TB in the host. On the contrary, harmful effects of CO on E. coli, P. aeruginosa, and S. aureus have been recognized, in which exposures to CO inhibit key enzymes of the electron transport chain required for bacterial respiration, resulting in microbial death [20], [21]. As Mtb resides in the CO2 rich atmosphere of the lung, it can utilize CO2 as a source of carbon and energy [22], [23], [24], [25], [26]. Mtb retains all the genes requisite to catabolize cholesterol to CO2 through the tricarboxylic acid cycle (TCA) [27]. Six autotrophic CO2 fixation pathways are known to date. Three of them (rTCA cycle, the reductive acetyl-CoA pathway and the 3-hydroxypropionate bicycle) were found in bacteria [28]. The existence of a key enzyme of the rTCA cycle, including fumarate reductase, ATP-citrate lyase β-chain, and α-ketoglutarate: ferredoxin oxidoreductase in Mtb suggests that a reductive TAC may operate to fix CO2 in TB [29], [30], [31]. However, a complete list of key enzymes of the rTCA cycle that most likely function in carbon fixation is unknown for Mtb. In the present study, two carbon fixation pathways [denoted as “carbon fixation in photosynthetic organisms” and “carbon fixation pathways in prokaryotes” in Kyoto Encyclopedia of Genes and Genomes (KEGG)] most likely involved in energy production and conversion of CO2 were discovered by mapping the protein sequences of all 23 strains of Mtb in KEGG database. Further, as CO2 is plentiful in the human lungs and the genes involved in the CO2 fixation pathways are primarily expressed during the dormant phase, inhibition of CO2 fixation could be used in the development of novel drug treatments against TB.
2. Materials and Methods
2.1. Data
The protein sequences of 23 completely sequenced strains of M. tuberculosis were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/genome/genomes/166). List of the studied strains were listed in Supplementary data: Table S1.
2.2. Identification of Putative Orthologous
The protein sequences of Mtb were separated according to their type of strain. An all-against-all protein sequence comparison was executed by using Basic Local Alignment Search Tool (BLAST) [32]. The best hit from every other genome was compared to proteins from the given genome. The pair of sequences that were the best match when either sequence was used as a query was defined as Best Bidirectional Hits (BBH), and the sequences in the pair were considered related. The relationship between the sequence pair was not estimated, if the best hit depended on only one direction. In addition to the above criterion, a sequence pair was considered to be related by BBH if the e-value from each BLAST comparison was less than 0.001 in both directions. Since in this methodology only comparisons between protein sequences from two separate genomes were executed, obvious paralogous genes were avoided. Furthermore, the introduction of the stringent e-value cutoff eliminated false matches. The output of this step was a table of putative orthologous genes.
2.3. Pathway Analysis
A complete set of protein sequences from the table of putative orthologous genes was annotated using KEGG Automatic Annotation Server (KAAS; http://www.genome.jp/kegg/kaas/) [33]. KAAS was completed using the BBH method. The identified genes involved in the relevant pathways were compared to reference pathways of Mycobacterium species to identify complete versus incomplete or orphan pathways. A pathway was labeled complete if all the genes in the reference pathway were found in the putative orthologs table, whereas a pathway was considered as an orphan if it was not found in the reference pathways of Mycobacterium species in KEGG database. In this study, two orphan pathways, i.e. “carbon fixation in photosynthetic organisms (ko00710)” and “carbon fixation pathways in prokaryotes (ko00720)” were discovered.
2.4. Enzyme Annotation
Protein sequences from both pathways were annotated for domain (http://pfam.sanger.ac.uk/) [34], molecular function (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi) [35], and metabolic pathway (http://www.genome.jp/kegg/). The relevant information was also retrieved from TDR target database V5 (http://tdrtargets.org) [36] and EMBL-EBI InterProScan-5 (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) [37].
2.5. Potential Targets
All proteins from the pathway were compared with the human proteome by using BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The proteins with no human orthologous genes, at query coverage ≥50 % and sequence identity ≥40 % were considered as probable candidates. The relevant information of known targets and related drugs were retrieved from the TDR target database (http://tdrtargets.org) [36] and the DrugBank database (http://www.drugbank.ca/) [38], [39], [40], [41], respectively. The essentiality of genes was calculated by BLAST against the Database of Essential Genes (http://www.essentialgene.org). The antigenic region (epitope) of proteins was predicted by an EMBOSS explorer (http://emboss.bioinformatics.nl/cgi-bin/emboss/antigenic). The transmembrane region and protein mass was predicted by TMHMM-V.2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and molecular weight calculator tool (http://www.sciencegateway.org/tools/proteinmw.htm), respectively.
3. Results and Discussion
3.1. Identification of Missing Genes and Enzymes for Carbon Fixation
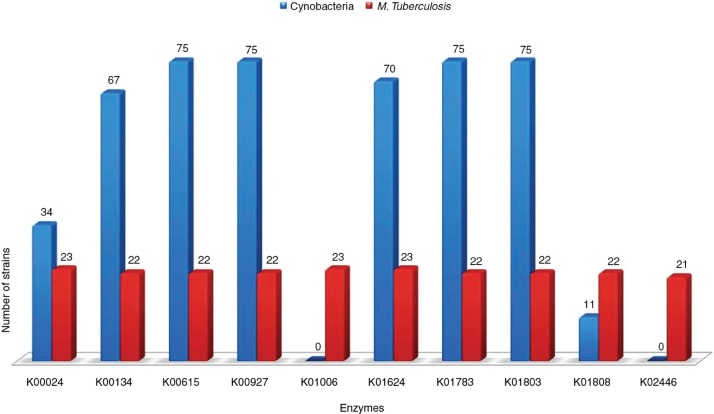
The present investigation identified and characterized common and orphan pathways using the complete set of protein sequences from the 23 strains of Mtb. A total of 90,737 unique protein entries were extracted using Perl script developed in-house (https://gist.github.com/amitkatiyar/). All-against-all comparison showed 7421 unique putative orthologs (bidirectional-best BLAST hits) containing approximately 2558 (35 %) proteins in >90 % of the tested strains. These 7421 unique putative orthologs were mapped to 136 annotated metabolic pathways comprising of 1769 KEGG orthologs (Supplementary data: Table S2). We found one or more proteins from 33 pathways that were formerly not reported in the KEGG for Mycobacteria. Among these pathways, two interesting pathways (i.e. ko00710: carbon fixation in photosynthetic organisms and ko00720: carbon fixation pathways in prokaryotes) belonging to class “energy metabolism” were selected. The extensive pathway analysis revealed a total of 222 and 983 proteins encoding for 10 (ko00710) and 32 (ko00720) enzymes, respectively, which were formerly not annotated for Mtb in KEGG (Table 1; Supplementary data: Table S3). These protein enzymes were re-annotated using Pfam domain and KEGG pathways. The results show that protein enzymes were correctly annotated and are involved in the rTCA cycle for CO2 fixation (Supplementary data: Table S4). These rTCA cycle-specific enzymes were found to be shared conserved among all the strains of TB (Table 2). However, the missing enzymes in a few of the observed strains may be due to issues with computational annotation. The enzyme acetyl-CoA C-acetyltransferase (EC2.3.1.9; K00626) contains the maximum number of orthologous genes (214) and more than seven paralogous genes in each tested strain. The CO2 fixation pathway enzymes of TB (data compiled from 23 strains) and cyanobacteria (data compiled from 75 strains) were compared that suggesting a close relationship between them (Supplementary data: Table S5). The result shows an identical pathway (ko0710) with eight shared enzymes between Cyanobacteria and TB (Figure 1). The remaining two key enzymes, namely pyruvate, orthophosphate dikinase (K01006; ppdK; EC 2.7.9.1) and fructose-1, 6- bisphosphatase II (K02446; glpX; EC 3.1.3.11) were only present in Mtb. Notably, we did not find pathways-00720 in Cyanobacteria as found in TB.
Table 1:
Carbon fixation enzymes of the reductive citric acid cycle (rTCA) in Mycobacterium tuberculosis.
| *KO | Enzyme description | *EC number |
|---|---|---|
| (A) ko00710: carbon fixation in photosynthetic organisms | ||
| K00024 | Malate dehydrogenase (MDH) | EC:1.1.1.37 |
| K00134 | Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | EC:1.2.1.12 |
| K00615 | Transketolase (TK) | EC:2.2.1.1 |
| K00927 | Phosphoglycerate kinase (PGK) | EC:2.7.2.3 |
| K01006 | Pyruvate,orthophosphate dikinase (ppdK) | EC:2.7.9.1 |
| K01624 | Fructose-bisphosphate aldolase, class II (FBA) | EC:4.1.2.13 |
| K01783 | Ribulose-phosphate 3-epimerase (rpe) | EC:5.1.3.1 |
| K01803 | Triosephosphate isomerase (TIM) | EC:5.3.1.1 |
| K01808 | Ribose 5-phosphate isomerase B (rpiB) | EC:5.3.1.6 |
| K02446 | Fructose-1,6-bisphosphatase II (glpX) | EC:3.1.3.11 |
| (B) ko00720: carbon fixation pathways in prokaryotes | ||
| K00024 | Malate dehydrogenase (MDH) | EC:1.1.1.37 |
| K00031 | Isocitrate dehydrogenase (IDH1) | EC:1.1.1.42 |
| K00031 | Isocitrate dehydrogenase (IDH2) | EC:1.1.1.42 |
| K00174 | 2-oxoglutarate ferredoxin oxidoreductase subunit alpha (korA) | EC:1.2.7.3 |
| K00175 | 2-oxoglutarate ferredoxin oxidoreductase subunit beta (korB) | EC:1.2.7.3 |
| K00239 | Succinate dehydrogenase flavoprotein subunit (sdhA) | EC:1.3.99.1 |
| K00240 | Succinate dehydrogenase iron-sulfur subunit (sdhB) | EC:1.3.99.1 |
| K00241 | Succinate dehydrogenase cytochrome b556 subunit (sdhC) | EC:1.3.99.1 |
| K00242 | Succinate dehydrogenase membrane anchor subunit (sdhD) | EC:1.3.99.1 |
| K00244 | Fumarate reductase flavoprotein subunit (frdA) | EC:1.3.99.1 |
| K00245 | Fumarate reductase flavoprotein subunit (frdB) | EC:1.3.99.1 |
| K00246 | Fumarate reductase subunit C (frdC) | EC:1.3.99.1 |
| K00247 | Fumarate reductase subunit D (frdD) | EC:1.3.99.1 |
| K00626 | Acetyl-CoA C-acetyltransferase (2.AA440) | EC:2.3.1.9 |
| K00925 | Acetate kinase (ackA) | EC:2.7.2.1 |
| K01006 | Pyruvate,orthophosphate dikinase (ppdK) | EC:2.7.9.1 |
| K01491 | Methylenetetrahydrofolate dehydrogenase (NADP+)//Methenyltetrahydrofolate cyclohydrolase (fold) | EC:1.5.1.5 EC:3.5.4.9 |
| K01679 | Fumarate hydratase, class II | EC:4.2.1.2 |
| K01681 | Aaconitate hydratase (ACO) | EC:4.2.1.3 |
| K01782 | 3-hydroxyacyl-CoA dehydrogenase//enoyl-CoA hydratase//3-hydroxybutyryl-CoA epimerase (fadJ) | EC:1.1.1.35// EC:4.2.1.17// EC:5.1.2.3 |
| K01847 | Methylmalonyl-CoA mutase (MUTB) AA750 | EC:5.4.99.2 |
| K01895 | Acetyl-CoA synthetase (ACSS) | EC:6.2.1.1 |
| K01902 | Succinyl-CoA synthetase alpha subunit (sucD) | EC:6.2.1.5 |
| K01903 | Succinyl-CoA synthetase beta subunit (sucC) | EC:6.2.1.5 |
| K01958 | Pyruvate carboxylase (PC) | EC:6.4.1.1 |
| K01963 | Acetyl-CoA carboxylase carboxyl transferase subunit beta (accD) | EC:6.4.1.2 |
| K01965 | Propionyl-CoA carboxylase alpha chain (PCCA) | EC:6.4.1.3 |
| K01966 | Propionyl-CoA carboxylase beta chain (PCCB) | EC:6.4.1.3 |
| K03518 | Carbon-monoxide dehydrogenase small subunit | EC:1.2.99.2 |
| K03519 | Carbon-monoxide dehydrogenase medium subunit | EC:1.2.99.2 |
| K03520 | Carbon-monoxide dehydrogenase large subunit | EC:1.2.99.2 |
| K05606 | Methylmalonyl-CoA/ethylmalonyl-CoA epimerase (MCEE) | EC:5.1.99.1 |
| K13788 | Phosphate acetyltransferase (PTA) | EC:2.3.1.8 |
These key enzymes were found in two pathways, namely ko00710# and ko00720#, containing 10 and 32 unique enzymes, respectively. The pathway analysis revealed the role of identifying enzymes in CO2 fixation. #ko00710: Carbon fixation in photosynthetic organisms; #ko00720: Carbon fixation pathways in prokaryotes; *KO: KEGG orthologs; *EC number: Enzyme commission number.
Table 2:
Comparative assessment of existing carbon fixation enzymes of rTCA cycle in 23 strains of Mycobacterium tuberculosis.
| *KO | Enzyme description | *EC | No of strains | Enzymes encoding genes in Mtb strains |
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 | S13 | S14 | S15 | S16 | S17 | S18 | S19 | S20 | S21 | S22 | S23 | ||||
| (A) ko00710: carbon fixation in photosynthetic organisms |
||||||||||||||||||||||||||
| K00024 | Malate dehydrogenase (MDH) | EC:1.1.1.37 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00134 | Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | EC:1.2.1.12 | 22 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00615 | Transketolase (TK) | EC:2.2.1.1 | 22 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00927 | Phosphoglycerate kinase (PGK) | EC:2.7.2.3 | 22 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01006 | Pyruvate,orthophosphate dikinase (ppdK) | EC:2.7.9.1 | 24 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01624 | Fructose-bisphosphate aldolase, class II (FBA) | EC:4.1.2.13 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01783 | Ribulose-phosphate 3-epimerase (rpe) | EC:5.1.3.1 | 22 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01803 | Triosephosphate isomerase (TIM) | EC:5.3.1.1 | 22 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01808 | Ribose 5-phosphate isomerase B (rpiB) | EC:5.3.1.6 | 22 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K02446 | Fructose-1,6-bisphosphatase II (glpX) | EC:3.1.3.11 | 20 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| (B) ko00720: carbon fixation pathways in prokaryotes |
||||||||||||||||||||||||||
| K00024 | Malate dehydrogenase (MDH) | EC:1.1.1.37 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00031 | Isocitrate dehydrogenase (IDH) | EC:1.1.1.42 | 23 | 2 | 3 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 |
| K00174 | 2-oxoglutarate ferredoxin oxidoreductase subunit alpha (korA) | EC:1.2.7.3 | 22 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00175 | 2-oxoglutarate ferredoxin oxidoreductase subunit beta (korB) | EC:1.2.7.3 | 22 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00239 | Succinate dehydrogenase flavoprotein subunit (sdhA) | EC:1.3.99.1 | 23 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| K00240 | Succinate dehydrogenase iron-sulfur subunit (sdhB) | EC:1.3.99.1 | 23 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| K00241 | Succinate dehydrogenase cytochrome b556 subunit (sdhC) | EC:1.3.99.1 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00242 | Succinate dehydrogenase membrane anchor subunit (sdhD) | EC:1.3.99.1 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00244 | Fumarate reductase flavoprotein subunit (frdA) | EC:1.3.99.1 | 21 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00245 | Fumarate reductase flavoprotein subunit (frdB) | EC:1.3.99.1 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00246 | Fumarate reductase subunit C (frdC) | EC:1.3.99.1 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00247 | Fumarate reductase subunit D (frdD) | EC:1.3.99.1 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K00626 | Acetyl-CoA C-acetyltransferase | EC:2.3.1.9 | 23 | 9 | 10 | 7 | 9 | 9 | 8 | 10 | 10 | 10 | 9 | 8 | 10 | 7 | 11 | 8 | 10 | 10 | 10 | 9 | 11 | 8 | 10 | 11 |
| K00925 | Acetate kinase (ackA) | EC:2.7.2.1 | 22 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01006 | Pyruvate,orthophosphate dikinase (ppdK) | EC:2.7.9.1 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01491 | Methylenetetrahydrofolate dehydrogenase (NADP+) / Methenyltetrahydrofolate cyclohydrolase (fold) | EC:1.5.1.5 EC:3.5.4.9 |
22 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01679 | Fumarate hydratase, class II | EC:4.2.1.2 | 20 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01681 | Aaconitate hydratase (ACO) | EC:4.2.1.3 | 19 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01782 | 3-hydroxyacyl-CoA dehydrogenase / enoyl-CoA hydratase / 3-hydroxybutyryl-CoA epimerase (fadJ) | EC:1.1.1.35 EC:4.2.1.17 EC:5.1.2.3 |
23 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01847 | Methylmalonyl-CoA mutase (MUTB) | EC:5.4.99.2 | 20 | 2 | 2 | 0 | 2 | 3 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| K01895 | Acetyl-CoA synthetase (ACSS) | EC:6.2.1.1 | 22 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01902 | Succinyl-CoA synthetase alpha subunit (sucD) | EC:6.2.1.5 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01903 | Succinyl-CoA synthetase beta subunit (sucC) | EC:6.2.1.5 | 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01958 | Pyruvate carboxylase (PC) | EC:6.4.1.1 | 21 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01963 | Acetyl-CoA carboxylase carboxyl transferase subunit beta (accD) | EC:6.4.1.2 | 21 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01965 | Propionyl-CoA carboxylase alpha chain (PCCA) | EC:6.4.1.3 | 20 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K01966 | Propionyl-CoA carboxylase beta chain (PCCB) | EC:6.4.1.3 | 22 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K03518 | Carbon-monoxide dehydrogenase small subunit | EC:1.2.99.2 | 20 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| K03519 | Carbon-monoxide dehydrogenase medium subunit | EC:1.2.99.2 | 19 | 1 | 0 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| K03520 | Carbon-monoxide dehydrogenase large subunit | EC:1.2.99.2 | 20 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| K05606 | Methylmalonyl-CoA/ethylmalonyl-CoA epimerase (MCEE) | EC:5.1.99.1 | 22 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| K13788 | Phosphate acetyltransferase (PTA) | EC:2.3.1.8 | 23 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
Strain-wise comparisons showed that most of the strains (except for a few strains that lack either one or two genes) contain genes encoding for respective enzymes in both pathways. *KO: KEGG orthologs; *EC number: Enzyme commission number; *Mycobacterium tuberculosis (strains): given below. S1: CCDC5180 uid161941; S2: H37Rv uid170532; S3: Haarlem3 NITR202 uid202216; S4: CDC1551 uid57775; S5: CCDC5079 uid161943; S6: RGTB423 uid162179; S7: CTRI 2 uid161997; S8: KZN 4207 uid83619; S9: CCDC5079_uid203790; S10: Beijing NITR203 uid197218; S11: EAI5 NITR206 uid202218; S12: F11 uid58417; S13: RGTB327 uid157907; S14: Erdman ATCC 35801 uid193763; S15: CAS NITR204 uid202217; S16: EAI5 uid212307; S17: KZN 605 uid54947; S18: UT205 uid162183; S19: H37Rv uid57777; S20: H37Ra uid58853; S21: Haarlem uid54453; S22: KZN 1435 uid59069; S23: uid185758.
Figure 1:
The comparison of CO2 fixation pathway enzymes (ko00710) between M. tuberculosis and Cyanobacteria. The comparison showed that Cyanobacteria shared eight enzymes with M. tuberculosis.
3.2. Evidence of CO2 Fixation by Reductive TCA Cycle
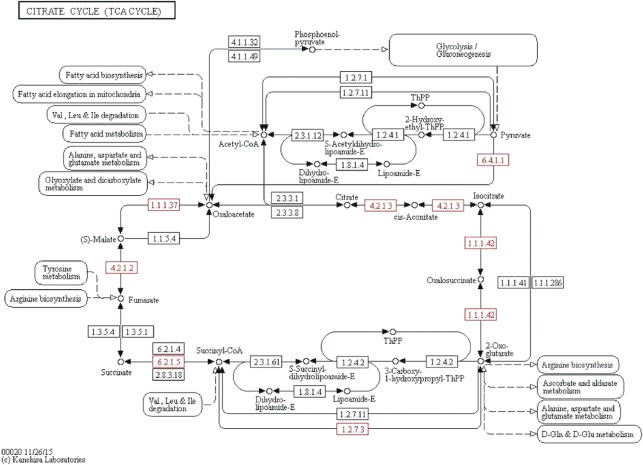
The citrate cycle (TCA cycle, Krebs cycle) is used by all aerobic organisms to produce energy by the oxidation of pyruvate. The TCA cycle is characterized as oxidative or reductive by the presence of three enzymes known as ATP-citrate lyase, 2-oxoglutarate synthase, and fumarate reductase [42], [43], [44]. The rTCA cycle reverses the reactions of the oxidative citric acid cycle by the replacement of three enzymes: the succinate dehydrogenase has to be replaced by the fumarate reductase, the NAD+-dependent 2-oxoglutarate dehydrogenase has to be replaced by the ferredoxin-dependent 2-oxoglutarat synthase and the citrate synthase has to be replaced by the ATP citrate lyase. The Mtb genome of 23 strains was annotated to encode a complete TCA cycle in this study. The results disclosed the presence of citrate synthase (EC 2.3.3.1), 2-oxoglutarate dehydrogenase (EC 1.2.4.2) and succinate dehydrogenase (EC 1.3.5.1) enzymes in Mtb (Figure 2). The findings indicate evidence of the existence of an oxidative TCA cycle in Mtb. Furthermore, the findings of fumarate reductase (EC 1.3.99.1), 2-oxoglutarate synthase (EC 1.2.7.3) and a homologue for citrate-oxaloacetate lyase (EC 4.1.3.6) enzymes showed evidence of an reduvtive TCA cycle for CO2 fixation in Mtb (Figure 2). Most of the enzymes (such as cis-aconitase, EC 4.2.1.3; isocitrate dehydrogenase, EC 1.1.1.42; succinyl-CoA ligase, EC 6.2.1.5; fumarate hydratase, EC 4.2.1.2 and malate dehydrogenase, EC 1.1.1.37) of the oxidative or reductive TCA cycle were common (Figure 2). These results are consistent with the previous hypothesis that Mycobacterium can activate the metabolic switch to anaerobic respiration where a complete or partial TCA cycle may operate in the reductive mode. This switch permits both carbon fixation and restoration to the balance of oxidative and reductive reactions in environmental changes, thus allowing the pathogen to survive, grow, and persist [31]. The first key enzyme of the rTCA pathway is fumarate reductase (Figure 2, reaction 3), which catalyzes the reduction of fumarate to succinate using Ubiquinol as an electron donor. We observed that the genome of Mtb encodes four distinct fumarate reductase (EC 1.3.99.1) namely, flavoprotein subunit A (frdA; K00244), subunit B (frdB; K00245), subunit C (frdC; K00246) and subunit D (frdD; K00247) (Table 1; Supplementary data: Table S4). The study also confirmed that the Mtb grnome encodes two distinct 2-oxoglutarate ferredoxin:oxidoreductase (OGOR; EC 1.2.7.3) enzymes, namely, subunit alpha (korA; K00174) and subunit beta (korB; K00175) (Table 1; Supplementary data: Table S4). The enzyme OGOR is the key enzyme of the rTCA cycle that fixes carbon dioxide [45], [46], [47]. The enzyme OGOR (Figure 2, reaction 5) catalyzes the reductive carboxylation of succinyl-CoA to 2-oxoglutarate. An anaerobic enzyme “ATP-citrate lyase” catalyzes the division of citrate to oxaloacetate and acetyl-coA. A homologue for citrate lyase subunit beta (citE; K01644; EC 4.1.3.6), synonym of citrate-oxaloacetate was observed in the genome of Mtb. Remarkably, we did not find the key activity of the pyruvate synthase (EC 1.2.7.1) in an rTCA cycle of Mtb. This may be due to a single or a set of alternative enzymes that can fill the missing reactions. Another important enzyme of the rTCA cycle is “isocitrate dehydrogenase or IDH” (Figure 2, reaction 6) that catalyzes the reversible conversion of isocitrate to 2-oxoglutarate in prokaryotes. Here, we observed that the genome of Mtb encodes two types of isocitrate dehydrogenase (EC 1.1.1.42), i.e. IDH1 (dimeric) and IDH2 (monomeric) as reported previously in Mtb [48]. Notably, all 23 strains of Mtb having IDH enzymes indicated their importance for the organism. Multiple sequence alignment revealed a closer similarity of isocitrate dehydrogenase (ICD)-1 from Mtb to eukaryotic NADP+-dependent ICDs, whereas ICD-2 from Mtb groups with bacterial ICDs (data not shown). We also explored the TDR database and observed the expression of many rTCA cycle-specific enzymes in dormant phase of TB (Table 3). The findings demonstrated that Mtb can activate reductive rTCA (during low oxygen tension or anaerobic environment) for the fixation of CO2 in TB. In support of this theory, several studies have been presented that Mtb could operate a reductive TCA half cycle under anaerobic situations, permitting it to metabolize glucose by generating succinate as an obligatorily secreted, fermentation product of fumarate, following reductive carboxylation of pyruvate or phosphoenolpyruvate to malate and/or oxaloacetate [49]. In addition, several mycobacterial strains including Mtb can grow on CO as the sole source of carbon and energy. A remarkable attribute of Mtb is its capability to survive with minimal growth inhibition [50] during the elevated CO concentration [30] by expressing CO resistance genes. Carbon monoxide dehydrogenase (CO-DH) is an enzyme catalyzing the oxidation of CO to carbon dioxide in Mycobacterium [4], [30]. Here, we observed that the genome of Mtb encodes three copies of the CO-DH (EC 1.2.99.2) enzymes, namely CO dehydrogenase-small subunit (K03518), medium subunit (K03519) and large subunit (K03520). However, the genome of three strains (RGTB423_uid162179; RGTB327_uid157907; UT205_uid162183) of Mtb did not encode for CO-DH enzyme. This might be due to lack of annotation or absence of CO resistance genes.
Figure 2:
Pictorial representation of reductive citric acid cycle for autotrophic CO2 fixation. The enzyme catalyzed reactions are shown by arrows. Enzyme activities: 1, malate dehydrogenase (EC 1.1.1.37); 2, fumarate hydratase (fumarase) (EC 4.2.1.2); 3, fumarate reductase; 4, succinyl-CoA synthetase (EC 6.2.1.5); 5, 2-oxoglutarate:ferredoxin oxidoreductase (EC 1.2.7.3); 6, isocitrate dehydrogenase (EC 1.1.1.42); 7, aconitate hydratase (aconitase) (EC 4.2.1.3); 8, ATP citrate lyase (EC 2.3.3.8); and 9, pyruvate: ferredoxin oxidoreductase (EC 1.2.7.1). Fdred, reduced ferredoxin. The enzymes observed in the genome of tuberculosis strains were highlighted with red color.
Table 3:
Carbon fixation enzymes accepted as a potential drug target in Mycobacterium tuberculosis.
| *KO | Enzyme description | $EC number | Human homologues (Query coverage/Sequence identity) | Gene expression (dormant phase) |
Epitopes/ Antigenicity | &Number of homologues in DEG | Trans domains | Associated compounds/Druggability | Low mass (<100 kDa) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower (%) | Mid (%) | Upper (%) | |||||||||
| (A) ko00710: carbon fixation in photosynthetic organisms |
|||||||||||
| K00024 | Malate dehydrogenase (MDH) | EC:1.1.1.37 | Human positive: 98/52 % | 00–20 | NA | NA | 13; 33.0 (67.0 %) | 3 | Absent | Nitrofurazone (E. coli) | 34.33 |
| K00134 | Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | EC:1.2.1.12 | Human positive: 97/51 % | 20–40 | NA | NA | 16; 72.2 (22.8 %) | 22 | Absent | No drug | 35.96 |
| K00615 | Transketolase (TK) | EC:2.2.1.1 | Human negative: 87/25 % | 00–20 | NA | NA | 30; 51.2 (48.8 %) | 16 | Absent | No drug | 79.97 |
| K00927 | Phosphoglycerate kinase (PGK) | EC:2.7.2.3 | Human negative: 98/38 % | 00–20 | NA | NA | 20; 82.5 (17.5 %) | 21 | Absent | No drug | 42.52 |
| K01006 | Pyruvate,orthophosphate dikinase (ppdK) | EC:2.7.9.1 | Human negative: 07/41 % | 00–20 | NA | 40–60 | 21; 53.1 (46.9 %) | 1 | Absent | No drug | 54.7 |
| K01624 | Fructose-bisphosphate aldolase, class II (FBA) | EC:4.1.2.13 | Human negative: 27/29 % | 00–20 | NA | 60–80 | 12; 16.6 (17.5 %) | 17 | Absent | No drug | 36.55 |
| K01783 | Ribulose-phosphate 3-epimerase (rpe) | EC:5.1.3.1 | Human positive: 95/44 % | 00–20 | 40–60 | NA | 11: 75.7 (24.3 %) | 14 | Absent | No drug | 24.24 |
| K01803 | Triosephosphate isomerase (TIM) | EC:5.3.1.1 | Human negative: 93/39 % | 20–40 | NA | NA | 11; 51.5 (48.5 %) | 18 | Absent | No drug | 27.41 |
| K01808 | Ribose 5-phosphate isomerase B (rpiB) | EC:5.3.1.6 | Human negative: 50/24 % | 60–80 | NA | 80–100 | 08; 82.5 (17.5 %) | 7 | Absent | No drug | 17.66 |
| K02446 | Fructose-1,6-bisphosphatase II (glpX) | EC:3.1.3.11 | Human negative: 35/26 % | 20–40 | 40–60 | NA | 13; 33.9 (66.1 %) | 3 | Absent | No drug | 39.74 |
| (B) ko00720: Carbon fixation pathways in prokaryotes |
|||||||||||
| K01782 | 3-hydroxyacyl-CoA dehydrogenase / Enoyl-CoA hydratase / 3-hydroxybutyryl-CoA epimerase (fadJ) | EC:1.1.1.35 EC:4.2.1.17 EC:5.1.2.3 | Human negative: 97/32 % | Unknown | Unknown | Unknown | 33;NA | 2 | Absent | No drug | 76.11 |
| K00024 | Malate dehydrogenase (MDH) | EC:1.1.1.37 | Human positive: 98/52 % | 0–20 | NA | NA | 13; 33.0 (67.0 %) | 3 | Absent | Nitrofurazone (E. coli) | 34.33 |
| K00031 | Isocitrate dehydrogenase (IDH2) | EC:1.1.1.42 | Human negative: 14/25 % | 20–40 | NA | 80–100 | 38; 87.2 (12.8 %) | 3 | Absent | No drug | 82.56 |
| Isocitrate dehydrogenase (IDH1) | Human positive: 97/66 % | 60–80 | 19; 68.4 (31.6 %) | 1 | 45.52 | ||||||
| K00175 | 2-oxoglutarate ferredoxin oxidoreductase subunit beta (korB) | EC:1.2.7.3 | Human negative: 12/32 % | Unknown | Unknown | Unknown | 17; NA | 4 | Absent | Nitrofurantoin (E. coli) | 46.76 |
| K00174 | 2-oxoglutarate ferredoxin oxidoreductase subunit alpha (korA) | EC:1.2.7.3 | Human negative: 11/30 % | Unknown | Unknown | Unknown | 29; NA | 3 | Absent | No drug | 69.68 |
| K03519 | Carbon-monoxide dehydrogenase medium subunit | EC:1.2.99.2 | Human negative: 56/24 % | NA | 40–60 | 60–80 | 15; 92.2 (7.8 %) | 2 | Absent | No drug | 31.56 |
| K03520 | Carbon-monoxide dehydrogenase large subunit | EC:1.2.99.2 | Human negative: 97/23 % | NA | 40–60 | 60–80 | 33; 45.2 (54.8 %) | 2 | Absent | No drug | 85.9 |
| K03518 | Carbon-monoxide dehydrogenase small subunit | EC:1.2.99.2 | Human negative: 86/39 % | 20–40 | NA | 60–80 | 06; 27.4 (72.6 %) | 1 | Absent | No drug | 17.22 |
| K00241 | Succinate dehydrogenase cytochrome b556 subunit (sdhC) | EC:1.3.99.1 | Human negative: 24/49 % | 20–40 | NA | 80–100 | 3; 5.5 (94.5 %) | Unknown | 3 | No drug | 16.3 |
| K00242 | Succinate dehydrogenase membrane anchor subunit (sdhD) | EC:1.3.99.1 | Human negative: 57/25 % | 20–40 | NA | 60–80 | 4; 6.1 (93.9 %) | 1 | 2 | No drug | 17.37 |
| K00246 | Fumarate reductase subunit C (frdC) | EC:1.3.99.1 | Human negative: 66/26 % | NA | NA | 60–80 | 4; 15.8 (84.2 %) | Unknown | 3 | No drug | 14.82 |
| K00247 | Fumarate reductase subunit D (frdD) | EC:1.3.99.1 | Human negative: 29/38 % | NA | 40–60 | 60–80 | 5; 52.5 (47.5 %) | Unknown | 3 | No drug | 13.75 |
| K00239 | Succinate dehydrogenase flavoprotein subunit (sdhA) | EC:1.3.99.1 | Human negative: 84/32 % | 20–40 | NA | NA | 24; 26.9 (73.1 %) | 17 | Absent | Thiabendazole (E. coli) | 70.69 |
| Human positive:-94/48 % | 22; 27.3 (72.7 %) | 64.83 | |||||||||
| K00244 | Fumarate reductase flavoprotein subunit (frdA) | EC:1.3.99.1 | Human negative: 88/37 % | 20–40 | NA | NA | 22; 27.3 (72.7 %) | 17 | Absent | Thiabendazole (E. coli) | 63.77 |
| K00240 | Succinate dehydrogenase iron-sulfur subunit (sdhB) | EC:1.3.99.1 | Human positive: 85/41 % | 20–40 | NA | NA | 10; 31.6 (68.4 %) | 8 | Absent | No drug | 29.34 |
| Human negative: 87/25 % | 10; 42.1 (57.9 %) | 28.63 | |||||||||
| K00245 | Fumarate reductase flavoprotein subunit (frdB) | EC:1.3.99.1 | Human negative: 89/38 % | 20–40 | NA | NA | 10; 31.6(68.4 %) | 8 | Absent | No drug | 27.24 |
| K01491 | Methylenetetrahydrofolate dehydrogenase (NADP+) / Methenyltetrahydrofolate cyclohydrolase (fold) | EC:1.5.1.5 EC:3.5.4.9 | Human positive: 91/40 % | 0–20 | NA | NA | 11; 36.2 (63.8 %) | 18 | Absent | No drug | 32.09 |
| K13788 | Phosphate acetyltransferase (PTA) | EC:2.3.1.8 | Human negative: 12/30 % | NA | 40–60 | 60–80 | 31; 66.8 (33.2 %) | 8 | Absent | No drug | 75.1 |
| K00626 | Acetyl-CoA C-acetyltransferase | EC:2.3.1.9 | Human negative: 04–99/21–44 % | Unknown | Unknown | Unknown | 03–38; NA | 2+1 | Absent | No drug | 8.8–53.57 |
| K00925 | Acetate kinase (ackA) | EC:2.7.2.1 | Human negative: 04–22/27–41 % | Unknown | 40–60 | Unknown | 17–38; 61.4 (38.6 %)-NA | 6–7 | Absent | No drug | 41.32–95.4 |
| K01006 | Pyruvate,orthophosphate dikinase (ppdK) | EC:2.7.9.1 | Human negative: 07/41 % | 0–20 | NA | 40–60 | 21; 53.1 (46.9 %) | 1 | Absent | No drug | 54.7 |
| K01679 | Fumarate hydratase, class II | EC:4.2.1.2 | Human negative: 33/30 % | NA | NA | 40–60 | 20; 48.0 (52.0 %) | 5 | Absent | No drug | 29.36 |
| K01681 | Aaconitate hydratase (ACO) | EC:4.2.1.3 | Human positive: 98/49 % | 20–40 | NA | 80–100 | 40; 51.6 (48.4 %) | 11 | Absent | No drug | 102.46 |
| K05606 | Methylmalonyl-CoA/ethylmalonyl-CoA epimerase (MCEE) | EC:5.1.99.1 | Human negative: 86/32 % | Unknown | Unknown | Unknown | 6;NA | 1 | Absent | No drug | 16.63 |
| K01847 | Methylmalonyl-CoA mutase (MUTB) | EC:5.4.99.2 | Human positive: 97/61 % | NA | NA | 80–100 | 38; 86.0 (14.0 %) | Unknown | Absent | No drug | 80.64 |
| Human negative: 73/28 % | 60–100 | 27; 57.7 (42.3 %) | 64.75 | ||||||||
| K01895 | Acetyl-CoA synthetase (ACSS) | EC:6.2.1.1 | Human positive: 95/46 % | NA | 40–60 | NA | 29; 62.3 (37.7 %) | Unknown | Absent | No drug | 71.48 |
| K01903 | Succinyl-CoA synthetase beta subunit (sucC) | EC:6.2.1.5 | Human negative: 96/37 % | 0–40 | NA | NA | 15; 33.4 (66.6 %) | 6 | Absent | No drug | 40.93 |
| K01902 | Succinyl-CoA synthetase alpha subunit (sucD) | EC:6.2.1.5 | Human positive: 95/48 % | 0–20 | NA | NA | 16; 91.3 (8.7 %) | 5 | Absent | No drug | 31.23 |
| K01958 | Pyruvate carboxylase (PC) | EC:6.4.1.1 | Human positive: 96/47 % | 0–40 | NA | NA | 54; 79.1 (20.9 %) | 24 | Absent | No drug | 124.34 |
| K01963 | Acetyl-CoA carboxylase carboxyl transferase subunit beta (accD) | EC:6.4.1.2 | Human negative: 81/25 % | Unknown | Unknown | Unknown | 26;NA | 36 | Absent | No drug | 51.78 |
| K01966 | Propionyl-CoA carboxylase beta chain (PCCB) | EC:6.4.1.3 | Human positive: 92/53 % | 0–40 | NA | NA | 26; 75.0 (25.0 %) | 38 | Absent | No drug | 60.38 |
| K01965 | Propionyl-CoA carboxylase alpha chain (PCCA) | EC:6.4.1.3 | Human negative: 99/39 % | Unknown | Unknown | Unknown | 33;NA | 32 | Absent | No drug | 71.13 |
The selected enzymes were verified by a number of attributes defined as good drug targets (e.g. non-human homologs, unique function, essentiality of genes, gene expression, etc.). Druggability analysis exposed that to this date, no drugs are available against the proposed drug targets. #ko00720: carbon fixation pathways in prokaryotes; *KO: KEGG orthologs; $EC number: Enzyme commission number; &DEG: Database of Essential Genes; NA: Not found.
3.3. Carbon Fixation Enzymes as New Anti-TB Targets
The discovery of new targets that are hard to repress by mutation and essential for the bacterial survival are the primary requirement in the drug discovery process [51]. In the present study, we focused on a number of parameters to confirm “carbon fixation enzymes” as a good antibiotic drug target in TB. The considered parameters included genes homologous to humans, genes essential to the pathogen growth and survival, genes expressed during adaptation to dormancy, chokepoint reaction, low molecular weight of a protein, presence or absence of transmembrane helices, and target publication(s) in PubMed. This study does not include drug findings against carbon fixation enzymes, therefore we neither checked for crystallographic structures nor performed homology modeling. The study is evaluated based on known information. Initially implemented BLAST analysis showed that the proposed targets of TB were non-homologous to human genes (Table 3). This reduces the chances of uninvited host-drug interactions and therefore avoids host toxicity [52]. Next, the proposed targets were examined for the gene essentiality to pathogen growth and survival using the “Database of Essential Genes (DEG).” As a result, we found essential homologues for 35 key enzymes commonly from the pathways ko00710 and ko00720 (Table 3; Supplementary data: Table S6). The three enzymes, namely propionyl-CoA carboxylase beta chain (K01966), acetyl-CoA carboxylase carboxyl transferase subunit beta (K01963) and propionyl-CoA carboxylase alpha chain (K01965) contain the highest number of homologues i.e. 38, 36 and 32, respectively. Targeting for any of these essential genes will prevent or kill bacterial growth. In the analysis, homologues for five enzymes (K00241; K00246; K00247; K01847 and K01895) were not observed, potentially due to lack of essential genes or gene annotation in database of essential genes. Furthermore, the “TDR target database” showed the higher expression (80–100 %) of ribose 5-phosphate isomerase B (rpiB; K01808; EC 5.3.1.6), isocitrate dehydrogenase (IDH2; K00031; EC 1.1.1.42), succinate dehydrogenase cytochrome b556 subunit (sdhC; K00241; EC 1.3.99.1), aconitate hydratase (ACO; K01681; EC 4.2.1.3), and methylmalonyl-CoA mutase (MUTB; K01847; EC 5.4.99.2) enzymes during TB adaptation to dormancy [53] (Table 3). The expression of these and other key enzymes during TB adaptation to dormancy signifies the need to survive in a dormant state. The proposed target genes were also selected by the uniqueness of their essential functions in the metabolome (metabolic chokepoint). We clustered “carbon fixation enzymes” by their unique enzyme commission (EC) number as it might perform unique reactions [54]. As a result, 20 clusters or unique chokepoint reactions were detected from ko00720 pathway of M. tuberculosis. Interestingly, we observed unique chokepoints reactions for all 10 enzymes from ko00710 pathway of M. tuberculosis (Table 1). Enzymes involved in unique essential chokepoint reactions are the good metabolic drug targets as their function cannot be compensated by another enzyme. In addition, the proposed targets also satisfied additional key properties of good drug targets [54] such as low mass [<100 kDa]; no transmembrane domain; and publication in PubMed (Table 3). The selected properties of the proposed targets will be useful due to the following reasons: (1) cloning efficiency is significantly higher at low molecular weight (<100 kDa) as compared with high molecular weight (<100 kDa), (2) protein with no transmembrane domain is easy to crystallize as compared to membrane proteins which are difficult to crystallize due to their heterogeneous nature. In addition, the detergent micelle or liposome used for solubilizing and stabilizing the membrane proteins can interfere with protein crystallization, (3) the publication on the target protein will help to identify unwanted off-target effects, molecular function and structure related information. These results confirmed “carbon fixation enzymes” as a good metabolic drug target of M. tuberculosis. Additionally, several studies have described the essentiality of carbon fixation enzymes for bacterial survival and therefore may be used as a potential target. For instance, six autotrophic pathways and ICD, a key regulatory enzyme (released during late exponential growth phase) in the citric acid cycle are used as autolysis markers [55]. Malate dehydrogenase and the ICD has been reported as a potential antigen for serodiagnosis [56], [57]. Similarly, Mtb requires the enzyme isocitrate lyase (ICL) for survival during nutrient starvation, growth and virulence in vivo. The essentiality of icl1 has been reported for slow growth on glycerol [25] and the multiplication of mtb in macrophages and mice [53], [54]. It has been also reported that physiologically significant levels of CO2 evade the growth arrest of Mycobacterium bovis BCG under persistent hypoxia [30]. Therefore, inhibition of CO2 fixation enzymes could support the development of novel drug treatments against TB.
4. Conclusions
In this study, we identified and characterized the common and orphan pathways in M. tuberculosis. We bioinformatically confirmed the existence of a complete list of genes by encoding the relevant enzymes of the reductive rTCA, most likely involved in energy production and conversion of CO2. The CO2 fixation pathway enzymes of TB and cyanobacteria were compared that suggesting a close relationship between them. The CO2 fixation enzymes were confirmed as a potential drug target by analyzing a number of attributes necessary to be a good bacterial target.
Supporting Information
Acknowledgments
We thank the Indian Council of Medical Research (ICMR) for supporting this work through the Second phase of Task Force Biomedical Informatics Center of ICMR.
Supplementary Material
The online version of this article offers supplementary material (jib-2017-0041).
Conflict of interest statement
Authors state no conflict of interest. All authors have read the journal’s publication ethics and publication malpractice statement available at the journal’s website and hereby confirm that they comply with all its parts applicable to the present scientific work.
References
- [1].Alexander PE, De P. The emergence of extensively drug-resistant tuberculosis (TB): TB/HIV coinfection, multidrug-resistant TB and the resulting public health threat from extensively drug-resistant TB, globally and in Canada. Can J Infect Dis Med Microbiol. 2007;18:289–91. doi: 10.1155/2007/986794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sethi S, Mewara A, Dhatwalia SK, Singh H, Yadav R, Singh K. et al. Prevalence of multidrug resistance in Mycobacterium tuberculosis isolates from HIV seropositive and seronegative patients with pulmonary tuberculosis in north India. BMC Infect Dis. 2013;13:137. doi: 10.1186/1471-2334-13-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lee S, Lee SH, Mok JH, Lee SJ, Kim KH, Lee JE. et al. Is multi-drug resistant tuberculosis more prevalent in HIV-infected patients in korea? Yonsei Med J. 2016;57:1508–10. doi: 10.3349/ymj.2016.57.6.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zacharia VM, Shiloh MU. Effect of carbon monoxide on Mycobacterium tuberculosis pathogenesis. Med Gas Res. 2012;2:30. doi: 10.1186/2045-9912-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Poole RK, Hughes MN. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol Microbiol. 2000;36:775–83. doi: 10.1046/j.1365-2958.2000.01889.x. [DOI] [PubMed] [Google Scholar]
- [6].Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–6. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- [7].Darwin KH, Nathan CF. Role for nucleotide excision repair in virulence of Mycobacterium tuberculosis. Infect Immun. 2005;73:4581–7. doi: 10.1128/IAI.73.8.4581-4587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shi S, Ehrt S. Dihydrolipoamide acyltransferase is critical for Mycobacterium tuberculosis pathogenesis. Infect Immun. 2006;74:56–63. doi: 10.1128/IAI.74.1.56-63.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Venugopal A, Bryk R, Shi S, Rhee K, Rath P, Schnappinger D. et al. Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe. 2011;9:21–31. doi: 10.1016/j.chom.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].da Silveira NJ, Bonalumi CE, Uchoa HB, Pereira JH, Canduri F, de Azevedo WF. DBMODELING: a database applied to the study of protein targets from genome projects. Cell Biochem Biophys. 2006;44:366–74. doi: 10.1385/cbb:44:3:366. [DOI] [PubMed] [Google Scholar]
- [11].Heberle G, de Azevedo WF. Bio-inspired algorithms applied to molecular docking simulations. Curr Med Chem. 2011;18:1339–52. doi: 10.2174/092986711795029573. [DOI] [PubMed] [Google Scholar]
- [12].de Azevedo WF. Protein targets for development of drugs against Mycobacterium tuberculosis. Curr Med Chem. 2011;18:1255–7. doi: 10.2174/092986711795029564. [DOI] [PubMed] [Google Scholar]
- [13].Gokhale K, Tilak B. Mechanisms of bacterial acetohydroxyacid synthase (AHAS) and specific inhibitors of Mycobacterium tuberculosis AHAS as potential drug candidates against tuberculosis. Curr Drug Targets. 2015;16:689–99. doi: 10.2174/1389450116666150416115547. [DOI] [PubMed] [Google Scholar]
- [14].Sharma R, Kaur A, Sharma AK, Dilbaghi N, Sharma AK. Nano-based anti-tubercular drug delivery and therapeutic interventions in tuberculosis. Curr Drug Targets. 2017;18:72–86. doi: 10.2174/1389450116666150804110238. [DOI] [PubMed] [Google Scholar]
- [15].Singh G, Kumar A, Maan P, Kaur J. Cell wall associated factors of Mycobacterium tuberculosis as major virulence determinants: current perspectives in drugs discovery and design. Curr Drug Targets. 2017;18:1904–18. doi: 10.2174/1389450118666170711150034. [DOI] [PubMed] [Google Scholar]
- [16].Tailleux L, Waddell SJ, Pelizzola M, Mortellaro A, Withers M, Tanne A. et al. Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PLoS One. 2008;3:e1403. doi: 10.1371/journal.pone.0001403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Waddell SJ, Butcher PD. Microarray analysis of whole genome expression of intracellular Mycobacterium tuberculosis. Curr Mol Med. 2007;7:287–96. doi: 10.2174/156652407780598548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 2004;84:218–27. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- [19].Boon C, Dick T. How Mycobacterium tuberculosis goes to sleep: the dormancy survival regulator DosR a decade later. Future Microbiol. 2012;7:513–8. doi: 10.2217/fmb.12.14. [DOI] [PubMed] [Google Scholar]
- [20].Desmard M, Davidge KS, Bouvet O, Morin D, Roux D, Foresti R. et al. A carbon monoxide-releasing molecule (CORM-3) exerts bactericidal activity against Pseudomonas aeruginosa and improves survival in an animal model of bacteraemia. FASEB J. 2009;23:1023–31. doi: 10.1096/fj.08-122804. [DOI] [PubMed] [Google Scholar]
- [21].Nobre LS, Al-Shahrour F, Dopazo J, Saraiva LM. Exploring the antimicrobial action of a carbon monoxide-releasing compound through whole-genome transcription profiling of Escherichia coli. Microbiology. 2009;155(Pt 3):813–24. doi: 10.1099/mic.0.023911-0. [DOI] [PubMed] [Google Scholar]
- [22].Schaefer WB, Cohn ML, Middlebrook G. The roles of biotin and carbon dioxide in the cultivation of Mycobacterium tuberculosis. J Bacteriol. 1955;69:706–12. doi: 10.1128/jb.69.6.706-712.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nishihara H. Studies on the metabolism of the tubercule bacillus with the use of radioactive substrates in the presence and absence of streptomycin. J Biochem. 1954;41:167–81. [Google Scholar]
- [24].Long ER, Anderson RJ, Rittenberg D, Karnovsky ML, Henderson HJ. The carbon metabolism of the tubercule bacillus. Am Rev Tuberc. 1955;71:609–15. doi: 10.1164/artpd.1955.71.5.609. [DOI] [PubMed] [Google Scholar]
- [25].Beste DJ, Bonde B, Hawkins N, Ward JL, Beale MH, Noack S. et al. 13C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog. 2011;7:e1002091. doi: 10.1371/journal.ppat.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Beste DJ, Noh K, Niedenfuhr S, Mendum TA, Hawkins ND, Ward JL. et al. 13C-flux spectral analysis of host-pathogen metabolism reveals a mixed diet for intracellular Mycobacterium tuberculosis. Chem Biol. 2013;20:1012–21. doi: 10.1016/j.chembiol.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC. et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci USA. 2007;104:1947–52. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hugler M. et al. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–60. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- [29].Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D. et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- [30].Park SW, Hwang EH, Park H, Kim JA, Heo J, Lee KH. et al. Growth of mycobacteria on carbon monoxide and methanol. J Bacteriol. 2003;185:142–7. doi: 10.1128/JB.185.1.142-147.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Srinivasan V, Morowitz HJ. Ancient genes in contemporary persistent microbial pathogens. Biol Bull. 2006;210:1–9. doi: 10.2307/4134531. [DOI] [PubMed] [Google Scholar]
- [32].Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- [33].Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–5. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR. et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Magarinos MP, Carmona SJ, Crowther GJ, Ralph SA, Roos DS, Shanmugam D. et al. TDR Targets: a chemogenomics resource for neglected diseases. Nucleic Acids Res. 2012;40:D1118–27. doi: 10.1093/nar/gkr1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–40. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P. et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D. et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–6. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A. et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–41. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–7. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shiba H, Kawasumi T, Igarashi Y, Kodama T, Minoda Y. The CO2 assimilation via the reductive tricarboxylic acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch Microbiol. 1985;141:198–203. [Google Scholar]
- [43].Schauder R, Widdel F, Fuchs G. Carbon assimilation pathways in sulfate-reducing bacteria II. Enzymes of a reductive citric acid cycle in the autotrophic Desulfobacter hydrogenophilus. Arch Microbiol. 1987;148:218–25. [Google Scholar]
- [44].Siebers B, Tjaden B, Michalke K, Dorr C, Ahmed H, Zaparty M. et al. Reconstruction of the central carbohydrate metabolism of Thermoproteus tenax by use of genomic and biochemical data. J Bacteriol. 2004;186:2179–94. doi: 10.1128/JB.186.7.2179-2194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yamamoto M, Arai H, Ishii M, Igarashi Y. Characterization of two different 2-oxoglutarate:ferredoxin oxidoreductases from Hydrogenobacter thermophilus TK-6. Biochem Biophys Res Commun. 2003;312:1297–302. doi: 10.1016/j.bbrc.2003.11.078. [DOI] [PubMed] [Google Scholar]
- [46].Yamamoto M, Arai H, Ishii M, Igarashi Y. Role of two 2-oxoglutarate:ferredoxin oxidoreductases in Hydrogenobacter thermophilus under aerobic and anaerobic conditions. FEMS Microbiol Lett. 2006;263:189–93. doi: 10.1111/j.1574-6968.2006.00415.x. [DOI] [PubMed] [Google Scholar]
- [47].Yamamoto M, Ikeda T, Arai H, Ishii M, Igarashi Y. Carboxylation reaction catalyzed by 2-oxoglutarate:ferredoxin oxidoreductases from Hydrogenobacter thermophilus. Extremophiles. 2010;14:79–85. doi: 10.1007/s00792-009-0289-4. [DOI] [PubMed] [Google Scholar]
- [48].Banerjee S, Nandyala A, Podili R, Katoch VM, Hasnain SE. Comparison of Mycobacterium tuberculosis isocitrate dehydrogenases (ICD-1 and ICD-2) reveal differences in coenzyme affinity, oligomeric state, pH tolerance and phylogenetic affiliation. BMC Biochemistry. 2005;6:20. doi: 10.1186/1471-2091-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Watanabe S, Zimmermann M, Goodwin MB, Sauer U, Barry CE, Boshoff HI. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shiloh MU, Manzanillo P, Cox JS. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe. 2008;3:323–30. doi: 10.1016/j.chom.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Terstappen GC, Reggiani A. In-silico research in drug discovery. Trends Pharmacol In Sci. 2001;22:23–6. doi: 10.1016/s0165-6147(00)01584-4. [DOI] [PubMed] [Google Scholar]
- [52].Freiberg C, Wieland B, Spaltmann F, Ehlert K, Brotz H, Labischinski H. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J Mol Microbiol Biotechnol. 2001;3:483–9. [PubMed] [Google Scholar]
- [53].Murphy DJ, Brown JR. Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect Dis. 2007;7:84. doi: 10.1186/1471-2334-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yeh I, Hanekamp T, Tsoka S, Karp PD, Altman RB. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res. 2004;14:917–24. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Andersen P, Askgaard D, Ljungquist L, Bennedsen J, Heron I. Proteins released from Mycobacterium tuberculosis during Growth. Infect Immun. 1991;59:1905–10. doi: 10.1128/iai.59.6.1905-1910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ohman R, Ridell M. Purification and characterisation of isocitrate dehydrogenase and malate dehydrogenase from Mycobacterium tuberculosis and evaluation of their potential as suitable antigens for the serodiagnosis of tuberculosis. Tuberc Lung Dis. 1996;77:454–61. doi: 10.1016/s0962-8479(96)90120-3. [DOI] [PubMed] [Google Scholar]
- [57].Florio W, Bottai D, Batoni G, Esin S, Pardini M, Maisetta G. et al. Identification, molecular cloning, and evaluation of potential use of isocitrate dehydrogenase II of Mycobacterium bovis BCG in serodiagnosis of tuberculosis. Clin Diagn Lab Immunol. 2002;9:846–51. doi: 10.1128/CDLI.9.4.846-851.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.