Abstract
Replication of nucleic acids in the absence of genetically encoded enzymes represents a critical process for the emergence of cellular life. Repeated separation of complementary RNA strands is required to achieve multiple cycles of chemical replication, yet thermal denaturation under plausible prebiotic conditions is impaired by the high temperatures required to separate long RNA strands and by concurrent degradation pathways, the latter accelerated by divalent metal ions. Here we show how the melting temperature of oligoribonucleotide duplexes can be tuned by changes in pH, enabling the separation of RNA strands at moderate temperatures. At the same time, the risk of phosphodiester bond cleavage is reduced under the acid denaturation conditions herein described, both in the presence and in the absence of divalent metal ions. Through a combination of ultraviolet and circular dichroism thermal studies and gel electrophoresis, we demonstrate the relevance of geological pH oscillations in the context of the RNA strand separation problem. Our results reveal new insights in the field of prebiotic chemistry, supporting plausible geochemical scenarios in which non-enzymatic RNA replication might have taken place.
RNA, or a close variant thereof, is considered the first genetic polymer that appeared on early Earth. Recent investigations have shown how RNA mononucleotides, along with other building blocks considered necessary for life, might have chemically formed from the interplay of plausible prebiotic reaction networks in a defined geochemical setting.1−6 However, the transition from the chemistry of RNA monomers to the biology of a self-replicating system has been challenged by the many problems encountered with multiple cycles of non-enzymatic RNA replication.7 Chemical copying of RNA oligonucleotides might have resulted either from the templated polymerization of activated ribonucleotides8−10 or from the templated ligation of shorter RNA fragments.11 In both cases, a dead-end, double-stranded product is generated, and separation of the template and daughter strand is required prior to a new round of replication. The plausibility of thermal denaturation of RNA is hampered by the high melting temperature of oligoribonucleotides, to the point that duplexes of >30 bp are considered impossible to melt under plausible prebiotic conditions.7 Concurrent RNA degradation is known to occur at elevated temperatures and is made worse by the presence of divalent metal ions12 (such as Mg2+), which are required as catalysts during RNA replication. From the observation that unnatural 2′,5′-linkages destabilize RNA duplexes,13 Szostak and co-workers suggested backbone heterogeneity as a possible solution to the strand separation problem.14 However, the instability of such linkages in a duplex environment15,16 and the need to separate strands of native RNA prompted us to interrogate plausible geochemical scenarios on early Earth in the search for additional solutions.
Our curiosity was piqued by a report describing the different pH values of NaCl solutions at the eutectic point and after thawing, in principle leading to pH variations of >5 units between the two phases.17 More recently, heat fluxes have been shown to support the establishment of stable pH gradients in a closed system, such as those that could have occurred inside rock pores on primordial Earth.18 In both cases, geological heating and freezing could have acted as natural energy sources/sinks to support the development of local pH gradients, thus opening the possibility of repetitive pH oscillations. As DNA is known to undergo reversible acid denaturation19−22 (presumably through protonation of GC base pairs and consequent formation of Hoogsteen base pairing), the melting temperature of RNA oligonucleotides is likely to be similarly influenced by the pH of the solution.23,24 Hence, we envisioned that natural pH fluctuations might have supported cycles of RNA strand separation and reannealing, circumventing the need for high temperatures and the undesirable degradation thereby incurred.
Here we report our investigation of the pH dependence of the melting temperature and stability of different RNA oligonucleotides and the relevance of such results in the context of the prebiotic RNA strand separation problem.
Our study initially focused on outlining the effect of acidic pH values on the temperature at which 50% of a 13mer RNA duplex is denatured [melting point (Tm)]. Ultraviolet (UV) thermal melting curves were measured by incubating stoichiometric amounts of oligonucleotide 1a and its complementary strand 1b [duplex 1, GC content of 53.8% (Table S1)] at different pH values (3.1–7.1) and monitoring the variation of their absorbance at 260 nm as a function of the temperature of the solution (Figure 1A,B). Being a tribasic acid (pKa1 = 3.13, pKa2 = 4.76, and pKa3 = 6.40) whose dissociation constants are little influenced by temperature changes,25 citrate was the buffer of choice, enabling the evaluation of melting point variations over a wide pH range and in the absence of buffer or temperature effects.
Figure 1.
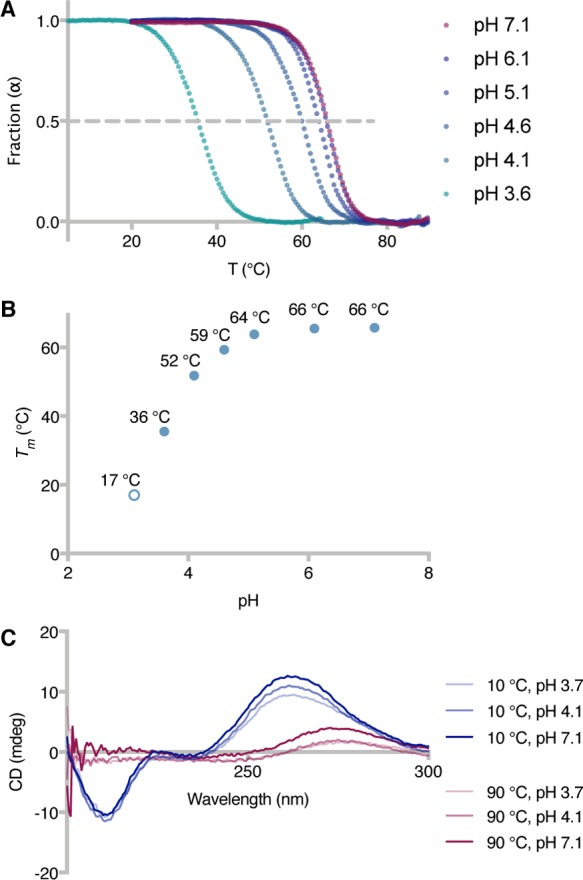
Modulation of RNA melting temperature by pH variations. (A) UV thermal melting curves of duplex 1 at different pH values (100 mM Na+, 10 mM citrate buffer, pH 3.6–7.1). Fraction of duplex RNA (α)32 vs temperature (T): α = 1 for double-stranded RNA, and α = 0 for single-stranded RNA. (B) Plot of duplex 1Tm vs solution pH, as determined from panel A. The Tm at pH 3.1 was calculated from the first-derivative maximum of the thermal melting curves (see the Supporting Information). (C) Circular dichroism spectra of duplex 1 at different pH and temperature values.
At pH 7.1, duplex 1 exhibited a melting point of 65.7 °C, and minimal variations were observed when the pH of the solution was decreased to either 6.1 or 5.1 (Tm = 65.5 or 63.8 °C, respectively). On the other hand, significant changes in the duplex Tm were registered at pH <4.6, with melting temperatures as low as 35.5 and 17.0 °C at pH 3.6 and 3.1, respectively. In parallel, we measured the CD thermal melting curves of duplex 1 to verify that the observed variations were the result of double-stranded to single-stranded transitions, ruling out possible effects arising from changes in the UV absorption of the nucleobases upon protonation (Figure 1C and Figures S1A–C and S2). The 10 °C CD spectra at pH 3.7, 4.1, and 7.1 all exhibited a negative couplet and a positive couplet around 210 and 260 nm, respectively, characteristic of double-stranded RNA structures.26 Upon heating, the 210 nm negative couplet progressively disappeared, together with a shift in the positive couplet maximum from 260 to 275 nm and a decrease in its intensity. These results confirm that denaturation of duplex 1 occurred upon heating over the entire pH range tested, and the melting temperatures determined by CD spectral changes were comparable with the data obtained in UV melting studies (Figure S1D). At pH 3.7 and 4.6, the intensity of the CD signal was slightly lower than the spectra at higher pH values. This effect is presumably caused by nucleobase protonation, and we do not exclude the presence of base pairing arrangements other than the standard Watson–Crick type.21,22
Additionally, we assessed the general effect of pH variations on the melting temperature of two new 13mer RNA duplexes, differing from 1 with respect to their GC content [30.8% GC content for duplex 2 and 69.2% GC content for duplex 3 (Table S1)]. As expected, changing the ratio of GC to AU base pairs affected the Tm values of both duplexes (Tm = 50.3 and 75.3 °C at pH 7.1 for duplexes 2 and 3, respectively), but the difference between the melting temperatures at pH 7.1 and 3.6 (ΔTm) was independent of the duplex employed [ΔTm = 30.2, 33.3, and 29.5 °C for duplexes 1–3, respectively (Table 1)].
Table 1. Tm and ΔTm Values of RNA Duplexesa.
| duplex | GC content (%) | Tm (°C) at pH 3.6 | Tm (°C) at pH 7.1 | ΔTm (°C) |
|---|---|---|---|---|
| 1 | 53.8 | 35.5 ± 0.5 | 65.7 ± 0.3 | 30.2 |
| 2 | 30.8 | 17.0 ± 0.0 | 50.3 ± 0.3 | 33.3 |
| 3 | 69.2 | 45.8 ± 0.3 | 75.3 ± 0.3 | 29.5 |
| 4 | 50.0 | 56.5 ± 0.5 | 84.2 ± 0.3 | 27.7 |
Values determined by UV thermal melting experiments (±standard deviation). Conditions: 100 mM Na+, 10 mM citrate buffer, pH 3.6 or 7.1.
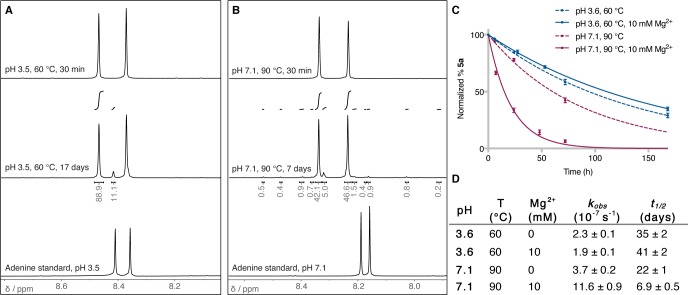
We next evaluated the potential of acidic pH values for the denaturation of duplexes that hardly melt under prebiotic conditions. Szostak and co-workers have previously shown that a 30mer RNA duplex containing four 2′,5′-linkages could be denatured at a temperature 15 °C lower than that of the corresponding naturally linked RNA duplex [50.0% GC content for duplex 4 (Table S1)].14 We thus measured the UV thermal curves of the fully 3′,5′-linked duplex 4 at pH 3.6 and 7.1 and found a remarkable 27.7 °C drop in its melting temperature upon exposure to acidic conditions [Tm = 56.5 and 84.2 °C at pH 3.6 and 7.1, respectively (Table 1)]. These results demonstrate how the melting temperature of RNA duplexes could be significantly altered upon changes in the environmental pH. Nevertheless, the prebiotic relevance of these results relies on whether moderate temperatures and low pH values would constitute an advantage over neutral pH and higher temperatures. A major concern in denaturing RNA duplexes is the increased risk of extensive phosphodiester bond cleavage. Furthermore, although less vulnerable than DNA,27−29 RNA might undergo a certain degree of depurination if exposed to acidic conditions. A direct comparison of the extent of degradation occurring during acidic or neutral RNA strand separation was thus needed. We investigated RNA depurination and phosphodiester bond cleavage following exposure to the conditions required to denature duplex 4 (either pH 3.5 and 60 °C or pH 7.1 and 90 °C, as determined from the UV melting experiments). As a model for RNA depurination, we studied the degradation of adenosine by 1H nuclear magnetic resonance (NMR) spectroscopy and found it to be highly stable under the acidic denaturation conditions (citrate buffer, pH 3.5, 60 °C), with only ∼11% of depurination after heating for 17 days (Figure 2A and Figure S3). By comparison, neutral denaturation conditions (citrate buffer, pH 7.1, 90 °C) resulted in ∼16% unspecific adenosine degradation after incubation for only 7 days (Figure 2B and Figure S4). In parallel, we estimated the extent of RNA backbone cleavage27,30 by monitoring the degradation of a FAM-labeled oligonucleotide [5a (Table S1)] by gel electrophoresis and fluorescence imaging. The average half-life of a phosphodiester bond (t1/2) was determined to be on the order of 35 days, when oligonucleotide 5a was incubated in citrate buffer at pH 3.6 and 60 °C (Figure 2C,D and Figure S5). Remarkably, inclusion of Mg2+ in the reaction mixture did not accelerate the rate of phosphodiester cleavage (t1/2 = 41 days). On the contrary, the t1/2 dropped to 22 days when oligonucleotide 5a was heated at 90 °C at neutral pH (citrate buffer, pH 7.1) and further decreased to 7 days in the presence of the divalent metal catalyst. Altogether, these data suggest that an acidic environment would be advantageous both for decreasing the melting temperature required to denature RNA duplexes and for decreasing the degree of concomitant RNA degradation. This is particularly true when divalent metal ions are to be present in the environment, the hydrolytic catalytic activity of which has long been considered a major obstacle to the preservation of RNA integrity at neutral pH. When these data are evaluated in the context of multiple cycles of non-enzymatic RNA replication, low pH values would therefore enable the separation of longer RNA duplexes with a lower degree of degradation, when compared to those characteristics under neutral conditions.
Figure 2.
Degradation under acidic or neutral denaturation conditions. (A) 1H NMR spectra [magnification of the H-C(2) and H-C(8) region] showing the extent of adenosine degradation at pH 3.5 and 60 °C. (B) Same as panel A at pH 7.1 and 90 °C. (C) Plot of the percentage of oligonucleotide 5a as a function of time, following incubation at either pH 3.5 and 60 °C or pH 7.1 and 90 °C, in the presence or absence of Mg2+. (D) Average (±standard deviation) hydrolysis rate constants (kobs) and phosphodiester bond half-lives (t1/2), as calculated from panel C.
Lastly, we sought to establish reversibility in the system upon exposure to repetitive pH oscillation cycles. Oligonucleotide 5a and its complement 5b [50.0% GC content for duplex 5 (Table S1)] were incubated in citrate buffer at pH 3.5, and the duplex Tm was measured by means of UV thermal melting [Tm = 47 °C (Figure 3)]. The pH of the solution was then increased to 6.8 by addition of Hepes buffer to the original mixture, and a new thermal curve was recorded (Tm = 68 °C). The use of FAM-labeled oligonucleotide 5a enabled us to estimate that 0.8% of RNA strands had degraded as a result of the initial UV thermal melting measurement at low pH. In parallel, we performed a control experiment, taking into account the duplex degradation and measuring the melting temperature of duplex 5 only after adjusting the pH of the solution to neutral (Tm = 68 °C). The Tm of duplex 5 at pH 6.8 was found to be identical in the two experiments, thus confirming the possibility of reverting the melting temperature of RNA by simply changing the pH of the solution. These results additionally rule out the chance of phosphodiester bond isomerization at acidic pH,31 as even a single 3′,5′- to 2′,5′-linkage transition would have led to a measurable decrease in the duplex Tm.14,16
Figure 3.
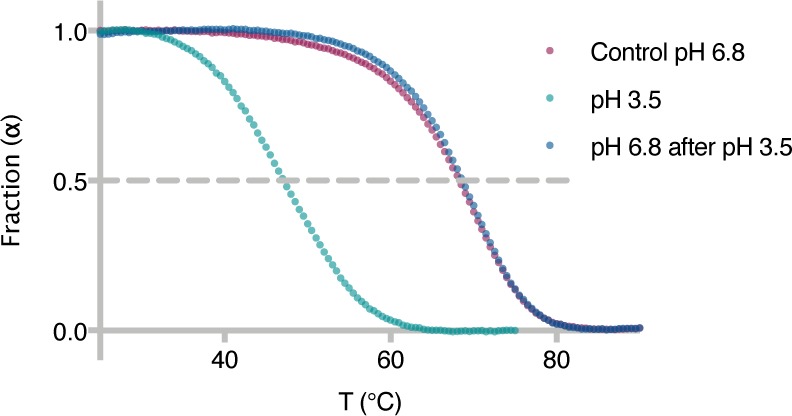
Reversibility in the system and absence of phosphodiester linkage isomerization. UV thermal melting curves of duplex 5 at pH 3.6 (1 M Na+, 10 mM citrate buffer) and after adjusting the pH to 6.8 (1 M Na+, 10 mM citrate buffer, 200 mM Hepes buffer) and comparison with a control experiment at pH 6.8 (1 M Na+, 10 mM citrate buffer, 200 mM Hepes buffer). α = 1 for double-stranded RNA, and α = 0 for single-stranded RNA.
In conclusion, this report demonstrates the possibility of reversibly tuning the melting temperature of RNA duplexes by simply altering the pH of the environment. Our results support the relevance of pH oscillations on early Earth, suggesting a model in which low pH values and moderate temperatures would have enabled the separation of RNA strands previously considered impossible to melt in a prebiotic setting. At the same time, RNA integrity is better preserved under the conditions required for acid rather than neutral denaturation, especially when magnesium ions are needed for subsequent replication steps. Therefore, the non-enzymatic replication of RNA oligonucleotides might have benefitted from natural geochemical environments in which the combination of pH and temperature oscillations (as could have occurred in rock pores18 or by freeze–thaw cycles17) would have supported multiple cycles of RNA strand separation and reannealing. For instance, freezing of brine solutions results in a pH increase of ≤5 units, the range of which apparently relies on nothing more than the initial pH of the solution.17 Thus, the transition from low pH and high temperature to high pH and low temperature required for RNA strand separation and replication cycles could have occurred in such natural environments. Nonetheless, RNA strand separation is only a small piece in the complex jigsaw that is the formation and evolution of RNA before the advent of life. Further investigations will be needed to provide explanations to the other many challenges in the field, including the possibility of preventing RNA strand invasion and inhibition in the course of multiple replication cycles.7
Acknowledgments
The authors thank all J.D.S. group members for fruitful discussions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.8b01080.
Materials and methods and supplementary figures and tables (PDF)
This work was supported by the Medical Research Council (Grant MC_UP_A024_1009) and a grant from the Simons Foundation (Grant 290362 to J.D.S.).
The authors declare no competing financial interest.
Supplementary Material
References
- Sutherland J. D. (2016) The origin of life-out of the blue. Angew. Chem., Int. Ed. 55, 104–121. 10.1002/anie.201506585. [DOI] [PubMed] [Google Scholar]
- Patel B. H.; Percivalle C.; Ritson D. J.; Duffy C. D.; Sutherland J. D. (2015) Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem. 7, 301–307. 10.1038/nchem.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stairs S.; Nikmal A.; Bučar D.-K.; Zheng S.-L.; Szostak J. W.; Powner M. W. (2017) Divergent prebiotic synthesis of pyrimidine and 8-oxo-purine ribonucleotides. Nat. Commun. 8, 15270. 10.1038/ncomms15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibard C.; Bhowmik S.; Karki M.; Kim E.-K.; Krishnamurthy R. (2017) Phosphorylation, oligomerization and self-assembly in water under potential prebiotic conditions. Nat. Chem. 10, 212–217. 10.1038/nchem.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritson D. J.; Battilocchio C.; Ley S. V.; Sutherland J. D. (2018) Mimicking the surface and prebiotic chemistry of early Earth using flow chemistry. Nat. Commun. 9, 1821. 10.1038/s41467-018-04147-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer P. B.; Xu J.; Thompson S. J.; Gillen E.; Sutherland J. D.; Queloz D. (2018) The origin of RNA precursors on exoplanets. Sci. Adv. 4, eaar3302 10.1126/sciadv.aar3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W. (2012) The eightfold path to non-enzymatic RNA replication. J. Syst. Chem. 3, 2. 10.1186/1759-2208-3-2. [DOI] [Google Scholar]
- Weimann B. J.; Lohrmann R.; Orgel L. E.; Schneider-Bernloehr H.; Sulston J. E. (1968) Template-directed synthesis with adenosine-5′-phosphorimidazolide. Science 161, 387. 10.1126/science.161.3839.387. [DOI] [PubMed] [Google Scholar]
- Inoue T.; Orgel L. E. (1982) Oligomerization of (guanosine 5′-phosphor)-2-methylimidazolide on poly(C): An RNA polymerase model. J. Mol. Biol. 162, 201–217. 10.1016/0022-2836(82)90169-3. [DOI] [PubMed] [Google Scholar]
- Li L.; Prywes N.; Tam C. P.; O’Flaherty D. K.; Lelyveld V. S.; Izgu E. C.; Pal A.; Szostak J. W. (2017) Enhanced nonenzymatic RNA copying with 2-aminoimidazole activated nucleotides. J. Am. Chem. Soc. 139, 1810–1813. 10.1021/jacs.6b13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler F. R.; Chan C. K. W.; Duffy C. D.; Gerland B.; Islam S.; Powner M. W.; Sutherland J. D.; Xu J. (2013) Prebiotically plausible oligoribonucleotide ligation facilitated by chemoselective acetylation. Nat. Chem. 5, 383–389. 10.1038/nchem.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butzow J. J.; Eichhorn G. L. (1965) Interactions of metal ions with polynucleotides and related compounds. IV. Degradation of polyribonucleotides by zinc and other divalent metal ions. Biopolymers 3, 95–107. 10.1002/bip.360030110. [DOI] [PubMed] [Google Scholar]
- Giannaris P. A.; Damha M. J. (1993) Oligoribonucleotides containing 2’,5′-phosphodiester linkages exhibit binding selectivity for 3′,5′-RNA over 3′,5′-ssDNA. Nucleic Acids Res. 21, 4742–4749. 10.1093/nar/21.20.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhart A. E.; Powner M. W.; Szostak J. W. (2013) Functional RNAs exhibit tolerance for non-heritable 2’-5′ versus 3′-5′ backbone heterogeneity. Nat. Chem. 5, 390–394. 10.1038/nchem.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher D. A.; McHale A. H. (1976) Hydrolytic stability of helical RNA: a selective advantage for the natural 3′,5′-bond. Proc. Natl. Acad. Sci. U. S. A. 73, 1149–1153. 10.1073/pnas.73.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani A.; Sutherland J. D. (2017) Non-enzymatic RNA backbone proofreading through energy-dissipative recycling. Angew. Chem., Int. Ed. 56, 6563–6566. 10.1002/anie.201703169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka N.; Tanaka M.; Okitsu K.; Bandow H. (2006) Rise in the pH of an unfrozen solution in ice due to the presence of NaCl and promotion of decomposition of gallic acids owing to a change in the pH. J. Phys. Chem. A 110, 10628–10632. 10.1021/jp0634095. [DOI] [PubMed] [Google Scholar]
- Keil L. M. R.; Möller F. M.; Kieß M.; Kudella P. W.; Mast C. B. (2017) Proton gradients and pH oscillations emerge from heat flow at the microscale. Nat. Commun. 8, 1897. 10.1038/s41467-017-02065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunville L. G.; Geiduschek E. P. (1960) DNA composition and stability to acid denaturation. Biochem. Biophys. Res. Commun. 2, 287–292. 10.1016/0006-291X(60)90186-8. [DOI] [Google Scholar]
- Bunville L. G.; Geiduschek E. P.; Rawitscher M. A.; Sturtevant J. M. (1965) Kinetics and equilibria in the acid denaturation of deoxyribonucleic acids from various sources. Biopolymers 3, 213–240. 10.1002/bip.360030301. [DOI] [Google Scholar]
- Courtois Y.; Fromageot P.; Guschlbauer W. (1968) Protonated polynucleotide structures. 3. An optical rotatory dispersion study of the protonation of DNA. Eur. J. Biochem. 6, 493–501. 10.1111/j.1432-1033.1968.tb00472.x. [DOI] [PubMed] [Google Scholar]
- Segers-Nolten G. M. J.; Sijtsema N. M.; Otto C. (1997) Evidence for Hoogsteen GC base pairs in the proton-induced transition from right-handed to left-handed poly(dG-dC)·poly(dG-dC). Biochemistry 36, 13241–13247. 10.1021/bi971326w. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Krishnamurthy R. (2009) Mapping the landscape of potentially primordial informational oligomers: oligo-dipeptides tagged with orotic acid derivatives as recognition elements. Angew. Chem., Int. Ed. 48, 8124–8128. 10.1002/anie.200904188. [DOI] [PubMed] [Google Scholar]
- To the best of our knowledge, investigations of RNA have been limited to homopolymer complexes in the form of poly(C)·poly(G) or poly(C)·poly(I) (which undergo the formation of triple-stranded helical structures upon protonation):; a Thiele D.; Guschlbauer W. (1969) Polynucléotides protonés. VII. Transitions thermiques entre differents complexes de l’acide polyinosinique et de l’acide polycytidylique en milieu acide. Biopolymers 8, 361–378. 10.1002/bip.1969.360080307. [DOI] [PubMed] [Google Scholar]; b Thiele D.; Guschlbauer W. (1971) Protonated polynucleotide structures. IX. Disproportionation of poly(G)·poly(C) in acid medium. Biopolymers 10, 143–157. 10.1002/bip.360100111.5544938 [Google Scholar]; Other reports have focused on RNA mismatches stabilization following protonation:; c Biała E.; Strazewski P. (2002) Internally mismatched RNA: pH and solvent dependence of the thermal unfolding of tRNAAla acceptor stem microhairpins. J. Am. Chem. Soc. 124, 3540–3545. 10.1021/ja0161305. [DOI] [PubMed] [Google Scholar]
- Stoll V. S.; Blanchard J. S. (1990) Buffers: principles and practice. Methods Enzymol. 182, 24–38. 10.1016/0076-6879(90)82006-N. [DOI] [PubMed] [Google Scholar]
- Ranjbar B.; Gill P. (2009) Circular dichroism techniques: biomolecular and nanostructural analyses- a review. Chem. Biol. Drug Des. 74, 101–120. 10.1111/j.1747-0285.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- Zoltewicz J. A.; Clark D. F.; Sharpless T. W.; Grahe G. (1970) Kinetics and mechanism of the acid-catalyzed hydrolysis of some purine nucleosides. J. Am. Chem. Soc. 92, 1741–1750. 10.1021/ja00709a055. [DOI] [PubMed] [Google Scholar]
- Garrett E. R.; Mehta P. J. (1972) Solvolysis of adenine nucleosides. I. Effects of sugars and adenine substituents on acid solvolyses. J. Am. Chem. Soc. 94, 8532–8541. 10.1021/ja00779a040. [DOI] [PubMed] [Google Scholar]
- Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature 362, 709–715. 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Kua J.; Bada J. L. (2011) Primordial ocean chemistry and its compatibility with the RNA world. Origins Life Evol. Biospheres 41, 553–558. 10.1007/s11084-011-9250-5. [DOI] [PubMed] [Google Scholar]
- Oivanen M.; Kuusela S.; Lönnberg H. (1998) Kinetics and mechanisms for the cleavage and isomerization of the phosphodiester bonds of RNA by Brønsted acids and bases. Chem. Rev. 98, 961–990. 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- Xu J.; Duffy C. D.; Chan C. K. W.; Sutherland J. D. (2014) Solid-phase synthesis and hybrization behavior of partially 2′/3′-o-acetylated RNA oligonucleotides. J. Org. Chem. 79, 3311–3326. 10.1021/jo5002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.