ABSTRACT
TAR DNA-binding protein 43 (TDP-43) is an RNA-binding protein that regulates transcription, translation and alternative splicing of mRNA. We have shown previously that null mutations of the Drosophila ortholog, Tar DNA-binding homolog (tbph), causes severe locomotion defects in larvae that are mediated by a reduction in the expression of a type II voltage-gated calcium channel, cacophony (cac). We also showed that TDP-43 regulates the inclusion of alternatively spliced exons of cacophony; tbph mutants showed significantly increased expression of cacophony isoforms lacking exon 7, a particularly notable finding as only one out of the 15 predicted isoforms lacks exon 7. To investigate the function of exon 7, we generated Drosophila mutant lines with a deletion that eliminates exon 7. This deletion phenocopies many defects in tbph mutants: a reduction in cacophony protein (Dmca1A) expression, locomotion defects in male and female third instar larvae, disrupted larval motor output, and also reduced activity levels in adult male flies. All these defects were rescued by expression of cacophony transcripts containing exon 7. By contrast, expression of a cacophony cDNA lacking exon 7 resulted in reduced cacophony protein levels and failed to rescue larval locomotion.
KEY WORDS: ALS, Model organism, Motor program, TDP-43, Dmca1A, Neurodegeneration
Summary: The voltage-gated calcium channel gene cacophony encodes multiple alternatively spliced variants. CRISPR/Cas9 genomic editing precisely eliminated a single cacophony exon in Drosophila, leading to larval crawling defects.
INTRODUCTION
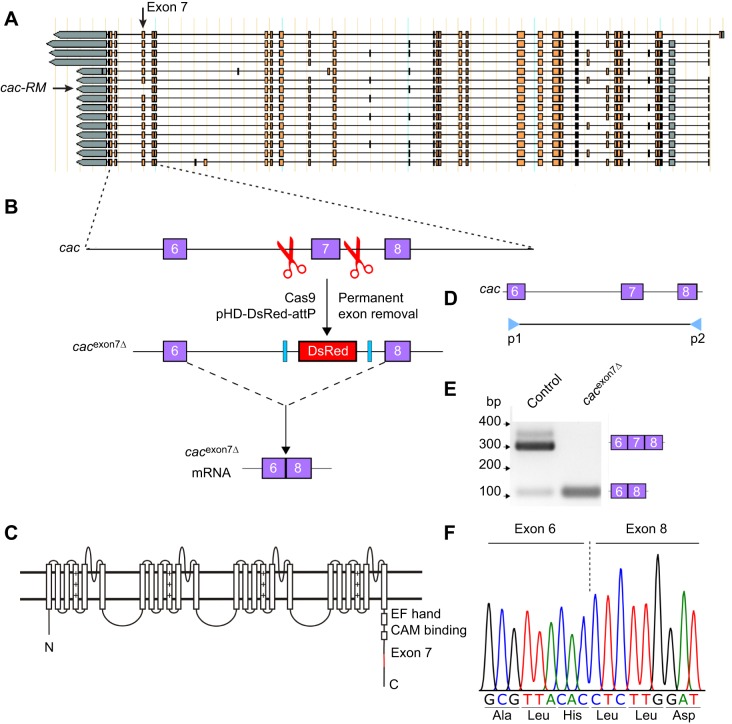
The amyotrophic lateral sclerosis (ALS)- and frontotemporal lobar degeneration (FTLD)-associated protein TAR DNA-binding protein 43 (TDP-43) is an RNA- and DNA-binding protein that regulates the expression and splicing of thousands of gene transcripts (Sephton et al., 2011; Hazelett et al., 2012). We have shown previously that loss of the Drosophila TDP-43 ortholog, tbph, caused a decrease in the expression of the primary type II voltage-gated channel, cacophony (Chang et al., 2014). Although tbph mutants showed a 50% reduction in cacophony protein (Dmca1A) expression, there was no decrease in total cacophony transcript levels (Chang et al., 2014). Of the cacophony transcripts expressed, there was a significant enrichment for transcripts lacking exon 7, which codes for a portion of the C-terminal cytoplasmic domain of the protein, suggesting that tbph functions to regulate the inclusion of specific cacophony exons (Chang et al., 2014). Of the 15 reported cacophony isoforms, only one, cac-RM, lacks exon 7, suggesting that this exon is of functional importance to the channel (Fig. 1A).
Fig. 1.
Generation of cacophony exon 7 deletion lines. (A) Predicted alternatively spliced cacophony transcripts from FlyBase, showing the position of exon 7 and that a single predicted isoform, cac-RM, lacks exon 7. (B) Schematic diagram showing the process of using CRISPR/Cas9 to delete exon 7 of cacophony. Cas9 was targeted to cut the genomic DNA on either side of exon 7, resulting in two double-stranded breaks, which were repaired via homologous recombination with a donor template containing DsRed, which was then incorporated into the genome. Splicing between exon 6 and 8 excludes the DsRed cassette, yielding cacexon7Δ mRNA. (C) Schematic diagram of the predicted cacophony protein, showing the location of exon 7 near the C-terminus. (D) Location of the primers in exon 6 and 8 used for RT-PCR to confirm the deletion of exon 7. (E) Representative DNA gel of the RT-PCR products from adult heads, showing a major 300 bp band that includes exon 7 and a minor 100 bp band lacking exon 7 from control animals. By contrast, RT-PCR from the cacexon7Δ line only yielded the 100 bp band. (F) Sequencing chromatogram resulting from the sequencing of the 100 bp band from the cacexon7Δ line showed that exon 6 splices directly to exon 8, excluding exon 7.
There is evidence that functional specificity of channels can be conferred by the differential expression of discreet channel isoforms (Lipscombe et al., 2013). The cacophony gene encodes the α-subunit of the Drosophila type II voltage-gated calcium channel, which is homologous to the vertebrate type II, N-type voltage-gated calcium channel (CaV2.2) (Kawasaki et al., 2002). The CaV2.2 channel has been implicated in synaptogenesis, regulation of gene expression, and neurotransmission at the neuromuscular junction (Brosenitsch and Katz, 2001; Kawasaki et al., 2004). The C-terminus of the gene coordinates channel inactivation, modulation by G-proteins, modulation by calmodulin (CAM; as there is a CAM-binding site), and protein–protein interactions that regulate activity and/or target the channel to specific cellular compartments (Gray et al., 2007). Inclusion and exclusion of specific exons of the channel are tissue specific and confer functional specificity. For example, in mice, exon 37a is expressed preferentially in the dorsal root ganglia, where it functions to increase the sensitivity of the N-type channel to the voltage-independent form of G-protein modulation (Gray et al., 2007). Therefore, natural variants of Cav2.2 can regulate the global activity of the channel within the context of the cellular milieu. Examples of disease-causing mutations that altered splicing of voltage-gated calcium channels include FTLD, Parkinsonism linked to chromosome 17, ALS, spinocerebellar ataxia 8, myotonic dystrophy and Timothy syndrome (Splawski et al., 2004; Cooper et al., 2009).
We have shown that loss of tbph caused disrupted larval locomotion, loss of motor burst pattern and coordination, and led to altered splicing and reduced levels of cacophony protein (Hazelett et al., 2012; Chang et al., 2014; Lembke et al., 2017). All of these effects were rescued by genetically restoring levels of cacophony. To further investigate the relationship between splicing of cacophony and motor burst rhythmicity and coordination, we specifically deleted the exon that appears to be the primary target of tbph, exon 7, using the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR associated gene 9) genome editing system (Gratz et al., 2014).
We found that targeted deletion of exon 7 reduced cacophony protein expression and caused locomotion defects in third instar larvae, similar to those reported in tbph mutants (Hazelett et al., 2012; Chang et al., 2014), but did not change the total expression of cacophony transcripts. Despite the fact that cacophony is the primary voltage-gated calcium channel at the neuromuscular junction (NMJ) and necessary for full evoked neurotransmission (Kawasaki et al., 2002, 2004), we found no consistent reduction in evoked release. Deletion of exon 7 also caused defects in larval locomotion and unpatterned, arrhythmic motor bursts similar to those of tbph mutants. These results also demonstrate a novel approach to assessing the function of specific exons in alternatively spliced genes: by precisely deleting specific exons, the contribution of these splice variants in physiology and development can be assessed at the whole-animal level.
MATERIALS AND METHODS
Fly stocks
All Drosophila stocks were reared at 25°C using standard procedures (Greenspan, 2004). The following fly strains were obtained from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu): D42-GAL4 motor neuron driver (w[*]; P{w[+mW.hs]=GawB}D42), OK6-GAL4 motor neuron driver (P{w[+mW.hs]=GawB}OK6), UAS-cacophony-EGFP (w[*]; P{w[+mC]=UAS-cac1-EGFP}786C), genomic duplication covering the cacophony gene (w[1118]; Dp(1;3)DC131, PBac{y[+mDint2] w[+mC]=DC131}VK00033). The duplication line carries a 77,150 bp genomic fragment on the third chromosome, which includes the cacophony locus and an additional 23,342 bp of flanking sequence. The cacexon7Δ mutants were crossed with w1118 at least four times to minimize any off-target effects caused by CRISPR/Cas9 (Lee et al., 2016) and to reduce differences in the genetic background between lines.
Generation of the cacexon7Δ mutants
A schematic diagram of the strategy used to generate flies in which exon 7 of cacophony was deleted is shown in Fig. 1. Fly embryos expressing the endonuclease Cas9 under the control of vas regulatory sequences (w[1118]; PBac{y[+mDint2]=vas-Cas9}VK00037/CyO, P{w[+mC]=Tb[1]}Cpr[CyO-A]) were injected by BestGene (Chino Hills, CA, USA) with two plasmids. One plasmid contained a donor template and was injected at a concentration of 500 ng µl−1. The donor template contained two regions, each of about 1 kb, of cacophony sequence from either side of exon 7, which were separated by sequence coding for a red fluorescent protein (DsRed) under the control of an eye-specific promoter. The other plasmid contained two guide RNAs and was injected at a concentration of 200 ng µl−1. The guide RNAs target Cas9 to sites on either side of exon 7 – 382 bp upstream and 562 bp downstream – resulting in two double-stranded breaks. This break was then repaired by homologous recombination using the donor template.
The donor template was generated by using PCR to amplify two homology arms that immediately flank exon 7 of cacophony and incorporate them into the pHD-DsRed-attP vector (Gratz et al., 2014). Since both homology arms did not perfectly flank exon 7, this incorporation resulted in a loss of 41 bp of intron on the 5′ end of exon 7 and a loss of 27 bp on the 3′ end of exon 7. About 1 kb of cacophony sequence was amplified from the P[acman] BAC CH321-64N05 library (Venken et al., 2009) for both the 5′ and the 3′ homology arms (using primers 5′ homology arm S and R and primers 3′ homology arm S and R, Table 1, respectively). The 5′ fragment was incorporated into the AarI site of the 5′ multiple cloning site (MCS) in the pHD-DsRed-attP vector and the 3′ fragment was incorporated into the SapI site of 3′ MCS in the pHD-DsRed-attP vector. Protospacer adjacent motif (PAM) sequences that would be used as targets by the guide RNAs were then removed from the homology arms using primers [5′ and 3′ PAM site-directed mutagenesis (SDM); Table 1] designed through the NEBaseChanger web tool. The mutated homology arms were then generated following the instructions of the Q5 site-directed mutagenesis kit (New England BioLabs, MA, USA) and confirmed by sequencing.
Table 1.
Sequences of the primers used in this study (5′–3′)
The guide RNA (gRNA) sequences were chosen using the Drosophila RNAi Screening Center CRISPR2 web tool (http://www.flyrnai.org/crispr2/). The two sequences selected were located in the introns between exons 7 and 8 and between exons 6 and 7 of cacophony. These two gRNAs (see Table 1, 5′ gRNA and 3′ gRNA) were cloned into two sibling vectors, pBFv-U6.2 and pBFv-U6.2B, respectively (Kondo and Ueda, 2013). Finally, these two independent gRNA cassettes were fused into a single plasmid for increased injection efficiency (Kondo and Ueda, 2013).
Generation of the UAS-cacophony-EGFP and UAS-cacexon7Δ-EGFP flies
Fly embryos with phiC31 (φC31) integration sites (y[1] w[67c23]; P{y[+t7.7]=CaryP}attP2) were injected by BestGene with one of two transformation plasmids. One plasmid contained a cacophony cDNA (cac1) fused to the EGFP coding sequence as described previously (Kawasaki et al., 2002), while the other was constructed with the same cac EGFP-fused coding sequence with a deletion to exclude exon 7 of cacophony. To generate cacophony sequence without exon 7, DNA was synthesized (Biomatik, Wilmington, DE, USA) at a length of 1515 bp using sequence from FlyBase as a reference with exon 7 removed. The synthesized sequence contained the restriction sites AgeI on the 5′ end and KpnI on the 3′ end, which were also present in cac1. The synthesized sequence was cut out of its cloning vector using AgeI and KpnI and shuttled into cac1. The final constructs were incorporated into the transformation vector pUASTattB (Bischof et al., 2007) and sequenced to confirm the lack of exon 7. The transformation constructs were prepared for injection using ZymoPURE Plasmid Mini Prep Kit (Zymo Research, Irvine, CA, USA) and injected at a concentration of 500 ng µl−1.
Genotyping of the cacexon7Δ mutants
Two sets of primers were designed to span a region that included the DsRed sequence at the 5′ end of cacophony and the 5′ homology arm (primers 5′ GT S and 5′ GT R; Table 1) and the region spanning DsRed at the 3′ end of cacophony and 3′ homology arm (primers 3′ GT S and 3′ GT R; Table 1). PCR was then performed using both sets of primers and Q5 high-fidelity polymerase (New England BioLabs). Both fragments were then TA-cloned for sequencing to confirm the expected sequence of the cacophony genomic region in which exon 7 was replaced by the DsRed cassette.
Real-time reverse transcription (RT)-PCR
Total RNA was extracted from either adult heads or larval CNS using Trizol (Life Technologies, Carlsbad, CA, USA) and cDNA synthesis was performed using SuperScript III (Life Technologies). To confirm that splicing took place as expected in the absence of exon 7, sequence from exon 6 to exon 8 was amplified with Q5 using primers cacEx6-8 S and cacEx6-8 R (see Table 1) and TA-cloned for sequencing. Real-time RT-PCR was used to determine the relative expression levels of cacophony in control flies and in the cacexon7Δ flies. Primer pairs against exon 23 and exon 24 of cacophony, and against EF2b as an internal control, were used for real-time RT-PCR reactions (see Table 1). Exons 23 and 24 were chosen because they are conserved in all predicted transcripts on FlyBase. Real-time RT-PCR was performed in a StepOne thermocycler (Life Technologies) using Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK). The PCR reaction conditions were 95°C for 10 min, followed by 40 cycles of 15 s at 95°C, 30 s at 63°C and 40 s at 72°C.
Delta Ct (ΔCt) values were calculated as the mean value of cacophony Ct minus the mean value of EF2b Ct. For larval CNS samples, a single technical replicate and 3 biological replicates were used and, for adult heads, 3 technical replicates and 4 biological replicates were processed. For each sample, the ΔCt for each biological replicate was calculated and normalized to the values of controls for relative expression levels.
Immunoblotting
The following antisera and antibodies (dilution; catalog number; source) were used for immunoblotting: rabbit anti-cacophony [1:4000; as previously described (Chang et al., 2014)], mouse anti-GAPDH (1:333; SC-365062; Santa Cruz, Dallas, TX, USA), chicken anti-GFP (1:2500; GFP-1020; Aves Labs, Inc., Tigard, OR, USA), mouse anti-rabbit H+L (1:2500; 211-005-109), goat anti-mouse H+L (1:2500; 115-005-146), goat anti-mouse peroxidase conjugate (1:5000; 115-035-003), rabbit anti-goat peroxidase conjugate (1:5000; 305-035-045) and goat anti-chicken peroxidase conjugate (1:10,000; 103-035-155) all from Jackson ImmunoResearch, West Grove, PA, USA.
Adult fly heads were dissected on a cold plate, homogenized in LDS loading buffer (Life Technologies) containing protease inhibitor (Roche Molecular Systems, Pleasanton, CA, USA) and allowed to incubate at room temperature for 45 min. TCEP was used to reduce samples at 75°C for 10 min, followed by centrifugation at 10,000 g for 15 min. The supernatant was kept, denatured at 75°C for an additional 10 min and proteins were fractionated using SDS-PAGE. Following transfer to a PVDF (polyvinylidene difluoride) membrane (Life Technologies), the membranes were blocked with 0.1% gelatin in PBST (PBS containing 0.05% Tween-20). Blots were then incubated in antisera against cacophony, GFP or GAPDH, followed by incubation with mouse anti-rabbit H+L or goat anti-mouse H+L antisera, and finally HRP-conjugated goat anti-mouse, HRP-conjugated rabbit anti-goat antisera or HRP-conjugated goat anti-chicken antisera as appropriate. The proteins were then detected with enhanced chemiluminescence DuoLuX (Vector laboratories, Inc., Burlingame, CA, USA). The relative cacophony levels were normalized to GAPDH levels using Fiji (Schindelin et al., 2012).
Longevity and behavioral assays
The cacexon7Δ mutants were outcrossed to control flies (described above) at least 4 times to minimize the effects of different genetic backgrounds and potential CRISPR off-target effects. The longevity of each line was assessed by collecting virgin males and females and placing them in separate vials of 10–15 animals in each vial. Vials were inspected for dead flies every other day and the remaining flies placed in new vials every 7 days.
To assess the climbing ability of adult flies, we carried out negative geotaxis assays on adult male flies as previously described (Gargano et al., 2005). Groups of 15 flies were placed in vials and the distance that individual flies climbed up the wall of a vial in 4 s was measured each week. Adult locomotory activity was assessed for 1-week-old individual male flies using an activity monitor (TriKinetics, Waltham, MA, USA) as previously described (Vanderwerf et al., 2015). Adult activity was logged as the total activity over the first 24 h the flies were in the recorder.
Larval locomotion was determined as previously described (Lembke et al., 2017). Third instar larvae were rinsed in PBS and placed on 2% agarose plates at room temperature. The crawling paths of the larvae were recorded for 5 min using a moticam 1000 connected to a PC and using the MIPlus07 software (Motic Images). The distance traveled in each video was traced and quantified using ImageJ software (http://imagej.nih.gov/ij/). The number of full posterior–anterior peristaltic waveforms was also recorded from these videos for each genotype.
Electrophysiological methods
Intracellular recordings were made from larval body wall muscle 6 in abdominal segment 3. Recordings were performed using glass microelectrodes as previously described (Lembke et al., 2017) and were performed at room temperature in extracellular HL3 saline containing (in mM): 70 NaCl, 5 KCl, 20 MgCl2, 10 NaHCO3, 115 sucrose, 5 trehalose, 5 HEPES and 1.0 CaCl2. Membrane potentials were recorded using an Axoclamp-2A amplifier (Molecular Devices) connected to a PC (Dell) and only recordings with resting membrane potential at or more negative than −55 mV were used. Excitatory junctional potentials (EJPs) were evoked by injection of current into severed axons, at 0.5 Hz, via a suction electrode and an A310 Accupulser (World Precision Instruments) through an isolation transformer. Miniature end plate potentials (mEPPs) were recorded over 3 min and analyzed using Mini Analysis 6.0.0.7 (Synaptosoft). The average single EJP amplitude of each recording was taken from 30 EPSPs, whose amplitudes were measured using Clampfit 10.2 software (Molecular Devices, Axon Instruments).
Motor activity recordings were made from peripheral nerves projecting from the second and seventh neuromeres of the intact CNS in third instar larvae as previously reported (Lembke et al., 2017). Nerves were suctioned en passant with a glass suction electrode in HL saline containing the following (in mmol ml–1): 70 NaCl, 5 KCl, 4 MgCl2, 10 NaHCO3, 115 sucrose, 5 trehalose, 5 HEPES and 1.8 CaCl2. Preparations were acutely incubated with 30 μmol ml–1 pilocarpine to stimulate fictive crawling and activate the motor program (Johnston and Levine, 1996). Recordings were done using an A-M Systems Differential AC Amplifier, digitized at 10 kHz, and stored with the Digidata 1440A digitizer as above. Recordings were made over 10 min and bandpass filtered (100 Hz to 10 kHz) using Clampex software (Molecular Devices). Autocorrelations and cross-correlations were computed for each trace as previously described (Lembke et al., 2017).
Experimental design and statistical analysis
GraphPad Prism 6 software was used to generate the graphs in this work. Data sets were analyzed for significance using one-way or two-way ANOVA followed by Dunnett's multiple comparison correction two-tailed t-test or Mantel–Cox log ranked test as stated in each figure legend. A 95% confidence interval was used to determine significance (P<0.05). See figure legends for sample sizes, P-values, and associated F- and t-values.
RESULTS
Deletion of exon 7 caused a decrease in cacophony protein expression but not total transcript levels
Our previous data showed that loss of tbph resulted in defective larval locomotion and reduction in the inclusion of exon 7 in cacophony (Hazelett et al., 2012; Chang et al., 2014). Furthermore, a causal relationship between cacophony and tbph-dependent larval locomotion was demonstrated by rescuing larval locomotion with increased expression of cacophony (Chang et al., 2014; Lembke et al., 2017). The cacophony cDNA used in these studies included exon 7, raising the question of whether larval locomotion required the specific inclusion of exon 7 or whether simply increased levels of cacophony were sufficient. To begin to address this question we used the CRISPR/Cas9 genome editing system to create a deletion covering exon 7 of cacophony (Fig. 1). Two plasmids were constructed, one that contained sequences complementary to the genomic sequence to direct Cas9-mediated cleavage of the genome on either side of exon 7 and the other containing complementary sequences on either side of exon 7, to direct homologous mediated recombination after exon 7 had been removed (Fig. 1B). This second plasmid also contained a cassette for eye-specific expression of DsRed flanked by splice acceptor and donor sequences (Gratz et al., 2014). Exon 7 codes for a 66-residue sequence at the C-terminal tail of cacophony (Fig. 1C). Drosophila embryos that expressed Cas9 in germline cells (Gratz et al., 2014) were injected with these two plasmids and 21 independent lines expressing DsRed were isolated. Two of these lines were homozygous viable, whereas the remaining 19 lines were homozygous lethal. We crossed the 19 lethal lines to a line containing a duplication that covers the genomic region of cacophony (DC131) and found that it rescued viability, indicating that the lethality locus was close to, or within, the cacophony locus. We then tested to see whether the lethality could be rescued by expressing a cacophony cDNA using the pan-neuronal Appl-GAL4 driver. We failed to observe any rescue of lethality, suggesting that the lethality was not due to loss of cacophony expression, but possibly due to off-target CRISPR effects, located nearby cacophony. For all the results described in this article, both of the homozygous viable lines gave similar results to each other and the results reported are for one of these lines. RT-PCR using primers that hybridized to exon 6 and 8 (Fig. 1D) using RNA from control animals generated a major 306 bp band expected for a product that included exon 7 and a minor band of 105 bp expected for a product that lacked exon 7. The mutant line, named cacexon7Δ, generated only this 105 bp product (Fig. 1E). When we sequenced this PCR product, the results showed that correct splicing between exon 6 and 8 had occurred, with a resulting sequence that was in frame and predicted to generate a full-length protein (Fig. 1F).
To determine whether the deletion had an impact on the overall transcription of cacophony or stability of the transcript, we used real-time RT-PCR with a primer pair spanning the intron between exon 24 and exon 23 (common to all cacophony isoforms) (Table 1). Using heads from adult males or females or whole larvae as a source of RNA showed that there was no difference in the levels of cacophony transcript between control or cacexon7Δ flies (Fig. 2A). We then examined the levels of cacophony protein, using immunoblots and an antibody specific to cacophony (Chang et al., 2014). Most of the different predicted isoforms of cacophony generate proteins with predicted sizes ranging from 212.1 to 212.6 kDa with cac-PM, the isoform lacking exon 7, having a predicted size of 204.9 kDa. These differences in size are not discernable on immunoblots and, as predicted, a single band with an apparent size of about 205 kDa was detected on immunoblots (Chang et al., 2014). Immunoblots of either adult heads or larval CNS from the cacexon7Δ line also showed a single band of this size (Fig. 2B). In addition, these blots showed that there was a reduction in the intensity of the band in the cacexon7Δ line compared with controls (Fig. 2B). We quantified the intensity of the bands, revealing a significant reduction in the level of cacophony protein detected in the mutant lines using either adult heads or larval CNS (Fig. 2B). After characterizing the molecular characteristics of the mutant line, we examined their phenotypes.
Fig. 2.
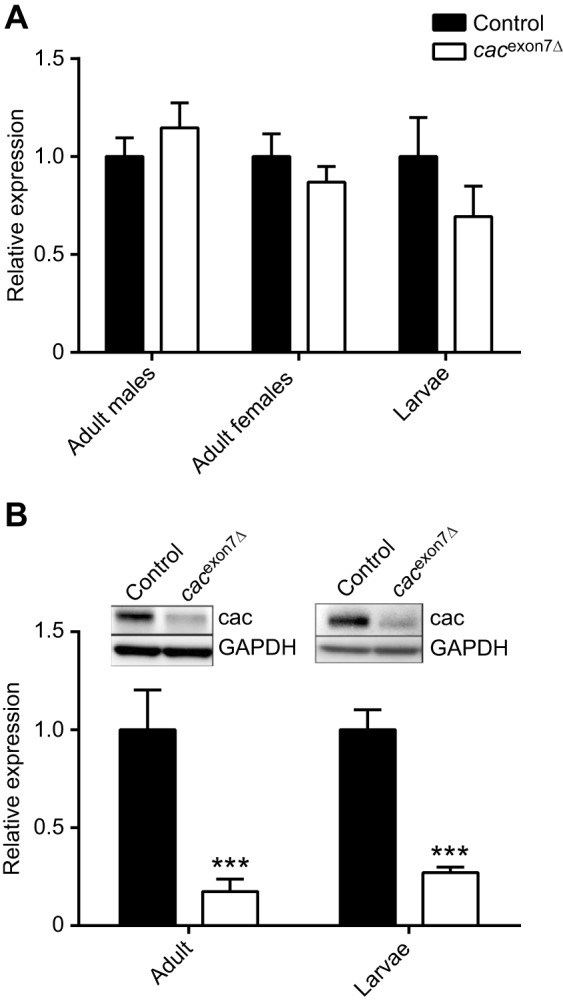
Quantification of cacophony transcripts and protein. (A) Real-time RT-PCR showed no significant difference in the levels of cacophony transcript between controls and the cacexon7Δ line for male or female heads or larval CNS. For each sample, primers against exons conserved across all predicted cacophony transcripts and against the loading control, EF2b, were used. The relative expression levels were calculated relative to EF2b and normalized to the relative expression in control flies. Mean and s.e.m. from four independent samples for adult heads and three independent samples for larval CNS are shown. Two-way ANOVA showed no significant differences between control and mutant lines (F2,16=1.62; P=0.23; P>0.05). (B) The levels of cacophony protein are reduced in cacexon7Δ mutants in both larvae and adults. Proteins from heads of adults or whole larvae were separated by PAGE, transferred to PVDF membrane and probed with cacophony protein antisera (Chang et al., 2014) and GAPDH as a loading control. The intensity of the cacophony immunoreactive band was quantified, divided by the intensity of its corresponding GAPDH band and normalized to the ratio obtained for the control sample. Mean and s.e.m. from four independent samples for adult heads and three independent samples for larvae are shown. Two-way ANOVA followed by Dunnett's multiple comparison test showed a significant reduction of cacophony protein levels in the cacexon7Δ mutant in both adults and larvae compared with controls (F1,12=42.7; ***P<0.001).
Deletion of exon 7 caused reduced locomotory activity in adults
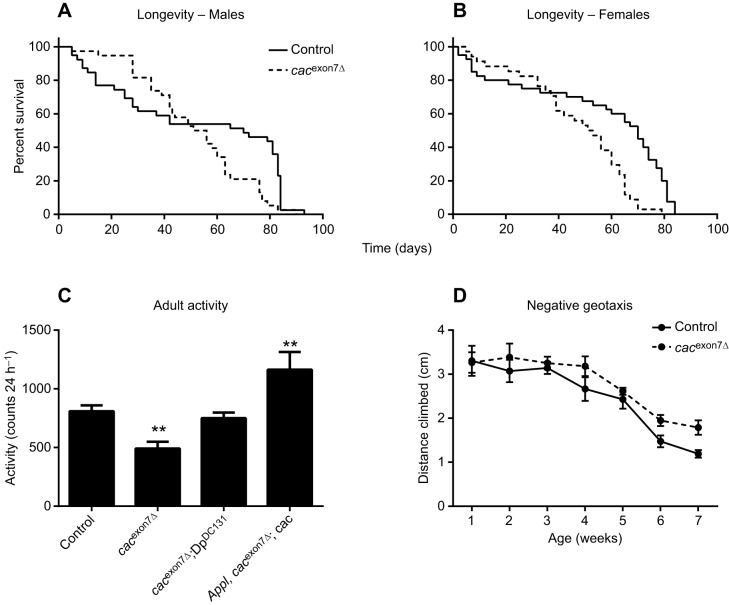
To determine whether there were any deleterious effects of this reduced level of cacophony, we assessed the longevity of cacexon7Δ flies. These data, shown in Fig. 3A and B, revealed that there was a slight, but statistically significant, reduction in the median lifespan in both male and female flies.
Fig. 3.
Adult phenotypes of the cacexon7Δ mutant. Survival plots of male (A) and female (B) adult flies, showing a slightly reduced median lifespan in both male and female flies: males: cacexon7Δ, 54 days (N=38), compared with controls, 70 days (N=39) (Mantel–Cox log ranked test, P=0.02); females: cacexon7Δ, 52 days (N=34), compared with controls, 70 days (N=40) (Mantel–Cox log ranked test, P=0.0002). (C) Activity levels of individual adult male flies. The level of activity over 24 h of the cacexon7Δ mutant (N=36) was significantly reduced compared with control flies (N=57). This reduced activity was restored with the presence of a genomic duplication (DpDC131) (N=31) or by expressing cacophony in all neurons using a pan-neuronal GAL4 driver (Appl) (N=21). The data were analyzed using one-way ANOVA followed by Dunnett's multiple comparison test. (F3,144=12.1; **P<0.01). (D) Negative geotaxis assay showed no significant difference in the performance of the cacexon7Δ mutant (N=9) compared to controls (N=8) (two-way ANOVA followed by Dunnett's multiple comparison test; P>0.05).
We also assessed the general activity levels of the cacexon7Δ flies in a bioactivity recorder, which showed that male cacexon7Δ flies had reduced levels of activity compared with controls. To confirm that this defect was due to changes in cacophony, we used a duplication line (DpDC131) that contained a duplication of a portion of the X chromosome that spans cacophony and includes the predicted promotor region. When we combined this duplication with the exon 7 deletion, the activity of the flies was restored to normal (Fig. 3C). We then used the pan-neuronal Appl-GAL4 driver and a UAS-cacophony construct that contained exon 7 (Kawasaki et al., 2004) to determine whether neuronal expression of exon 7 (+) cacophony was required for normal adult activity. The results shown in Fig. 3C showed that this was sufficient to restore the levels of activity in the cacexon7Δ flies. To assess the climbing ability of adult flies, we performed negative geotaxis assays by measuring the distance they climbed in 4 s. The results of this assay are shown in Fig. 3D: cacexon7Δ flies performed as well as control flies and showed similar levels of reduction in performance with age. In addition to assessing the adult phenotypes, we also examined phenotypes in larvae.
Deletion of exon 7 caused locomotion defects in third instar larvae
We had previously shown that tbph mutants show cacophony-dependent defective larval locomotion (Chang et al., 2014; Lembke et al., 2017). We therefore tested whether the cacexon7Δ mutants showed similar defects in larval locomotion. Total distance crawled was measured in third instar larvae and both male and female mutant larvae showed decreased crawling distance (Fig. 4A). To confirm that this defect was due to changes in cacophony, we again used the DpDC131 duplication line. When we combined this duplication with the cacexon7Δ mutants, the distance crawled by the larvae was restored to normal (Fig. 4B). We then used a variety of cell-specific GAL4 drivers and a UAS-cacophony construct that contained exon 7 (Kawasaki et al., 2004) to determine which neurons required exon 7 (+) cacophony for normal larval locomotion. Broad motor neuron GAL4 drivers (D42 and OK6) were sufficient to fully rescue the total distance crawled by larvae (Fig. 4B). We also used a cholinergic driver (Cha-GAL4) to drive expression of cacophony in sensory neurons and interneurons, which failed to rescue the crawling defect (Fig. 4B). Our previous studies (Lembke et al., 2017) identified a GAL4 driver line (R75C03) that expressed in two pairs of neurons in the brain that, when used to express cacophony in tbph mutants, was capable of fully rescuing the larval crawling defects. When we used this line to drive cacophony in the cacexon7Δ mutant larvae, we also measured a significant increase in crawling distance, although it did not fully rescue the defect (Fig. 4B).
Fig. 4.
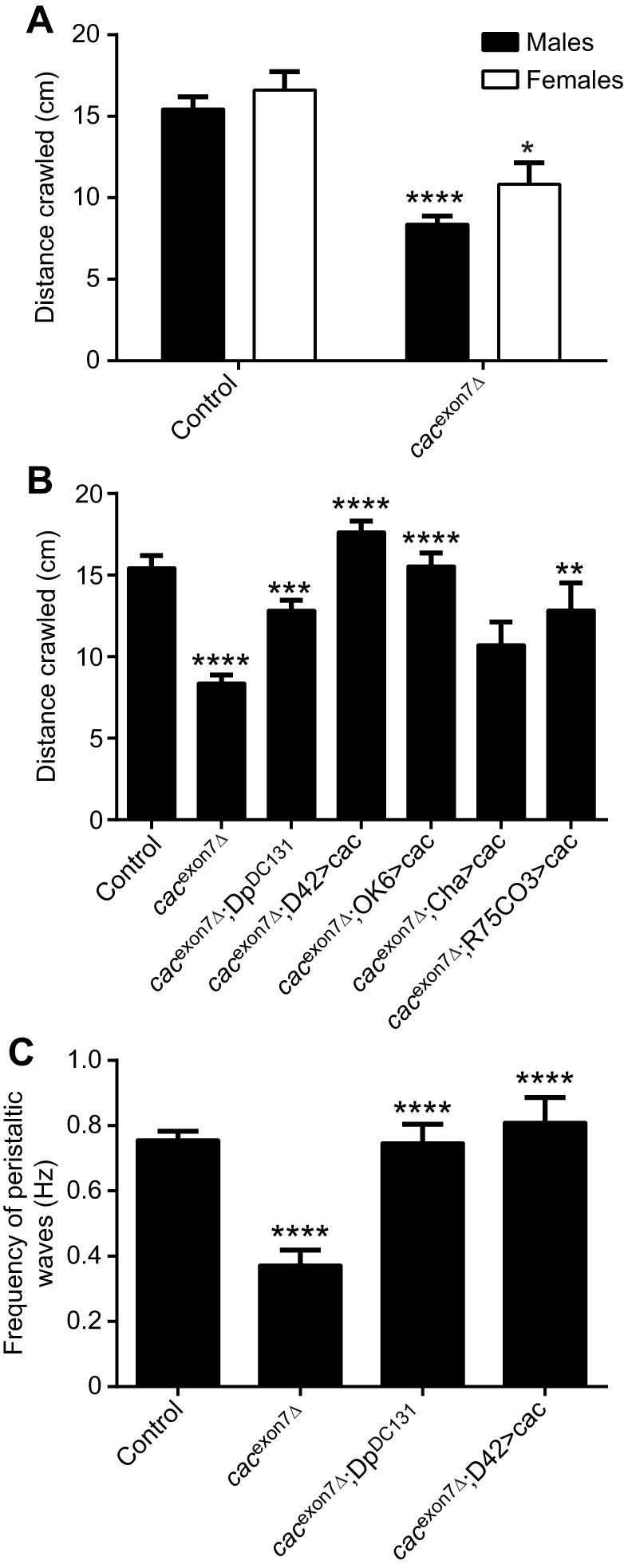
Larval locomotion is disrupted in the cacexon7Δ mutant. Third instar larvae were allowed to crawl across plain agar plates and the total distance traveled in 5 min was measured. (A) The total distance crawled in 5 min was significantly reduced in the cacexon7Δ mutant in both male (N=53 for controls and N=38 for mutants) and female (N=8 for controls and N=10 for mutants) larvae (two-way ANOVA followed by Sidak's multiple comparisons test, F2105=31.0; *P<0.05, ****P<0.0001). (B) Restoration of crawling in male larvae by expression of cacophony in motor neurons. The distance crawled by the cacexon7Δ mutants (N=38) was significantly reduced compared with controls (N=53) and was significantly increased by the presence of a genomic duplication (DpDC131; N=32) or by expressing cacophony in motor neurons using either the D42 (N=32) or OK6 (N=20) GAL4 drivers. No significant increase was seen with a cholinergic driver (Cha-GAL4; N=17) and a significant rescue was seen using the R75CO3 driver (N=19). One-way ANOVA followed by Dunnett's multiple comparisons test; F6,204=15.3; **P<0.01, ***P<0.001, ****P<0.0001. (C) The frequency of peristaltic waves in male larvae was significantly reduced in cacexon7Δ mutants (N=9) compared with controls (N=19) and was significantly increased by the presence of a genomic duplication (DpDC131) (N=10) or by expressing cacophony in motor neurons using the D42 GAL4 driver (N=10). One-way ANOVA followed by Dunnett's multiple comparisons test; F3,44=13.9; ****P<0.0001.
Larval crawling is a highly stereotyped behavior consisting of posterior to anterior peristaltic waves that travel the length of the larva (Fox et al., 2006; Inada et al., 2011). To analyze in more detail the locomotion defect of the deletion lines, we measured the frequency of the peristaltic waves. As expected, the cacexon7Δ mutant larvae showed a significant reduction in the frequency of peristaltic waves, which was rescued with the DpDC131 duplication and expression of cacophony using the D42 motor neuron driver (Fig. 4C). These defects in larval locomotion suggested defects at the NMJ, which we also examined.
Synaptic neurotransmission at the NMJ
Drosophila cacophony is necessary for evoked neurotransmission at the NMJ (Kawasaki et al., 2004; Lee et al., 2014) and the reduced levels of cacophony protein in the exon 7 deletion lines suggested that there could also be defects in neurotransmission at the NMJ. To examine whether there were any defects in the cacexon7Δ mutants, we recorded evoked and spontaneous transmission at the larval NMJ of body wall muscle 6 (Fig. 5). Surprisingly, when we measured the EJP amplitudes of the cacexon7Δ mutants, we found that there was no change in the EJP amplitude when compared to controls (Fig. 5A,B).
Fig. 5.
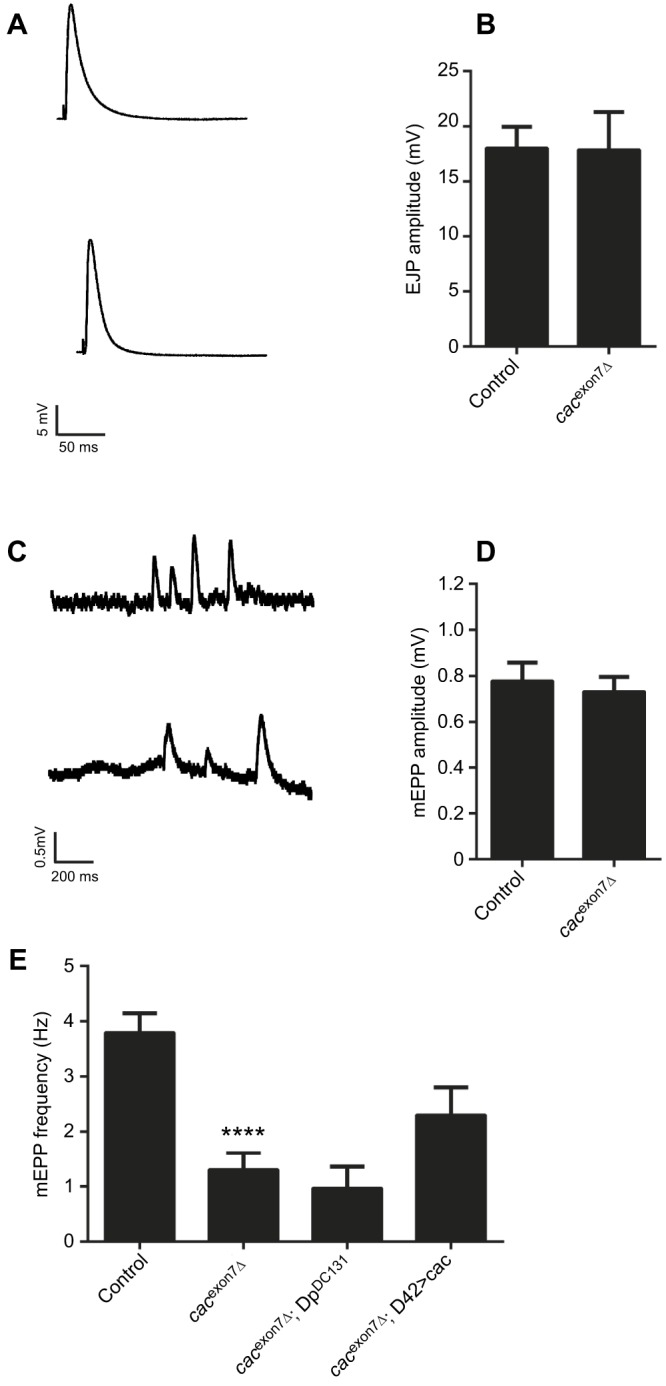
Synaptic physiology at the larval NMJ in the cacexon7Δ mutant. (A) Representative examples of excitatory junctional potentials (EJPs) from control (top) and cacexon7Δ mutant (bottom) larvae. (B) EJP amplitude was unchanged in cacexon7Δ mutants (N=11) compared to controls (N=17) (two-tailed t-test, t=0.04 d.f.=26; P>0.05). (C) Representative example of miniature end plate potentials (mEPPs) in control (top) and cacexon7Δ mutant (bottom) larvae. (D) The mEPP amplitude was unchanged in the cacexon7Δ mutant (N=11) compared to controls (N=17) (two-tailed t-test, t=0.04 d.f.=26; P>0.05). (E) The mEPP frequency was significantly reduced in the cacexon7Δ mutant (N=11) compared with controls (N=17) and was not rescued by either the DpDC131 duplication (N=5) or by expressing cacophony in motor neurons using the D42 GAL4 driver (N=9) (one-way ANOVA followed by Dunnett's multiple comparisons test, F3,38=11.2; ****P<0.0001). All experiments were carried out in 1 mmol l−1 calcium using male larvae.
We had previously shown that reduction in endogenous cacophony protein at the larval NMJ caused reduced frequencies of spontaneous neurotransmitter release (Lembke et al., 2017), and we also examined spontaneous neurotransmission by measuring the amplitude and frequency of mEPPs (Fig. 5C–E). The mEPP amplitude was unchanged in cacexon7Δ mutants, but the frequency was significantly reduced (Fig. 5D,E). We then examined the effect of incorporating the DpDC131 duplication and also of expressing cacophony cDNA in all motor neurons, and found that neither the duplication nor expressing cacophony in all motor neurons increased the frequency of mEPPs, suggesting that the reduction in mEPP frequency was not due to the deletion of exon 7 from cacophony (Fig. 5E). The absence of major defects in synaptic transmission suggested defects in the motor output of the nervous system.
Motor pattern output
The cacexon7Δ mutant animals showed a reduction in the frequency of peristaltic waves (Fig. 4C). This phenotype suggested that there was also defective motor output from the CNS (Fox et al., 2006; Lembke et al., 2017), and we therefore monitored the motor output from the CNS in semi-intact larvae.
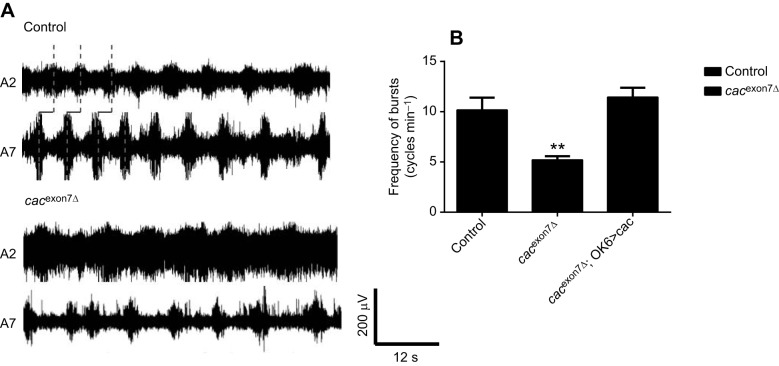
Focal extracellular recordings were made en passant from intact peripheral nerves projecting to muscle 6/7 in abdominal segment 2 (A2) and abdominal segment 7 (A7), as previously described (Lembke et al., 2017) (Fig. 6A). In the presence of pilocarpine, the cacexon7Δ deletion line show a significant reduction in the frequency of non-random motor bursts (Fig. 6A,B), which was rescued by driving cacophony in motor neurons using the OK6-GAL4 driver (Fig. 6B), suggesting that deletion of exon 7 significantly affects motor burst frequency.
Fig. 6.
The motor output of cacexon7Δ mutants was disrupted. (A) Representative examples of the motor output shows that control larvae exhibit regular, patterned motor bursts that progress from abdominal segment 7 (A7) and abdominal segment 2 (A2). By contrast, recordings from the cacexon7Δ mutants show irregular and poorly defined bursts. (B) Frequency of motor bursts. An autocorrelation analysis was performed on each recording to test whether the bursting showed non-randomness. The cacexon7Δ mutants showed non-random bursting at a frequency significantly reduced compared with control larvae, which was rescued by expressing cacophony in motor neurons using the OK6 GAL4 driver (one-way ANOVA followed by Dunnett's multiple comparison test; F2,18=12.8; **P<0.01). All recordings were done in HL3.1 saline solution containing 1.8 mmol l−1 calcium and 30 μmol l–1 pilocarpine. Recordings were taken from A2 and A7 motor nerves. Data represent the mean and s.e.m. of 7 animals for each genotype.
Exon 7 is required for full protein expression and restoration of larval locomotion
Deletion of exon 7 caused reduced cacophony protein levels and defective larval locomotion. To determine whether these two phenotypes are causally related, we generated two new UAS-cacophony lines. One was generated using the same cacophony cDNA used to make the original UAS-cac1 line (Kawasaki et al., 2002) and the other was constructed using the same cDNA as starting material but engineered to delete exon 7. Both constructs were generated in plasmids containing attB sites for site-specific integration into the genome using the phiC3 system to ensure equivalent expression levels (Bischof et al., 2007). We first crossed both lines with a pan-neuronal GAL4 driver and immunoblotted adult heads with an anti-GFP antibody to determine whether equivalent levels of cacophony-EGFP protein were present (Fig. 7A). Both the UAS-cac (exon 7+) line and the original UAS-cac1 line resulted in a robust GFP signal, whereas there was a notably lower intensity signal from the UAS-cac (exon 7−) line.
Fig. 7.
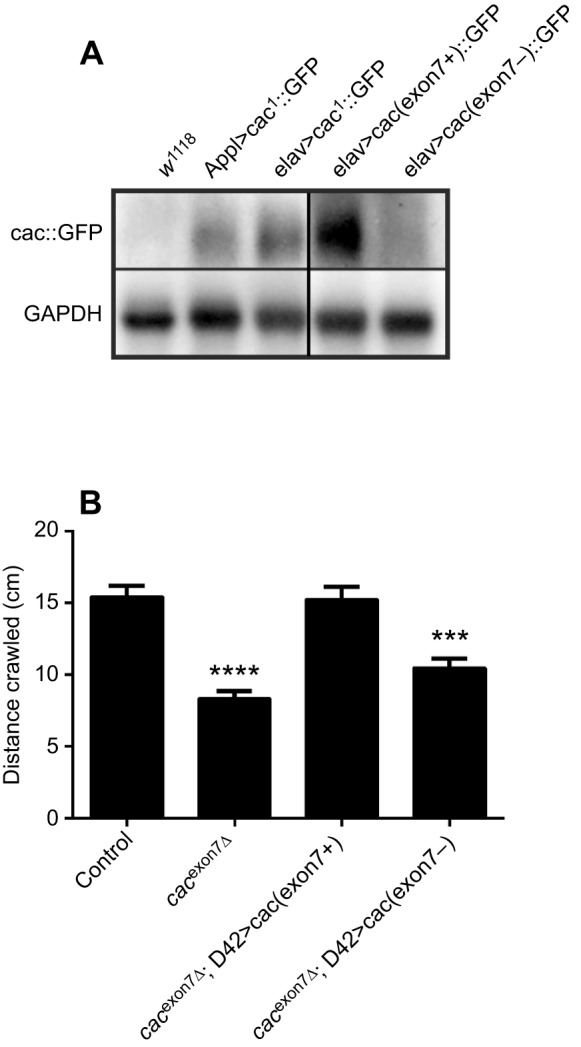
Expression of a UAS-cacophony transgene lacking exon 7 failed to rescue larval crawling and resulted in low cacophony protein expression. (A) Immunoblots using anti-GFP antisera show reduced levels of cacophony-GFP in flies expressing a UAS-cacophony transgene lacking exon 7 [elav>cac(exon7−)::GFP] compared with flies expressing a UAS-cacophony transgene containing exon 7 [elav>cac(exon7+)::GFP] or flies expressing the original cac1 transgene (Appl>cac1::GFP or elav>cac1::GFP). (B) Expression of the UAS-cacophony transgene lacking exon 7 in motor neurons using the D42-GAL4 driver was unable to rescue the crawling defects in cacexon7Δ mutants, whereas expression of the UAS-cacophony transgene containing exon 7 fully rescued crawling. Data represent the mean and s.e.m. of at least 10 animals and were analyzed using one-way ANOVA followed by Dunnett's multiple comparison test (F3,114=23.5; ***P<0.01, ****P<0.0001).
To test whether the presence of exon 7 in the cDNA had any effect on larval crawling, we drove its expression in motor neurons using the D42-GAL4 motor neuron driver. The larval locomotion of the cacexon7Δ mutant was rescued by cac(exon7+), whereas driving cac(exon7−) expression with D42-GAL4 failed to rescue crawling, suggesting that normal larval movement depended on the expression of exon 7.
DISCUSSION
Drosophila cacophony is a member of the type II voltage-gated calcium channel family, member of which act to regulate many neuronal processes, including synaptic neurotransmission at the NMJ, synaptogenesis and synaptic homeostasis, as well as regulation of gene expression (Catterall, 2011). Here, we describe the generation of a new cacophony mutant fly line that has a single exon deletion, which eliminates exon 7 in the coding region of the protein. The cacophony gene has multiple splice variants and, in wild-type animals, exon 7 is included in all but one predicted splice variant. RT-PCR showed that, in wild-type larvae, approximately 50% of the total level of cacophony mRNA contains exon 7 (Chang et al., 2014), whereas, in adult heads from wild-type animals, almost all of the cacophony transcripts include exon 7 (Fig. 1E). This would have suggested that any phenotypes resulting from the loss of exon 7 would be more severe in adults compared with larvae. Surprisingly, however, we did not detect any dramatic phenotypes in adults: their survival and longevity were similar to control animals and negative geotaxis was unaffected. The only significant effect was that the total level of locomotory activity was reduced in a cacophony-dependent manner. Interestingly, larval locomotion was also reduced in cacexon7Δ animals, which could be rescued by the expression of cacophony in motor neurons.
Our rationale for developing these mutants was our finding that null mutations in tbph, the Drosophila ortholog of the ALS- and FTLD-associated gene, TARDBP, caused a reduction in the incorporation of exon 7 in cacophony transcripts and a corresponding cacophony-dependent reduction in larval locomotion (Chang et al., 2014). In addition to both tbph and cacexon7Δ mutants showing defective larval locomotion, in both mutants there is also a reduction in the levels of cacophony protein, but no change in the total levels of cacophony transcript (Chang et al., 2014; Fig. 2).
Given the similarity of the cacophony-dependent effects in both tbph and cacexon7Δ mutants, we decided to investigate the causes of the defects in larval locomotion in cacexon7Δ mutants in more detail. We first examined synaptic transmission and, despite there being a reduced level of cacophony protein, there was no reduction in evoked neurotransmitter release. This was unexpected as another hypomorphic cacophony allele, cacTS2, led to a dramatic reduction in EJP amplitude (Kawasaki et al., 2002; Macabuag and Dolphin, 2015). However, it is consistent with our previous report that decreased endogenous cacophony protein expression in tbph mutants did not display reduced evoked neurotransmission (Lembke et al., 2017). This suggests that there is sufficient cacophony protein present at the NMJ for normal evoked transmission. We also found that the frequency of spontaneous mEPPs was significantly reduced in cacexon7Δ mutants, although this could not be restored by the expression of cacophony in motor neurons. This was particularly noteworthy as tbph mutants also exhibited a cacophony-dependent reduction in the frequency of mEPPs and, also, no effect on EJP amplitude was observed (Lembke et al., 2017). Although there are no reports that there is a reduction in mEPP frequency in the cacTS2 allele, studies on genes that lead to a reduction in cacophony protein at active zones showed no effect on mEPP frequency, although they also found a reduction in EJP amplitude (Kittel et al., 2006; Graf et al., 2012). Nevertheless, we have previously shown that pharmacological blockage of cacophony protein also led to reduced frequency of mEPPs (Lembke et al., 2017). These results seem to suggest that there is not a simple relationship between the levels of cacophony protein in the pre-synaptic terminal and evoked and spontaneous transmitter release.
Similarly, both tbph and cacexon7Δ mutants exhibited a disrupted larval motor program. We previously reported that tbph mutant larvae displayed cacophony-dependent unpatterned motor bursts and loss of coordination between body wall segments (Lembke et al., 2017). In the current study we also showed that cacexon7Δ mutants produced disrupted motor output (Fig. 6B), which could be restored by the motor neuron expression of cacophony. However, the phenotype did not appear to be as severe as in the tbph mutants as we still detected patterned output in the cacexon7Δ mutants (Fig. 6A).
To better understand the relationship between the presence of exon 7, cacophony protein levels and defects in larval locomotion, we generated two additional UAS constructs: one containing the same cacophony cDNA used for all previous rescues and the other containing an identical cDNA, with the exception that the 201 bp coding for exon 7 was deleted. Both constructs were targeted to the same insertion positions in the genome to ensure comparable levels of expression. Immunoblot analysis showed that, despite similar expression levels, cacophony protein levels were dramatically reduced in flies expressing the exon7− cDNA (Fig. 7A). Not surprisingly, when expressed in motor neurons in cacexon7Δ mutants, the cac(exon7+) cDNA fully rescued the larval locomotion defects, whereas cac(exon7−) cDNA failed to restore larval locomotion. These data suggest that the loss of exon 7 directly leads to reduced levels of protein, and that the reduced levels of cacophony protein in cacexon7Δ mutants is responsible for the phenotypes observed.
A significant unanswered question is the specific molecular and/or cellular function of exon 7. The data presented here clearly show that loss of exon 7 leads to reduced levels of cacophony protein and defective behavior. Exon 7 codes for a 66-residue portion of the cytoplasmic C-terminus of the channel. Although the C-terminus of cacophony protein contains a calcium/calmodulin binding domain and an EF hand, these are not included in exon 7. Exon 7 is highly conserved across insect cacophony orthologs; however, it is not conserved at the amino acid level in mammalian Cav type II channels. A possible clue to the function of exon 7 comes from studies of alternatively spliced forms of Cav2.2, which has two alternative exons in a similar location to exon 7 in cacophony. Alternative splicing of these exons in Cav2.2 regulates neuronal trafficking via the presence or absence of an AP-1 binding motif (Macabuag and Dolphin, 2015). Although exon 7 of cacophony does not appear to contain an AP-1 binding motif, there is an AP-1 binding motif (YPTL) nearby in exon 6. It is possible that the presence or absence of exon 7 modulates the ability of AP-1 binding and hence regulates trafficking of cacophony, and that defective trafficking of cacophony leads to increased degradation. Further experiments are needed to test this model directly.
One of the most striking findings in this study is the close parallel of larval phenotypes seen in both cacexon7Δ and tbph mutants. We had previously shown that TBPH could bind to cacophony transcripts (Chang et al., 2014), and had assumed that this was required for normal levels of translation and also for the correct inclusion of exon 7, and that these were two separate actions. In light of our current data showing that the independent deletion of exon 7 also results in reduced cacophony protein, we would suggest a simpler model in which reduced inclusion of exon 7 in tbph mutants is sufficient to lead to a reduction in cacophony protein and that this is sufficient to lead to the larval phenotypes observed in the tbph mutants.
Acknowledgements
We thank members of the Morton lab, especially Dr Jer-Cherng Chang, for critical comments on the manuscript and helpful discussions during the course of this work. We also thank Richard Ordway and Andrew Frank for providing UAS-cac plasmids, protocols and helpful advice in the cloning of cacophony cDNAs. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.M.L., D.B.M.; Methodology: K.M.L., A.D.L.; Formal analysis: D.B.M.; Investigation: K.M.L., A.D.L., J.A., D.B.M.; Writing - original draft: K.M.L., D.B.M., A.D.L.; Writing - review & editing: K.M.L., D.B.M., A.D.L.; Visualization: D.B.M.; Supervision: D.B.M.; Project administration: D.B.M.; Funding acquisition: D.B.M.
Funding
This study was supported by grants from NINDS (NS071186) and the Amyotrophic Lateral Sclerosis Association (23VU14). Deposited in PMC for release after 12 months.
References
- Bischof J., Maeda R. K., Hediger M., Karch F. and Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312-3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosenitsch T. A. and Katz D. M. (2001). Physiological patterns of electrical stimulation can induce neuronal gene expression by activating N-type calcium channels. J. Neurosci. 21, 2571-2579. 10.1523/JNEUROSCI.21-08-02571.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall W. A. (2011). Voltage-gated calcium channels. Cold Spring Harb. Perspect Biol. 3, a003947 10.1101/cshperspect.a003947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.-C., Hazelett D. J., Stewart J. A. and Morton D. B. (2014). Motor neuron expression of the voltage-gated calcium channel cacophony restores locomotion defects in a Drosophila, TDP-43 loss of function model of ALS. Brain Res. 1584, 39-51. 10.1016/j.brainres.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. A., Wan L. and Dreyfuss G. (2009). RNA and disease. Cell 136, 777-793. 10.1016/j.cell.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox L. E., Soll D. R. and Wu C.-F. (2006). Coordination and modulation of locomotion pattern generators in Drosophila larvae: effects of altered biogenic amine levels by the tyramine beta hydroxylase mutation. J. Neurosci. 26, 1486-1498. 10.1523/JNEUROSCI.4749-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargano J. W., Martin I., Bhandari P. and Grotewiel M. S. (2005). Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Exp. Gerontol. 40, 386-395. 10.1016/j.exger.2005.02.005 [DOI] [PubMed] [Google Scholar]
- Graf E. R., Valakh V., Wright C. M., Wu C., Liu Z., Zhang Y. Q. and DiAntonio A. (2012). RIM promotes calcium channel accumulation at active zones of the Drosophila neuromuscular junction. J. Neurosci. 32, 16586-16596. 10.1523/JNEUROSCI.0965-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M. and O'Connor-Giles K. M. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961-971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray A. C., Raingo J. and Lipscombe D. (2007). Neuronal calcium channels: splicing for optimal performance. Cell Calcium 42, 409-417. 10.1016/j.ceca.2007.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan R. J. (2004). Fly Pushing, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Hazelett D. J., Chang J.-C., Lakeland D. L. and Morton D. B. (2012). Comparison of parallel high-throughput RNA sequencing between knockout of TDP-43 and its overexpression reveals primarily nonreciprocal and nonoverlapping gene expression changes in the central nervous system of Drosophila. G3 2, 789-802. 10.1534/g3.112.002998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada K., Kohsaka H., Takasu E., Matsunaga T. and Nose A. (2011). Optical dissection of neural circuits responsible for Drosophila larval locomotion with halorhodopsin. PLoS ONE 6, e29019 10.1371/journal.pone.0029019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. M. and Levine R. B. (1996). Crawling motor patterns induced by pilocarpine in isolated larval nerve cords of Manduca sexta. J. Neurophysiol. 76, 3178-3195. 10.1152/jn.1996.76.5.3178 [DOI] [PubMed] [Google Scholar]
- Kawasaki F., Collins S. C. and Ordway R. W. (2002). Synaptic calcium-channel function in Drosophila: analysis and transformation rescue of temperature-sensitive paralytic and lethal mutations of cacophony. J. Neurosci. 22, 5856-5864. 10.1523/JNEUROSCI.22-14-05856.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki F., Zou B., Xu X. and Ordway R. W. (2004). Active zone localization of presynaptic calcium channels encoded by the cacophony locus of Drosophila. J. Neurosci. 24, 282-285. 10.1523/JNEUROSCI.3553-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel R. J., Wichmann C., Rasse T. M., Fouquet W., Schmidt M., Schmid A., Wagh D. A., Pawlu C., Kellner R. R., Willig K. I. et al. (2006). Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312, 1051-1054. 10.1126/science.1126308 [DOI] [PubMed] [Google Scholar]
- Kondo S. and Ueda R. (2013). Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715-721. 10.1534/genetics.113.156737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Ueda A. and Wu C.-F. (2014). Distinct roles of Drosophila cacophony and Dmca1D Ca 2+ channels in synaptic homeostasis: Genetic interactions with slowpoke Ca 2+ -activated BK channels in presynaptic excitability and postsynaptic response. Dev. Neurobiol. 74, 1-15. 10.1002/dneu.22120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. M., Cradick T. J., Fine E. J. and Bao G. (2016). Nuclease target site selection for maximizing on-target activity and minimizing off-target effects in genome editing. Mol. Ther 24, 475-487. 10.1038/mt.2016.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembke K. M., Scudder C. and Morton D. B. (2017). Restoration of motor defects caused by loss of Drosophila TDP-43 by expression of the voltage-gated calcium channel, Cacophony, in central neurons. J. Neurosci. 37, 9486-9497. 10.1523/JNEUROSCI.0554-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D., Andrade A. and Allen S. E. (2013). Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim. Biophys. Acta. 1828, 1522-1529. 10.1016/j.bbamem.2012.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macabuag N. and Dolphin A. C. (2015). Alternative splicing in Ca(V)2.2 regulates neuronal trafficking via adaptor protein complex-1 adaptor protein motifs. J. Neurosci. 35, 14636-14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton C. F., Cenik C., Kucukural A., Dammer E. B., Cenik B., Han Y., Dewey C. M., Roth F. P., Herz J., Peng J. et al. (2011). Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem 286, 1204-1215. 10.1074/jbc.M110.190884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I., Timothy K. W., Sharpe L. M., Decher N., Kumar P., Bloise R., Napolitano C., Schwartz P. J., Joseph R. M., Condouris K. et al. (2004). Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell 119, 19-31. 10.1016/j.cell.2004.09.011 [DOI] [PubMed] [Google Scholar]
- Vanderwerf S. M., Buck D. C., Wilmarth P. A., David L. L., Sears L. M., Morton D. B. and Neve K. A. (2015). Role for Rab10 in methamphetamine-induced behavior. PloS ONE 10, e0136167 10.1371/journal.pone.0136167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., Carlson J. W., Schulze K. L., Pan H., He Y., Spokony R., Wan K. H., Koriabine M., de Jong P. J., White K. P. et al. (2009). Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6, 431-434. 10.1038/nmeth.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]