ABSTRACT
Tumors are composed of heterogeneous phenotypes, each having different sensitivities to the microenvironment. One microenvironment characteristic – matrix stiffness – helps to regulate malignant transformation and invasion in mammary tumors, but its influence on oral squamous cell carcinoma (OSCC) is unclear. We observed that, on stiff matrices, a highly invasive OSCC cell line (SCC25) comprising a low E-cad to N-cad ratio (InvH/E:NL; SCC25) had increased migration velocity and decreased adhesion strength compared to a less invasive OSCC cell line (Cal27) with high E-cad to N-cad ratio (InvL/E:NH; Cal27). However, InvL/E:NH cells acquire a mesenchymal signature and begin to migrate faster when exposed to prolonged time on a stiff niche, suggesting that cells can be mechanically conditioned. Owing to increased focal adhesion assembly, InvL/E:NH cells migrated faster, which could be reduced when increasing integrin affinity with high divalent cation concentrations. Mirroring these data in human patients, we observed that collagen organization, an indicator of matrix stiffness, was increased with advanced disease and correlated with early recurrence. Consistent with epithelial tumors, our data suggest that OSCC cells are mechanically sensitive and that their contribution to tumor progression is mediated in part by this sensitivity.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Cancer, Extracellular matrix, Elasticity, Collagen, Hydrogel
Highlighted Article: Matrix stiffness modulates OSCC cell migration similarly to that of other epithelial carcinomas but by using a different focal-adhesion-mediated mechanism.
INTRODUCTION
Oral squamous cell carcinoma (OSCC) is the most common oral cancer in the USA (Markopoulos, 2012) and originates from epithelial cells, whose genetic mutations induce them to lose polarity, invade adjacent connective tissue and even metastasize to distant tissues (Leemans et al., 2018). With the loss of polarity, cells undergo an epithelial-to-mesenchymal transition (EMT) and acquire a migratory phenotype (Smith et al., 2013; Nieto et al., 2016; Lamouille et al., 2014). EMT is orchestrated by the expression and/or nuclear localization of several families of transcriptional factors (Yao et al., 2017; Kong et al., 2015; Fan et al., 2013), including TWIST, SNAIL and ZEB. One change accompanying EMT in OSCC cells is a switch from epithelial to mesenchymal adhesion proteins, i.e. E-cadherin (E-cad) to N-cadherin (N-cad) (Angadi et al., 2016). However, EMT is a dynamic, transitional process and cells at different stages of the process can co-exist within the same tumor. In addition to tumor heterogeneity, the tumor microenvironment (TME) is often involved in this process and can even modulate cancer progression. Tumors, cancer-associated fibroblasts (CAFs), the immune system and the extracellular matrix (ECM) often interact in reciprocal ways that promote subpopulations to grow, become aggressive and spread (Hanahan and Coussens, 2012; Bissell and Hines, 2011). Although many TME properties have been studied extensively in a variety of tumor types, the influence of TME on OSCC remains relatively understudied, despite its prevalence and, especially, in the context of how TME physical properties regulate OSCC cell behavior.
OSCC is clinically observed as an ulcer with irregular, elevated and indurated margins (Scully and Porter, 2001). As with mammary (Levental et al., 2009), ovarian (McKenzie et al., 2018), head and neck, esophageal, and colorectal cancer (Conklin et al., 2011; Hanley et al., 2016), OSCC tumors present clinically as a region that is stiffer than normal counterpart tissue (Scully and Porter, 2001). As such, stiff matrix in vitro triggers an invasive phenotype in mammary epithelial cells and increases migration in an epithelial ovarian cancer cell line (McKenzie et al., 2018; Wei et al., 2015; Paszek et al., 2005). Similar stiffness sensitivities and tumor stromal changes have been found in 2D cell culture for lung (Tilghman et al., 2010), prostate (Moazzem Hossain et al., 2014) and hepatocellular (Yangben et al., 2013) carcinomas. The ubiquitous nature of stiffness-mediated cell behavior changes begs the question of how cells sense ECM properties, such as stiffness. Mechano-sensing often occurs through a complex series of structures, beginning with focal adhesions (FAs) that directly connect cells to the ECM through integrins and, ultimately, to the cytoskeleton and nucleus (Holle and Engler, 2011). Positive feedback between these structures promotes FA formation and maturation, force generation, migration or invasion, and the expression and translocation of EMT and YAP/TAZ transcription factors to the nucleus (Nardone et al., 2017). These factors often have co-regulators that control localization, such as with TWIST1 whose cytoplasmic partner G3BP2 regulates its translocation to the nucleus and induction of an invasive phenotype (Wei et al., 2015). Yet, all of these signals are transient, as cancer cells often transition back and forth between epithelial and mesenchymal states, raising the question of whether OSCCs have mechanical memory. After being exposed to a stiff niche, mammary epithelial cells migrated faster and showed YAP-dependent increases in actomyosin expression, even when the second niche was softer (Nasrollahi et al., 2017). Studies also often focus on cell–matrix interactions in the absence of cell–cell connections. Recent work both in normal (Sunyer et al., 2016; Xi et al., 2017) and transformed epithelia (Lintz et al., 2017), suggests that cell sheets sense a combination of matrix and cell stiffness to direct migration; but whether OSCC cells respond individually or collectively to stiffness differences, or whether they have a mechanical memory is unclear.
Thus, to better understand stiffness responses in OSCC, we examined stiffness-mediated responses in four OSCC cell lines with a range of epithelial and invasive phenotypes, as well as tumor-recurrence-free survival time of OSCC patients, assessed by collagen organization as a surrogate for stiffness (Conklin et al., 2011; Hanley et al., 2016; Wei et al., 2015). We found that the epithelial phenotype appears plastic when cells conditioned within a stiff niche present EMT-like responses; focal adhesions; moreover, specifically, integrin activation, appears to be crucial in the regulation of this response. At the clinical level, increases in stiffness, as measured by enhanced collagen organization, appears to correlate with advanced disease and shorter recurrence-free survival time. Together, this suggest that the progression of oral cancers, as with other epithelial tumors, is mechanically sensitive.
RESULTS
Increased EMT marker expression and invasion correlate in oral squamous cell carcinomas
Oral cancer cells – which have epithelial origins – exhibit a spectrum of EMT marker expression and the ability to localize those markers to the nucleus. To illustrate this, protein and mRNA expression from four OSCC cell lines were analyzed; SCC9 and SCC25 cell lines had higher N-cadherin (N-cad) to E-cadherin (E-cad) ratios compared to those in Cal27 and FaDu cell lines (Fig. 1A), indicating that SCC cell lines have a more mesenchymal-like phenotype. Accordingly, mRNA analyses of EMT transcription factors, e.g. Zeb1, Zeb2, Snail1, Snail2 and Twist, also showed that SCCs cell lines have higher expression of all EMT markers analyzed (Fig. 1B). Cal27 and SCC25 cells are also morphologically distinct from each other (Fig. 1C). Thus, to correlate EMT marker expression and morphological differences with the level of invasiveness of the cell lines, we carried out organotypic cultures, for which cells were cultivated on top of a fibroblast-embedded collagen matrix for 21 days in an air–liquid interface. Cells with more E-cad than N-cad, e.g. Cal27, did not invade the collagen matrix compared to SCC9 – a line with more N-cad than E-cad, which invades the collagen matrix by day 21 (Fig. 1D). The latter resembles histopathological specimens of patients diagnosed with OSCC, where there is significant stromal invasion (Angadi et al., 2016; Colley et al., 2011). On the basis of these differences, we subsequently divided cell lines according to invasiveness and EMT protein expression, e.g. less invasive with high E-cad to N-cad ratio (InvL/E:NH; Cal27) and highly invasive with low E-cad to N-cad ratio (InvH/E:NL; SCC25).
Fig. 1.
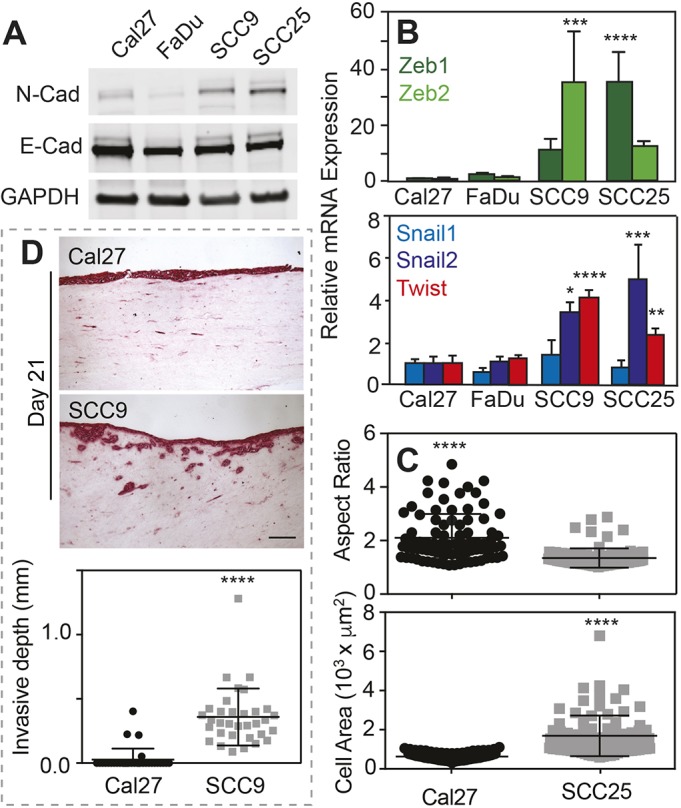
Invasive oral squamous cell carcinoma cell lines have increased N-cadherin and EMT marker expression. (A) Western blot of indicated cell lines (Cal27, FaDu, SCC9, SCC25) and proteins (N-Cad, E-cad, GADPH). (B) Relative mRNA expression of the indicated EMT transcription factors for the indicated cell lines. n=4 samples for each plot. *, **, *** and **** represent P<0.05, 0.01, 0.001 and 0.0001, respectively, for one-way ANOVA and Tukey's multiple comparison test. (C) Plot of aspect ratio (major to minor cell axes) and cell area for the indicated cell lines. ****P<0.001 for two-tailed Welch's t-test. n=86 and 87 for Cal27 and SCC25, respectively, from triplicate experiments. (D) Histological sections (top) of a collagen gel invasion assay stained using PicroSirius Red, of cells cultured at an air-liquid interface. The indicated cell lines were cultured on the collagen gels for 21 days prior to staining. Plot showing the invasion depth (bottom). n=3 samples and ****P<0.001 for two-tailed Welch's t-test. Scale bar is 50 μm.
InvL/E:NH cell migration is initially stiffness insensitive
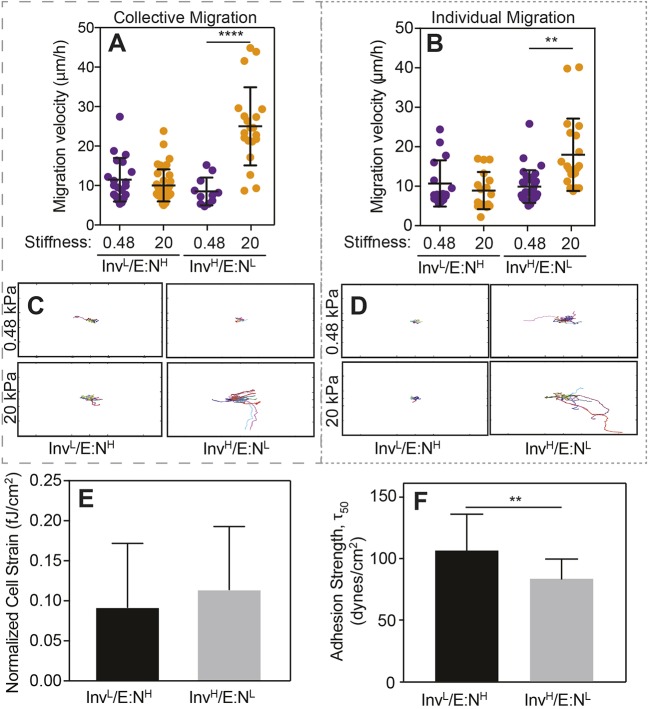
Although well-studied mammary and ovarian cancer cell lines exhibit EMT, and become invasive at increased stiffness (McKenzie et al., 2018; Wei et al., 2015; Paszek et al., 2005), such correlation is not yet clear for oral cancer. Thus, we analyzed the migration profile of InvL/E:NH and InvH/E:NL cells after they had been plated on collagen-coated hydrogels for 12 h with two different stiffness – a soft (0.48 kPa) matrix, close to healthy tongue stiffness (Brown et al., 2015; Cheng et al., 2011), and a stiff (20 kPa) matrix that represents the degree of stiffening and stromal remodeling present in other cancers (McKenzie et al., 2018; Wei et al., 2015; Paszek et al., 2005). We observed that InvL/E:NH cells tended to migrate in cell clusters, especially on the soft substrate, and InvH/E:NL cells tended to migrate individually, which resembles epithelial and mesenchymal cell behavior, respectively (Fig. S1) (Friedl and Alexander, 2011). This behavior also appeared to be independent of substrate stiffness. Although morphology or mode of migration did not differ with respect to stiffness, we found that InvH/E:NL cells had significantly increased collective and individual migration velocity (Fig. 2A,B), and were more processive on stiff substrates relative to InvL/E:NH cells (Fig. 2C,D). Interestingly, InvL/E:NH cells appeared to be stiffness insensitive with respect to migration velocity and directionality, whereas InvH/E:NL cells were fastest on stiff substrates when migrating collectively (P<0.01 between collective and single migration on 20 kPa substrates). In all cases, migration appeared to occur through random walk, based on a lack of difference in either the angle between migration steps (i.e. orientation correlation coefficient; Engler et al., 2004) or the persistence index (i.e. path length divided by total displacement; Fig. S2).
Fig. 2.
InvL/E:NH cell migration is stiffness insensitive. (A,B) Collective (A) and individual (B) cell migration for the indicated cell lines on soft (purple) and stiff (orange) substrates. Each point represents the average velocity of an individual cell. For panel A, n=20, 40, 20 and 20 cells for the groups from triplicate experiments. For panel B, n=16, 17, 31, and 20 cells for the groups from triplicate experiments. **P<0.01 and ****P<0.0001 for Welch's t-test comparisons of velocities as a function of stiffness for each cell line. (C,D) Rose plots of collective (C) and individual (D) cell migration pathways for the indicated cell lines on soft (top) and stiff (bottom) substrates over a 24 h period. n=4 for each plot. Plot size is 1 mm2. (E) Average cell strain normalized to cell area for the indicated cell line. n=42 (InvL/E:NH) or 17 (InvH/E:NL) cells from triplicate experiments. fJ, femtojoules. (F) Average adhesion strength for the indicated cell lines exposed to shear stress that caused 50% of the population to detach, i.e. τ50. n=9 for both groups. **P<0.01 for Welch's t-test.
To determine what underlying functional differences drive these observations, we assessed how strongly cells pulled on their surroundings when migrating and how well cells adhered to their surroundings, by using traction force microscopy (Munevar et al., 2001) and spinning disk assays (Fuhrmann et al., 2017), respectively. We found no difference between the contractile forces between these lines (Fig. 2E). However, we did observe lower adhesion strength for InvH/E:NL cells relative to their InvL/E:NH counterparts (Fig. 2F), consistent with previous reports that metastatic cell lines have lower adhesion strength (Fuhrmann et al., 2017). Together, these data suggest that the lower adhesion strength of InvH/E:NL cells can produce labile adhesions primed to create faster migrating cells. Furthermore, our data further suggest two questions: (1) how stable is each population and (2) are focal adhesion differences driving this phenomenon?
InvL/E:NH cells exhibit increased EMT and migration after prolonged exposure to a stiff niche
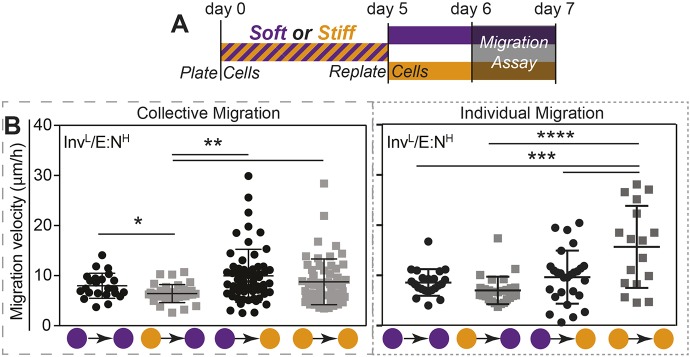
Although InvL/E:NH cells were initially stiffness insensitive, we next asked whether they remain insensitive to stiffness and EMT when cultivated for a prolonged period of time in a stiff niche. After 5 days in a stiff niche, absolute mRNA expressions of five EMT transcription factors were generally higher in InvL/E:NH cells. However, although higher, none were statistically similar to those in InvH/E:NL cells, which appeared stiffness insensitive, albeit at significantly higher expression (Fig. 3A). In addition to marker expression, we assessed E-cad and N-cad protein expression after 5 days, finding that culturing InvL/E:NH cells in a stiff niche for 5 days was sufficient to induce a 2.5-fold increase in N-cad expression, consistent with Twist-mediated activation of N-cadherin expression (Hao et al., 2012). Again, InvH/E:NL cells expressed significant levels of N-cad but did not exhibit a trend with stiffness (Fig. 3B). Since EMT marker expression changed, we next determined whether that change impacted cell migration. After cultivation for 5 days in a soft or stiff niche, InvL/E:NH cells were re-plated onto either soft or stiff niches and migration velocity was analyzed (Fig. 4A). When challenged with combinations of soft and stiff, neither combination exhibited a significant change in migration. However, whereas all groups migrated faster when re-plated on stiff matrix, independently of mode of migration or initial stiffness conditions, single-cell migration for cells initially on stiff matrix was significantly faster compared to cells initially cultured on soft matrix (Fig. 4B). This behavior was not observed for collective migration, as the initial seeding conditions did not affect migration speed after re-plating on stiff gels. This is unlike other epithelial lineages that collectively migrate faster (Sunyer et al., 2016; Xi et al., 2017). Together these data suggest that InvL/E:NH cells can be induced by stiffness, much like mammary epithelial cells (Wei et al., 2015), in order to begin to express EMT markers and exhibit behaviors consistent with a stable mesenchymal state. However, while mammary cells initially cultured on stiff matrix maintain their phenotype independent of their secondary matrix (Nasrollahi et al., 2017), InvL/E:NH cells exhibited a dual phenotype: cells initially cultured on stiff and re-plated on soft matrix did not appear to exhibit memory, whereas those re-plated on stiff matrix did appear to be primed for migration. Furthermore, this behavior is only exhibited when cells migrate in a single, mesenchymal manner.
Fig. 3.
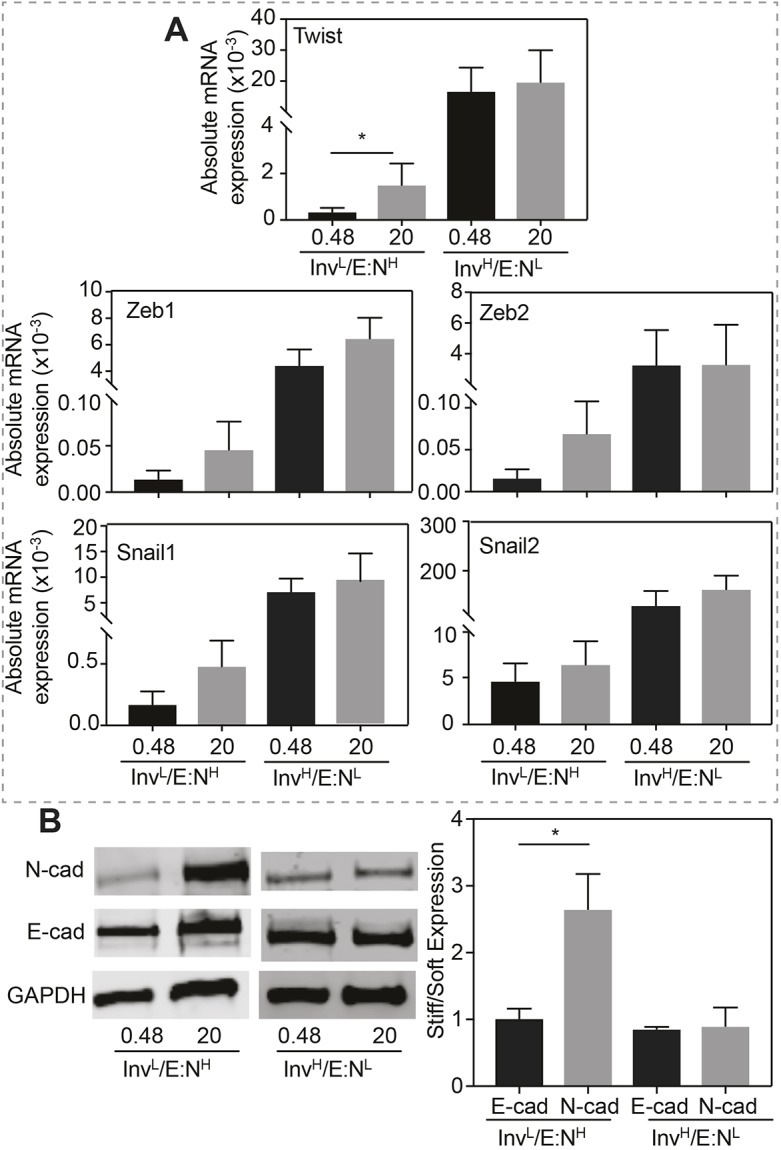
Prolonged exposure to stiff substrates induces InvL/E:NH cells to express EMT markers. (A) Absolute mRNA expression for the indicated transcription factors, normalized to GAPDH, is plotted for the indicated cells cultured on soft or stiff substrates. n=5, 5, 9 and 8 samples for each condition from triplicate experiments. *P<0.05 for Welch's t-test. (B) Western blots (left panel) and average expression ratio (in kPa, right panel) of N-cadherin, E-cadherin and GAPDH for the indicated cell lines. For each protein, the average expression ratio on stiff to soft substrate is plotted. n=4 for each protein. *P<0.05 for Welch's t-test relative to the other stiffness or cadherin.
Fig. 4.
InvL/E:NH cells exhibit ‘memory’ after prolonged exposure to a stiff niche. (A) Schematic of experimental design for cell commitment and migration assay. (B) Collective (left) and individual (right) migration was monitored for InvL/E:NH cells on soft–stiff substrate combinations as indicated (soft=purple; stiff=orange). All data were analyzed using one-way ANOVA with Tukey's multiple comparisons test with *P<0.05, **P<0.01. n=23, 35, 61 and 58 cells for collective migration, and n=22, 27, 27 and 17 cells for individual migration from triplicate experiments.
Adhesion of InvL/E:NH cells is modulated by prolonged exposure to a stiff niche
To understand whether focal adhesion differences drive the gradual conversion of InvL/E:NH cells from epithelial to mesenchymal behaviors, e.g. low to high migration, we assessed the extent to which the substrate can imprint on the focal adhesion assembly of a cell. As previously, InvL/E:NH cells were cultured on soft or stiff substrates, replated, and focal adhesion assembly was assessed by paxillin staining on stiff substrates (Fig. 5A). Individual cells cultured initially on soft substrate – which migrate less than their counterparts continuously cultured on stiff substrate, had less adhesive areas (Fig. 5B). These data suggest that, regarding oral carcinomas, larger adhesions can enable cells to migrate faster, which is unlike their mammary counterparts (Fuhrmann et al., 2017) perhaps due to faster single-cell migration (Fig. 4). To normalize substrate-induced adhesive differences in InvL/E:NH cells (Fig. 2F), we cultured cells on continuously stiff and mixed stiffness substrates in high-cation media (cell-specific DMEM plus 2.5 mM MgCl2, see Materials and Methods), which has previously been shown to activate integrins and modulate adhesion independent of adhesion area (Fuhrmann et al., 2017); a high concentration of cations is also present in tumors relative to stroma (Seltzer et al., 1970, 1970). Whereas cells on continuously stiff substrate migrated faster in media containing normal cation levels (see Materials and Methods) compared with cells on mixed substrates (soft-to-stiff), high-cation media reduced migration differences for InvL/E:NH cells on continuously stiff and mixed substrates (Fig. 5C). These data suggest that InvL/E:NH cell migration after EMT requires larger but labile adhesions to adequately bind the stiffer ECM in the tumor and stroma.
Fig. 5.
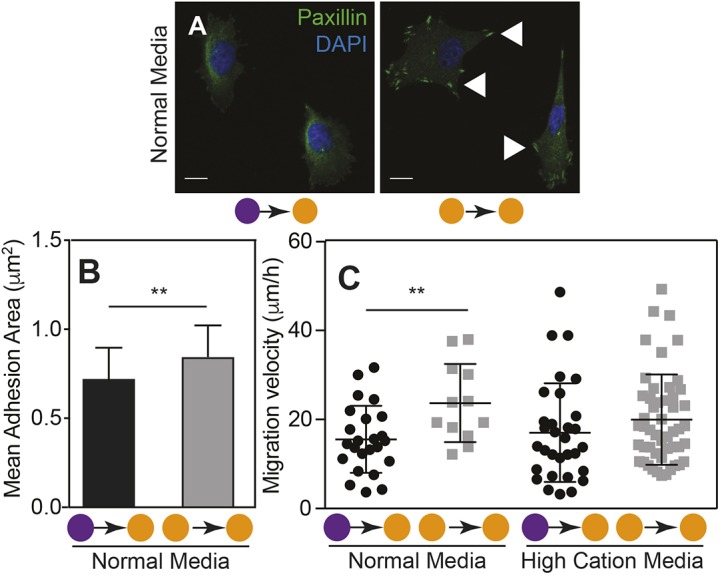
Long-term conditioning in a stiff niche increases the adhesion area in InvL/E:NH cells. (A) Images of InvL/E:NH cells on soft-to-stiff substrate (left) and stiff-to-stiff substrate (right) stained for paxillin (green) and nuclei (blue). Arrowheads indicate assembled focal adhesions. Scale bars: 10 μm. (B) Plot of the mean adhesive area of InvL/E:NH cells cultured in normal medium on soft-to-stiff or stiff-to-stiff substrate (n=39 or 51, respectively). **P<0.01 for Welch's t-test comparisons. (C) Individual cell migration velocity is plotted for cells as outlined in Fig. 4A. Each point represents the average velocity of an individual cell. Cells were cultured on soft-to-stiff or stiff-to-stiff substrate in normal or high-cation medium as indicated. n=24, 12, 31 and 50 cells for each condition. **P<0.01 for Welch's t-test comparisons of velocities as a function of stiffness for each line. Soft=purple; stiff=orange.
Increasing collagen organization predicts poor outcome in OSCC patients
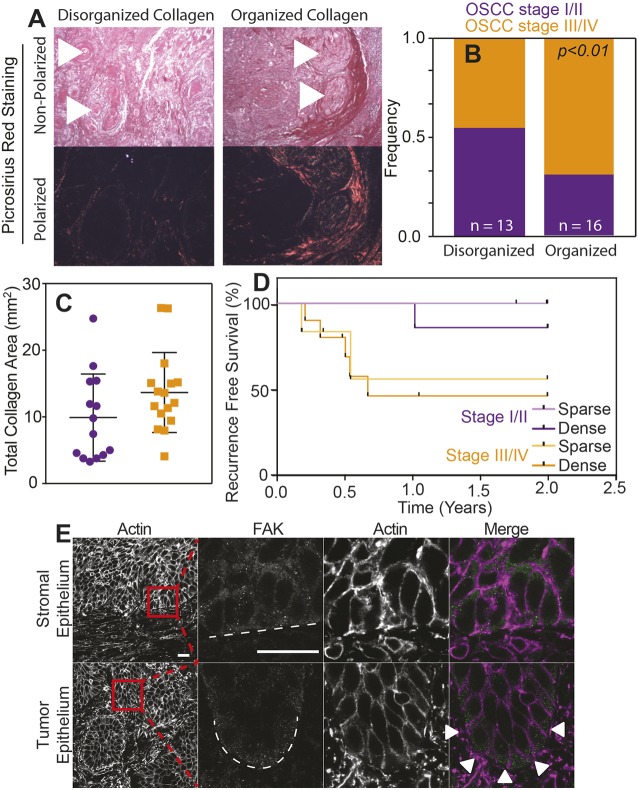
Collagen organization has previously been used as a surrogate for in vivo tumor rigidity (Wei et al., 2015) and correlates with poor outcome regarding a variety of tumors (Conklin et al., 2011; Hanley et al., 2016). Thus, we examined tumor sections of 29 OSCC patients (Table S1) by polarized PicroSirius Red staining to determine whether the margins that surrounded oral carcinomas had aligned collagen (Fig. 6A, arrowheads), which is indicative of stiffer tissue (Acerbi et al., 2015). We found that organized collagen surrounding tumor margins was associated with higher clinical TN stage (Fig. 6B), as well as more collagen present within OSCCs and tumor stroma (Fig. 6C). When using both tumor severity and collagen amount within the tumor to stratify patient outcomes, recurrence within the first 2-year period was most prevalent in patients having a higher TN stage, with organized and densely packed collagen surrounding tumor margins (Fig. 6D), and was below average patient outcomes (Liu et al., 2013). Concordant InvL/E:NH cell behavior on stiff matrix that shows phenotype conversion and enhanced migration agree with these results, and demonstrate that large, mesenchymal-like tumors are supported by highly organized collagen that facilitates stromal invasion. Moreover when TN stage III/IV tumors are stained for intracellular proteins that would respond to more densely packed collagen, we found that an adhesion-associated protein, e.g. focal adhesion kinase (FAK), exhibited asymmetric distribution compared to stromal regions (Fig. 6E), which is also consistent with activated adhesive complexes that result in differential migration of tumor cells in culture (Fig. 5).
Fig. 6.
Higher collagen organization of tumor correlates with poorer outcome for OSCC patients. (A) PicroSirius Red staining of histological sections of tumors with the indicated collagen organization. The same image was taken under polarized and non-polarized (brightfield) light (bottom and top images, respectively). Arrowheads indicate the location of selected keratin pearls. (B) Frequency of clinical TN stages I/II (purple) vs III/IV (orange) for OSCC relative to collagen organization within the tumor. Number of patients in each group were n=13 and n=16. P<0.05 for Welch's t-test comparing collagen organization and tumor stages. (C) Plotted collagen area of PicroSirius Red-stained tumor tissue of clinical TN stages I/II (purple) vs III/IV (orange) observed under polarized light. (D) Survival rate without tumor recurrence in percent of OSCC patients. Data of patients with tumors of the same TN stage but with above (dense) or below (sparse) average total collagen area were pooled. Kaplan–Meier plot of recurrence of recurrence-free survival for patients in the indicated categories on the basis of histological assessment. By using a log-rank (Mantel–Cox) test, P<0.05 was assessed and compared between dense stage III/IV and sparse stage I/II tumor tissue. n=6, 7, 6 and 10 for sparse stage I/II, dense stage I/II, sparse stage III/IV and dense stage III/IV tumor tissue, respectively. (E) Histological staining for actin and focal adhesion kinase (FAK) of representative stomal and OSCC tumor epithelium. The last three panels in each row show the magnification of the boxed area in the first panel. Dashed lines indicate the edge of each indicated feature; arrowheads indicate FAK polarization to the tumor edge. Scale bars: 20 μm.
DISCUSSION
Cancer cells are exceedingly diverse within a tumor, having different phenotypes and, sometimes, even expressing different oncogenes (Navin et al., 2011; Greaves and Maley, 2012). As a consequence, subsets of cancer cells can be exceedingly influenced by the tumor microenvironment, inducing a change in phenotype and migration away from the primary mass. By contrast, stromal cells can be influenced by the tumor, i.e. cancerized, to help remodel the niche (Curtius et al., 2018). What results is a tumor-adjacent stroma composed of many new biological, chemical and physical signals (Quail and Joyce, 2013) that support metastasis. As we described earlier, this dynamic, reciprocal interaction has been well-studied in vitro, specifically for the influence that niche stiffness has on mammary (Levental et al., 2009; Wei et al., 2015; Paszek et al., 2005), ovarian (McKenzie et al., 2018), lung (Tilghman et al., 2010), prostate (Moazzem Hossain et al., 2014) and hepatocellular (Yangben et al., 2013) carcinomas but not for OSCCs. Our data demonstrated that cells comprising different levels of invasiveness respond differently to matrix stiffness. Whereas InvL/E:NH cells remain surprisingly plastic, and able to increase migration and EMT marker expression gradually over time as directly by matrix stiffness, while their counterpart InvH/E:NL cells appear more mesenchymal and adopt migratory phenotypes rapidly when the substrate permits. These observations are consistent with OSCC progression observed in patients, where advanced stages of disease have more organized collagen around keratin pearls, which can trigger a more-invasive phenotype and increased disease recurrence.
Tumor stiffness is correlated with increased EMT and invasion in breast cancer (Wei et al., 2015). As observed clinically for OSCC, lesions have a stiffened margin (Scully and Porter, 2001). Thus, we hypothesized that this exerts influence in cancer progression, although tumor stiffness was not explicitly measured from OSCC patient samples. Consistent with the concept that different tumor subset can exhibit different stiffness responses, we found that two OSCC cell lines comprising different E-cad expression – indicating that they are at different stages of EMT – were initially more (InvH/E:NL) or less (InvL/E:NH) migratory on a stiff 20 kPa substrate. Migration speed is inversely correlated to adhesion strength in mammary cancer cells (Fuhrmann et al., 2017) and, consistent with this, we found that the more migratory InvH/E:NL cells also exhibited lower adhesion strength. Once chronically exposed to stiff substrates, InvL/E:NH cells did acquire a more-mesenchymal state, with increased EMT marker expression and migration speed but increased mean adhesion area (Fuhrmann et al., 2017). This is in contrast to invasive mammary cancer cells, which have smaller, labile adhesions relative to their non-malignant counterparts (Fuhrmann et al., 2017). Artificially enhancing integrin affinity reduced migration speed of stiffness-conditioned InvL/E:NH cells, suggesting that, despite their size, they can still be distributed in a manner labile enough to facilitate higher migration speed. Despite their trends regarding stiffness, these OSCC cell lines in particular exhibit smaller adhesions than other SCC cell lines (Hoshino et al., 2012) and metastatic mammary lines (Fuhrmann et al., 2017), which may still enable adhesion to be labile and migration to be effective. Intracellular connections to the molecular clutch and cytoskeleton proteins (Case and Waterman, 2015; Elosegui-Artola et al., 2018) might also be more dynamic in OSCC cells and help facilitate their invasion. Matrix stiffness can assemble (Levental et al., 2009; Paszek et al., 2005; Provenzano et al., 2009) and polarize them in stiff 3D matrix (Mekhdjian et al., 2017) so, even if InvH/E:NL cells assembled robust adhesions, they may be primed for migration based on their directionality. Interestingly, OSCC patients with a poor prognosis had an increase in FAK expression within the invasive front of tumors (Flores et al., 2018), further suggesting that polarity of the adhesive machinery and not just its amount in a cell contributes to tumor invasion. Thus, it would appear that tumor stiffness modulates EMT and prime cells based on the environment and cell state to trigger an increase in migratory behavior.
Cancer invasion, similar to that of neoplastic cells, requires a reciprocal relationship between cells and ECM that helps to dictate whether cells adopt collective or individual migration modes (Friedl and Alexander, 2011), which each allows migration at varying speeds. In a niche that resembles a basement membrane, malignant epithelia grow collectively, without protrusions; but in stromal gels containing type I collagen, faster single-cell migration is preferred (Nguyen-Ngoc et al., 2012). We found that, whereas invasive OSCCs are faster compared with their non-invasive counterparts that were, initially, substrate insensitive, collective migration of InvH/E:NL cells was faster than single-cell migration. Epithelial sheets can efficiently use durotaxis (Sunyer et al., 2016; Xi et al., 2017), i.e. migration guided by rigidity gradients, but it would appear that for OSCC cells, a collective mode of migration is more effective, as they are more mesenchymal-like. A stiffer niche enhances nuclear translocation of YAP (Taubenberger et al., 2016) and signals (Calvo et al., 2013) that promote focal adhesion assembly (Nardone et al., 2017), but weaker InvH/E:NL cell-matrix adhesion might indicate that invasive OSCC cells migrate together and slower than cells of other tumors, whose preferred mode of migration is as single cells (Nguyen-Ngoc et al., 2012). However, unlike Nguyen-Ngoc and co-workers, we have only explored OSCC migration on type I collagen substrates, but ECM composition directly modulates migration (Ramos Gde et al., 2016) and how processive cells can be (Montenegro et al., 2017). Thus, our observation of faster collective migration could be limited by ECM ligand, which – although type I collagen is the dominant ligand in tumor-adjacent stroma (Pickup et al., 2014) – could nonetheless alter the ratio of cells that migrate individually compared with those migrating collectively.
Stiff tissue observed by collagen organization has already been associated with poor survival in breast, head and neck, esophageal, and colorectal cancer (Conklin et al., 2011; Hanley et al., 2016), so one possible means of assessing the impact of stiffness, migration mode and mechanism on oral carcinomas is to directly examine them. By using PicroSirius Red staining (Drifka et al., 2016), we found that collagen organization correlated with the advanced stages of disease and resulted in early recurrence of the disease. An important caveat about most in vitro systems, including ours, is that it lacks in supporting stromal cells, such as cancer-associated fibroblasts (CAFs). CAFs can modify tumor ECM by increased collagen deposition and alignment that, as a consequence, turns the niche into a stiffer microenvironment (Pankova et al., 2016). With CAFs being present in the indurated tumor margins and capable of remodeling the niche, we suggest that our model, in which InvL/E:NH cells are mechanically conditioned by their niche to undergo EMT and increase their migration, occurs in patients. While these patient observations will benefit from additional studies, it is safe to conclude that InvL/E:NH cells can be mechanically conditioned with a stiff matrix, and that this drives EMT, increases migratory velocity and affects focal adhesion assembly in a manner that, at least partially, reflects the advanced stages of OSCC lesions in vivo.
MATERIALS AND METHODS
Cell culture
OSCC cell lines were a kind gift from Akihiro Sakai, University of California San Diego (UCSD). Mycoplasma testing was performed at the UCSD Stem Cell Core Facility using PCR in November 2017. Cal-27 cells were cultivated in DMEM high glucose (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gemni Bio) and 1% penicillin/streptomycin (Gibco); FaDu, SCC-9 and SCC-25 cells were maintained in DMEM/F12 with 15 mM HEPES (Teknova) supplemented with 10% FBS, 1% penicillin/streptomycin, and 400 ng/ml of hydrocortisone (Sigma). Cells were maintained in incubator at 37°C with 5% CO2. For cell culture in high-cation medium, 2.5 mM of MgCl2 was added to the above-described media.
Real-time PCR
RNA was extracted from cells using Trizol and total RNA quantity and quality was checked using absorbance (A280/A260). cDNA was generated by adding random hexamer primers and Super Script III reverse transcriptase (Thermo) to 2 μg of RNA. Quantitative PCR was performed (45 cycles, 95°C for 15 s followed by 60°C for 1 min) using a 7900HT Fast Real-Time PCR System (Thermo) with the primer sets described below and SYBR Green Supermix (Bio-Rad Laboratories). All quantification cycle (Cq) values were normalized to GAPDH and a fibronectin standard was used to analyze absolute RNA expression. Experiments were performed in biological and technical triplicates using the CFX Connect (Bio-Rad) real-time PCR detection system. Human primer sequences were used as follows: Twist1 (5′-TGCATGCATTCTCAAGAGGT-3′, 5′-CTATGGTTTTGCAGGCCAGT-3′), Snail1 (5′-CTAGCGAGTGGTTCTTCTG-3′, 5′-CTGCTGGAAGGTAAACCTCTG-3′), Snail2 (5′-ATGAGGAATCTGGCTGCTGT-3′, 5′-CAGGAGAAAATGCCTTTGGA-3′), Zeb1 (5′-GCCAATAAGCAAACGATTCTG-3′, 5′-CTTGTCTTTCATCCTGATTTCC-3′), Zeb2 (5′-CAGTCCAGACCAGTATTCCT-3′, 5′-GCAATTCTCCCTGAAATCCT-3′), GAPDH (5′-TCGACAGTCAGCCGCATCTTC-3′, 5′-ACCAAATCCGTTGACTCCGAC-3′).
Western blotting
Cells were lysed using RIPA buffer (50 mM HEPES, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1% Triton, 10% glycerol, 25 mM sodium deoxycholate, 0.1% SDS) supplemented with Roche Complete Protease Inhibitor (Sigma) and PhosSTOP (Sigma). Total protein was quantified with Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Cell lysates (20 µg) were loaded and separated in 4-12% Bis-Tris Gels and MES running buffer (50 mM MES, 50 mM Tris base, 1 mM EDTA, 0.1% w/v SDS). Proteins were transferred to nitrocellulose membranes using the iBlot semi-dry transfer system (Invitrogen). Membranes were blocked with 5% Sea Block blocking buffer (Thermo Fisher Scientific) in Tris-buffered saline with Tween (TBS-T, 150 mM NaCl, 15 mM Tris-HCl, 20 mM Tris base, 0.1% Tween) for 1 h. Membranes were then immunoassayed for E-cadherin (1:1000, Cell Signaling, 24E10), N-cadherin (1:1000, Abcam, ab76011) and GAPDH (1:7500, Abcam, ab8245) overnight at 4°C. After washing three times for 5 min in TBS-T, membranes were incubated with Alexa Fluor 680 donkey anti-mouse (1:10,000, Invitrogen, A10038) and Alexa Fluor 790 donkey anti-rabbit (1:10,000, Invitrogen, A11374) for 2 h. Membranes were washed three times for 5 min in TBS-T before imaging. Images were acquired using the LI-COR Odyssey CLx imaging system detection system and Image Studio Lite was used to analyze and quantify the bands. Values for each protein were normalized to the loading control.
Fabrication of polyacrylamide hydrogels
Polyacrylamide hydrogels (PAAGs) were made on No. 1 12 mm and 25 mm glass coverslips that had been methacrylated by first oxidizing the surface through UV/ozone exposure (BioForce Nanosciences) followed by functionalization with 20 mM 3-(trimethoxysilyl)propyl methacrylate (Sigma-Aldrich, cat # 440159) in ethanol. A polymer solution containing either 3%/0.06% acrylamide/bis-acrylamide (Fisher) for 0.48 kPa hydrogels or 8%/0.264% for 20 kPa hydrogels, 1% v/v of 10% ammonium persulfate (Fisher), and 0.1% v/v of N,N,N′,N′-tetramethylethylenediamine (VWR) was prepared. 15 μl for 12 mm coverslips and 30 μl for 25 mm coverslips of hydrogel solution was sandwiched between a functionalized coverslip and a dichlorodimethylsilane-treated glass slide and polymerized for 15 min. Hydrogels were incubated in 0.2 mg/ml sulfo-SANPAH (Fisher, cat # 22589) in sterile 50 mM HEPES pH 8.5, activated with UV light (wavelength 350 nm, intensity 4 mW/cm2) for 10 min, washed three times in HEPES, and then incubated in 150 μg/ml collagen solution (Corning) overnight at 37°C.
Migration assay
OSCC cells were plated on either 0.48 or 20 kPa PAAGs for 12 h and were then imaged with a Nikon Eclipse Ti-S microscope equipped with a motorized temperature- and CO2-controlled stage. Cells were imaged at 10× in brightfield at multiple positions every 15 min for 20 h. For long-term conditioning in a soft or stiff niche, cells were cultivated for 5 days in either soft or stiff PAAGs, then trypsinized and plated onto soft and stiff hydrogel to analyze migration as mentioned. For experiments using high-cation medium, 2.5 mM of magnesium chloride was added before image acquisition. For analysis of migration parameters, the nucleus of each migratory cell was used as a reference point to track each cell with the ‘Manual Tracking’ plugin on ImageJ. Migration was considered to be in single-cell mode when a cell did not touch any other cell during its migration movement. Migration was considered to be in collective-cell mode when cells migrated in a group of two or more cells.
Traction force microscopy
Traction force microscopy was performed as previously described (Holle et al., 2013). Briefly, polyacrylamide hydrogels (2 kPa, 4%/0.1% acrylamide/bis-acrylamide) were fabricated as described above but with the addition of 2% v/v 568/605 fluorescent 0.2 μm FluoSpheres (ThermoFisher) to the polymer solution. This stiffness was selected due to its optimal deformability resulting in improved resolution of traction forces (Holle et al., 2013). After coating with collagen type I as described above, cells were plated and allowed to attach overnight at 37C and 5% CO2. Images were obtained of single cells followed by the microspheres underneath them with a 60× water immersion objective using a Nikon Eclipse TI-S microscope equipped with a CARV II confocal system (BD Biosciences), motorized stages with a Cool-Snap HQ camera (Photometrics) controlled by Metamorph (Molecular Devices). Cells were released with 10% Triton X-100 and the same microsphere positions were acquired. Bead displacements were determined by using a particle image velocimetry script in MATLAB (The MathWorks, Natick, MA) and normalized to the cell area − codes will be provided by the corresponding author upon request.
Cell-adhesion strength assay
Glass coverslips (25 mm, Fisher Scientific, St Louis, MO) were sonicated in ethanol and pure water before incubation with 5 mg/cm2 collagen type I (rat-tail, Corning) for 60 min at room temperature. Cells were allowed to attach for 24 h at 37°C and 5% CO2 in media containing high-cation levels. Coverslips were then mounted on a custom-built spinning-disk device and dipped into temperature-controlled spinning buffer (37°C). The spinning buffer was phosphate-buffered saline [PBS; without Mg2+ and Ca2+ (Cellgro, Manassas, VA)] supplemented with 4.5 mg/ml dextrose. Once immersed in spinning buffer, coverslips were spun for 5 min at defined angular velocities; cells were fixed with 3.7% formaldehyde immediately after spinning. Quantification of adhesion strength was used according to previous publications (Fuhrmann et al., 2017; Fuhrmann and Engler, 2015). Briefly, coverslips were stained with Hoechst 33342 (Invitrogen, H3570) and imaged at 10× magnification on a Nikon (Melville, NY) Ti-S microscope (∼1000 individual images stitched together with Metamorph 7.6 software and custom macros) and analyzed using a custom-written MATLAB (The MathWorks, Natick, MA) program. Cell densities, as a function of radial position and, subsequently, shear, were stored and combined with other measurements, e.g. those obtained at different RPMs. A sigmoidal decay fit was used to quantify adhesion strength – codes will be provided by the corresponding author upon request.
Immunofluorescence
OSCC cells were directly fixed on the PAAGs (4% paraformaldehyde, 15 min, RT), washed with PBS, permeabilized (0.1% Tween-20, 20 min, RT) and blocked (10% goat serum, 1% BSA, 0.1% Tween-20 and 0.3 M glycine, 30 min, RT). Paxillin (1:250, Abcam, ab32084) primary antibody was incubated overnight at 4°C, washed 3× with blocking buffer, and incubated with Alexa Fluor 488 goat anti-rabbit secondary antibody for 2 h at room temperature (1:500, Invitrogen, A11008). Finally, nuclei were stained with Hoechst 33342 for 15 min at room temperature (1:7500, Invitrogen, H3570), washed with PBS and distilled water. Coverslips were mounted on slides with Fluormount-G (SouthernBiotech) and sealed on the edges using nail polish. Images were obtained by using a LSM780 confocal microscope with a 63× objective and Zeiss software. Images were analyzed using ImageJ analysis software.
OSCC organotypic culture
Rat-tail-derived collagen type I (Corning) was used to produce a 3D matrix with a final concentration of 1.8 mg/ml according to the manufacturer’s instructions. Briefly, rat-tail collagen type I was added on ice to 10× DMEM and the pH adjusted with 0.1 M NaOH to 7. 1×105 primary fibroblasts were embedded in the matrix, as approved by the Ethical Committee of UFRGS (CAE#59124916.6.0000.5327). On top of the matrix, 5×105 of OSCC cells were added. Once cells had reached confluence, the matrix was lifted to create an air–liquid interface. The system was cultivated for 21 days, formalin fixed and paraffin embedded, and then sectioned. Sections were deparafinized and re-hydrated, followed by staining with hematoxylin and eosin. Pictures were taken with an Olympus CX41 microscope coupled to a QColor 5 digital camera (Olympus) at 20× magnification and invasion depth was analyzed using ImageJ software.
Tumors and PicroSirius Red staining
Tumor cells of 29 patients (Table S1) were included in this study, as approved by the Institutional Review Board of the Irmandade da Santa Casa de Misericórdia de Porto Alegre (ISCMPA) under protocol 2.324.217. All patient samples were obtained from the ISCMPA tissue bank and only de-identified patient samples were used. For each sample, only biometric patient information was obtained to create the Kaplan–Meier plot. Formalin-fixed and paraffin embedded 5-μm sections were deparafinized, re-hydrated and followed by staining with PicroSirius Red staining according to manufacturer's protocol (EasyPath). Polarized microscope (BEL photonics) coupled with a camera device (Bioptika) was used at 4× magnification to acquire images. Per patient sample, ten fields were acquired. The scoring rubric (which was defined prior to blind scoring) was defined as ‘organized collagen’ in tumors comprising prominent linearized collagens fibers in the margins of the tumors, or as ‘disorganized collagen’ in tumors comprising either short collagen fibers of a high degree of circularity or low/no PicroSrius Red staining. The total collagen area (mm2) was measured in the polarized images using ImageJ software.
Statistical analysis
All experiments were performed with at least three biological replicates, n indicates the number of technical replicates. Bar graphs represent the mean+ standard deviation (+s.d.). Box and whisker graphs represent the median and extend to the 25% and 75% quartiles. Sample size was determined on the basis of previously published studies in which similar assays were performed (Pickup et al., 2014). We did not exclude any data. No degree of randomization or blind scoring was performed. Statistical differences among two groups were tested using two-tailed Welch's t-test, and differences amongst more than two groups were analyzed with one-way analysis of variance (ANOVA) test followed by Tukey's multiple comparison test. To compare survival distributions of two-patient populations, a log-rank (Mantel-Cox) test was used. Statistical analyses were performed using Graphpad Prism software, with the threshold for significance level set at P<0.05.
Supplementary Material
Acknowledgements
The authors thank Dr Akihiro Sakai of UC San Diego for providing several of the cell lines and reagents used in this work.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: B.F.M., A.J.E., M.L.L.; Methodology: B.F.M., A.K., J.K.P., V.G.Z., A.J.E., M.L.L.; Validation: A.K., J.K.P., V.G.Z., M.D.M., A.J.E., M.L.L.; Formal analysis: B.F.M., A.K., J.K.P.; Investigation: B.F.M., A.K., J.K.P., V.G.Z., M.D.M.; Resources: B.F.M., V.G.Z., A.J.E., M.L.L.; Writing - original draft: B.F.M., A.K., J.K.P., A.J.E., M.L.L.; Visualization: A.K., J.K.P., A.J.E., M.L.L.; Supervision: A.J.E., M.L.L.; Project administration: B.F.M., A.J.E., M.L.L.; Funding acquisition: A.J.E., M.L.L.
Funding
The authors declare no competing interests. Funding for this work was provided by the National Institutes of Health (grant numbers: R01CA206880 to A.J.E., R21CA217735 to A.J.E., T32AR060712 to A.K. and F32HL126406 to J.K.P.) as well as by the National Science Foundation (grant number: 1463689 to A.J.E.) and the Graduate Research Fellowship program (A.K.). Additional fellowship support was provided the Brazilian Federal Agency for Support and Evaluation of Graduate Education award (grant number: 88881.135357/2016-01 to B.F.M.) and the ARCS/Roche Foundation Scholarship Award Program in Life Sciences (A.K.) Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.224360.supplemental
References
- Acerbi I., Cassereau L., Dean I., Shi Q., Au A., Park C., Chen Y. Y., Liphardt J., Hwang E. S. and Weaver V. M. (2015). Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 7, 1120-1134. 10.1039/C5IB00040H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angadi P. V., Patil P. V., Angadi V., Mane D., Shekar S., Hallikerimath S., Kale A. D. and Kardesai S. G. (2016). Immunoexpression of epithelial mesenchymal transition proteins E-cadherin, β-catenin, and N-cadherin in oral squamous cell carcinoma. Int. J. Surg. Pathol. 24, 696-703. 10.1177/1066896916654763 [DOI] [PubMed] [Google Scholar]
- Bissell M. J. and Hines W. C. (2011). Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 17, 320-329. 10.1038/nm.2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. C., Cheng S., McKenzie D. K., Butler J. E., Gandevia S. C. and Bilston L. E. (2015). Tongue stiffness is lower in patients with obstructive sleep apnea during wakefulness compared with matched control subjects. Sleep 38, 537-544. 10.5665/sleep.4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo F., Ege N., Grande-Garcia A., Hooper S., Jenkins R. P., Chaudhry S. I., Harrington K., Williamson P., Moeendarbary E., Charras G. et al. (2013). Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15, 637-646. 10.1038/ncb2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case L. B. and Waterman C. M. (2015). Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17, 955-963. 10.1038/ncb3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Gandevia S. C., Green M., Sinkus R. and Bilston L. E. (2011). Viscoelastic properties of the tongue and soft palate using MR elastography. J. Biomech. 44, 450-454. 10.1016/j.jbiomech.2010.09.027 [DOI] [PubMed] [Google Scholar]
- Colley H. E., Hearnden V., Jones A. V., Weinreb P. H., Violette S. M., MacNeil S., Thornhill M. H. and Murdoch C. (2011). Development of tissue-engineered models of oral dysplasia and early invasive oral squamous cell carcinoma. Br. J. Cancer 105, 1582-1592. 10.1038/bjc.2011.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin M. W., Eickhoff J. C., Riching K. M., Pehlke C. A., Eliceiri K. W., Provenzano P. P., Friedl A. and Keely P. J. (2011). Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178, 1221-1232. 10.1016/j.ajpath.2010.11.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtius K., Wright N. A. and Graham T. A. (2018). An evolutionary perspective on field cancerization. Nat. Rev. Cancer 18, 19-32. 10.1038/nrc.2017.102 [DOI] [PubMed] [Google Scholar]
- Drifka C. R., Loeffler A. G., Mathewson K., Mehta G., Keikhosravi A., Liu Y., Lemancik S., Ricke W. A., Weber S. M., Kao W. J. et al. (2016). Comparison of picrosirius red staining with second harmonic generation imaging for the quantification of clinically relevant collagen fiber features in histopathology samples. J. Histochem. Cytochem. 64, 519-529. 10.1369/0022155416659249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola A., Trepat X. and Roca-Cusachs P. (2018). Control of mechanotransduction by molecular clutch dynamics. Trends Cell Biol. 28, 356-367. 10.1016/j.tcb.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Engler A. J., Griffin M. A., Sen S., Bonnemann C. G., Sweeney H. L. and Discher D. E. (2004). Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J. Cell Biol. 166, 877-887. 10.1083/jcb.200405004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C. C., Wang T. Y., Cheng Y. A., Jiang S. S., Cheng C.-W., Lee A. Y.-L. and Kao T.-Y. (2013). Expression of E-cadherin, Twist, and p53 and their prognostic value in patients with oral squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 139, 1735-1744. 10.1007/s00432-013-1499-9 [DOI] [PubMed] [Google Scholar]
- Flores A. P. C., Dias K. B., Hildebrand L. C., Oliveira M. G., Lamers M. L. and Sant'ana Filho M. (2018). Focal adhesion kinases in head and neck squamous cell carcinoma. J. Oral Pathol. Med. 47, 246-252. 10.1111/jop.12674 [DOI] [PubMed] [Google Scholar]
- Friedl P. and Alexander S. (2011). Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147, 992-1009. 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Fuhrmann A. and Engler A. J. (2015). The cytoskeleton regulates cell attachment strength. Biophys. J. 109, 57-65. 10.1016/j.bpj.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann A., Banisadr A., Beri P., Tlsty T. D. and Engler A. J. (2017). Metastatic state of cancer cells may be indicated by adhesion strength. Biophys. J. 112, 736-745. 10.1016/j.bpj.2016.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M. and Maley C. C. (2012). Clonal evolution in cancer. Nature 481, 306-313. 10.1038/nature10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. and Coussens L. M. (2012). Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 21, 309-322. 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- Hanley C. J., Noble F., Ward M., Bullock M., Drifka C., Mellone M., Manousopoulou A., Johnston H. E., Hayden A., Thirdborough S. et al. (2016). A subset of myofibroblastic cancer-associated fibroblasts regulate collagen fiber elongation, which is prognostic in multiple cancers. Oncotarget 7, 6159-6174. 10.18632/oncotarget.6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L., Ha J. R., Kuzel P., Garcia E. and Persad S. (2012). Cadherin switch from E- to N-cadherin in melanoma progression is regulated by the PI3K/PTEN pathway through Twist and Snail. Br. J. Dermatol. 166, 1184-1197. 10.1111/j.1365-2133.2012.10824.x [DOI] [PubMed] [Google Scholar]
- Holle A. W. and Engler A. J. (2011). More than a feeling: discovering, understanding, and influencing mechanosensing pathways. Curr. Opin. Biotechnol. 22, 648-654. 10.1016/j.copbio.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holle A. W., Tang X., Vijayraghavan D., Vincent L. G., Fuhrmann A., Choi Y. S., Del Álamo J. C. and Engler A. J. (2013). In situ mechanotransduction via vinculin regulates stem cell differentiation. Stem Cells 31, 2467-2477. 10.1002/stem.1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino D., Jourquin J., Emmons S. W., Miller T., Goldgof M., Costello K., Tyson D. R., Brown B., Lu Y., Prasad N. K. et al. (2012). Network analysis of the focal adhesion to invadopodia transition identifies a PI3K-PKCα invasive signaling axis. Sci. Signal. 5, ra66 10.1126/scisignal.2002964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y. H., Syed Zanaruddin S. N., Lau S. H., Ramanathan A., Kallarakkal T. G., Vincent-Chong V. K., Wan Mustafa W. M., Abraham M. T., Abdul Rahman Z. A., Zain R. B. et al. (2015). Co-expression of TWIST1 and ZEB2 in oral squamous cell carcinoma is associated with poor survival. PLoS ONE 10, e0134045 10.1371/journal.pone.0134045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S., Xu J. and Derynck R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178-196. 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans C. R., Snijders P. J. F. and Brakenhoff R. H. (2018). The molecular landscape of head and neck cancer. Nat. Rev. Cancer 18, 269-282. 10.1038/nrc.2018.11 [DOI] [PubMed] [Google Scholar]
- Levental K. R., Yu H. M., Kass L., Lakins J. N., Egeblad M., Erler J. T., Fong S. F. T., Csiszar K., Giaccia A., Weninger W. et al. (2009). Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891-906. 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintz M., Muñoz A. and Reinhart-King C. A. (2017). The mechanics of single cell and collective migration of tumor cells. J. Biomech. Eng. 139 10.1115/1.4035121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.-H., Chen H.-J., Wang P.-C., Chen H.-S. and Chang Y.-L. (2013). Patterns of recurrence and second primary tumors in oral squamous cell carcinoma treated with surgery alone. Kaohsiung J. Med. Sci. 29, 554-559. 10.1016/j.kjms.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Lydiatt W. M., Patel S. G., O'sullivan B., Brandwein M. S., Ridge J. A., Migliacci J. C., Loomis A. M. and Shah J. P. (2017). Head and Neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J. Clin. 67, 122-137. 10.3322/caac.21389 [DOI] [PubMed] [Google Scholar]
- Markopoulos A. K. (2012). Current aspects on oral squamous cell carcinoma. Open Dent. J. 6, 126-130. 10.2174/1874210601206010126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenzie A. J., Hicks S. R., Svec K. V., Naughton H., Edmunds Z. L. and Howe A. K. (2018). The mechanical microenvironment regulates ovarian cancer cell morphology, migration, and spheroid disaggregation. Sci. Rep. 8, 7228 10.1038/s41598-018-25589-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhdjian A. H., Kai F., Rubashkin M. G., Prahl L. S., Przybyla L. M., Mcgregor A. L., Bell E. S., Barnes J. M., Dufort C. C., Ou G. et al. (2017). Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix. Mol. Biol. Cell 28, 1467-1488. 10.1091/mbc.e16-09-0654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazzem Hossain M., Wang X., Bergan R. C. and Jin J.-P. (2014). Diminished expression of h2-calponin in prostate cancer cells promotes cell proliferation, migration and the dependence of cell adhesion on substrate stiffness. FEBS Open Biol. 4, 627-636. 10.1016/j.fob.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro C. F., Casali B. C., Lino R. L. B., Pachane B. C., Santos P. K., Horwitz A. R., Selistre-De-Araujo H. S. and Lamers M. L. (2017). Inhibition of αvβ3 integrin induces loss of cell directionality of oral squamous carcinoma cells (OSCC). PLoS ONE 12, e0176226 10.1371/journal.pone.0176226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munevar S., Wang Y.-L. and Dembo M. (2001). Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys. J. 80, 1744-1757. 10.1016/S0006-3495(01)76145-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardone G., Oliver-De La Cruz J., Vrbsky J., Martini C., Pribyl J., Skládal P., Pešl M., Caluori G., Pagliari S., Martino F. et al. (2017). YAP regulates cell mechanics by controlling focal adhesion assembly. Nat. Commun. 8, 15321 10.1038/ncomms15321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrollahi S., Walter C., Loza A. J., Schimizzi G. V., Longmore G. D. and Pathak A. (2017). Past matrix stiffness primes epithelial cells and regulates their future collective migration through a mechanical memory. Biomaterials 146, 146-155. 10.1016/j.biomaterials.2017.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N., Kendall J., Troge J., Andrews P., Rodgers L., Mcindoo J., Cook K., Stepansky A., Levy D., Esposito D. et al. (2011). Tumour evolution inferred by single-cell sequencing. Nature 472, 90-94. 10.1038/nature09807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc K.-V., Cheung K. J., Brenot A., Shamir E. R., Gray R. S., Hines W. C., Yaswen P., Werb Z. and Ewald A. J. (2012). ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc. Natl. Acad. Sci. USA 109, E2595-E2604. 10.1073/pnas.1212834109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M. A., Huang R. Y.-J., Jackson R. A. and Thiery J. P. (2016). EMT: 2016. Cell 166, 21-45. 10.1016/j.cell.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Pankova D., Chen Y., Terajima M., Schliekelman M. J., Baird B. N., Fahrenholtz M., Sun L., Gill B. J., Vadakkan T. J., Kim M. P. et al. (2016). Cancer-associated fibroblasts induce a collagen cross-link switch in tumor stroma. Mol. Cancer Res. 14, 287-295. 10.1158/1541-7786.MCR-15-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek M. J., Zahir N., Johnson K. R., Lakins J. N., Rozenberg G. I., Gefen A., Reinhart-King C. A., Margulies S. S., Dembo M., Boettiger D. et al. (2005). Tensional homeostasis and the malignant phenotype. Cancer Cell. 8, 241-254. 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Pickup M. W., Mouw J. K. and Weaver V. M. (2014). The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 15, 1243-1253. 10.15252/embr.201439246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano P. P., Inman D. R., Eliceiri K. W. and Keely P. J. (2009). Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 28, 4326-4343. 10.1038/onc.2009.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D. F. and Joyce J. A. (2013). Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423-1437. 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos Gde O., Bernardi L., Lauxen I., Sant'ana Filho M., Horwitz A. R. and Lamers M. L. (2016). Fibronectin modulates cell adhesion and signaling to promote single cell migration of highly invasive oral squamous cell carcinoma. PLoS ONE 11, e0151338 10.1371/journal.pone.0151338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully C. and Porter S. (2001). Oral cancer. West J. Med. 174, 348-351. 10.1136/ewjm.174.5.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltzer M. H., Rosato F. E. and Fletcher M. J. (1970). Serum and tissue calcium in human breast carcinoma. Cancer Res. 30, 615-616. [PubMed] [Google Scholar]
- Smith A., Teknos T. N. and Pan Q. (2013). Epithelial to mesenchymal transition in head and neck squamous cell carcinoma. Oral Oncol. 49, 287-292. 10.1016/j.oraloncology.2012.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyer R., Conte V., Escribano J., Elosegui-Artola A., Labernadie A., Valon L., Navajas D., Garcia-Aznar J. M., Munoz J. J., Roca-Cusachs P. et al. (2016). Collective cell durotaxis emerges from long-range intercellular force transmission. Science 353, 1157-1161. 10.1126/science.aaf7119 [DOI] [PubMed] [Google Scholar]
- Taubenberger A. V., Bray L. J., Haller B., Shaposhnykov A., Binner M., Freudenberg U., Guck J. and Werner C. (2016). 3D extracellular matrix interactions modulate tumour cell growth, invasion and angiogenesis in engineered tumour microenvironments. Acta Biomater. 36, 73-85. 10.1016/j.actbio.2016.03.017 [DOI] [PubMed] [Google Scholar]
- Tilghman R. W., Cowan C. R., Mih J. D., Koryakina Y., Gioeli D., Slack-Davis J. K., Blackman B. R., Tschumperlin D. J. and Parsons J. T. (2010). Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS ONE 5, e12905 10.1371/journal.pone.0012905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S. C., Fattet L., Tsai J. H., Guo Y., Pai V. H., Majeski H. E., Chen A. C., Sah R. L., Taylor S. S., Engler A. J. et al. (2015). Matrix stiffness drives epithelial mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol. 17, U678-U306. 10.1038/ncb3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W., Sonam S., Beng Saw T., Ladoux B. and Teck Lim C. (2017). Emergent patterns of collective cell migration under tubular confinement. Nat. Commun. 8, 1517 10.1038/s41467-017-01390-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yangben Y., Wang H., Zhong L., Chiang M. Y., Tan Q., Singh G. K., Li S., Yang L. et al. (2013). Relative rigidity of cell-substrate effects on hepatic and hepatocellular carcinoma cell migration. J. Biomater. Sci. Polym. Ed. 24, 148-157. 10.1163/156856212X627856 [DOI] [PubMed] [Google Scholar]
- Yao X., Sun S., Zhou X., Zhang Q., Guo W. and Zhang L. (2017). Clinicopathological significance of ZEB-1 and E-cadherin proteins in patients with oral cavity squamous cell carcinoma. Onco Targets Ther. 10, 781-790. 10.2147/OTT.S111920 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.