Abstract
Studies of laboratory strains of Saccharomyces cerevisiae have uncovered signaling pathways involved in mating, including information-processing strategies to optimize decisions to mate or to bud. However, lab strains are heterothallic (unable to self-mate), while wild yeast are homothallic. And while mating of lab strains is studied using cycling haploid cells, mating of wild yeast is thought to involve germinating spores. Thus, it was unclear whether lab strategies would be appropriate in the wild. Here, we have investigated the behavior of several yeast strains derived from wild isolates. Following germination, these strains displayed large differences in their propensity to mate or to enter the cell cycle. The variable interest in sex following germination was correlated with differences in pheromone production, which were due to both cis- and trans-acting factors. Our findings suggest that yeast spores germinating in the wild may often enter the cell cycle and form microcolonies before engaging in mating.
INTRODUCTION
The molecular and genetic tractability of the budding yeast, Saccharomyces cerevisiae, has made it a premier model organism for the study of many aspects of biology (Botstein et al., 1997; Botstein and Fink, 2011). Among these, the yeast mating pathway has yielded paradigm-setting discoveries concerning signal transduction, pheromone biogenesis, regulation of gene expression, and the cell biology of chemotropism and cell–cell fusion (Arkowitz, 2009; Michaelis and Barrowman, 2012; Merlini et al., 2013; Atay and Skotheim, 2017). While molecular mechanisms have been well studied in the laboratory, surprisingly little is known about S. cerevisiae mating outside the lab. Recent work has emphasized the importance of studying the life history of S. cerevisiae as well as other model organisms in order to properly interpret laboratory findings (Knop, 2006; Boynton and Greig, 2014; Liti, 2015). Here we consider ways that mating may differ in the lab and in the wild.
Lab strains of yeast can be grown as diploids, or they can be maintained as haploids of either mating type (MATa or MATα). In the lab, mating assays are conducted under “orgy” conditions (Hartwell, 1973) in which large numbers of a and α cells first encounter each other when they are abruptly mixed together by the investigator. Haploids secrete small peptide pheromones, a-factor and α-factor, which trigger a number of changes to prepare cells for mating (Alvaro and Thorner, 2016). Successful mating involves arrest in the G1 phase of the cell cycle, polarization of growth toward the mating partner, and expression of numerous genes that enhance cell–cell adhesion and eventual fusion of the cells and nuclei to form a diploid zygote.
Successful mating is not guaranteed: for example, the nonmotile haploid cells may be too far apart, or another partner may mate with the intended target. Recent studies have revealed a sophisticated decision-making system that appears optimized for mating success. Cells in liquid media can optimize their decision to mate or to proliferate by detecting the ratio of opposite-sex partners to same-sex competitors (Banderas et al., 2016). Cells on solid media detecting suboptimal pheromone levels undergo a “chemotropic growth” program in which they arrest transiently and grow toward the mating partner, but then reenter the cell cycle and form a bud in the direction of growth (Erdman and Snyder, 2001; Hao et al., 2008). This response places the resulting daughter cell closer to the pheromone source, presumably increasing the chances that it will mate. In the following cell cycle, the cells retain and exploit a memory of the fact that they had previously responded to the pheromone: the original “mother” cell no longer arrests in response to the same low levels of pheromone (Caudron and Barral, 2013), while the daughter cell is more likely to arrest in response to low pheromone levels (Doncic et al., 2015). Thus, cells with better chances of mating are primed to mate, while cells unlikely to mate invest their energies more productively in vegetative growth.
These decision-making processes were uncovered using lab strains, and the behaviors were interpreted to optimize outcomes under specific lab mating conditions. However, lab strains differ from wild strains in an important respect: the ability to switch mating type. After producing its first daughter, a haploid mother cell switches mating type in the next cell cycle, while its daughter does not (Haber, 2012). This means that the next cell cycle will yield cells of both mating types in close proximity, providing an opportunity to mate in a process termed “haplo-selfing” (Figure 1A). Owing to the presence of mating-type switching, wild yeast strains do not proliferate stably as haploids. Almost all wild S. cerevisiae isolates (environmental and clinical) are diploid.
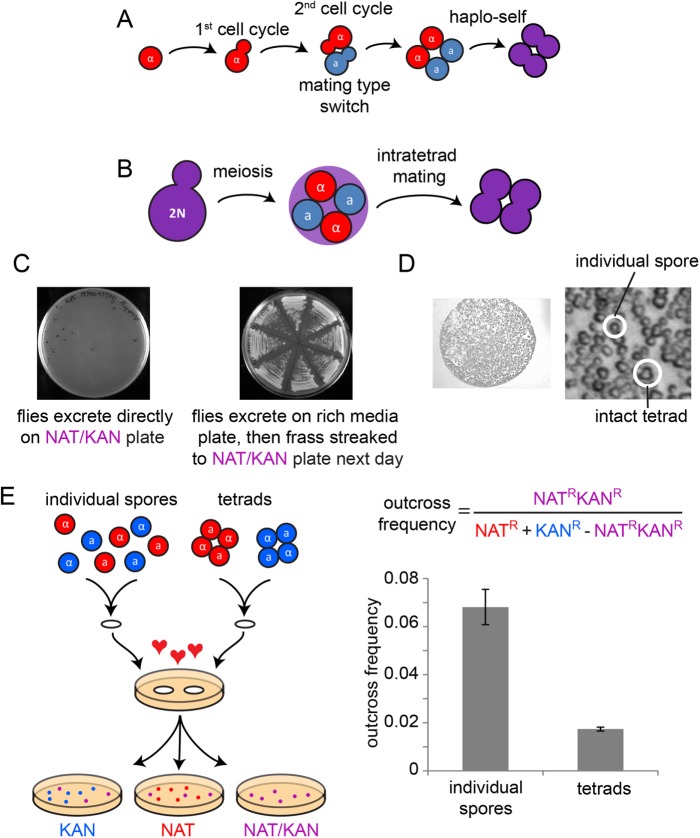
FIGURE 1:
Yeast mating scenarios. (A) Cartoon depiction of mating type switching allowing haplo-selfing. (B) Cartoon depiction of intratetrad mating. (C) Left: flies were fed tetrads that carried either NAT or KAN resistance markers (DLY19740 and DLY19741), and then allowed to excrete on a NAT/KAN double drug plate for 1 h. Excretion spots were marked on the cover of the plate, but showed no growth after 2 d. Right: after feeding, flies were allowed to excrete on a plate without drugs for 1 h. The next day, excretion spots showed yeast growth that was then streaked onto a NAT/KAN double drug plate. (D) Excretion spots show a mix of intact tetrads and isolated spores. (E) Either individual spores or intact tetrads with drug resistance markers (indicated by blue or red) were placed on filters where cells germinated and mated overnight. Then cells were recovered from the filters and plated to identify the rate of outcrossing. Separated spores showed a higher outcross frequency than intact tetrads (mean ± SEM of three plates, t test, p < 0.006).
The circumstances under which mating occurs in the wild are not well understood. Unfavorable nutritional conditions initiate a program of meiosis and sporulation, whereby a diploid cell generates a tetrad with four haploid spores in an ascus (Neiman, 2011). Spores are metabolically dormant and have tough outer cell walls; they can survive harsh conditions and prolonged starvation. Upon exposure to fermentable sugars, spores germinate, awaken metabolic pathways, swell, and break the outer cell wall (Herman and Rine, 1997; Joseph-Strauss et al., 2007). If a partner of the opposite mating type is available (e.g., a sibling spore in a tetrad), germinating spores can mate to regenerate a diploid (Taxis et al., 2005; Figure 1B). This “intratetrad mating” would differ from that examined in lab-based mating assays because the germinating spore has a physiology different from that of a haploid cell grown for many generations in rich media, and because there would be only one or two available partners to choose from within the tetrad.
If no suitable partner were present when a spore germinated, the haploid cell would enter the cell cycle, form a bud, and then switch mating type in the next cycle. Even if no mating partner were close by, mating-type switching guarantees that a partner will soon become available immediately adjacent to the cell. In that respect, it is unclear whether the decision-making strategies uncovered for lab strains would make sense for wild strains of yeast.
To better understand the mating behavior of wild yeast, we examined the events following germination of spores (either in tetrads or on their own) from various wild and lab strains. Depending on the strain, the frequency of intratetrad mating varied widely. For several strains, germinating spores and their progeny chose to bud instead of mate with a nearby potential partner, generating microcolonies with haploid cells of both sexes (due to mating-type switching). Mating subsequently took place in the microcolony context. Pheromone production upon germination was variable between strains, probably contributing to the variable interest in mating. Our findings suggest that mating in the wild may often occur in the context of mixed mating-type microcolonies, a context similar to that of mating in the lab. Our findings have implications for decision making and outbreeding.
RESULTS
Mating of germinating cells in insect frass
In the wild, insects are thought to provide dispersal vectors for yeast (Gilbert, 1980; Reuter et al., 2007). Insects carry yeast cells on their legs, and insects eat yeast cells. In a remarkable recent study, diploid yeast cells were fed to wasps that were then induced to hibernate. Over the ensuing weeks, the yeast underwent meiosis, sporulation, germination, and mating, all in the digestive tract of the hibernating wasp (Stefanini et al., 2016), providing one potential wild yeast mating scenario. Although diploid yeast cells would likely be digested following ingestion by active (nonhibernating) insects, yeast spores were shown to survive passage through the digestive tracts of fruit flies (Reuter et al., 2007; Coluccio et al., 2008). Flies were also shown to increase the frequency of yeast outbreeding (Reuter et al., 2007), suggesting that spores that survive digestion might subsequently germinate and mate in fly frass.
We fed Drosophila melanogaster flies tetrads from two diploid strains of yeast, each homozygous for a different drug resistance marker. When the flies excreted directly onto solid media containing both drugs, no cells grew (Figure 1C), indicating that no detectable mating took place within the gut of the fly. However, if the flies excreted on solid media lacking drugs, and the frass was streaked onto media containing both drugs the next day, many outbred colonies grew. This supports the idea that wild yeast might mate following spore germination in fly frass.
Although the flies were fed tetrads, digestive enzymes within the fly gut have been shown to separate sibling spores (Reuter et al., 2007). Imaging the fly frass, we found a mix of intact tetrads and isolated spores (Figure 1D). It was suggested, based on indirect evidence, that isolated spores would be more likely than spores in tetrads to engage in outbreeding (Reuter et al., 2007). Consistent with that hypothesis, we found using a quantitative mating assay that nonsibling mating was more common when starting with isolated spores than when starting with tetrads (Figure 1E). These findings support the idea that in the wild, yeast spores or tetrads might be deposited in a new and nutrient-rich environment following excretion by flies, and that mating occurs following spore germination in fly frass. However, it was not clear whether the germinating spores were the cells that actually mated, or whether mating followed one or more haploid vegetative cycles.
Mating behavior of germinating spores in tetrads
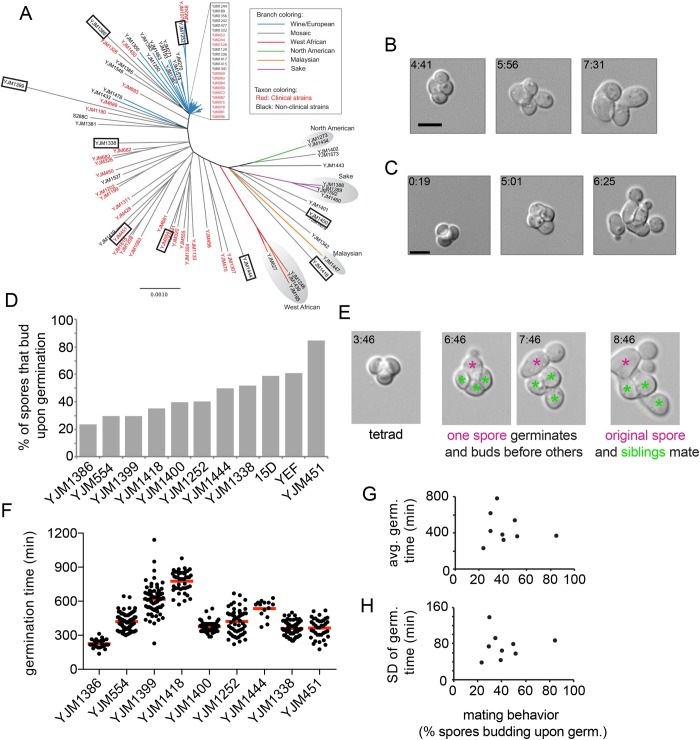
To study the mating behavior of wild yeast strains, we selected a set of nine genetically diverse strains from the 100-genomes collection (Strope et al., 2015; Figure 2A), derived from isolates collected from different environments (Supplemental Table 1). These homothallic strains are homozygous diploids derived by mating-type switching and haplo-selfing. Diploids were incubated in potassium acetate media to induce sporulation, and tetrads were placed on microscope slabs with rich media and imaged by time-lapse microscopy to observe behavior following germination. Upon germination in a tetrad, spores either mated with a sibling (Figure 2B) or entered the cell cycle and formed a bud (Figure 2C). Under these conditions, many germinating spores entered the cell cycle even if a suitable mating partner was present. Quantification of the percentage of spores that mated or budded following germination revealed that even in the wild strain with the highest propensity to mate, around 20% of spores entered the cell cycle rather than mate (Figure 2D). The mating behavior was variable between wild strains, but intratetrad mating was generally much lower than in a previously described lab strain (Taxis et al., 2005). This does not appear to represent a general difference between wild and lab strains, because two S288C-derived lab strains (labeled 15D and YEF) showed lower mating propensities than most of the wild strains (Figure 2D).
FIGURE 2:
Intratetrad mating behavior following germination. (A) Genetically diverse wild strains used in this study. Image adapted from Strope et al. (2015). (B) Germinating tetrad spores from YJM1418 demonstrating intratetrad mating where all four spores mate (time after plating on rich media indicated in h:min). (C) Germinating tetrad spores from YJM1338 demonstrating all four spores entering cell cycle and producing buds rather than mating. (D) Quantification of mating behavior upon germination (n > 36 spores per strain). (E) Germinating tetrad spores from YJM1418 demonstrating one spore germinating and producing a bud prior to the germination of other spores in the same tetrad. Fourth panel shows the original early germinator mating with an intratetrad partner in its second cell cycle, as well as the other two spores mating. (F) Tetrad spores were separated from one another and then placed on an agarose slab containing glucose. Germination time was defined as the time between exposure to glucose and the first time point with a visible bud (n > 14 for each strain). (G, H) Relation between mating behavior from B and average germination time (G) or interspore variability in germination time (H) (assessed using the SD). Scale bar, 5 μm.
One possible explanation for the failure of sibling spores to mate following germination is a difference in the germination times of the spores. If one spore germinated significantly before its potential mating partners, the first spore would have no available partners until after it had undergone one or more cell cycles, and perhaps haplo-selfing after mating-type switching (Figure 2E). To assess germination timing in the cell populations, we digested tetrads to yield single spores and imaged them. Germination times were indeed variable, both between spores from the same strain and between strains (Figure 2F). However, there was no obvious correlation between intratetrad mating and germination time (either average time or variability between spores: Figure 2, G and H). Thus, while asynchrony in germination may lower the efficiency of intratetrad mating, it does not appear to be a dominant factor in explaining the different rates of intratetrad mating between strains.
Mating behavior of isolated germinating spores
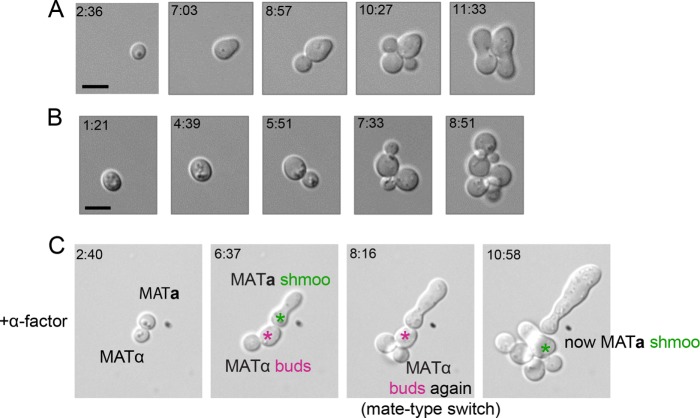
As individual spores might be more prevalent than intact tetrads in fly frass, we next separated spores from each other and imaged the events following germination. With no partner nearby, a germinating spore would be expected to haplo-self (Figure 1A). Indeed, some spores showed this behavior (Figure 3A), but we also observed formation of microcolonies that deferred mating (Figure 3B). These findings are consistent with the deferred mating observed with tetrads.
FIGURE 3:
Isolated spore mating behavior following germination. (A) Germinating spore from YJM1399 demonstrating haplo-selfing. (B) Germinating spore from YJM451 demonstrating formation of a microcolony through repeated budding. (C) Two isolated spores from YJM1399 germinating in the presence of 2 μM α-factor. The top spore arrests and forms a shmoo, and the bottom spore enters the cell cycle twice and then arrests and forms a shmoo, indicating that it has undergone a mating-type switch. Scale bar, 5 μm.
In principle, deferred mating might result from inefficient mating-type switching, so that opposite mating-type cells do not coexist until larger microcolonies have formed. To detect mating-type switching from MATα to MATa, we germinated isolated spores in the presence of saturating α-factor. Some spores, presumably MATa, never entered the cell cycle and instead formed long mating projections. Other germinating spores, presumably MATα, entered the cell cycle and budded. After two budding cycles, these cells arrested and formed projections, indicating that they had switched mating type (Figure 3C). This suggests that mating type switching occurs as predicted following spore germination, so that germinating spores generate microcolonies with cells of both mating types. Thus, the delayed mating we observed is probably due to a lack of interest in mating upon germination. We conclude that interest in sex following germination is a variable trait in wild yeast and that many wild mating events are likely to involve cycling haploid cells rather than germinating spores.
Pheromone sensitivity and production in germinating spores
Potential reasons for a difference among wild yeast strains in the propensity to mate include differences in either the production of pheromones or the reception of pheromones derived from a potential mating partner. For the following set of experiments, we initially focused on strains YJM451 (low propensity to mate; clinical isolate) and YJM1399 (high propensity to mate; cherry tree isolate).
To assess sensitivity to pheromone, spores from each strain were germinated on slabs containing different concentrations of exogenous α-factor. At 2 μM α-factor, 50% of spores (presumably the MATa spores) from both strains germinated, arrested, and formed shmoos (Figure 4A). At 100 nM α-factor, a mixed response was observed, with slightly under half of the germinating spores elongating (presumably due to a transient G1 arrest) before budding (Figure 4B). For YJM451, 42% of spores elongated (n = 265), while for YJM1399, 47% of spores elongated (n = 244). For the spores that elongated, we quantified the time from germination (the first noticeable expansion of the spore) to budding, reflecting the duration of the G1 arrest. YJM451 cells arrested for 201 ± 41 min (mean ± SD), while YJM1399 cells arrested for 283 ± 67 min, indicating a modest but statistically significant (p < 0.001) difference in mean pheromone sensitivity that could contribute to the difference in mating behavior.
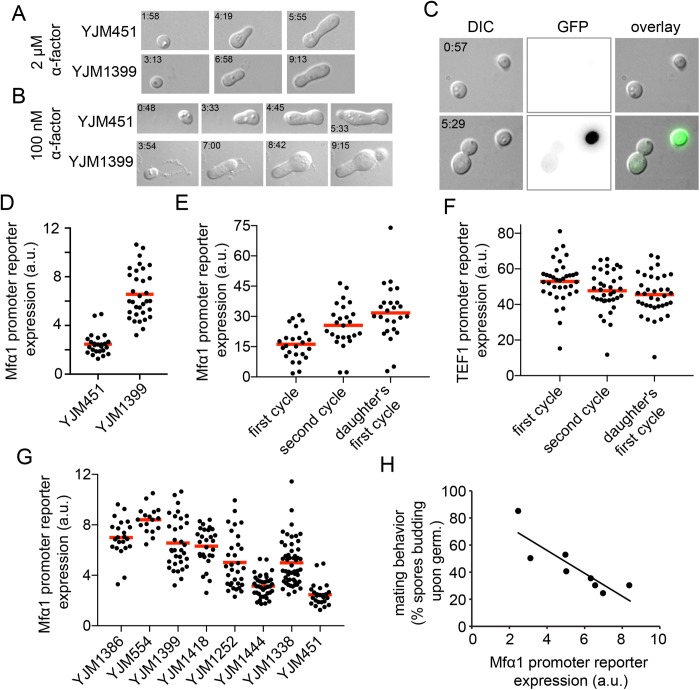
FIGURE 4:
Pheromone sensitivity and pheromone production in germinating spores. Spores of strains YJM451 and YJM1399 were isolated from tetrads and germinated in the presence of 2 μM α-factor (A) or 100 nM α-factor (B). (C) Two isolated spores from DLY20288. The top-right spore (presumed MATα) shows expression of the Mfα1 reporter following germination, whereas the bottom-left spore (presumed MATa) does not. (D) Spores from DLY20922 and DLY21020 (containing the Mfα1 reporter integrated into YJM451 and YJM1399) were isolated from tetrads and imaged during germination. Expression of the Mfα1 reporter was determined just before bud emergence (see Materials and Methods). (E) Spores from DLY22729 (a lab strain in the YEF background) carrying the Mfα1 reporter were treated as in D, and expression was determined just before bud emergence during the first cell cycle, the second cell cycle, and the first cell cycle of the daughter. The second cell cycle and the daughter signals were each significantly higher than the first cell cycle (t test, p < 0.002). (F) Spores from DLY18930 carrying the TEF1 reporter were treated as in E. (G) Spores from the indicated wild strains carrying the Mfα1 reporter were treated as in D (note that data in D are also included in G). (H) Correlation between the percentage of spores that bud following germination (from Figure 2D) and the averaged Mfα1 reporter expression (from G; Pearson’s correlation, R2 = 0.74, p < 0.01).
To assess production of pheromone, we integrated a fluorescent reporter at the MFα1 locus (replacing the ORF for pheromone production). As expected, ∼50% of germinating spores (presumably MATα) expressed the reporter (Figure 4C), and we quantified the reporter intensity following germination, 8 min before detection of the first bud. Reporter expression was significantly higher for strain YJM1399 than for strain YJM451 (Figure 4D). Thus, the higher propensity of strain YJM1399 to mate may be due, at least in part, to the higher transcription rate of MFα1.
Interestingly, we found that in our lab strain, expression of the MFα1 reporter was higher in the second than in the first cell cycle following germination (Figure 4E). Moreover, daughter cells made by the germinated mother expressed higher levels of the reporter in their first cell cycles than the germinating mothers had in their first cycles (Figure 4E). In all cases, the reporter expression level was measured at the time point preceding bud emergence. In contrast to MFα1, expression from a control TEF1 promoter was similar in the first and second cell cycles following germination, and in daughter cells (Figure 4F). These findings suggest that pheromone production ramps up over the first two (and perhaps more) cycles following germination. If the magnitude and timing of such a ramp-up were variable between strains, that might lead to the observed variability in propensity to mate immediately following germination.
To assess the degree to which mating propensity is correlated with MFα1 expression, we integrated the reporter at the MFα1 locus for the entire set of strains. MFα1 expression showed significant variability between strains (Figure 4G) and was well correlated with intratetrad mating propensity (R2 = 0.74, p < 0.01; Figure 4H). This correlation suggests that variability in the amount of pheromone produced at the time of germination may explain some of the variability in the mating behavior of wild strains.
Basis for variability in pheromone production in wild strains
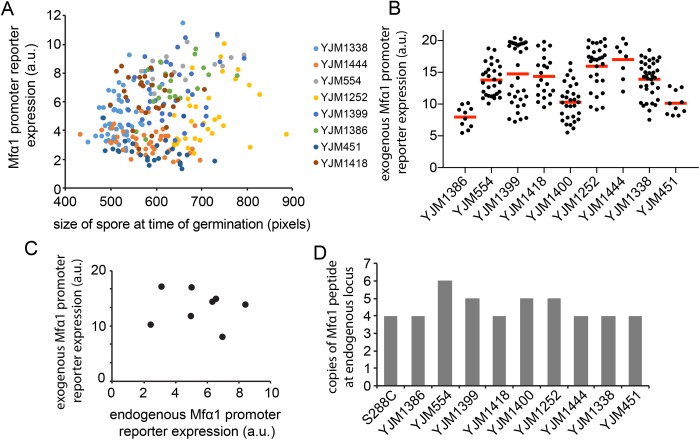
A potential explanation for variable pheromone production could be variability in the synthetic capacity of germinating spores. In particular, there was considerable variation in the size of the spores, and we speculated that larger spores might produce more pheromone. However, we did not detect any correlation between cell size and MFα1 reporter expression, either within or between strains (Figure 5A). Thus, it seems likely that variable MFα1 expression is due to cis- or trans-acting factors affecting the promoter. To test whether trans-acting factors were the dominant contributor to the variable expression, we integrated a reporter from a lab strain driven by the MFα1 promoter into each wild strain. The different wild strains exhibited differences in reporter expression at the time of germination (Figure 5B), indicating that trans-acting factors vary between the strains. Surprisingly, comparison of MFα1 expression from the trans gene and from the endogenous locus revealed little correlation (Figure 5C), suggesting that cis-acting alterations also play a role in MFα1 expression. Examination of the genome sequences revealed several polymorphisms in the MFα1 promoter (Supplemental Figure S1), consistent with that possibility.
FIGURE 5:
Basis for differences in pheromone production between strains. (A) Relation between spore size and Mfα1 reporter expression in germinating spores. Each dot is one cell, color-coded by strain. (B) Mfα1 reporter driven by an exogenous lab strain Mfα1 promoter shows variable expression in different strains. (C) Relation between average endogenous Mfα1 reporter expression and average exogenous Mfα1 reporter expression. (D) Copy number of Mfα1 peptides at the endogenous locus of the different wild strains (Strope et al., 2015).
We note that pheromone production depends on more than just MFα1 transcription. There are two genes encoding α-factor (MFα1 and MFα2), and pheromone production involves proteolytic processing of the primary protein product, which encodes four copies of the pheromone peptide in lab strains. We found that the number of pheromone peptide copies in MFα1 varied (up to six copies) among the wild strains we examined (Figure 5D; Strope et al., 2015), presumably contributing to variation in pheromone secretion. In sum, these data suggest that pheromone expression is variable in wild strains, due to both cis- and trans-acting differences between strains.
DISCUSSION
The yeast mating pathway is one of the best-understood information-processing systems in eukaryotic biology. Recent work has highlighted the decision-making capabilities of the pheromone response pathway, revealing behaviors that have been interpreted as optimizing the mating potential of the cells under imagined scenarios motivated by consideration of lab strains that are heterothallic (incapable of mating-type switching and therefore haplo-selfing; Erdman and Snyder, 2001; Paliwal et al., 2007; Hao et al., 2008; Caudron and Barral, 2013; Doncic et al., 2015; Banderas et al., 2016). However, most yeasts in the wild are homothallic, so mating scenarios in the wild may differ significantly. Thus, it is important to understand the likely situations that the information-processing capabilities of this pathway were evolved to accommodate. Our primary finding is that many yeast strains derived from wild isolates show a low propensity to mate upon germination. This behavior has significant implications, as detailed below.
One consequence is that mating in the wild may often occur in the context of mixed–mating type microcolonies rather than between germinating spores in a tetrad. That scenario is similar to lab mating protocols, supporting conclusions about yeast information-processing systems drawn from lab studies.
A second consequence of the delayed interest in sex is that germinating spores would undergo a few cell cycles as haploids before mating. This provides an opportunity for natural selection to purge deleterious mutations and enrich for beneficial mutations.
Another consequence is to reduce the reproductive isolation that might arise as a result of asynchronous spore germination. Our work suggests that in many wild strains, there is considerable spore-to-spore variability in germination time (e.g., strain YJM1399 in Figure 2F). Differences in germination time have been proposed to promote reproductive isolation, leading to speciation (Murphy and Zeyl, 2012). However, delayed propensity to mate upon germination means that mating would often occur between haploid cells in microcolonies, rather than between germinating spores themselves. That would allow opportunities to mate between progeny of spores that germinated at different times.
A final consequence concerns opportunities for outbreeding. The high levels of heterozygosity observed in some wild diploid yeasts (Liti et al., 2009; Muller and McCusker, 2009; Magwene et al., 2011; Kelly et al., 2012; Strope et al., 2015) suggest that outbreeding occurs with appreciable frequency, particularly in human-associated environments. Delayed interest in sex could lead to a greater frequency of outbreeding. If germinating spores were the mating entity, then unrelated spores would have to find themselves in very close proximity (and germinate synchronously) in order to mate. However, if a haploid microcolony were formed by one or both spores, cells at the periphery of each microcolony would have the opportunity to mate to genetically different partners, yielding outbred diploids.
MATERIALS AND METHODS
Yeast strains and plasmids
Standard molecular genetics procedures were used for strain construction. Yeast strains, plasmids, and construction details are listed in the Supplement. Wild yeast strains were originally isolated and characterized in Strope et al. (2015).
Sporulation conditions and spore isolation
Prior to sporulation, yeast was grown to near saturation in liquid YEPD-rich media at 30°C. Approximately 8 ml of saturated culture was washed briefly with water and resuspended in 8 ml of 2% KAC media (2% potassium acetate supplemented with 0.004% adenine, 0.003% histidine, 0.004% uracil, 0.008% leucine, and 0.003% tryptophan). Cultures were kept at 30°C for 10 d and then moved to 4°C.
To separate spores from tetrads, 1 ml of sporulated cells was washed with water, resuspended in 20–30 μl of lyticase solution (2.72% lyticase powder, 1 M sorbitol, 100 mM PIPES, pH 6.5), and incubated for 20 min at room temperature. Spores were then resuspended in 1 ml of water and sonicated at 50% amplitude for ∼20 s. Samples were heated at 55°C for 30 min to kill any remaining diploid cells and then stored at 4°C.
Feeding yeast to flies
Sporulated yeast cultures (1 ml) were washed with water and resuspended in the residual water. A sample of this paste (5 μl) was pipetted on a 30-mm grape plate, and 50 w1118 flies were then incubated in the plate for 6 h. Fifteen flies were then transferred to a 10-cm YEPD plate or a YEPD plate supplemented with clonNAT and kanamycin for 1 h. After the flies were removed, excrement spots were noted, and the next day, the spots from the YEPD plate were streaked onto a YEPD plate with both drugs.
Filter mating assay
Quantitative, filter-based mating assays were essentially as performed in Hartwell (1973). Spores from each genotype (5 × 105; tetrads were counted as four) were mixed and filtered onto triplicate nitrocellulose filters, which were placed on a YEPD agarose plate. After 4 h, cells were washed in phosphate-buffered saline and sonicated in 50-ml conical tubes. One hundred fifty cells from each filter were plated in triplicate on YEPD plates supplemented with clonNAT or kanamycin, and 2000 cells were plated on YEPD plates with both drugs. Outcross frequency was calculated as the percentage of recovered cells that were resistant to both drugs (NATRKANR/(NATR+ KANR – NATRKANR)), where NATR is the number of NAT-resistant colonies (whether or not they are also KAN-resistant) and KANR is the number of KAN-resistant colonies (whether or not they are also NAT-resistant).
Live cell imaging
From 100 to 200 μl of spores (see Sporulation conditions and spore isolation) were washed once with water and then resuspended in ∼8 μl of medium (2% dextrose in complete synthetic medium). Samples of cells (0.75 μl) were mounted onto a 2% agarose slab with the same medium, sealed with petroleum jelly, and kept in a humidity chamber at 30°C until imaging.
Live cell imaging was performed on a Zeiss Axio Observer essentially as described in Howell et al. (2012). Differential interference contrast (DIC) images were acquired every 3 min using the autofocus function in the MetaMorph software. If fluorescence images were also being acquired, then DIC images with autofocus were acquired every 8 min and fluorescence images were taken every 16 min. GFP images were acquired with 800 EM gain, 200 ms exposure, and 20 z-steps that were 0.42 μm apart. GFP quantification of MFα1 reporters was performed on the images acquired at the time point just before budding was visualized. The fluorescence z-stack was maximum-projected and the mean fluorescence value was recorded. For experiments using each strain’s endogenous MFα1 promoter or TEF1 promoter, mean fluorescence values were normalized against Bem1-GFP fluorescence in DLY19805 that was germinated and imaged concurrently.
Supplementary Material
Acknowledgments
We thank D. Kiehart for the inspiration to investigate fly frass as a potential yeast dating scene, and for enthusiastic pointers on how to collect and identify fly frass. We thank P. Magwene for the wild yeast strains and for helpful advice throughout the project. We thank M. McMurray, N. Buchler, B. Errede, A. Gladfelter, D. Kiehart, P. Magwene, J. McCusker, and members of the D.J.L. lab for thoughtful discussion and critical reading of the manuscript. Special thanks are due to the D. Fox lab for supplying flies and helping with fly techniques and to D. McClure for help with data analysis. This work was funded by National Institutes of Health/National Institute of General Medical Sciences Grants GM103870 and GM122488 to D.J.L.
Abbreviations used:
- DIC
differential interference contrast
- GFP
green fluorescent protein
- ORF
open reading frame
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0528) on October 24, 2018.
REFERENCES
- Alvaro CG, Thorner J. (2016). Heterotrimeric G protein-coupled receptor signaling in yeast mating pheromone response. J Biol Chem , 7785–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkowitz RA. (2009). Chemical gradients and chemotropism in yeast. Cold Spring Harb Perspect Biol , a001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atay O, Skotheim JM. (2017). Spatial and temporal signal processing and decision making by MAPK pathways. J Cell Biol , 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banderas A, Koltai M, Anders A, Sourjik V. (2016). Sensory input attenuation allows predictive sexual response in yeast. Nat Commun , 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Fink GR. (2011). Yeast: an experimental organism for 21st century biology. Genetics , 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Chervitz SA, Cherry JM. (1997). Yeast as a model organism. Science , 1259–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton PJ, Greig D. (2014). The ecology and evolution of non-domesticated Saccharomyces species. Yeast , 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron F, Barral Y. (2013). A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Cell , 1244–1257. [DOI] [PubMed] [Google Scholar]
- Coluccio AE, Rodriguez RK, Kernan MJ, Neiman AM. (2008). The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS One , 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncic A, Atay O, Valk E, Grande A, Bush A, Vasen G, Colman-Lerner A, Loog M, Skotheim JM. (2015). Compartmentalization of a bistable switch enables memory to cross a feedback-driven transition. Cell , 1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S, Snyder M. (2001). A filamentous growth response mediated by the yeast mating pathway. Genetics , 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DG. (1980). Dispersal of yeasts and bacteria by Drosophila in a temperate forest. Oecologia , 135–137. [DOI] [PubMed] [Google Scholar]
- Haber JE. (2012). Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics , 33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG. (2008). Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell , 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. (1973). Synchronization of haploid yeast cell cycles, a prelude to conjugation. Exp Cell Res , 111–117. [DOI] [PubMed] [Google Scholar]
- Herman P, Rine J. (1997). Yeast spore germination: a requirement for Ras protein activity during re-entry into the cell cycle. EMBO J , 6171–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AS, Jin M, Wu CF, Zyla TR, Elston TC, Lew DJ. (2012). Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell , 322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph-Strauss D, Zenvirth D, Simchen G, Barkai N. (2007). Spore germination in Saccharomyces cerevisiae: global gene expression patterns and cell cycle landmarks. Genome Biol , R241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AC, Shewmaker FP, Kryndushkin D, Wickner RB. (2012). Sex, prions, and plasmids in yeast. Proc Natl Acad Sci USA , E2683–E2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M. (2006). Evolution of the hemiascomycete yeasts: on life styles and the importance of inbreeding. Bioessays , 696–708. [DOI] [PubMed] [Google Scholar]
- Liti G. (2015). The fascinating and secret wild life of the budding yeast S. cerevisiae. Elife , 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. (2009). Population genomics of domestic and wild yeasts. Nature , 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwene PM, Kayıkçı Ö, Granek JA, Reininga JM, Scholl Z, Murray D. (2011). Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc Natl Acad Sci USA , 1987–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L, Dudin O, Martin SG. (2013). Mate and fuse: how yeast cells do it. Open Biol , 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis S, Barrowman J. (2012). Biogenesis of the Saccharomyces cerevisiae pheromone a-factor, from yeast mating to human disease. Microbiol Mol Biol Rev , 626–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller LAH, McCusker JH. (2009). Microsatellite analysis of genetic diversity among clinical and nonclinical Saccharomyces cerevisiae isolates suggests heterozygote advantage in clinical environments. Mol Ecol , 2779–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy HA., Zeyl CW. (2012). Prezygotic isolation between Saccharomycescerevisiae and Saccharomycesparadoxus through differences in mating speed and germination timing. Evolution (NY) , 1196–1209. [DOI] [PubMed] [Google Scholar]
- Neiman AM. (2011). Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics , 737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal S, Iglesias PA, Campbell K, Hilioti Z, Groisman A, Levchenko A. (2007). MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature , 46–51. [DOI] [PubMed] [Google Scholar]
- Reuter M, Bell G, Greig D. (2007). Increased outbreeding in yeast in response to dispersal by an insect vector. Curr Biol , 81–83. [DOI] [PubMed] [Google Scholar]
- Stefanini I, Dapporto L, Berná L, Polsinelli M, Turillazzi S, Cavalieri D. (2016). Social wasps are a Saccharomyces mating nest. Proc Natl Acad Sci USA , 2247–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS, McCusker JH. (2015). The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res , 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Keller P, Kavagiou Z, Jensen LJ, Colombelli J, Bork P, Stelzer EHK, Knop M. (2005). Spore number control and breeding in Saccharomyces cerevisiae: a key role for a self-organizing system. J Cell Biol , 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.