Abstract
Methionine availability during overall amino acid limitation metabolically reprograms cells to support proliferation, the underlying basis for which remains unclear. Here we construct the organization of this methionine-mediated anabolic program using yeast. Combining comparative transcriptome analysis and biochemical and metabolic flux-based approaches, we discover that methionine rewires overall metabolic outputs by increasing the activity of a key regulatory node. This comprises the pentose phosphate pathway (PPP) coupled with reductive biosynthesis, the glutamate dehydrogenase (GDH)-dependent synthesis of glutamate/glutamine, and pyridoxal-5-phosphate (PLP)-dependent transamination capacity. This PPP-GDH-PLP node provides the required cofactors and/or substrates for subsequent rate-limiting reactions in the synthesis of amino acids and therefore nucleotides. These rate-limiting steps in amino acid biosynthesis are also induced in a methionine-dependent manner. This thereby results in a biochemical cascade establishing a hierarchically organized anabolic program. For this methionine-mediated anabolic program to be sustained, cells co-opt a “starvation stress response” regulator, Gcn4p. Collectively, our data suggest a hierarchical metabolic framework explaining how methionine mediates an anabolic switch.
INTRODUCTION
Cell growth is expensive and is therefore tightly coordinated with the intrinsic cellular metabolic state. In general, the metabolic costs incurred during growth and proliferation come from two well-studied phenomena. First, to successfully complete division, a cell makes substantial metabolic investments: to replicate its genome, as well as synthesize building blocks like amino acids, lipids, nucleotides, and other macromolecules (Nelson and Cox, 2017). Second, the process of protein synthesis required for growth itself consumes large amounts of energy (Warner, 1999; Warner et al., 2001), as the translational output of cells increases (Jorgensen and Tyers, 2004). Understanding such changes in cellular metabolic state coupled to biosynthetic outputs, in the context of commitments to cell growth and proliferation, is now the focus of several studies (Brauer et al., 2008; Dechant and Peter, 2008; Boer et al., 2010; Broach, 2012; Ljungdahl and Daignan-Fornier, 2012). For example, one context where there is intense interest in understanding metabolic alterations enabling growth is in cancers, where the identification of the biosynthetic and metabolic requirements for cell growth, incuding during phenomena such as the Warburg effect, are areas of current interest (Warburg, 1956; DeBerardinis et al., 2008; Tong et al., 2009; Vander Heiden et al., 2009). Yet metabolic rewiring is often complex. Therefore, understanding how specific “sentinel” metabolites can function directly as growth signals, and identifying the necessary steps by which such metabolites can metabolically reprogram cells, has been challenging.
However, simple, tractable cellular models can be used to dissect and deconvolute such complex phenomena. Studies using Saccharomyces cerevisiae have been instrumental in identifying dedicated, conserved strategies utilized by eukaryotic cells to integrate metabolic state with growth (Gray et al., 2004; Brauer et al., 2008; Boer et al., 2010; Cai et al., 2011; Slavov and Botstein, 2011; Broach, 2012; Ljungdahl and Daignan-Fornier, 2012; Xu et al., 2013; Mulleder et al., 2016; Ye et al., 2017). In such reductive studies using yeast, preferred carbon or nitrogen sources are typically limited (thereby slowing overall growth). Subsequently, specific factors are reintroduced individually or in combination. This reconstitutes minimal components required for metabolically reprogramming cells and thereby allows the precise dissection of mechanistic events. Such approaches have discovered several novel nutrient sensing systems, mechanisms, or transcriptional programs by which growth outputs are controlled by the build-up and utilization of specific metabolites (Tu et al., 2005, 2007; Boer et al., 2008, 2010; Brauer et al., 2008; Laxman and Tu, 2010; Laxman et al., 2013; Cai et al., 2011; Klosinska et al., 2011; Slavov and Botstein, 2013; Sutter et al., 2013; Xu et al., 2013; Ye et al., 2017).
Interestingly, some amino acids directly function as anabolic signals, potently activating growth pathways independent of their roles as nitrogen or carbon sources. For example, leucine and glutamine activate the target of rapamycin (TOR) pathway directly (González and Hall, 2017; Wolfson and Sabatini, 2017). In this context, studies from diverse organisms suggest that methionine is a strong growth signal or “growth metabolite” (Sugimura et al., 1959; Breillout et al., 1990; Troen et al., 2007; Cavuoto and Fenech, 2012; Sutter et al., 2013; Lee et al., 2014, 2016; Gu et al., 2017). The most direct evidence for methionine as a growth signal comes from studies in yeast. When S. cerevisiae are shifted from complex, amino acid–replete medium with lactate as the carbon source to a minimal medium with the same carbon source, the addition of methionine alone (likely through its metabolite S-adenosylmethionine [SAM]) strongly promotes growth and proliferation (Wu and Tu, 2011; Laxman et al., 2013, 2014b; Sutter et al., 2013; Ye et al., 2017). Thus, even during otherwise overall nutrient limitation, methionine can induce proliferation. Despite these advances, a fundamental question regarding methionine as a growth signal remains unanswered. What is the biochemical logic of the methionine-mediated anabolic program (i.e., how does methionine result in an anabolic reprogramming)? Further, what mechanisms regulate this methionine-mediated anabolic rewiring, even in overall amino acid limiting conditions? We address these related questions in this study.
Here, using a minimal, reconstitutive system in yeast, we uncover how methionine uniquely rewires cellular metabolism to an anabolic state, even in otherwise amino acid–limited conditions. We find that methionine activates very specific metabolic nodes to mediate this anabolic reprogramming. When these nodes are coincidently activated, they further induce a cascade of dependent metabolic processes leading to the overall biosynthesis of amino acids and nucleotides. For sustaining this anabolic program in the presence of methionine, cells co-opt Gcn4p, a mediator of a nutrient stress/survival response. Collectively, these results position methionine at the apex of an overall anabolic network and suggest an overarching, hierarchically organized metabolic logic to explain how methionine availability results in metabolic rewiring and controlling cellular metabolic state.
RESULTS
Methionine mediates a transcriptional rewiring reflecting a “growth state”
When wild-type, prototrophic yeast cells are shifted from a complex, amino acid–rich medium with lactate as the sole carbon source (rich medium, RM) to a synthetic minimal medium containing nitrogen base and lactate (minimal medium, MM), they show a significant lag phase and slower growth. Supplementation with all 20 standard amino acids restores growth after this nutrient downshift (Sutter et al., 2013; Laxman et al., 2014b). Importantly, methionine supplementation alone substantially increases growth (Figure 1A). This growth rescue by supplementing a single amino acid is comparable to (in our hands) or even better than (Sutter et al., 2013) adding all 18 other nonsulfur amino acids (nonSAAs) together. Collectively, this suggested that despite otherwise overall amino acid limitation, methionine availability alone can increase proliferation. However, an inherent paradox emerges from these observations. For increased proliferation, a cell requires an increased supply of anabolic precursors. Methionine itself is not a good “nutrient source” (i.e., it is a poor carbon or nitrogen source), unlike the combined pool of other nonSAAs. Therefore, simply adding methionine alone cannot create a nutrient-replete medium. If this is so, then how do cells build-up precursors for anabolism in the presence of methionine, when other amino acids are limiting? In other words, how might methionine mediate a complete switch to an anabolic state and resolve this apparent metabolic supply problem? We reasoned that dissecting the methionine-mediated overall transcriptional response might provide insight into a possible anabolic program mediated by methionine, revealing any logic inherent within. This could then explain any core metabolic response dependent on methionine that drives proliferation.
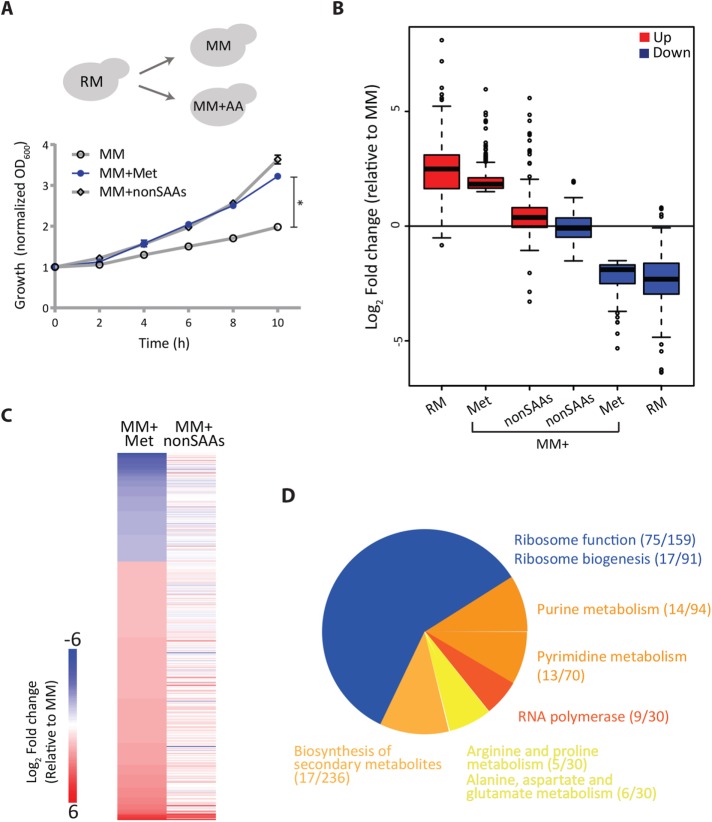
FIGURE 1:
Methionine mediates a transcriptional rewiring reflecting a “growth state.” (A) Methionine and cell proliferation during amino acid limitation. Shown are growth profiles of WT cells grown in rich medium (RM) and shifted to minimal medium (MM) with or without the indicated amino acid supplements (2 mM each; nonSAAs indicates all the nonsulfur amino acids except tyrosine). The growth profile with methionine is in blue (n = 4). (B) Global trends of gene expression in RM and methionine supplemented MM. The boxplot shows fold changes in gene expression levels of two gene classes (up- or down-regulated) relative to MM for cells grown in different amino acid combinations (RM, MM + Met, MM + nonSAAs). The gene classes were defined as those genes that had a significant change (up in red, Met-induced; down in blue, Met-repressed) in MM + Met relative to MM. Also see Supplemental File E1 for gene lists. (C) Effect of methionine on a global transcriptional response in cells. The heat map shows differentially expressed genes in cells grown in MM plus methionine compared with MM (left column), with cells grown in MM plus nonSAAs compared with MM (right column). Also see Supplemental file E1 for gene lists and Supplemental Figures 2 and 3 for related volcano plots and cladograms. (D) GO-based analysis of the methionine-induced genes. The pie chart depicts the processes grouped by GO analysis for the up-regulated transcripts between MM plus methionine and MM set. Numbers in the bracket indicate the number of genes from the query set/ total number of genes in the reference set for the given GO category. Also see Supplemental File E2 for GO annotations and Supplemental Figure 6 for related GO groupings.
We first addressed how methionine reprograms cells into an anabolic state, focusing on elucidating early transcriptional events, even before the overall proliferation is observed. We performed comprehensive RNA-seq analysis on distinct sample sets of wild-type cells: 1) RM grown or cells shifted to 2) MM for 2h, 3) MM + Met for 2h (Met set), and 4) MM + nonSAAs for 2h. Transcript reads from the biological replicates showed exceptional correlation across all conditions (Pearson correlation coefficient, R ≥ 0.99) (Supplemental Figure 1). Setting a stringent cut-off, we initially considered differentially expressed genes with ≥log2 1.5-fold changes (i.e., ∼2.8-fold change) and a p value cut-off <10-4 for further analysis. We initially compared global transcription trends in wild-type (WT) cells growing in RM, MM + Met, or MM + nonSAAs to MM, with the main focus on what happens when methionine is the sole variable (i.e., MM and MM + Met). We first examined overall global gene expression trends in these conditions (compared with MM), looking at the distribution of the most induced or down-regulated genes (Figure 1B and Supplemental Figure 2). Here we compared the global gene expression trends (broad trends of up- or down-regulated genes) exhibited by cells in MM + Met to cells grown in RM or MM + nonSAAs, all relative to MM (i.e., we compared the fold-change in expression levels of both up-/down-regulated genes in MM + Met, to the same genes in RM or MM + nonSAAs, all baselined to these gene expression levels in MM) (Figure 1B). Notably, the overall MM + Met gene expression profile very closely resembled the signature of cells in RM, in contrast to the cells in MM + nonSAAs (which were nearly indistinguishable from MM) (Figure 1B and Supplemental Figures 2 and 3). This suggests that methionine is perceived by cells as a stronger anabolic cue than all other nonsulfur amino acids combined. Methionine is sufficient to switch cells into a transcriptional state resembling that of rapidly proliferating cells in RM (which is complex, amino acid–rich medium ideal for growth).
We next more closely examined the overall transcriptional response unique to methionine by comparing transcriptomes of cells growing in MM versus MM + Met (i.e., the only variable being methionine). This comparison identified 372 genes, of which 262 genes were up-regulated in the Met set (Figure 1C and Supplemental file E1). Using gene ontology (GO), these genes were grouped into related processes (Figure 1D, Supplemental file E2). Given that there is an eventual growth increase in MM + Met, we observed a grouping suggesting a transcriptional induction of genes related to the core translational machinery. Additionally, GO also grouped multiple induced genes into “nucleotide metabolism,” that is, under “purine/pyrimidine” or “nucleobase and nucleotide metabolism,” along with the “biosynthesis of secondary metabolites” (Figure 1D). The induction of all of these processes would be expected for any cell in a “growth/proliferative” state, since proliferation relies on increased translation and replication. Unsurprisingly therefore, this GO grouping of induced genes showed a signature of a cell in a “proliferative state.”
Methionine uniquely induces the PPP-GDH-PLP node
However, this grouping does not satisfactorily resolve the metabolic supply problem highlighted earlier, which is the goal in this study. While GO-based analyses are very effective to explain entire transcriptional programs, GO relies on enriching pathway terms relying on multiple genes within a pathway to be overrepresented. In contrast, for metabolic changes, it is standard knowledge that entire metabolic pathways need not be regulated, especially at the level of transcripts. This is because substantial metabolic regulation relies on cofactor and substrate availability, as well as by controlling only key nodes or “rate-limiting steps” in metabolism (Nelson and Cox, 2017). Sometimes, therefore, if a metabolic transformation is mediated by the induction of select, specific nodes, GO analysis alone may not fully reveal this, and a secondary analysis is required. Therefore, we took an additional approach to manually rebuild connections and groupings of only the metabolism related transcripts that are induced by methionine, based on known metabolic reaction requirements. In this additional reconstruction, we broke down the analysis into two components as follows: 1) whether the regulated biochemical step, and its subsequent product, was critical for multiple other biosynthetic reactions (i.e., if it was a major substrate or cofactor generating step) and 2) whether the protein encoded by the gene regulated a known “bottleneck” or rate-limiting metabolic step in a general anabolic pathway. For this second point, we considered bottleneck steps as the following: the first enzyme in a metabolic pathway typically commits the fate of the entry-point metabolite, and the enzyme itself is often regulated by end-product inhibition (Nelson and Cox, 2017). Similarly, the last enzyme in a pathway forms the end product, which can be an entry point (and therefore rate limiting) for a downstream pathway (Nelson and Cox, 2017). We reasoned that by this secondary analysis, it may be possible to better organize a possible biochemical hierarchy. In this analysis, we therefore did not particularly emphasize on the number of genes in a single pathway that are induced (which is one major consideration in GO-based pathway enrichment). Our reasoning was that these bottleneck nodes may not be picked up in the first analysis, especially if some metabolic pathways are composed of very few limiting steps.
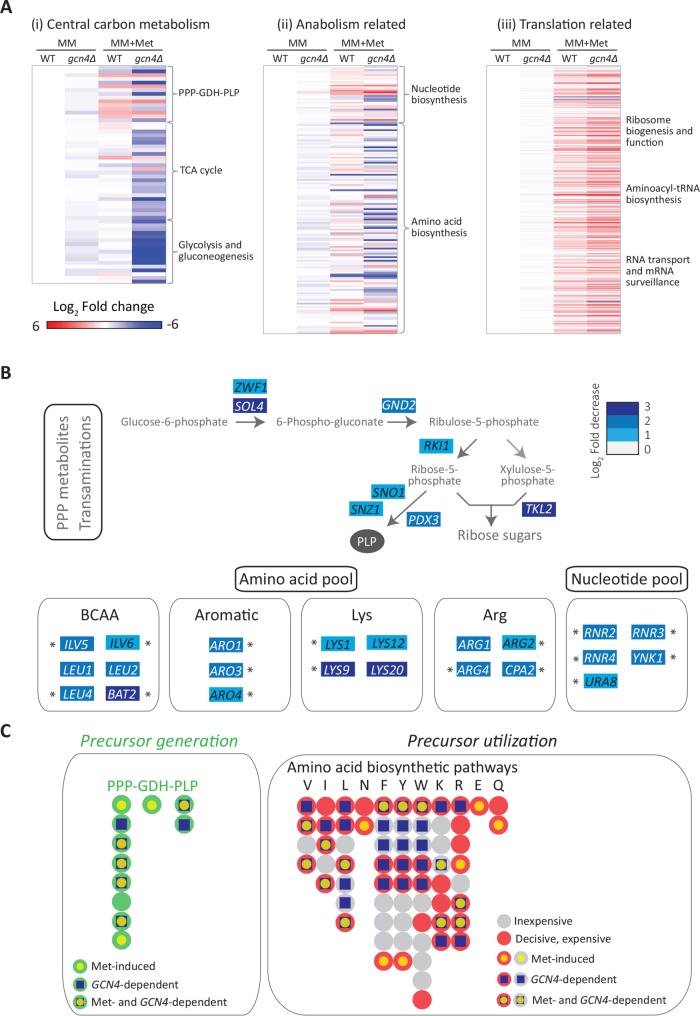
Through this secondary reconstruction, we identified and recompartmentalized the metabolic response regulated by methionine into a group of key biochemical reaction nodes, as described. First, for the category 1 grouping, that is, the substrate and cofactor producing metabolic reactions, we considered “central carbon/carbohydrate metabolism” as a single class. Only a few genes involved in classical “central carbon/carbohydrate metabolism” were up-regulated in the presence of methionine, and, strikingly, none of them group to glycolysis, the tricarboxylic (TCA) cycle, or gluconeogenesis (Figure 2A). However, the genes encoding three key enzymes of the pentose phosphate pathway (PPP) (GND1, RKI1, and TKL1), which regulate four steps in the PPP, were strongly induced in the presence of methionine. Furthermore, two other genes (SOL3 and TAL1), which control two other steps in the PPP, were also induced by methionine (at just below the log2 1.5-fold (∼2.8-fold) stringent cut-off limit we had set) (Figure 2A). More specifically, Gnd1p catalyzes the last step in the oxidative arm of the PPP, generating nicotinamide adenine dinucleotide phosphate (NADPH) and producing ribulose-5-phosphate. RKI1, TKL1, and TAL1 are three of the four genes of the nonoxidative arm of the PPP, which make ribose-5-phosphate and other critical intermediates. Additionally, HXK2, encoding hexokinase, which converts glucose to glucose-6-phosphate was induced. While this is not even classified under the PPP by GO grouping, glucose-6-phosphate is the substrate for the first, rate-limited step of the PPP, and so we included it under the PPP in our grouping (Figure 2A). In this grouping, these targets were not picked up simply by random chance. Of the 11 genes comprising the eight steps of the PPP, six (HXK2, SOL3, GND1, RKI1, TKL1, and TAL1) are induced by methionine. Thus, the PPP arm of central carbon metabolism was significantly transcriptionally induced by methionine (p = 10-4, Fisher’s exact test), while for any of the other central carbon metabolic pathways, any pathway level induction was not significant. Second, we noted that a regulator essential for the formation of pyridoxal-5-phosphate (PLP) (encoded by SNO1) was strongly induced by methionine (Figure 2A). PLP is a central cofactor, required for all transamination reactions (Nelson and Cox, 2017), but notably does not get a GO grouping, because it does not fall in a large pathway/group. Third, the transcript of Gdh1p, which regulates the key nitrogen assimilation reaction, resulting in the formation of glutamate (and GLN1, which is further required to make glutamine), was highly induced in methionine (Figure 2A). This reaction requires NADPH (which is itself produced in the PPP) and importantly is also critical for the subsequent formation of all other amino acids and nucleotides (Figure 2A). Thus, from this grouping, we observe that the PPP, glutamate dehydrogenase (GDH), and PLP pathways were collectively strongly and significantly induced by methionine (p = 1.2 × 10-6, Fisher’s exact test) (Figure 2B, also see Supplemental Figures 4 and 5). Notably, the reactions in these pathways produce metabolic precursors that are typically simultaneously utilized in other biosynthetic reactions (as described in the subsequent section and illustrated in Figure 2B). We therefore grouped these together as a single PPP-GDH-PLP metabolic node. We hypothesized that this PPP-GDH-PLP node could be central for all the subsequent, downstream anabolic outputs.
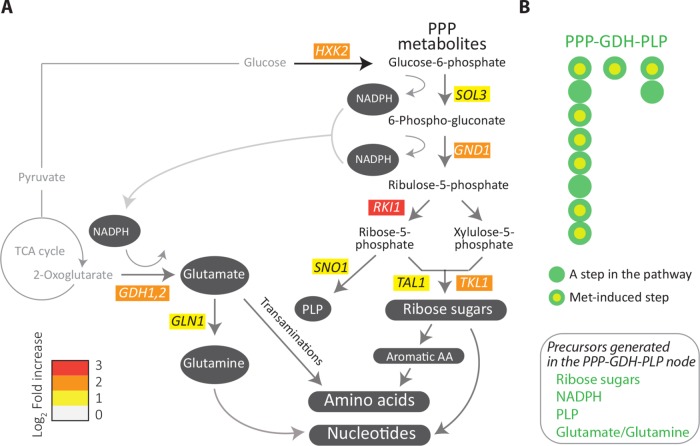
FIGURE 2:
Methionine uniquely induces the PPP-GDH-PLP node. (A) Metabolic pathways induced by methionine. Illustration of the results of a manual regrouping of the methionine responsive genes into their relevant metabolic pathways, restricted only to central carbon metabolism, and subsequent anabolic processes. The pathway map includes individual genes in central carbon metabolism which are induced by methionine (indicating the fold changes in gene expression). The color bar indicates the fold increase in gene expression. (B) A bird’s-eye view description of the PPP-GDH-PLP node regulated by methionine. Each bead (or filled circle) represents a step in the pathway (see details in Supplemental Figures 4 and 5). Methionine-induced steps are shown with a yellow fill at the centre of the circle for the given step. Precursors generated through this node are shown in the inset; p = 1.2 × 10–6 (Fisher’s exact test) for the methionine-dependent induction of the PPP-GDH-PLP node.
Methionine sets up a hierarchical metabolic response leading to anabolism
We next analyzed our transcriptome data for the other metabolic genes up-regulated by methionine, which would be considered anabolic genes. These could be grouped broadly under “amino acid biosynthesis,” “nucleotide synthesis,” and “oxidoreduction/transamination” categories. Further, here we focused on the substrates or cofactors required for the functions of these pathways. Notably, only some genes in each of these large, multistep, multienzyme pathways were induced in the presence of methionine. However, when these induced genes were organized by their substrate or cofactor requirements, we noted that essentially all enzymes encoded by these genes utilized either a PPP intermediate/product, and/or NADPH, and/or glutamate/glutamine, or combinations of all of these (Figure 3A). These substrates or cofactors thus are all derived from the earlier defined PPP-GDH-PLP node (Figure 3A). We further examined the steps in the respective biosynthetic pathways that these genes regulated (coming from Figure 3A). For this, we categorized all steps in the amino acid biosynthesis pathway as either the rate-limiting/initiation step and/or final step in the production of that amino acid. Here, we organized them based on the use of precursors or cofactors, derived from this PPP-GDH-PLP node (Figure 3B and Supplemental Figures 4 and 5). Strikingly, we observed that the methionine-induced genes in these pathways regulated only the most critical, rate-limiting, or costly steps in amino acid biosynthesis (p = 7.4 × 10-5, Fisher’s exact test) but had no significant role (p = 0.517, Fisher’s exact test) in regulating the other genes in the pathway that encode enzymes for inexpensive steps (Figure 3B). Finally, we note that these methionine-induced genes in the “amino acid biosynthesis” bin do not just broadly represent all amino acid biosynthesis but synthesize what are considered conventionally to be the costliest amino acids to synthesize (Barton et al., 2010). These are the aromatic amino acids, the branched-chain amino acids, and lysine (which is also overrepresented in ribosomal and core translational machinery proteins [Laxman et al., 2013]).
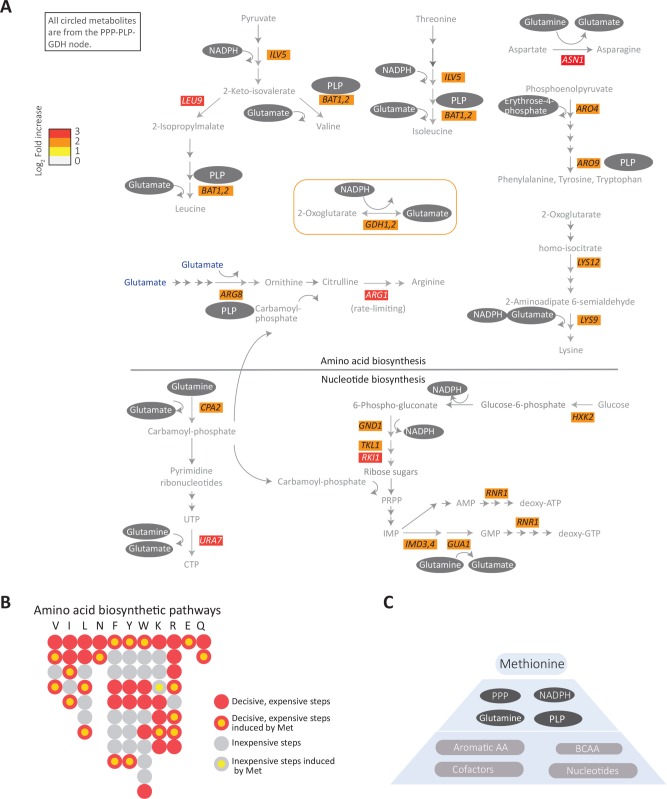
FIGURE 3:
Methionine sets up a hierarchical metabolic response leading to anabolism. (A) Grouping of the methionine-induced genes, focusing on amino acid and nucleotide metabolism. The schematic shows the methionine-responsive genes in various amino acid and nucleotide biosynthesis pathways, along with their fold changes in gene expression (indicated by the color bar). The substrates/cofactors produced by the PPP-GDH-PLP node (see Figure 2) for the individual steps in these pathways is also mapped on to the schematic. Note that all gene products induced by methionine in these pathways use PPP intermediates, NADPH, PLP and/or glutamine/glutamate (indicated within gray ovals) in their biochemical reactions. (B) A bird’s-eye view description of the amino acid biosynthesis steps regulated by methionine, with metabolically expensive or inexpensive steps indicated. Each bead (or filled circle) represents a step in the pathway (prepared according to the individual amino acid pathways shown at https://pathway.yeastgenome.org/; details in Supplemental Figures 4 and 5). A step is considered expensive (red) when it is either the entry point or the exit point or if it involves ATP utilization or reduction. All the rest of the steps are considered inexpensive (gray). Methionine-induced steps are shown with a yellow fill at the centre of the circle for the given step. p = 7.4e-5 (Fisher’s exact test) for methionine-dependent induction of genes encoding the critical, rate-limiting, or costly steps in amino acid biosynthesis (not significant for the inexpensive steps). (C) A proposed hierarchical organization of the methionine-mediated anabolic remodeling. Methionine induces expression of genes in the PPP-GDH-PLP node, which provides precursors for the key steps in the biosynthesis of all other amino acids and nucleotides, and these steps are also directly induced by methionine.
Finally, nucleotide biosynthesis involves very elaborate, multistep, multienzyme pathways. Methionine does not induce large groups of genes in nucleotide biosynthesis. However, we find that the methionine-dependent, up-regulated genes related to nucleotide synthesis, functionally utilize substrates or cofactors from the PPP-GDH-PLP node (Figure 3A). Notably, RNR1, which encodes the key enzyme in converting ribonucleotides to deoxyribonucleotides (and hence the critical hub for DNA synthesis), is strongly up-regulated on methionine addition (Figure 3A), while most other steps in this pathway (which are not rate limiting) are not regulated by methionine. Rnr1p activity requires reduced thioredoxin, which itself is NADPH dependent.
Separately, as a control, we expanded this analysis and compared the methionine response to minimal medium supplemented with all the other nonSAAs. Here the overall metabolic grouping or organization (for methionine-induced genes) remained unchanged (Supplemental Figure 2), since the nonSAA response resembles the MM response. The nonSAAs do not induce the PPP-GDH-PLP node (or subsequent dependent anabolic nodes). Thus, the methionine-dependent induction of the PPP-GDH-PLP node, and the subsequent dependent pathways, is unique to the presence of methionine in the medium. Finally, in MM+nonSAAs, the few highly induced genes (compared with RM) function in methionine- (and sulfur-amino acid) related biosynthesis or salvage (Supplemental Figure 6 and Supplemental File E1) and not additional reactions. This further substantiates our overall observations for the role of methionine as an “anabolic signal.”
Collectively, these data and organization of a methionine-dependent transcriptional program suggest not just a general transcriptional remodeling to a “growth state” but also a deeper hierarchical organization of an anabolic program (Figure 3C). In this putative hierarchical organization, methionine induces genes regulating the PPP, key transamination reactions, and the synthesis of glutamine/glutamate (the PPP-GDH-PLP node). These three processes directly allow critical steps in synthesis of the costliest amino acids and nucleotides. The key, limiting steps in these subsequent synthesis reactions are themselves induced by methionine, collectively setting up a structured anabolic program (Figure 3C). These data thus uniquely position methionine as an anabolic cue.
The anabolic program induced by methionine requires GCN4
How might methionine mediate this very specific transcriptional response to induce these metabolic nodes and genes? We reasoned that there could be a methionine-dependent activation of a transcriptional regulator(s) that controls this metabolic axis, including amino acid and nucleotide biosynthetic genes. Further, the methionine effect was most strongly observed in conditions of overall amino acid limitation. While there is currently no known methionine-dependent transcriptional regulator that can control these metabolic nodes, there is in fact a well-known master regulator of general amino acid biosynthesis. The conserved transcription factor Gcn4p (Atf4 in mammals) is a transcriptional activator, primarily controlling the amino acid biosynthetic genes during amino acid starvations and amino acid imbalance (Hinnebusch, 2005). However, the activity of Gcn4p has been mainly studied during starvation as a “stress response” regulator and not in contexts involving increased proliferation. We wondered whether a possible connection between methionine and Gcn4p might exist in conditions where increased proliferation is observed despite overall amino acid limitation. We first monitored the amounts of endogenous Gcn4p (chromosomally tagged with a C-terminal human influenza hemagglutinin [HA] epitope) after a shift to MM, with and without supplementation of different amino acids including methionine. Interestingly, Gcn4p amounts increased substantially specifically on methionine supplementation alone (when other amino acids were not supplemented), compared with either MM or MM supplemented with all 18 other nonSAAs (Figure 4A). This observation was further independently confirmed using immunofluorescence-based experiments (Supplemental Figure 7A). We therefore asked whether GCN4 was necessary for the increased growth on methionine supplementation. Notably, the gcn4Δ cells did not show any increased growth in methionine supplemented medium but instead grew comparably to WT cells in MM (Figure 4B). As controls, in all other conditions (lacking methionine, or in RM), the growth of gcn4Δ cells was indistinguishable from the WT cells (Supplemental Figure 7B). Collectively, these data suggest that GCN4 is required for the methionine-mediated growth in otherwise amino acid–poor conditions. We therefore hypothesized that the methionine-dependent transcriptional response, particularly that of the core anabolic program defined earlier, might be mediated by Gcn4p. To address the role for GCN4 in this anabolic response, we carried out a comparison of transcriptomes of gcn4Δ cells grown in RM, MM, MM + Met, and MM + nonSAAs with wild-type cells grown in the respective conditions.
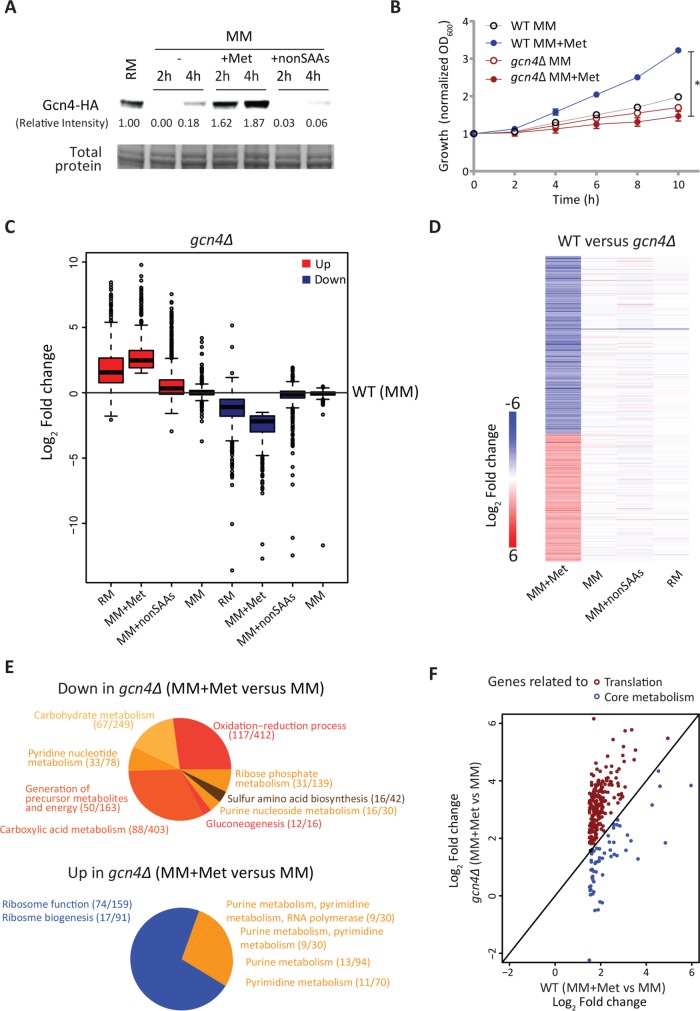
FIGURE 4:
The anabolic program induced by methionine requires GCN4. (A) Gcn4p is induced by methionine. Gcn4p amounts were detected by Western blotting of WT cells (expressing Gcn4p with an HA epitope, tagged at the endogenous locus) shifted from RM to MM or MM supplemented with the indicated combinations of amino acids. A representative blot is shown (n = 3). Also see Supplemental Figure 7A. (B) GCN4 is necessary for methionine-mediated increased growth. WT and gcn4Δ cells were shifted from RM to MM with or without methionine supplementation and growth was monitored. Also see Supplemental Figure 7B (n = 4). (C) Trends of gene expression in RM and methionine supplemented MM in gcn4Δ cells. Gene expression levels of transcripts in gcn4Δ cells grown in RM or shifted to MM or MM plus methionine or MM plus nonSAAs were compared with only the WT MM set. Also see Supplemental File E1 for related gene lists and Supplemental Figure 8 for related volcano plots. (D) Global transcriptional response in the absence of GCN4. The heat map shows differentially expressed genes (log2 1.5-fold change; p < 10-4) between WT and gcn4Δ cells in the respective growth conditions. Also see Supplemental File E1 for related gene lists and Supplemental Figure 8 for related volcano plots. (E) GO-based analysis of the methionine-responsive genes in gcn4Δ cells. A pie chart showing the processes grouped by GO analysis for the up-regulated and down-regulated transcripts between MM + methionine and MM sets in the gcn4Δ background. Numbers in the bracket indicate the number of genes from the query set/total number of genes in the reference set for the given GO category. Also see Supplemental file E2 for related GO groupings. (F) The methionine-induced metabolic program requires GCN4. Categorization of the GCN4-dependent transcripts in the presence of methionine, as related to metabolism, or translation. The expression level of the methionine-responsive transcripts related to metabolism and translation in WT set (MM plus methionine vs. MM) was compared with the gcn4Δ background. The genes related to the metabolic steps described in Figures 1 and 2 are marked with blue circles, while genes related to ribosome biogenesis and function are marked with red circles. Also see Supplemental Figure 9 for the list of GCN4-dependent genes related to metabolism, picked up in this analysis. In all panels, data shows mean ± SD. *p < 0.05.
We first examined global trends of gene expression (similar to those in Figure 1B) in the absence of GCN4 and compared those to the WT set. Here the baseline was the gene expression of WT cells in MM. The transcriptional response in all conditions excluding MM + Met (i.e., MM, RM, or nonSAA) was almost unaffected in gcn4Δ cells compared with WT cells (Figure 4C). However, in MM + Met, the absence of GCN4 revealed a strong transcriptional response. Notably, if gene expression trends of only the group of genes induced in methionine (similar to the analysis done in Figure 1B for the WT set) were considered, that is, the genes induced in gcn4Δ cells in methionine, then the overall global gene expression trends in the MM + Met even more strongly resembled the RM set than that of WT cells in this condition (Figure 4C). This paradoxically suggested that in the absence of GCN4, supplementing methionine invokes an even stronger transcriptional response in cells. To address this, we first compared transcriptomes of WT cells with gcn4Δ cells, under the same combination of conditions used earlier and analyzed our data with the same filters used in the preceding section. The overall changes between transcriptomes of WT versus gcn4Δ cells are shown (Figure 4D, Supplemental Figure 8, and Supplemental File E1). In all conditions except methionine, WT and gcn4Δ cells showed very similar gene expression profiles (Figure 4D). Strikingly, only in the presence of methionine do WT and gcn4Δ cells show a contrast in gene expression. This suggests a strong regulatory role for GCN4 only in the presence of methionine (see detailed analysis in the next section).
To better understand what component of the methionine response was directly induced in a GCN4-dependent manner, we organized the subset of genes induced and down-regulated in methionine (compared with MM) in the gcn4Δ cells by function. When grouped using GO, we found that genes generally related to “central carbon metabolism” and “nucleotide metabolism” were down-regulated in the presence of methionine in the gcn4Δ background (top panel in Figure 4E, Supplemental file E2). Further, in the presence of methionine, a large number of genes (∼200) were highly induced in the gcn4Δ cells (even more than in WT cells in methionine), and these grouped into the general groups of “ribosome/translation” and “nucleotide synthesis” (bottom panel in Figure 4E and Supplemental File E2). This representation was more striking than that seen in WT cells (Figure 1D), suggesting an even stronger methionine-dependent induction of translation related genes in cells lacking GCN4.
All these data revealed that cells lacking GCN4 showed a very strong transcriptional response in MM + Met, with aspects of the signature of induced transcripts similar to, but stronger than, WT cells in MM + Met. Note, however, that (as shown earlier in Figure 4B) GCN4 was essential for the growth induction due to methionine. These data therefore paradoxically suggested that while the overall “growth signature” response due to methionine remained (in the induced genes), a subset of genes down-regluated in cells lacking GCN4 may be pivotal for this growth outcome. We next considered only the set of genes induced by methionine in WT cells and grouping them into two bins, one representing the genes related to ribosome function and translation and the other the core metabolism related genes. Relative amounts of these transcripts were compared with cells lacking GCN4 (only in the MM + Met condition). Strikingly, transcripts of all genes that mapped to ribosome/translation function (from Figure 1D) were higher in gcn4Δ cells compared with WT cells in the presence of methionine (Figure 4F). In contrast, transcript amounts of every gene mapping to core metabolism were decreased in a GCN4-dependent manner (Figure 4F; see Supplemental Figure 9). Thus, this reorganization suggested that in the presence of methionine, the induction of only the core anabolic program requires GCN4.
In methionine-rich medium the absence of GCN4 results in an anabolic failure
The earlier comparison (Figure 4F) was limited, considering only the highly induced genes in WT cells in the presence of methionine. For a more unbiased comparision, we therefore more systematically examined the entire transcriptomes of WT and gcn4Δ cells in only the MM + Met condition. In this condition, both sets have methionine present, and therefore the only variable is GCN4. Here, we binned the transcriptomes into the following large groups: 1) genes involved in central carbon metabolism (which includes the PPP-GDH-PLP node, the TCA cycle, glycolysis, and gluconeogenesis), 2) amino acid and nucleotide biosynthesis (general anabolism), and 3) translation related genes (Figure 5A). Through this analysis, two clear trends emerged. First, transcripts in the PPP-GDH-PLP node were significantly decreased in GCN4-deficient cells (p = 4.2 × 10–3, Fisher’s exact text). Further, the entire central carbon metabolism node was strongly and significantly decreased in GCN4-deficient cells, compared with WT cells, in the MM + Met condition (Figure 5A) (p = 2.2 × 10–16, Fisher’s exact test). Next, the entire anabolic arm was strongly and significantly decreased in GCN4-deficient cells, compared with WT cells, only in the MM + Met condition (p = 4.1 × 10–8, Fisher’s exact test), with the amino acid biosynthesis steps being particularly significantly down-regulated (p = 4.7 × 10–10) (Figure 5A). In contrast, transcript amounts of genes related to translation/ribosome biogenesis were increased in the absence of GCN4 (Figure 5A) (p = 2.2 × 10–16). Together, these analyses revealed that in methionine-rich conditions, the absence of GCN4 results in a collapse of the entire anabolic program. This further explains why, despite the increase in translation-related transcripts, cells lacking GCN4 cannot sustain proliferation in the presence of methionine.
FIGURE 5:
In methionine-rich medium the absence of GCN4 results in an anabolic failure. (A) Global transcriptional response in the presence of methionine in WT cells or cells lacking GCN4. The heat maps show transcript abundances of 1) genes involved in central carbon metabolism (including the PPP-GDH-PLP node), 2) anabolism (including amino acid biosynthesis), and 3) translation related processes in the respective growth conditions and genetic backgrounds. Note: compared with WT cells, the loss of GCN4 shows little effect in MM. In cells supplemented with methionine, cells lacking GCN4 have a strongly reduced central carbon metabolism component (p = 2.2 × 10–16) and anabolic component (p = 4.1 × 10-8) and increased translation component (p = 2.2 × 10–16) (Fisher’s exact tests). (B) GCN4 is required for the metabolic program due to methionine. Grouping of the GCN4-dependent genes based on the defined PPP-GDH-PLP–dependent metabolic node. The schematic shows the GCN4-dependent genes (comparison of MM plus methionine set between WT and gcn4Δ) in the PPP, amino acid, and nucleotide biosynthesis pathways, along with fold changes in gene expression. The arrows marked blue in the PPP are the steps down-regulated in gcn4Δ cells. The rate-limiting steps in the pathway are marked by an asterisk. (C) A bird’s-eye view depiction of the methionine-induced genes and the GCN4-dependent genes (in the presence of methionine) mapped onto pathways that either generate biosynthetic precursors or pathways that utilize these precursors. The left panel shows steps induced by methionine in WT cells and the GCN4 dependence (in methionine medium) for the PPP-GDH-PLP node. The right panel shows the critical, expensive steps or the inexpensive steps in amino acid biosynthesis pathways (details in Supplemental Figures 4 and 5), with a mapping of these as methionine-induced and/or GCN4 dependent. Each bead (or filled circle) represents a step in the pathway (prepared as shown in Figure 3B). For the given step, methionine-induced steps are shown with a yellow fill at the center of the circle, and GCN4-dependent steps are shown with a blue square at the center of the circle. Left panel, p = 4.2 × 10–3 (Fisher’s exact test) for GCN4-dependent genes that control the PPP-GDH-PLP node. Right panel, p = 7 × 10–3 (Fisher’s exact test) for GCN4-dependent genes encoding the critical, rate-limiting steps in amino acid biosynthetic pathways.
For more clarity, we examined the GCN4-dependent transcripts that regulated the PPP-GDH-PLP node or specific steps in amino acid biosynthesis (Figure 5B). This is the metabolic hierarchy we have described earlier. The genes from the PPP and transamination reactions were strongly down-regulated in gcn4Δ cells compared with WT cells in methionine (Figure 5B). However, the transcript of the Gdh1 enzyme (required for glutamate synthesis) was only methionine but not GCN4 dependent (Figure 5B). Further, the transcripts of methionine-induced genes encoding multiple branched-chain or aromatic amino acid and lysine and arginine biosynthesis were also GCN4 dependent. Finally, while most nucleotide biosynthesis genes were not GCN4 dependent, the entire RNR complex (which is critical for the NTP to dNTP conversion, required for DNA synthesis) was GCN4 dependent (Figure 5B). We mapped these methionine-induced and GCN4-dependent genes onto the metabolic hierarchy defined earlier (the PPP-GDH-PLP node), and the amino acid biosynthetic pathways (Figure 5C and Supplemental Figures 4 and 5). In this mapping, we found that the loss of GCN4 significantly down-regulates the PPP-GDH-PLP node (p = 4.2 × 10-3, Fisher’s exact test). This also includes a down-regulation of genes that were not induced in WT cells by methionine. However, if only the genes in this node induced in WT in the presence of methionine are considered (i.e., the overlapping set), then their down-regulation in cells lacking GCN4 remains significant (p = 3 × 10-2, Fisher’s exact test). For the amino acid biosynthetic pathways alone, in the presence of methionine, the absence of GCN4 results in a significant down-regulation of this entire arm (4.7 × 10-10, Fisher’s exact test). If only the critical steps in these pathways that are induced in WT cells in the presence of methionine are considered (the overlapping set), then, again, the loss of GCN4 results in a significant down-regulation of these steps (p = 7.8 × 10-6, Fisher’s exact test). Collectively, these data reveal that in the presence of methionine, GCN4 is required for the induced hierarchical anabolic program, and its absence in methionine-rich medium results in an anabolic collapse.
The PPP-GDH-PLP nodal enzymes are methionine dependent and largely GCN4 dependent
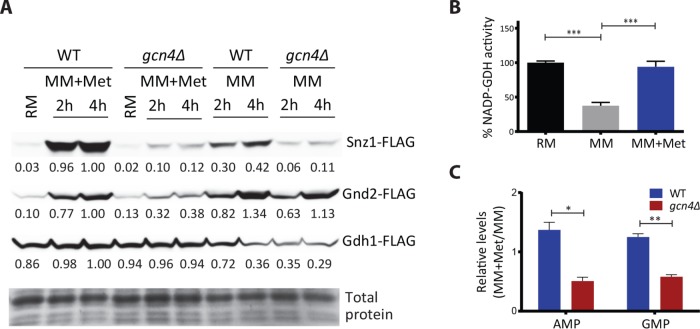
We next biochemically estimated the protein amounts of three transcripts that represent the PPP-GDH-PLP node. These are Snz1p, Gnd2p, and Gdh1p. Snz1p is required for pyridoxal phosphate (PLP) biosynthesis (Dong et al., 2004), which is essential for all transamination reactions (Nelson and Cox, 2017). As illustrated earlier in Figure 2A, PLP biosynthesis itself requires the PPP intermediate erythrose-4-phosphate as a substrate. Gnd2p is the key NADPH generating enzyme in the oxidative branch of the PPP. Gdh1p consumes NADPH and makes glutamate from 2-ketoglutarate. We measured amounts of these three proteins from WT and gcn4Δ cells growing in MM and MM + methionine (Figure 6A). Notably, Snz1p and Gnd2p showed a strongly methionine- and GCN4-dependent induction (Figure 6A and Supplemental Figure 10). Gdh1p was strongly induced by methionine but was not dependent on GCN4 (Figure 6A). However, since this activity itself is related to the PPP and PLP node, we measured in vitro Gdh1p activity (NADP-GDH activity) in lysates from cells growing in MM or with methionine and found that overall Gdh1p activity was higher in cells growing with methionine (Figure 6B). These data collectively suggest that the effective output of this PPP-GDH-PLP node in the presence of methionine should be GCN4 dependent. Here an expected final readout of this biochemical coupling are changes in steady-state nucleotide amounts, also predicting a GCN4 dependence in methionine-replete conditions. Comparing relative amounts of nucleotides in wild-type and gcn4Δ cells, we noted decreased nucleotide amounts in gcn4Δ cells in the presence of methionine (Figure 6C). Collectively, these data support our proposed paradigm of a coupled induction of the PPP-GDH-PLP node by methionine, leading to increased amino acid and nucleotide synthesis.
FIGURE 6:
The PPP-GDH-PLP nodal enzymes are methionine dependent and largely GCN4 dependent. (A) Snz1p, Gnd2p, and Gdh1p amounts in WT or gcn4Δ cells, with methionine in the medium as the variable. WT and gcn4Δ cells expressing FLAG-tagged Snz1p or Gnd2p or Gdh1p were shifted from RM to MM or MM plus methionine, and amounts of these proteins were detected by Western blotting. A representative blot is shown in each case (n = 2). Also see Supplemental Figure 10. (B) NADP-dependent glutamate dehydrogenase activity with methionine in the medium as a variable. Crude extracts of WT cells grown in RM and shifted to MM or MM plus methionine were analyzed for intracellular biosynthetic NADP–glutamate dehydrogenase activity (n = 4). (C) Relative nucleotide amounts in the presence of methionine in WT or gcn4Δ cells. WT and gcn4Δ cells grown in RM were shifted to MM (4 h) with and without methionine, and the relative amounts of AMP and guanosine 5’-monophosphate (GMP) from metabolite extracts of the respective samples were measured by LC-MS/MS (n = 2 biological replicates, with technical replicates). In all panels, data indicate mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
Methionine increases amino acid biosynthesis
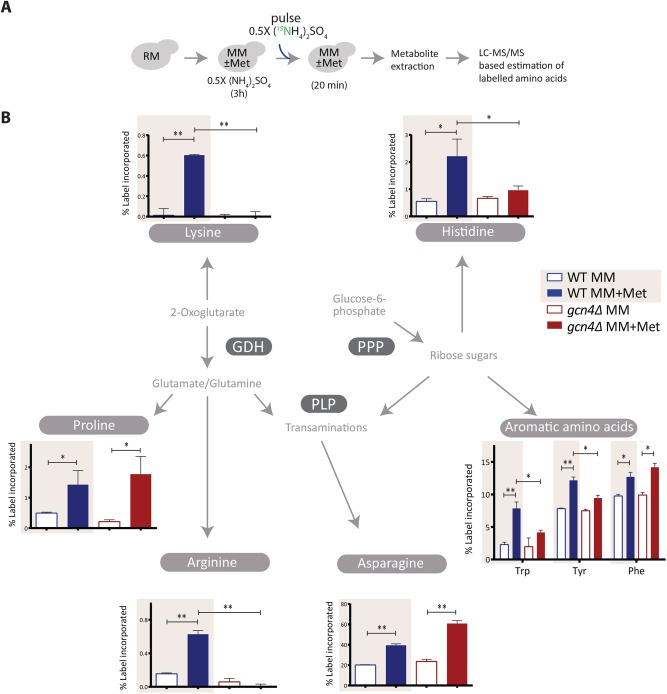
Steady-state metabolite measurements alone do not definitively show this metabolic coupling, since any steady-state metabolite measurement (as in Figure 6C) cannot directly distinguish synthesis from consumption. Therefore, to directly address this hierarchical anabolic program, we resorted to a stable-isotope pulse labeling and an LC-MS/MS-based approach to first directly measure the new synthesis of amino acids. To WT or gcn4Δ cells in the respective medium with or without methionine, we pulsed 15N-labeled ammonium sulfate and measured the 15N incorporation into amino acids (Figure 7A and Supplemental Table 2) before an effective steady-state of labeled amino acid synthesis and consumption was reached (Supplemental Figure 11). This permits the detection of newly synthesized amino acids, which will incorporate the 15N label. We observed that biosynthesis of all the aromatic amino acids, lysine, histidine, proline, arginine, and asparagine, is strongly dependent on methionine presence (Figure 7B). For technical reasons, we could not measure label incorporation into branched-chain amino acids. Notably, the label immediately (∼20 min) percolated in asparagine and aromatic amino acid biosynthesis and showed a very strong methionine dependence (Figure 7B). Asparagine, proline, and phenylalanine biosynthesis were methionine dependent even in gcn4Δ cells, pointing toward possible GCN4-independent influences of methionine. For all the other amino acids measured, the biosynthesis was both methionine as well as GCN4 dependent (Figure 7B). These data directly indicate that methionine availability controls the key nodes around the PPP-GDH-PLP axis, thereby generating the amino acid pool required for proliferation and that this is largely regulated by GCN4.
FIGURE 7:
Methionine increases amino acid biosynthesis. (A) A schematic showing the experimental design of 15N pulse-labeling experiment to measure amino acid biosynthetic flux. Cells were shifted to MM with and without methionine and maintained for 3 h, 15N-ammonium sulfate was pulsed into the medium, and the indicated, labeled metabolites were measured. (B) Methionine increases amino acid biosynthesis in a GCN4-dependent manner. 15N label incorporation into newly synthesized amino acids in WT and gcn4Δ cells was measured, as shown in A. For all the labeled moieties, fractional abundance of the label was calculated. Also see Supplemental Figure 11 for the label incorporation kinetics experiment and Supplemental Table 2 for mass spectrometry parameters (n = 2 biological replicates, with technical replicates). In all panels, data indicate mean ± SD. *p < 0.05, **p < 0.01.
Methionine increases nucleotide biosynthesis
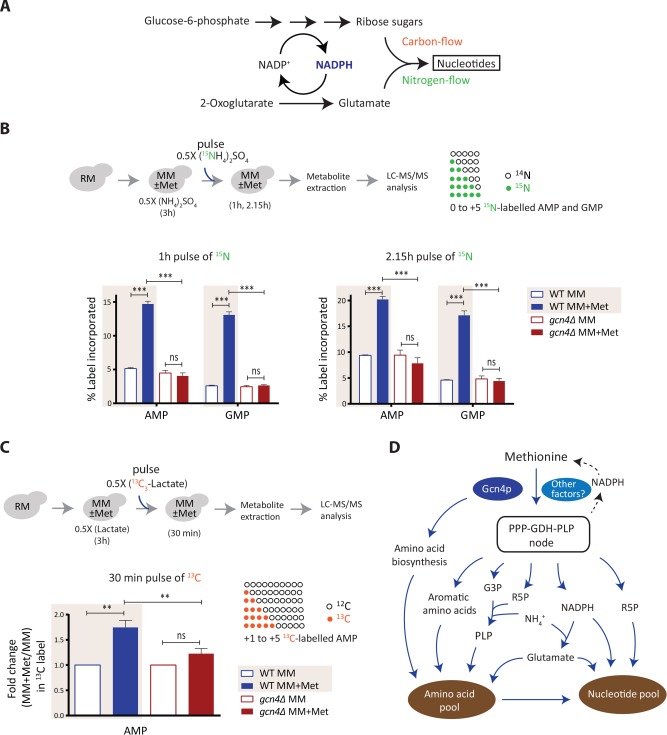
Given that the PPP and amino acid biosynthesis are directly regulated by methionine and GCN4, and the PPP metabolites and amino acids together couple to nucleotide synthesis, tuning this node should alter the rates of nucleotide biosynthesis. In principle, this will reflect the overall flux through the coupled steps of the PPP, glutamate/glutamine synthesis, and the use of intermediates from amino acid biosynthetic pathways for carbon and nitrogen assimilation into nucleotides (Figure 8A). Here the carbon skeleton of nucleotides comes partly from the PPP, the nitrogen base is derived from glutamine/glutamate and aspartate, and glutamate synthesis is itself coupled to NADPH from the PPP (Figure 8A). We therefore adopted a direct estimation of methionine- and GCN4-dependent increases in nucleotide synthesis (similarly to the approach in Figure 7), predicting increased de novo nucleotide synthesis due to methionine. To this end, using a stable-isotope-based nitrogen or carbon pulse labeling approach, coupled to targeted LC-MS/MS-based measurement of nucleotides, we separately measured the incorporation of the nitrogen and carbon label into nucleotides, as illustrated in Figure 8, B and C. We observed a strong increase in 15N-labeled nucleotides on the addition of methionine, in ∼1 h (Figure 8B and Supplemental Table 2). Furthermore, this methionine-mediated incorporation of 15N label in nucleotides was entirely GCN4 dependent (Figure 8B and Supplemental Figure 12).
FIGURE 8:
Methionine increases nucleotide biosynthesis. (A) Schematic showing carbon and nitrogen inputs in nucleotide biosynthesis and their coupling to the PPP/NADPH metabolism. (B) Methionine increases nucleotide biosynthesis in a GCN4-dependent manner. The WT and gcn4Δ cells treated and pulse-labeled with 15N ammonium sulfate as illustrated in the top panel. For all the labeled moieties, fractional increase of the incorporated label was calculated to measure newly synthesized AMP and GMP (also see Supplemental Figure 12 for cytidine 5’-monophosphate [CMP] and uridine 5’-monophosphate [UMP]) (n = 3). (C) Methionine enhances carbon flux into AMP biosynthesis. An experimental setup similar to that in B was employed, using 13C-lactate for carbon labeling. Label incorporation into nucleotides (from +1 to +5) was accounted for calculations. (Note: GMP could not be estimated because of MS/MS signal interference from unknown compounds in the metabolite extract) (n = 2 biological replicates, with technical replicates.) (D) A model illustrating how methionine triggers an anabolic program leading to cell proliferation. Methionine promotes the synthesis of PPP metabolites, PLP, NADPH, and glutamate (up-regulated genes in the pathways are shown in blue), which directly feed into nitrogen metabolism. As a result, methionine activates biosynthesis of amino acids and nucleotides, allowing the cells to grow in amino acid limiting medium. GCN4 is required to sustain this anabolic program. In all panels data indicate mean ± SD. ns: nonsignificant difference, **p < 0.01, ***p < 0.001.
Monitoring carbon flux is extremely challenging in a nonfermentable carbon source like lactate (as compared with glucose), given the difficulties of following the labeled carbon molecules. Despite that, like the 15N-labeling experiments described above, a similar experimental design was adopted to measure the 13C-label incorporation into adenosine 5’-monophosphate (AMP) (Figure 8C and Supplemental Table 2). We observed a significant increase in 13C-labeled AMP on the addition of methionine, and this methionine-dependent incorporation of 13C label in AMP was not observed in cells lacking Gcn4p (Figure 8C). Collectively, these data show a methionine- and GCN4-dependent increase in de novo synthesis of nucleotides, coupling carbon and nitrogen flux that is dependent on the PPP and glutamate synthesis. Note that the overall kinetics of incorporation of label are entirely in line with the predicted hierarchy. Increased amino acid labels (shown in Figure 7) were seen in ∼20-min postlabeled ammonium sulfate addition, while the nucleotide label increase occurs in ∼1 h, subsequent to the observed amino acid label increase. Thus, we directly demonstrate first the synthesis of new amino acids, and the subsequent synthesis of nucleotides, in a methionine- and GCN4-dependent manner.
DISCUSSION
In this study, we show how methionine drives cellular proliferation by metabolically rewiring cells to an anabolic state, even under otherwise amino acid–limited conditions. We propose a regulated, hierarchical activation of metabolic processes by methionine, which leads to overall anabolism. We also present a mechanism of how methionine mediates this anabolic program.
Starting with a global transcriptome analysis (Figure 1), we systematically build the underlying metabolic foundations of a methionine-mediated anabolic switch. Methionine mediates a global transcriptional remodeling in cells, which controls the anabolic program (Figures 1–3). To understand the core metabolic logic within this transcriptional response, we emphasized control points at rate- or resource-limiting biochemical steps, instead of exclusively relying on GO-based organization. The organizational metabolic logic that could be constructed was striking (Figures 2 and 3). First, methionine positively regulates the PPP (Figure 2). The PPP provides the pentose sugar backbones for nucleotides, along with reducing equivalents (NADPH), which can allow reductive biosynthesis for a variety of anabolic molecules (Nelson and Cox, 2017). Further, for amino acid and nucleotide synthesis, the outputs of pyridoxal phosphate-dependent transaminations (Eliot and Kirsch, 2004), as well as glutamate synthesis by glutamate dehydrogenase (GDH) is critical. Methionine directly induced this PPP-GDH-PLP node (Figure 2). This node provides the necessary substrates and/or cofactors for all the subsequent metabolic steps induced by methionine. Furthermore, in these subsequent metabolic steps (the synthesis of amino acids and nucleotides), methionine induces the expression of genes that control the most rate limiting or final steps (Figure 3). Notably, essentially every one of these (methionine regulated) steps use cofactors or intermediates from the PPP-GDH-PLP node (Figures 2 and 3). Thus, in this constructed metabolic hierarchy, methionine sits on top and controls the PPP-GDH-PLP node, which subsequently drives the generation of anabolic precursors, as illustrated in the schematics in Figures 3C and 8D.
The methionine-dependent growth, and the increased activity in these defined metabolic nodes (i.e., the overall anabolic program), requires GCN4 to be sustained (Figures 4 and 5). Gcn4p is traditionally understood as a regulator of amino acid biosynthesis during starvation (Natarajan et al., 2001). Indeed, many of the GCN4 targets picked up in our study compare well with the landmark study of GCN4 targets (Natarajan et al., 2001) (see Supplemental Figure 13 and Supplemental Table 3). A role for GCN4 in the presence of methionine in synchronously controlling this PPP-GDH-PLP node, coupled to amino acid and nucleotide biosynthesis, and thus sustaining anabolism has not been previously appreciated. Further, when methionine is present, in otherwise amino acid–limited conditions, the absence of GCN4 results in a collapse of the anabolic program, and therefore cells cannot sustain proliferation. This role for GCN4 in supporting proliferation is in contrast to its well-studied role in supporting survival during starvation, allowing a restoration of amino acid levels. Interestingly, GCN4 is required to sustain only the anabolic program induced by methionine and not for the induction of the translation machinery (as seen in Figures 4 and 5). The induction of translation due to methionine might be through other mechanisms, including activation of the TOR pathway (Sutter et al., 2013; Laxman et al., 2014a). Thus, there seems to be some separation of the methionine sensing machinery, the factors controling the translational response, and the effectors of the anabolic program (Gcn4p), suggesting a multicomponent transcriptional program to achieve overall methionine-dependent proliferation. Identifying other components of this overall proliferative program will be obvious directions of future studies.
A combination of rigorous biochemical and metabolic flux-based analysis using stable-isotopes directly demonstrate this hierarchical coupling of the PPP, NADPH utilization, and transamination reactions (in both nitrogen assimilation and carbon assimilation). This is shown first in the increased synthesis of aromatic and branch-chain amino acids and next in the synthesis of nucleotides in a methionine- and GCN4-dependent manner (Figures 7 and 8). Collectively, our data permit the construction of an overall structured hierarchy of metabolic events, mediated by methionine, to set up an anabolic program.
The central role of the PPP in anabolism is now textbook knowledge (Nelson and Cox, 2017). Yet, this importance of the PPP in mediating an anabolic rewiring is now being appreciated due to the association of the PPP in cancer metabolism (Cairns et al., 2011; Patra and Hay, 2014). While many anabolic transformations require contributions from the PPP, the metabolic cues regulating the PPP (and coupling to other processes) are not immediately obvious. Additionally, these studies ignore or underplay coincident but necessary metabolic events for proliferation, emphasizing only single-pathway metabolic reactions. Our study directly addresses how methionine (and possibly its downstream metabolite SAM) acts as an anabolic signal for cells through setting up of a hierarchical program, with the co-incident PPP-GDH-PLP node being critically important. This striking role of methionine regulating an anabolic program seems analogous to another central metabolite, acetyl-CoA, which is better known to determine cellular decisions toward growth (Cai et al., 2011; Shi and Tu, 2013; Comerford et al., 2014; Mariño et al., 2014; Pedro and Madeo, 2015; Krishna and Laxman, 2018). Correlations can be made from our observations to known roles of methionine in cancer cell metabolism, and metazoan growth. The earliest observations of methionine as a proliferative cue in some cancers dates back to the 1950s (Sugimura et al., 1959; Breillout et al., 1990; Cavuoto and Fenech, 2012), and several types of cancer cells are addicted to methionine (Halpern et al., 1974; Stern and Hoffman, 1986; Guo et al., 1993; Lu and Epner, 2000; Poirson-Bichat et al., 2000; Kokkinakis et al., 2001; Cellarier et al., 2003; Cavuoto and Fenech, 2012; Clarke et al., 2016). Other, distinct studies show that Drosophila fed on methionine-rich diets exhibit rapid growth, high fecundity, and shorter lifespans (Troen et al., 2007; Lee et al., 2014, 2016), all hallmarks of what a “growth signal” will do. Studies from yeast show that methionine inhibits autophagy and regulates the TORC1 to boost growth (Laxman et al., 2013, 2014b; Sutter et al., 2013). One of the earliest-known cell-cycle entry checkpoints is linked to methionine (Unger and Hartwell, 1976). Further, on sulfate (and thereby methionine) starvation, cells arrest their growth to promote survivability (Boer et al., 2008) and transform their proteome to preferentially express proteins containing fewer cysteine/methionine residues to save sulfur (Fauchon et al., 2002). There are other, less appreciated, connections of methionine metabolism and the PPP. Yeast cells lacking ZWF1 (encoding glucose-6-phosphate dehydrogenase, the first enzyme in the PPP) exhibit methionine auxotrophy (Thomas et al., 1991), and methionine supplementation also increases the oxidative stress tolerance of zwf1Δ (Campbell et al., 2016). Despite these studies highlighting a critical role of methionine, a hierarchical logic explaining the organizational principles of the anabolic program mediated by methionine has thus far been elusive. Our study provides this.
We observe that while the growth rate in MM + Met and MM + nonSAAs is similar, distinct metabolic adaptations operate when cells are limited for either methionine or all other nonSAAs. Methionine addition has a unique anabolic response targeted toward synthesis of all other amino acids and nucleotides, whereas expression profile in MM + nonSAAs is largely similar to MM with one striking difference, which is the up-regulation of genes involved in methionine biosynthesis. Although the nonSAAs, particularly glycine, aspartate, and glutamine, can feed into anaplerotic as well as nucleotide biosynthetic pathways, the proliferation rate of cells in MM + nonSAAs is lower than MM + Met + nonSAAs (Supplemental Figure 7B), further indicating that cells perceive methionine availability as a strong growth signal and its absence as a bottleneck for growth.
Our use of a “less-preferred” carbon source, lactate, has helped reveal regulatory phenomena otherwise hidden in glucose and amino acid–rich laboratory conditions, where a surfeit of costly metabolic resources (for example, unlimited PPP intermediates) are present. Tangentially, several recent reports emphasize the importance of lactate as a carbon source in rapidly proliferating cells (Kennedy et al., 2013; Faubert et al., 2017; Hu et al., 2017), and our observations might inform how proliferation is achieved in these conditions. Furthermore, the Gcn4p ortholog in mammals, Atf4, play important roles in cancer cell proliferation (Bi et al., 2005; Ye et al., 2010; Palam et al., 2015), where many cancers continue to grow in apparently poor nutrient environments. In methionine-rich (but otherwise amino acid limiting) conditions, Gcn4p/Atf4 might function to promote growth and not just help cells recover from nutrient stress. A separate question will be to understand how Gcn4p is itself regulated under these otherwise amino acid–limited conditions by methionine. Note that our studies would not have been possible without using prototrophic (“wild-type”) yeast strains to study responses to amino acids. Typically, studies utilize laboratory strains derived from an auxotrophic backgrounds (e.g., S288C/BY4741), which require supplemented uracil, histidine, leucine, and methionine for survival (Sherman, 1991; Brachmann et al., 1998; Pronk, 2002; Corbacho et al., 2011; Gerashchenko and Gladyshev, 2014; Alam et al., 2016), and where therefore overall amino acid homeostasis is severely altered. This precludes systematic experiments with amino acid limitation, such as those in this study.
We close by suggesting a possible metabolic cost-based hypothesis for what might make methionine a strong growth cue. The de novo synthesis of methionine and its immediate metabolites (notably SAM) is exceptionally costly in terms of NADPH molecules invested (Thomas and Surdin-Kerjan, 1997; Mampel et al., 2005; Kaleta and Schäuble, 2013; Sutter et al., 2013). Cells require at least six molecules of NADPH to reduce sulfur and synthesize a single molecule of methionine. Since biology has tied multiple anabolic processes to reductive biosynthesis (dependent on NADPH from the PPP), the availability of methionine might be an ancient signal to represent a metabolic state where reductive equivalents are sufficiently available for all other reductive biosynthetic processes as a whole.
MATERIALS AND METHODS
Yeast strains and growth media
The prototrophic CEN.PK strain (WT) was used in all experiments (van Dijken et al., 2000). Strains with gene deletions or chromosomally tagged proteins (at the C-terminus) were generated as described. Strains used in this study are listed in Supplemental Table 1.
The growth media used in this study are RM (1% yeast extract, 2% peptone, and 2% lactate) and MM (0.17% yeast nitrogen base without amino acids, 0.5% ammonium sulfate, and 2% lactate). All amino acids were supplemented at 2 mM. NonSAAs refers to the mixture of all standard amino acids (2 mM each) except methionine, cysteine, and tyrosine.
The indicated strains were grown in RM with repeated dilutions (∼36 h), and the culture in the log phase (absorbance at 600 nm of ∼1.2) was subsequently switched to MM, with or without addition of the indicated amino acids. For growth curves, the RM acclimatized cultures were used and diluted in a fresh medium with the starting absorbance of ∼0.2 and the growth was monitored at the indicated time intervals.
Western blot analysis
Approximately 10 OD600 cells were collected from respective cultures, pelleted, and flash-frozen in liquid nitrogen until further use. The cells were resuspended in 400 μl of 10% trichloroacetic acid and lysed by bead-beating three times (30 s of beating and then 1 min of cooling on ice). The precipitates were collected by centrifugation, resuspended in 400 μl of SDS–glycerol buffer (7.3% SDS, 29.1% glycerol, and 83.3 mM Tris base), and heated at 100°C for 10 min. The supernatant after centrifugation was treated as the crude extract. Protein concentrations from extracts were estimated using bicinchoninic acid assay (Thermo Scientific). Equal amounts of samples were resolved on 4 to 12% Bis–Tris gels (Invitrogen). Coomassie blue–stained gels were used as loading controls. Western blots were developed using the antibodies against the respective tags. We used the following primary antibodies: monoclonal FLAG M2 (Sigma) and HA (12CA5, Roche). Horseradish peroxidase–conjugated secondary antibodies (mouse and rabbit) were obtained from Sigma. For Western blotting, standard enhanced chemiluminescence reagents (GE Healthcare) were used. ImageJ was used for quantification.
Immunofluorescence measurements
Yeast cells were fixed with 3.7% formaldehyde, washed, and resuspended in spheroplasting buffer (40 mM potassium phosphate buffer, pH 6.5; 0.5 mM MgCl2; 1.2 M sorbitol). Spheroplasts were prepared by zymolyase (MP Biomedicals; 08320921) treatment and spread on a slide pretreated with 50 μl of 1 mg/ml polylysine (Sigma-Aldrich; P6407). Gcn4-HA was stained with the mouse monoclonal anti-HA (12CA5) primary antibody (Roche; 11583816001) and Alexa Fluor 488-conjugated Goat anti-Mouse IgG (H + L) secondary antibody (Thermofisher; A32723). DNA was stained with 1 μg/ml 4’,6-diamidino-2-phenylindole (DAPI) for 2 min, washed, and mounted in Fluoromount-G (Southern Biotech; 0100-01). The cells were imaged using Olympus FV1000 confocal microscope.
RNA-seq analysis
Total RNA from yeast cells was extracted using hot acid phenol method (Collart and Oliviero, 2001). The quality of RNA was checked on Bioanalyzer using an RNA 6000 Nano kit (Agilent), and the libraries were prepared using the TruSeq RNA library preparation kit V2 (Illumina). The samples were sequenced on Illumina platform HiSeq2500. The raw data are available with NCBI-SRA under the accession number SRP101768. Genome and the annotation files of S. cerevisiae S288C strain were downloaded from the Saccharomyces Genome Database (SGD; www.yeastgenome.org/). The 100-mer, single-end reads obtained from RNA sequencing experiments were mapped to the S288C genome using Burrows Wheeler Aligner (Li and Durbin, 2009). Mapped reads with the mapping quality of ≥20 were used for the further analysis. The number of reads mapped to each gene was calculated, and the read count matrix was generated. The read count matrix was fed into EdgeR, a Bioconductor package used for analyzing differential gene expression (Robinson et al., 2010). Genes which are differentially expressed by at least threefold with the p value of <0.0001 were considered for further analysis. Normalized gene expression was calculated by dividing the number of reads by the gene length and the total number of reads for those samples, then dividing each of these values with the mode of its distribution (Srinivasan et al., 2013). Normalized expression levels of the genes between the replicates are well correlated with the Pearson correlation coefficient (R) values more than 0.99 (see Supplemental Figure 1). Mapping of genes to the related pathways and gene ontology analysis were carried out using public databases such as Yeastcyc (Caspi et al., 2014), GeneCodis (Carmona-Saez et al., 2007; Nogales-Cadenas et al., 2009; Tabas-Madrid et al., 2012), and SGD (Dwight et al., 2002).
Metabolite extractions and measurements by LC-MS/MS
Cells were grown in RM for ∼36 h and transferred to MM with and without methionine for the indicated time. After incubation, cells were rapidly harvested, and metabolite was extracted as described earlier (Tu et al., 2007). Metabolites were measured using the liquid chromatography (LC)-mass spectrometry (MS) methods described earlier (Laxman et al., 2014b; Walvekar et al., 2018). Standards were used for developing multiple reaction monitoring methods on Thermo Scientific TSQ Vantage triple stage quadrupole mass spectrometer or Sciex QTRAP 6500. For the positive-polarity mode, metabolites were separated using a Synergi 4μ Fusion-RP 80A column (150 × 4.6 mm, Phenomenex) on Agilent’s 1290 infinity series UHPLC system coupled to mass spectrometer. Buffers used for separation were: buffer A: 99.9% H2O/0.1% formic acid and buffer B: 99.9% methanol/0.1% formic acid (flow rate, 0.4 ml/min; T = 0 min, 0% B; T = 3 min, 5% B; T = 10 min, 60% B; T = 10.1 min, 80% B; T = 12 min, 80% B; T = 14 min, 5% B; T = 15 min, 0% B; T = 20 min, stop). For the negative-polarity mode, metabolites were separated using a Luna HILIC 200A column (150 × 4.6 mm, Phenomenex). Buffers used for separation were as follows: buffer A, 5 mM ammonium formate in H2O, and buffer B, 100% acetonitrile (flow rate: 0.4 ml/min; T = 0 min, 95% B; T = 1 min, 40% B; T = 7 min, 10% B; T = 11 min, 1% B; T = 13 min, 95% B; T = 17 min, stop). The area under each peak was calculated using either AB SCIEX MultiQuant software 3.0.1 or Thermo Xcalibur software 2.2 SP1.48 (Qual and Quan browsers).
15N- and 13C-based metabolite labeling experiments
For detecting 15N-label incorporation in amino acids and nucleotides, 15N-ammonium sulfate with all nitrogens labeled (Sigma-Aldrich) was used. For 13C-labeling experiment, 13C-lactate with all carbons labeled (Cambridge Isotope Laboratories) was used. In the labeling experiments, 0.5X refers to 0.25% ammonium sulfate or 1% lactate. All the parent/product masses measured are enlisted in Supplemental Table 2. Amino acid measurements were done in the positive-polarity mode. For all the nucleotide measurements, release of the nitrogen base was monitored in the positive-polarity mode. For the 13C-label experiment, the phosphate release was monitored in the negative-polarity mode. Under these conditions, the nitrogen base release cannot be monitored here, as the nitrogen base itself has carbon skeleton, which will complicate the analysis. The HPLC and MS/MS protocol was similar to those explained above.
GDH assays
Glutamate dehydrogenase activity was measured as described in Doherty (1970), with some modifications. Yeast cells were lysed by bead beating in lysis buffer (100 mM potassium phosphate buffer, pH 7; 5% glycerol; 1 mM phenylmethylsulfonyl fluoride (PMSF); 0.1% Tween-20; 1 mM EDTA; 1 mM 2-mercaptoethanol). NADP-dependent activity was measured by monitoring oxidation of NADPH (assay buffer: 100 mM Tris–HCl, pH 7.2; 10 mM 2-ketoglutarate, pH adjusted to 7.2; 100 mM ammonium chloride; 0.1 mM NADPH) at 340 nm. Protein concentrations from extracts were estimated using bicinchoninic acid assay (Thermo Scientific). One enzyme unit corresponds to the amount of enzyme required to oxidize one µmol of NADPH min-1 at room temperature.
Statistical analysis
In most experiments, Student’s t test or Fisher’s exact test was applied for calculating the p values (as indicated). Wherever necessary, other tests were applied and indicated accordingly.
Supplementary Material
Acknowledgments
We acknowledge Dhananjay Shinde, Padma Ramakrishnan, and the NCBS/inStem/C-CAMP mass spectrometry facility for LC-MS/MS support and Avadheesh Pandit and the NCBS/inStem/C-CAMP next-generation sequencing facility for assistance with library preparation. We thank Utpal Banerjee, Mark Sharpley, Marco Foiani, Christopher Bruhn, Krishnamurthy Natarajan, Arati Ramesh, and P. J. Bhat for critical comments on this article. This work was supported by a Wellcome Trust-DBT IA intermediate fellowship (IA/I/14/2/501523), grant BT/PR13446/COE/34/30/2015 from the Department of Biotechnology, Government of India, and inStem/DBT institutional support to S.L. A.S.W., R.G., and R.S acknowledge bridging fellowships (from inStem), and A.S.W., R.G., and R.S. are supported by Department of Science and Technology and Science and Engineering Research Board (DST-SERB) national postdoctoral fellowships (PDF/2015/000225, PDF/ 2016/000416, and PDF/2016/001877, respectively).
Abbreviations used:
- AMP
adenosine 5’-monophosphate
- CMP
cytidine 5’-monophosphate
- GDH
glutamate dehydrogenase
- GMP
guanosine 5’-monophosphate
- GO
gene ontology
- HA
human influenza hemagglutinin
- LC
liquid chromatography
- Met
methionine
- MM
minimal medium
- MS
mass spectrometry
- NADPH
nicotinamide adenine dinucleotide phosphate
- nonSAAs
nonsulfur amino acids
- PLP
pyridoxal-5-phosphate
- PPP
pentose phosphate pathway
- RM
rich medium
- SAM
S-adenosylmethionine
- TCA
tricarboxylic acid
- TOR
target of rapamycin
- UMP
uridine 5’-monophosphate
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0515) on October 24, 2018.
REFERENCES
- Alam MT, Zelezniak A, Mülleder M, Shliaha P, Schwarz R, Capuano F, Vowinckel J, Radmanesfahar E, Krüger A, Calvani E, et al. (2016). The metabolic background is a global player in Saccharomyces gene expression epistasis. Nat Microbiol , 15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MD, Delneri D, Oliver SG, Rattray M, Bergman CM. (2010). Evolutionary systems biology of amino acid biosynthetic cost in yeast. PLoS One , e11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia J, Raleigh J, et al. (2005). ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J , 3470–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D. (2008). Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci USA , 6930–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. (2010). Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol Biol Cell , 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast , 115–132. [DOI] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. (2008). Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell , 352–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breillout F, Antoine E, Poupon MF. (1990). Methionine dependency of malignant tumors: a possible approach for therapy. J Natl Cancer Inst , 1628–1632. [DOI] [PubMed] [Google Scholar]
- Broach JR. (2012). Nutritional control of growth and development in yeast. Genetics , 73–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. (2011). Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell , 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RRA, Harris IS, Mak TWT. (2011). Regulation of cancer cell metabolism. Nat Rev Cancer , 85–95. [DOI] [PubMed] [Google Scholar]
- Campbell K, Vowinckel J, Keller MA, Ralser M. (2016). Methionine metabolism alters oxidative stress resistance via the pentose phosphate pathway. Antioxid Redox Signal , 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. (2007). GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol , R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, et al. (2014). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res , D459–D471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavuoto P, Fenech MF. (2012). A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat Rev , 726–736. [DOI] [PubMed] [Google Scholar]
- Cellarier E, Durando X, Vasson MP, Farges MC, Demiden A, Maurizis JC, Madelmont JC, Chollet P. (2003). Methionine dependency and cancer treatment. Cancer Treat Rev , 489–499. [DOI] [PubMed] [Google Scholar]
- Clarke CJ, Berg TJ, Birch J, Ennis D, Mitchell L, Cloix C, Campbell A, Sumpton D, Nixon C, Campbell K, et al. (2016). The initiator methionine tRNA drives secretion of type II collagen from stromal fibroblasts to promote tumor growth and angiogenesis. Curr Biol , 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Oliviero S. (2001). Preparation of yeast RNA. In: Current Protocols in Molecular Biology, Hoboken, NJ: John Wiley & Sons. [DOI] [PubMed] [Google Scholar]
- Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, Walters H, Tantawy MN, Fu A, Manning HC, et al. (2014). Article acetate dependence of tumors. Cell , 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbacho I, Teixidó F, Velázquez R, Hernández LM, Olivero I. (2011). Standard YPD, even supplemented with extra nutrients, does not always compensate growth defects of Saccharomyces cerevisiae auxotrophic strains. Antonie Van Leeuwenhoek , 591–600. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Sayed N, Ditsworth D, Thompson CB. (2008). Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev , 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R, Peter M. (2008). Nutrient signals driving cell growth. Curr Opin Cell Biol , 678–687. [DOI] [PubMed] [Google Scholar]
- Doherty D. (1970). l-Glutamate dehydrogenases (yeast). Methods Enzymol , 850–856. [Google Scholar]
- Dong Y-X, Sueda S, Nikawa J-I, Kondo H. (2004). Characterization of the products of the genes SNO1 and SNZ1 involved in pyridoxine synthesis in Saccharomyces cerevisiae. Eur J Biochem , 745–752. [DOI] [PubMed] [Google Scholar]
- Dwight SS, Harris MA, Dolinski K, Ball CA, Binkley G, Christie KR, Fisk DG, Issel-Tarver L, Schroeder M, Sherlock G, et al. (2002). Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic Acids Res , 69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliot AC, Kirsch JF. (2004). Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem , 383–415. [DOI] [PubMed] [Google Scholar]
- Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, et al. (2017). Lactate metabolism in human lung tumors. Cell , 358–371. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J. (2002). Sulfur sparing in the yeast proteome in response to sulfur demand mire. Mol Cell , 713–723. [DOI] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. (2014). Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res , e134–e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A, Hall MN. (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J , e201696010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. (2004). “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev , 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Orozco JM, Saxton RA, Condon KJ, Liu GY, Krawczyk PA, Scaria SM, Harper JW, Gygi SP, Sabatini DM. (2017). SAMTOR is an S -adenosylmethionine sensor for the mTORC1 pathway. Science , 813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HY, Herrera H, Groce A, Hoffman RM. (1993). Expression of the biochemical defect of methionine dependence in fresh patient tumors in primary histoculture. Cancer Res , 2479–2483. [PubMed] [Google Scholar]
- Halpern BC, Clark BR, Hardy DN, Halpern RM, Smith RA. (1974). The effect of replacement of methionine by homocystine on survival of malignant and normal adult mammalian cells in culture. Proc Natl Acad Sci USA , 1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. (2005). Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol , 407–450. [DOI] [PubMed] [Google Scholar]
- Hu X, Chao M, Wu H. (2017). Central role of lactate and proton in cancer cell resistance to glucose deprivation and its clinical translation. Signal Transduct Target Ther , 16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M. (2004). How cells coordinate growth and division. Curr Biol , R1014–R1027. [DOI] [PubMed] [Google Scholar]
- Kaleta C, Schäuble S. (2013). Metabolic costs of amino acid and protein production in Escherichia coli. Biotechnol J , 1105–1114. [DOI] [PubMed] [Google Scholar]
- Kennedy KM, Scarbrough PM, Ribeiro A, Richardson R, Yuan H, Sonveaux P, Landon CD, Chi J-T, Pizzo S, Schroeder T, et al. (2013). Catabolism of exogenous lactate reveals it as a legitimate metabolic substrate in breast cancer. PLoS One , e75154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosinska MM, Crutchfield CA, Bradley PH, Rabinowitz JD, Broach JR. (2011). Yeast cells can access distinct quiescent states. Genes Dev , 336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkinakis DM, Hoffman RM, Frenkel EP, Wick JB, Han Q, Xu M, Tan Y, Schold SC. (2001). Synergy between methionine stress and chemotherapy in the treatment of brain tumor xenografts in athymic mice. Cancer Res , 4017–4023. [PubMed] [Google Scholar]
- Krishna S, Laxman S. (2018). A minimal “push–pull” bistability model explains oscillations between quiescent and proliferative cell states. Mol Biol Cell , 2243–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Shi L, Tu BP. (2014b). Npr2 inhibits TORC1 to prevent inappropriate utilization of glutamine for biosynthesis of nitrogen-containing metabolites. Sci Signal , ra120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Tu BP. (2014a). Methionine is a signal of amino acid sufficiency that inhibits autophagy through the methylation of PP2A. Autophagy , 386–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. (2013). Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell , 416–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Tu BP. (2010). Systems approaches for the study of metabolic cycles in yeast. Curr Opin Genet Dev , 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BC, Kaya A, Gladyshev VN. (2016). Methionine restriction and life-span control. Ann NY Acad Sci , 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BC, Kaya A, Ma S, Kim G, Gerashchenko MV, Yim SH, Hu Z, Harshman LG, Gladyshev VN. (2014). Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat Commun , 3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics , 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl PO, Daignan-Fornier B. (2012). Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics , 885–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Epner DE. (2000). Molecular mechanisms of cell cycle block by methionine restriction in human prostate cancer cells. Nutr Cancer , 123–130. [DOI] [PubMed] [Google Scholar]
- Mampel J, Schröder H, Haefner S, Sauer U. (2005). Single-gene knockout of a novel regulatory element confers ethionine resistance and elevates methionine production in Corynebacterium glutamicum. Appl Microbiol Biotechnol , 228–236. [DOI] [PubMed] [Google Scholar]
- Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al. (2014). Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell , 710–725. [DOI] [PubMed] [Google Scholar]
- Mulleder M, Calvani E, Alam MT, Wang RK, Eckerstorfer F, Zelezniak A, Ralser M. (2016). Functional metabolomics describes the yeast biosynthetic regulome. Cell , 553–565. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. (2001). Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol , 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Cox M. (2017). Principles of Biochemistry, New York: W. H. Freeman. [Google Scholar]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. (2009). GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res , W317–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palam LR, Gore J, Craven KE, Wilson JL, Korc M. (2015). Integrated stress response is critical for gemcitabine resistance in pancreatic ductal adenocarcinoma. Cell Death Dis , e1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra KC, Hay N. (2014). The pentose phosphate pathway and cancer. Trends Biochem Sci , 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedro MB, Madeo F. (2015). Review acetyl coenzyme A: a central metabolite and second messenger. Cell Metab , 805–821. [DOI] [PubMed] [Google Scholar]
- Poirson-Bichat F, Gonçalves RA, Miccoli L, Dutrillaux B, Poupon MF. (2000). Methionine depletion enhances the antitumoral efficacy of cytotoxic agents in drug-resistant human tumor xenografts. Clin Cancer Res , 643–653. [PubMed] [Google Scholar]
- Pronk JT. (2002). Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol , 2095–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics , 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (1991). Getting started with yeast. Methods Enzymol , 3–21. [DOI] [PubMed] [Google Scholar]
- Shi L, Tu BP. (2013). Acetyl-CoA induces transcription of the key G1 cyclin CLN3 to promote entry into the cell division cycle in Saccharomyces cerevisiae. Proc Natl Acad Sci USA , 7318–7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Botstein D. (2011). Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol Biol Cell , 1997–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Botstein D. (2013). Decoupling nutrient signaling from growth rate causes aerobic glycolysis and deregulation of cell size and gene expression. Mol Biol Cell , 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Chandraprakash D, Krishnamurthi R, Singh P, Scolari VF, Krishna S, Seshasayee ASN. (2013). Genomic analysis reveals epistatic silencing of “expensive” genes in Escherichia coli K-12. Mol Biosyst , 2021–2033. [DOI] [PubMed] [Google Scholar]
- Stern PH, Hoffman RM. (1986). Enhanced in vitro selective toxicity of chemotherapeutic agents for human cancer cells based on a metabolic defect. J Natl Cancer Inst , 629–639. [DOI] [PubMed] [Google Scholar]
- Sugimura T, Birnbaum SM, Winitz M, Greenstein JP. (1959). Quantitative nutritional studies with water-soluble, chemically defined diets. VIII. The forced feeding of diets each lacking in one essential amino acid. Arch Biochem Biophys , 448–455. [DOI] [PubMed] [Google Scholar]
- Sutter BM, Wu X, Laxman S, Tu BP. (2013). Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell , 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. (2012). GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res , W478–W483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Cherest H, Surdin-Kerjan Y. (1991). Identification of the structural gene for glucose-6-phosphate dehydrogenase in yeast. Inactivation leads to a nutritional requirement for organic sulfur. EMBO J , 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y. (1997). Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev , 503–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB. (2009). The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev , 32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troen AM, French EE, Roberts JF, Selhub J, Ordovas JM, Parnell LD, Lai C-Q. (2007). Lifespan modification by glucose and methionine in Drosophila melanogaster fed a chemically defined diet. Age (Dordr) , 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. (2005). Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science , 1152–1158. [DOI] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. (2007). Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci USA , 16886–16891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger MW, Hartwell LH. (1976). Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. Proc Natl Acad Sci USA , 1664–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB, Vander Heiden MG, Cantley LC, Thompson CB. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science , 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin MLF, Heijnen JJ, Hoare M, Lange HC, et al. (2000). An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol , 706–714. [DOI] [PubMed] [Google Scholar]
- Walvekar A, Rashida Z, Maddali H, Laxman S. (2018). A versatile LC-MS/MS approach for comprehensive, quantitative analysis of central metabolic pathways [version 1; referees: 2 approved]. Wellcome Open Res , 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. (1956). On the origin of cancer cells. Science , 309–314. [DOI] [PubMed] [Google Scholar]
- Warner JR. (1999). The economics of ribosome biosynthesis in yeast. Trends Biochem Sci , 437–440. [DOI] [PubMed] [Google Scholar]
- Warner JR, Vilardell J, Sohn JH. (2001). Economics of ribosome biosynthesis. Cold Spring Harb Symp Quant Biol , 567–574. [DOI] [PubMed] [Google Scholar]
- Wolfson RL, Sabatini DM. (2017). The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab , 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tu BP. (2011). Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol Biol Cell , 4124–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-F, Létisse F, Absalan F, Lu W, Kuznetsova E, Brown G, Caudy AA, Yakunin AF, Broach JR, Rabinowitz JD. (2013). Nucleotide degradation and ribose salvage in yeast. Mol Syst Biol , 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Sutter BM, Wang Y, Kuang Z, Tu BP. (2017). A metabolic function for phospholipid and histone methylation. Mol Cell , 180–193. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. (2010). The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J , 2082–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.