Abstract
Heat shock factor 1 is the master transcriptional regulator of molecular chaperones and binds to the same cis-acting heat shock element (HSE) across the eukaryotic lineage. In budding yeast, Hsf1 drives the transcription of ∼20 genes essential to maintain proteostasis under basal conditions, yet its specific targets and extent of inducible binding during heat shock remain unclear. Here we combine Hsf1 chromatin immunoprecipitation sequencing (seq), nascent RNA-seq, and Hsf1 nuclear depletion to quantify Hsf1 binding and transcription across the yeast genome. We find that Hsf1 binds 74 loci during acute heat shock, and these are linked to 46 genes with strong Hsf1-dependent expression. Notably, Hsf1’s induced DNA binding leads to a disproportionate (∼7.5-fold) increase in nascent transcription. Promoters with high basal Hsf1 occupancy have nucleosome-depleted regions due to the presence of “pioneer factors.” These accessible sites are likely critical for Hsf1 occupancy as the activator is incapable of binding HSEs within a stably positioned, reconstituted nucleosome. In response to heat shock, however, Hsf1 accesses nucleosomal sites and promotes chromatin disassembly in concert with the Remodels Structure of Chromatin (RSC) complex. Our data suggest that the interplay between nucleosome positioning, HSE strength, and active Hsf1 levels allows cells to precisely tune expression of the proteostasis network.
INTRODUCTION
The cellular response to thermal stress is directed by a transcriptional program whose basic components—sequence-specific activator, cis-response DNA element, and core target genes—have been conserved since the last common ancestor in the eukaryotic lineage (Verghese et al., 2012). Heat shock factor 1 (Hsf1), master activator of the heat shock response, is a winged helix-turn-helix transcription factor that recognizes DNA sequence motifs (heat shock elements) located upstream of genes encoding chaperones and other cytoprotective heat shock proteins (HSPs) (Wu, 1995; Gomez-Pastor et al., 2018). HSPs maintain cellular protein homeostasis (proteostasis) and are required in higher concentrations under stressful conditions to contend with unfolded cytosolic and nucleoplasmic proteins. This is principally achieved via Hsf1-mediated transcriptional up-regulation of the genes encoding these proteins. In addition to its evolutionarily conserved role, mammalian HSF1 contributes to oncogenesis by driving distinct transcriptional programs in both tumor cells and their supporting stroma (Dai et al., 2007; Mendillo et al., 2012; Scherz-Shouval et al., 2014). HSF1 function also has been linked to normal development, neurodegenerative disease, and aging (Neef et al., 2011; Li et al., 2017). Thus, gaining a deeper understanding of Hsf1 biology may inform development of novel approaches to modulate human HSF1.
Hsf1 is subject to multiple layers of regulation, with two shared across multiple phyla. Under nonstressful conditions, Hsf1 exists primarily as a non-DNA-binding monomer in either nucleus or cytoplasm. In response to thermal or other proteotoxic stress, the protein trimerizes and acquires the capacity for high-affinity DNA binding. In higher eukaryotes, the Hsp90 and Hsp70 chaperones (and their cochaperones) as well as the TRiC/CCT cochaperone complex have been suggested as direct repressors of Hsf1 under basal conditions (Abravaya et al., 1992; Zou et al., 1998; Neef et al., 2014). According to this model, the chaperones are titrated away on heat shock by unfolded or misfolded proteins, allowing Hsf1 to trimerize, bind its cognate HSEs, and transactivate HSP genes. Although certain details of Hsf1 regulation differ between metazoans and budding yeast (Sorger and Nelson, 1989; Liu et al., 1997), it was recently shown that Hsp70 binds yeast Hsf1 and negatively regulates it. Hsp70 transiently releases Hsf1 in response to thermal stress and then rebinds on exposure to sustained stress, thereby constituting a two-component feedback loop (Zheng et al., 2016; Krakowiak et al., 2018). Hsf1 has also been suggested to be the downstream effector of various signaling cascades. In the case of yeast, phosphorylation positively tunes Hsf1’s transactivation of target genes independent of chaperone regulation (Zheng et al., 2016).
Hsf1-regulated, heat shock-responsive genes have served as a paradigm for understanding basic mechanisms of transcription. For example, the existence of paused RNA polymerase II (Pol II) at the 5′-end of genes was first identified at Drosophila HSP70 (Rougvie and Lis, 1988). Hsf1-regulated genes in Saccharomyces cerevisiae use both SAGA and TFIID pathways for activation, although SAGA plays a dominant role (Ghosh and Pugh, 2011; de Jonge et al., 2017; Vinayachandran et al., 2018). In addition, Mediator has been shown to be a key coactivator of Hsf1-driven transcription in both metazoans and yeast (Park et al., 2001; Fan et al., 2006; Singh et al., 2006; Kim and Gross, 2013). Dynamic genewide eviction of nucleosomes and their subsequent redeposition has been observed at HSP genes in both yeast and Drosophila (Zhao et al., 2005; Petesch and Lis, 2008; Kremer and Gross, 2009). Recently, Hsf1-regulated yeast genes were observed to undergo striking alteration in their local structure and engage in highly specific cis- and trans-intergenic interactions on their transcriptional activation, coalescing into discrete intranuclear foci (Chowdhary et al., 2017, 2018).
In budding yeast, the Msn2 and Msn4 gene-specific transcription factors drive transcription of a large set of genes (200–300) in response to a variety of environmental stresses, including heat, oxidative, osmotic, and salt stress (Gasch et al., 2000; Elfving et al., 2014). Msn2/Msn4-regulated genes include several HSP genes, but the Msn2/Msn4 regulon is by and large distinct from that controlled by Hsf1. Nonetheless, the identity of the genes whose acutely induced expression is under the direct control of Hsf1 remains unclear. Prior global localization studies of Hsf1 have either lacked resolution or did not evaluate Hsf1 occupancy beyond the basal or chronically induced states (Lee et al., 2002; Hahn et al., 2004; Solis et al., 2016; de Jonge et al., 2017).
Here we investigate the link between Hsf1 occupancy, its function, and the epigenetic determinants underpinning its genome localization under basal, acute, and chronic heat shock states. Our results reveal that Hsf1 binds to a core set of 43 loci under control conditions and that the vast majority of these and 31 others are occupied at substantially higher levels following heat shock; of these 74 bound loci, 46 are associated with genes whose transcriptional activation is Hsf1 dependent. Additionally, our analysis reveals a central role played by preset nucleosome positioning and Remodels Structure of Chromatin (RSC) complex in regulating yeast’s dynamic transcriptional response to heat shock.
RESULTS
Yeast Hsf1 binds inducibly to the vast majority of its target genomic sites
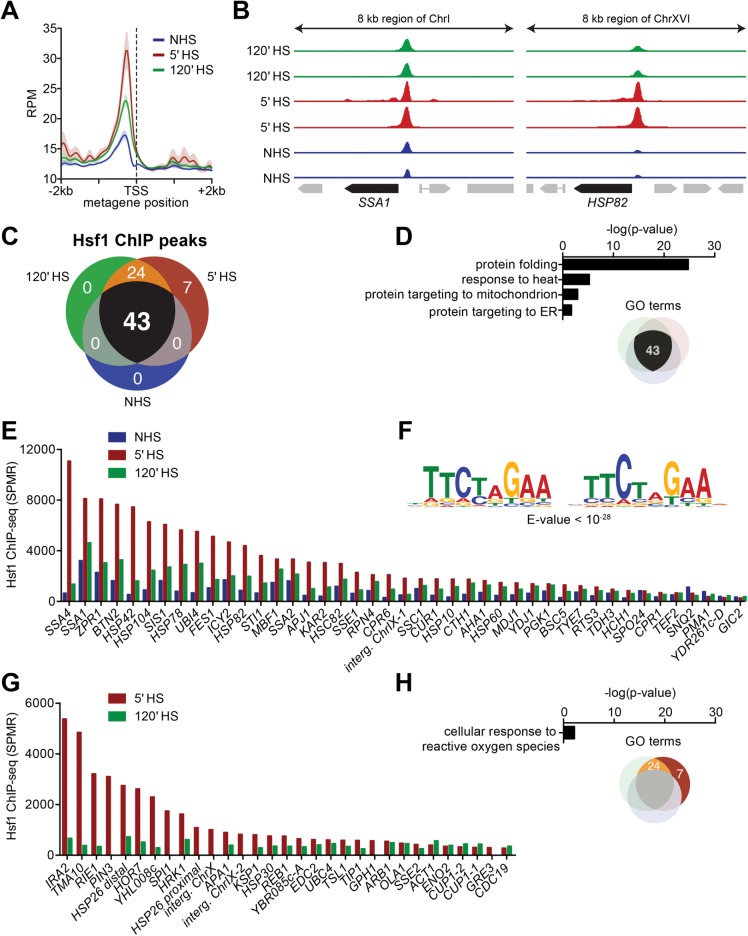
To investigate Hsf1 DNA binding genomewide, we performed chromatin immunoprecipitation sequencing (ChIP-seq) to determine where and how strongly it binds under basal (non–heat shock [NHS]), acute heat shock (HS), and chronic heat shock states. To circumvent problems typically encountered when quantifying ChIP-seq data that combine dynamic protein binding with high coverage over a small genome, we performed parallel IPs using both anti-Hsf1 serum as well as preimmune serum at all time points, and we generated paired-end sequencing data to determine the full sequence of the captured fragments (see Materials and Methods). Analytically, we subtracted signal from the matched preimmune samples, used only properly paired reads, and allowed for duplicate reads when quantifying fragment pileups. Such an approach revealed that Hsf1 binding could be detected under NHS conditions (30°C) and was centered ∼200 base pairs upstream of the transcription start site (TSS) (Figure 1A). In cells exposed to acute heat shock (30° to 39°C shift for 5 min), Hsf1 occupancy increased at least fourfold genomewide and remained elevated in cells chronically exposed (2 h) to the higher temperature. Replicate locus-specific and genomewide occupancy profiles are provided in Figure 1B and Supplemental Figure S1A, which illustrate both the specificity and reproducibility of our ChIP-seq analysis.
FIGURE 1:
Hsf1 ChIP-seq reveals differential basal and heat-shock-inducible binding across the Hsf1 regulon. (A) Metagene plot of Hsf1 ChIP-seq signal genomewide with respect to the TSS under NHS conditions and following 5-min and 120-min HS. Strain BY4741 was used for this and all other Hsf1 ChIP-seq assays in this work. RPM, reads per million mapped reads. (B) IGV browser images of the SSA1 and HSP82 loci showing Hsf1 ChIP-seq signal in two biological replicates under NHS conditions and following 5-min and 120-min HS. The y-axes are normalized to the maximum displayed signal in the 5-min time point. Preimmune serum ChIP samples (not shown) were used for peak calling (see Materials and Methods). (C) Venn diagram showing the number of Hsf1 ChIP peaks that surpassed the background cutoff in both biological replicates under each condition. (D) Gene ontology (GO) term enrichment values for genes immediately downstream of the 43 ChIP peaks. (E) Normalized Hsf1 ChIP signal at the 43 peaks identified under all three conditions; represent the mean of two biological replicates. SPMR, signal per million mapped reads. (F) Consensus motif identified under Hsf1 ChIP peaks detected under all conditions. (G) Normalized Hsf1 ChIP signal at the 31 peaks detected only under HS conditions; presented as in E. (H) GO term enrichment values for the 31 Hsf1 ChIP peaks detected only under HS conditions.
Two sets of Hsf1 genomic targets were identified. A core set, composed of 43 loci, was occupied under all conditions, with occupancy typically increased in cells exposed to thermal stress (Figure 1, C and E). A second set, composed of 31 loci, showed subthreshold occupancy under NHS conditions but was inducibly occupied following acute HS; of these, 24 remained Hsf1 bound in cells chronically exposed to thermal stress (Figure 1, C and G). We performed locus-specific chromatin immunoprecipitation (ChIP) to validate examples of each (Supplemental Figure S1B and unpublished data). The core set of Hsf1 targets was enriched for proteostasis components in the cytosol, endoplasmic reticulum (ER), and mitochondria (Figure 1D). Notably, within these 43 core targets, we observed strong inducible binding with a broad range of inducible levels—up to 50-fold at select sites (Figure 1E). Indeed, all but five core targets exhibited elevated occupancy. Thus, yeast Hsf1 inducibly binds 69/74 of its genomic targets on heat shock.
Analysis of the regions occupied by Hsf1 pulled out a sequence of 20 base pairs composed of tandem inverted repeats of NTTCT as the most enriched motif (Figure 1F). This motif is consistent with previous characterization of HSEs from other organisms (Xiao et al., 1991; Leach et al., 2016; Vihervaara et al., 2017). Among the heat-shock-only Hsf1 binding sites, there was a modest enrichment for oxidative stress-responsive genes (Figure 1H) but no enrichment for an alternative binding motif.
DNA-bound Hsf1 is differentially active during basal and acute heat shock states
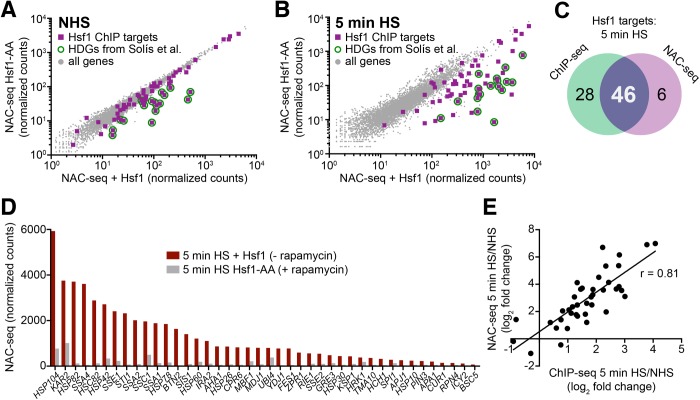
As previous genomewide studies (Lee et al., 2002; Hahn et al., 2004; Eastmond and Nelson, 2006; Solis et al., 2016; de Jonge et al., 2017) failed to define the Hsf1-dependent repertoire of target genes during acute heat shock, we were interested in knowing whether genes with strong heat shock-inducible and heat shock-only binding were in fact dependent on Hsf1 for their transcriptional induction. To test this, we deployed a yeast strain in which we could rapidly induce nuclear export of Hsf1 using the Anchor-Away system (Haruki et al., 2008; Solis et al., 2016). Analysis of genomewide transcription rates under NHS and 5 min HS states using nascent mRNA sequencing (NAC-seq; see Materials and Methods) revealed that the basal transcription of 18 genes was Hsf1 dependent (Figure 2A), consistent with previous observations (Solis et al., 2016), while heat shock-induced transcription of this set of genes plus an additional 34 were also Hsf1 dependent (Figure 2, B–D; Supplemental Table S1). Most of these genes (46/52) were also occupied by Hsf1 in 5 min heat-shocked cells (Figure 2C), arguing that this set of genes, derived from both core and inducible categories (Figure 1, E and G), is directly regulated by Hsf1. The remaining six genes exhibited subthreshold Hsf1 occupancy. While not meeting our stringent cut-off (see Materials and Methods), these six, which include the Hsp70-encoding SSA3 gene, may be direct targets of Hsf1 as well (see Supplemental Figure S1B). The 28 Hsf1 targets whose transcription was Hsf1 independent are largely composed of highly expressed housekeeping genes (Supplemental Figure S2) that are presumably controlled by multiple gene-specific, functionally redundant activators. Notably, for the 46 genes corroborated by both ChIP-seq and NAC-seq, induced DNA binding by Hsf1 led to a disproportionate increase in nascent transcription during the first 5 min of heat shock (typically 7.5-fold and in certain cases nearly 20-fold [Figure 2E]) (see Discussion).
FIGURE 2:
NAC-seq coupled with Hsf1-Anchor Away reveals genes dependent on Hsf1 for their basal and induced transcription. (A) NAC-seq counts transcriptome-wide under NHS conditions in the presence and absence of nuclear Hsf1 using the Hsf1 Anchor Away system (Hsf1-AA). Rapamycin (1 µM) was added for 45 min to deplete Hsf1 from the nucleus. Purple squares are genes with Hsf1 ChIP peaks; green circles show Hsf1 targets identified previously under NHS conditions (Solis et al., 2016). Genes with Hsf1 ChIP peaks above background (≥250 SPMR) and whose expression was significantly reduced by rapamycin treatment (p < 0.01; two-tailed t test) were designated as Hsf1-dependent genes (HDGs). Eighteen fell into this category: AHA1, BTN2, CUR1, CPR6, FES1, HCH1, HSC82, HSP104, HSP42, HSP78, HSP82, MBF1, MDJ1, SIS1, SSA1, SSA2, STI1, and YDJ1. (B) Analysis, representation, and designations as in A, except following 5 min HS. (C) Venn diagram comparing Hsf1 ChIP-seq gene targets and NAC-seq targets following a 5-min HS. ChIP-seq gene targets were derived from 74 total peaks, three of which were intergenic, two of which were linked to a single gene (HSP26), and four of which were linked to Hsf1-dependent, bidirectionally transcribed genes (see Figure 3, B and C). (D) NAC-seq counts for shared ChIP-seq/NAC-seq Hsf1 targets following a 5-min HS in the presence and absence of nuclear Hsf1. (Note: NAC-seq counts represent total nascent transcription and are not normalized for gene length.) Only one gene from the four Hsf1-dependent bidirectional genes is shown. (E) Scatter plot showing the correlation between HS-inducible Hsf1 DNA binding and HS-inducible transcription of shared ChIP-seq/NAC-seq Hsf1 targets. ChIP-seq signals represent the mean of two biological replicates.
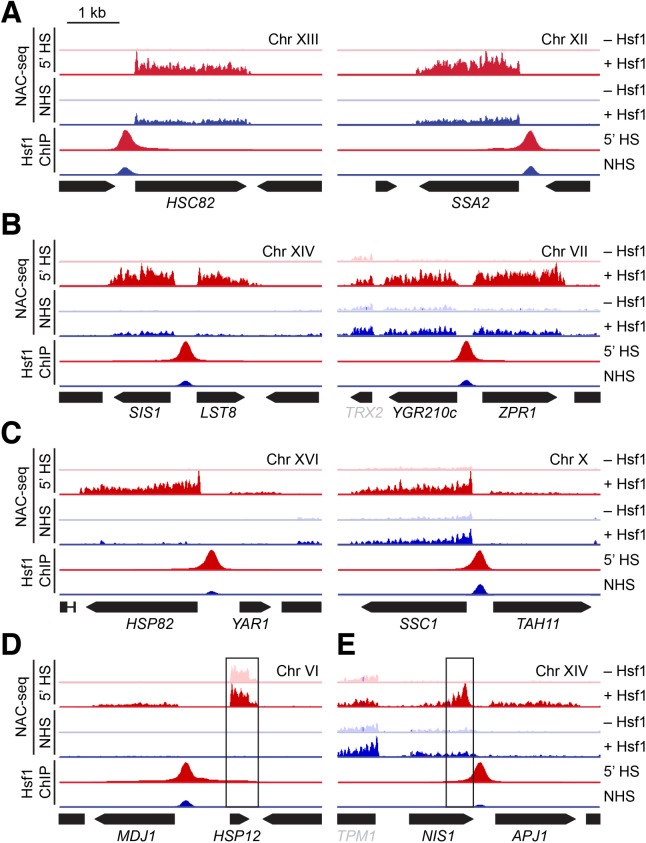
Examination of individual Hsf1-dependent genes revealed surprising variation with respect to their genomic arrangement and transcriptional response. Some, such as HSC82 and SSA2, were exclusively activated by Hsf1 and, as revealed by Anchor Away, entirely dependent on it for their expression (Figure 3A; compare NAC-seq tracks –/+ Hsf1 [see also Figure 2D]). Others, such as genes within the bidirectional pairs SIS1-LST8 and YGR210C-ZPR1, were symmetrically activated and Hsf1 dependent (Figure 3B). However, the gene pairs HSP82-YAR1 and SSC1-TAH11 were asymmetrically activated, with one gene strongly induced while the other only weakly, yet both members were Hsf1 dependent (Figure 3C). In the case of YAR1, the Pol II transcript began well upstream of the gene, suggesting that this heat-shock-inducible RNA might be noncoding. Since in each case the gene more strongly activated by Hsf1 was positioned closer to the Hsf1 site, the most parsimonious explanation is that proximity of the HSE to the core promoter/TSS dictates robustness of Hsf1 activation. The bidirectionally transcribed MDJ1–HSP12 gene pair is striking for the strong Hsf1 dependence of one gene, MDJ1, but not of the other, HSP12 (Figure 3D; Supplemental Table S1). Indeed, HSP12 appears to be primarily regulated by the alternative thermal stress-responsive activators Msn2 and Msn4 (Gasch et al., 2000). Also notable is the tandemly oriented NIS1–APJ1 gene pair. While both genes are heat shock induced and Hsf1 dependent, NIS1 is transcribed in the antisense direction (Figure 3E). Thus, Pol II transcripts arising from YAR1 and NIS1 are consistent with the idea that Hsf1 directs the expression of heat-shock-inducible lncRNAs.
FIGURE 3:
Hsf1 stimulates both unidirectional and bidirectional transcription. (A) IGV browser images of 5-kb windows at Hsf1-dependent loci. Tracks show NAC-seq and Hsf1 ChIP-seq under NHS and 5-min HS conditions; NAC-seq was conducted in the presence and absence of nuclear Hsf1 using the Hsf1-AA system. ChIP-seq and NAC-seq tracks were normalized to the maximum displayed value for each locus in the 5-min heat-shock sample. NAC-seq tracks represent both sense and anti-sense transcription yet for simplicity are shown above the line for each gene. Hsf1 was anchored away with 1 µM rapamycin for 45 min. (B) As in A but for loci that show near-stoichiometric, Hsf1-dependent bidirectional transcription. (C) As in B but for loci that show substoichiometric, Hsf1-dependent transcription of the nonchaperone gene. (D) As in C but for the MDJ1/HSP12 locus on chromosome VI. Although both MDJ1 and HSP12 are induced by heat shock, only MDJ1 is Hsf1 dependent. (E) As in C but for the NIS1/APJ1 locus on chromosome XIV. Demonstrates heat-shock- and Hsf1-dependent bidirectional transcription in the sense direction for APJ1 and in the antisense direction for NIS1.
A preset chromatin state poises Hsf1 for constitutive occupancy of core target loci
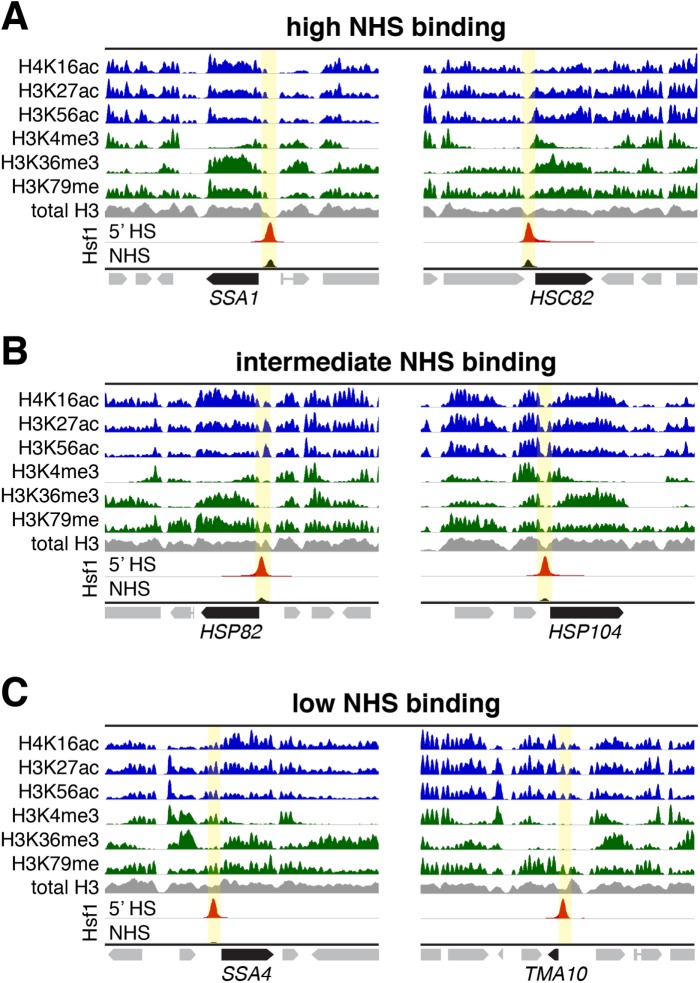
As we observed a broad spectrum of Hsf1 binding across the genome under both basal and heat-inducing conditions, we wished to know whether one or more properties of the preset chromatin landscape correlated with this behavior. We examined these at three classes of Hsf1 target genes: 1) those with strong Hsf1 binding under NHS conditions (“High NHS Binding”), 2) those with intermediate Hsf1 binding under NHS conditions (“Intermediate NHS Binding”), and 3) those whose Hsf1 binding was only detectable following 5 min HS (“Low NHS Binding”).
High NHS Binding genes such as SSA1, SSA2, and HSC82 displayed prominent nucleosome-free regions (NFRs) at the site of Hsf1 binding (Figure 4A and Supplemental Figure S3A; highlighted in yellow). In addition, nucleosomes flanking and/or downstream of these NFRs were enriched in histone marks linked to transcription, including acetylated H2A, H3, H4; H3K4me3; H3K36me3; and H3K79me (Li et al., 2007). While these marks are consistent with the strong basal transcription of these genes (Supplemental Table S1), their enrichment varied between genes. The preset chromatin landscape of Intermediate NHS Binding genes, epitomized by HSP78, HSP82, and HSP104 was similar although the breadth of the NFR was noticeably reduced (Figure 4B and Supplemental Figure S3B). This may reflect the smaller proportion of cells with Hsf1 prebound upstream of these genes. Finally, the preset chromatin of Low NHS Binding genes, exemplified by SSA4, HSP26, and TMA10, either lacked an NFR altogether (SSA4) or showed a greatly muted NFR (TMA10, HSP26) (Figures 4C and Supplemental Figure S3C). Instead, the upstream regions of these genes were enriched in histone acetylation marks, while classic methylation marks of transcription (H3K4me3, H3K36me36, and H3K79me3) were depleted. Absence of the latter is consistent with the fact that these genes express at very low levels in NHS cells (Supplemental Table S1). Notably, no class displayed enrichment of the Htz1 (H2A.Z) variant, contrary to models which posit that this histone is enriched at nucleosomes flanking promoter-associated NFRs (Hartley and Madhani, 2009).
FIGURE 4:
Hsf1’s constitutive occupancy correlates with preset accessible chromatin, while its heat-inducible occupancy correlates with preexisting histone acetylation and partial nucleosome occupancy. (A) ChIP-seq profiles of the indicated covalently modified histones, total H3 and Hsf1 over 10 kb windows of representative High NHS Binding loci (high Hsf1 occupancy under control conditions). Location of Hsf1 binding peak is highlighted in yellow. All tracks except Hsf1 were normalized to their own maximum displayed signal and are from publicly available data sets (NHS state; see Materials and Methods). Hsf1 tracks depict both NHS and 5-min HS states and are normalized to the 5-min HS sample (this study). (B) As in A, except depicted are Intermediate Hsf1 Binding loci. (C) As in A, except depicted are Low Hsf1 Binding loci.
The foregoing analysis suggests that a primary determinant of Hsf1 occupancy in NHS cells is whether the upstream region of a target gene is preassembled into stable chromatin. A powerful model to test this idea is the HSP82 upstream activation sequence (UAS), which is occupied by Hsf1 under basal conditions (Figure 1E and Supplemental Figures S1B and S8A) while also partially assembled into chromatin, as inferred from histone ChIP and ChIP-seq assays (Figures 4B and 5A and Supplemental Figure S8B).
FIGURE 5:
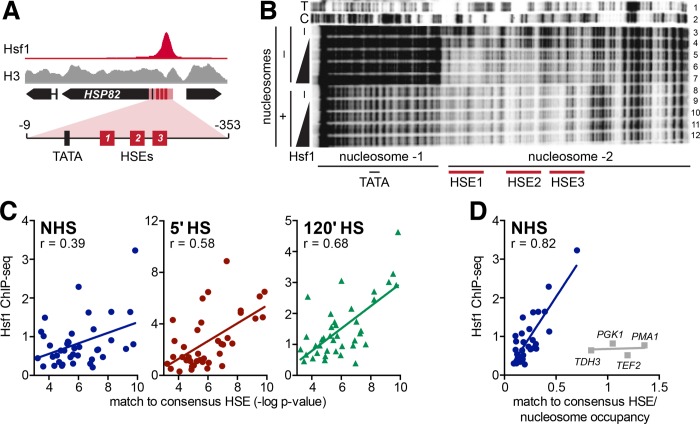
Hsf1 DNA binding is impeded by nucleosomes, both in vitro and in vivo. (A) Hsf1 and H3 ChIP-seq signal at HSP82 under NHS conditions, normalized to their maximum displayed values. The region used for nucleosome reconstitution and DNase I footprinting is highlighted. (B) DNA corresponding to the HSP82 upstream region depicted in A (spanning –9 to –353 [ATG = +1]) and 32P-end labeled on the upper strand was either reacted directly with GST-Hsf1 (lanes 4–7) or following its reconstitution into a dinucleosome (lanes 9–12). Reconstitution was achieved using a 1:1 (wt/wt) HeLa histone: DNA ratio and salt dilution, followed by purification over a glycerol gradient (see Supplemental Figure S4). Both naked DNA and chromatin templates were challenged with increasing amounts of recombinant Hsf1 (lanes 3 and 8 are -Hsf1 controls) and then subjected to DNase I digestion. DNA was purified and electrophoresed on an 8% sequencing gel. (C) Scatter plots of Hsf1 ChIP-seq signal as a function of the strength of the HSE for the NHS, 5-min HS, and 120-min HS states. HSE strength was determined by MEME as a p value corresponding to how well the binding site beneath the summit of each ChIP peak matched the consensus HSE motif (Figure 1F). ChIP-seq signals represent the mean of two biological replicates. (D) As in C, but here each p value was divided by the H3 ChIP-seq signal below the summit of the Hsf1 peak under NHS conditions (H3 ChIP-seq data from Qiu et al. [2016]). Outliers (gray) consist of Hsf1-independent housekeeping genes.
To directly test the ability of yeast Hsf1 to bind nucleosomal DNA, we reconstituted the upstream region of HSP82 into chromatin using purified HeLa core histones (Supplemental Figure S4; see Materials and Methods). Strikingly, this procedure resulted in the assembly of a dinucleosome composed of one nucleosome centered over the core promoter and a second over the UAS (HSEs1-3) and surrounding region, as revealed by DNase I footprinting (Figure 5B, compare lane 8 with lane 3). The nucleosome over the core promoter appears to be rotationally phased as revealed by the chromatin-specific, ∼10-base DNase I cleavage periodicity (Sollner-Webb et al., 1978). Equally striking, this footprinting pattern resembles that obtained by DNase I genomic footprinting of an inactivated allele of HSP82 (hsp82-ΔHSE1) to which Hsf1 cannot bind and whose upstream region is occupied by two strongly positioned nucleosomes (Gross et al., 1993; Venturi et al., 2000). When we challenged this template with recombinant Hsf1, no Hsf1-dependent DNase I footprint could be detected (Figure 5B, lanes 9–12), in contrast to naked DNA, where protection was evident over the entire UASHS (Figure 5B, lane 3 vs. lanes 5–7). Thus, although recombinant Hsf1 is capable of cooperative, high-affinity binding to the HSP82 UAS (Figure 5B and Erkine et al. [1999]), it cannot bind to the same DNA sequence when it is preassembled into a stable dinucleosome.
Genomewide Hsf1 occupancy is largely dictated by accessible, high-quality HSEs
While the above analysis suggests that preexisting chromatin state plays an important role in dictating Hsf1 occupancy, other parameters, such as the quality of the HSE, may also play a role. To test this, we quantified how well the putative Hsf1-bound sequence under each ChIP peak matched the HSE consensus sequence we derived from all ChIP peaks (TTCTAGAAnnTTCTAGAA; Figure 1F) and compared this score (Supplemental Figure S1C) with the amount of Hsf1 ChIP signal. We found little correlation under NHS conditions, but improved correlation as a function of time during heat shock (120′ > 5′ > NHS) (Figure 5C). Strikingly, when the quality of the HSE was considered in conjunction with nucleosome density as determined by total histone H3 ChIP-seq signal, a strong correlation was found under NHS conditions between this parameter (HSE quality/nucleosome density) and Hsf1 occupancy (Figure 5D, r = 0.82). That is, 64% of the variance in Hsf1 ChIP signal across the genome in the control state can be accounted for by HSE quality/nucleosome density alone.
Pioneer transcription factors are enriched within upstream regions of genes with high levels of constitutively bound Hsf1
How are nucleosome-free or nucleosome-depleted regions formed at Hsf1 binding sites? In mammals, “pioneer” transcription factors, epitomized by FoxA1 and related proteins, have been shown to potentiate the subsequent binding of gene-specific activators through their ability to invade repressive chromatin and create locally accessible regions (reviewed in Zaret and Carroll [2011] and Zaret et al. [2016]). The DNA binding proteins Rap1, Reb1, and Abf1 (sometimes termed General Regulatory Factors) have similar qualities: abundant, sequence-specific, constitutively bound to DNA and frequently located within nuclease-hypersensitive, open chromatin (Buchman et al., 1988; Chasman et al., 1990; Bai et al., 2011; Ganapathi et al., 2011). We therefore asked whether the genomewide localization of Rap1, Reb1, and Abf1 in the NHS state showed any correspondence to that of Hsf1. As shown in Supplemental Figure S5A, there is a striking enrichment of these factors overlapping or in close proximity to the nucleosome-depleted, high-NHS binding Hsf1 sites of SSA1 and HSC82. This relationship also exists with certain Intermediate NHS Binding Genes (e.g., HSP82) although not with others (e.g., HSP104) (Supplemental Figure S5B). It is particularly evident at strongly expressed housekeeping genes such as TEF2 and TDH3 (Supplemental Figure S5D); at such genes, Hsf1’s contribution to transcription is modest, especially under NHS conditions (Supplemental Table S1). In contrast, the Low NHS Binding Genes SSA4 and HSP26 were located in Rap1/Reb1/Abf1 deserts with comparatively high H3 occupancy (Supplemental Figure S5C). Overall, there was a modest but significant correlation between the amount of pioneer factor bound within 1 kb upstream of the 43 core Hsf1 targets (R2 = 0.26; p = 0.0008) (Supplemental Figure S5E). Together, this analysis suggests that pioneer factor binding is an important factor distinguishing high Hsf1 NHS binding targets from the low NHS binding, heat shock-inducible targets.
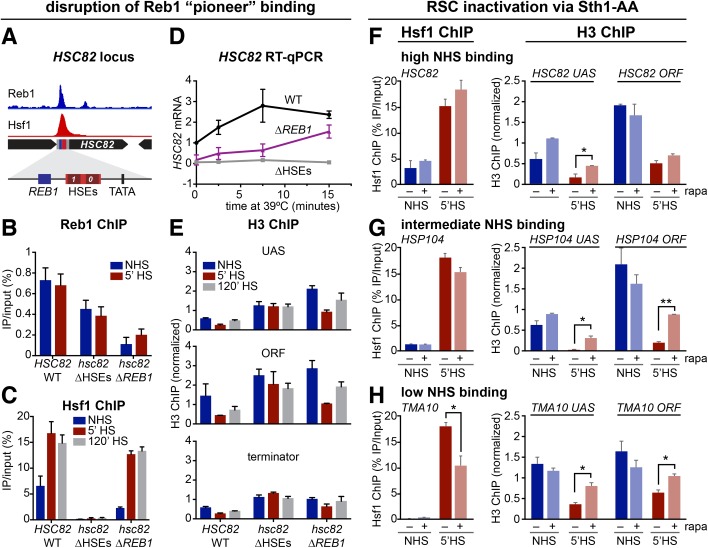
Reb1 potentiates open chromatin, Hsf1-transactivation, and Hsf1-mediated nucleosome displacement
To more directly address the role of pioneer factors in fostering Hsf1 binding and activity, we investigated the effect of mutating a high-affinity Reb1 binding site upstream of HSC82, creating a mutant allele termed hsc82-∆REB1, and compared it to an isogenic mutant bearing a dual substitution of the two HSEs (termed hsc82-∆HSEs) (Erkine et al., 1996). Hsf1 ChIP-seq and Reb1 ChEC-seq suggest high-level, overlapping occupancy of Reb1 and Hsf1 at HSC82 under control conditions (Figure 6A) (Zentner et al., 2015). Locus-specific ChIP confirmed this and, moreover, revealed that Reb1 occupancy was modestly diminished by a chromosomal substitution of HSE0 and HSE1 that obviates Hsf1 binding (Figure 6, B and C). Similarly, chromosomal substitution of the Reb1 site, resulting in strongly reduced Reb1 occupancy (Figure 6B), negatively impacted Hsf1 binding (Figure 6C). Interestingly, mutation of the Reb1 site diminished hsc82 expression greather than sevenfold in nonstressed cells and up to fivefold in acutely stressed ones (Figure 6D), and this corresponded to a pronounced increase (three- to fourfold) in nucleosome occupancy within the UAS and coding region of the gene. Thus, the ability of Hsf1 to remodel chromatin was impaired at hsc82-∆REB1. Likewise, obviating Hsf1 binding led to an increase in H3 occupancy over both UAS and gene coding regions and additionally suppressed the eviction of histones that takes place during heat shock (Figure 6E). Collectively, the data suggest that pioneer factors functionally cooperate with Hsf1 at select genes. In the absence of pioneer factor binding, other factors, including an increase in available Hsf1 (through chaperone titration as described above), are still able to drive nucleosome displacement on acute heat shock (schematically summarized in Figure 7).
FIGURE 6:
The pioneer factor Reb1 enables Hsf1 binding to a high NHS binding target while RSC cooperates with Hsf1 to displace nucleosomes during heat shock. (A) Browser shot showing Reb1 ChEC-seq (Zentner et al., 2015) and Hsf1 ChIP-seq signal under NHS conditions at a 5-kb window around the HSC82 locus. Both tracks are normalized to their maximum displayed values. Expanded view of the HSC82 promoter shows the locations of the Reb1 binding site, HSEs, and TATA box (centered at –249, –193, and –138, respectively; ATG = +1). (B) Reb1-myc9 ChIP-qPCR analysis of the hsc82 promoter under NHS and 5-min HS conditions in isogenic HSC82, hsc82-∆HSEs, and hsc82-∆REB1 cells conducted as described under Materials and Methods. The hsc82-∆HSEs allele bears multiple point substitutions within HSE0 and HSE1 while hsc82-∆REB1 bears a 10-base-pair chromosomal substitution of the Reb1 binding site (Erkine et al., 1996). Shown are means + SD (N = 2 biological replicates; qPCR = 4). (C) Hsf1 ChIP analysis of the hsc82 UAS region, conducted and analyzed as in B. (D) Quantification of HSC82 mRNA expression level by RT-qPCR over a heat-shock time course. Plotted are means ± SD (N = 2; qPCR = 4). (E) Histone H3 ChIP analysis of the HSC82 promoter, midcoding region (open reading frame [ORF]), and 3′-UTR-terminator region. H3 ChIP signals were normalized to those detected at a nontranscribed locus, ARS504, which served as an internal recovery control. (F–H) Hsf1 and H3 ChIP at HSC82, HSP104, and TMA10 (as indicated) under NHS and 5-min HS conditions, in the presence and absence of the RSC catalytic subunit, Sth1 (rapa - and rapa +, respectively). Conditional nuclear depletion of Sth1 was achieved using an Sth1-AA strain that was pretreated with 1 µM rapamycin (rapa) for 2.5 h. Analysis and display as in B. *, p < 0.05; **, p < 0.01, two-tailed t test.
FIGURE 7:
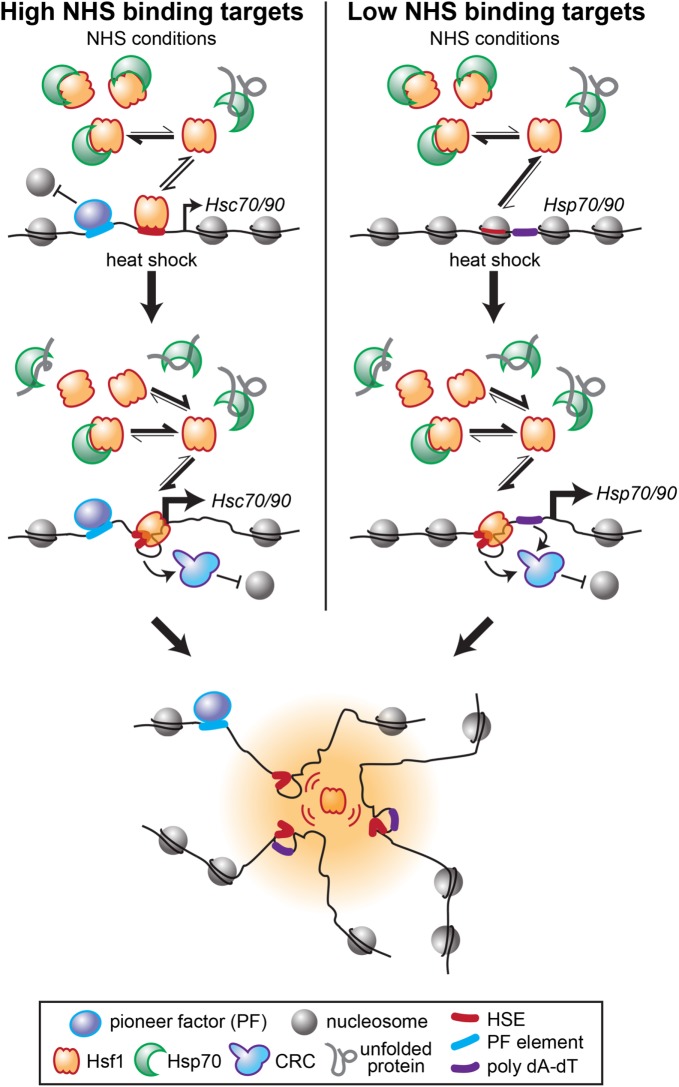
Model for differential basal and inducible Hsf1 binding across the genome. Target genes with high levels of Hsf1 binding under non-heat-shock conditions (High NHS binding targets) have nucleosome-free (or depleted) regions upstream of the TSS due to the presence of pioneer factors. Such NFRs are occupied by the small fraction of DNA binding-competent Hsf1 present, stimulating basal transcription of linked HSC70/90 genes (e.g., SSA1, SSA2, HSC82). These genes nonetheless show approximately twofold increases in Hsf1 binding on acute heat shock when the bulk of Hsf1 is liberated from repressive association with Hsp70. This inducible binding further restructures the loci and drives increased transcription. In contrast, Low NHS binding targets (e.g., SSA4, HSP26, TMA10) lack proximal pioneer factor binding, and Hsf1 binding sites are occluded by nucleosomes. The fraction of free Hsf1 available under NHS conditions is insufficient to invade the chromatin at these sites. On heat shock, the large increase in DNA-binding competent Hsf1 allows Hsf1 to cooperatively bind its cognate HSEs, and along with poly(dA-dT) tracts that help recruit CRCs, to restructure the loci and drive high-level transcription. Active Hsf1 then drives looping and coalescence of its target loci into transcriptionally active foci that may form phase-separated assemblies.
RSC facilitates nucleosome depletion and Hsf1 binding within pioneer factor deserts
Finally, we asked whether the abundant, ATP-dependent chromatin remodeling complex (CRC), RSC (Cairns et al., 1996), plays a substantive role in creating a favorable chromatin template for Hsf1 binding and HS-dependent chromatin remodeling (Zhao et al., 2005; Zhang et al., 2014). RSC has been shown to remodel a nucleosome covering Gal4 binding sites, rendering these sites accessible and, as a consequence, the linked GAL1 and GAL10 genes responsive to Gal4 activation (Floer et al., 2010). Using Anchor Away as above, we conditionally depleted Sth1, the catalytic subunit of RSC, from the nuclei of Sth1-FRB tagged cells and then subjected the cells to a subsequent acute heat shock (or not) and evaluated the effect on Hsf1 and H3 occupancy at representative High, Intermediate, and Low NHS Binding genes. Sth1 perturbation minimally affected Hsf1 and H3 occupancy at the High NHS Binding gene, HSC82, although it did lead to an increase in H3 occupancy at the Intermediate NHS Binding gene, HSP104 (Figure 6, F and G). Interestingly, at the Low NHS Binding gene, TMA10, both Hsf1 and H3 occupancy were significantly affected (Figure 6H). These results suggest that RSC plays an increasingly important, nonredundant role in Hsf1-mediated regulation of genes whose preset chromatin structure is antagonistic to Hsf1 occupancy and function.
DISCUSSION
Yeast Hsf1 inducibly binds DNA genomewide in response to thermal stress
Results presented here indicate that Hsf1 binds heat-inducibly to the vast majority of its transcriptional targets (69/74). While seemingly contrary to original claims that yeast Hsf1 constitutively binds HSEs, based on EMSA (Sorger et al., 1987), genetic analysis (Jakobsen and Pelham, 1988), or genomic footprinting (Gross et al., 1990), in fact our data validate that a small fraction (∼25%) of Hsf1 constitutively binds select HSEs even under NHS conditions. This is consistent with the ability of yeast Hsf1 to trimerize under cell-free conditions (Sorger and Nelson, 1989). Isolation of cell lysates or nuclei (prerequisite for EMSA and genomic footprinting) require spheroplasting, a procedure that induces the heat shock response (Adams and Gross, 1991), thereby providing a plausible explanation for the earlier claims. In vivo dimethyl sulfate footprinting analysis provided initial evidence for inducible Hsf1 binding in S. cerevisiae (Giardina and Lis, 1995).
Hsf1’s occupancy under NHS conditions, while correlating poorly with the quality of the bound HSE, shows a striking correlation to a related parameter, quality of HSE/nucleosome occupancy (r = 0.82). Consistent with this, recombinant Hsf1 cannot bind HSEs assembled into a stable nucleosome, even one that has been reconstituted with hyperacetylated histones (A.M.E. and D.S.G., unpublished observations). Thus, other factors must come into play, and we demonstrate that a preset, nucleosome-free (or depleted) chromatin structure typifies the upstream regions of High NHS Binding genes. One attribute underpinning the NFRs of high Hsf1 occupancy promoters is the presence of constitutively bound pioneer factors Rap1, Reb1, and Abf1. These abundant, DNA-binding proteins are overrepresented at such genes, while unrepresented at Low NHS Binding genes. They may work through recruitment of CRCs that mediate the nucleosome-free state (Krietenstein et al., 2016). Alternative sequence-specific factors may serve an analogous role at High NHS Binding genes such as ZPR1 with low occupancy of Rap1, Reb1, and Abf1 (Supplemental Figure S5E).
At Low NHS Binding Genes, relocalization of RSC that occurs on heat shock (Vinayachandran et al., 2018) may be enhanced by the presence of poly(dA-dT) motifs (Lorch et al., 2014) that are significantly enriched within the NFRs of these genes (Supplemental Figure S6). Given that our evidence argues against a dominant role for RSC, Hsf1 likely recruits multiple CRCs (Zhao et al., 2005; Shivaswamy and Iyer, 2008; Krietenstein et al., 2016). In addition, heat shock–activated Hsf1 triggers widespread changes in the genome that may contribute to its ability to occupy nucleosomal genomic sites and expand its regulon. Hsf1-target genes engage in frequent intergenic (both cis- and trans-) interactions during acute heat shock; such coalescence is strictly dependent on Hsf1 and encompasses High NHS, Intermediate NHS, and Low NHS genes (Chowdhary et al., 2017, 2018). HSP gene coalescence may be indicative of liquid–liquid phase separation postulated to underlie transcriptional control in higher eukaryotes (Hnisz et al., 2017). Thus, it could be hypothesized that the high local concentration of Hsf1 liberated from Hsp70 on heat shock would enable Hsf1 to gain a toehold at inducible targets and begin to recruit Mediator and other coactivators (many of which, like Hsf1 itself, harbor intrinsically disordered regions), driving coalescence among targets. These “Hsf1 bodies” could serve as a platform to recruit RSC and other CRCs through protein–protein interactions centered at coalesced UAS/promoters (see model in Figure 7).
DNA-bound Hsf1 exists in multiple, functionally distinct states
An important implication of our study is that DNA-bound Hsf1 significantly differs in its ability to stimulate transcription of its targets depending on the proteotoxic state of the cell. Under control conditions, a core set of 43 genes is detectably occupied by Hsf1. Following acute HS, most of these 43 are occupied at substantially higher levels (genomewide Hsf1 occupancy increased ≥4-fold). Nascent transcription of these genes increased even more, nearly 30-fold. Thus, DNA-bound Hsf1 exists in a far more active state (∼7.5-fold) in acutely stressed cells. A possible basis for this is that the Hsp70 chaperone, shown to bind Hsf1 in whole cell extracts isolated from NHS cells but not from those exposed to 5 min HS (Zheng et al., 2016; Krakowiak et al., 2018), is also associated with DNA-bound Hsf1 under NHS but not acute HS conditions. During more chronic exposures to stress, nascent transcription is reduced (Solis et al., 2016), and this may be accompanied by Hsp70 rebinding to DNA-bound Hsf1 (Zheng et al., 2016).
Related to the above, Hsf1-dependent transcription varies from one promoter to another under a particular condition, even after normalization of ChIP-seq signal (Supplemental Figure S7). What might account for this? Undoubtedly, quality of core promoter elements contributes. However, additional factors might also come into play, such as pioneer factors and/or chromatin remodeling complexes, that may either enhance or suppress the transcription-stimulating activity of DNA-bound Hsf1. One way this could be accomplished is by facilitating the association/disassociation of Hsp70. Especially intriguing is the hyperactive state of Hsf1 bound upstream of the essential basal targets HSC82 and SSA2 (Supplemental Figure S7A).
Quality of HSE strongly correlates with Hsf1 binding in heat-shock-induced cells
Earlier work on Hsf1 suggested that its binding avidity and transcriptional activity were linked to the presence of various types of HSEs, termed perfect, gap, step, and direct repeat (reviewed in Sakurai and Takemori [2007]). However, we have found that only when the role of chromatin is lessened—for example, following nucleosome disassembly that accompanies heat shock (Zhao et al., 2005; Kremer and Gross, 2009)—does a strong correlation exist between Hsf1 occupancy and quality of the HSE (defined as match to the consensus; Figure 5C). Indeed, 7/8 perfect HSEs located within promoter regions were occupied by Hsf1 following acute heat shock, while a far lower fraction of suboptimal HSEs were similarly occupied (e.g., 15/82 TTCnnGAAnnTTC motifs and 13/110 GAAnnTTCnnGAA motifs; Supplemental Table S2). It is thus likely that extent of match to the consensus HSE, rather than presence of gap, step, or direct repeat HSEs per se, dictates Hsf1 binding avidity in vivo.
A mutual antagonism exists between Hsf1 and nucleosomes under NHS conditions
A striking observation is that the HSP82 upstream regulatory region can be reconstituted into two sequence-positioned nucleosomes using only core histones and DNA. This chromatin structure, which recapitulates the dinucleosome present within the upstream region of hsp82-ΔHSE1 (Gross et al., 1993; Venturi et al., 2000) and resembles the one reconstituted on a similar DNA sequence outfitted with p53 binding sites (Laptenko et al., 2011), may actively antagonize Hsf1 binding. Indeed, under NHS conditions, Hsf1 binding is virtually undetectable at hsp82-ΔHSE1; instead, there is a three- to fourfold increase in histone H3 occupancy (Supplemental Figure S8). At HSP82+ under NHS conditions, Hsf1 exists in a dynamic equilibrium with chromatin, as suggested by the cohabitation of Hsf1 with H2A, H2B, H3, and H4 (this study and Zhao et al. [2005]), creating a chromatin structure hypersensitive to DNase I cleavage (Gross et al., 1990). Following acute heat shock, Hsf1’s cooperative binding of HSEs (Erkine et al., 1999), coupled with its recruitment of CRCs, HATs, and other coactivators (Kremer and Gross, 2009; Kim and Gross, 2013), leads to a 40-fold increase in nascent transcription and eviction of histones that extends across the gene (Supplemental Table S1; Supplemental Figure S8B) (Zhao et al., 2005). Thus, the chromatin state of the HSP82 promoter is subject to a dynamic remodeling process even under NHS conditions that is largely directed by Hsf1 itself.
Relationship to previous genomewide studies
It is instructive to compare findings reported here to previous genomewide studies of Hsf1 occupancy. Using ChIP microarray analysis, Thiele, Iyer and colleagues reported that S. cerevisiae Hsf1 binds 210 genomic sites, 165 of which were located upstream of distinct open reading frames and the majority of which were heat-inducibly bound by Hsf1 (Hahn et al., 2004). Our ChIP-seq analysis, by contrast, identified only 74 genomic sites occupied by Hsf1, 69 of which were inducibly occupied and 46 of which were located upstream of genes whose heat-induced transcription was Hsf1 dependent. One explanation for the fewer genomic targets identified in our analysis is that we incorporated a high threshold for peak calls. This led to the exclusion of six genes likely occupied by Hsf1 since their heat-shock-induced transcription is Hsf1 dependent. However, this fact alone cannot entirely explain the disparity between the data sets, since only 31/43 core Hsf1 targets and 14/31 inducible targets identified here were also identified in the earlier ChIP-chip analysis. Holstege and colleagues examined Hsf1 occupancy in cells maintained under control, nonstressful conditions. Using ChIP-seq coupled with comparative dynamic transcription using Hsf1-AA strains, they identified 21 Hsf1 targets under NHS conditions (de Jonge et al., 2017); we identify these same 21 genes using a combination of ChIP-seq, NAC-seq, and Anchor Away, plus four others (HSP82, SSA2, SSA4, and UBI4).
Several previous studies examined the interplay of nucleosomes and Hsf1 binding. In contrast to our observations, Brown and colleagues reported little loss of histone over the UAS/promoter regions of Hsf1-bound loci on heat shock of the pathogenic yeast Candida albicans (Leach et al., 2016). Why this is the case is unclear but might be related to differential recruitment of CRCs. Similarly to findings reported here, Drosophila HSF occupancy was found to be strongly linked to preset chromatin that is nucleosome depleted, enriched in active histone marks, and whose landscape has prebound pioneer factors (Guertin and Lis, 2010). Most prominent of the pioneer factors identified was GAGA factor, whose roles include opening of promoter chromatin and facilitating the recruitment and pausing of Pol II (Fuda et al., 2015). Following its heat-shock-induced binding, Drosophila HSF drives striking remodeling of chromatin (Petesch and Lis, 2008), as we have observed for yeast Hsf1. HSF also functions to release Pol II into the body of highly activated HSP genes (Duarte et al., 2016). Likewise, in mammalian cells, heat shock activation of HSF1 target genes is regulated at the level of Pol II pause release (Mahat et al., 2016), and HSF1 binding to both promoters and enhancers is accompanied by a marked increase in H4 acetylation (Vihervaara et al., 2017).
These recent studies additionally suggest that only a fraction of all heat-shock-inducible genes in insects and mammals are under control of HSF1 (Duarte et al., 2016; Mahat et al., 2016). Likewise, in yeast, we observed that only 10% of heat-shock-inducible genes (i.e., those exhibiting greater than or equal to threefold increase in transcription) were Hsf1 dependent. Nonetheless, in each organism, the HSF1 regulon encompasses the HSP genes that encode chaperones and cochaperones, consistent with coevolution of the transcription factor, its cognate cis-response element, and a core set of target genes.
Conclusions
The results presented here point to three clear conclusions: 1) Yeast Hsf1, like metazoan HSFs, inducibly binds to its target promoters across the genome during acute heat shock, contrary to popular models that presume the existence of a fundamental difference in DNA-binding behavior. 2) There are a small number of essential chaperone genes to which Hsf1 binds strongly under NHS conditions; these genes have NFRs due to the presence of pioneer factors that expose strong HSEs. 3) Hsf1 expands its target regulon during heat shock by cooperating with CRCs to reveal HSEs occluded by nucleosomes, concomitant with driving its target genes—dispersed throughout the genome—into a coalesced, potentially phase-separated, state (Chowdhary et al., 2017, 2018). Together, these mechanisms allow cells to dynamically control the breadth and magnitude of the heat shock response to tune the proteostasis network according to need.
MATERIALS AND METHODS
Yeast strains
Strains AJ1001, AJ1002, and AJ1003 are derivatives of SLY101, HSE102, and GRF200, respectively, in which REB1 was C-terminally tagged with the Myc9 epitope as previously described (Kim and Gross, 2013). Similarly, ACY101 is a derivative of HHY212 (Haruki et al., 2008) in which STH1 was C-terminally tagged with the FRB domain. All other strains have been previously described (see Table 1).
TABLE 1:
Yeast strains.
| Strain name | Genotype | Source or reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| BY4742-Hsf1-AA | BY4741; MATα tor1-1 fpr1Δ RPL13A-FKBP12::NAT MET15+ HSF1-FRB-yEGFP::KAN-MX | F. Holstege, University Medical Center Utrecht, The Netherlands |
| SLY101 | MATα ade- can1-100 cyh2r his3-11,15 leu2-3,112 trp1-1 ura3 | Lee and Gross, 1993 |
| HSE102 | SLY101; hsc82-ΔHSEs | Erkine et al., 1996 |
| GRF200 | SLY101; hsc82-ΔREB1 | Erkine et al., 1996 |
| AJ1001 | SLY101; REB1-MYCx9::TRP1 | This study |
| AJ1002 | HSE102; REB1-MYCx9::TRP1 | This study |
| AJ1003 | GRF200; REB1-MYCx9::TRP1 | This study |
| KEY102 | SLY101; hsp82-ΔHSE1 | Gross et al., 1993 |
| KEY105 | SLY101; hsp82-ΔHSE1· | Gross et al., 1993 |
| DPY1283 | MATα tpk1/2/3-as TOR1-1 fpr1∆::NAT RPL13A-FKBP::TRP1 HSF1-FRB::HIS3 TEF2pr-mKate-4xHSE-EmGFP::URA3 RPB3-FLAG::KAN | Solis et al., 2016 |
| HHY212 | MATa tor1-1 fpr1::loxP-LEU2-loxP RPL13A-2 × FKBP12::loxP-TRP1-loxP ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 | Haruki et al., 2008 |
| ACY101 | HHY212; STH1-FRB::HIS3 | This study |
Chromatin immunoprecipitation
ChIP experiments were performed as previously described (Kim and Gross, 2013) except as noted below. Mid–log cell cultures (350–500 ml), pregrown in YPDA medium (yeast extract-peptone-dextrose supplemented with 0.002% adenine) at 30°C, were used in both rapamycin (final concentration 1 μg/ml) and heat-shock time-course experiments. To effect a heat shock, an equal volume of prewarmed YPDA (50°–55°C) was added to each culture, instantaneously raising the temperature to 39°C. This was maintained thereafter in a shaking water bath. Aliquots (50 ml) were removed at each time point to which formaldehyde was added to a final concentration of 1% at the temperature of the treatment (30°C for NHS and 39°C for HS). Cells were harvested, washed, and resuspended in 250 μl lysis buffer and lysed with vigorous shaking in the presence of glass beads (∼300 mg) at 4°C for 30 min. Cell lysates were then transferred to 1.5-ml TPX tubes and sonicated at 4°C using a Diagenode Biorupter Plus (50 cycles with 30 s pulses), generating chromatin fragments with a mean size of ∼200–300 base pairs. TPX tubes were centrifuged to clarify supernatants that were then brought up to 2000 μl using ChIP lysis buffer. To perform immunoprecipitation, the equivalent of 500–800 μg chromatin protein (typically 200–400 μl) was incubated with one of the following antibodies: 1 μl of anti-Hsf1 (Erkine et al., 1996), 2.5 μl of anti-Myc (Santa Cruz Biotechnology), and 1 μl of anti-H3 globular domain (Abcam; ab1791).
Immunoprecipitated DNA was resuspended in 60 μl TE (10 mM Tris-HCl, 0.5 mM EDTA, pH 8); 2 μl was used in quantitative PCR (qPCR) with Power SYBR Green PCR Master Mix (Applied Biosystems 4367660) on an Applied Biosystems 7900HT Real-Time PCR system. DNA was quantified using a standard curve specific for each amplicon, and background signal arising from beads alone was subtracted. Background was determined by signal arising from incubating an equivalent volume of chromatin extract with Protein A or Protein G–Sepharose beads (GE Healthcare Life Sciences; CL-4B or 4 Fast Flow, respectively). To normalize for variation in yield of chromatin extracts, in certain cases input was used. Briefly, 40 μl was removed from the 2000 μl chromatin lysate isolated as described above, and the volume was brought up to 400 μl. Formaldehyde-induced cross-links were reversed, and DNA was deproteinized as for ChIP samples. Purified Input DNA was dissolved in 300 μl TE, and 2 μl was removed for qPCR. The signal arising from this represents 2% of total input chromatin. In the case of H3 ChIP, signal at a test locus was normalized to that obtained at ARS504 as described (Zhang et al., 2014). Primers used for detection and quantification of genomic loci in ChIP and Input DNAs are listed in Supplemental Table S3.
ChIP sequencing
ChIP-Seq experiments were performed as above except as noted below. BY4741 mid–log cell cultures (600 ml) were used for each heat-shock time-course sample (0-, 5-, and 120-min HS). Cells were harvested as 50-ml aliquots, with each pellet washed and resuspended in 250 μl lysis buffer and lysed with vigorous shaking in the presence of ∼300 mg glass beads at 4°C for 30 min. Cell lysates were then transferred to 1.5-ml TPX tubes and sonicated at 4°C using a Diagenode Biorupter Plus (60 cycles with 30-s pulses). This procedure generates chromatin fragments with a mean size of 100–250 base pairs. TPX tubes were centrifuged to clarify supernatants that were pooled and brought up to 3000 μl using ChIP lysis buffer. To perform immunoprecipitation, 100 μl was incubated with 4 μl of anti-Hsf1 antiserum (Erkine et al., 1996). As a control, 4 μl preimmune serum were used for each of the three time points. Five separate IPs were conducted for both the Hsf1 ChIP and the preimmune ChIP using 40 μl Protein A–Sepharose beads for each. Following purification of the IP and deproteinization, each ChIP DNA was suspended in 60 μl TE; the five samples were combined into one 300-μl pooled sample and quantified using the Qubit Fluorimeter assay. ChIP DNA (5 ng) was used to generate barcoded libraries using NEBNext Ultra DNA Library Prep Kit for Illumina (NEB E7370). Libraries were sequenced on lllumina Mi-Seq and NextSeq 500 instruments located in the LSUHSC Core Research Facility. Raw data are deposited on GEO; see Data access below.
Hsf1 ChIP-seq analysis
Reads were aligned to the SacCer3 build of the yeast genome using Bowtie2 (Langmead and Salzberg, 2012). We used MACS2 to call and quantify peaks, filtering for properly paired reads and controlling for nonspecific binding using condition-matched preimmune serum ChIP-seq data. We generated a signal file that outputs fragment pileups per million reads in bedgraph format (signal per million mapped reads; SPMR). It was important that we allowed MACS2 (Zhang et al., 2008) to model the expected value of duplicate reads (–keep dup auto) due the compact size of the genome and the high coverage of the data set. Only peaks that had SPMR summit values above the background threshold of 250 were designated Hsf1 targets. Motif enrichment analysis was performed using Multiple EM for Motif Elicitation (MEME)-ChIP (Ma et al., 2014). The MACS command used for a generic sample/preimmune pair that had been fragmented to 180 base pairs:
macs2 callpeak -t SAMPLE.bam -c PREIMMUNE.bam -f BAMPE -g 1.2e7 -n SAMPLE -p 1e-3 –nomodel -B –SPMR –extsize 180 –keep-dup auto
We note that the algorithm calls a single motif under each peak (a contiguous sequence that has the closest match to consensus) and thus does not reveal existence of ancillary HSEs. These do, in fact, exist and can contribute to the cooperative binding of Hsf1 (e.g., HSP82 [Erkine et al., 1999]). We also note that when visualized using the Integrative Genomics Viewer (IGV) (Robinson et al., 2011), small values often appear stretched and poorly reflect quantitative SPMR values (Figures 1B, 3, and 4 and Supplemental Figures S2 and S5, A–D). These are qualitative unless one zooms out so each peak is only in one pixel per bin.
Analysis of previously published NHS data sets
Histone modification and Htz1 ChIP-seq data were obtained from GSE61888 (Weiner et al., 2015). Histone H3 data are from GSE74787 (Qiu et al., 2016). Pioneer factor ChEC-seq data are from GSE67453 (Zentner et al., 2015). As above, data were visualized using the IGV.
Nascent transcript sequencing
Nascent mRNA was enriched by purifying RNA that coprecipitates with RNA Pol II-component Rpb3. Hsf1 Anchor-Away cells expressing Rpb3-FLAG (strain DPY1283) were grown at 30°C to OD600 = 0.8 in YPD, treated ± 1 µM rapamycin for 45 min, and then harvested after a 5-min heat shock at 39°C (or mock-treated for an additional 5-min at 30°C [NHS sample]). Cells were collected and lysed in a coffee grinder as described (Zheng et al., 2016) and resuspended in 1 ml IP buffer (20 mM HEPES, pH 7.4, 110 mM KOAc, 0.5% Triton X-100, 0.1% Tween-20, 10 mM MnCl2, 1X Complete mini EDTA-free protease inhibitor, 50 U/ml SuperaseIn). DNA was digested with 150 U/ml RNase-free DNase for 20 min on ice and samples were centrifuged at 20,000 × g. Clarified lysate was added to 50 µl prewashed anti-FLAG M2 magnetic beads and incubated for 2 h at 4°C. Beads were washed four times with 1 ml IP buffer and eluted in 2 × 50 µl of 3xFLAG peptide (0.5 mg/ml) in IP buffer. RNA was purified using the Qiagen miRNeasy kit and libraries were prepared using the NEB Next Ultra RNA kit. Illumina sequencing libraries were sequenced at the Whitehead Genome Technology Core. Reads were aligned using Tophat, quantified with HTSeq-Count and normalized using DESeq2. A compilation of genomewide NAC-seq data is provided in Supplemental Table S1. Raw data are deposited at GEO; see Data access.
Reverse transcription–quantitative PCR (RT-qPCR)
Cells were cultivated in 600- to 650-ml mid–log cultures, and 50-ml aliquots were removed for each heat-shock time point and treated with 1/100th volume of 2 M sodium azide to terminate transcription. Total RNA was extracted using phenol-chloroform and purified using an RNeasy kit (Qiagen 74204). Purified RNA (0.5–2 μg) and random primers were used in each cDNA synthesis using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems 4368814). Synthesized cDNA was diluted 1:20, and 5 µl of diluted cDNA was added to each 20 µl real-time PCR. Relative cDNA levels were quantified by the delta delta (∆∆) Ct method. The Pol III transcript SCR1 was used as a normalization control for quantification of HSP mRNA levels. PCR primers used to detect cDNAs are provided in Supplemental Table S3.
Nucleosome reconstitution and Hsf1 in vitro binding assay
For nucleosomal reconstitution of the HSP82 upstream regulatory region, we amplified a 345-base-pair region spanning –353 to –9 (where +1 = ATG start codon) by PCR using a plasmid (KEM101) harboring the HSP82 EcoRI fragment (–1359 to +1543) as template. The forward primer was end labeled and gel purified. Reconstitution was achieved using the salt dilution method. Briefly, a 20-µl reaction containing 5 µg (2 × 106 cpm) of PCR fragment, 5 µg of unlabeled HaeIII/MspI-digested lambda DNA, and 10 µg HeLa histones were incubated in 15 mM Tris–HCl, pH 7.5, 5 M NaCl, 0.2 mM EDTA, 0.2 mM PMSF for 20 min at 37°C and then 5 min at room temperature (RT). At 10-min intervals thereafter, 10 µl of 15 mM Tris–HCl, pH 7.5, 0.2 mM EDTA, and 0.2 mM PMSF were added 18 times to a final volume of 200 µl. This mixture was then loaded onto a 7.5–35% glycerol gradient (3.75 ml total volume) and spun at 33,500 rpm in a Beckman L8 rotor for 15 h at 4°C. The gradient was fractionated into 25 equal volumes and 5 µl from each fraction were electrophoresed on a 5% polyacrylamide Tris–borate–EDTA gel. The fraction containing pure dinucleosomes (no free DNA, no chromatin aggregates) was used for footprinting (see Supplemental Figure S4).
To conduct DNase I footprinting, we challenged the template, either purified dinucleosome or free DNA, with recombinant Hsf1. GST-Hsf1, purified from Escherichia coli as previously described (Erkine et al., 1999), was preincubated with ∼125 ng dinucleosomal template (50,000 cpm) in 50 μl of 15 mM Tris–HCl, pH 7.5, 50 mM KCl, 4 mM MgCl2 for 20 min at RT (0, 0.5, 1.5, 4.5, 13.5 ng GST-Hsf1 were used; see Figure 5B). In parallel, ∼125 ng of naked DNA was similarly incubated with GST-Hsf1. A 1/100 dilution of DNase I (5 µl; Sigma D7291; 100 U/ µl stock) was then added to each mixture, and digestion was allowed to proceed for 2 min. It was terminated by addition of EDTA to 5 mM.
Data access
ChIP-seq and NAC-seq data sets can be accessed using GEO accession number GSE117653.
Supplementary Material
Acknowledgments
We thank Amol Kainth for critical reading of the manuscript and assistance with data analysis; Paula Polk (LSUHSC Core Research Facility) for assistance with ChIP-seq; Tom Volkert and the Whitehead Institute Genome Technology Core for assistance with NAC-seq; Apeng Chen for strain construction; Jodi Taylor for assistance with data mining; and Frank Holstege for yeast strain BY4742-Hsf1-AA. This work was supported by an NIH Early Independence Award (DP5 OD017941) to D.P., a Feist-Weiller Postdoctoral Grant to J.A., and grants from the National Science Foundation (MCB-1518345) and National Institutes of Health (GM 128065) to D.S.G.
Abbreviations used:
- Ac
acetylation
- EMSA
electrophoretic mobility shift assay
- GEO
Gene Expression Omnibus
- GO
gene ontology
- HS
heat shock
- HSE
heat shock element
- Hsf1
heat shock factor 1
- NHS
non–heat shock
- me
methylation
- MEME
Multiple EM for Motif Elicitation
- NFR
nucleosome-free region
- rapa
rapamycin
- RPM
reads per million mapped reads
- RSC
Remodels Structure of Chromatin
- RT
room temperature
- SPMR
signal per million mapped reads
- TE
Tris–EDTA
- TSS
transcription start site
- UAS
upstream activation sequence
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-06-0353) on October 17, 2018.
REFERENCES
- Abravaya K, Myers MP, Murphy SP, Morimoto RI. (1992). The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev , 1153–1164. [DOI] [PubMed] [Google Scholar]
- Adams CC, Gross DS. (1991). The yeast heat shock response is induced by the conversion of cells to spheroplasts and by potent transcriptional inhibitors. J Bacteriol , 7429–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Ondracka A, Cross FR. (2011). Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol Cell , 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman AR, Lue NF, Kornberg RD. (1988). Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol , 5086–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. (1996). RSC, an essential, abundant chromatin-remodeling complex. Cell. , 1249–1260. [DOI] [PubMed] [Google Scholar]
- Chasman DI, Lue NF, Buchman AR, LaPointe JW, Lorch Y, Kornberg RD. (1990). A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev , 503–514. [DOI] [PubMed] [Google Scholar]
- Chowdhary S, Kainth AS, Gross DS. (2017). Heat shock protein genes undergo dynamic alteration in their three-dimensional structure and genome organization in response to thermal stress. Mol Cell Biol , e00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary S, Kainth AS, Pincus DP, Gross DS. (2018). Heat Shock Factor 1 drives intergenic association of its target gene loci upon heat shock. bioRxiv 400499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. (2007). Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell , 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge WJ, O’Duibhir E, Lijnzaad P, van Leenen D, Groot Koerkamp MJ, Kemmeren P, Holstege FC. (2017). Molecular mechanisms that distinguish TFIID housekeeping from regulatable SAGA promoters. EMBO J , 274–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte FM, Fuda NJ, Mahat DB, Core LJ, Guertin MJ, Lis JT. (2016). Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev , 1731–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond DL, Nelson HC. (2006). Genome-wide analysis reveals new roles for the activation domains of the Saccharomyces cerevisiae heat shock transcription factor (Hsf1) during the transient heat shock response. J Biol Chem , 32909–32921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N, Chereji RV, Bharatula V, Bjorklund S, Morozov AV, Broach JR. (2014). A dynamic interplay of nucleosome and Msn2 binding regulates kinetics of gene activation and repression following stress. Nucleic Acids Res , 5468–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkine AM, Adams CC, Diken T, Gross DS. (1996). Heat shock factor gains access to the yeast HSC82 promoter independently of other sequence-specific factors and antagonizes nucleosomal repression of basal and induced transcription. Mol Cell Biol , 7004–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkine AM, Magrogan SF, Sekinger EA, Gross DS. (1999). Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol Cell Biol , 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Chou DM, Struhl K. (2006). Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol , 117–120. [DOI] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. (2010). A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell , 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Guertin MJ, Sharma S, Danko CG, Martins AL, Siepel A, Lis JT. (2015). GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet , e1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi M, Palumbo MJ, Ansari SA, He Q, Tsui K, Nislow C, Morse RH. (2011). Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res , 2032–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell , 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Pugh BF. (2011). Sequential recruitment of SAGA and TFIID in a genomic response to DNA damage in Saccharomyces cerevisiae. Mol Cell Biol , 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Lis JT. (1995). Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol Cell Biol , 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R, Burchfiel ET, Thiele DJ. (2018). Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol , 4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Adams CC, Lee S, Stentz B. (1993). A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J , 3931–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, English KE, Collins KW, Lee S. (1990). Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J Mol Biol , 611–631. [DOI] [PubMed] [Google Scholar]
- Guertin MJ, Lis JT. (2010). Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet , e1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR. (2004). Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol , 5249–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. (2009). Mechanisms that specify promoter nucleosome location and identity. Cell. , 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. (2008). The anchor-away technique, rapid, conditional establishment of yeast mutant phenotypes. Mol Cell , 925–932. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. (2017). A phase separation model for transcriptional control. Cell , 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen BK, Pelham HRB. (1988). Constitutive binding of yeast heat shock factor to DNA in vivo. Mol Cell Biol , 5040–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Gross DS. (2013). Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and Mediator Tail subunits Med15 and Med16. J Biol Chem , 12197–12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakowiak J, Zheng X, Patel N, Feder ZA, Anandhakumar J, Valerius K, Gross DS, Khalil AS, Pincus D. (2018). Hsf1 and Hsp70 constitute a two-component feedback loop that regulates the yeast heat shock response. eLife , e31668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer SB, Gross DS. (2009). SAGA and Rpd3 chromatin modification complexes dynamically regulate heat shock gene structure and expression. J Biol Chem , 32914–32931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, Korber P. (2016). Genomic nucleosome organization reconstituted with pure proteins. Cell , 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods , 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Beckerman R, Freulich E, Prives C. (2011). p53 binding to nucleosomes within the p21 promoter in vivo leads to nucleosome loss and transcriptional activation. Proc Natl Acad Sci USA , 10385–10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MD, Farrer RA, Tan K, Miao Z, Walker LA, Cuomo CA, Wheeler RT, Brown AJ, Wong KH, Cowen LE. (2016). Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans. Nat Commun , 11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Gross DS. (1993). Conditional silencing: The HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol Cell Biol , 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. (2002). Transcriptional regulatory networks in Saccharomyces cerevisiae. Science , 799–804. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. (2007). The role of chromatin during transcription. Cell , 707–719. [DOI] [PubMed] [Google Scholar]
- Li J, Labbadia J, Morimoto RI. (2017). Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol , 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Liu PC, Santoro N, Thiele DJ. (1997). Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J , 6466–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. (2014). Role of DNA sequence in chromatin remodeling and the formation of nucleosome-free regions. Genes Dev , 2492–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Noble WS, Bailey TL. (2014). Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat Protoc , 1428–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. (2016). Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell , 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. (2012). HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell , 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Jaeger AM, Gomez-Pastor R, Willmund F, Frydman J, Thiele DJ. (2014). A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep , 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Jaeger AM, Thiele DJ. (2011). Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat Rev Drug Discov , 930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Werner J, Kim JM, Lis JT, Kim Y-J. (2001). Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol Cell , 9–19. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. (2008). Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell , 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Chereji RV, Hu C, Cole HA, Rawal Y, Clark DJ, Hinnebusch AG. (2016). Genome-wide cooperation by HAT Gcn5, remodeler SWI/SNF, and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation. Genome Res , 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. (2011). Integrative genomics viewer. Nat Biotechnol , 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. (1988). The RNA polymerase II molecule at the 5’ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell , 795–804. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Takemori Y. (2007). Interaction between heat shock transcription factors (HSFs) and divergent binding sequences: binding specificities of yeast HSFs and human HSF1. J Biol Chem , 13334–13341. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Santagata S, Mendillo ML, Sholl LM, Ben-Aharon I, Beck AH, Dias-Santagata D, Koeva M, Stemmer SM, Whitesell L, Lindquist S. (2014). The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell , 564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Iyer VR. (2008). Stress-dependent dynamics of global chromatin remodeling in yeast: A dual role for SWI/SNF in the heat shock stress response. Mol Cell Biol , 2221–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Erkine AM, Kremer SB, Duttweiler HM, Davis DA, Iqbal J, Gross RR, Gross DS. (2006). A functional module of yeast Mediator that governs the dynamic range of heat-shock gene expression. Genetics , 2169–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis EJ, Pandey JP, Zheng X, Jin DX, Gupta PB, Airoldi EM, Pincus D, Denic V. (2016). Defining the essential function of yeast Hsf1 reveals a compact transcriptional program for maintaining eukaryotic proteostasis. Mol Cell , 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B, Melchior W, Jr, Felsenfeld G. (1978). DNAase I, DNAase II and staphylococcal nuclease cut at different, yet symmetrically located, sites in the nucleosome core. Cell , 611–627. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Lewis MJ, Pelham HRB. (1987). Heat shock factor is regulated differently in yeast and HeLa cells. Nature , 81–84. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Nelson HCM. (1989). Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell , 807–813. [DOI] [PubMed] [Google Scholar]
- Venturi CB, Erkine AM, Gross DS. (2000). Cell cycle-dependent binding of yeast heat shock factor to nucleosomes. Mol Cell Biol , 6435–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J, Abrams J, Wang Y, Morano KA. (2012). Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev , 115–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara A, Mahat DB, Guertin MJ, Chu T, Danko CG, Lis JT, Sistonen L. (2017). Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun , 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayachandran V, Reja R, Rossi MJ, Park B, Rieber L, Mittal C, Mahony S, Pugh BF. (2018). Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock. Genome Res , 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hsieh TH, Appleboim A, Chen HV, Rahat A, Amit I, Rando OJ, Friedman N. (2015). High-resolution chromatin dynamics during a yeast stress response. Mol Cell , 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. (1995). Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol , 441–469. [DOI] [PubMed] [Google Scholar]
- Xiao H, Perisic O, Lis JT. (1991). Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell , 585–593. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. (2011). Pioneer transcription factors: establishing competence for gene expression. Genes Dev , 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Lerner J, Iwafuchi-Doi M. (2016). Chromatin scanning by dynamic binding of pioneer factors. Mol Cell , 665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Kasinathan S, Xin B, Rohs R, Henikoff S. (2015). ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo. Nat Commun , 8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao L, Anandhakumar J, Gross DS. (2014). Uncoupling transcription from covalent histone modification. PLoS Genetics , e1004202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol , R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Herrera-Diaz J, Gross DS. (2005). Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol Cell Biol , 8985–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Krakowiak J, Patel N, Beyzavi A, Ezike J, Khalil A, Pincus D. (2016). Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife , e18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. (1998). Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell , 471–480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.