Abstract
The nuclear pore complex (NPC) protein Nup2 plays interphase nuclear transport roles and in Aspergillus nidulans also functions to bridge NPCs at mitotic chromatin for their faithful coinheritance to daughter G1 nuclei. In this study, we further investigate the interphase functions of Nup2 in A. nidulans. Although Nup2 is not required for nuclear import of all nuclear proteins after mitosis, it is required for normal G1 nuclear accumulation of the NPC nuclear basket–associated components Mad2 and Mlp1 as well as the THO complex protein Tho2. Targeting of Mlp1 to nuclei partially rescues the interphase delay seen in nup2 mutants indicating that some of the interphase defects in Nup2-deleted cells are due to Mlp1 mislocalization. Among the inner nuclear membrane proteins, Nup2 affects the localization of Ima1, orthologues of which are involved in nuclear movement. Interestingly, nup2 mutant G1 nuclei also exhibit an abnormally long period of extensive to-and-fro movement immediately after mitosis in a manner dependent on the microtubule cytoskeleton. This indicates that Nup2 is required to limit the transient postmitotic nuclear migration typical of many filamentous fungi. The findings reveal that Nup2 is a multifunctional protein that performs diverse functions during both interphase and mitosis in A. nidulans.
INTRODUCTION
The distinguishing feature of eukaryotes is the compartmentalization of the genetic material into a nucleus by the nuclear envelope (NE). The NE contains nuclear pore complexes (NPCs) that mediate nuclear–cytoplasmic transport as well as inner and outer nuclear membrane proteins that make contact with the chromatin and cytoskeleton, respectively (Hetzer, 2010). The NE of animal cells additionally contains proteins called lamins that make up the nuclear lamina conferring tensile strength on the nuclear periphery to resist mechanical forces (Dahl et al., 2004). In organisms lacking lamins such as yeast, inner nuclear membrane (INM) proteins perform lamin-like functions (King et al., 2008; Gonzalez et al., 2012) and enable nuclear architecture to resist cytoskeletal forces by anchoring chromatin to the nuclear periphery (King et al., 2008; Schreiner et al., 2015).
Regulated bidirectional transport between the nucleus and the cytoplasm is mediated via NPCs (Strambio-De-Castillia et al., 2010). The overall NPC structure is conserved from yeast to vertebrates and is made up of ∼30 different proteins called nucleoporins (abbreviated as Nups) present in multiple copies (Rout et al., 2000; Alber et al., 2007). Nups can be classified based on their location within the NPC structure and include Nups residing on the nuclear side that are connected by a distal ring to form a basket-like structure (Grossman et al., 2012). The core scaffolding member of this NPC basket is the conserved translocated promoter region (TPR) protein in vertebrates, the orthologue of Mlp1 and Mlp2 in budding yeast (Krull et al., 2010; Niepel et al., 2013). The nuclear basket also possesses the FG-repeat Nups Nup153 and Nup50 in vertebrates that are homologous to Nup60 and Nup2 in budding yeast (Guan et al., 2000; Hood et al., 2000; Rout et al., 2000; Makise et al., 2012). In addition to these proteins, the nuclear basket is the docking site for Mad1 and Mad2 in yeast, fungi, plants, and vertebrates (Campbell et al., 2001; Ikui et al., 2002; Iouk et al., 2002; De Souza et al., 2009; Ding et al., 2012). MAD (mitotic arrest deficient) proteins locate to interphase NPCs but locate to kinetochores during mitosis to mediate the spindle assembly checkpoint (SAC) response that ensures fidelity of chromosome segregation (Foley and Kapoor, 2013). Several studies indicate that the nuclear basket–associated proteins play roles in transcription, RNA biogenesis, SUMO homeostasis, and chromatin organization (Strambio-De-Castillia et al., 2010).
Nucleocytoplasmic transport is executed by nuclear transporters (NTRs) known as karyopherins/importins/exportins (Stewart, 2007). The NTRs recognize specific signatures on their cargoes called the nuclear localization sequence (NLS) or the nuclear export sequence to be transported into or out of nuclei, respectively (Stewart, 2007). The best studied type of NLS, also referred to as the “classical NLS,” contains either a single cluster of basic amino acids such as lysine and arginine (monopartite NLS) as found in the SV40 T-antigen NLS (Kalderon et al., 1984) or two clusters of basic amino acids separated by a spacer (bipartite NLS) such as in the nucleoplasmin NLS (Dingwall et al., 1988). The classical NLS sequences are specifically recognized by importin α in the importin α:β heterodimer that docks them through the nuclear pores to release them into the nucleoplasm with the help of karyopherin release factors (KaRFs; Gilchrist et al., 2002).
One such KaRF is Nup2 and its mammalian orthologue Nup50 (Guan et al., 2000; Dilworth et al., 2001; Gilchrist et al., 2002). Nup50 is essential for embryonic development in mice (Smitherman et al., 2000) and for viability in the filamentous fungus Aspergillus nidulans (Osmani et al., 2006). Nup2 is a highly mobile nucleoporin with low residence times at NPCs (Denning et al., 2001; Dilworth et al., 2001; Rabut et al., 2004) and exists in an additional nucleoplasmic pool in flies and vertebrates (Kalverda et al., 2010; Buchwalter et al., 2014). The structure of Nup2 is conserved from yeast to vertebrates with an N-terminal region that binds importin α, a C-terminal region that binds RanGTP, and a central FG-rich region (Booth et al., 1999; Lindsay et al., 2002; Matsuura et al., 2003; Matsuura and Stewart, 2005). Nup2 is targeted to nuclear pores via Nup60 in budding yeast (Denning et al., 2001), Nup153 in mammals (Makise et al., 2012), and NupA in A. nidulans (Markossian et al., 2015). Nup2 performs various functions in the nuclear transport cycle such as accelerating nuclear import rates, recycling importin α back into the cytoplasm, and in nuclear protein export (Booth et al., 1999; Guan et al., 2000; Hood et al., 2000; Gilchrist et al., 2002; Lindsay et al., 2002; Matsuura et al., 2003; Matsuura and Stewart, 2005; Makise et al., 2012; Finn et al., 2013). Nup2 also interacts with active genes in the nucleoplasm, participates in gene regulation, and controls the epigenetic state of chromatin by preventing spreading of repressive marks (Ishii et al., 2002; Dilworth et al., 2005; Schmid et al., 2006; Kalverda et al., 2010; Buchwalter et al., 2014). In A. nidulans and mammalian cells, Nup2 locates to mitotic chromatin (Osmani et al., 2006; Dultz et al., 2008) and Nup2 is required for normal mitotic progression in A. nidulans (Markossian et al., 2015). Recently identified roles of Nup2 include chromosome organization during meiosis (Chu et al., 2017).
We recently reported that A. nidulans Nup2 is required for faithful mitotic NPC segregation by bridging NPCs to segregating chromatin via a central targeting domain (Suresh et al., 2017). Nup2 null mutants inherit fewer NPCs after mitosis and accumulate misplaced NPC proteins in the cytoplasm (Suresh et al., 2017). Because Nup2 mutants have reduced NPC number, we sought to understand the consequence of the lack of Nup2 on interphase progression. We previously reported a defect in the G1 nuclear accumulation of Mad1 in nup2 mutants (Markossian et al., 2015). In this work, we report that lack of Nup2 delays interphase progression and leads to defects in the G1 nuclear accumulation of specific classical NLS-bearing protein cargoes including the NPC basket proteins Mlp1, Mad2 and the nucleoplasmic protein Tho2. Furthermore, the conserved INM protein Ima1 involved in nuclear movement (Borrego-Pinto et al., 2012) fails to locate normally in nup2 mutants. This is accompanied by prolonged G1 nuclear movements after mitosis that are dependent on the microtubule cytoskeleton. Together, we have shown that Nup2 is a multifunctional protein that performs diverse functions that contribute to normal NPC basket formation, nuclear positioning, and transport in A. nidulans.
RESULTS
Nup2 is required for the normal localization of the nuclear pore basket–associated proteins Mad2 and Mlp1 in G1 after completion of mitosis
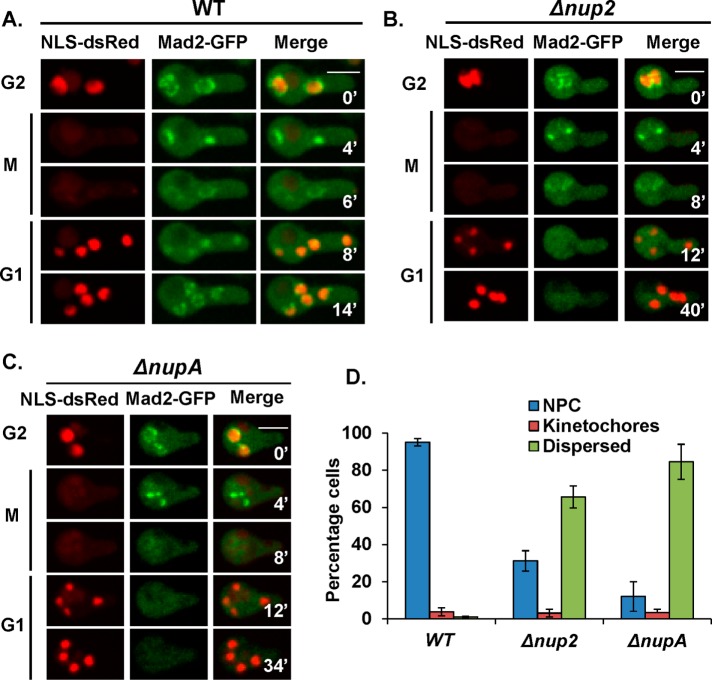
A. nidulans Nup2 is dispensable for rapid reimport of NLS-DsRed protein during mitotic exit, a nuclear transport marker used to monitor nuclear import that contains a NLS from the APSES transcription regulator StuA (Toews et al., 2004; Markossian et al., 2015). NLS-DsRed is released from nuclei following NPC disassembly during early mitosis and is efficiently reimported in G1 after NPCs are reassembled (Figure 1A). Although dispensable for NLS-DsRed import, Nup2 is required for the postmitotic G1 nuclear import of the NPC basket–associated protein Mad1 (Markossian et al., 2015). Because Mad1 resides with Mad2 at the nuclear basket (Lee et al., 2008; De Souza et al., 2009; Ding et al., 2012), we investigated the nuclear localization of Mad2-GFP without Nup2 function. In interphase wild-type (WT) cells, Mad2-GFP locates to NPCs (Figure 1A) but then transitions to kinetochores during mitosis to mediate the SAC (Waters et al., 1998; Shah et al., 2004; Musacchio and Salmon, 2007; De Souza et al., 2009). Once the SAC is satisfied, Mad2-GFP disperses from nuclei during late mitosis and gets reimported into G1 nuclei and then incorporated back to NPCs (Figure 1A). In cells lacking Nup2 (Figure 1B) or NupA function (Figure 1C), we found a marked defect in Mad2-GFP cell cycle–specific locations. Although Mad2-GFP located normally to interphase NPCs and mitotic kinetochores, it failed to accumulate normally into postmitotic G1 nuclei but instead remained dispersed in the cytoplasm for an extended period without Nup2 or NupA function (Figure 1, B and C).
FIGURE 1:
Nup2 and NupA facilitate the G1 nuclear import of the nuclear basket protein Mad2. Time-lapse microscopy of Mad2-GFP and NLS-DsRed in (A) WT (strain SGS282), (B) Δnup2 (SGS285-H), and (C) ΔnupA (SGS287-H). (D) Quantitation of the mislocalization of Mad2 in asynchronously growing WT, Δnup2, and ΔnupA cells (n = 375 cells). Scale bar, ∼5 μm.
In normal growing cultures a high population of WT cells (95%) has Mad2-GFP at NPCs (Figure 1D) reflective of the long interphase duration compared with the short mitosis in A. nidulans (Bergen and Morris, 1983). A small percentage of WT cells (∼5%) have Mad2-GFP at mitotic nuclear locations reflective of those in mitosis (Figure 1D). A very low percentage of cells have Mad2-GFP dispersed from nuclei when it is transiently released from nuclei during mitotic exit before nuclear transport is reestablished during G1 (Figure 1D). These ratios were significantly altered without Nup2 or its targeting partner NupA (Markossian et al., 2015), with a large increase in the percentage of cells with dispersed Mad2-GFP and a corresponding decrease in the percentage of cells with NPC-localized Mad2 (Figure 1D). In Nup2-deleted cells, although the interphase location of Mad2 is affected, its mitotic kinetochore localization and ability to signal the SAC response (Foley and Kapoor, 2013) was unaffected as revealed by an active SAC in cells without Nup2 (Markossian et al., 2015). This indicates that Nup2 and NupA are required for the timely import and/or incorporation of Mad2 to NPCs after mitosis in G1.
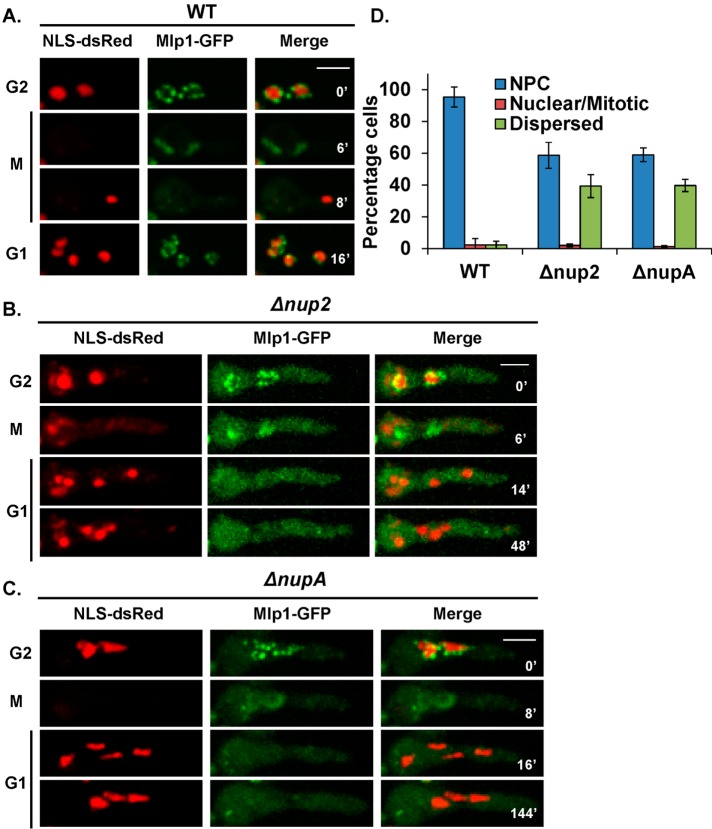
Nup2 is required for normal nuclear localization of the nuclear basket–associated proteins Mad1 and Mad2 and both proteins are tethered to the NPCs via a third protein, Mlp1 (De Souza et al., 2009). We therefore analyzed the localization of Mlp1 without Nup2 or without NupA. Mlp1 localizes to interphase NPCs, transitions onto the mitotic spindle matrix during mitosis, then disperses into the cytoplasm and is immediately imported and incorporated into G1 nuclei concomitant with NLS-DsRed in WT cells (Figure 2A). However, without Nup2 (Figure 2B) or NupA (Figure 2C), although Mlp1 located normally during G2 and mitosis, it failed to accumulate into G1 nuclei and remained cytoplasmic. Quantitation in a population of cells, as described for Mad2 (Figure 1D), indicated that Mlp1-GFP remained dispersed in most of the cells in a population suggestive of a long delay in its NPC incorporation (Figure 2D). Mlp1-GFP was eventually incorporated to NPCs (Supplemental Figure S1A) and almost all cells located Mlp1 to NPCs before entering mitosis (Supplemental Figure S1B). We have previously reported that Nup2 mutants exhibit a range of nuclear sizes and shapes with nuclear chromatin appearing in unequal masses (Osmani et al., 2006). These defects were also recapitulated in NupA mutants (Figure 2C) in which nuclei characteristically underwent dynamic changes in shape due to constant pulling forces as can be seen in Supplemental Movie 1, which tracks Mlp1-GFP and NLS-DsRed in a ∆nupA cell. Together, these data show that Nup2 and NupA are required for the timely nuclear localization of the nuclear basket proteins Mlp1 and Mad2 during G1.
FIGURE 2:
Nup2 and NupA facilitate the postmitotic nuclear import of the NPC basket protein Mlp1. Time-lapse microscopy of Mlp1-GFP and NLS-DsRed in (A) WT (SGS102), (B) Δnup2 (SGS121-H), and (C) ΔnupA cells (SGS105-H). (D) Quantitation of the mislocalization of Mlp1 in asynchronously growing WT, Δnup2, and ΔnupA cells (n = 450 cells). Scale bar, ∼5 μm.
Movie S1.
△nupA cell expressing Mlp1-GFP and NLS-DsRed going through mitosis. Images were collected every 8 minutes.
While analyzing the defects in the nuclear accumulation of Mad2 and Mlp1 in Nup2-deleted cells, we observed that interphase was longer without Nup2 function. Calculation of interphase timing by measuring the time between consecutive mitosis when NLS-DsRed disperses revealed that interphase is significantly prolonged in Nup2-deleted cells (Supplemental Figure S1C). Whereas WT cells spend around 200 min in interphase at room temperature (23–25°C), Nup2-deleted cells spend, on average, 450 min in interphase (Supplemental Figure S1C). Together, these studies indicate that Nup2 is required for normal postmitotic nuclear localization of nuclear basket–associated proteins and for normal interphase progression timing.
Retargeting Mlp1 to interphase nuclei without Nup2 partly rescues the interphase delay but not the mitotic delay
Mlp1 is a key element of the nuclear pore basket that performs various interphase functions including mRNA export and surveillance, and regulation of SUMO protein dynamics (Galy et al., 2004; Zhao et al., 2004). We considered the possibility that lack of Mlp1 at NPCs during interphase might therefore contribute to some Nup2 deletion defects. To address this, we first determined the impact of Mlp1 on cell cycle duration in A. nidulans. It was previously reported that Mlp1-deleted cells undergo several nuclear divisions but cannot grow to form a colony (De Souza et al., 2009). The localizations of Nup2 (unpublished data) and NupA are largely unaffected without Mlp1 (Supplemental Figure S2B). We could therefore monitor mitosis by following NupA-GFP and NLS-DsRed. NupA is the targeting partner for Nup2 and, like Nup2, locates to interphase NPCs and mitotic chromatin (Markossian et al., 2015). We found that Mlp1 deletion led to a prolonged presence of NupA on chromatin and a prolonged period with dispersed NLS-DsRed indicating that progression through mitosis is delayed (Supplemental Figure S2, A and B). Quantitation of mitotic timing revealed a mitotic delay in around 50% of cells that varied between 11 and 50 min in contrast to <10′ for WT cells at room temperature (Supplemental Figure S2C). Measurement of interphase timing by monitoring two consecutive rounds of NLS-DsRed dispersal indicated a modest increase in interphase duration without Mlp1 (Supplemental Figure S2D). These results support the conclusion that Mlp1 is important for normal rates of cell cycle progression in A. nidulans.
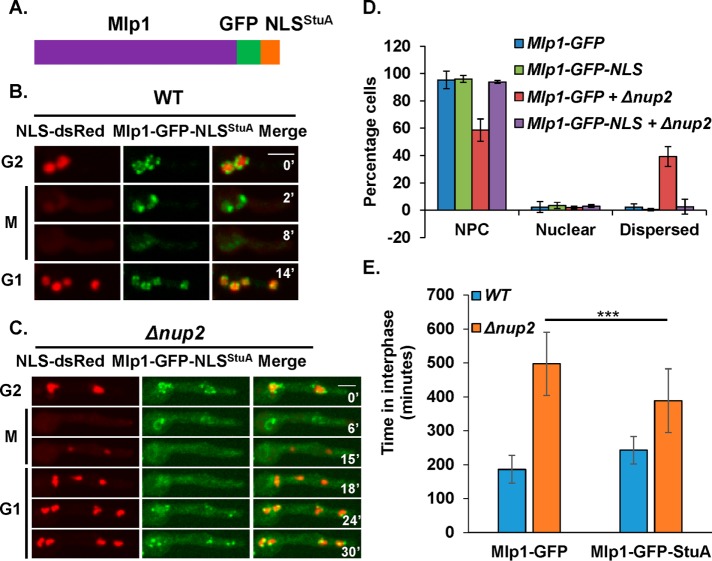
To determine whether Mlp1 mislocalization contributes to Nup2-deletion phenotypes in interphase, we targeted Mlp1 into interphase nuclei without Nup2. This was achieved by fusing the endogenous copy of Mlp1 with the NLS sequence of StuA (Suelmann et al., 1997) that is independent of Nup2 for its nuclear import (Markossian et al., 2015). NLSStuA was fused in frame with endogenous Mlp1-GFP at its carboxy terminus to enable visualization of the fusion protein by live cell microscopy (Figure 3A). In WT cells, Mlp1-GFP-NLSStuA behaved like Mlp1-GFP and located to NPCs in interphase, nuclear structures during mitosis, dispersed later in mitosis, and was reimported and incorporated into NPCs in G1 (Figure 3B). In the absence of Nup2, although Mlp1-GFP-NLSStuA behaved like Mlp1-GFP during G2 and M, there was a difference between the two proteins during G1 (Figure 3C). Whereas Mlp1-GFP failed to return to G1 nuclei (Figure 2B), Mlp1-GFP-NLSStuA located to nuclei much quicker in G1 (Figure 2B vs. Figure 3C). Although there was a delay in the nuclear import of Mlp1-GFP-NLSStuA in G1 compared with StuA-NLS-DsRed, this delay was negligible compared with the delay seen for Mlp1-GFP. In a cell population, the percentage of cells with dispersed Mlp1-GFP were rescued to WT levels in Mlp1-GFP-NLSStuA cells (Figure 3D) indicating that the interphase NPC location of Mlp1 was largely restored without Nup2 by the NLS addition.
FIGURE 3:
Fusion of Mlp1 with the StuA NLS rescues its interphase nuclear targeting in Nup2-deleted cells. (A) Cartoon depicting the chimeric protein construct of Mlp1 fused C-terminally with GFP followed by the StuA-NLS. Live cell confocal images of Mlp1-GFP-NLSStuA and NLSStuA-DsRed in (B) WT cells (SGS311) and (C) Δnup2 cells (SGS306-H) during G2-M-G1 transitions. (D) Quantitation of cells based on the localization of Mlp-GFP-NLSStuA and Mlp1-GFP in asynchronously growing WT and Δnup2 cells (n = 250 and 450 cells respectively, for WT and Δnup2 cells). (E) Time spent in interphase in Mlp1-GFP and Mlp-GFP-NLSStuA cells with or without Nup2. p value < 0.001 (n = 20 mitoses each). Scale bar, ∼5 µm.
Mlp1 plays interphase roles and its absence leads to an interphase delay (Supplemental Figure S2D). Therefore, restoration of the interphase location of Mlp1 in Nup2-deleted cells might at least in part rescue the interphase delay. Consistent with this expectation, Nup2-deleted cells with Mlp1-GFP-NLSStuA spent significantly less time in interphase compared with Nup2-deleted cells with Mlp1-GFP (Figure 3E). Despite partial rescue in interphase delay in Nup2-deleted cells when Mlp1 location was corrected, no rescue in colony growth (Supplemental Figure S3A) or mitotic timing was observed (Supplemental Figure S3B). The results indicate that Nup2 contributes to normal interphase progression partly by ensuring normal interphase localization of Mlp1.
Nup2 is required for normal G1 nuclear accumulation of specific nucleoplasmic cargoes
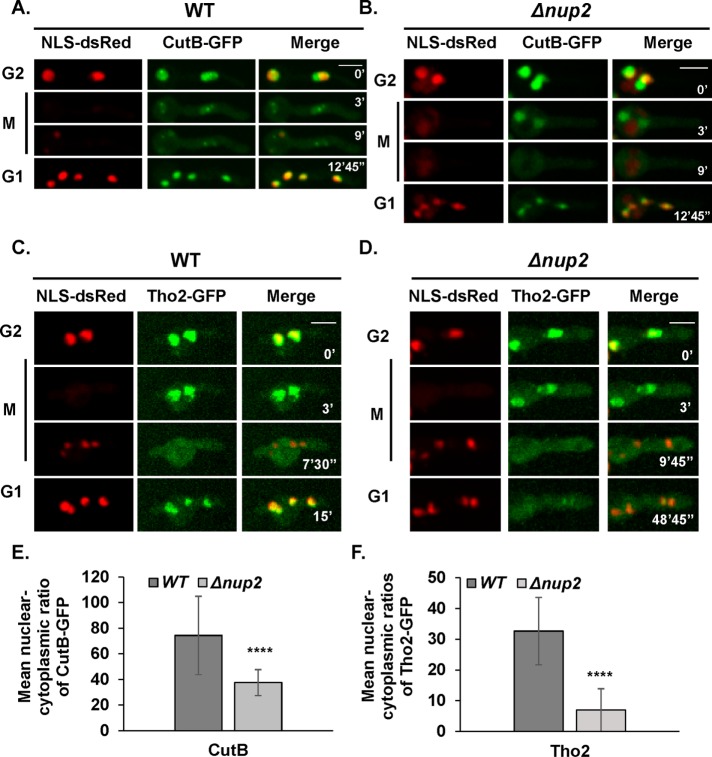
Nup2 deletion affects the nuclear localization of the nuclear basket–associated proteins Mad1, Mad2, and Mlp1. Mad1 is imported via the importin α/β pathway in yeast (Scott et al., 2005) and Mlp1 is imported via the same pathway in vertebrates (Ben-Efraim et al., 2009). Nup2 could therefore potentially affect nuclear import of specific proteins that are associated with NPCs. We therefore wished to determine whether Nup2-affected proteins extend beyond NPC proteins. In yeast and human cells, Nup2 interacts with importin α to increase the rate of cargo disassociation via the canonical importin α/β pathway (Gilchrist et al., 2002; Lindsay et al., 2002; Matsuura et al., 2003; Matsuura and Stewart, 2005). In A. nidulans, Nup2 purifies with importins α and β1 as well as NupA (Markossian et al., 2015). A. nidulans has four nuclear transport pathways that are essential based on knockout analysis of karyopherin genes (Markina-Inarrairaegui et al., 2011). Among these pathways, we expected that Nup2 might function through the importin α/β pathway and identified potential cargo proteins that interact with importins α and/or β. The candidates were identified using affinity purifications of importin α and β followed by mass spectroscopy analysis (A.O. and S.O., unpublished data). We identified two importin α/β interacting proteins encoded by the loci AN2226 and AN5694 (Supplemental Table S3). AN2226 encodes the predicted orthologue of the THO complex protein Tho2 involved in transcription elongation in Saccharomyces cerevisiae (Piruat and Aguilera, 1998). AN5694 encodes a ribonucleotidyltransferase protein called CutB involved in mRNA degradation in A. nidulans (Morozov et al., 2012). Tho2 and CutB were endogenously tagged with GFP at the carboxy terminus and their localization during the cell cycle examined. Both proteins contain predicted classical NLS sequences (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). CutB-GFP located to nuclei during interphase as expected (Figure 4A). During mitosis, CutB-GFP largely dispersed from nuclei albeit with different kinetics compared with NLS-DsRed (Figure 4A). During mitosis in WT cells, CutB located to focal structures and this concentration was compromised in Nup2 null mutants (Figure 4, A and B). What these structures represent is currently unknown. After completion of mitosis, CutB-GFP located back to interphase nuclei in WT cells (Figure 4A). In the absence of Nup2, the localization of CutB-GFP was mildly affected in G1 (Figure 4B). In an interphase cell population, measurement of the nuclear–cytoplasmic ratio of CutB-GFP revealed an approximate twofold difference between WT and Δnup2 cells (Figure 4E). Similar to CutB-GFP, Tho2-GFP located to interphase nuclei (Figure 4C). During mitosis Tho2-GFP dispersed from nuclei later than NLS-DsRed in WT and Δnup2 cells (Figure 4, C and D). Although Tho2-GFP reaccumulated into WT nuclei in G1 similar to NLS-DsRed, it remained largely cytoplasmic in Δnup2 cells (Figure 4, C and D). In a cell population, measurement of the nuclear–cytoplasmic ratio of Tho2-GFP indicated an approximate fivefold reduction in Δnup2 cells (Figure 4F). Together, these results indicate that Nup2 differentially impacts the interphase nuclear accumulation of importin α/β interacting nuclear proteins while having little to no effect on the nuclear marker protein NLS-DsRed (Markossian et al., 2015).
FIGURE 4:
Nup2 affects G1 nuclear localization of different proteins to various degrees. Live cell imaging of CutB-GFP in relation to NLS-DsRed during interphase and mitosis in (A) WT (SGS206) and (B) Δnup2 cells (SGS290-H). (C) Live cell imaging of Tho2-GFP in relation to NLS-DsRed during interphase and mitosis in WT (SGS212) and (D) Δnup2 cells (SGS263-H). Quantitation of the nucleo/cytoplasmic ratios of (E) CutB-GFP and (F) Tho2-GFP signal intensities in interphase cells indicating differential effects without Nup2. p value < 0.0001 for E and F (n = 25 nuclei each). Scale bar, ∼5 μm.
We next examined whether the G1 nuclear accumulation of Mlp1 and Tho2 is dependent on the transport functions of Nup2, which are known to involve its conserved N- and C-terminal importin α and RanGTP-binding domains, respectively (Booth et al., 1999; Lindsay et al., 2002; Matsuura et al., 2003; Matsuura and Stewart, 2005). These domains were removed from endogenous Nup2 (referred to as Nup2ΔΔ) and the truncated protein is, perhaps surprisingly, known to be functional (Suresh et al., 2017). The G1 localization of Mlp1 and Tho2 was examined in Nup2ΔΔ cells and found to be mildly affected when compared with cells without Nup2 function (Figure 5, A and B, compared with Figures 2B and 4D, respectively). Measurement of interphase timing in Nup2ΔΔ cells indicated a slight delay (Figure 5C) compared with Δnup2 mutants (Supplemental Figure S1C). These studies indicate that Nup2 affects G1 nuclear accumulation of proteins in a manner largely independent of its conserved importin α and Ran-binding domains involved in accelerating the rate of nuclear transport in yeast and vertebrate cells.
FIGURE 5:
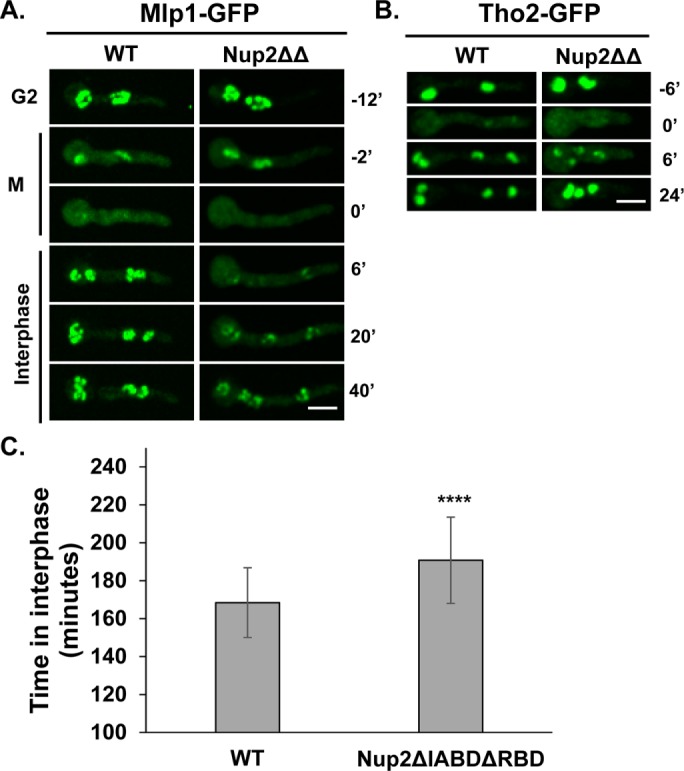
Localization of Mlp1 and Tho2 is largely unaffected in Nup2ΔIABDΔRBD-GFP cells. Localization of (A) Mlp1-GFP (SGS337 and SGS90) and (B) Tho2-GFP (SGS340 and SGS193) in Nup2ΔIABDΔRBD cells in comparison to WT cells during G2-M-G1 transitions. (C) Interphase timing in Nup2ΔIABDΔRBD cells based on consecutive cycles of mitotic NLS-DsRed dispersal in comparison to WT cells. p value < 0.0001 (n = 50 mitoses each for control and mutants). Scale bar, ∼5 µm.
Nup2 is required for the normal localization of the INM protein Ima1
Because Nup2 deletion leads to specific nuclear accumulation defects in NPC-associated as well as nucleoplasmic proteins, we next examined whether the localization of INM proteins is affected without Nup2 function. Some INM proteins bear classical NLS sequences and are imported through nuclear pores via the importin α/β pathway involving Nup2 in budding yeast (King et al., 2006). Previous analyses showed that the localization of the INM protein MtgA (Chemudupati et al., 2016), the fission yeast orthologue of Bqt4 involved in anchoring telomeres to the NE (Chikashige et al., 2009), is unaffected without Nup2 function (Markossian et al., 2015). The localization of the INM protein Src1 (Liu et al., 2015) was also unaffected without Nup2 (unpublished data).
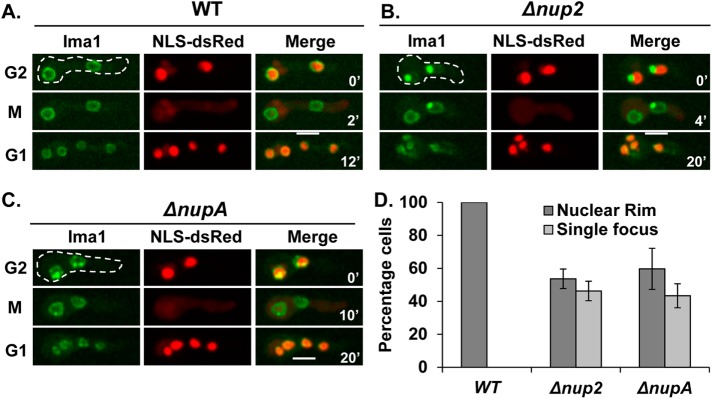
Ima1 is an integral INM protein that helps connect the nucleus to the cytoskeleton along with the other INM proteins Lem2 and Man1 (King et al., 2008; Hiraoka et al., 2011). The orthologue of vertebrate Ima1, termed Samp1, is involved in nuclear movement (Borrego-Pinto et al., 2012). Interestingly, without Nup2 function, we observed a defect in the localization of Ima1 within the NE. Ima1-GFP localizes uniformly around the nuclear periphery in WT cells during interphase and mitosis (Figure 6A). However, when Nup2 or NupA was deleted, Ima1 concentrated into one or two foci at the nuclear periphery during interphase. It then notably redistributed throughout the NE during mitosis (Figure 6, B and C). Quantitation of a cell population, based on the localization of Ima1-GFP, revealed a significant penetrance of this defect (Figure 6D). To determine whether microtubules contribute to the focal accumulation of Ima1-GFP in Nup2-deleted cells, the cells were treated with benomyl and we followed the localization of Ima1-GFP. The focal accumulation of Ima1-GFP in Nup2-deleted cells was found to be MT-independent (Supplemental Figure S4). Together, these results indicate that Nup2 functions to ensure normal localization of the integral INM protein Ima1 during interphase.
FIGURE 6:
Nup2 is required for normal interphase localization of the INM protein Ima1. Localization of the INM protein Ima1-GFP in relation to NLS-DsRed in (A) WT (HA449) cells, (B) Δnup2 cells (SGS159-H), and (C) ΔnupA cells (SM106-H) during G2-M-G1 transitions. (D) Quantitation of cells based on the localization of Ima1-GFP along the nuclear rim vs. at a concentrated focus in WT, Δnup2, and ΔnupA cells (n = 387, 158, and 186 cells, respectively). Scale bar, ∼5 µm.
Nup2 mutants exhibit increased microtubule-dependent nuclear movement during G1
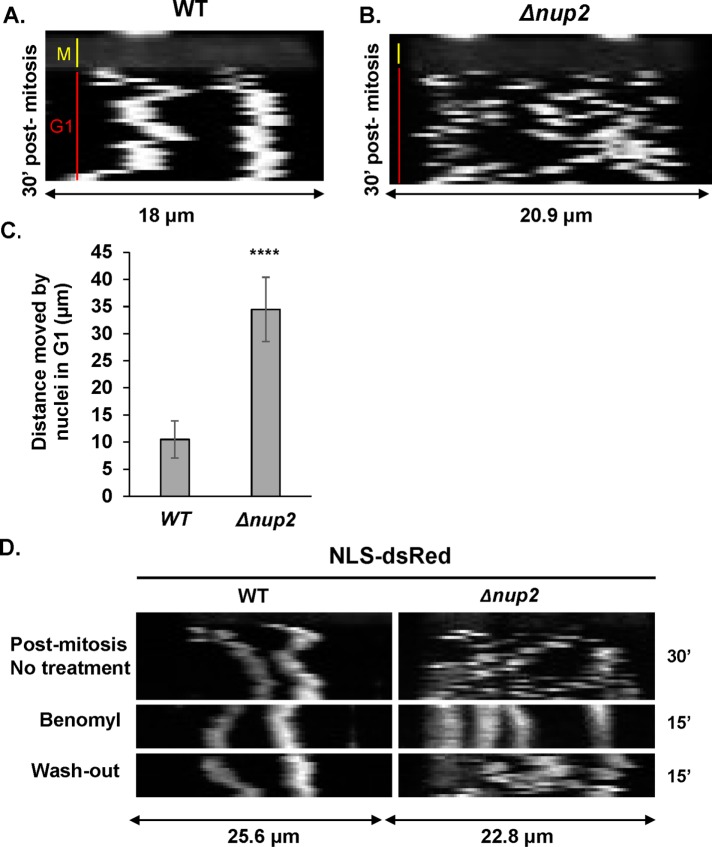
A. nidulans cells contain evenly spaced nuclei in a shared cytoplasm. However, a prior study revealed that in WT cells, postmitotic G1 daughter nuclei undergo transient bidirectional rapid movement along hyphae for a short duration before being more stably positioned evenly apart from each other (Fox et al., 2002). Similar transient (ca. 5–15 min) postmitotic nuclear migrations have also been reported for other fungi (reviewed in Aist and Morris [1999]). Fox et al. tracked GFP-tagged BimG, a type 1 phosphatase that locates to nuclei and spindle pole bodies, to observe the rapid nuclear movements after completion of mitosis (Fox et al., 2002, in their Supplemental file 2 movie). By monitoring NLS-DsRed we were also able to track mitosis (dispersal of NLS-DsRed from nuclei) as well as postmitotic G1 nuclear movements (NLS-DsRed within nuclei after mitosis). By generating kymographs of cells completing mitosis we could track G1 nuclei and observed nuclei moving immediately after mitosis before they became less mobile (Figure 7A). Interestingly, Nup2 mutants exhibited prolonged postmitotic nuclear movement that extended for more than 30 min after mitosis (Figure 7B). In Nup2 null mutants, unlike WT cells, it was difficult to track specific individual nuclei through time because of their higher mobility and tendency to travel past each other. Therefore, we have presented kymographs of an entire cell with two nuclei in this case (Figure 7B). Quantitation of the total distance moved by nuclei for identical time periods postmitosis indicated a significant increase in the distance moved by nuclei in Δnup2 in comparison to WT cells (Figure 7C).
FIGURE 7:
Nup2 is required to limit microtubule-dependent nuclear mobility after mitosis in early G1. Kymographs of NLS-DsRed during mitosis (yellow line) and for 30 min into G1 (red line) in (A) WT cells (SGS197) and (B) Δnup2 cells (SGS159-H). (C) Quantitation of total distance moved by nuclei in G1 analyzed for 15 min at 1 min intervals (n = 10 mitoses each for WT and Δnup2 cells. p value < 0.0001). (D) Kymographs of NLS-DsRed 30 min postmitosis followed by addition of 2.4 µg/ml benomyl for 15 min and after benomyl washout for 15 min.
We next examined whether the excessive movement of postmitotic G1 nuclei was dependent on the microtubule cytoskeleton. Treatment of postmitotic G1 cells with the microtubule depolymerizing drug benomyl had little effect on the movement of G1 nuclei of WT cells (Figure 7D). However, in Nup2-deleted cells, addition of benomyl stopped the extensive nuclear movement seen during postmitotic G1 nuclei (Figure 7D). However, upon removal of benomyl, allowing repolymerization of microtubules (Horio and Oakley, 2005), the atypical nuclear movements resumed in the Nup2-deleted cells (Figure 7D). These results indicate that Nup2 is required to limit the normal postmitotic microtubule-dependent nuclear movements transiently triggered after completion of mitosis in A. nidulans.
DISCUSSION
Nup2 is an essential NPC protein in A. nidulans with roles in interphase and mitosis (Matsuura et al., 2003; Matsuura and Stewart, 2005; Markossian et al., 2015; Suresh et al., 2017). We recently reported that Nup2 mediates NPC segregation by bridging NPCs to chromatin during mitosis such that nup2 mutants inherit fewer NPCs in their NE after completion of mitosis (Suresh et al., 2017). Notably, however, if the mitotic NPC segregation defect is corrected in Nup2 null cells, by providing an artificial chromatin-NPC bridge, cells still remain inviable (Suresh et al., 2017) indicating that additional essential Nup2 functions exist. In the current study, we investigated the consequences of the loss of Nup2 during interphase and report that Nup2 is required for the nuclear accumulation of specific proteins after mitosis as well as for stable postmitotic nuclear positioning in G1.
Nup2 was previously shown to be required for the timely G1 accumulation of the NPC basket–associated protein Mad1 although Nup2 is not required for the nuclear accumulation of NLS-DsRed (Markossian et al., 2015). Extending this analysis to other nuclear pore basket–associated proteins, we find that the Mad1-anchoring protein Mlp1, as well as Mad2, also do not accumulate normally in G1 nuclei without Nup2. In yeast and vertebrate cells, the tethering of Mlp1 to NPCs is dependent on Nup153 (NupA is the likely A. nidulans Nup153 functional orthologue; Hase and Cordes, 2003). However, our studies in A. nidulans indicate that Nup2 and NupA are involved in the nuclear import of Mlp1, Mad1, and Mad2. In A. nidulans, Mlp1 acts as a tether for Mad1 and Mad2 at the NPC basket within nuclei (De Souza et al., 2009) and in the absence of Mlp1, both Mad1 and Mad2 are transported into nuclei but then fail to locate to NPCs and remain in the nucleoplasm (De Souza et al., 2009). This indicates that their nuclear accumulation is not dependent on NPC tethering. In contrast, when Nup2 is deleted neither Mad1 nor Mad2 locates within nuclei after mitosis but instead remain in the cytoplasm for an extended duration. The data indicate that Nup2 likely plays a role in the nuclear transport of Mlp1, Mad1, and Mad2 but not for the import of NLS-DsRed. This suggests Nup2 might preferentially affect nuclear import of NPC-associated proteins. However, analysis of two additional nuclear proteins provides evidence that Nup2 affects the nuclear accumulation of different proteins to different degrees. Although the conventional nuclear transport roles of Nup2 are mediated via its N- and C-terminal domains, lack of these domains in A. nidulans does not drastically impact the nuclear accumulation of the proteins affected in the absence of Nup2. These findings suggest that Nup2 promotes nuclear accumulation of specific proteins in G1 via mechanisms that are independent of its conserved IABD and RBD domains and could potentially involve effective mitotic NPC inheritance (Suresh et al., 2017).
Because Mlp1 is the key architectural element of the nuclear basket (Krull et al., 2010; Niepel et al., 2013), lack of nuclear Mlp1 in Nup2-deleted cells compromises the NPC basket structure. mRNA surveillance and export proceeds with docking of mRNPs onto the nuclear basket of NPCs (Zhang et al., 2002; Zhao et al., 2004; Green et al., 2003; Saroufim et al., 2015; Schneider et al., 2015). Hence, it is likely that aspects of RNA biology are defective in the absence of Nup2, although this remains to be directly tested. Interestingly, some interphase defects caused by the mislocalization of Mlp1 to the cytoplasm in the Δnup2 null mutants are somewhat fixable as indicated by a rescue in interphase delay by artificial targeting of Mlp1 to NPCs without Nup2. We have previously shown that deletion of Mlp1 does not impact the SAC response in A. nidulans (De Souza et al., 2009). Consistent with that observation, we find that in the absence of Mlp1, cells undergo a delay before anaphase with Nup2 and NupA locating throughout unseparated mitotic chromatin. Therefore, it is likely that Mlp1 performs yet unidentified early mitotic roles in A. nidulans.
In addition to NPC proteins, Nup2 is required for the G1 nuclear accumulation of other proteins that purify with importins α/β. Although the examined proteins CutB and Tho2 possessed classical NLS sequences, Tho2 was more sensitive for its nuclear import to lack of Nup2 function. Therefore, there might be other determinants in proteins in addition to their NLS sequences that dictate the Nup2 dependence of their nuclear import as previously reported for Nup124 (Dang and Levin, 2000; Varadarajan et al., 2005; Sistla et al., 2007). Nuclear protein transport dependencies on specific Nups have been observed before which could also contribute to differential import of a subset of proteins (Balasundaram et al., 1999; Dang and Levin, 2000; Varadarajan et al., 2005; Sistla et al., 2007; Walde et al., 2012).
Another consideration to take into account for understanding the varying effects of Nup2 deletion on nuclear import of different proteins is that NLS sequences with varied strengths have previously been noted (Hodel et al., 2001; Kakar et al., 2007). In yeast and humans, Nup2 contributes to the recycling of importin-α from the nucleus to the cytoplasm, making it available for further import complex formation (Solsbacher et al., 2000; Matsuura and Stewart, 2005). If importin-α becomes limiting in the cytoplasm without Nup2, one can imagine a scenario in which only high-affinity NLS sequence-bearing proteins would manage to bind the available limited importin α and get imported preferentially over weaker NLS-bearing proteins. Moreover, the abnormal occurrence of peripheral FG Nups in the cytoplasm without Nup2 (Suresh et al., 2017) could lead to their binding of importin β, further limiting the importin α/β complex available for nuclear import. In the future, it might be informative to examine the localization of importin α and β to determine whether their localizations are altered by lack of Nup2.
Another nuclear protein that is mislocalized in the absence of Nup2 is the INM protein Ima1. As an integral INM protein containing multiple transmembrane domains, Ima1 remains at the NE throughout interphase and during mitosis in WT cells. However, in the absence of Nup2 or NupA, the levels of Ima1 around the NE are reduced and one to two foci of the protein instead accumulate at the nuclear periphery. Most noticeably, when cells enter mitosis, Ima1 levels at the foci drop while the levels around the NE increase. Upon completion of mitosis and entry into interphase Ima1 remains at the NE but no longer locates at foci. Ima1 subsequently accumulates at the NE foci before the next mitosis. These Ima1 locations in Nup2 and NupA null cells might be explained if the ability of Ima1 to locate to the INM during mitosis is linked to the opening of NPCs that occurs during A. nidulans mitosis. In this scenario, the absence of Nup2 or NupA might potentially prevent nuclear import of Ima1 such that when NPCs open during mitosis Ima1 can then enter nuclei to the INM through the partially open mitotic NPCs. Once it has entered the mitotic nuclei, it remains there after mitosis while newly synthesized Ima1 protein fails to enter nuclei and so again accumulates at foci. In this model, Nup2 and NupA would be involved in the nuclear import of Ima1. Some INM proteins are transported to the INM after integrating into the endoplasmic reticulum membrane through the nuclear pore membrane into the INM (King et al., 2006; Meinema et al., 2011). Therefore, if INM proteins fail to be transported though NPCs to the INM, they typically remain located within the endoplasmic reticulum (ER) (King et al., 2006; Meinema et al., 2011). However, rather than occurring in the ER without Nup2/A function, Ima1 locates to foci at the nuclear periphery. At this time, we cannot explain why Ima1 accumulates at such foci in the absence of Nup2/A. Further studies are required to identify the exact location of the Ima1 foci and to better understand whether this defective location is in fact caused by defects in nuclear transport of Ima1 in the absence of Nup2 and NupA. Interestingly, a recent study found that the INM proteins Heh1 and Heh2 in S. cerevisiae possess highly potent NLS sequences that can bind the importin β–binding domain of importin α in the absence of importin β (Lokareddy et al., 2015). In such a scenario when the import complexes lack importin β, RanGTP binding would be insufficient for cargo release. Importantly, Nup2 displaces Heh2 from importin by competing for the same binding site on importin α, facilitating cargo release in the nucleoplasm. Therefore, the presence of noncanonical, high-affinity NLS sequences could also contribute to differential nuclear protein import defects in the absence of Nup2. However, deletion of the N- and C-terminal regions of Nup2 have minimal effect on protein import suggesting that the central region of Nup2 might be involved in cargo release.
Another unexpected phenotype observed in Nup2 null mutants was excessive nuclear movement during G1 and lack of stable nuclear positioning in the cytoplasm as is typical of WT cells. In A. nidulans, and some other filamentous fungi, nuclei are evenly spaced within interphase hyphae via microtubules and the motor protein dynein. During mitosis, as daughter nuclei are forming, they are moved via the mitotic spindle apparatus, most noticeably during telophase. However, once nuclei exit mitosis and enter G1 they undergo additional transient increased movements (Fox et al., 2002), which can involve them passing each other moving both toward and away from the growing tip of the cell, mediated by microtubules. Because cellular compartmentalization occurs by septation after mitosis in filamentous fungi, nuclear mixing involving transient rapid movements up and down hyphae perhaps provides a mechanism for cells to maintain a heterogeneous nuclear population within compartments. Consistent with reported studies (Fox et al., 2002), we found that WT nuclei exhibit transient rapid movements before being more steadily positioned in the G1 cytoplasm after mitosis (Figure 7). However, Nup2 mutant nuclei continue to move along hyphae, in both directions, for a prolonged duration. Interphase nuclear movement and positioning is known to be an active microtubule-dependent process in A. nidulans (Oakley and Morris, 1980). The rapid nuclear movement in cells lacking Nup2 was abolished when microtubules are depolymerized, indicating that the transient rapid nuclear movements seen in G1 are also microtubule based, similar to WT cells. How cells regulate the transition from rapid two-way nuclear movements in early G1 to having nuclear movement regulated to ensure less mobile and evenly spaced locations for the rest of interphase remains to be determined. However, our analysis indicates Nup2 and NupA functions are required for cells to complete this transition. To test whether the excessive G1 nuclear movements in Nup2 nulls are a consequence of the mitotic NPC segregation defect, we utilized cells where this defect is fixed by an artificial NPC-chromatin bridge as described previously (Suresh et al., 2017). Interestingly, analyses of the nuclei in Nup2 null cells in which the NPC segregation defect is fixed revealed that they still moved their nuclei excessively over a 15 min period after mitosis (27.7 ± 7.5 μm compared with WT cells 10.5 ± 3.4 μm). In addition, in a population, WT cells are rarely seen to exhibit nuclear movements in interphase, whereas Nup2 null cells exhibited abnormal nuclear movements irrespective of whether mitotic NPC segregation proceeded normally or not. Therefore, the nuclear movement phenotypes seen in Nup2 nulls is independent of defects in NPC segregation. Interestingly, Samp1, the mammalian orthologue of the INM protein Ima1, is involved in nuclear migration and positioning (Borrego-Pinto et al., 2012). Although deletion of Ima1 in A. nidulans has no obvious effect on nuclear movement (unpublished data), it is difficult to rule out the possibility that mislocalized Ima1 in Nup2-deleted cells could contribute to defective nuclear positioning. To test whether mislocalization of Ima1-GFP could contribute to nuclear positioning defects, we performed a double deletion of Ima1 and Nup2 to track nuclear movement and positioning. However, the double-deleted cells were unable to grow past the swollen spore stage prohibiting further analysis.
Taken together, our studies reveal that Nup2 performs diverse functions to ensure normal nuclear functions and behavior and cell cycle progression. Although studies in budding yeast and human cells have highlighted the importance of Nup2 in the nuclear transport cycle, work in the filamentous fungus A. nidulans reveals additional intricacies in the functions of Nup2 during nuclear transport and beyond. It is interesting to note that Nup2 is involved in NPC inheritance in A. nidulans indicating further studies are warranted to determine if specific defects in nuclear biology caused by lack of Nup2 or NupA represent a global response to reduced NPC number.
MATERIALS AND METHODS
General techniques
Classical genetic techniques, media preparation, culture, and transformation of A. nidulans were done as described previously (Pontecorvo et al., 1953; Osmani et al., 2006; Markossian et al., 2015). The heterokaryon rescue technique was used to study the phenotypes of the essential genes nup2, nupA, and mlp1 as described previously (Osmani et al., 2006). Strains used in the study are listed in Supplemental Table 1, and the oligos used in the study are listed in Supplemental Table 2. All experiments were repeated at least three times and at least two independent transformants were examined for every experiment.
Imaging analysis
Cells were grown and imaged live as described previously (Markossian et al., 2015). A spinning disk confocal system (UltraVIEW Vox CSUX1 system; PerkinElmer) with 440-, 488-, 515-, and 561-nm solid state lasers and dual back-thinned EM CCD cameras (C9100-13; Hamamatsu Photonics) on a microscope (Ti-E; Nikon) with the dual camera mode utilizing a 60×/1.4 NA Plan-Apochromat objective lens (Nikon) was used. The distance between imaging planes was 0.8 μm. The microscopy images are represented as maximum intensity projections.
Quantitation analysis
Analysis of microscopy data was performed using the following software: 1) Volocity (PerkinElmer), 2) UltraVIEW (PerkinElmer), and 3) ImageJ (National Institutes of Health). Microsoft Excel (Microsoft) was used for plotting graphs and statistical analysis. A paired, two-tailed t test was performed to test for statistical significance. Data points represent the mean ± SD. Localization analysis of Mad2-GFP, Mlp1-GFP, and Mlp1-GFP-NLSStuA in nup2 or nupA deletions was performed as described previously for Mad1-GFP (Markossian et al., 2015). Mitotic cells were identified based on the dispersal of the NLS-DsRed signal from nuclei and the localization of Mad2-GFP as a nuclear focus and Mlp1-GFP/ Mlp1-GFP-NLSStuA as nuclear signals. For measurement of Ima1 mislocalization, cells with a focus of Ima1-GFP at the nuclear periphery were measured as defective. The nuclear periphery was inferred from the NLS-DsRed signal within nuclei. For measuring interphase timing, long-term time-lapse images were captured at 10-min intervals. NLS-DsRed dispersal was counted as mitosis and the time between two consecutive mitoses was calculated as the time spent in interphase. Typically, the time between first and second or second and third mitoses was measured and used for the analysis. For measurement of mitotic timing, the time for which the NLS-DsRed signal was dispersed from nuclei in the cell was measured. For analysis of Δmlp1 cells, a mitotic time of 10 min or lower was counted as normal mitosis; higher than 10 min was counted as delayed mitosis. For measuring nuclear and cytoplasmic localization of CutB-GFP and Tho2-GFP, nuclei were defined using the nuclear NLS-DsRed signal and the mean green fluorescence intensity in the defined red voxels was measured. For measuring mean cytoplasmic intensity, regions of interest (ROIs) were drawn in the cytoplasm and the average fluorescence intensity was measured. Mean background intensities were measured using the same ROIs in the field but adjacent to the cells of interest and subtracted from the nuclear and cytoplasmic intensity values. A ratio was then obtained using the background-corrected values. For measurement of nuclear movement, nuclei were tracked using the nuclear NLS-DsRed signal. The total distance moved by G1 nuclei for 15 min after mitosis followed at 1 min intervals was measured using Volocity software and plotted. Kymographs were generated from movies collected at 1 min intervals following the localization of NLS-DsRed in WT and mutants. Benomyl was added at a concentration of 2.4 µg/ml to depolymerize microtubules (Ovechkina et al., 1999).
Supplementary Material
Acknowledgments
We are grateful to the members of the Osmani lab for feedback and the undergraduate students Kimberlee Hunt and Leymaan Abdurehman for help with experiments. This work was supported by a grant from the National Institutes of Health (Grant no. GM042564) to S.A.O.
Abbreviations used:
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- IABD
importin-α binding domain
- INM
inner nuclear membrane
- KaRF
karyopherin release factors
- MT
microtubule
- NE
nuclear envelope
- NPC
nuclear pore complex
- NLS
nuclear localization sequence or signal
- NTR
nuclear transport receptor
- Nup
nucleoporin
- RBD
Ran-binding domain
- SAC
spindle assembly checkpoint
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-04-0223) on October 24, 2018.
REFERENCES
- Aist JR, Morris NR. (1999). Mitosis in filamentous fungi: how we got where we are. Fungal Genet Biol , 1–25. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait T, et al (2007). The molecular architecture of the nuclear pore complex. Nature , 695–701. [DOI] [PubMed] [Google Scholar]
- Balasundaram D, Benedik MJ, Morphew M, Dang VD, Levin HL. (1999). Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1. Mol Cell Biol , 5768–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim I, Frosst PD, Gerace L. (2009). Karyopherin binding interactions and nuclear import mechanism of nuclear pore complex protein Tpr. BMC Cell Biol , 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen LG, Morris NR. (1983). Kinetics of the nuclear division cycle of Aspergillus nidulans. J Bacteriol , 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth JW, Belanger KD, Sannella MI, Davis LI. (1999). The yeast nucleoporin Nup2p is involved in nuclear export of importin α/Srp1p. J Biol Chem , 32360–32367. [DOI] [PubMed] [Google Scholar]
- Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, Mattaj IW, Gomes ER. (2012). Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci (Pt 5), 1099–1105. [DOI] [PubMed] [Google Scholar]
- Buchwalter AL, Liang Y, Hetzer MW. (2014). Nup50 is required for cell differentiation and exhibits transcription-dependent dynamics. Mol Biol Cell , 2472–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Chan GK, Yen TJ. (2001). Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci (Pt 5), 953–963. [DOI] [PubMed] [Google Scholar]
- Chemudupati M, Osmani AH, Osmani SA. (2016). A mitotic nuclear envelope tether for Gle1 also impacts nuclear and nucleolar architecture. Mol Biol Cell , 3757–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, Haraguchi T, Hiraoka Y. (2009). Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J Cell Biol , 413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DB, Gromova T, Newman TAC, Burgess SM. (2017). The nucleoporin Nup2 contains a meiotic-autonomous region that promotes the dynamic chromosome events of meiosis. Genetics , 1319–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Kahn SM, Wilson KL, Discher DE. (2004). The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci (Pt 20), 4779–4786. [DOI] [PubMed] [Google Scholar]
- Dang VD, Levin HL. (2000). Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p. Mol Cell Biol , 7798–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA. (2009). Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol Biol Cell , 2146–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D, Mykytka B, Allen NP, Huang L, Al B, Rexach M. (2001). The nucleoporin Nup60p functions as a Gsp1p–GTP-sensitive tether for Nup2p at the nuclear pore complex. J Cell Biol , 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. (2001). Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol , 1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Chait BT, Wozniak RW, Aitchison JD. (2005). The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol , 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Muthuswamy S, Meier I. (2012). Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Mol Biol , 203–216. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Robbins J, Dilworth SM, Roberts B, Richardson WD. (1988). The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J Cell Biol , 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. (2008). Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol , 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn EM, DeRoo EP, Clement GW, Rao S, Kruse SE, Kokanovich KM, Belanger KD. (2013). A subset of FG-nucleoporins is necessary for efficient Msn5-mediated nuclear protein export. Biochim Biophys Acta , 1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. (2013). Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol , 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox H, Hickey PC, Fernandez-Abalos JM, Lunness P, Read ND, Doonan JH. (2002). Dynamic distribution of BIMG(PP1) in living hyphae of Aspergillus indicates a novel role in septum formation. Mol Microbiol , 1219–1230. [DOI] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell , 63–73. [DOI] [PubMed] [Google Scholar]
- Gilchrist D, Mykytka B, Rexach M. (2002). Accelerating the rate of disassembly of karyopherin∙cargo complexes. J Biol Chem , 18161–18172. [DOI] [PubMed] [Google Scholar]
- Gonzalez Y, Saito A, Sazer S. (2012). Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus , 60–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DM, Johnson CP, Hagan H, Corbett AH. (2003). The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc Natl Acad Sci USA , 1010–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman E, Medalia O, Zwerger M. (2012). Functional architecture of the nuclear pore complex. Annu Rev Biophys , 557–584. [DOI] [PubMed] [Google Scholar]
- Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L. (2000). Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol , 5619–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase ME, Cordes VC. (2003). Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell , 1923–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW. (2010). The nuclear envelope. Cold Spring Harb Perspect Biol , a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Maekawa H, Asakawa H, Chikashige Y, Kojidani T, Osakada H, Matsuda, Haraguchi T. (2011). Inner nuclear membrane protein Ima1 is dispensable for intranuclear positioning of centromeres. Genes Cells , 1000–1011. [DOI] [PubMed] [Google Scholar]
- Hodel MR, Corbett AH, Hodel AE. (2001). Dissection of a nuclear localization signal. J Biol Chem , 1317–1325. [DOI] [PubMed] [Google Scholar]
- Hood JK, Casolari JM, Silver PA. (2000). Nup2p is located on the nuclear side of the nuclear pore complex and coordinates Srp1p/importin-α export. J Cell Sci (Pt 8), 1471–1480. [DOI] [PubMed] [Google Scholar]
- Horio T, Oakley BR. (2005). The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol Biol Cell , 918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikui AE, Furuya K, Yanagida M, Matsumoto T. (2002). Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J Cell Sci (Pt 8), 1603–1610. [DOI] [PubMed] [Google Scholar]
- Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. (2002). The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol , 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. (2002). Chromatin boundaries in budding yeast: the nuclear pore connection. Cell , 551–562. [DOI] [PubMed] [Google Scholar]
- Kakar M, Davis JR, Kern SE, Lim CS. (2007). Optimizing the protein switch: altering nuclear import and export signals, and ligand binding domain. J Control Release , 220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. (1984). A short amino acid sequence able to specify nuclear location. Cell (3 Pt 2), 499–509. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. (2010). Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell , U360–U381. [DOI] [PubMed] [Google Scholar]
- King MC, Drivas TG, Blobel G. (2008). A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell , 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. (2006). Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature , 1003–1007. [DOI] [PubMed] [Google Scholar]
- Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. (2010). Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J , 1659–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Sterling H, Burlingame A, McCormick F. (2008). Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev , 2926–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Plafker K, Smith AE, Clurman BE, Macara IG. (2002). Npap60/Nup50 is a tri-stable switch that stimulates importin-α:β-mediated nuclear protein import. Cell , 349–360. [DOI] [PubMed] [Google Scholar]
- Liu HL, Osmani AH, Osmani SA. (2015). The inner nuclear membrane protein Src1 is required for stable post-mitotic progression into G1 in Aspergillusnidulans. PLoS One , e0132489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokareddy RK, Hapsari RA, van Rheenen M, Pumroy RA, Bhardwaj A, Steen A, Veenhoff LM, Cingolani G. (2015). Distinctive properties of the nuclear localization signals of inner nuclear membrane proteins Heh1 and Heh2. Structure , 1305–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS. (2012). The Nup153-Nup50 protein interface and its role in nuclear import. J Biol Chem , 38515–38522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markina-Inarrairaegui A, Etxebeste O, Herrero-Garcia E, Araujo-Bazan L, Fernandez-Martinez J, Flores JA, Osmani, SA, Espeso EA. (2011). Nuclear transporters in a multinucleated organism: functional and localization analyses in Aspergillusnidulans. Mol Biol Cell , 3874–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markossian S, Suresh S, Osmani AH, Osmani SA. (2015). Nup2 requires a highly divergent partner, NupA, to fulfill functions at nuclear pore complexes and the mitotic chromatin region. Mol Biol Cell , 605–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M. (2003). Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J , 5358–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Stewart M. (2005). Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J , 3681–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FA, Kralt A, van den Bogaart G, Lusk CP, Poolman B, Veenhoff LM. (2011). Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science , 90–93. [DOI] [PubMed] [Google Scholar]
- Morozov IY, Jones MG, Gould PD, Crome V, Wilson JB, Hall AJ, Rigden DJ, Caddick MX. (2012). mRNA 3′ tagging is induced by nonsense-mediated decay and promotes ribosome dissociation. Mol Cell Biol , 2585–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. (2007). The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol , 379–393. [DOI] [PubMed] [Google Scholar]
- Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C. (2013). The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell , 3920–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Morris NR. (1980). Nuclear movement is beta–tubulin-dependent in Aspergillusnidulans. Cell , 255–262. [DOI] [PubMed] [Google Scholar]
- Osmani AH, Davies J, Liu HL, Nile A, Osmani SA. (2006). Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillusnidulans. Mol Biol Cell , 4946–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, Oakley BR, Osmani SA. (2006). Identification and analysis of essential Aspergillusnidulans genes using the heterokaryon rescue technique. Nat Protoc , 2517–2526. [DOI] [PubMed] [Google Scholar]
- Ovechkina YY, Pettit RK, Cichacz ZA, Pettit GR, Oakley BR. (1999). Unusual antimicrotubule activity of the antifungal agent spongistatin 1. Antimicrob Agents Chemother , 1993–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruat JI, Aguilera A. (1998). A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J , 4859–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. (1953). The genetics of Aspergillus nidulans. Adv Genet , 141–238. [DOI] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. (2004). Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol , 1114–1121. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol , 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saroufim MA, Bensidoun P, Raymond P, Rahman S, Krause MR, Oeffinger M, Zenklusen D. (2015). The nuclear basket mediates perinuclear mRNA scanning in budding yeast. J Cell Biol , 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. (2006). Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell , 379–391. [DOI] [PubMed] [Google Scholar]
- Schneider M, Hellerschmied D, Schubert T, Amlacher S, Vinayachandran V, Reja R, Pugh BF, Clausen T, Köhler A. (2015). The nuclear pore-associated TREX-2 complex employs mediator to regulate gene expression. Cell , 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SG, King MC. (2015). The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun , 7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW. (2005). Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell , 4362–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. (2004). Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol , 942–952. [DOI] [PubMed] [Google Scholar]
- Sistla S, Pang JV, Wang CX, Balasundaram D. (2007). Multiple conserved domains of the nucleoporin Nup124p and its orthologs Nup1p and Nup153 are critical for nuclear import and activity of the fission yeast Tf1 retrotransposon. Mol Biol Cell , 3692–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smitherman M, Lee K, Swanger J, Kapur R, Clurman BE. (2000). Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol Cell Biol , 5631–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J, Maurer P, Vogel F, Schlenstedt G. (2000). Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin α. Mol Cell Biol , 8468–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. (2007). Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol , 195–208. [DOI] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. (2010). The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol , 490–501. [DOI] [PubMed] [Google Scholar]
- Suelmann R, Sievers N, Fischer R. (1997). Nuclear traffic in fungal hyphae: in vivo study of nuclear migration and positioning in Aspergillus nidulans. Mol Microbiol , 757–769. [DOI] [PubMed] [Google Scholar]
- Suresh S, Markossian S, Osmani AH, Osmani SA. (2017). Mitotic nuclear pore complex segregation involves Nup2 in Aspergillus nidulans. J Cell Biol , 2813–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toews MW, Warmbold J, Konzack S, Rischitor P, Veith D, Vienken K, Vinuesa C, Wei H, Fischer R. (2004). Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr Genet , 383–389. [DOI] [PubMed] [Google Scholar]
- Varadarajan P, Mahalingam S, Liu P, Ng SB, Gandotra S, Dorairajoo DS, Balasundaram D. (2005). The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr. Mol Biol Cell , 1823–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walde S, Thakar K, Hutten S, Spillner C, Nath A, Rothbauer U, Wiemann S, Kehlenbach RH. (2012). The nucleoporin Nup358/RanBP2 promotes nuclear import in a cargo- and transport receptor-specific manner. Traffic , 218–233. [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. (1998). Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol , 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ. (2002). Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol , 6498–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wu CY, Blobel G. (2004). Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J Cell Biol , 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.