Summary
Pichinde virus (PICV) is a nonpathogenic arenavirus with a bi-segmented RNA genome (L and S segments) that encodes four viral genes. We have developed a reverse genetics system to generate recombinant tri-segmented PICV (rP18tri) that packages three RNA segments (L, S1, and S2) and can encode up to two foreign genes. Using influenza virus HA and NP as model antigens, we show that the rP18tri vector can induce strong humoral and cell mediated immunity, which further increases upon a booster dose. We propose that this novel rP18tri vector can be developed into a useful vaccine platform for other antigens, particularly when strong cellular immunity and prime-boost vaccination strategy are desired.
Keywords: viral vaccine vector, Pichinde virus, arenavirus, vaccine
1. Introduction
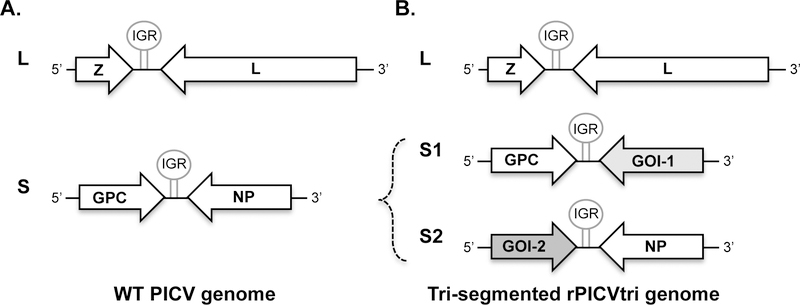
Pichinde virus (PICV) is a non-pathogenic enveloped RNA virus within the Arenaviridae family. PICV was first isolated from its natural hosts Oryzomys albigularis (rice rats) in the Pichinde valley of Colombia, South America (1), and is not known to cause disease in humans or animals. The sero-prevelance is very low even among humans living or working in close association with habitats of infected rodents (1). Therefore, there is virtually no preexisting immunity against PICV in the general population. This is in contrast to another prototypic arenavirus, lymphocytic choriomeningitis virus (LCMV), which has a global distribution with up to 5% seroprevalence in human populations. The PICV genome consists of two RNA genome segments, the long (L) and short (S) segments. Each segment encodes two genes in the opposite orientation, (Fig. 1A). The L segment encodes the small RING-domain containing matrix protein Z and a large L polymerase protein. The S segment encodes the glycoprotein GPC and the nucleoprotein NP. All 4 genes are essential for the basic life cycle of the virus. The 15-kDa Z protein mediates virus budding and regulates viral RNA synthesis. The L protein (~200 kDa) is the RNA-dependent RNA polymerase (RdRp) that is required for viral RNA transcription and replication. The GPC glycoprotein precursor is post-translationally cleaved into three subunits, stable signal peptide (SSP), GP1, and GP2, which together form a tripartite complex that mediates receptor binding and membrane fusion. The NP protein encapsidates the viral genomic RNAs and plays an essential role in viral RNA synthesis. In addition, the NP and Z proteins have been found to play important roles in suppressing the host innate immune responses (2).
Fig. 1.
Genomic organization of PICV and tri-segmented rPICVtri. (A) Wild-type (WT) PICV genome consists of two genomic RNA segments (L and S), each encoding two viral genes in opposite orientation. (B) Tri-segmented recombinant PICV (rPICVtri) genome consists of three RNA segments, L, S1, and S2. S1 encodes GPC and the gene of interest (GOI-1). S2 encodes GOI-2 and NP. GOI, gene-of-interest; IGR, intergenic region.
Arenaviruses target dendritic cells (DCs) and macrophages early in the infection, therefore these viruses are considered a potential vaccine vectors (3–6). Following a strategy first developed by de la Torre group for LCMV (4), we have recently generated a tri-segmented PICV (P18 strain) vaccine vector (rP18tri) that can simultaneously express dual foreign antigens in addition to its own genes (Fig. 1B). The rP18tri-based viruses are attenuated in vitro and in vivo, and can effectively induce strong T cell and humoral responses with limited anti-vector neutralizing antibodies (7). Therefore this novel rP18tri vector exhibits multiple features of an ideal live viral vector including safety, immunogenicity, robust antigen-encoding capacity, targeting antigen-presenting cells (APCs), and the lack of a strong anti-vector immunity.
2. Materials
2.1. Plasmids:
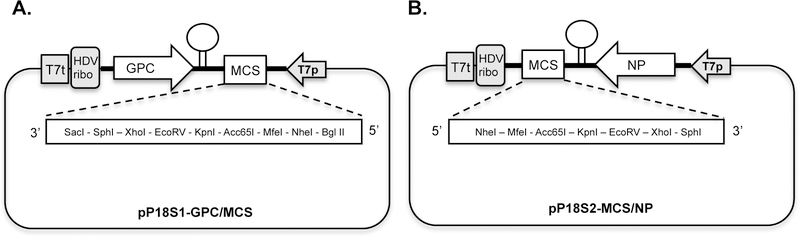
pP18S1-GPC/MCS: This plasmid (Fig. 2A) expresses the antigenomic strand of the rP18 S1 segment under control of the T7 promoter. The rP18 S1 segment encodes the glycoprotein GPC and a multiple-cloning-site (MCS) that replaces the NP gene and allows the convenient cloning of a gene of interest. The stem-loop structure represents the intergenic region (IGR) of viral RNA. The hepatitis delta virus ribozyme (HDVribo) sequence, which immediately follows the P18S1 RNA segment and preceeds the T7 terminator sequence (T7t), is used to generate authentic PICV S RNA ends (8).
pP18S2-MCS/NP: This plasmid (Fig. 2B) expresses the antigenomic strand of the rP18 S2 segment under control of the T7 promoter. The rP18 S2 segment encodes the nucleoprotein NP and a multiple-cloning-site (MCS) that replaces the GPC gene and allows the convenient cloning of a gene of interest. The stem-loop structure represents the intergenic region (IGR) of viral RNA. The hepatitis delta virus ribozyme (HDVribo) sequence, which immediately follows the P18S1 RNA segment and preceeds the T7 terminator sequence (T7t), is used to generate authentic PICV S RNA ends (8).
pPolI-HA and pPolI-NP plasmids: These plasmids express the segment 4 (encoding Hemagglutinin HA) and the segment 5 (encoding nucleoprotein NP), respectively, of influenza A virus H1N1 laboratory strain A/PR8, and were obtained from Drs. Brownlee and Fodor (Oxford University, UK).
pP18L: This plasmid expresses the full-length antigenomic strand of the rP18 L segment under the control of the T7 promoter. The hepatitis delta virus ribozyme (HDVribo) sequence, which immediately follows the P18S1 RNA segment and preceeds the T7 terminator sequence (T7t), is used to generate authentic PICV S RNA ends (8).
Fig. 2.
Vector maps of plasmids encoding S1 and S2 RNA segments. Plasmids pP18S1-GPC/MCS (A) and pP18S2-MCS/NP (B) are based on the PICV P18 reverse genetics system plasmid pP18S, which encodes the full-length P18 S RNA segment, followed by hepatitis delta virus ribozyme sequence (HDVribo), under a T7 promoter. Genomic organization of the rP18tri vector expressing eGFP together with influenza virus A/PR8HA (rP18tri-G/H) orNP(rP18tri-G/P). IAV, influenza virus; IGR, intergenic region MCS, multiple cloning sites.
2.2. Primer sequences
HA Forward: ggtgctagcATGAAGGCAAACCTACTGGTCCTG (Nhe I site is underlined)
HA Reverse: gcaggtaccTCAGATGCATATACTGCACTGC (Kpn I site is underlined)
NP Forward: ggtgctagcATGGCGTCCCAAGGCACCAAAC (Nhe I site is underlined)
NP Reverse: gcaggtaccTTAATTGTCGTACTCCTCTGC (Kpn I site is underlined)
2.3. Molecular cloning:
Phusion DNA polymerase (New England Biolabs, MA)
Qiagen gel purification kit
Restriction enzymes Nhe I and Kpn I
Qiagen nucleotide removal kit
Antarctic Phosphatase (New England Biolabs, MA)
T4 DNA ligase (New England Biolabs, MA)
Qiagen mini-prep kit
2.4. Cell lines:
Baby hamster kidney (BHK-21) cells
BSRT7–5 cells (obtained from Dr. K-K Conzelmann, Ludwig-Maximilians-Universität, Germany) are BHK-21 cells stably expressing the T7 RNA polymerase.
African green monkey kidney (Vero) cells
Madin-Darby canine kidney (MDCK) cells
2.5. Cell culture reagents
BHK-21 cells are grown in Dulbecco’s modified Eagle medium (DMEM with high glucose) supplemented with 10% fetal bovine serum (FBS) and 50 μg/ml penicillin-streptomycin.
BSRT7–5 cells are grown in Minimal essential medium (MEM) supplemented with 10% FBS, 1 μg/ml Geneticin, and 50 μg/ml penicillin-streptomycin.
Vero cells are grown in MEM supplemented with 10% FBS and 50 μg/ml penicillin-streptomycin.
MDCK cells are grown in MEM supplemented with 10% FBS and 50 μg/ml penicillin-streptomycin.
2.6. Transfection reagents
Opti-MEM
Lipofectamine-2000
2.7. Plaque assay
First overlay: 0.6 ml of 2% agar and 2.4 ml of complete MEM medium supplemented with 10% FBS and 50 μg/ml penicillin-streptomycin.
Second overlay: 0.4 ml of 2% agar, 1.6 ml of complete MEM medium, 120 μl of neutral red dye stock solution (0.33% stock concentration)
2.8. Mouse experiment
Six- to eight-week old female C57BL/6 mice were obtained from Charles River Laboratories.
Influenza A virus A/PR8 is a laboratory mouse-adapted strain that causes lethal infection in mice (9) and was obtained from Dr. Richard W. Compans (Emory University, GA).
Isoflurane is used to sedate mice prior to intranasal (IN) inoculation.
3. Methods
3.1. Cloning of antigen genes into the PICV-based rP18tri vectors
PCR amplification of antigen genes: Influenza A virus HA and NP genes are used as model antigens to explain the construction and usage of the rP18tri-based vaccine candidates. HA and NP open-reading-frames (ORFs) were amplified from the pPolI-HA and pPolI-NP plasmids, respectively, with the 5’ primers containing the NheI site (HA Forward and NP Forward) and the 3’ primers containing the KpnI site (HA Reverse and NP Reverse), in a 50-μl reaction that contains 1× Phusion HF buffer, 50 ng of plasmid DNA, 25 pmol of each primer, 10 uM dNTPs, and 1 unit of Phusion DNA polymerase. The PCR conditions were 98°C for 30 sec, and 30 cycles of 98°C for 10 sec, 55°C for 15 sec, and 72°C for 30 sec. The 1.7-kb and 1.5-kb PCR products for HA and NP, respectively, were purified from agarose gel slices using Qiagen gel purification kit and digested with NheI and KpnI in a 40-μl reaction that contains 1× CutSmart Buffer, 1 μl of NheI (10 Units), and 1 μl of KpnI-HF (20 Units), at 37°C for 4 h (See Note 1). The digested PCR fragments were purified using Qiagen nucleotide removal kit.
Digestion of the viral vector plasmids. One microgram of each of the pP18S1-GPC/MCS and pP18S2-MCS/NP plasmids (Fig. 2) was digested with NheI and KpnI in a 30-μl reaction that contains 1× CutSmart Buffer, 1 μl of NheI (10 Units) and 1 μl of KpnI-HF (20 units), at 37°C for 4 h (See Note 1). At the end of the restriction enzyme reaction, 3 μl of 10× Antarctic Phosphatase Buffer and 1 μl of Antarctic Phosphatase were added and incubated at 37°C for 1 h followed by a 5-min incubation at 70°C to inactivate the enzyme.
Ligation and transformation. The NheI/KpnI-treated HA PCR fragment was ligated into the digested pP18S1-GPC/MCS vector, while the NheI/KpnI-treated NP PCR fragment was ligated with the digested pP18S2-MCS/NP vector, at an insert-to-vector ratio of 3 to 1. The 10-μl ligation reaction contains 1 μl of T4 ligase buffer, 1 μl of 10mM dNTPs, and 1μl of T4 DNA ligase (400 Units) (See Note 2). After incubating at room temperature for 2 h (See Note 3), 5 μl of the ligation reaction was used to transform 100-μl of E. coli competent Stbl2 cells that were plated on LB-Amp plate (See Note 4). Plasmid DNAs were extracted from the bacterial cultures using the Qiagen mini-prep kit and screened by restriction enzyme digestion using the NheI and KpnI-HF enzymes. The positive clones were verified by sequencing and named pP18S1-GPC/H and pP18S2-P/NP, respectively.
3.2. Generation of recombinant tri-segmented PICVs from plasmid transfection
Recombinant tri-segmented PICVs are generated by simultaneously transfecting 3 plasmids encoding the L, S1, and S2 RNA segments (Fig. 3) into BHK-21 cells that constitutively express the T7 RNA polymerase. Supernatants were collected from the transfected cells and detected for infectious viruses by plaque assaying on Vero cells. Single plaques were picked from plates and amplified in BHK-21 cells in order to prepare viral vaccine stocks (Fig. 3).
Fig. 3.
Steps to generate the tri-segmented rP18tri-based vaccine stocks. BSR-T7 cells are transfected with 3 plasmids encoding the L, S1, and S2 RNA segments. Supernatants from the transfected cells are detected for infectious viruses by plaque assay. Virus stocks are prepared by growing up plaque-purified viruses in BHK-21 cells.
Seed 4×105 BSRT7–5 cells (See Note 5) per well in 6-well plate with antibiotics-free culture medium and incubate the cells overnight at 37°C and 5% CO2.
The following day, change to fresh medium 1 h prior to transfection.
Mix 250 μl of Opti-MEM (See Note 6) with three plasmids, pP18L, pP18S1-GPC/MCS containing gene of interest, and pP18S2-MCS/NP containing gene of interest, each at 1 μg, and incubate for 5 min at room temperature.
Mix 250 μl of Opti-MEM with 7 μl of Lipofectamine-2000 and incubate for 5 min at room temperature.
Mix Opti-MEM/plasmids with Opti-MEM/lipofectamine and further incubate the mixture for 30 min at room temperature before adding to the cells.
After 4-h incubation at 37°C, replace the transfection medium with antibiotics-free culture medium.
Incubate the cells at 37°C and 5% CO2. Collect 100 μl of supernatants daily from 2 to 5d post-transfection for plaque assay in order to determine whether viable tri-segmented rP18tri-HA/NP virus has been generated (See Note 7).
3.3. Plaque assay
Seed 3×105 Vero cells per well in six-well plates and incubate overnight at 37°C and 5% CO2.
Conduct 10-fold serial dilutions (normally from 101 to 106 dilutions) of the collected supernatants in MEM medium (See Note 8).
Aspirate medium from the cells and add 500 μl of the appropriate dilution to each well. Gently rock the plate to make sure that the cell monolayer is evenly covered with the viral dilution.
Incubate the infected cells for 1 h at 37°C and 5% CO2; rock the plate every 10 minutes.
Prepare the first overlay and keep in 55°C water bath. Melt the 2% agar by microwave and equilibrate to 75°C before adding to the complete MEM medium.
Remove the infection medium and immediately add 3ml of the first overlay per well. After the agar has solidified, incubate cells at 37°C and 5% CO2 for 4 days.
Add 2 ml of the second overlay per well and, after agar has solidified, incubate cells at 37°C and 5% CO2 for 1 day.
Count plaques and calculate viral titers as plaque-forming unit (PFU)/ml.
Proceed to the plaque purification step.
3.4. Plaque Purification
Add 1 ml of MEM medium to 1.5-ml microtubes.
Choose an isolated plaque that is well separated from other plaques in the plaque assay plate (See Note 9).
Use a P1000 pipette to insert a 1-ml pipette tip into the agar straight down to the plaque and gently aspirate a small volume of materials.
Pipet up and down to dispense the collected agar into a 1-ml MEM in a microtube.
Vortex at high speed. In a microcentrifuge, spin at 200 g for 5 min. Collect the 1-ml supernatants and store at −80°C, or immediately infect BHK-21 cells for viral stock preparation.
3.5. Preparation of the rP18tri-based viral vaccine stocks
Seed 8×105 BHK-21 cells in a 10-cm culture dish and incubate overnight at 37°C and 5% CO2.
Aspirate the culture medium, add 1 ml of supernatants from a single plaque (See Note 10), incubate for 1 h at 37°C and 5% CO2, rock plate every 15 min.
Add another 7 ml of fresh culture medium to the infected cells, further incubate for 48 h at 37°C and 5% CO2.
Collect supernatants, centrifuge at 200 g for 5 min to remove debris, filter supernatants through a 0.45 μm filter (Millipore, MA).
Aliquot and store viral stocks at −80°C (See Note 11).
To make concentrated viral stocks, gently dispense 40 ml of viral sample on top of 5 ml of 20% sucrose cushion and centrifuge the sample at 13,000 g for 2 h in a SW28 rotor (Beckman Coulter, CA). Resuspend the pellet in 500 μl of PBS. Aliquot and store at −80°C.
Quantify viral titer by plaque assaying as described in 3.3 (See Note 12).
3.6. Immunization of mice with rP18tri-HA/NP in a lethal flu-mouse model
Inoculation of C57BL/6 mice with recombinant viruses is conducted in a biosafety cabinet (See Note 13).
For intraperitoneal (IP) inoculation, use a 25-gauge needle and 1-ml syringe to inject 1×105 pfu (See Note 14) of rP18tri-HA/NP in 60 μl volume into the peritoneum of the mouse. For intramuscular (IM) inoculation, restrain the mouse in a restrainer, use a 25-gauge needle and 1-ml syringe to inject 1×105 pfu of rP18tri-HA/NP in 50 μl volume into the thigh muscle. A booster dose can be given 21 days later.
Monitor the mouse body weight and other disease signs daily (See Note 15).
For blood collection, 100-μl of blood is taken from facial vein using a 21-gauge needle and 1-ml syringe (See Note 16). Blood cells were collected at 7 days after each dose in heparinized tubes and used immediately for analysis of T cell response (7). Blood was collected at 14 days after each dose into 1.5-ml microtubes. After blood clotting at 37°C for 1 h, collect the serum by centrifugation at 6,000 rpm for 10 min in a microfuge. Serum can be stored at −20°C until being used for analysis of neutralizing antibodies (7).
For a challenge study, immunized mice were inoculated intranasally (IN) with 10× 50% mouse-lethal dose (MLD50) of influenza A virus A/PR8 (See Note 13). Mice were anesthetized with 5% isoflurane in the anesthesia machine. Hold the anesthetized mouse (See Note 17) in one hand, tilt the mouse backward slightly, and use a P200 pipet to inoculate 50 μl of A/PR8 virus into the right and left nostrils (9). Place the mouse back into the cage (See Note 18). Monitor mouse body weight and disease signs daily up to 14 dpi. Mice are euthanized when reaching the predetermined terminal points (See Note 19).
Acknowledgments
This work was supported by NIH grants R21AI094133 and R01AI083409 to Y.L, and R01AI093580 to H.L.
Footnotes
Notes
The restriction enzyme digestion reactions can be kept overnight in a 37°C water bath.
A vector-alone ligation reaction should be conducted as a negative control.
The ligation reaction can be carried out at either ambient (room) temperature or 16°C for variable lengths of time from 30 min to overnight.
We prefer to propagate PICV reverse genetics plasmids in Stbl2 cells at 30°C, partly because arenavirus genome contains the high GC-rich intergenic region (IGR) that can be responsible for genomic instability if amplified in conventional competent cells (e.g., DH5α) and at higher temperature (e.g., 37°C).
In order to increase transfection efficiency, use low passaged BSRT7–5 cells. Higher passaged cells may limit transfection efficiency and thereby reduce virus yield.
For plasmid transfection, use fresh aliquots of Opti-MEM. Avoid repeated opening of Opti-MEM bottle in order to avoid its oxidation.
All work with recombinant viruses should be performed in a biosafety cabinet. Centrifugation of recombinant viruses is conducted in a sealed rotor. Proper personal protection equipment (PPE) such as lab coats, gloves, and toe-covered shoes are required.
For serially diluting the virus, add 900 μ medium to 6 tubes. Transfer 100 μ of the PICV stock virus to the first tube containing 900 μ of medium. Vortex the mixture for 15 sec and transfer 100 μ of the diluted virus to the next tube.
We normally pick single plaques from samples that were collected at the earliest time points after plasmid transfection. Viruses collected at earlier times are less likely to contain spontaneous mutations.
High-titer viral stocks are generated from plaque-purified virus.
Store virus stocks in small aliquots to avoid repeated freeze-thaw cycles.
Titers of the rP18tri-based viral vaccines can vary from 105 to 107 pfu/ml.
Inoculation of mice with viruses should be conducted in a biosafety cabinet inside an ABSL-2 facility. Proper PPEs include disposable long-sleeved gown, surgical mask, hair cover, shoe cover, and gloves.
Lower dose of viral vaccines such as 1,000 pfu was found to provide protective immunity in the lethal influenza-mouse model (7).
Mice immunized with the rP18tri-based vaccine vectors can be downgraded from ABSL-2 to ABSL-1 facility 3 days after inoculation, as these vaccine vectors are severely attenuated in vitro and in vivo (7).
Maximal volume of blood collected from one mouse per week is 0.5% of the mouse’s body weight.
Anesthesia is confirmed by toe pinching.
Mice infected with influenza A virus A/PR8 were housed in an ABSL-2 facility.
Endpoints are determined using an established scoring system (9). Hunched posture is 3 points, ruffled fur is 3 points, not eating or drinking 2 points, greater than 20% body weight loss 10 points, neurological symptoms (hind-limb paralysis) 10 points. When the total points exceed 16, the animal should be euthanized.
References
- 1.Trapido H, Sanmartin C (1971) Pichinde virus, a new virus of the Tacaribe group from Colombia. Am J Trop Med Hyg 20:631–641 [PubMed] [Google Scholar]
- 2.Meyer B and Ly H (2016) Inhibition of Innate Immune Responses Is Key to Pathogenesis by Arenaviruses. J Virol 90:3810–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flatz L, Hegazy AN, Bergthaler A, Verschoor A, Claus C, Fernandez M, et al. (2010) Development of replication-defective lymphocytic choriomeningitis virus vectors for the induction of potent CD8+ T cell immunity. Nat Med 16, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emonet SF, Garidou L, McGavern DB, and de la Torre JC (2009) Generation of recombinant lymphocytic choriomeningitis viruses with trisegmented genomes stably expressing two additional genes of interest. Proc Natl Acad Sci U S A 106, 3473–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popkin DL, Teijaro JR, Lee AM, Lewicki H, Emonet S, de la Torre JC, et al. (2011) Expanded potential for recombinant trisegmented lymphocytic choriomeningitis viruses: protein production, antibody production, and in vivo assessment of biological function of genes of interest. J Virol 85, 7928–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ortiz-Riano E, Cheng BY, Carlos de la Torre J, and Martinez-Sobrido L (2013) Arenavirus reverse genetics for vaccine development. J Gen Virol 94, 1175–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhanwani R, Zhou Y, Huang Q, Verma V, Dileepan M, Ly H, et al. (2015) A novel live Pichinde virus-based vaccine vector induces enhanced humoral and cellular immunity upon a booster dose. J Virol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan S, McLay SL, Wang J, Kumar N, Ly H, and Liang Y (2009) Development of infectious clones for virulent and avirulent pichinde viruses: a model virus to study arenavirus-induced hemorrhagic fevers. J Virol 83, 6357–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuoka Y, Lamirande EW, and Subbarao K (2009) The mouse model for influenza. Curr Protoc Microbiol Chapter 15, Unit 15G 13.. [DOI] [PubMed] [Google Scholar]