Abstract
Objective
To characterize the genetic and clinical features of patients with autosomal dominant adult-onset demyelinating leukodystrophy (ADLD) carrying duplication and deletion upstream of lamin B1 (LMNB1).
Methods
Ninety-three patients with adult-onset leukoencephalopathy of unknown etiology were genetically analyzed for copy numbers of LMNB1 and its upstream genes. We examined LMNB1 expression by reverse transcription-qPCR using total RNA extracted from peripheral leukocytes. Clinical and MRI features of the patients with ADLD were retrospectively analyzed.
Results
We identified 4 patients from 3 families with LMNB1 duplication. The duplicated genomic regions were different from those previously reported. The mRNA expression level of LMNB1 in patients with duplication was significantly increased. The clinical features of our patients with LMNB1 duplication were similar to those reported previously, except for the high frequency of cognitive impairment in our patients. We found 2 patients from 1 family carrying a 249-kb genomic deletion upstream of LMNB1. Patients with the deletion exhibited relatively earlier onset, more prominent cognitive impairment, and fewer autonomic symptoms than patients with duplication. The presence of cerebellar symptoms and lesions may be characteristic in our patients with the deletion compared with the previously reported family with the deletion. Magnetic resonance images of patients with the deletion exhibited a widespread distribution of white matter lesions including the anterior temporal region.
Conclusions
We identified 4 Japanese families with ADLD carrying duplication or deletion upstream of LMNB1. There are differences in clinical and MRI features between the patients with the duplication and those with the deletion upstream of LMNB1.
Autosomal dominant adult-onset demyelinating leukodystrophy (ADLD; OMIM #169500) is a slowly progressive adult-onset leukoencephalopathy that predominantly affects the cerebral white matter. The onset of ADLD is typically in the fourth and fifth decade of life.1 Patients with ADLD are clinically characterized by early development of autonomic symptoms such as bladder and/or bowel impairment and orthostatic hypotension. Autonomic symptoms usually precede or occur together with other accompanying clinical features such as pyramidal signs and cerebellar ataxia. Cognitive impairment is observed in some patients.2,3 Characteristic MR images include diffuse and symmetrical lesions in the cerebral white matter and cerebellar peduncles. The periventricular rim adjacent to the lateral ventricle in the cerebral white matter is spared or less affected.1
Duplication of lamin B1 (LMNB1) encoding lamin B1 has been identified as a cause of ADLD.4 To date, 26 pedigrees with ADLD carrying LMNB1 duplication have been reported from different ethnic backgrounds.3,5,6 The regions of duplication commonly include the entire LMNB1, but differed in size among the pedigrees, ranging from 128 to 475 kb.7 In 2015, it was reported that a deletion of 660 kb in the region upstream of LMNB1 was identified as the cause of ADLD in an Italian pedigree.8 In both mutations, i.e., duplication and deletion upstream of LMNB1, the expression mRNA level of LMNB1 was elevated.4,7,8 The key mechanism involved in the ADLD pathogenesis seems to be lamin B1 overproduction; however, it has not been fully understood how lamin B1 overproduction causes demyelination, leading to ADLD.
In this study, we analyzed the copy number variation (CNV) of LMNB1 and the upstream genes to identify LMNB1-related ADLD in families with adult-onset leukoencephalopathies of unknown etiology. By this analysis, we identified 4 ADLD pedigrees including 3 families with LMNB1 duplication and 1 family with the upstream deletion of LMNB1. We here report the genetic and clinical characteristics of Japanese families with LMNB1-related ADLD.
Methods
Standard protocol approvals, registrations, and patient consents
This study was conducted in accordance with the Helsinki declaration and approved by the Institutional Review Board of Niigata University. Written informed consent was obtained from the patients or their caregivers.
Patients
One-hundred ten patients clinically suspected of having an adult-onset leukoencephalopathy, whose etiologies have not been determined, were referred to our institute for genetic analysis between September 2015 and April 2017. Genetic analysis was performed by PCR-based Sanger sequencing analysis of genes including colony-stimulating factor 1 receptor (CSF1R), AARS2, NOTCH3, and SNORD118. By this analysis, 11 patients who were found to harbor CSF1R mutations, 2 patients with AARS2 mutations, 2 patients with NOTCH3 mutations, 1 patient with SNORD118 mutations, and 1 patient with neuronal intranuclear inclusion disease were excluded. The remaining 93 patients were included in this study.
Genetic analysis
Genomic DNA was extracted from peripheral leukocytes using a QIAamp DNA Blood Maxi kit (QIAGEN, Hilden, Germany). CNV was analyzed by real-time PCR assay using TaqMan probes (Thermo Fischer Scientific, Waltham, MA) designed for exons 3 (Hs02537023_cn), 6 (Hs00696436_cn), and 10 (Hs00579415_cn) of LMNB1. TaqMan probes were also designed for exon 5 of phosphorylated adaptor for RNA export (PHAX) (Hs0092512_cn), exon 8 of aldehyde dehydrogenase 7 family member A1 (ALDH7A1) (Hs00222439_cn), and exon 2 of GRAM domain containing 3 (GRAMD3) (Hs01106540_cn) to examine the CNV of the upstream genomic region of LMNB1. The amount of a PCR product was calculated on the basis of the threshold cycle (Ct), namely, the cycle in which fluorescence was detected above the baseline on an ABI PRISM 7900HT instrument (Applied Biosystems, Waltham, MA). We analyzed the results using CopyCaller Software v2.0 (Applied Biosystems). The control DNA CEPH 1347-02 was used as a reference genomic DNA sample, and the endogenous control was calculated by TaqMan copy number reference assay. The range of genomic CNVs around LMNB1 was examined by microarray-based copy number profiling using an Affymetrix CytoScan HD array (Thermo Fischer Scientific).
LMNB1 expression assay
Total RNA was extracted from patients with LMNB1-related ADLD using a PAXgene blood RNA kit (QIAGEN). RNA integrity number (RIN) was determined using Bioanalyzer 2100. Complementary DNA was synthesized by SuperScript IV VILO Master Mix (Thermo Fischer Scientific) using RNA showing a RIN score of 7 or higher. mRNA expression level was analyzed by real-time PCR assay using 2 TaqMan probes (Hs01059207_m1, Hs01059210_m1) (Thermo Fischer Scientific) designed for LMNB1. The expression of LMNB1 was normalized to that of ACTB (Hs99999903_m1) or TBP (Hs99999910_m1). The amount of the PCR product was calculated on the basis of Ct, the cycle where fluorescence was detected above the baseline on an ABI PRISM 7900HT instrument (Applied Biosystems). We analyzed the result by a comparative Ct method, in which the average of 7 control subjects (age, 53.1 ± 17.8 years, mean ± SD) was set to 1. Data are presented as median ± SD. Statistical analysis was performed with the Mann–Whitney U test.
Clinical assessment
We retrospectively analyzed the clinical characteristics of 6 patients from 4 families with LMNB1-related ADLD. We examined their sex, ages at onset and examination, initial symptoms, and the presence or absence of clinical symptoms including autonomic nervous dysfunction, pyramidal tract signs, and cerebellar ataxia using medical records. Cognitive functions were evaluated by Mini-Mental State Examination or Montreal Cognitive Assessment. Brain MRI with T1-weighted imaging (T1WI), T2-weighted imaging, fluid-attenuated inversion recovery (FLAIR), diffusion-weighted imaging (DWI), and apparent diffusion coefficient (ADC) was performed on the 6 patients with ADLD.
Results
LMNB1 duplication
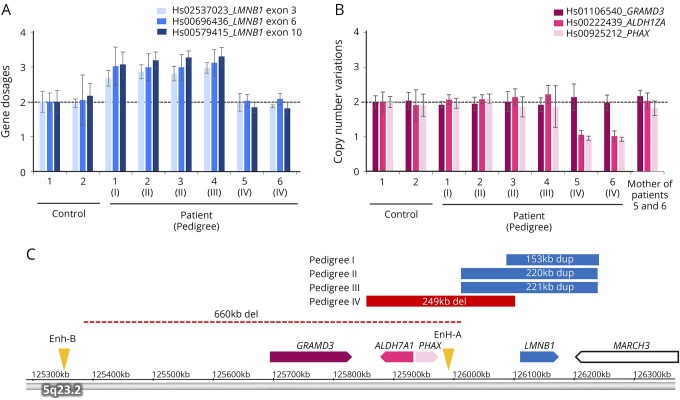
We examined the CNV of LMNB1 in 93 patients with adult-onset leukoencephalopathy of unknown etiology by TaqMan-based real-time PCR assay. The gene dosage of LMNB1 was increased approximately by 1.5-fold in 4 patients from 3 families (pedigrees I–III, figure 1A). This suggests that these patients were heterozygous for LMNB1 duplication.
Figure 1. Analysis of CNVs of LMNB1 and its upstream region.
(A) Gene dosages for exons 3, 6, and 10 of LMNB1 were determined by TaqMan-based real-time PCR assay. The copy numbers of 3 exons of LMNB1 in patients 1–4 were increased by approximately 1.5-fold compared with control subjects, suggesting the presence of duplication of LMNB1 in these patients. (B) The copy number variations of regions upstream of LMNB1 including GRAMD3, ALDH7A1, and PHAX were determined by TaqMan-based real-time PCR assay. The copy numbers of ALDH7A1 and PHAX were decreased approximately by half in patients 5 and 6, suggesting the presence of the upstream deletion of LMNB1. (C) The genomic regions of duplication (blue) and deletion (red) were analyzed using an Affymetrix CytoScan HD array and are shown on the basis of information obtained from the UCSC genome browser (assembly GRCh37/hg19). The regions of duplication were 153 kb in pedigree I, 220 kb in pedigree II, and 221 kb in pedigree III. The deletion upstream of LMNB1 in a previous report is shown by a dotted line.8 The positions of original enhancer A (Enh-A) and alternative enhancer B (Enh-B) for LMNB1 are indicated by arrowheads. ALDH7A1 = aldehyde dehydrogenase 7 family member A1; CNV = copy number variation; GRAMD3 = GRAM domain containing 3; LMNB1 = lamin B1; PHAX = phosphorylated adaptor for RNA export.
Next, we determined the genomic region of the duplication by microarray-based copy number profiling using an Affymetrix CytoScan HD array. Consistent with the results of quantitative real-time PCR assay, the gene dosage of the entire region of LMNB1 was increased (figure 1C). In addition to LMNB1, the gene dosage of the partial region of MARCH3 was also increased. The regions of duplication were 153 kb (hg19 chr5: 126,086,500–126,239,452) in pedigree I, 220 kb (126,012,578–126,232,143) in pedigree II, and 221 kb (126,012,161–126,233,043) in pedigree III.
Deletion upstream of LMNB1
A previous study showed a 660-kb deletion upstream of LMNB1 as a plausible cause of ADLD in an Italian family.8 Thus, we examined the CNV of GRAMD3, ALDH7A1, and PHAX, which are located upstream of LMNB1 in 93 patients. By this analysis, we identified the deletion upstream of LMNB1 in 2 patients in pedigree IV (figure 1B). The range of deletion was determined to be 249 kb (hg19 chr5: 125,855,011-126,103,996), which includes PHAX and ALDH7A1 (figure 1C).
mRNA expression of LMNB1 in blood of patients
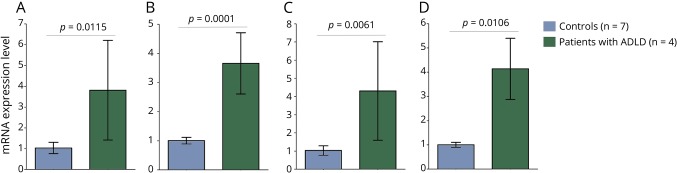
To ascertain whether duplication and deletion upstream of LMNB1 found in this study result in alternation of mRNA expression of LMNB1, we performed quantitative real-time reverse transcription-PCR assay using RNA extracted from peripheral leukocytes of the patients. We analyzed 2 amplicons detecting the LMNB1 transcript using 2 control transcripts of ACTB and TBP. The relative expression levels of LMNB1 mRNA in 4 patients with LMNB1 duplication were significantly increased in comparison with those of controls (figure 2). The LMNB1 mRNA expression level in the blood of patient 5 with the deletion was comparable to those of control subjects and his unaffected mother (data not shown).
Figure 2. LMNB1 mRNA expression.
The relative mRNA expression level of LMNB1 in patients with LMNB1 duplication (n = 4) and control subjects (n = 7) was determined using RNA extracted from peripheral blood by quantitative RT-PCR assay. qRT-PCR was performed using primer pairs spanning exons 6 and 7 (A and B) or exons 9 and 10 (C and D) of LMNB1. mRNA expression level of LMNB1 was normalized to those of ACTB (A and C) and TBP (B and D). The average value of control subjects was set to 1. Error bars indicate standard deviation. The statistical significance of difference was examined by the Mann–Whitney U test. LMNB1 = lamin B1; RT = reverse transcription.
Clinical characteristics
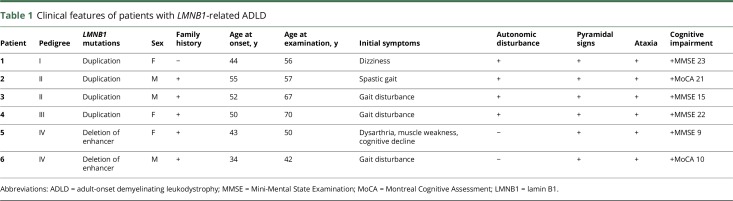
Details of clinical presentations of the patients with LMNB1-related ADLD are summarized in table 1. Familial occurrence was observed in 3 pedigrees (figure e-1, links.lww.com/NXG/A126). Patient 1 was apparently sporadic. Her father who died at the age of 71 years and mother at the age of 85 years did not develop ADLD. The mean age at onset of the patients with LMNB1 duplication was 50.3 years, ranging from 44 to 55 years. The most frequent initial symptom was gait disturbance. Subsequently, pyramidal signs, ataxia, and autonomic symptoms such as orthostatic hypotension, dysuria, and constipation were observed in all the patients with LMNB1 duplication. Notably, cognitive impairment was recognized in all the patients with the duplication. Reversible exacerbation with exposure to hot water bath or high fever was observed in patients 1, 2, 4, and 5.
Table 1.
Clinical features of patients with LMNB1-related ADLD
The ages at onset in patients with the deletion upstream of LMNB1 were 43 and 34 years. These patients showed pyramidal signs, ataxia, and prominent cognitive impairment. In contrast to the patients with duplication, patients with the deletion did not show apparent autonomic symptoms.
MRI characteristics
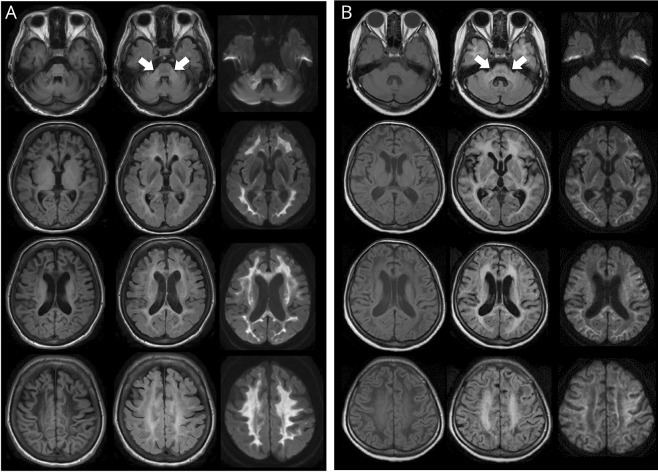
MR images of each patient are shown in figure 3 and figure e-2 (links.lww.com/NXG/A126). All the patients showed bilateral hyperintensities in the cerebral white matter and middle cerebellar peduncles (MCP) visualized by FLAIR (figure 3). As previously described, the periventricular white matter was spared or less affected.1,9 T1WI and FLAIR showed that the affected cerebral white matter appeared to be replaced by fluid (figure 3, left and middle panels). The lesions of the cerebral and superior cerebellar peduncles were detectable by FLAIR in patient 1. Patients with the deletion showed a more widespread distribution of white matter lesions extending to the anterior temporal region than patients with the duplication (figure 3B). DWI showed hyperintensity signals in the white matter and MCP, particularly in patients with the duplication (figure 3, A and B, right panel). ADC values were increased or normal in affected lesions of the cerebral white matter (figure e-3).
Figure 3. MRI findings in patients with ADLD with duplication and those with deletion upstream of LMNB1.
(A) Findings of MRI with T1WI (left panel), FLAIR (middle panel), and DWI (right panel) of patient 1 with LMNB1 duplication at the age of 56 years. Arrows point to the MCP lesion. (B) Findings of MRI with T1WI (left panel), FLAIR (middle panel), and DWI (right panel) of patient 5 with the upstream deletion of LMNB1 at the age of 50 years. ADLD = adult-onset demyelinating leukodystrophy; DWI = diffusion-weighted imaging; FLAIR = fluid-attenuated inversion recovery; MCP = middle cerebellar peduncle; LMNB1 = lamin B1.
Discussion
We here report Japanese families with LMNB1-related ADLD carrying duplication or deletion upstream of LMNB1. The duplicated genomic regions in these Japanese patients were different from those of the reported families with duplication including the Japanese K4975 family.4,7,10 The genomic regions of duplication in pedigrees II and III were similar, suggesting that they may share a common founder of the duplication. We showed that the LMNB1 mRNA expression level was significantly elevated, as determined by the analysis using RNA extracted from peripheral blood leukocytes from the patients with LMNB1 duplication (figure 2). Consistent with our results, previous reports have shown that the levels of lamin B1 protein were increased in leukocytes or brains of patients with LMNB1 duplication.4,7,9,11,12 Thus, lamin B1 overproduction seems to be a key mechanism underlying the pathogenesis of ADLD. However, the pathomechanism by which LMNB1 mRNA overexpression causes ADLD has not been fully elucidated.
Lamin B1 is a component of nuclear lamina and ubiquitously expressed in all cells with nuclei. Lamin B1 plays a role in regulating gene expression during DNA replication.13 It has been demonstrated that transgenic mice overexpressing LMNB1 showed age-dependent demyelination similar to ADLD.14,15 Comprehensive mRNA expression analysis in these mice revealed decreased mRNA expression levels of genes involved in the synthesis of lipid and cholesterol, which are the major components of myelin.14 This suggests that maintenance and repair of myelin may be impaired by lamin B1 overproduction, leading to the development of ADLD.
Previous studies have shown that autonomic symptoms precede or occur together with gait disturbance and motor symptoms in patients with LMNB1 duplication.1,16–19 The most frequent initial symptom was gait disturbance in our patients with duplication. Thus, it should be noted that patients with ADLD may initially show motor symptoms. Cognitive impairment was observed in all the patients with the duplication in this study, although the frequency of cognitive impairment was reported to be 63% in patients with the duplication.2,3
The characteristics of MR images in previous reports showed changes in the cerebral white matter and middle cerebellar peduncles, which were similarly observed in our patients with the duplication.1,9 T1WI and FLAIR revealed the white matter degeneration, which is replaced by fluid in our patients (figure 3). These findings are similarly observed in patients with vanishing white matter disease (VWMD).20 Patients 5 and 6 were initially suspected as having VWMD, and genetic testing of eIF2B was performed with negative results. Thus, differential diagnosis between ADLD and VWMD may be required in patients with such MRI findings.
We identified new patients with ADLD in pedigree IV carrying the deletion upstream of LMNB1. The region of the deletion in this study was 249 kb, which was narrower than that of the 660-kb deletion found in the previous Italian family (figure 1C).8 It was demonstrated that the LMNB1 mRNA expression level was increased in brains of patients with the deletion.8 The deletion in the Italian pedigree contains a putative enhancer region (Enh-A) regulating LMNB1 expression and the insulator between PHAX and ALDH7A1 (figure 1C). They speculated that another upstream enhancer (Enh-B) may alternatively work and enhance LMNB1 expression if the Enh-A and the insulator are deleted. Findings in our patients with the deletion supported this notion because the 188-kb deleted region shared by our patients and the previously reported patients commonly included the insulator and Enh-A. However, the mRNA expression level of LMNB1 derived from peripheral leukocytes did not increase in our patient with the deletion. The reason why the mRNA expression level was not apparently altered in our patient may be explained by the difference in tissues used for mRNA examination. The alternative enhancer (Enh-B) was reported to work in a forebrain-specific manner8; thus, its deletion may not cause the overexpression of LMNB1 in peripheral blood in patients. The mRNA expression level of LMNB1 may be altered in brain tissues, especially in oligodendrocytes. It would be important to analyze the LMNB1 mRNA expression level in brains of patients with the deletion when the autopsied brain samples become available.
In previous reports, the patients with the deletion were clinically characterized by later age at onset (age at onset: 47.2 ± 6.4 years) and the absence of autonomic symptoms at onset and cerebellar ataxia, as compared with the patients with duplication.21,22 Similarly, our patients with the deletion also lacked autonomic symptoms. In contrast to a previous report,22 our patients with the deletion showed relatively severe cognitive decline and the presence of cerebellar ataxia (table e-1, links.lww.com/NXG/A126). Cerebellar lesions were also noticeable on MR images of our patients (figure 3B). The presence of cerebellar symptoms and the cerebellar lesions revealed by MRI may be characteristic in our patients with the deletion because the Italian family lacked cerebellar symptoms and rarely exhibited cerebellar lesions on MRI.22 A novel MR finding of this study is that the cerebral white matter lesions extended to the anterior temporal region in patients with the deletion. The temporal white matter lesions were also observed in other white matter diseases including cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy and cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy.23,24 ADLD should be considered as a differential diagnosis for patients exhibiting anterior temporal lobe white matter lesions on MRI.
In this study, we identified 6 patients with ADLD of 93 patients with adult-onset leukoencephalopathy of unknown etiology. There were differences in clinical and MRI features between patients with ADLD with duplication and those with deletion upstream of LMNB1. The characteristic clinical and imaging features in patients with LMNB1-related ADLD may provide the clue for efficient molecular diagnosis in patients with adult-onset leukoencephalopathies.
Glossary
- ADC
apparent diffusion coefficient
- ADLD
adult-onset demyelinating leukodystrophy
- ALDH7A1
aldehyde dehydrogenase 7 family member A1
- CSF1R
colony-stimulating factor 1 receptor
- CNV
copy number variation
- DWI
diffusion-weighted imaging
- FLAIR
fluid-attenuated inversion recovery
- GRAMD3
GRAM domain containing 3
- LMNB1
lamin B1
- MCP
middle cerebellar peduncle
- PHAX
phosphorylated adaptor for RNA export
- RIN
RNA integrity number
- T1WI
T1-weighted imaging
- VWMD
vanishing white matter disease
Author contributions
N. Mezaki: drafting of the manuscript, study concept, acquisition of data, and analysis of data. T. Miura: study concept, acquisition of data, and analysis of data. K. Ogaki, M. Eriguchi, Y. Mizuno, K. Komatsu, H. Yamazaki, N. Suetsugu, S. Kawajiri, R. Yamasaki, T. Ishiguro, T. Konno, H. Nozaki, K. Kasuga, Y. Okuma, J.-I. Kira, and H. Hara: acquisition of data. O. Onodera: acquisition of data and study supervision. T. Ikeuchi: drafting of the manuscript, study concept, interpretation of data, and obtained the funding.
Study funding
Supported in part by grant-in-aid (JP16H01331 and 26117506 to T.Ikeuchi) from the Japan Society for the Promotion of Science, a grant-in-aid for Research on Intractable Disease from the Japanese Ministry of Health, Labour and Welfare, Japan (12103055 to T.Ikeuchi and O.O.), a grant-in-aid (18kk0205009 to T.Ikeuchi) from Japan Agency for Medical Research and Development, and a grant from the Tsubaki Memorial Foundation (to N.M.).
Disclosure
N. Mezaki has received foundation and society research support from the Tsubaki Memorial Foundation and JA Niigata Kouseiren grant. T. Miura, K. Ogaki, M. Eriguchi, Y. Mizuno, K. Komatsu, H. Yamazaki, N. Suetsugi, and S. Kawajiri report no disclosure. R. Yamasaki serves on the editorial board of Clinical and Experimental Neuroimmunology. T. Ishiguro, T. Konno, and H. Nozaki report no disclosure. K. Kasuga has received research support from the Japan Society for the Promotion of Science. Y. Okuma reports no disclosures. J.-I. Kira has served on the editorial boards of Multiple Sclerosis, BMC Medicine, Journal of the Neurological Sciences, Multiple Sclerosis and Related Disorders, PLoS One, Acta Neuropathologica Communications, and Clinical and Experimental Neuroimmunology; has provided consultancy for Biogen Idec Japan; and has received research support from governmental entities the Ministry of Health, Labour and Welfare, Japan Agency for Medical Research and Development, and the MEXT KAKENHI program. H. Hara has received funding for travel from Novartis and for speaker honoraria from Eisai and has received Grant-in-Aid for Scientific Research. O. Onodera has received funding for speaker honoraria from Kyowa Hakko Kirin Co., Ltd., Bristol-Myers Squibb, Ono Pharmaceutical Co., Ltd., Mitsubishi Tanabe Pharma, Takeda, Daiichi-Sankyo, Fujifilm, Sanofi, and FP-Pharma and has received governmental entity research support for Scientific Research from a Grant-in-Aid from the Research Committee for Hereditary Cerebral Small Vessel Diseases, and from the Ministry of Health, Labour and Welfare of Japan. T. Ikeuchi has served on the editorial board of Parkinsonism and Related Disorders; has acted as a consultant for Janssen Pharmaceuticals Inc. and Chugai Pharmaceutical Co. Ltd.; serves on the speakers' bureaus of Daiichi Sankyo, Eisai, Ono Pharmaceutical Co. Ltd., Novartis, and Takeda; and has received governmental entity research support from KAKENHI and the Ministry of Health. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Finnsson J, Sundblom J, Dahl N, Melberg A, Raininko R. LMNB1-related autosomal-dominant leukodystrophy: clinical and radiological course. Ann Neurol 2015;78:412–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwankhaus JD, Katz DA, Eldridge R, Schlesinger S, McFarland H. Clinical and pathological features of an autosomal dominant, adult-onset leukodystrophy simulating chronic progressive multiple sclerosis. Arch Neurol 1994;51:757–766. [DOI] [PubMed] [Google Scholar]
- 3.Sandoval-Rodríguez V, Cansino-Torres MA, Sáenz-Farret M, Castañeda-Cisneros G, Moreno G. Autosomal dominant leukodystrophy presenting as Alzheimer's-type dementia. Mult Scler Relat Disord 2017;17:230–233. [DOI] [PubMed] [Google Scholar]
- 4.Padiath QS, Saigoh K, Schiffmann R, et al. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat Genet 2006;38:1114–1123. [DOI] [PubMed] [Google Scholar]
- 5.Nahhas N, Sabet Rasekh P, Vanderver A, Padiath Q. Autosomal Dominant Leukodystrophy With Autonomic Disease. Seattle: GeneReviews (R); 1993. [Google Scholar]
- 6.Dai Y, Ma Y, Li S, et al. An LMNB1 Duplication caused adult-onset autosomal dominant leukodystrophy in Chinese family: clinical manifestations, neuroradiology and genetic diagnosis. Front Mol Neurosci 2017;10:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giorgio E, Rolyan H, Kropp L, et al. Analysis of LMNB1 duplications in autosomal dominant leukodystrophy provides insights into duplication mechanisms and allele-specific expression. Hum Mutat 2013;34:1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giorgio E, Robyr D, Spielmann M, et al. A large genomic deletion leads to enhancer adoption by the lamin B1 gene: a second path to autosomal dominant adult-onset demyelinating leukodystrophy (ADLD). Hum Mol Genet 2015;24:3143–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melberg A, Hallberg L, Kalimo H, Raininko R. MR characteristics and neuropathology in adult-onset autosomal dominant leukodystrophy with autonomic symptoms. AJNR Am J Neuroradiol 2006;27:904–911. [PMC free article] [PubMed] [Google Scholar]
- 10.Asahara H, Yoshimura T, Suda S, Furuya H, Kobayashi T. A Japanese family with probably autosomal dominant adult-onset leukodystrophy. Rinsho Shinkeigaku 1996;36:968–972. [PubMed] [Google Scholar]
- 11.Schuster J, Sundblom J, Thuresson AC, et al. Genomic duplications mediate overexpression of lamin B1 in adult-onset autosomal dominant leukodystrophy (ADLD) with autonomic symptoms. Neurogenetics 2011;12:65–72. [DOI] [PubMed] [Google Scholar]
- 12.Columbaro M, Mattioli E, Maraldi NM, et al. Oct-1 recruitment to the nuclear envelope in adult-onset autosomal dominant leukodystrophy. Biochim Biophys Acta 2013;1832:411–420. [DOI] [PubMed] [Google Scholar]
- 13.Tang CW, Maya-Mendoza A, Martin C, et al. The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J Cel Sci 2008;121:1014–1024. [DOI] [PubMed] [Google Scholar]
- 14.Rolyan H, Tyurina YY, Hernandez M, et al. Defects of lipid synthesis are linked to the age-dependent demyelination caused by lamin B1 overexpression. J Neurosci 2015;26:12002–12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heng MY, Lin ST, Verret L, et al. Lamin B1 mediates cell-autonomous neuropathology in a leukodystrophy mouse model. J Clin Invest 2013;123:2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brussino A, Vaula G, Cagnoli C, et al. A novel family with Lamin B1 duplication associated with adult-onset leukoencephalopathy. J Neurol Neurosurg Psychiatry 2009;80:237–240. [DOI] [PubMed] [Google Scholar]
- 17.Dos Santos MM, Grond-Ginsbach C, Aksay SS, et al. Adult-onset autosomal dominant leukodystrophy due to LMNB1 gene duplication. J Neurol 2012;259:579–581. [DOI] [PubMed] [Google Scholar]
- 18.Eldridge R, Anayiotos CP, Schlesinger S, et al. Hereditary adult-onset leukodystrophy simulating chronic progressive multiple sclerosis. N Engl J Med 1984;311:948–953. [DOI] [PubMed] [Google Scholar]
- 19.Brunetti V, Ferilli MA, Nociti V, Silvestri G. Teaching neuroImages: autosomal dominant leukodystrophy in a sporadic case. Neurology 2014;83:e121. [DOI] [PubMed] [Google Scholar]
- 20.van der Knaap MS, Pronk JC, Scheper GC. Vanishing white matter disease. Lancet Neurol 2006;5:413–423. [DOI] [PubMed] [Google Scholar]
- 21.Quattrocolo G, Leombruni S, Vaula G, et al. Autosomal dominant late-onset leukoencephalopathy. Clinical report of a new Italian family. Eur Neurol 1997;37:53–61. [DOI] [PubMed] [Google Scholar]
- 22.Brussino A, Vaula G, Cagnoli C, et al. A family with autosomal dominant leukodystrophy linked to 5q23.2-q23.3 without lamin B1 mutations. Eur J Neurol 2010;17:541–549. [DOI] [PubMed] [Google Scholar]
- 23.Chabriat H, Joutel A, Dichgans M, Tournier-Lasserve E, Bousser MG. Cadasil Lancet Neurol 2009;8:643–653. [DOI] [PubMed] [Google Scholar]
- 24.Nozaki H, Sekine Y, Fukutake T, et al. Characteristic features and progression of abnormalities on MRI for CARASIL. Neurology 2015;85:459–463. [DOI] [PubMed] [Google Scholar]