Abstract
Objective:
Non-invasive prenatal screening (NIPS) utilizes circulating cell-free DNA (cfDNA) to screen for fetal genetic abnormalities. NIPS is the first widely-available prenatal screen to assess genotypic sex. Most pediatricians have limited familiarity with NIPS technology and potential etiologies of discordant results. Increased familiarity may provide diagnostic insight and improve clinical care.
Study Design:
We reviewed all patients with discordant genotypic fetal sex assessed by cfDNA and neonatal phenotypic sex referred to our medical center.
Result:
Four infants with discordant cfDNA result and phenotypic sex were identified. Etiologies include vanishing twin syndrome, difference of sexual development, sex chromosome aneuploidy and maternal chimerism.
Conclusion:
We present four cases illustrating potential etiologies of discordant cfDNA result and postnatal phenotypic sex. Unanticipated cfDNA results offer the perinatologist a unique opportunity for early diagnosis and targeted treatment of various conditions, many of which may not have otherwise been detected in the perinatal period.
INTRODUCTION
Since its clinical introduction in 2011, maternal screening utilizing circulating cell-free DNA (cfDNA), also referred to as non-invasive prenatal screening (NIPS), has revolutionized prenatal screening for fetal chromosome disorders1–4. NIPS is a highly sensitive prenatal screening modality that screens common aneuploidies (typically 13, 18, 21 and sex chromosome aneuploidies). Unlike maternal serum analyte screening, such as the quadruple, integrated or sequential screen, NIPS routinely assesses a fetal sex chromosome complement, providing parents with an early prediction of fetal sex. Although highly sensitive, it should always be emphasized that NIPS is a screening platform. As such, any NIPS result that is abnormal, ambiguous, or discrepant to fetal or postnatal phenotype requires further evaluation and diagnostic testing5–8.
Cell-free DNA exists in human circulation from various sources, including cell apoptosis and pathogen breakdown. In pregnancy, the fraction of non-maternal, placental cfDNA increases with gestational age and rapidly decreases after delivery of the placenta9. After 9–10 weeks gestation, placental cfDNA is of sufficient quantity in maternal circulation that it can be obtained via routine maternal phlebotomy and assessed on a NIPS platform. NIPS platforms employ various methods; however, all exploit placental cell-free DNA circulating in maternal plasma as a surrogate for the fetus. Various factors influence the capacity of NIPS to obtain an accurate and/or conclusive result, including the fraction of circulating cfDNA of placental origin (the “fetal fraction”), maternal body mass index (BMI), and increased risk of anueploidy7. The sensitivity of NIPS varies by the chromosome assessed but is overall very high (female: 95.4–97.5%; male: 99.1%)6,10,12.
Prior to NIPS, the only widely-available screening for fetal sex was a mid-gestation fetal anatomic ultrasound, which assessed phenotypic sex11. Although routinely performed, ultrasound assessment of the fetal genitalia is an optional component of the fetal anatomic ultrasound. As such, parents may decline assessment. If fetal phenotypic sex cannot be evaluated during anatomic ultrasound, repeat ultrasound may not be offered. Prior to NIPS, fetal genotypic assessment required a diagnostic procedure with chromosomal assessment and was typically performed for an alternative indication such as maternal age or fetal anomaly10. In contrast, NIPS routinely screens a sex chromosome complement, providing a highly sensitive and specific result to a large group of women who likely would not have otherwise undergone a diagnostic procedure and therefore not have received fetal genotypic sex assessment. In the United States, most patients undergoing cfDNA screening elect to be informed of predicted fetal sex. However, some commercial laboratories offer the ability to opt-in or -out of knowing this information in which case this result would not be included in the clinical report.
Given the relatively recent clinical introduction of NIPS, many pediatricians are less familiar with this prenatal screening modality, the important ways in which it differs from maternal serum screening and the potential etiologies of unanticipated results. An understanding of this screening modality and its limitations may lead to earlier pediatric diagnosis and impact medical management. Herein, we describe possible etiologies of discordant fetal NIPS result and postnatal phenotypic sex. Based on review of cases evaluated at our center, we developed a suggested clinical and laboratory evaluation workflow for when this unanticipated result presents postnatally.
METHODS
All subjects referred to our medical center for the indication of discordant NIPS from postnatal phenotype from January 2014 to June 2016 were reviewed. Available medical records describing NIPS result, prenatal care, postnatal care, and clinical evaluation of mother and infant were reviewed and results were collated. Case series was considered IRB exempt.
RESULTS
Four infants were identified with a fetal sex reported on NIPS that was discordant from postnatal phenotypic sex (Table 1). In each case, NIPS resulted as normal male. All patients declined prenatal diagnostic chromosomal assessment. Phenotypic fetal sex was not assessed by ultrasound in 2 of 4 cases, either by parental request or inability to visualize. Postnatal evaluation varied widely from phenotypic examination only to postnatal karyotype with referral to specialist care. Given the wide-variability in observed clinical practice in the patients in this study, postnatal evaluation guidelines were developed with input from obstetrics, medical genetics, pediatrics, genetic counseling and pathology (Figures 1&2).
Table 1:
Clinical summary of cases 1–4
| CASE 1 | CASE 2 | CASE 3 | CASE 4 | |
|---|---|---|---|---|
| NIPS Platform | Progenity verifi | Sequenom MaterniT21 Plus | Illumina Verinata | NK |
| Chromosomes assessed | 21, 18, 13, X and Y | 21, 18, 13, X and Y | 21, 18, 16, 13, 9, X and Y | NK |
| Gestational age NIPS performed | 11w, 6d; 22w, 4d | 11w, 4d | 17w, 5d | 12w, 4d |
| Fetal Fraction | NK | NK | NK | NK |
| NIPS sex result | Male (x2) | Male | Male | Male |
| Age discordant NIPS -phenotypic sex appreciated (method) | 21–22w (ultrasound) | Birth (physical exam) | Birth (autopsy) | 21w4d (ultrasound) |
| Prenatal diagnostic testing | No | No | No | No |
| Pregnancy outcome | Term, AGA | Term, AGA | Demise, 21w1d | Term, AGA |
| Post-natal physical examination of external genitalia | Normal female | Normal external female genitalia, no clitoromegaly, palpable center gonad | Normal female internal and external genitalia | Normal female |
| Post-natal karyotype result | 46,XX [60/60], 500 band level | 46,XY [30/30], 550–600 band level. | mos 45,X[18]/46,X,i(Y)(q10)[2] | NK |
| Post-natal FISH | NK | SRY positive, all cells | SRY negative, i(Y)(q10) | NK |
| Final diagnosis | Co-twin demise | 46,XY DSD | Sex chromosome mosaicism | Maternal chimera (iatrogenic) |
KEY: AGA=average (size) for gestational age; d=days;w=weeks; yo=years old; DOL=days of life; DNA=deoxyribonucleic acid; DSD=disorder of sexual development; NK=not known (either not performed or results unavailable); NIPS=non-invasive prenatal screening; SRY=sex-determining region Y;
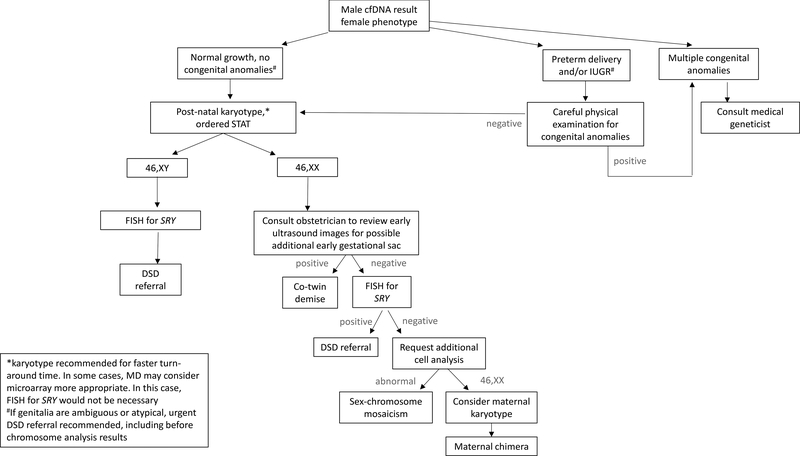
Figure 1:
Recommended clinical evaluation when cfDNA predicts male fetal sex and postnatal physical examination is consistent with female genitalia.
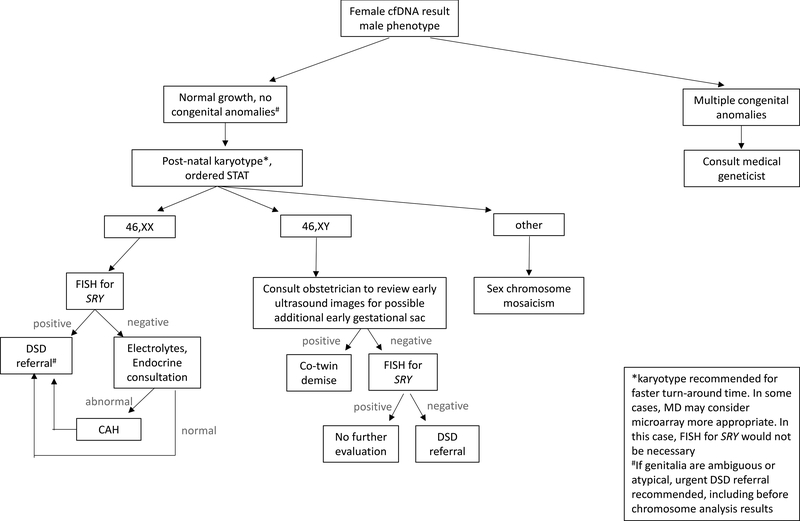
Figure 2:
Recommended clinical evaluation when cfDNA predicts female fetal sex and postnatal phenotype is consistent with male genitalia.
Case 1 was a term infant born after an uncomplicated delivery. NIPS performed at 11 weeks gestation was normal (i.e. no aneuploidy detected) and consistent with a male fetus (Table 1). Mid-gestation ultrasound examination suggested female external genitalia and a single amniotic sac. Amniocentesis was declined. NIPS was repeated at 22 weeks for the small possibility of a lab error, but again, results were consistent with a genetic male. Postnatal peripheral blood chromosome analysis was 46,XX [60/60] at a 500-band level. Although maternal chimerism was considered a potential cause, maternal karyotype was declined. Early co-twin demise of a male fetus (vanishing twin syndrome) was considered the most likely etiology.
Case 2 was the second child to non-consanguineous parents born at term after an uncomplicated delivery. NIPS performed at 11 weeks was normal and consistent with a male fetus. External genitalia were not assessed at the mid-gestation ultrasound due to fetal positioning. Postnatal physical examination was consistent with normal female external genitalia. After some confusion regarding the clinical relevance of this discrepancy, the infant was referred to a multidisciplinary Differences of Sex Development (DSD) clinic for full evaluation at 11 days of age. Specialized DSD physical examination revealed normal female external genitalia, no clitoromegaly, and a palpable gonad in the left inguinal canal. Laboratory evaluation did not suggest an endocrinopathy. Pelvic ultrasound showed a possible Mullerian structure. Postnatal peripheral blood karyotype was normal 46,XY and the patient was diagnosed with non- syndromic 46,XY DSD.
Case 3 was referred to Maternal-Fetal Medicine at 17 weeks gestation due to fetal hydrops. Fetal sex was not assessed on anatomic ultrasound. The pregnant patient declined amniocentesis with diagnostic chromosomal assessment but elected NIPS. NIPS results were normal and consistent with a male fetus. Unfortunately, fetal hydrops progressed and resulted in demise at 21 weeks’ gestation. Fetal autopsy identified normal internal and external female genitalia. Postnatal karyotype was 45,X[18]/46,X,i(Y)(q10)[2], consistent with mosaic Turner syndrome with an iso-dicentric Y-chromosome (Table 1).
Case 4 was a 26-year-old primigravida with a complicated medical history including congenital unilateral renal agenesis and renal transplant from a male donor. NIPS performed at 12 weeks was normal and consistent with a male fetus (Table 1). Mid-gestation ultrasound and postnatal examination were consistent with female external genitalia. The mother declined pre- and post-natal diagnostic chromosome analysis. Circulating cfDNA from the (male) transplanted kidney was considered the most likely source of discordant NIPS and phenotype. Of note, transplant recipients are no longer candidates for several NIPS platforms.
DISCUSSION
Given the swift clinical uptake of cfDNA screening and ongoing technological advancement, many pediatricians have limited understanding of the limitations of NIPS and potential etiology of discordant results. This report aims to review known potential causes of discordant cfDNA result and phenotypic sex as well as provide a diagnostic process for the pediatrician to evaluate this complex clinical presentation.
As seen in Case 1, vanishing twin syndrome is the most common cause of discordant NIPS result and phenotypic sex. After a co-twin demise, the fetus may be resorbed into the maternal circulation, incorporated into the placenta, “mummified” into a fetus papyraceus, or rarely, resorbed by the surviving twin to create a human chimera13. After a demise, placental cfDNA may persist for weeks in maternal circulation as the fetus and placenta are resorbed. As NIPS cannot distinguish between a viable and non-viable fetus, results may appear discordant if the co-twin demise is of the opposite genetic sex8,14,15. Even if an early gestational sac or fetal remnant is identified, it is unlikely that the sex of the co-twin can be phenotypically assessed15. Therefore, diagnostic chromosomal assessment to confirm genetic sex is recommended for the surviving neonate and could be considered for the placenta or co-twin remnant, if identified.
Differences of sex development (DSD) are highly complex conditions, as illustrated by Case 2. From a medical, social and ethical standpoint, these conditions are best managed by specialized teams. A DSD may represent an isolated condition, such as complete androgen insensitivity syndrome, or be part of a more involved syndrome, such as congenital adrenal hyperplasia or Smith-Lemli-Opitz syndrome16–20. Given the high complexity of DSD conditions and the potential for life- threatening complications, urgent referral is appropriate for any neonate whose chromosome complement (including NIPS) differs from the phenotypic external and/or internal genitalia or in whom the genitalia appear abnormal (Figures 1&2).
If noted prenatally, diagnostic testing via amniocentesis and referral to a maternal-fetal medicine specialist and/or medical geneticist with DSD expertise can be offered. If diagnostic testing is declined prenatally, postnatal chromosomal microarray or karyotype with fluorescence in situ hybridization (FISH) for SRY and urgent referral to a multi-disciplinary DSD clinical team is recommended. A discordant NIPS result may provide an opportunity for early diagnosis of a DSD that might otherwise escape detection until childhood, puberty, or even adulthood8,18–20. Communication between the obstetrician and pediatrician is essential; a discordant NIPS result may be the only early indication of a DSD, particularly for DSD conditions that lead to sex reversal.
Sex chromosome aneuploidies with or without mosaicism are an important possible cause of a discordant NIPS result and postnatal phenotype, with significant implications for medical management11,21,22. When NIPS detects a Y-chromosome component, it is considered definitively non-maternal, assumed to be of placental origin and genetic sex is often interpreted as male. However, placental cfDNA is only a small fraction of cfDNA in maternal circulation; sex chromosome mosaicism may not be adequately detected, as reflected in NIPS positive predictive values of approximately 55%23. Individuals with a mosaic monosomy X with a Y-chromosome component genotype can have significant phenotypic variability, ranging from normal external female genitalia with systemic features of Turner syndrome to normal phenotypic male20,22,24–26. Sex chromosome mosaicism can be difficult to detect by physical examination; postnatal chromosome analysis is recommended if sex chromosome mosaicism is suspected based on discordant NIPS result or otherwise.
A chimera refers to an individual with two unique cell lines that cannot be derived from a single cell line. This can occur spontaneously, as when one twin resorbs the other, or iatrogenically, as with a solid organ or stem cell transplantation. As a viable transplanted organ undergoes physiologic cell turnover, it sheds apoptotic cells into maternal circulation, creating an additional source of non-maternal cfDNA, as illustrated by Case 43,27.
Spontaneous chimeras are considered extremely rare, although the incidence is likely under-recognized28. Maternal chimerism was considered for Case 1, given the fetal fraction was high and nearly identical between NIPS drawn at 11 weeks and a repeat at 22 weeks (Table 1).
Although maternal karyotype was declined, and this possibility could not be definitively excluded, maternal chimera was empirically considered less likely than a co-twin demise. To our knowledge, spontaneous maternal chimerism has never been noted as a cause of discordant NIPS. Theoretically, suggestive factors may include an unusually large fetal fraction or a consistent fetal fraction on repeat testing (if performed), with the “fetal fraction” for the Y-chromosome actually representing maternal chimerism.
In addition, other etiologies to consider when NIPS result and phenotypic sex are discordant include intrauterine growth restriction (IUGR), preterm birth (PTB), a genetic syndrome and laboratory error. IUGR and PTB may cause male under-virilization due to one of several proposed mechanisms, including perturbation of the hypothalamic-pituitary-adrenal axis, maternal malnutrition and unidentified environmental factors30–31. Abnormal genitalia is a component of numerous multi-system genetic syndromes, although describing each here is beyond the scope of this report16,17,20,25. Referral to a clinical geneticist is recommended if multiple congenital anomalies are identified and/or if a genetic syndrome is suspected. A sample or laboratory handling error may result in a discordant NIPS result and phenotype but should be a diagnosis of exclusion. It is incorrect to presume that a previous male pregnancy (delivery or miscarriage) can explain the discrepancy between a male NIPS result and the findings of typical postnatal female external genitalia. Although intact fetal cells can persist in maternal circulation for decades, their quantity is significantly below the threshold of detection by current NIPS methods32.
A limitation of this case series is the inability to calculate a prevalence of discordant cfDNA genotype and postnatal phenotype. The majority of women with uncomplicated pregnancies in our region are followed by their local obstetrics provider. In each of the cases presented, the patient was referred to our regional, tertiary medical centers for the indication of discordant NIPS and post-natal phenotype or other complication requiring a higher level of care or expertise not available locally. This prevents us from knowing the total number of women who received NIPS during the study period and from determining a prevalence of discordant fetal sex results. Our case series aims to increase awareness of potential etiologies of discordant results. Further study into prevalence of this clinical presentation is warranted.
In conclusion, NIPS is the first non-invasive prenatal screening platform to routinely evaluate genetic sex. Given the relatively recent introduction of this methodology, many pediatricians have limited exposure to NIPS, and subsequently, unanticipated results. Clinical recognition of the potential etiologies of discordant phenotypic and genotypic results and recommended clinical evaluation provides an opportunity for early diagnosis and may be used to guide medical management.
Figure 3.
Differential Diagnosis: discordant cfDNA result and phenotypic sex
ACKNOWLEDGMENTS
Thank you to Lauren H. Brown, MS; Sheila Weiss, MS; Patricia Fechner, MD; Linda Ramsdell, MS; Margaret Shnorhavorian, MD; Elizabeth McCauley, PhD; Anne-Marie Amies-Oelschlager, MD and Jane Hitti, MD for their contributions to the patient’s clinical care, enthusiasm for this case series and interesting discussions. Funding support provided by the National Institutes of Health NIH5T32GM007454 (H.M.B.).
Abbreviations
- cfDNA
cell-free DNA
- NIPS
non-invasive prenatal screening
- DSD
differences of sexual development
- AGA
average (size) for gestational age
- SRY
sex-determining region Y
- DNA
deoxyribonucleic acid
- IUGR
Intrauterine growth restriction
- PTB
preterm birth
Footnotes
Conflict of Interest: The authors have no conflicts of interest to disclose. No honorarium, grant, or other form of payment was given to any of the authors to produce this manuscript.
REFERENCES
- 1.Bianchi DW, Parsa S, Bhatt S, Halks-Miller M, Kurtzman K, Sehnert AJ, et al. Fetal sex chromosome testing by maternal plasma DNA sequencing: clinical laboratory experience and biology. Obstet Gynecol. 2015; 125: 375–82. [DOI] [PubMed] [Google Scholar]
- 2.Wapner RJ, Babiarz JE, Levy B, Stosic M, Zimmermann B, Sigurjonsson S, et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol. 2015; 212: 332.e1–9. [DOI] [PubMed] [Google Scholar]
- 3.Neufeld-Kaiser WA, Cheng EY, Liu YJ. Positive predictive value of non-invasive prenatal screening for fetal chromosome disorders using cell-free DNA in maternal serum: independent clinical experience of a tertiary referral center. BMC Med. 2015; 13: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snyder MW, Simmons LE, Kitzman JO, Coe BP, Henson JM, Daza RM, et al. Copy- number variation and false positive prenatal aneuploidy screening results. N Engl J Med. 2015; 372: 1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol 2015; 45: 249–266. [DOI] [PubMed] [Google Scholar]
- 6.Devaney SA, Palomaki GE, Scott JA, Bianchi DW. Noninvasive fetal sex determination using cell-free fetal DNA: a systematic review and meta-analysis. JAMA 2011; 306: 627–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daley R, Hill M, Chitty LS. Non-invasive prenatal diagnosis: progress and potential. Arch Dis Child Fetal Neonatal Ed. 2014; 99: F426–30. [DOI] [PubMed] [Google Scholar]
- 8.Richardson EJ, Scott FP, McLennan AC. Sex discordance identification following non- invasive prenatal testing. Prenat Diagn. 2017; 37: 1298–1304. [DOI] [PubMed] [Google Scholar]
- 9.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997; 350: 485–487. [DOI] [PubMed] [Google Scholar]
- 10.Colmant C, Morin-Surroca M, Fuchs F, Fernandez H, Senat MV. Non-invasive prenatal testing for fetal sex determination: is ultrasound still relevant? Eur J Obstet Gynecol Reprod Biol. 2013; 171: 197–204. [DOI] [PubMed] [Google Scholar]
- 11.American Institute of Ultrasound in Medicine. AIUM practice guideline for the performance of obstetric ultrasound examinations. J Ultrasound Med. 2013;32:1083–101 [DOI] [PubMed] [Google Scholar]
- 12.Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. 2017; 50: 302–314. [DOI] [PubMed] [Google Scholar]
- 13.Shin SY, Yoo HW, Lee BH, Kim KS, Seo EJ. Identification of the mechanism underlying a human chimera by SNP array analysis. Am. J. Med. Genet 2012; 158: 2119–2123. [DOI] [PubMed] [Google Scholar]
- 14.Masala M, Saba L, Zoppi MA, Puddu R, Picciau A, Capponi V, et al. Pitfalls in noninvasive fetal RhD and sex determination due to a vanishing twin. Prenat Diagn. 2015; 35: 506–8. [DOI] [PubMed] [Google Scholar]
- 15.Curnow KJ, Wilkins-Haug L, Ryan A, Kirkizlar E, Stosic M, Hall MP, et al. Detection of triploid, molar, and vanishing twin pregnancies by a single-nucleotide polymorphism- based noninvasive prenatal test. Am J Obstet Gynecol. 2015; 212: 79.e1–9. [DOI] [PubMed] [Google Scholar]
- 16.Chitty LS, Chatelain P, Wolffenbuttel KP, Aigrain Y. Prenatal management of disorders of sex development. J Pediatr Urol. 2012; 8: 576–84. [DOI] [PubMed] [Google Scholar]
- 17.Baetens D, Mladenov W, Delle Chiaie B, Menten B, Desloovere A, Iotova V, et al. , Extensive clinical, hormonal, and genetic screening in a large consecutive series of 46,XY neonates and infants with atypical sexual development. Orphanet Journal of Rare Diseases. 2014; 9: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franasiak JM, Yao X, Ashkinadze E, Rosen T, Scott RT Jr. Discordant embryonic aneuploidy testing and prenatal ultrasonography prompting androgen insensitivity syndrome diagnosis. Obstet Gynecol. 2015; 125: 383–6. [DOI] [PubMed] [Google Scholar]
- 19.Mansfield N, Boogert T, McLennan A. Prenatal diagnosis of a 46,XX male following noninvasive prenatal testing. Clin Case Rep. 2015; 3: 849–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parisi MA, Ramsdell LA, Burns MW, Carr MC, Grady RE, Gunther DF, et al. A Gender Assessment Team: experience with 250 patients over a period of 25 years. Genet Med. 2007; 9: 348–57. [DOI] [PubMed] [Google Scholar]
- 21.Coyle D, Kutasy B, Han Suyin K, Antao B, Lynch SA, McDermott MB, et al. Gonadoblastoma in patients with 45,X/46,XY mosaicism: A 16-year experience. J Pediatr Urol. 2016; 12: 283.e1–283.e7. [DOI] [PubMed] [Google Scholar]
- 22.Tosson H, Rose SR, Gartner LA. Description of children with 45,X/46,XY karyotype. Eur J Pediatr. 2012; 171: 521–529.2. [DOI] [PubMed] [Google Scholar]
- 23.Neufeld-Kaiser WA, Cheng EY, Liu YJ. Positive predictive value of non-invasive prenatal screening for fetal chromosome disorders using cell-free DNA in maternal serum: independent clinical experience of a tertiary referral center. BMC Med. 2015; 13: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Siu VM. Is there a correlation between the proportion of cells with isodicentric Yp at amniocentesis and phenotypic sex? Prenat Diagn. 2010; 3: 839–44. [DOI] [PubMed] [Google Scholar]
- 25.Iruretagoyena JI, Grady M, Shah D. Discrepancy in fetal sex assignment between cell free fetal DNA and ultrasound. J Perinatol. 2015; 35: 229–30. [DOI] [PubMed] [Google Scholar]
- 26.Colindres JV, Axelrad M, McCullough L, Smith EO, Huang GO, Tu DD, et al. Evidence- Based Management of Patients with 45,X/46,XY Gonadal Dysgenesis and Male Sex Assignment: from Infancy to Adulthood. Pediatr Endocrinol Rev. 2016; 13: 585–601. [PubMed] [Google Scholar]
- 27.Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A. 2011; 108: 6229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Souter VL, Parisi MA, Nyholt DR, Kapur RP, Henders AK, Opheim KE, et al. A case of true hermaphroditism reveals an unusual mechanism of twinning. Hum Genet. 2007; 121: 179–85. [DOI] [PubMed] [Google Scholar]
- 29.de Andrade Machado Neto F, Moreno Morcillo A, Trevas Maciel-Guerra A, Guerra- Junior G. Idiopathic male pseudohermaphroditism is associated with prenatal growth retardation. Eur J Pediatr. 2005; 164: 287–91. [DOI] [PubMed] [Google Scholar]
- 30.Lek N, Miles H, Bunch T, Pilfold-Wilkie V, Tadokoro-Cuccaro R, Davies J, et al. Low frequency of androgen receptor gene mutations in 46 XY DSD, and fetal growth restriction. Arch Dis Child. 2014; 99: 358–61. [DOI] [PubMed] [Google Scholar]
- 31.Wu G, Bazer FW, Cudd TA, Meininger CJ, Spencer TE. Maternal nutrition and fetal development. J Nutr. 2004; 134: 2169–72. [DOI] [PubMed] [Google Scholar]
- 32.Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci U S A. 1996; 93: 705–8. [DOI] [PMC free article] [PubMed] [Google Scholar]