Abstract
Admixture and polyploidization are major recognized eukaryotic genome evolutionary processes. Their impacts on genome dynamics vary among systems and are still partially deciphered. Many banana cultivars are triploid (sometimes diploid) interspecific hybrids between Musa acuminata (A genome) and M. balbisiana (B genome). They have no or very low fertility, are vegetatively propagated and have been classified as “AB,” “AAB,” or “ABB” based on morphological characters. We used NGS sequence data to characterize the A versus B chromosome composition of nine diploid and triploid interspecific cultivars, to compare the chromosome structures of A and B genomes and analyze A/B chromosome segregations in a polyploid context. We showed that interspecific recombination occurred frequently between A and B chromosomes. We identified two large structural variations between A and B genomes, a reciprocal translocation and an inversion that locally affected recombination and led to segregation distortion and aneuploidy in a triploid progeny. Interspecific recombination and large structural variations explained the mosaic genomes observed in edible bananas. The unprecedented resolution in deciphering their genome structure allowed us to start revisiting the origins of banana cultivars and provided new information to gain insight into the impact of interspecificity on genome evolution. It will also facilitate much more effective assessment of breeding strategies.
Keywords: banana, chromosomes, genome evolution, interspecific hybrids, Musa spp, polyploidy
Introduction
Interspecific hybridization through admixture and/or allopolyploidization is widely acknowledged as being a major genome evolutionary process (Pennisi 2013; Wu et al. 2014; Van de Peer et al. 2017). The short-term consequences of these events on genome dynamics differ between systems (Van de Peer et al. 2017; Wendel et al. 2018) and may depend on the level and type of differentiation between the partners involved. In the long term, surviving polyploidy events have evolved toward diploidization through still only partially understood mechanisms (Freeling 2017; Pelé et al. 2018). In the mid-term, asexual propagation has been suggested as a way to overcome the low level of fertility resulting from destabilized karyotypes (Freeling 2017; Hojsgaard 2018). Many crops such as banana are derived from interspecific hybridization, they are interesting models for studying admixture and allopolyploidization. Banana has been selected through domestication for seedless fruits, this unique feature is a result of low fertility and parthenocarpy.
Wild bananas originated from South-East Asia, where they diverged into species and subspecies following geographical isolation in distinct continental regions and islands (Janssens et al. 2016). Cultivars are derived from natural hybridization between wild diploid Musa species and subspecies fostered by human migrations (Perrier et al. 2011). They are triploid, sometimes diploid, seedless, and parthenocarpic clones selected by early farmers that were dispersed through centuries of vegetative propagation. Musa acuminata (A genome, 2n = 2x = 22) is involved in all the known cultivars and is present in the southern part of the distribution range, whereas M. balbisiana (B genome, 2n = 2x = 22) contributes to several cultivars and originates from North-East India to South-East China (Simmonds and Shepherd 1955). Divergence between these two species has been estimated between 4.6 and 20 Ma (Lescot et al. 2008; Janssens et al. 2016). M. acuminata has been divided into several subspecies based on morphological features, geographical distribution and partial interfertility. Cultivars have been classified in genomic groups based on their morphological similarities to wild diploid species and their ploidy level (Cheesman 1947; Simmonds and Shepherd 1955), with the main groups being conventionally referred to as “AA,” “AB,” “AAA,” “AAB,” and “ABB.” They were further classified into subgroups based on agronomic characteristics. Almost 40% of global banana production concerns A/B interspecific triploid cultivars, including the Plantain cooking banana group (classified as “AAB”) that represents ∼18% of total banana production and is mainly grown in West Africa and Central/South America.
The origin and the domestication process of banana interspecific cultivars is still a matter of investigation. It is generally recognized that M. balbisiana plants were transported southward via human migrations to regions where hybridization with M. acuminata occurred (Perrier et al. 2011). Based on the reported peculiar organelle transmission in Musa, with the paternal inheritance of mitochondrial genomes and maternal inheritance of chloroplasts (Fauré et al. 1994; Carreel et al. 2002), a scenario regarding the formation of triploid “AAB” and “ABB” has been proposed. A primary interspecific A/B hybrid produced a 2n gamete that was fertilized by M. acuminata (AA), resulting in an “AAB” progeny, or by M. balbisiana (BB), resulting in an “ABB” progeny. The nature and ploidy of the primary A/B hybrid are still unresolved and the occurrence of one or more backcrosses in the origin of edible banana cultivars has been evoked (De Langhe et al. 2010). The involvement of one or several steps in cultivar formation and the extent of interspecific recombination would have an impact on their chromosome composition. Genomic in situ hybridization (GISH) studies have begun revising the conventional classification by revealing that the “ABB” clone Pelipita had 8A and 25B chromosomes and that “AAB” plantain had 21A and 12B (D’Hont et al. 2000).
Chromosome pairing at meiosis is generally regular (bivalent) in wild diploid banana species but was found to be altered with the frequent formation of univalent and multivalent chromosomes in interspecific (AB) or intersubspecific (AA) diploid hybrids (Fauré et al. 1993; Shepherd 1999; Hippolyte et al. 2010; Jeridi et al. 2011). These analyses suggested that Musa species and subspecies differed in their chromosome structure, resulting in disturbed meioses in the hybrids and contributing to their loss of fertility (Dodds and Simmonds 1948; Shepherd 1999; Jeridi et al. 2012). Martin et al. (2017) recently described such a large chromosome structural variation (LSV) in banana in the form of a large reciprocal translocation between M. acuminata chromosomes 1 and 4. This translocation was found in the wild subspecies M. acuminata spp. malaccensis from which it is suggested to have emerged. It was shown to generate high segregation distortion in progenies obtained from heterozygous accessions. In A/B polyploid hybrids, homologous chromosome pairing was generally assumed, although recent GISH (Jeridi et al. 2011) and genetic mapping (Noumbissié et al. 2016) studies have provided clues that challenge this idea. The first evidence of LSV between the A and B genomes was reported based on chromosome segregation in an interspecific progeny (Noumbissié et al. 2016).
Musa breeding is complicated by the fact that cultivated bananas are generally triploid and have very low fertility levels, a condition that ensures edibility. Enhanced knowledge of the chromosome constitution of interspecific banana cultivars and its consequence on chromosome segregation would help gain greater insight into Musa evolution and domestication and more generally genome evolution in allopolyploids, while also ultimately benefitting banana breeding programs.
The present study aimed to characterize the genome structure of diploid, triploid, and tetraploid A/B interspecific banana accessions and to study the impact of these genome structures on chromosome segregation and recombination. We thus developed methods to exploit resequencing data and used them to: 1) characterize the A/B genome structure of nine interspecific banana accessions; 2) compare the global chromosome organization of M. acuminata and M. balbisiana (A and B genomes); and 3) analyze the impact of interspecificity on chromosome segregation and recombination in the progeny of an interspecific “AAAB” tetraploid breeding accession.
Results
Substantial Deviations in Banana Cultivar A/B Chromosome Constitutions from the Conventional Classification
We analyzed the A versus B chromosome constitution of nine interspecific banana hybrid accessions: two diploid cultivars, six triploid cultivars from four subgroups (Plantain, Silk, Mysore, and Pelipita), as well as one tetraploid breeding accession. These accessions, together with nine M. acuminata and three M. balbisiana representatives, were sequenced through WGS or RadSeq and sequence reads were mapped onto the M. acuminata reference genome V2 (Martin et al. 2016). We first compared the read coverage of each SNP position to the mean coverage value along chromosomes so as to verify the euploid chromosome constitution of the accessions (i.e., 2, 3, or 4 sets of each chromosome for the 2x, 3x, and 4x accessions, respectively). All accessions had the expected euploid chromosome constitution (supplementary fig. S1, Supplementary Material online). We then selected M. acuminata or M. balbisiana-specific SNPs as the ones being present versus absent in all representatives of these species (diallelic SNPs only). Their coverage ratio was analyzed along each of the 11 basic chromosomes to determine the A versus B homeologous chromosome constitution (fig. 1 and supplementary fig. S1, Supplementary Material online).
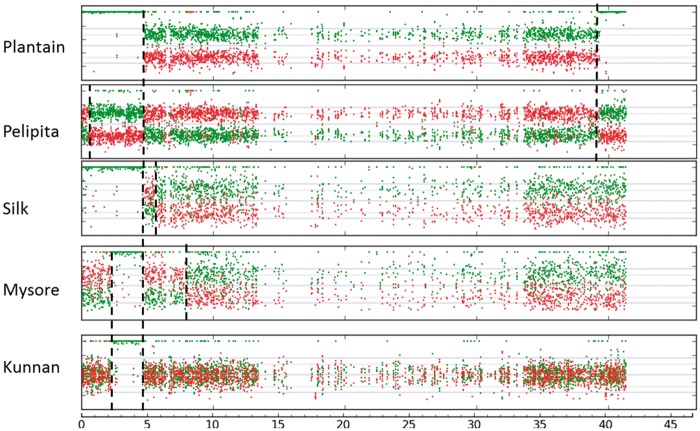
Fig. 1.
Mosaic genome structure of A/B interspecific banana cultivars. Coverage ratios for alleles specific to the A genome (green dots) and to the B genome (red dots) are plotted along the 11 pseudochromosomes of the Musa acuminata reference genome. Open boxes indicate chromosome segments that differ from the conventional genomic classification (“AB,” “ABB,” “AAB”). The scale is indicated (in Mb) at the bottom of each figure.
For the two diploid cultivars, the results showed an AB chromosome constitution for all chromosome sets except for a few chromosomal segments that displayed an AA constitution. Two such fragments were observed in the Kunnan accession: a 2.4-Mb fragment within chromosome 9 and a fragment of ∼6 Mb at one end of chromosome 8. For the Safet Velchi accession, one AA fragment of 7 Mb was observed at one end of chromosome 6 (fig. 1).
For cultivar Pelipita, conventionally classified as “ABB,” the results were consistent with a global ABB chromosome constitution, but several notable exceptions were detected (fig. 1). The entire chromosomes 2 and 11 and fragments of various lengths of chromosomes 6, 7, and 10 (from several megabases to almost the entire chromosome) displayed a BBB constitution. In addition, two segments of chromosome 9 (one on each arm) displayed an AAB constitution. The five accessions conventionally classified as “AAB” from the Silk, Plantain, and Mysore subgroups showed an AAB chromosome constitution for most of the chromosome sets, but here again exceptions were noted (fig. 1). The two cultivars from the Silk subgroup, that is, Figue Pomme Géante and Figue Pomme Naine, displayed an identical chromosome composition (fig. 1 and supplementary fig. S2, Supplementary Material online). They had one segment on chromosome 9 with an ABB constitution and four segments located in the distal regions of chromosomes 2, 3, 4, and 9 with an AAA constitution. The two Plantain cultivars, that is, French Clair and Red Yade (fig. 1 and supplementary fig. S2, Supplementary Material online), displayed an identical chromosome composition with six segments of chromosomes 4, 6, 8, 9, and 10 showing an AAA composition. In addition, the entire Plantain chromosome 7 displayed an ABB composition. Cultivar Pisang Ceylan (Mysore subgroup, AAB) showed five chromosome segments displaying an ABB composition in chromosomes 4, 7, 8, and 9 and four segments with an AAA composition in chromosomes 3, 4, 9, and 11 (fig. 1).
For the CRBP39 tetraploid breeding accession derived from a cross between the “AAB” plantain French Clair and an AA accession (M53), most chromosome sets showed an AAAB constitution (supplementary fig. S3, Supplementary Material online). Exceptions concerned the same chromosome segments as its Plantain parent French Clair. In addition, most of chromosome 2 displayed an AABB composition and the entire chromosome 10 displayed an AAAA composition.
All interspecific banana cultivars studied here, showed a mosaic A/B chromosome structure that we interpret as resulting from A/B recombination. The observed uniform SNP read coverage ratio along chromosomes excluded deletion events or aneuploidy as alternative interpretation. Likewise, the differences between CRBP39 and “AAB” Plantain could be explained by two A/B recombination events during the meiosis of French Clair (supplementary fig. S3, Supplementary Material online).
Identification of Two Large Structural Variations between Musa A and B Genomes
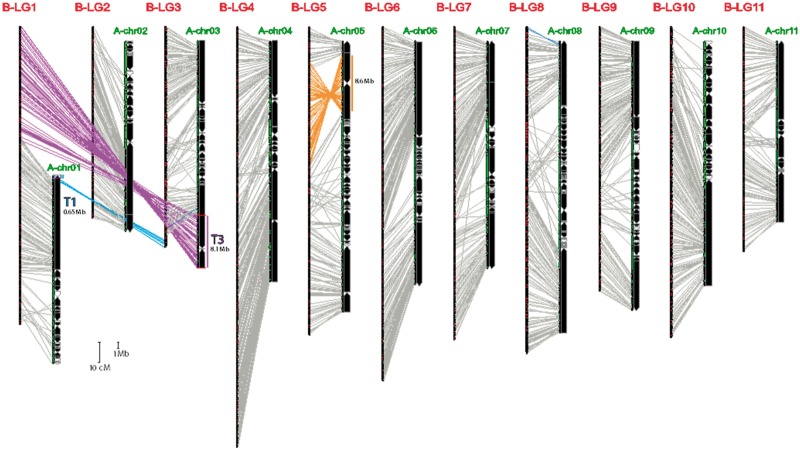
We compared the global chromosome structure conservation (i.e., synteny) between the M. balbisiana accession Pisang Klutuk Wulung (PKW) and the M. acuminata accession DH-Pahang through the construction a dense genetic map for the B genome and comparison with the A genome reference sequence assembly. A self-progeny of 105 individuals from the M. balbisiana PKW accession was genotyped by RadSeq. A total of 10,074 heterozygous markers were selected and grouped at LOD 9 into 11 linkage groups of 580–1,370 markers (supplementary table S2, Supplementary Material online). A very low level of segregation distortion uniformly spread along chromosomes was observed, with two linkage groups corresponding to chromosomes 10 and 11 that were more affected than other LGs (supplementary fig. S4, Supplementary Material online).
Markers from the B genetic map were aligned on the A reference sequence, revealing global synteny and collinearity conservation between M. balbisiana (PKW) and M. acuminata (DH-Pahang) for most chromosome arms (fig. 2). For these regions, >95% of markers belonging to a B linkage group were aligned to a single A reference chromosome (supplementary table S2, Supplementary Material online). Two major exceptions were observed: a reciprocal translocation involving chromosome 1 and chromosome 3 and an inversion on chromosome 5 (fig. 2). On the M. acuminata physical map, the reciprocal translocation involved a small 0.65-Mb segment on chromosome 1 (referred to as T1 for translocated fragment from chromosome 1) and a large 8.5-Mb segment of chromosome 3 (referred to as T3). The inversion involved a large 9-Mb segment on chromosome 5A. In pericentromeric regions, the comparison was complicated by the low precision of the genetic and physical maps resulting from the low recombination rate and high repeat content (fig. 2).
Fig. 2.
Comparison of the genetic map of the Musa balbisiana PKW accession (genome B) with the M. acuminata (genome A) reference sequence assembly. The 11 linkage groups of PKW (genome B) are represented on the left (scale bar: 10 cM) and the genome A reference sequence assembly on the right with black or open arrows representing oriented or nonoriented scaffolds, respectively (scale bar: 1 Mb). Markers from each LG of the B genetic map that were aligned to one A reference chromosome are connected in gray. Spindles of >10 markers highlighting large structural variations between A and B genomes are connected in different colors: orange for a large inversion on chromosome 5 and blue or purple for a large reciprocal translocation involving chromosomes 1 (T1) and 3 (T3).
Characterization of Interspecific A and B Chromosome Recombination and Segregation Patterns in an “AAAB”×AA Progeny
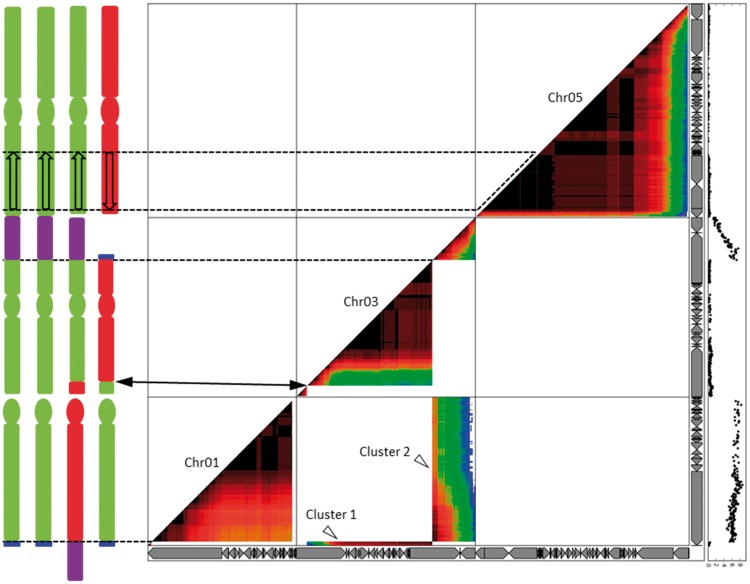
A progeny of 185 individuals from the “AAAB” (CRBP39)×AA cross was genotyped by Radseq. We selected 20,824 SNP markers specific to the B genome and projected their pairwise associations in the progeny on the M. acuminata reference assembly (supplementary fig. S5, Supplementary Material online). The markers were well distributed along the 11A reference chromosomes except for chromosomes or chromosome segments for which no B homeologs were present in CRBP39 (i.e., chromosome 10 and parts of chromosomes 4, 6, 8, and 9). In chromosome 3, we observed a genetic linkage breakpoint that we tentatively interpreted as corresponding to an A/B recombination during the formation of CRBP39 (fig. 3), which did not result in a change in the allelic ratio.
Fig. 3.
Pairwise association of B genome specific SNPs in AAAB×AA progeny projected on chromosomes 1, 3, and 5 of the A genome reference assembly. Chromosomes 1, 3, and 5 of the Musa acuminata reference genome assembly are represented by horizontal and vertical gray arrow boxes (each one representing a scaffold). The marker linkage/pairwise association is represented by a warm-cool color gradient from dark red to blue for strong linkage and weak linkage, respectively. The black curve on the right represents segregation distortion (calculated as the log10P value of the χ2 test to assess the deviation from the expected Mendelian segregation ratio). Two clusters of B markers linking two independent A reference chromosomes corresponding to the reciprocal translocation detected in PKW (fig. 2) are observed (Clusters 1 and 2, open arrowheads). No recombination was detected (dash line) in the region corresponding to the inversion in PKW as compared with the A reference genome. The putative karyotype of the tetraploid CRBP39 is indicated on the left. Vertical bars represent chromosome arms, circles represent centromeric regions and A/B chromosome segments are in green and red, respectively. Structural variations are indicated in the chromosome bodies (translocated fragments: T1 = blue, T3 = purple; inverted fragment = open arrow) and connected to the dot plot with black lines. In chromosome 3, the black arrows indicate a breakpoint in genetic linkage, potentially corresponding to an A/B recombination breakpoint in the CRBP39 chromosome.
Interestingly, clusters of markers of the B genome projected on reference chromosomes 1A and 3A were highly linked with a linkage intensity similar to that observed for physically close markers belonging to a single reference chromosome (fig. 3). Cluster 1 connected markers projected on a 1-Mb distal region of chromosome 1 to markers projected on most of chromosome 3 (fig. 3). Cluster 2 connected markers projected on the rest of chromosome 1 to markers projected on an 8-Mb distal region of chromosome 3 (fig. 3). A high level of segregation distortion was observed in markers corresponding to the 1B and 3B chromosomes (fig. 3). In addition, strong linkage—suggesting an absence or a very low level of interspecific recombination—was observed in the region of chromosome 5 found inverted in PKW compared with M. acuminata (fig. 3).
The most parsimonious interpretation of these results is that the B genome involved in the origin of “AAB” Plantain and thus of CRBP39 bears the same large structural variations as PKW (i.e., a large reciprocal translocation involving chromosomes 1 and 3 and a large chromosome inversion on chromosome 5) as compared with the M. acuminata reference genome assembly.
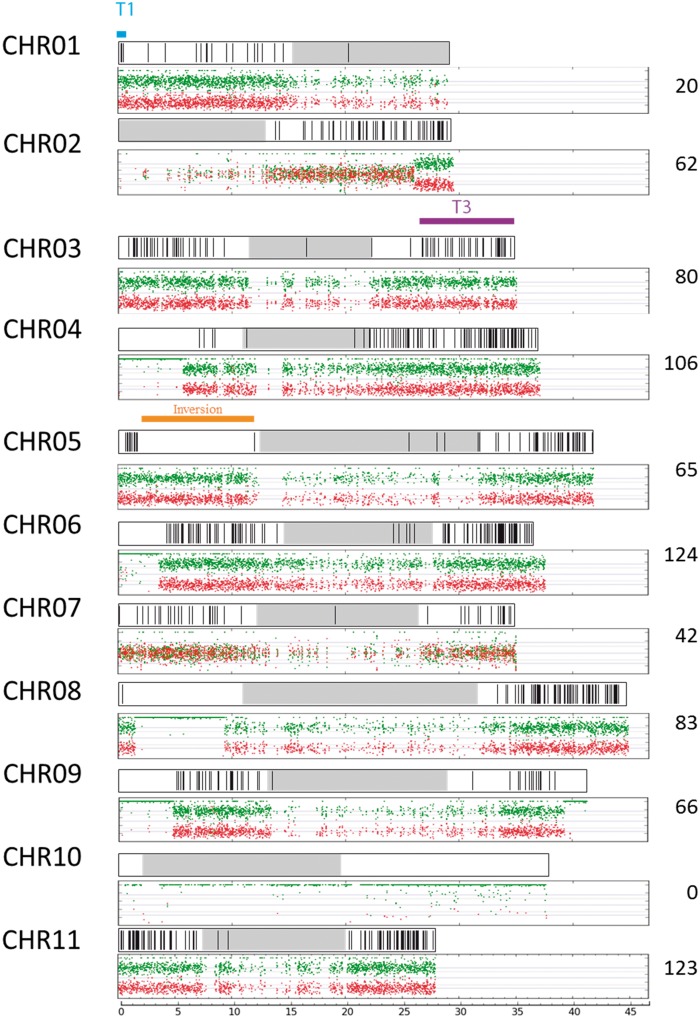
Chromosomes resulting from recombination between A and B genomes in the tetraploid accession CRBP39 were identified by comparing the A/B chromosomal composition of each of the 185 progeny individuals to that of CRBP39. The detected interspecific recombination events are shown along the 11A reference chromosomes (fig. 4).
Fig. 4.
Interspecific A/B recombination detected in the CRBP39 “AAAB” progeny. The A/B mosaic genome structure of the 11 chromosome sets of the parent CRBP39 “AAAB” are represented based on the A reference chromosome assembly (bottom box) (see fig. 1 legend). Interspecific recombination breakpoints (black vertical lines) recorded in the 180 progenies from CRBP39×Pahang are plotted on the A reference chromosome assembly (top box). The total number of breakpoints per chromosome recorded in the 185 progenies is indicated on the right. Large structural variations between the A and B genome are indicated in blue and purple for reciprocal translocation (T1, T3) and orange for inversion.
The results revealed that interspecific recombination occurred on all chromosomes for which A and B homeologs were present and generally widespread on the chromosome arms. As expected, fewer recombination events were observed in pericentromeric regions. One noteworthy exception concerned the region of chromosome 5 which was inverted between A and B genomes and for which no A/B chromosome recombinant was observed (fig. 4) although recombinants were detected at both ends of the chromosome 5 arms. This was consistent with the higher linkage intensity for markers projected on this segment (fig. 3). In addition, a lower number of interspecific recombinations was detected for chromosomes 1 and 3, which are involved in the 1/3 chromosome translocation distinguishing A and B genomes. However, recombination breakpoints were detected within the T1 and T3 segments, showing that pairing occurred between chromosome 1A and the T1 fragment translocated on chromosome 3B and also between chromosome 3A and the T3 translocated fragment on chromosome 1B. In regions with an AABB chromosome constitution, such as the entire chromosome 7, fewer A/B recombination events were observed compared with AAAB regions (fig. 4). Statistically, the number of A/B recombinations in an AABB configuration should be increased by one sixth relative to an AAAB configuration. When comparing the A/B recombination level observed on chromosome 7 (AABB) to that observed for chromosome 11 (AAAB) (the only chromosome class with an entire AAAB structure and no structural variations), a two-third reduction in recombination was observed, suggesting some degree of preferential pairing between homologs.
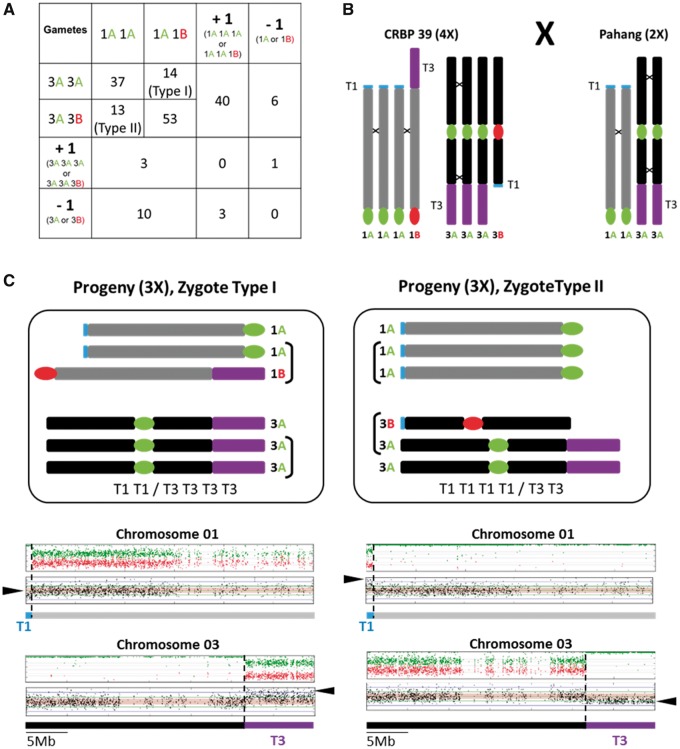
We then analyzed the euploid versus aneuploid nature of all progeny individuals by examining the Radseq sequencing coverage along chromosomes. Among the 185 individuals of the progeny, only 39% had three complete homeologous chromosomes per chromosome set (2n = 3x = 33). All other individuals exhibited various degrees of aneuploidy (table 1). For 73 plants, a single chromosome set showed aneuploidy, whereas for 40 plants, two or more chromosome sets were concerned (35 plants with two chromosomes sets, 4 plants with three chromosome sets, and 1 plant with four chromosome sets). Aneuploidy mainly affected the three chromosome sets that differed by an LSV between A and B genomes with 44% of the progeny showing aneuploidy for chromosome 1, 25% for chromosome 3 and 8% for chromosome 5 (table 1). Aneuploidy involving chromosome 5 was mainly a gain of one chromosome (12 plants), whereas only three plants showed a chromosome loss. Aneuploidy involving chromosome 1 and/or chromosome 3 concerned 95 plants and was of a distinct type (fig. 5A). A total of 27 plants contained the expected 2n = 3x = 33 chromosomes but had an altered ratio for the T1 and T3 chromosome segments involved in the reciprocal translocation (fig. 5B). Two types of segmental aneuploidy resulted from particular combinations of A and B chromosome 1 and chromosome 3 that led to various copy numbers for segments T1 and T3. Type I corresponded to a chromosome constitution of 1AAB/3AAA in the progeny, resulting in a T1T1T3/T3T3T3 constitution (fig. 5C). Type II corresponded to a chromosome constitution of 1AAA/3AAB in the progeny, resulting in a T1T1T1/T3T3T1 constitution (fig. 5C). In addition, 71 plants showed a gain or loss of an entire chromosome 1 or 3 (54 plants for chromosomes 1 and 17 plants for chromosome 3) (table 1). In these plants, various combinations of chromosomes 1A, 1B, 3A, and 3B were observed, resulting in various ratios of segments T1 and T3.
Table 1.
Chromosomal Characteristics of Aneuploidy Recorded in the Progeny.
| Chromosome | CHR01 | CHR02 | CHR03 | CHR04 | CHR05 | CHR06 | CHR07 | CHR08 | CHR09 | CHR10 | CHR11 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Complete chromosome gain | 7 | 1 | 4 | 0 | 12 | 0 | 0 | 2 | 2 | 0 | 2 | 38 |
| Complete chromosome loss | 47 | 3 | 13 | 1 | 3 | 1 | 0 | 0 | 0 | 0 | 4 | 63 |
| Partial gain | 13 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 27 |
| Partial loss | 14 | 0 | 15 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 15 |
| Percent of progeny affected | 43.8 | 2.2 | 24.8 | 0.5 | 8.1 | 0.5 | – | 1.6 | 1.1 | – | 3.2 | 61.1 |
Note.—The number of plants with complete or partial chromosome aneuploidy is indicated for each chromosome set with their respective proportions in the progeny.
Fig. 5.
Aneuploidy pattern involving chromosomes 1 and 3 observed in CRBP39 “AAAB”×Pahang “AA” progeny. (A) Number of progeny observed resulting from gametes with distinct combinations of homeologous chromosomes 1 (rows) and chromosomes 3 (columns). Aneuploid gametes with complete gain (+1) or loss (−1) are indicated. (B) Schematic representation of the homeologous chromosome constitution of the progeny parents for chromosomes 1 and 3. (C) Schematic representation of the homeologous chromosome constitution for chromosomes 1 and 3 for Type I and Type II progeny individuals (top boxes). A/B mosaic chromosome structure (see fig. 1 legend) and chromosomal segment coverage with the read-depth of each SNP marker plotted along the chromosomes for Type I and Type II progeny individuals (bottom boxes). T1 and T3 correspond to the reciprocal translocation differentiating chromosome 1 and 3 in A and B genomes. Arrows point to observed changes in allele ratios and read coverage.
Discussion
A/B Chromosome Recombination Is Frequent in Interspecific Bananas but Segregation Is Locally Affected by Large Structural Variations between A and B Genomes
We demonstrated, when analyzing “AAAB” CRBP39 progeny, that interspecific recombination between A and B genomes occurred all along the chromosomes but was locally affected by large structural variations. Recombinations were observed in segments with three A copies and one B copy, that is, in which the B segment could only pair with an A segment, but also in segments with two A and two B copies. Although in this latter case some degree of preferential pairing between homologs might have occurred.
Comparison of the chromosome structure of M. balbisiana, represented by the Pisang Klutuk Wulung (PKW) accession, and M. acuminata, represented by the DH-Pahang accession, revealed the presence of two large structural variations: one paracentric inversion on chromosome 5 and one reciprocal translocation involving chromosomes 1 and 3. Our analysis of chromosome segregation in “AAAB” CRBP39 progeny provided strong evidence that the B genome of CRBP39, and its Plantain parent French Clair, have the same chromosomal structures as the M. balbisiana PKW accession. M. acuminata has been divided, based on chromosome pairing configurations in hybrids between subspecies, into “translocation groups” with one standard group and six other groups differing from the standard group by one to three translocations (Shepherd 1999; Martin et al. 2017). This classification partially overlapped the subspecies classification. For the A/B comparison, we used the M. acuminata DH-Pahang accession, a M. acuminata spp. malaccensis accession derived from the standard A chromosome structure group (D’Hont et al. 2012; Martin et al. 2017). Within M. balbisiana, no clear subdivisions have been revealed and this species is still considered to be less genetically diverse (Swangpol et al. 2007; Gayral et al. 2010). Previous chromosome pairing studies in a synthetic A/B hybrid involving an A genome from the standard group suggested the presence of one translocation and probably an inversion (Shepherd 1999). In addition, we showed that the M. balbisiana accession involved in the origin of plantains, which is believed to be different from PKW (Hippolyte et al. 2012), has the same chromosome structure as the M. balbisiana PKW accession. So far, these data (although limited) are in favor of a common chromosome structure within M. balbisiana.
Structural variations are recognized as being major actors of speciation processes by reducing fertility (Stathos and Fishman 2014; Quach et al. 2016) and recombination in hybrids, thus limiting gene flow (Rieseberg 2001). Inversions have been widely studied, particularly in Drosophila and sunflower where their role in reducing recombination between alleles that independently increase fitness has been demonstrated (Rieseberg et al. 1995; Rieseberg 2001; Hoffmann et al. 2004; Hoffmann and Rieseberg 2008). In crops, segregation distortion and reduced fertility induced by structural heterozygosity have been reported (Chen et al. 2015; Samans et al. 2017), which is generally a constraint for breeding programs. In banana, the presence of a heterozygous reciprocal translocation between chromosome 1 and chromosome 4 in a diploid M. acuminata accession has generated high chromosome segregation distortion and reduced recombination (Martin et al. 2017).
In the “AAAB” CRBP39 progeny, the large structural variations between A and B genomes had a major impact on chromosome segregation, locally resulting in a very high level of segregation distortion, a high proportion of aneuploids in progenies (61%), and low fertility. The progeny resulted from pollination of thousands of flowers and the use of embryo rescue techniques, clearly indicating that drastic selection processes had occurred at the gametic and/or zygotic level, resulting in a reduction of fertility. The observed aneuploids included a gain and loss of one copy of chromosomes but also a gain or loss of one copy of the chromosome segment involved in the reciprocal translocation differentiating A and B genomes (segmental aneuploid). In the progeny of a diploid M. acuminata accession structurally heterozygous for the chromosome 1/4 translocation, no aneuploid offsprings were observed (Martin et al. 2016). More generally, aneuploidy has not been reported in diploid banana progenies (Fauré et al. 1993; Hippolyte et al. 2010). Our finding of many aneuploids showed that the double chromosome content of the gametes generated by the tetraploid CRBP39 accession partially mitigated the detrimental effects of unbalanced gametes. In other systems, aneuploidy has also been observed in early stage polyploids (Chester et al. 2012; Zhang et al. 2013). Further investigations are needed to determine whether they represent ephermer byproducts or contribute to polyploid evolution (Matsushita et al. 2012; Wu Y et al. 2018).
The progeny we analyzed was derived from plantain cultivars that are believed to have their A chromosomes contributed by M. acuminata spp. banksii (Perrier et al. 2011; Hippolyte et al. 2012). This subspecies belongs to the standard A chromosome structure group (Shepherd 1999; Martin et al. 2017). Other interspecific cultivars involved other M. acuminata subspecies and could thus have additional differences in their chromosomal organization compared with the B genome, for example, Figue Pomme and Safet Velchi have one set of A genome chromosomes 1 and 4, which differ from the standard structure by a reciprocal translocation (Martin et al. 2017). In interspecific progenies derived from these accessions, these structural variations could induce additional chromosome recombination and segregation disturbances in other genome regions.
A New Mosaic Image of the Genome Structure of A/B Interspecific Cultivars That Will Help Elucidate Their Origin
The interspecific genome structure of all nine accessions studied here deviated from the conventional classification based on morphological resemblance to wild M. acuminata and M. balbisiana (Simmonds and Shepherd 1955). For this classification, 15 morphological descriptors were scored and the overall score range was used to group edible banana varieties into distinct genomic classes (“AA,” “AB,” “AAA,” “AAB,” “ABB”). The conventional classification has been questioned by De Langhe et al. (2010) based on wide deviations from the expected scoring range for some accessions and on the transmission patterns of chloroplast and mitochondrial markers, and also by D’Hont et al. (2000) based on GISH experiments. Note that GISH only allows assignment of an A versus B origin to centromeric/pericentromeric parts of banana chromosomes that have a high number repetitive elements. The methodology developed enabled us to refine these results at whole chromosome scale. Here, we showed that all nine accessions studied had a mosaic interspecific genome structure, implying that interspecific recombination occurred during the crosses that led to these accessions.
Within each genomic group, banana cultivars were subdivided into subgroups based on agro-morphological characteristics. Banana clones have been multiplied by vegetative propagation for hundreds of years. It is thus believed that the agro-morphological variations observed within subgroups result from genetic or epigenetic somatic mutations. Molecular genetic diversity studies have generally confirmed these subgroups but exceptions to genetic homogeneity have also been found (Carreel et al. 1994; Hippolyte et al. 2012; Sardos et al. 2016). Within some subgroups such as African Plantains, accessions are highly genetically similar, which suggests a common origin from a single genotype (Noyer et al. 2005; Sardos et al. 2016). The two clones of the Plantain subgroup that we analyzed had an identical mosaic genome structure, which is in line with this hypothesis. The two clones of the Silk subgroup we analyzed also had an identical mosaic genome structure. This was expected since they represent size variants of the same “Figue Pomme” accession (Figue Pomme Géante and Figue Pomme Naine). Recently, based on DArT analysis, the Silk subgroup was divided into two sets of clones (Sardos et al. 2016). We may here have a representative structure of one of the two Silk types but further analyses on a larger number of accessions are needed to confirm this. The possibility we described here to precisely characterize the mosaic interspecific structure of cultivars will help to better define cultivar subgroups and gain further insight into their origins.
Insight into the Origin a Few 2× and 3× Interspecific Hybrid Subgroups
Diploid Interspecific Hybrids
Edible diploid AB hybrids are rare and confined to India (De Langhe et al. 2010). The only two documented cases are Kunnan accessions and the Ney Poovan subgroup, including Safet Velchi (Stover and Simmonds 1987; De Langhe et al. 2010; Christelová et al. 2017). Both diploid AB hybrids have a M. acuminata chloroplast cytotype (transmitted maternally) and mitochondrial cytotype (transmitted paternally) (Carreel et al. 1994; Boonruangrod et al. 2008). They were predicted to have resulted from an initial AA×BB cross followed by an AB×AA backcross, which would explain their M. acuminata cytotype. Here, we showed that these two hybrids had distinct A/B mosaic genome structures and thus were not derived from a single original genotype through somaclonal variation, thus confirming recent DArT and SSR marker results (Sardos et al. 2016; Christelová et al. 2017). In both of them, we found a balanced A and B homeologous chromosome constitution, with the exception of one or two small chromosome segments without any B homeolog contribution. If these AB diploids resulted from a single AB×AA backcross with random segregation of A and B chromosomes, then a more imbalanced A and B homeologous chromosome constitution could be expected. One hypothesis to explain the observed A/B mosaic patterns is that these AB diploids are the result of a cross between an AA accession and a BB accession already introgressed with some A genome. The small number and size of the observed AA fragments suggest that the introgressed BB accessions resulted from multiple backcrosses following the original A/B cross. This hypothesis is in favor of the involvement of introgressed secondary hybrids through backcrossing, as suggested by De Langhe et al. (2010), but with more backcrossing events in founding populations than initially proposed.
Plantain Subgroup and Pelipita (Philippine Origin)
Plantains have Musa acuminata spp. banksii chloroplast and mitochondrial cytotypes (Carreel et al. 2002). The mosaic genome structure observed here showed that Plantain formation must have involved at least one A/B interspecific hybrid meiosis. The probable scenario is thus an original cross between M. acuminata spp. banksii and M. balbisiana that produced a primary A/B hybrid. This hybrid was subsequently fertilized by M. acuminata spp. banksii, which explains why the two Plantain A genomes were found to be highly similar in our analysis (data not shown) and previous reports (Perrier et al. 2011; Hippolyte et al. 2012).
The mosaic genome structure of Pelipita is compatible with an origin involving a primary A/B hybrid crossed with M. balbisiana. Pelipita chloroplast and mitochondrial cytotypes originate from Musa acuminata spp. banksi and Musa balbisiana, respectively (Carreel et al. 2002; Boonruangrod et al. 2008). A scenario with a female A/B hybrid fertilized by M. balbisiana pollen is plausible. In addition, our data suggest that Plantain and Pelipita share an interspecific recombination breakpoint on chromosome 9 that could reflect a common ancestor (fig. 6). For both Plantains and Pelipita, a question remains on whether the primary A/B hybrid was a diploid that produced an unreduced gamete or a triploid that produced an AB gamete. The second option was proposed by De Langhe et al. (2010) based on the apparent absence of edible AB hybrids in their area of origin.
Fig. 6.
Focus on A/B mosaic structure of chromosome 9 in the studied cultivated banana accessions. Coverage ratios for alleles specific to the A genome (green dots) and to the B genome (red dots) are plotted along the 11 pseudochromosomes of the Musa acuminata reference genome. Vertical dash lines indicate interspecific recombination breakpoints. The scale is indicated (in Mb) at the bottom of each figure.
Silk and Mysore Subgroup (Indian Origin)
The interspecific mosaic genome structure showed, as for Plantain, that the origin of Figue Pomme Géante/Figue Pomme Naine (Silk) and Pisang Ceylan (Mysore) must have involved at least one A/B interspecific hybrid meiosis. The fact that more interspecific recombination breakpoints were observed in Pisang Ceylan may imply a higher recombination rate in the original A/B hybrid or, more likely, additional meiosis steps (i.e., crosses) were involved in its origin. The same question remains as to whether the primary A/B hybrid was a diploid that produced an unreduced gamete or a triploid that produced an AB gamete. However, despite their low fertility because diploid AB hybrids exist in India, the region of origin of the Silk and Mysore subgroups, the diploid AB hybrid option is more likely in this case (De Langhe et al. 2010). It was found, based on an RFLP marker study, that Silk accessions Figue Pomme and Figue Pomme Naine had M. acuminata chloroplast and mitochondrial cytotypes from the same cytotype group as the Safet Velchi and Kunnan diploids (Carreel et al. 2002). Boonruangrod et al. (2008) found identical PCR–RFLP-based cytotypes for Figue Pomme Géante (ITC0769) and Kunnan (ITC1034). A phylogenetic proximity between Silk accessions and the diploid Kunnan group was also noted using SSR markers (Christelová et al. 2017). In addition, our data showed that Figue Pomme, Pisang Ceylan, and Kunnan shared an interspecific recombination breakpoint on chromosome 9, and the last two showed a second common interspecific recombination breakpoint also on chromosome 9, suggesting that these accessions might share a common ancestor (fig. 6).
Note that the interspecific cultivars we analyzed, except the diploid Safet Velchi, shared some common recombined interspecific breakpoints on chromosome 9. It would be worth assessing the possible importance of these regions of chromosome 9 in the evolution/domestication of interspecific cultivars. Interestingly, all five triploid cultivars we studied had a euploid chromosome constitution (2n = 3x = 33). If the primary interspecific hybrids leading to these cultivars were polyploids, we could have expected some aneuploidy considering the substantial aneuploid rate observed in the CRBP39 progeny and in other plantain-derived progenies (Vuylsteke et al. 1993; Osuji et al. 1997). Furthermore, in “AAB”×AA crosses, Vuylsteke et al. (1993) observed very few individuals resulting from “AAB” 2x gametes. These observations suggest the involvement of diploid primary interspecific hybrids that would have produced euploid 2n gametes.
Conclusion
We developed a method that exploits sequencing data to help precisely unravel the interspecific mosaic genome structure of banana accessions. This new access to the mosaic structure of banana cultivar genomes should considerably facilitate the interpretation of phylogenetic, genetic, and functional studies and will help in refining banana subgroup composition and origin. Accordingly, we have presented data that supports the implication of BB genotypes already introgressed through several generations with A genome segments in the origin of some interspecific cultivars. Compared with most other crops for which seed production is crucial, the banana domestication process includes sterility as one major criterion for cultivar selection. Current hypothesis was that partially fertile edible cultivars were selected from the initial interspecific or intersubspecific primary hybrid pool. These vegetatively propagated hybrids then contributed to some rare breeding events with wild relatives that are considered to have evolved independently because of their fertility. However, the observed mosaic genome of diploid AB cultivars suggests that other scenarios with more hybridization steps between wild and selected compartments may have produced introgressed populations that contributed to current cultivars. Scenarios with a complex admixture history have been proposed during the domestication of annual crops such as maize (Hufford et al. 2013) and rice (Wang et al. 2017), but also for clonally propagated crops such as apple (Cornille et al. 2012), grape (Myles et al. 2011), and Citrus (Wu GA et al. 2018) in which traces of widespread ancient introgression have been found.
We showed that M. balbisiana and M. acuminata chromosomes often recombine in interspecific hybrids. This should facilitate recombination of traits between these two species in breeding programs, while also offering the possibility to eliminate endogenous viruses such as the banana streak virus integrated in B genomes (Chabannes et al. 2013; Noumbissié et al. 2016). However, we also showed that the presence of two large chromosomal structural variations between M. balbisiana and M. acuminata chromosomes generated a locally high level of segregation distortion and aneuploidy, which complicates genetic analysis and breeding. The new possibility described here to precisely characterize progenies and their parents regarding their genome structure (mosaic, structural heterozygote, and aneuploid vs. euploid) will enhance studies on much more efficient banana breeding strategies.
More generally, our results contribute to a better understanding of the complex phase following allopolyploidization, which varies among systems but is generally subjected to disturbed meiosis (Pelé et al. 2018). We showed that homoelogous recombinations are frequent in Musa but locally affected by the large chromosomal variations (LSV) that differentiate the parental species. In chromosomal regions involved in these LSVs, a high impact on meiosis was observed with a drastic reduction in viable recombination products and a high proportion of aneuploids.
Materials and Methods
Plant Material
Interspecific cultivars (Kunnan, AB, PT-BA-00163; Safet Velchi, AB, PT-BA-00359; Figue Pomme Naine, AAB, PT-BA-00087; Figue Pomme Géante, AAB, PT-BA-00086; Pisang Ceylan, AAB, PT-BA-00286; Red Yade, AAB, PT-BA-00355; Pelipita, ABB, PT-BA-00275) as well as representatives of M. acuminata (Niyarma Yik PT-BA-00242; Banksii PT-BA-00024; Waigu PT-BA-00412; Hawain2 PT-BA-00113; Tomolo ITC1187; Long Tavoy PT-BA-00178; and Selangor PT-BA-00363) and M. balbisiana (Pisang Klutuk Wulung [PKW] PT-BA-00302, Cameroun PT-BA-00018, Honduras PT-BA-00019, and Lal Velchi PT-BA-00172) were obtained from the CRB-Plantes Tropicales Antilles CIRAD-INRA collection. A self-progeny of 105 individuals from the M. balbisiana accession “Pisang Klutuk Wulung” was produced through embryo rescue at the CIRAD research station in Guadeloupe. A population of 185 hybrids between the CRBP39 female parent and the wild AA diploid Pahang (M. acuminata spp. malaccensis, CARBAP collection number NY09) male parent was produced through embryo rescue at the Centre Africain de Recherche sur Bananiers et Plantains (CARBAP) in Cameroon (Noumbissié et al. 2016). CRBP39 is a tetraploid breeding accession resulting from crosses between the Plantain cultivar French Clair (AAB genome) and the M53 accession (AA genome). Leaves were used for DNA extraction.
Genotyping, SNP Calling, and Data Filtering
Sequencing and Read Processing
Self-progeny of M. balbisiana “PKW” was genotyped using the RadSeq technology (Peterson et al. 2012). Library construction, sequencing, genotype calling, and data filtering were performed at BGI Tech Solutions (Hong Kong). About 62 Gb of clean sequence from the M. balbisiana population produced a total of 19,585 SNPs that were heterozygous in the PKW parent with a minimum number of alternative allele reads >3.
For the CRBP39×Pahang progeny, Musa acuminata diversity, Musa balbisiana diversity, and other Musa interspecific cultivars, sequencing libraries were constructed using either genotyping-by-sequencing or whole genome shotgun (WGS) approaches. Sequencing was performed at BGI Tech Solutions (Hong Kong), GeT-PlaGe (Toulouse, France), or Genoscope (Evry, France) facilities (supplementary table S1, Supplementary Material online).
For all accessions except the PKW self-population, reads were processed as follows: 1) reads were aligned to the Musa reference A genome version 2 (Martin et al. 2016) using BWA v0.7.15 with the mem algorithm (Li 2013). Reads aligning at several positions were removed using samtools v1.3 (Li et al. 2009). 2) For WGS libraries, redundant reads were removed using MarkDuplicate from Picard Tools v2.7.0. 3) Reads were locally realigned around indels using the IndelRealigner tool of GATK v3.3 package (McKenna et al. 2010). 4) For each accession, at each covered position, all mapping bases with a mapping quality ≥10 were counted with the bam-readcount program (https://github.com/genome/bam-readcount). The complete process described here was performed using the process_reseq_1.0.py custom python script available at (https://github.com/SouthGreenPlatform/vcfHunter).
Variant Calling
Variant sites were then filtered according to the following criteria: 1) only data with a coverage between 10 and 100,000 reads were considered; 2) for each accession, alleles supported by at least three reads and with a frequency ≥0.05 were kept as variant; 3) sites with at least one variant were kept for variant calling; and 4) for each selected site and each accession, a genotype was called based on the maximum likelihood of all possible genotypes calculated based on a binomial distribution assuming a sequencing error rate of 0.005. The resulting variant calling file was generated in VCF format. The complete process described here was performed using the VcfPreFilter.1.0 custom python script available at (https://github.com/SouthGreenPlatform/vcfHunter).
SNP Filtering
The VCF file was sequentially filtered according to the following parameters: 1) indel variant sites were removed; 2) genotype calls based on <10-fold coverage and <3-fold coverage for the minor allele were converted to missing data; 3) variant sites with >5% missing genotype calls were filtered out; 4) only biallelic sites were kept; and 5) SNP clusters identified using GATK v3.3 and defined as at least two SNPs in a 2-base window were removed. A total of 148,329 SNPs were obtained in the final VCF data set. Data filtering was performed using the vcfFilter.1.0.py custom python script available at (https://github.com/SouthGreenPlatform/vcfHunter).
Genome Ratio Analysis and Coverage Analysis
The complete filtered VCF file was used to assign the allele origin to M. acuminata or M. balbisiana, to plot the A/B genome allele coverage ratio along chromosomes and to calculate normalized site coverage along chromosomes. Two sets of three M. balbisiana accessions and nine M. acuminata accessions were used to assign SNPs to A and B groups, respectively. For each allele, at each position, alleles were assigned to a species if present in all representatives of the species and absent from the other species. Both homozygous and heterozygous positions were considered. If ambiguities remained (i.e., an allele is present with different frequencies in both A and B species standards), the SNP allele was discarded and not taken into account for further analysis. For each allele assigned to a species, the allele ratio was calculated as the number of sequencing reads supporting the allele divided by the total number of reads covering the position for the concerned accession. The allele ratio was then plotted along Musa reference genome V2 chromosomes (Martin et al. 2016). Ratios of 50%: 50% of A versus B specific SNPs revealed a balanced AB or AABB chromosome constitution. A ratio of 66% A:33% B or 33% A:66% B revealed an AAB or an ABB chromosome constitution, respectively. A ratio of 75% A: 25% B revealed an AAAB chromosome constitution. A ratio of 0% A:100% B and 0% B:100% A revealed no chromosome A and no chromosome B, respectively. To calculate the normalized site coverage along chromosomes, each SNP position coverage (i.e., number of reads recorded at this position) was divided by the mean coverage of the accession (i.e., mean number of reads of all positions of the accession) and this value was plotted along chromosomes. The SNP distribution along chromosomes was verified: SNP assigned to either A or B genomes were well spread along chromosomes with a decrease in SNP frequency for pericentromeric regions (supplementary fig. S6, Supplementary Material online). Plots for A/B ratio and sequencing coverage were generated and analyzed for all accessions including the CRBP39 lineage (the triploid AAB French Clair, tetraploid AAAB CRBP39, and the entire triploid progeny). For the AA and BB accessions used to assign specific SNPs, SNPs were well spread along the chromosomes and we did not observe segments with no specific SNPs that could have indicated gene flows between the two species (supplementary fig. S7, Supplementary Material online). A/B genome ratios plotted along chromosomes which identify the A/B mosaic landscape were visually scanned to detect breakpoints in the A and B ratio distribution. Breakpoints at the boundaries of T1 and T3 fragments corresponding to reciprocal translocation were not recorded as recombination points. A hotspot of recombination breakpoints located at the beginning of chromosome 3 probably resulting from an A/B recombination point already present in the CRBP39 genome was not recorded (see Results section). The approximate coordinates of the breakpoints were graphically recorded.
The complete process described here was performed using vcf2allPropAndCov.py and vcf2allPropAndCovByChr.py custom python scripts available at (https://github.com/SouthGreenPlatform/vcfHunter).
Genetic Mapping
Musa balbisiana Genetic Map Construction
After the filtering steps (no missing data in the progeny, at least two genotypes in the progeny with at least five individuals in the minor class), a total of 10,074 SNPs were retained to build a Musa balbisiana genetic map. The genetic map was constructed using the ML mapping algorithm in JoinMap 4.1 (Stam 1993; Van Ooijen 2006) with standard parameters. To compare the A/B genomes of Musa, markers from the M. balbisiana genetic map were aligned along the M. acuminata reference sequence V2 (Martin et al. 2016) using BWA (Li 2013). Marker locations in the M. acuminata reference sequence were compared with their locations in the M. balbisiana genetic map using SpiderMap software (JF Rami, unpublished).
CRBP39 Genome B Marker Linkage Analysis
Markers in the CRBP39×Pahang triploid progenies were selected based on their segregation ratio in the progeny: 1:1 ratio for simple dose markers and 1:5 ratio for double dose markers. Marker deviation from the expected ratio was accepted based on a χ2 test with a minimal P value of 1e-10 and 1e-7 for simple and double doses, respectively. From these markers, a total of 20,824 alleles identified by the genome ratio analysis and assigned to the B genome were selected. In addition, 213 markers were removed from the data set because their segregation did not match that of the surrounding markers. A subset was generated corresponding to the selection of one marker every six markers ordered along the Musa acuminata reference sequence V2. Markers of the subset were used to calculate simple matching pairwise dissimilarity (i.e., a measure of the observed recombination frequency between pairs of markers) and marker segregation. These statistics were used to generate a dotplot showing the marker pairwise distance along chromosomes associated with a linear plot showing marker segregation distortion. The process described here was performed using vcf2popNew.1.0.py, RecombCalculatorDDose.py, and Draw_dot_plot.py custom python scripts available at (https://github.com/SouthGreenPlatform/vcfHunter).
Availability of Supporting Data
VCF files and mapping matrix are available in the download section of the Banana Genome Hub (http://banana-genome-hub.southgreen.fr).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank CRB Plantes Tropicales Antilles CIRAD-INRA Guadeloupe for providing the plant material. This work was supported by the Centre de coopération Internationale en Recherche Agronomique pour le Développement (CIRAD). The authors are grateful for financial support from the CGIAR Research Program on Roots, Tubers, and Bananas (RTB), the French Government “Investissement d’Avenir” FRANCE GENOMIQUE (ANR-10-INBS-09) and the Agropolis Fondation “GenomeHarvest project” (ID 1504-006) through the “Investissements d’avenir” programme (Labex Agro: ANR-10-LABX-0001-01). This work was supported by the CIRAD—UMR AGAP HPC Data Center of the South Green Bioinformatics platform (http://www.southgreen.fr/).
Author Contributions
F.C.B. and A.D.H. jointly designed the study. F.S., S.R., D.Y., and R.H. designed and performed the genetic crosses, embryo rescue, and handled the progenies. C.H. performed progeny DNA extractions. A.L. and M.R. produced or made available the sequencing data. G.M. and F.C.B. designed and performed the bioinformatic analysis. F.C.B., N.Y., A.D.H., and G.M. interpreted the data. F.C.B. and A.D.H. wrote the article, while N.Y. and G.M. edited the article.
References
- Boonruangrod R, Desai D, Fluch S, Berenyi M, Burg K.. 2008. Identification of cytoplasmic ancestor gene-pools of Musa acuminata Colla and Musa balbisiana Colla and their hybrids by chloroplast and mitochondrial haplotyping. Theor Appl Genet. 118(1):43–55. [DOI] [PubMed] [Google Scholar]
- Carreel F, de Leon DG, Lagoda P, Lanaud C, Jenny C, Horry JP, du Montcel HT.. 2002. Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome 45(4):679–692. [DOI] [PubMed] [Google Scholar]
- Carreel F, Fauré S, De Leon DG, Lagoda PJL, Perrier X, Bakry F, Du Montcel HT, Lanaud C, Horry J-P.. 1994. Evaluation de la diversité génétique chez les bananiers diploïdes (Musa sp.). Genet Sel Evol. 26(Suppl):125s–136s. [Google Scholar]
- Chabannes M, Baurens F-C, Duroy P-O, Bocs S, Vernerey M-S, Rodier-Goud M, Barbe V, Gayral P, Iskra-Caruana M-L.. 2013. Three infectious viral species lying in wait in the banana genome. J Virol. 87(15):8624–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman EE. 1947. Classification of the bananas. I. The genus Ensete Horan and the genus Musa L. Kew Bulletin 2(2):97–117. [Google Scholar]
- Chen H, Khan MKR, Zhou Z, Wang X, Cai X, Ilyas MK, Wang C, Wang Y, Li Y, Liu F.. 2015. A high-density SSR genetic map constructed from a F2 population of Gossypium hirsutum and Gossypium darwinii. Gene 574(2):273–286. [DOI] [PubMed] [Google Scholar]
- Chester M, Gallagher JP, Symonds VV, Silva AVC, da Mavrodiev EV, Leitch AR, Soltis PS, Soltis DE.. 2012. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc Natl Acad Sci U S A. 109(4): 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christelová P, Langhe ED, Hřibová E, Čížková J, Sardos J, Hušáková M, Houwe IV, den Sutanto A, Kepler AK, Swennen R.. 2017. Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers Conserv. 26(4):801–824. [Google Scholar]
- Cornille A, Gladieux P, Smulders MJM, Roldán-Ruiz I, Laurens F, Cam BL, Nersesyan A, Clavel J, Olonova M, Feugey L.. 2012. New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genet. 8(5):e1002703.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Langhe E, Hribová E, Carpentier S, Dolezel J, Swennen R.. 2010. Did backcrossing contribute to the origin of hybrid edible bananas? Ann Bot. 106(6): 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hont A, Denoeud F, Aury J-M, Baurens F-C, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, et al. 2012. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488(7410):213–217. [DOI] [PubMed] [Google Scholar]
- D’Hont A, Paget-Goy A, Escoute J, Carreel F.. 2000. The interspecific genome structure of cultivated banana, Musa spp. revealed by genomic DNA in situ hybridization. Theor Appl Genet. 100(2):177–183. [Google Scholar]
- Dodds KS, Simmonds NW.. 1948. Sterility and parthenocarpy in diploid hybrids of Musa. Heredity 2(Pt 1):101–117. [DOI] [PubMed] [Google Scholar]
- Fauré S, Noyer J-L, Carreel F, Horry J-P, Bakry F, Lanaud C.. 1994. Maternal inheritance of chloroplast genome and paternal inheritance of mitochondrial genome in bananas (Musa acuminata). Curr Genet. 25(3): 265–269. [DOI] [PubMed] [Google Scholar]
- Fauré S, Noyer J-L, Horry J-P, Bakry F, Lanaud C, de León DG.. 1993. A molecular marker-based linkage map of diploid bananas (Musa acuminata). Theor Appl Genet. 87(4): 517–526. [DOI] [PubMed] [Google Scholar]
- Freeling M. 2017. Picking up the ball at the K/Pg boundary: the distribution of ancient polyploidies in the plant phylogenetic tree as a spandrel of asexuality with occasional sex. Plant Cell 29(2):202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayral P, Blondin L, Guidolin O, Carreel F, Hippolyte I, Perrier X, Iskra-Caruana M-L.. 2010. Evolution of endogenous sequences of banana streak virus: what can we learn from banana (Musa sp.) evolution? J Virol. 84(14):7346–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippolyte I, Bakry F, Seguin M, Gardes L, Rivallan R, Risterucci A-M, Jenny C, Perrier X, Carreel F, Argout X, et al. 2010. A saturated SSR/DArT linkage map of Musa acuminata addressing genome rearrangements among bananas. BMC Plant Biol. 10:65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippolyte I, Jenny C, Gardes L, Bakry F, Rivallan R, Pomies V, Cubry P, Tomekpe K, Risterucci AM, Roux N.. 2012. Foundation characteristics of edible Musa triploids revealed from allelic distribution of SSR markers. Ann Bot. 109(5):937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Sgro C, Weeks A.. 2004. Chromosomal inversion polymorphisms and adaptation. Trends Ecol Evol. 19(9):482–488. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH.. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 39:21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojsgaard D. 2018. Transient activation of apomixis in sexual neotriploids may retain genomically altered states and enhance polyploid establishment. Front Plant Sci. 9:230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford MB, Lubinksy P, Pyhäjärvi T, Devengenzo MT, Ellstrand NC, Ross-Ibarra J.. 2013. The genomic signature of crop-wild introgression in maize. PLoS Genet. 9(5):e1003477.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens SB, Vandelook F, De Langhe E, Verstraete B, Smets E, Vandenhouwe I, Swennen R.. 2016. Evolutionary dynamics and biogeography of Musaceae reveal a correlation between the diversification of the banana family and the geological and climatic history of Southeast Asia. New Phytol. 210(4):1453–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeridi M, Bakry F, Escoute J, Fondi E, Carreel F, Ferchichi A, D’Hont A, Rodier-Goud M.. 2011. Homoeologous chromosome pairing between the A and B genomes of Musa spp. revealed by genomic in situ hybridization. Ann Bot. 108(5):975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeridi M, Perrier X, Rodier-Goud M, Ferchichi A, D’Hont A, Bakry F.. 2012. Cytogenetic evidence of mixed disomic and polysomic inheritance in an allotetraploid (AABB) Musa genotype. Ann Bot. 110(8):1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Piffanelli P, Ciampi AY, Ruiz M, Blanc G, Leebens-Mack J, da Silva FR, Santos CMR, D’Hont A, Garsmeur O, et al. 2008. Insights into the Musa genome: syntenic relationships to rice and between Musa species. BMC Genomics 9:58.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Baurens F-C, Droc G, Rouard M, Cenci A, Kilian A, Hastie A, Doležel J, Aury J-M, Alberti A, et al. 2016. Improvement of the banana “Musa acuminata” reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genomics 17:243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Carreel F, Coriton O, Hervouet C, Cardi C, Derouault P, Roques D, Salmon F, Rouard M, Sardos J, et al. 2017. Evolution of the banana genome (Musa acuminata) is impacted by large chromosomal translocations. Mol Biol Evol. 34(9): 2140–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita SC, Tyagi AP, Thornton GM, Pires JC, Madlung A.. 2012. Allopolyploidization lays the foundation for evolution of distinct populations: evidence from analysis of synthetic Arabidopsis allohexaploids. Genetics 191(2):535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Boyko AR, Owens CL, Brown PJ, Grassi F, Aradhya MK, Prins B, Reynolds A, Chia J-M, Ware D, et al. 2011. Genetic structure and domestication history of the grape. Proc Natl Acad Sci U S A. 108(9):3530–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noumbissié GB, Chabannes M, Bakry F, Ricci S, Cardi C, Njembele J-C, Yohoume D, Tomekpe K, Iskra-Caruana M-L, D’Hont A, et al. 2016. Chromosome segregation in an allotetraploid banana hybrid (AAAB) suggests a translocation between the A and B genomes and results in eBSV-free offsprings. Mol Breed. 36(4):38–52. [Google Scholar]
- Noyer J-L, Causse S, Tomekpe K, Bouet A, Baurens F-C.. 2005. A new image of plantain diversity assessed by SSR, AFLP and MSAP markers. Genetica 124(1):61–69. [DOI] [PubMed] [Google Scholar]
- Osuji J, Vuylsteke D, Ortiz R.. 1997. Ploidy variation in hybrids from interploid 3x X 2x crosses in Musa. Tropicultura 15(1):37–39. [Google Scholar]
- Pelé A, Rousseau-Gueutin M, Chèvre A-M.. 2018. Speciation success of polyploid plants closely relates to the regulation of meiotic recombination. Front Plant Sci. 9:907 doi: 10.3389/fpls.2018.00907. Available from: https://www.frontiersin.org/articles/10.3389/fpls.2018.00907/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. 2013. More genomes from denisova cave show mixing of early human groups. Science 340(6134):799.. [DOI] [PubMed] [Google Scholar]
- Perrier X, De Langhe E, Donohue M, Lentfer C, Vrydaghs L, Bakry F, Carreel F, Hippolyte I, Horry J-P, Jenny C, et al. 2011. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc Natl Acad Sci U S A. 108(28):11311–11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE.. 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7(5):e37135.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach AT, Revay T, Villagomez DAF, Macedo MP, Sullivan A, Maignel L, Wyss S, Sullivan B, King WA.. 2016. Prevalence and consequences of chromosomal abnormalities in Canadian commercial swine herds. Genet Sel Evol. 48:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol. 16:8. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Linder CR, Seiler GJ.. 1995. Chromosomal and genic barriers to introgression in Helianthus. Genetics 141(3):1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samans B, Chalhoub B, Snowdon RJ.. 2017. Surviving a genome collision: genomic signatures of allopolyploidization in the recent crop species Brassica napus Plant Genome 10. doi:10.3835/plantgenome2017.02.0013. [DOI] [PubMed]
- Sardos J, Perrier X, Doležel J, Hřibová E, Christelová P, Van den houwe I, Kilian A, Roux N.. 2016. DArT whole genome profiling provides insights on the evolution and taxonomy of edible Banana (Musa spp.). Ann Bot. 118(7):1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd K. 1999. Cytogenetics of the genus Musa. International Network for the Improvement of Banana and Plantain, Montpellier, France.
- Simmonds NW, Shepherd K.. 1955. The taxonomy and origins of the cultivated bananas. Bot J Linn Soc. 55(359):302–312. [Google Scholar]
- Stam P. 1993. Construction of integrated genetic linkage maps by means of a new computer package: join Map. Plant J. 3(5):739–744. [Google Scholar]
- Stathos A, Fishman L.. 2014. Chromosomal rearrangements directly cause underdominant F1 pollen sterility in Mimulus lewisii–Mimulus cardinalis hybrids. Evolution 68(11):3109–3119. [DOI] [PubMed] [Google Scholar]
- Stover R, Simmonds N.. 1987. Bananas. 3rd ed London: Longrnans; p. 27–45. [Google Scholar]
- Swangpol S, Volkaert H, Sotto R, Seelanan T.. 2007. Utility of selected non-coding chloroplast DNA sequences for lineage assessment of Musa interspecific hybrids. Jounal of Biochemistry and Molecular Biology 40(4):577–587. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K.. 2017. The evolutionary significance of polyploidy. Nat Rev Genet. 18(7):411.. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW. 2006. JoinMap® 4. Software for the calculation of genetic linkage maps in experimental populations. Popul. Kyazma BV, Wagening. Neth. 33(10.1371). [Google Scholar]
- Vuylsteke DR, Swennen RL, Ortiz R.. 1993. Development and performance of balck sigatoka-resistant tetraploid hybrids of plantain Musa spp., AAB group). Euphytica 65(1):33–42. [Google Scholar]
- Wang H, Vieira FG, Crawford JE, Chu C, Nielsen R.. 2017. Asian wild rice is a hybrid swarm with extensive gene flow and feralization from domesticated rice. Genome Res. 27(6):1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Lisch D, Hu G, Mason AS.. 2018. The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr Opin Genet Dev. 49:1–7.29438956 [Google Scholar]
- Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, Perrier X, Ruiz M, Scalabrin S, Terol J, et al. 2014. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol. 32(7):656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GA, Terol J, Ibanez V, López-García A, Pérez-Román E, Borredá C, Domingo C, Tadeo FR, Carbonell-Caballero J, Alonso R, et al. 2018. Genomics of the origin and evolution of Citrus. Nature 554(7692):311–316. [DOI] [PubMed] [Google Scholar]
- Wu Y, Sun Y, Sun S, Li G, Wang J, Wang B, Lin X, Huang M, Gong Z, Sanguinet KA, et al. 2018. Aneuploidization under segmental allotetraploidy in rice and its phenotypic manifestation. Theor Appl Genet. 131(6):1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bian Y, Gou X, Zhu B, Xu C, Qi B, Li N, Rustgi S, Zhou H, Han F, et al. 2013. Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proc Natl Acad Sci U S A. 110(9): 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
VCF files and mapping matrix are available in the download section of the Banana Genome Hub (http://banana-genome-hub.southgreen.fr).