Abstract
Gene body methylation (gbM) is typically characterized by DNA methylation in the CG context within coding regions and is associated with constitutive genes that have moderate to high expression levels. A recent study discovered the loss of gbM in two plant species (Eutrema salsugineum and Conringia planisiliqua), illustrating that gbM is not necessary for survival and reproduction. The same paper stated there was no detectable effect of gbM loss on gene expression (GE). Here, we reanalyzed the GE data and accounted for experimental variability in expression level estimates. We show that the loss of gbM in E. salsugineum is associated with a small but highly significant decrease in GE relative to the closely related species Arabidospis thaliana. Our results are consistent with various evolutionary analyses that suggest gbM has a function, perhaps as a homeostatic effect on GE.
Keywords: gene body methylation, Brassicaceae, gene expression
Since its first genomic characterization in Arabidopsis thaliana (Cokus et al. 2008; Lister et al. 2008), gene body methylation (hereafter gbM) has been a puzzling phenomenon. Typically, plants methylate cytosines in three contexts: CG, CHG, and CHH (where H = A, C, or T). When all three contexts are methylated, gene expression (hereafter GE) is silenced. In contrast, A. thaliana gbM is characterized by elevated methylation levels in the CG context, but neither in the CHG nor in the CHH context of coding regions. This high CG methylation does not silence genes. In fact, gbM genes tend to be intermediately expressed (Zhang et al. 2006; Lister et al. 2008) and expressed across more tissues than Łunmethylated (UM) genes (Lister et al. 2008; Kawakatsu et al. 2016). Importantly, not all genes have gbM; only 20% of A. thaliana genes contain CG methylation above background levels (Takuno and Gaut 2012).
The puzzling aspect about gbM is its function. Some argue that gbM must have some function (Zilberman 2017), based on four observations rooted in evolutionary analyses. First, gbM levels are highly conserved between orthologs (Takuno and Gaut 2013; Seymour et al. 2014), even across ∼300 My of land plant evolution (Takuno et al. 2016). Second, gbM genes tend to evolve more slowly than UM genes (Takuno and Gaut 2012, 2013) with lower levels of polymorphism (Takuno et al. 2017), consistent with their enrichment for important functions (Zhang et al. 2006; Takuno and Gaut 2012). Third, most gbM genes are not particularly GC rich, suggesting that elevated CG methylation levels are not a simple consequence of cytosine availability (Takuno and Gaut 2013). In fact, gbM is maintained against mutational biases that decrease GC content over time (Takuno and Gaut 2013). Finally, gbM status seems to have a modest effect on GE, based on an evolutionary comparison between closely related Arabidopsis species (Takuno et al. 2017). This comparison focused on a small subset of orthologs that did not have conserved gbM levels between species. It revealed that gbM genes tend to be more highly expressed than their UM orthologous counterpart, but the overall trend was not convincingly significant. In another study comparing A. thaliana Swedish accessions, gbM genes were found to be more heavily methylated in northern latitudes, which was associated with a higher expression level (Dubin et al. 2015).
Other studies have suggested that gbM has no function and is instead a by-product of transposition and/or methylation pathways (Roudier et al. 2009; Teixeira and Colot 2009; Kawakatsu et al. 2016; Bewick and Schmitz 2017). This argument is consistent with observations that: 1) the loss of gbM in an A. thaliana mutant does not substantially alter GE (Roudier et al. 2009; Bewick et al. 2016) and 2) gbM variation across A. thaliana accessions does not strongly affect GE (Kawakatsu et al. 2016). Two recent papers have seemingly strengthened the argument against gbM functionality, because they found that two flowering plants (Eutrema salsugineum and Conringia planisiliqua) have no gbM throughout their genome. The identification of these species is important for two reasons. First, they have provided important insights into the mechanism that produces gbM, which had been mysterious (Bewick et al. 2017). Second, they provide prima facie evidence against gbM function, because these two plant species seem to exist happily without it. In support of this argument, Bewick et al. (2016) compared GE between genes that have gbM in A. thaliana and their UMŁ orthologs in E. salsugineum. They concluded that the two sets of genes had similar transcription levels, again suggesting that gbM has no function.
Here, we use the data of Bewick et al. (2016) to revisit GE analyses and to test whether an effect of gbM can be detected. We began by defining gbM genes and UM genes in A. thaliana (see Materials and Methods) using the same statistical approach and thresholds as Bewick et al. (2016). We then identified 12,189 orthologous UM genes in E. salsugineum, based on best-hits between species, again using the same methods as Bewick et al (2016). These ortholog pairs were then separated into two groups (table 1) for inclusion in GE analyses. Group 1 consists of 4,221 ortholog pairs that changed gbM status between species—that is, the gene was gbM in A. thaliana and UM in E. salsugineum. Group 2 includes 7,968 ortholog pairs that did not change gbM status, because they were UM in both species. These Groups 2 genes can be viewed as a “control” set and are necessary for comparative analyses.
Table 1.
Definition of the Two Gene Groups.
| Ortholog Pairs | Methylation in A. thaliana | Methylation in E. salsugineum | Change in Methylation Status | Number of Orthologous Genes |
|---|---|---|---|---|
| Group 1 | gbM | UM | Yes | 4,221 |
| Group 2 | UM | UM | No | 7,968 |
Note.—The first group consists of genes that have changed methylation status between A. thaliana and E. salsugineum. The second group has conserved methylation status as UM genes.
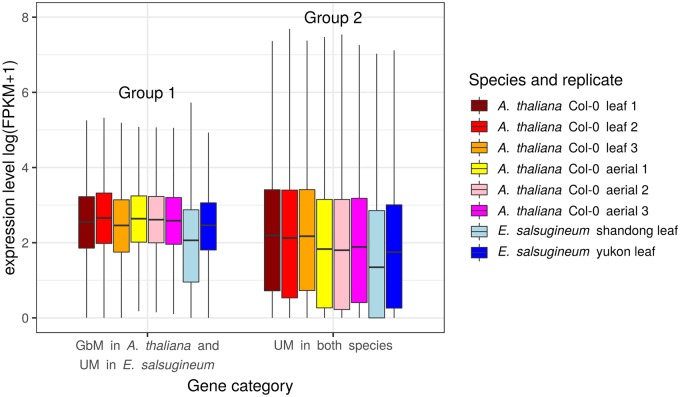
Given these two gene groups, we gathered GE data from Bewick et al. (2016) and contrasted expression levels between A. thaliana and E. salsugineum for Group 1 orthologs (fig. 1 ). These orthologs differed significantly in GE, with lower expression in E. salsugineum (one-sided Wilcoxon test W = 95218000, P-value < 2.2e–16). However, we also found significantly lower expression between the full data sets of 26,248 E. salsugineum genes compared with the 27,066 A. thaliana genes (Wilcoxon one-sided W = 3913700000, P-value < 2.2e–16). We concluded that these results are likely due to inherent experimental biases that cause global differences in expression levels between the two species. This global difference in GE between the two species may be one reason why Bewick et al. (2016) did not find a link between gbM and GE.
Fig 1.
Expression levels (log scale) for Arabidospis thaliana and Eutrema salsugineum in two gene Groups defined in table 1. The boxplot shows the median, the hinges are the first and third quartiles (the 25th and 75th percentiles) and the whiskers extend from the hinge to the largest or smallest value no further than 1.5 times the interquartile range (distance between the first and third quartiles).
To account for the global species difference in expression, which is (again) likely due to experimental biases since all genes were affected, we applied a linear model with mixed effects to the expression data (see Materials and Methods). In addition to accounting for GE differences between species, the model can address a specific hypothesis: if gbM modulates GE, we expect the 4,221 Group 1 genes to exhibit more substantial differences in GE between species than the 7,968 Group 2 genes (table 1). After applying the model, we found that the average expression in A. thaliana for Group 2 genes is 2.08 FPKM on a log scale (the intercept in table 2). The same genes (Group 2) were expressed at significantly lower levels in E. salsugineum (0.32 FPKM less on a log scale, on average; the species effect in table 2). In A. thaliana, Group 1 genes were significantly more expressed than Group 2 genes (0.49 FPKM more on a log scale on average; the Group effect in table 2), consistent with the known high to intermediate expression level of gbM genes. Finally, after taking into account the species effect, Group 1 orthologs have significantly lower expression levels in E. salsugineum compared with A. thaliana (the interaction effect in table 2). Although the change in expression level is small (–0.14 FPKM in log scale), the fact that it is significant shows that this difference is consistent across genes in the data set. The results also hold after excluding 1,321 lowly expressed genes. Therefore, we conclude that the loss of gbM in E. salsugineum Group 1 genes is associated with a small but significant decrease in expression level relative to the same genes which are gbM in A. thaliana.
Table 2.
Results of the Linear Regression Model with Mixed Effects (see Materials and Methods for details).
| Estimate | 95% Confidence Interval | t-Value | P-value | |
|---|---|---|---|---|
| Average expression in A. thaliana for Group 2 genes (intercept) | 2.08 | 1.96–2.21 | ||
| Difference in expression between E. salsugineum and A. thaliana for Group 2 genes (species effect) | –0.32 | –0.33 to –0.31 | –55.373 | <0.001 |
| Difference in expression between Group 1 and Group 2 genes in A. thaliana (Group effect) | 0.49 | 0.44–0.54 | 18.67 | <0.001 |
| Additional difference in expression between E. salsugineum and A. thaliana for Group 1 genes (interaction effect) | –0.14 | –0.16 to –0.13 | –15.63 | <0.001 |
Note.—See table 1 for the definition of gene Groups and for gene numbers. For each fixed effect of the model and their interaction, the estimated average change in expression level is shown in log scale, along with the 95% confidence interval, t-value, and P-value. The first line (intercept) shows the average expression (FPKM in log scale) for one species and one gene Group; subsequent lines show differences in expression observed with that intercept and whether the differences are significant.
Our result is important for at least two reasons. First, by utilizing E. salsugineum, a species naturally devoid of gbM, we have studied the effect of gbM loss from 4,221 Group 1 genes simultaneously. A previous paper took a similar approach but could study only hundreds of genes (Takuno et al. 2017). It is worth noting, however, that both studies have detected a modest but consistent association between gbM and GE, with a trend toward higher expression levels for gbM genes. Second, by contrasting the GE effects between two groups—that is, genes that did and did not differ in gbM status (table 1)—we have effectively controlled for experimental effects. We suspect that our conclusions differ from Bewick et al. (2016) in part due to the use of Group 2 as a control to contrast to Group 1 genes.
We believe these results to bolster the argument, which is largely based on evolutionary analyses, that gbM has some function, specifically with regard to effects on GE. Although the loss of gbM in E. salsugineum is associated with only a small reduction in GE, the effect is significant and notable across 4,221 genes, even if the effect is small at the scale of individual genes. We note that natural selection can act on functional genomic features that have small effects on fitness, such as codon usage bias (Duret and Mouchiroud 1999), as long as the species effective population size (Ne) is large enough for selection to act efficaciously. One reason why E. salsugineum lost gbM, whereas most angiosperms have preserved it, could be the relatively small Ne of this species and its specialized halophytic niche (Wang et al. 2018).
The question remains, however, as to the exact nature and mechanism linking gbM and GE. Zilberman (2017) argues persuasively that the primary effect of gbM is likely to be homeostatic, by (for example) preventing aberrant transcription within genes or restricting access to histone H2A.7, which is associated with gene responsiveness. If the effect of gbM is to better regulate GE, it may have been missed up to now due to technical limitations. Indeed, current estimates of GE are averaged across thousands of cells, and because the effect of gbM is small, it may be necessary to consider single cell approaches to better understand it (Ji et al. 2015; Zilberman 2017). Whatever the function, the effect of gbM is modest enough to be difficult to detect on experimental time-scales, but strong enough to be conserved among plant orthologs.
Materials and Methods
Data Set
We used data from Bewick et al. (2016) and Niederhuth et al. (2016) from five Brassicaceae species: A. thaliana, Arabidopsis lyrata, Brassica rapa, Capsella rubella, and E. salsugineum (supplementary table S1, Supplementary Material online). The methylation data consisted of the methylation status of each cytosine based on a binomial test (Lister et al. 2008). The RNA-seq data were reported by Bewick et al (2016) as the FPKM level for each gene for six A. thaliana replicates (three leaf and three aerial tissue) and two E. salsugineum replicates from leaf tissue (supplementary table S1, Supplementary Material online).
gbM Inference
For each gene, the methylation state was inferred using the same approach as Bewick et al. (2016) and Niederhuth et al. (2016), for details see Supplementary Material.
Analysis of Expression and Methylation Levels
GE levels were analyzed using a linear regression model with mixed effects using the R package lme4 (Bates et al. 2015). FPKM expression levels were log transformed. To account for interGE variability, a random gene effect was included in the model (see eq. 1). Similarly, a random tissue effect was included (eq. 1) to account for variability in expression between leaf and aerial RNA-seq data. The aim of this model was to compare the expression level of gbM genes in A. thaliana to genes that lost gbM in E. salsugineum, while taking into account any global difference in expression levels between the two species. In order to achieve this, we defined a fixed effect called “gene Group” (table 1). A species fixed effect was also used in the model to account for global differences in expression levels between the two species. The resulting model was written to examine an effect of gene Group on expression level after taking the species effect into account—that is,
| (1) |
Significance for fixed effects and their interaction were determined by comparing the fit of the full model to nested models that first removed the interaction and then removed one effect at a time. P-values for each effect and their interaction were computed via Wald-statistics approximation using sjPlot R package (Lüdecke 2018).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the European Molecular Biology Organization Long Term Fellowship Award EMBO ALTF 775‐2017 and by NSF grant IOS-1542703 to B.S.G. We are thankful to Shohei Takuno and two anonymous reviewers for commenting and improving the manuscript.
References
- Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw [Internet]. Available from: https://www.jstatsoft.org/article/view/v067i01, last accessed August 8, 2018.
- Bewick AJ, Ji L, Niederhuth CE, Willing E-M, Hofmeister BT, Shi X, Wang L, Lu Z, Rohr NA, Hartwig B. 2016. On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci U S A. 113(32):9111–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Niederhuth CE, Ji L, Rohr NA, Griffin PT, Leebens-Mack J, Schmitz RJ. 2017. The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 18(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Schmitz RJ. 2017. Gene body DNA methylation in plants. Curr Opin Plant Biol. 36:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Natur. 452(7184):215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin MJ, Zhang P, Meng D, Remigereau M-S, Osborne EJ, Paolo Casale F, Drewe P, Kahles A, Jean G, Vilhjálmsson B et al. 2015. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. Elif. 4:e05255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci U S A. 96(8):4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Neumann DA, Schmitz RJ. 2015. Crop epigenomics: identifying, unlocking, and harnessing cryptic variation in crop genomes. Mol Plant. 8(6):860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Huang S-SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y et al. 2016. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cel. 166(2):492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cel. 133(3):523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüdecke D. 2018. sjPlot—data visualization for statistics in social science. Available from: https://zenodo.org/record/1310947.
- Niederhuth CE, Bewick AJ, Ji L, Alabady MS, Kim KD, Li Q, Rohr NA, Rambani A, Burke JM, Udall JA et al. 2016. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17(1):194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Teixeira FK, Colot V. 2009. Chromatin indexing in Arabidopsis: an epigenomic tale of tails and more. Trends Genet. 25(11):511–517. [DOI] [PubMed] [Google Scholar]
- Seymour DK, Koenig D, Hagmann J, Becker C, Weigel D. 2014. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 10(11):e1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Gaut BS. 2012. Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol. 29(1):219–227. [DOI] [PubMed] [Google Scholar]
- Takuno S, Gaut BS. 2013. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci U S A. 110(5):1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Ran J-H, Gaut BS. 2016. Evolutionary patterns of genic DNA methylation vary across land plants. Nat Plants. 2:15222. [DOI] [PubMed] [Google Scholar]
- Takuno S, Seymour DK, Gaut BS. 2017. The evolutionary dynamics of orthologs that shift in gene body methylation between Arabidopsis species. Mol Biol Evol. 34(6):1479–1491. [DOI] [PubMed] [Google Scholar]
- Teixeira FK, Colot V. 2009. Gene body DNA methylation in plants: a means to an end or an end to a means? EMBO J. 28(8):997–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-J, Hu Q-J, Guo X-Y, Wang K, Ru D-F, German DA, Weretilnyk EA, Abbott RJ, Lascoux M, Liu J-Q. 2018. Demographic expansion and genetic load of the halophyte model plant Eutrema salsugineum. Mol Ecol. 27(14):2943–2955. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW-L, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cel. 126(6):1189–1201. [DOI] [PubMed] [Google Scholar]
- Zilberman D. 2017. An evolutionary case for functional gene body methylation in plants and animals. Genome Biol. 18(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.