Abstract
Forkhead box protein 3 (Foxp3) is indispensable for the development of CD4+CD25+ regulatory T cells (Tregs). Here we analyzed three prominent evolutionary conserved regions (ECRs) upstream of the transcription start site of the human FOXP3 gene. We show that ECR2 and ECR3 fragments derived from positions −1.3 to −2.0 kb and −5.0 to −6.0 kb, respectively, display basal transcriptional activity. Reporter constructs derived from ECR1, located between −0.6 and +0.23 kb and thus the most proximal ECR in respect of transcription initiation, remained almost inactive. However, ECR1 was transactivated by the NF-κB subunit p65 in HEK 293 cells. In Jurkat and primary T cells, in addition to p65, a second stimulus delivered by either T-cell receptor stimulation or addition of PMA was needed. Co-expression of IκBα inhibited p65-mediated FOXP3 proximal promoter transactivation, and the NF-κB inhibitor curcumin reduced Foxp3 neoexpression in IL-2/CD3/CD28/TGF-β stimulated PBMCs. Moreover, proximal FOXP3 promoter transactivation was inhibited by Foxp3 and the SP transcription factor family member SP3. Thus, the human proximal FOXP3 promoter is controlled by activation through the TCR involving PKC and the NF-κB subunit p65 and by inhibition through a negative feedback loop and SP3.
Keywords: T cells, Transcription factors, Tolerance/suppression/anergy
1. Introduction
Among the lymphoid lineage, a small population of T-lymphocytes is described as regulatory T cells (Tregs) that maintain peripheral immune tolerance and thus play a major role in immune responses and prevention of autoimmunity (Shevach, 2004). Foxp3, a member of the forkhead/windged-helix family of transcriptional regulators is indispensable for the development as well as execution of the suppressive function of Tregs in mice and humans and remains the most distinct marker for this T-cell subset: mutations in the human FOXP3 gene cause X-linked human IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome) and XLAAD (X-linked autoimmunity-allergic dysregulation syndrome) (Bennett et al., 2001; Chatila et al., 2000; Wildin et al., 2002). A two base pair frame shift in the murine FOXP3 gene leads to a truncated Foxp3 protein (lack of the forkhead domain) and to the autoimmune scurfy phenotype in male mice (Brunkow et al., 2001). Further, ectopic expression of Foxp3 in non-Treg cells and even in Jurkat T cells induced a Treg phenotype (Hori et al., 2003; Kim et al., 2007).
The forkhead domain of Foxp3 was shown to be required for nuclear localization and DNA binding (Schubert et al., 2001). Various genes are regulated on the transcriptional level by Foxp3 (Marson et al., 2007; Zheng et al., 2007). For instance, IL-2, IL-4 and IFN-γ transcription is repressed, whereas IL-2 receptor α-chain (CD25), glucocorticoid-induced TNF receptor family related protein (TNFRSF18, GITR), the co-stimulatory molecule cytotoxic T-lymphocyte antigen 4 (CTLA-4) and integrin alpha E chain (CD103) are activated (Hori et al., 2003). One of the mechanisms underlying transcriptional control of these genes by Foxp3 might be in the physical interaction with AML1 that inhibits AML1 to activate IL-2 and IFN-gamma gene expression through binding to their respective promoters (Ono et al., 2007).
Foxp3 expression is not entirely restricted to Tregs in humans: TGF-β and TCR signaling induced Foxp3 expression in CD4+CD25− T cells (Chen et al., 2003; Tran et al., 2007), and Morgan et al. (2005) demonstrated that Foxp3 mRNA transcription can be induced with PHA or CD3/CD28 antibodies in human CD25− PBMCs and CD8+ PBMCs within 24 to 40 h after stimulation. However, transient Foxp3 expression does not result in a regulatory phenotype (Wang et al., 2007). Long-term hyporeactive or suppressive phenotypes require stabilized expression of Foxp3 (Williams and Rudensky, 2007) mediated by epigenetic control through CpG demethylation and histone methylation (Floess et al., 2007).
Because of the importance of Foxp3 for development and peripheral function of Tregs, it is instrumental to understand FOXP3 gene regulation. Brunkow and colleagues cloned a 30.8 kb genomic fragment that contained the entire murine FOXP3 gene along with ~18 kb of the 5′ sequence and ~4 kb of the 3′ sequence. Within this sequence they found a distal, non-coding exon (named exon −2b) ~6.1 kb upstream of the first coding exon of the murine FOXP3 gene. In this region a potential promoter was predicted by GENESCAN (Brunkow et al., 2001). Mantel et al. and Ouaked et al. investigated a further promoter region: −1657 to +176 in respect to transcription initiation of the human FOXP3 gene. There these authors described proximal promoter elements i.e. a transcription start site, TATA box, GC box, CAAT box and transcription factor binding sites (Mantel et al., 2006; Ouaked et al., 2009). Other regulatory, in particular highly conserved enhancer elements that bind either NFAT and Smad3, or STAT5, CREB and ATF, respectively, were reported within “intron zero”, a region of approximately 6 kb that interrupts the 5′ untranslated region (Bassuny et al., 2003; Burchill et al., 2007; Floess et al., 2007; Kim and Leonard, 2007; Mantel et al., 2006; Tone et al., 2008; Zorn et al., 2006). Recently, it has been shown by several groups that Foxp3 expression and thymic Treg development requires TCR signaling via the NF-κB subunit c-Rel (Isomura et al., 2009; Long et al., 2009; Ruan et al., 2009). However, how NF-κB exerts its Foxp3 regulating effect is still a matter of debate. According to Long et al. prerequisite for Foxp3 expression is binding of a c-Rel-p50 complex to the promoter distal enhancer in intron zero (Long et al., 2009). In contrast Ruan et al. (2009) argue that a c-Rel-p65 complex at the Foxp3 promoter initiates an enhanceosome further consisting of NFAT, Smad, and CREB. In contrast Jana et al. (2009) have also reported recently that c-Rel does not bind at all to the Foxp3 promoter and they attribute to p65 and p50 rather an inhibitory effect on Foxp3 expression. Because these studies were all done with inbred mice, these differences are highly challenging, in particular in respect of comparison with and translation to humans. In addition to experimental conditions or cell types in different activation/differentiation stages, etc., that might underlie the differences in the mouse studies, human Tregs further vary in several molecular aspects to mouse ones (Ziegler, 2006). For instance, Foxp3 seems to be a more consistent marker for functional Tregs in mice than in humans where it appears more as a T-cell activation marker (Morgan et al., 2005; Tran et al., 2007), humans express a second isoform (Allan et al., 2005), and while expression of IL-35 by mouse Tregs is important for suppression, IL-35 is not even expressed by human Tregs (Bardel et al., 2008).
Here we aimed to get insight into the regulation of human Foxp3 and studied three prominent evolutionary conserved regions (ECRs) contained in a ~6 kb spanning region upstream of the human FOXP3 transcription start site. By using a 525 base pair promoter fragment derived from ECR1, the region closest to the FOXP3 transcription start site, we identified the NF-κB subunit p65 as a critical transactivator of the proximal Foxp3 promoter. Further, p65-mediated proximal Foxp3 promoter transactivation was inhibited by Foxp3 indicating control of Foxp3 expression by a negative feedback mechanism.
2. Material and methods
2.1. Bioinformatics analysis
For sequence alignments we used the “UCSC Genome Bioinformatics” tools (http://genome.cse.ucsc.edu) (Kent et al., 2002). Identification of ECRs was performed by the bioinformatics program “ECR-Browser” (http://ecrbrowser.dcode.org) (Ovcharenko et al., 2004). Transcription factor binding sites were analyzed by “CONSITE” (http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite) (Sandelin et al., 2004).
2.2. Construction of promoter fragments
Genomic DNA was isolated from peripheral blood of a human male donor and Jurkat T cells according to the manual provided with the Genomic DNA Purification Kit # K0519 (Fermentas, St. Leon-Rot, Germany). The genomic DNA was used as a template to amplify FOXP3 promoter fragments by PCR.
The ECR1_525 fragment was obtained from the blood genomic DNA by use of the ProofStart™ DNA Polymerase Kit (Quiagen, Vienna, Austria). The ECR1_525 amplification product was cloned into the Kpn I/Bgl II restriction sites of the luciferase reporter vector pGL3-Basic (Promega, Madison WI, USA). As a control, we generated a pGL3-Basic plasmid with an inverse orientated ECR1_525 insert.
To perform reporter assay experiments in T-cell lines we modified the plasmid pd2EGFP-N1 (Clontech, Mountain View CA, USA) expressing destabilized enhanced green fluorescent protein (d2EGFP) by removing the CMV promoter (=pd2EGFP-N1-ΔCMV). It was generated by restriction digestion with Ase I and Nhe I and filling the sticky ends by using T4-polymerase reaction. The Kpn I/Bgl II fragment of pGL3-ECR1_525 was subcloned into Kpn I/Bgl II and Kpn I/BamH I digested pd2EGFP-N1-ΔCMV vectors resulting in correct and inverse orientation of the FOXP3 promoter fragments.
All other FOXP3 promoter fragments (ECR1_855, ECR2_1072, ECR1+2_2008, ECR1+2_2208 and ECR3_1474) were amplified via PCR from genomic Jurkat DNA by the use of Extensor PCR Master Mix 2 (AB-GENE, Epsom, UK). All amplification products were cloned into pGEM-T-Easy (Promega). The inserts were isolated by EcoR I digestion and cloned into pd2EGFP-N1-ΔCMV resulting in correct and inverse orientated FOXP3 promoter plasmids. The pGL3-FOXP3 promoter constructs were generated from the corresponding pd2EGFP-N1-ΔCMV-FOXP3 promoter plasmids by a cloning step via the Bgl II/Kpn I restriction sites. Mutagenesis of the NF-κB binding site was performed by using the QuikChange® XL Site-Directed Mutagenesis Kit (STRATAGENE, Cedar Creek, Texas, USA).
Sequencing of all vectors and the production of oligonucleotides were carried out by MWG Biotech AG, Martinsried, Germany. The primers used for generation of the different constructs and the site-directed mutagenesis were: ECR1_525 (−499 to +26): Forward primer (Kpn I site): 5′-AATAGGTACCACATAGAGCTTCAGATTCTC-3′. Reverse primer (Bgl II site): 5′-TATAAGATCTCTGACAGAAAAGGATCAGCC-3′. ECR1_855 (−629 to +226): Forward primer: 5′-CCCTCGCCGGGTAGTTCAAGCAAT-3′. Reverse primer: 5′-CTTCACCTTTAAGTCTTCTGCCATT-3′. ECR2_1072 (−2333 to −1261): Forward primer: 5′-CAGGTGCTGGGGCAGGGAACAAT-3′. Reverse primer: 5′-GCTTTCTTTGGTGCTGGGCTTTGA-3′. ECR1+2_2008 (−1982 to +26): Forward primer: 5′-CTGTCCAACACACCCAAGCCATTT-3′. Reverse primer: 5′-TATAAGATCTCTGACAGAAAAGGATCAGCC-3′. ECR1+2_2208 (−1982 to +226): Forward primer: 5′-CTGTCCAACACACCCAAGCCATTT-3′. Reverse primer: 5′-CTTCACCTTTAAGTCTTCTGCCATT-3′. ECR3_1474 (−6315 to −4841): Forward primer: 5′-CCATCACTCTTTGACTGACTGACT-3′. Reverse primer: 5′-CATTAGCTGTCTGCTTCTGTTTTC-3′. Mutagenesis of the NF-κB binding site within ECR1: Forward primer: 5′-GAGAGAGAGAAAAAAAAAACTATGATAAGGCCCCCCCACCCCG-3′. Reverse primer: 5′-CGGGGTGGGGGGGCCTTATCATAGTTTTTTTTTTCTCTCTCTC-3′.
2.3. Cell culture
The human embryonic kidney (HEK) cell line 293 was cultured in DMEM (GIBCO, Invitrogen, Lofer, Austria) supplemented with 10% fetal bovine serum (SANOVA Pharma GesmbH, Vienna, Austria). Jurkat E6.1 T cells were cultured in RPMI 1640 medium (GIBCO) supplemented with 10% fetal bovine serum. Before usage, the media were supplemented by adding 2 mM glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin (all from Invitrogen).
PBMCs were isolated from blood of healthy donors by standard density-gradient centrifugation using Lymphoprep (Nycomed, Oslo, Norway). CD4+ T cells were purified from the PBMCs by depletion of CD8-, CD14-, CD16-, CD19-, CD20- and CD56-positive cells using mouse anti-human mAbs (CD8 mAb MEM-87, CD14 mAb MEM-18, CD16 mAb MEM-154, CD19 mAb WIJ09, CD20 mAb MEM-97, CD56 mAb MEM-188, all kindly provided by Dr. Vaclav Horejsi, Institute of Molecular Genetics, Academy of Sciences of the Czech Republic, Prague, Czech Republic) and magnetic sorting using goat anti mouse IgG coated microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). The resulting CD4+ T-cell population was separated into CD4+CD25− and CD4+CD25+ T cells by using anti-CD25 magnetic microbeads (Miltenyi Biotec). For stimulation, cells were cultured with 10 ng/ml phorbol 12-myristate 13-acetate (PMA; Sigma–Aldrich, Vienna, Austria), 1 µM ionomycin (Sigma–Aldrich), 1 µg/ml phytohemagglutinin (PHA; Murex HA16 #RE-30852801, Biomedica, Vienna, Austria), 1 µg/ml plate-bound CD3 mAb OKT3 (Ortho Pharmaceuticals, Raritan, NJ, USA) or 500 ng/ml soluble CD28 mAb Leu28 (Becton Dickinson, Franklin Lakes, USA) alone or in combination. Curcumin was obtained from Sigma–Aldrich.
2.4. Immunofluorescence analysis
Foxp3 staining was conducted by using the Alexa Fluor® 647 labeled mouse anti-human Foxp3 monoclonal antibody clone 259D/C7 from Becton Dickinson according to the manufacturer’s manual. PE-conjugated monoclonal anti-human CD25 clone 4E3, purchased from Miltenyi Biotec, was incubated with the cells for 30 min on ice. Antibody staining of cells was evaluated by using a LSR II flow cytometer from Becton Dickinson.
2.5. Luciferase reporter gene assays
Cells were seeded into 24-well plates the day prior to transfection using the calcium phosphate method. Data normalization was achieved by co-transfection of 0.3 µg pRC-CMV-βGal plasmid to each sample. We used the luciferase reporter gene assay high sensitivity kit from Roche Diagnostics Wien GmbH (Vienna, Austria). In brief, harvested cells were resuspended in 150 µl luciferase assay lysis buffer provided with the kit. Debris was removed by centrifugation. For measurement of luciferase activity we used 100 µl of supernatant in a Berthold Lumat LB 9501 device (Berthold Technologies GmbH, Vienna, Austria). Thirty microliters of the supernatant of each sample was used for the β-galactosidase assay using ortho-nitrophenyl β-d-galactopyranoside (ONPG, Sigma–Aldrich) as substrate. The protein concentration of each sample was measured by the BIO-RAD protein assay (Vienna, Austria) using 5 µl of the supernatant.
2.6. Fluorescence-based reporter assays
Jurkat cells (5 × 106) were mixed with up to 5 µg plasmid DNA and 100 µl ATP free “cytomix-buffer” according to van den Hoff et al. (1992). Human primary T cells (5 × 106) were mixed with up to 5 µg plasmid DNA and 100 µl “enhanced cytomix-buffer”, which was also devoid of ATP but contained in addition 1.25% DMSO and 100 mM sucrose. Electroporation of Jurkat and primary T cells was performed in 2 mm cuvettes using an “AMAXA™ Nucleofector” device (AMAXA GmbH, Cologne, Germany). Immediately after electroporation, cells were washed with RPMI 1640 medium and cultured as described. Dead cells were excluded by 7-amino-actinomycin D (7-AAD) staining and SSC/FSC properties to assess d2EGFP expressing cells among the living population. In some experiments, DsRed expression vector was co-electroporated to distinguish between transfected and untransfected cells and thus to determine d2EGFP expression just among transfected cells. Because of the spectral overlap of DsRed and 7-AAD we assessed in the respective experiments the living population due to the FSC/SSC properties only. Flow cytometry was performed using a LSR II flow cytometer (Becton Dickinson).
3. Results
3.1. The upstream region of the human FOXP3 gene contains three ECRs
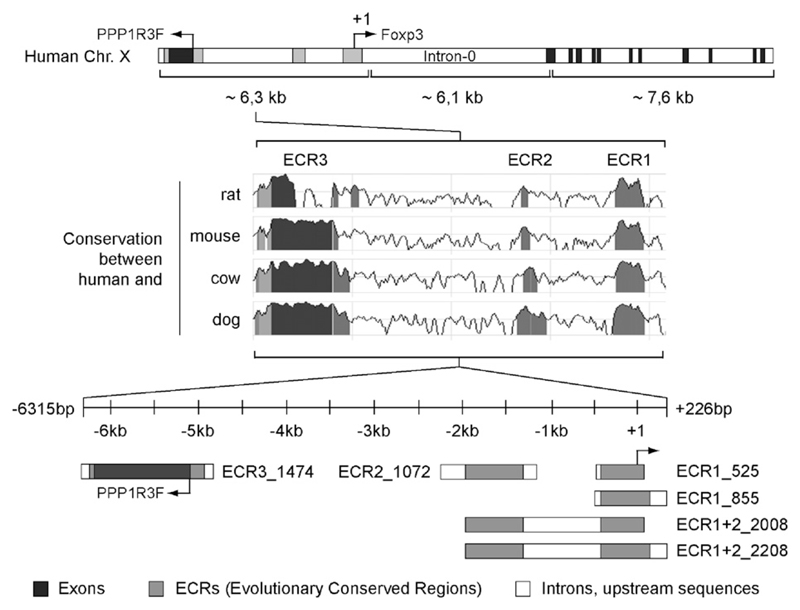
To identify sequences that may contain regulatory elements controlling the human FOXP3 gene we screened the 6.3 kb stretch upstream of the 5′-non-coding exon for ECRs with more than 85% identity/>100 base pairs between the human, mouse, rat, cow and dog FOXP3 loci. We found three main ECRs (for simplicity we named them ECR1 to 3). ECR1 comprises a stretch of ~0.4 to ~0.5 kb, which was described to contain the proximal FOXP3 promoter (Mantel et al., 2006). ECR2 is situated 1.3–2.0 kb upstream of the transcription start site. The utmost distant part of ECR2 (−1.85 to −2.0 kb) shows the most conserved sequences followed by an extended stretch of descending conservation. ECR3 occurs as a large conserved region between −5 and −6 kb, which is located in close proximity to the protein phosphatase 1 regulatory (inhibitor) subunit 3F (PPP1R3F) gene with opposite orientation to Foxp3 transcription.
Fig. 1 illustrates the human FOXP3 gene, transcription start site, ECRs and promoter fragments derived from the respective ECRs. The ECR1_525 (−499 to +26) promoter fragment covers the complete ECR1 stretch, in contrast to the ECR1_855 construct (−629 to +226), which contains additional flanking sequences. ECR2_1072 (−2333 to −1261) comprises the complete ECR2 including additional up- and downstream sequences. Furthermore, two fragments were amplified containing both ECR1 and ECR2: ECR1+2_2208 (−1982 to +226) and ECR1+2_2008 (−1982 to +26). ECR3_1474 was amplified from the region −6315 to −4841; it contains the potential promoter region ECR3, which includes the first exon and a small part of the first intron of PPP1R3F.
Fig. 1.
Schematic overview of the human Foxp3 locus, the identified ECRs and the amplified promoter fragments. The upper section of the figure shows the organization of the FOXP3 gene on chromosome X. The gene PPP1R3F (NM_033215) is located upstream. The three ECRs on the upstream region of human Foxp3 (indicated in gray) were identified based on the DNA sequence similarity between the human, rat, mouse, cow and dog FOXP3 loci using the bioinformatics program ‘ECR-Browser’ (database May’04/hg17; human Chr. X: 48883883–48863781; conservation: minimum length 100 base pairs, 85% identity). The cloned promoter fragments containing the identified ECRs are illustrated in the lower section. The black areas correspond to exons.
3.2. Basal transcriptional activities of the cloned ECR fragments
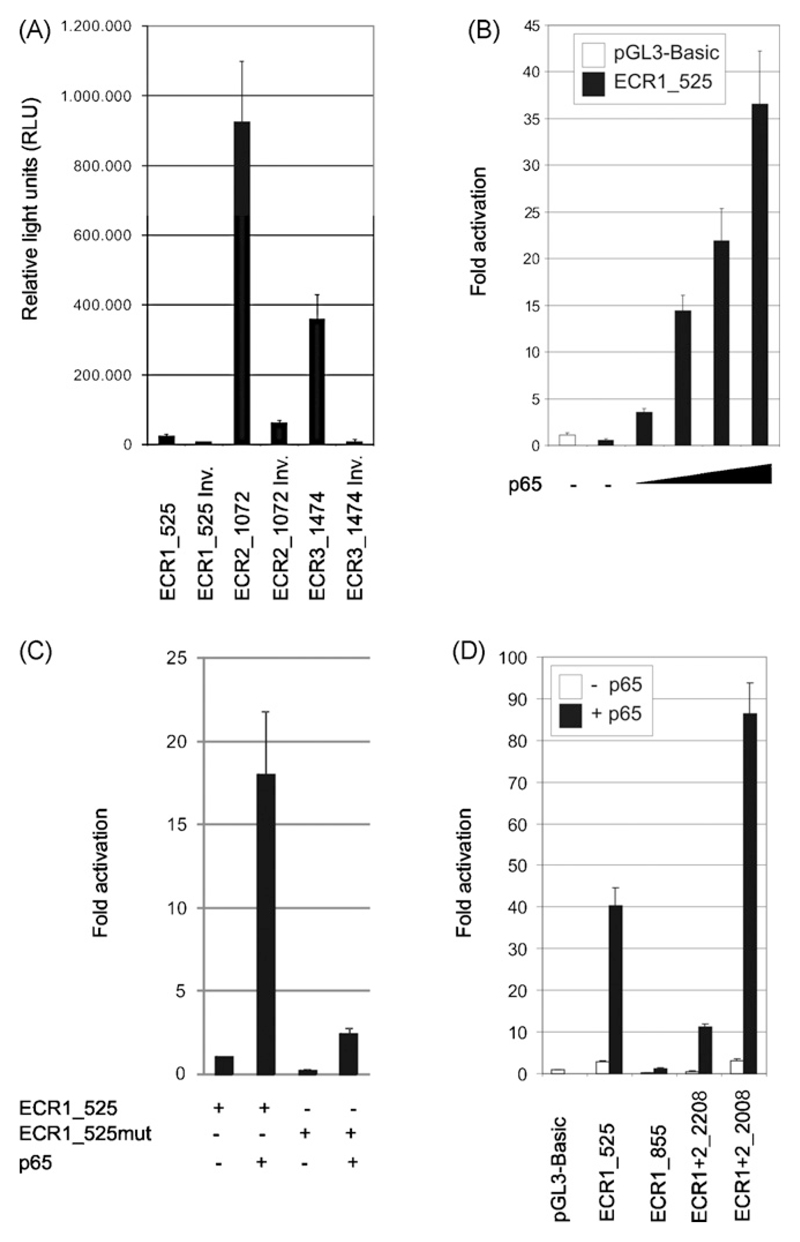
We cloned the different correct and inverse orientated promoter fragments into the pGL3-basic vector and transfected the resulting luciferase reporter plasmids into HEK 293 cells to investigate their basal transcriptional activity. Measurement of luciferase expression revealed low reporter activity of ECR1. In contrast, ECR2 showed high reporter activity suggesting an important upstream element that might contribute to FOXP3 gene regulation. Moreover, we observed basal transcriptional activity directed towards FOXP3 within the ECR3 fragment that overlaps with the first exon of the human PPP1R3F gene. Strikingly, PPP1R3F is transcribed in the opposite direction of FOXP3. The inverse orientated ECR3 fragment (ECR3_INV) corresponding to the transcriptional direction of PPP1R3F showed only negligible luciferase expression (Fig. 2A).
Fig. 2.
Transcriptional activities and p65-mediated transactivation of the cloned promoter fragments in HEK 293 cells analyzed by luciferase reporter assays. (A) The cells were transiently transfected with the indicated correct and inverse ECR-luciferase reporter constructs. After 40 h of cultivation, the basal transcriptional activity was determined. (B) HEK 293 cells were transiently co-transfected with 0.5 µg pGL3-Basic or proximal FOXP3 reporter construct pGL3-ECR1_525 plus 0.0, 0.3, 0.6, 0.9 or 1.2 µg of p65 cDNA. After 40 h of culture, cells were harvested and luciferase expression was measured. (C and D) Transient co-transfection of HEK 293 cells with 1 µg of indicated FOXP3 reporter constructs with or without 1 µg p65 cDNA. After 24 h of culture, cells were harvested and luciferase expression was measured. Each experiment is representative for three independent ones, performed in triplicates. Bars represent means of triplicates; the SD is indicated.
3.3. Transactivation of the proximal human FOXP3 promoter by NF-κB subunit p65
Based on literature and bioinformatics analysis (see Fig. S3), we selected several transcription factor candidates for ECR1 transactivation in a coexpression/luciferase reporter assay. We tested NFATc2/p and SP transcription factors recently published to control the proximal human FOXP3 promoter (Mantel et al., 2006). Further, we analyzed AML-1, ternary complex factor Elk-1, ETS domain family member PU.1, EGR-1, Foxp3, NF-κB family members p65 and cRel; for reviews see (Hayden and Ghosh, 2004; Sharrocks, 2001). We also included STAT5a because an inducible form of STAT5, when transiently activated in IL-2 knockout mice, increased CD4+CD25+ T-cell levels (Antov et al., 2003). In addition to these transcription factors, we selected the following controls: EAPP, E2F1, E2F4, GATA1, STAT1 (Harigae, 2006; Levy and Darnell, 2002; Novy et al., 2005; Trimarchi and Lees, 2002). As shown in Table 1A, only NF-κB subunit p65 transactivated ECR1_525 in HEK 293 cells significantly.
Table 1. Transactivation of ECR1_525 by different transcription factors.
| Transcription factor | A. Fold activation of luciferase reporter in HEK 293 cells | B. Percentage of d2EGFP expressing Jurkat T cells |
|
|---|---|---|---|
| Unstimulated | PMA/iono | ||
| / | 1.1 ± 0.2 | 1.1 | 2 |
| AML1a | 1 ± 0.2 | 1 | 2.7 |
| AML1b | 2.8 ± 0.3 | 1.2 | 1.9 |
| EAPP | 1.7 ± 0.1 | 1 | 2.5 |
| E2F1 | 0.3 ± 0.1 | 1.4 | 1.8 |
| E2F4 | 2.5 ± 0.4 | 0.9 | 2.2 |
| EGR1 | 2.1 ± 0.5 | 1.4 | 2.7 |
| Elk1 | 4.3 ± 0.1 | 0.7 | 1.5 |
| Foxp3 | 1.6 ± 0.2 | 0.6 | 2 |
| GATA1 | 4.7 ± 0.4 | 1.4 | 4.5 |
| NFATc1a | 1 ± 0.2 | 1.8 | 5.1 |
| NFATc2/p | 1.4 ± 0.4 | 1.2 | 3.1 |
| PU.1 | 2.7 ± 0.3 | 2.5 | 3 |
| p65 | 24.4 ± 3.9 | 3.5 | 10.9 |
| cRel | 1.6 ± 0.1 | 1.3 | 2.1 |
| STAT1 | 0.6 ± 0.1 | 1.5 | 2.1 |
| STAT5a | 1.2 ± 0.4 | 2.3 | 4.1 |
| SP1 | 1.3 ± 0 | 1,8 | 2.3 |
| SP3 | 1.4 ± 0.4 | 3 | 1.5 |
(A) HEK 293 cells were transiently co-transfected with 1 µg ECR1_525 luciferase reporter plasmid alone or in combination with 2 µg of expression vectors encoding the indicated transcription factors. Luciferase expression was measured after 24 h. The data are the mean value ± SD of three independent experiments. (B) Jurkat T cells were electroporated using 2 µg ECR1_525 d2EGFP reporter plasmid in combination with 1 µg of either one of the indicated transcription factor cDNA or as control empty pCIneoHA vector. Samples were divided and treated for 22 h with or without PMA/ionomycin. Prior to flow cytometry measurement, the cells were treated with 1 µg/ml 7-AAD to distinguish dead cells from the living population.
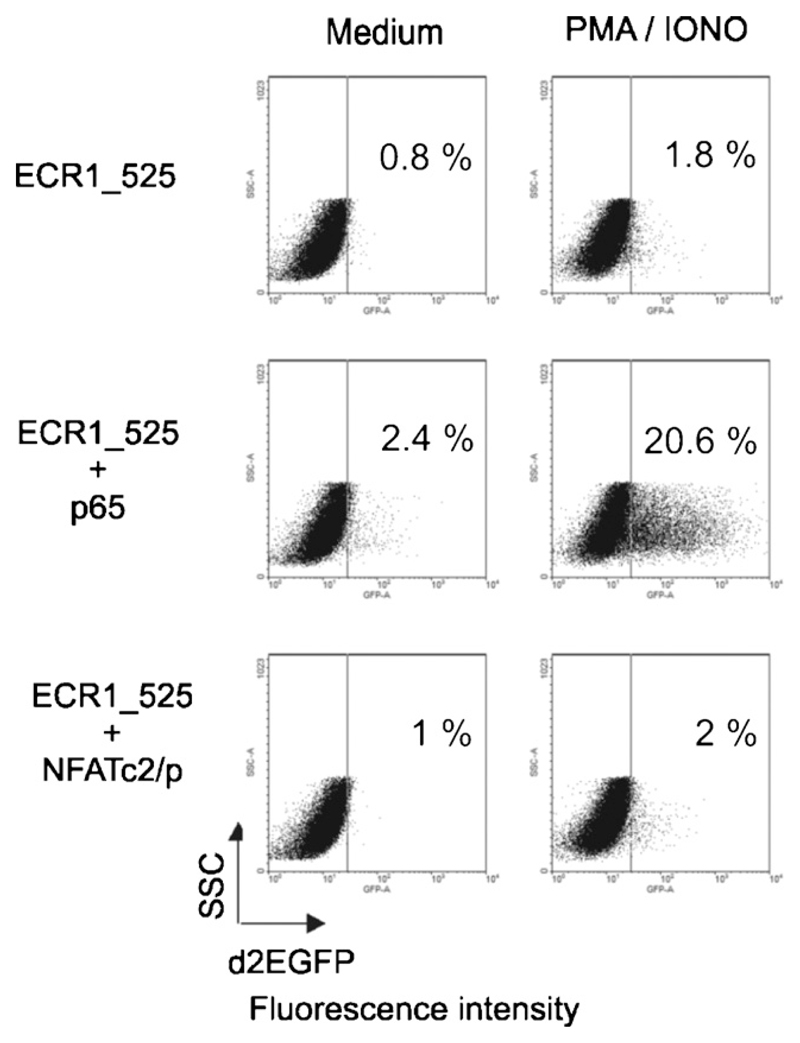
We next tested whether this holds also true in T cells. For this purpose, we established a reporter assay based on d2EGFP expression to measure reporter activity by flow cytometry. Briefly, we replaced the CMV promoter of the pd2EGFP-N1 vector by the ECR1_525 promoter fragment and transferred the resulting plasmids into Jurkat T cells by electroporation. After 22h of cultivation, the cells were harvested and flow cytometry was performed. Compared to the transcription factor screen in HEK 293 cells, we again found p65 as the most striking transactivator of the proximal promoter region in Jurkat T cells. Furthermore, only the transactivation ability of p65 was enhanced by PMA/ionomycin stimulation of the Jurkat T cells (Table 1B).
The specificity of transactivation of ECR1_525 by p65 was elaborated by a dose dependence and mutation analysis. First, cotransfection of ECR1_525 with increasing amounts of p65 resulted in a dose dependent transactivation: ECR1_525 alone (0.5 fold activation), +0.3 µg p65 cDNA (3.5 fold activation), +0.6 µg p65 cDNA (14.4 fold activation), +0.9 µg p65 cDNA (21.9 fold activation), +1.2 µg p65 cDNA (36.6 fold activation) (Fig. 2B). Second, partial mutation of the potential NF-κB binding site 5′-GAGAACCCCC-3′ (position −157/−148) to 5′-GATAAGGCCC-3′ (=ECR1_525mut) reduced p65-mediated ECR1 transactivation (Fig. 2C). Furthermore, we compared the transactivation ability of p65 on each fragment containing the proximal human FOXP3: ECR1_525 (−499/+26), ECR1_855 (−629/+226), ECR1+2_2208 (−1982/+226) and ECR1+2_2008 (−1982/+26). In contrast to ECR1_525, p65 could not transactivate ECR1_855 (Fig. 2D). This result suggested at least one transcriptional repressor-binding site either within the sequence stretches −629/−499 and/or +26/+226. To identify these sites, we analyzed ECR1+2_2208 that contains the complete ECR1_855 element plus ECR2 including the linker region −629/−499. This construct showed a fourfold lower luciferase activity upon p65 co-transfection compared to the ECR1_525 promoter fragment. Truncation of the region +26/+226 resulted in ECR1+2_2008, which showed in comparison to ECR1_525 a doubled transcriptional activity with p65. Thus, the stretch +26/+226 contains at least one transcriptional repressor-binding site (Fig. 2D).
Because of the high basal reporter activity within ECR2 and ECR3 (Fig. 2A) we assumed that these ECR regions contain (i) an alternative transcription start site and/or (ii) an enhancer element that acts on ECR1. Therefore, we generated additional reporter constructs to investigate potential enhancer function of ECR2 and ECR3. For this purpose ECRs were cloned in correct or inverse orientation downstream of the luciferase gene of empty pGL3-Basic, pGL3-ECR1_525 and pGL3-ECR1_855 plasmids. Then we checked the constructs in CD3 stimulated Jurkat T cells. However, we observed no enhancer activity of the downstream cloned ECR2 or ECR3 (supplementary Fig. S1). Further, compared to HEK 293 cells, the basal activity of ECR3 was reduced in Jurkat T cells (compare Fig. S1 and Fig. 2A).
3.4. p65-Mediated transactivation of ECR1 in Jurkat T cells requires PKC
Because p65 barely transactivated ECR1_525 in resting but well in PMA/ionomycin stimulated Jurkat T cells (Table 1B), we analyzed whether PMA or ionomycin signaling alone is able to induce the proximal FOXP3 promoter in combination with p65. We tested also other activation agents including CD3 and CD28 mAbs and PHA. Table 2 summarizes the results of three independent experiments and Fig. 3 shows flow cytometry profiles of a typical experiment of the triplicate data sets summarized in Table 2. One can see that PMA is critical for transactivation of the proximal FOXP3 promoter ECR1_525 by p65. Also signaling via CD3 activated the ECR1_525 promoter fragment in p65 transfected cells significantly. However, ionomycin, CD28 or PHA alone did not transactivate ECR 525, indicating that activation of the protein kinase C via CD3 or PMA stimulation is required for the induction of the proximal FOXP3 promoter by NF-κB/p65. In agreement with this finding PKC0 was found essential for TCR-mediated NF-κB activation (Li et al., 2005), and mice deficient in PKCθ harbor CD4+ T cells with reduced Foxp3 mRNA levels and low numbers of CD4+Foxp3+Treg cells (Gupta et al., 2008; Schmidt-Supprian et al., 2004).
Table 2. Activation dependent transactivation of ECR1-525 in Jurkat T cells.
| Treatment | ECR1_525+control | ECR1_525+p65 | ECR1_525+NFATc2/p |
|---|---|---|---|
| Medium | 1% (±0.2) | 1.2% (±1.1) | 0.7% (±0.5) |
| PMA | 1.5% (±0.6) | 13.4% (±3.1) | 1.5% (±0.5) |
| Iono | 0.6% (±0.2) | 0.3% (±1.1) | 1.3% (±1.8) |
| PHA | 1.3% (±1.1) | 4.4% (±2.7) | 1% (±1.0) |
| CD3 | 0.8% (±0.5) | 5.9% (±1.4) | 0.6% (±0.4) |
| CD28 | 0.8% (±0.4) | 2.1% (±1.5) | 0.7% (±0.7) |
| PMA/iono | 2.5% (±1.5) | 20.7% (±2.6) | 2.3% (±1.0) |
| PMA/PHA | 2.7% (±0.6) | 14.5% (±3.6) | 2.5% (±1.1) |
| CD3/CD28 | 1.2% (±0.4) | 6.3% (±1.6) | 0.7% (±0.1) |
Jurkat T cells were electroporated using 2 µg ECR1_525 in combination with 1 µg of either p65 cDNA, NFATc2/p cDNA or as a control empty pCIneoHA vector. After electroporation, cells were divided and treated for 22 h with the indicated stimuli. Prior to flow cytometry measurement, the cells were stained with 1 µg/ml 7-AAD to distinguish dead cells from the living population. The table summarizes the percentages of d2EGFP expressing cells and the respective SD of three independent experiments.
Fig. 3.
Transactivation of ECR1 in Jurkat T cells. Flow cytometry profiles of Jurkat T cells transfected with the pd2EGFP reporter under control of the proximal FOXP3 promoter ECR1_525. pd2EGFP expression was also analyzed upon co-transfection with p65 or NFATc2/p, and PMA/ionomycin stimulation. The dot blots show side scatter (SSC)/d2EGFP properties of life gated cells. This experiment is representative for three independent experiments, which are compiled in Table 2.
We display here also the effect of NFATc2/p that was earlier reported to be a key factor for transactivation of the proximal FOXP3 promoter (Mantel et al., 2006). In our assays NFATc2/p did not transactivate ECR1_525 neither in resting nor activated Jurkat T cells (Fig. 3 and Table 2). This finding correlates with Tone et al., who found poor binding of NFAT (position −406 to −306) in the Foxp3 promoter. According to that report the major contribution of NFAT to Foxp3 expression is activation of the enhancer at position +2137 to +2158 (Tone et al., 2008).
3.5. ECR1 transactivation in human CD4+CD25− T cells
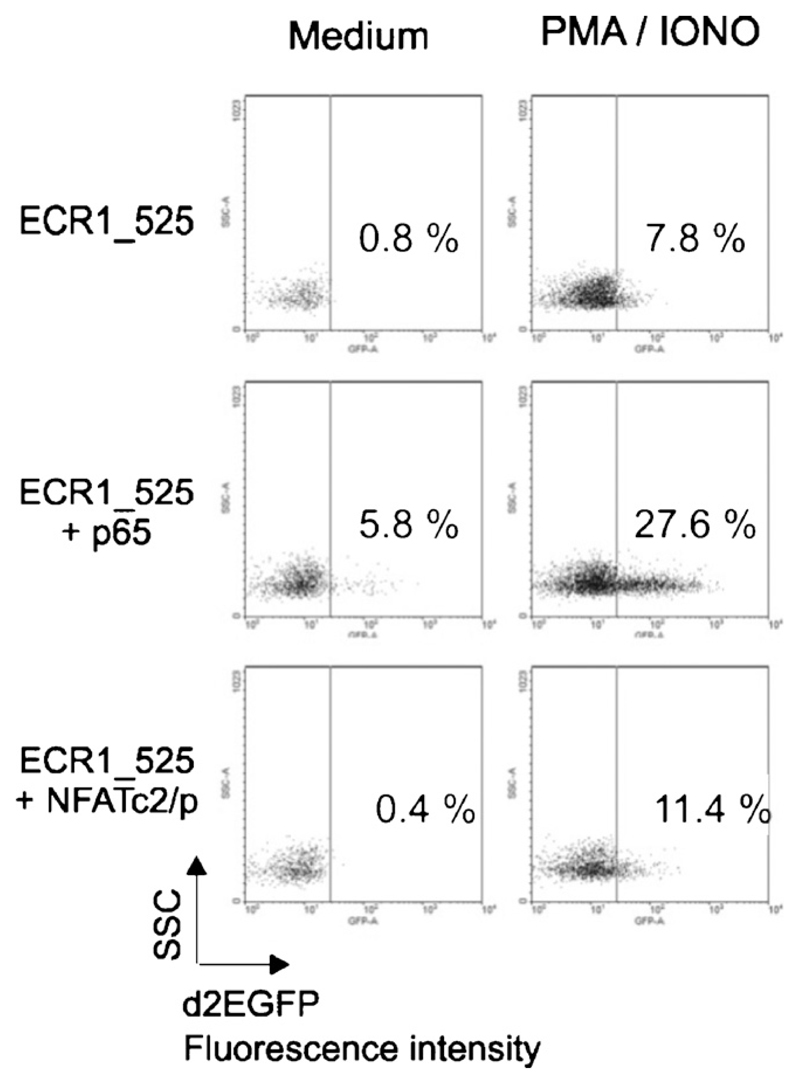
Next, we analyzed ECR1 transactivation in primary human CD4+CD25− T cells isolated from peripheral blood. We coelectroporated the cells with the proximal promoter fragment ECR1_525 plus vectors encoding p65 or NFATc2/p and cultured them with or without PMA/ionomycin for 24h. In contrast to Jurkat cells (Fig. 3, Tables 1 and 2), PMA/ionomycin stimulation alone transactivated ECR1 significantly (7.8% d2EGFP positive cells). Furthermore, we observed a slight p65-mediated transactivation of ECR1_525 in unstimulated cells (5.8% d2EGFP positive cells), which was dramatically enhanced upon PMA/ionomycin stimulation (27.6% d2EGFP positive cells). In contrast to p65, the transcription factor NFATc2/p alone failed to transactivate ECR1_525 but had a slight synergistic transactivatory effect upon PMA/ionomycin stimulation (Fig. 4).
Fig. 4.
ECR1 transactivation mediated by p65 or NFATc2/p in primary human T-lymphocytes. CD4+ CD25− T-lymphocytes were electroporated with 2 µg ECR1_525 pd2EGFP reporter in combination with 2 µg cDNA expressing the indicated molecules and 1 µg pDsRed expression vector. After electroporation, the cells were rested over night in RPMI 1640 medium without FCS. The next day the samples were divided, cultured in IL-2 (30 U/ml) and 10% FCS conditioned RPMI 1640 medium, and stimulated with or without 10 ng/ml PMA and 1 µM ionomycin for 24h. Dot blots show d2EGFP expressing cells among the transfected cells, which were identified and gated by DsRed expression. This experiment is representative for two independent ones.
3.6. NF-κB inhibitor curcumin stalls Foxp3 expression
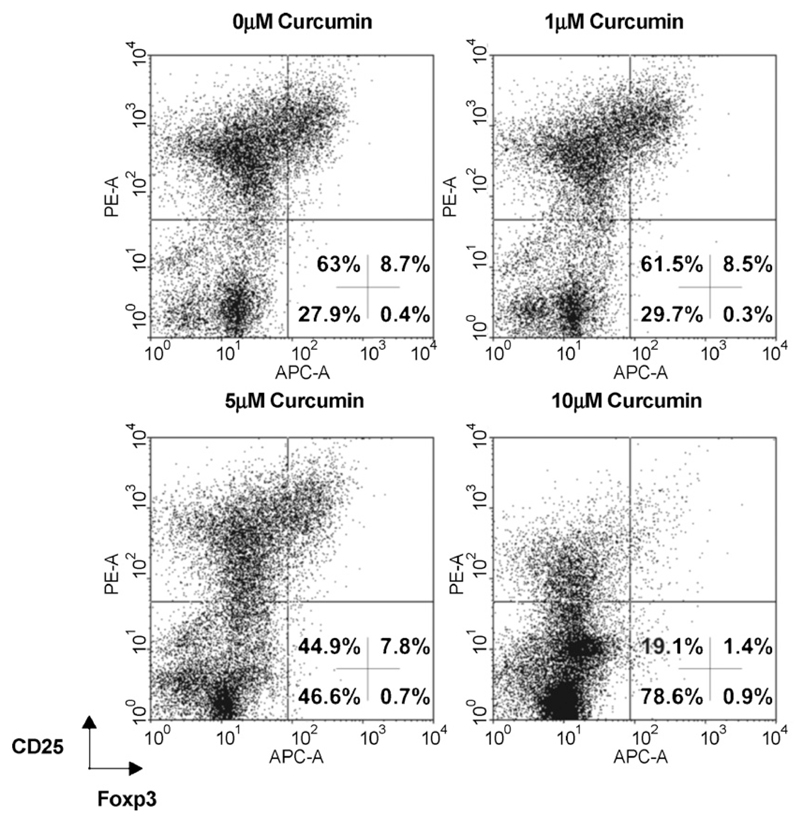
Curcumin is a potent inhibitor of PMA-mediated NF-κB activation (Ranjan et al., 2004) and in agreement we found that curcumin reduced p65-mediated ECR1 transactivation in Jurkat T cells (supplementary Fig. S2). Based on this observation we asked whether curcumin also inhibits the expression of endogenous Foxp3 in primary T cells. In order to address this question, we stimulated PBMCs for 48 h with IL-2, CD3, CD28 and TGF-β to induce Foxp3 expression and treated the cells with increasing concentrations of curcumin. This procedure markedly reduced expression of Foxp3 in the activated CD25 positive T blasts (Fig. 5).
Fig. 5.
Curcumin inhibits endogenous Foxp3 expression in primary human T cells. PBMCs were isolated from human blood and stimulated with IL-2 (50 U/ml), CD3 (5 µg/ml OKT3 plate-bound), CD28 (0.5 µg/ml LEU28 soluble), TGF-β (5 ng/ml) and treated with indicated amounts of curcumin. After 48 h of culture, we analyzed Foxp3 and CD25 expression by flow cytometry.
3.7. p65-Mediated transactivation of the proximal Foxp3 promoter is repressed by Foxp3 and SP3
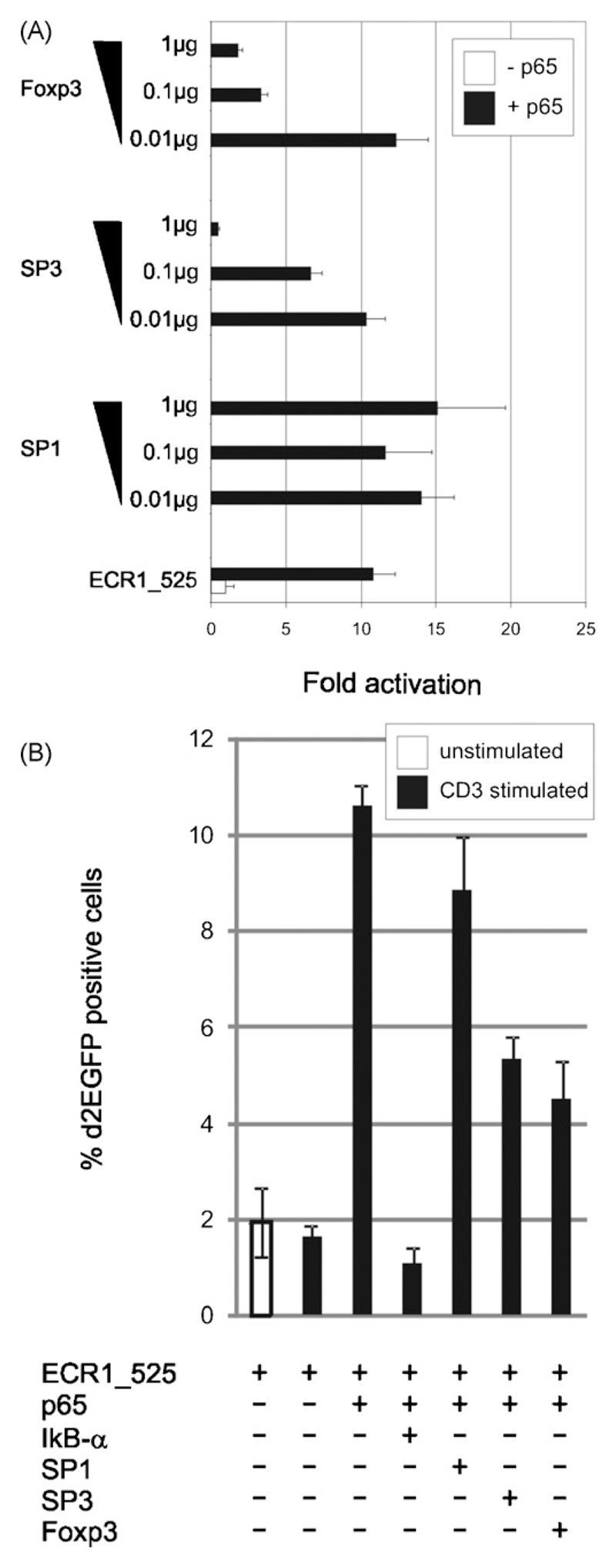
One of the dominant effects of Foxp3 on target genes is to suppress their activation (Marson et al., 2007). Therefore, we tested the potential negative regulatory function of Foxp3 on its own expression. Indeed, overexpression of Foxp3 inhibited p65-mediated transactivation of the proximal FOXP3 promoter fragment ECR1_525 in both HEK 293 cells (Fig. 6A) and Jurkat T cells (Fig. 6B).
Fig. 6.
Foxp3 and SP3 but not SP1 repress p65-mediated ECR1_525 reporter activity. (A) Transient co-transfection of HEK 293 cells with 1 µg ECR1_525 luciferase reporter plasmid plus 1 µg p65 and increasing concentrations of SP1, SP3 or Foxp3 cDNAs. Luciferase activity was measured after 24 h. Bars represent triplicates; standard deviations are indicated. (B) Jurkat T cells were electroporated with 1 µg ECR1_525 pd2EGFP reporter and 1 µg pDsRed expression vector alone or in combination with 1 µg of the indicated transcription factor cDNAs. After electroporation the samples were cultured for 24 h in the presence or absence of 1 µg/ml platebound CD3 antibody OKT3. Bar graph shows the percentage of d2EGFP expressing cells among the transfected cells, which were identified and gated by DsRed expression. Bars represent triplicates; standard deviations are indicated.
It was shown that SP transcription factors are able to bind to proximal FOXP3 promoter elements and that mutation of these binding sites reduces reporter gene expression (Mantel et al., 2006). Based on this finding, we tested the influence of transcription factor SP1 and SP3 on the proximal FOXP3 promoter. SP3 was able to repress p65-mediated ECR1_525 transactivation in a dose dependent manner in HEK 293 cells, in contrast to SP1, which had no effect (Fig. 6A). We obtained the same results when we used Jurkat T cells (Fig. 6B). The NF-κB repressor IκB-α was co-expressed as a control (Fig. 6B).
4. Discussion
In this work we cloned three evolutionary conserved regions (ECR1, ECR2 and ECR3), which are located upstream of the transcription start site of the human FOXP3 gene. In HEK 293 cells, ECR1, which comprises the proximal FOXP3 promoter, showed only low basal transcriptional activity indicating that its induction is strictly controlled. In contrast, high basal reporter activity was observed for ECR2 and ECR3 suggesting a potential regulator function of these sequence stretches for FOXP3 gene expression.
Indeed, by using constructs spanning ECR1 and ECR2, we identified a region upstream of −629 (most probably located within ECR2), that enhanced the transcriptional activity of the proximal FOXP3 promoter (Fig. 2D). ECR3 is located in close proximity to the open reading frame of PPP1R3F. Therefore, we originally assumed that it controlled this FOXP3 unrelated gene. However, ECR3 showed poled reporter transactivation in direction to FOXP3 (Fig. 2A). Thus, both ECR2 and ECR3 possess basal transcriptional activity in direction to the FOXP3 gene.
To further analyze whether ECR2 or ECR3 enhances transcriptional activity of ECR1, we cloned either ECR2 or ECR3 downstream of an ECR1-controlled luciferase reporter gene. However, this reporter was enhanced neither by ECR2 nor ECR3 (supplementary Fig. S1). Therefore, at the moment we do not have entirely conclusive data on the putative enhancer activity of the distal ECRs 2 or 3 on the proximal FOXP3 promoter in ECR1. In this respect one has to mention that Tone et al. did not observe reporter activity for the upstream regions −5551 to −4288 and −2402 to −1730 in the murine FOXP3 gene that partially correspond to the human regions ECR3 and ECR2, respectively. However, they identified enhancers downstream of the Foxp3 transcription start site (Tone et al., 2008).
When screening for regulators of ECR1 we identified the NF-κB subunit p65 as a critical factor; all other tested transcription factors failed or just weakly transactivated (Table 1). We confirmed p65-mediated ECR1 transactivation by performing various experiments using different cells (Figs. 2–4). Further, the addition of the NF-κB inhibitor curcumin reduced endogenous Foxp3 expression in primary peripheral blood T cells (Fig. 5), and overexpression of IκB-αblunted p65-mediated ECR1 transactivation. We also allocated the p65 binding site to the sequence stretch −157 to −148 in ECR1.
However, the human proximal FOXP3 promoter is controlled also by factors other than p65. This is indicated by a repressor activity, which we identified within the ECR1 stretch +26 to +226. This finding is corroborated by an earlier study reporting important regulatory elements within the region −245 to +176 (Mantel et al., 2006). This paper describes also binding sites for NFAT within the proximal FOXP3 promoter region and discusses the relevance of NFAT binding to the Foxp3 promoter by the sensitivity of FOXP3 induction to cyclosporine A (Mantel et al., 2006). However, in our hands, NFATc2/p overexpression had negligible potential to transactivate ECR1 in HEK 293 and Jurkat T cells; we only observed a moderate synergistic effect of NFATc2/p on transactivation of ECR1 in PMA/ionomycin stimulated primary CD4+CD25− T cells. In this respect, we would like to stress that cyclosporine A inhibits the nuclear translocation of p65 due to interference with degradation of IKBa and IKBb (Marienfeld et al., 1997). The relevance of NF-κB versus NFAT for Foxp3 expression is further corroborated by CD4+CD25++GITR++ cells from NFATc2/c3 deficient mice that display normal Treg function and unchanged Foxp3 expression levels (Bopp et al., 2005), while mice deficient in PKC, CARMA1 or Bcl10, that are upstream signaling molecules for NF-κB activation, display a substantial reduction or absence of Tregs (Barnes et al., 2009; Gupta et al., 2008; Medoff et al., 2009; Molinero et al., 2009; Schmidt-Supprian et al., 2004). The same Treg defect holds true for mice bearing a conditional deletion of IKKβ (Schmidt-Supprian et al., 2003). Finally, recently several independent reports provide convincing evidence that the NF-κB subunit c-Rel is critical for Foxp3 expression in mice (Isomura et al., 2009; Long et al., 2009; Ruan et al., 2009).
NF-κB could also be the key in the identified negative feedback regulation of Foxp3 (Fig. 6A and B). This mechanism could be executed by physical interaction of Foxp3 with NF-κB that was shown to result in inhibition of NF-κB activity (Bettelli et al., 2005). As a further potent repressor of p65-mediated ECR1 transactivation we discovered SP3 (Fig. 6A and B). How SP3 functions is not clear at the moment. On the BCAT-2 promoter, SP1 and SP3 transcription factors can bind to the same DNA motifs resulting in SP3-mediated repression of SP1-dependent promoter transactivation (Hagen et al., 1994). Interestingly, on the Fas promoter, SP1 is bound to the κB-SP1 site. The NF-κB p50-p65 heterodimer can occupy this site to induce the Fas promoter (Chan et al., 1999). On the FOXP3 promoter the identified SP binding site (Mantel et al., 2006) has poor SP1 binding affinity (Tone et al., 2008). Whether this site can be easier occupied by SP3 is subject of further assays as well as the role of the potential human regulatory FOXP3 elements ECR2 and ECR3 in specific transcription of Foxp3 for human Treg differentiation and function.
Supplementary Material
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.molimm.2010.04.002.
Acknowledgments
We thank Dr. Thomas Decker (University of Vienna, Vienna, Austria) for pRC-STAT1, Dr. Robert A. Hipskind (Institut de Génétique Moléculaire de Montpellier, Montpellier, France) for pCDNA3-HA-Elk-1, Dr. Rainer de Martin and DI Dr. Ulrike Resch (Medical University of Vienna, Vienna, Austria) for providing expression vectors encoding cJun, cRel, p65 and IκBα cDNA, Dr. David G. Roodman (University of Pittsburgh Cancer Institute, Pittsburgh, PA, USA) for pcDNA3-AML-1a and pcDNA3-AML-1b, Dr. Johann Rotheneder (Medical University of Vienna, Vienna, Austria) for the plasmids pCIneoHA, pCIneoHA-EAPP, pCIneoHA-E2F1, pCIneoHA-E2F4, pCIneoHA-EGR-1, pCIneoHA-SP1 and pCIneoHA-SP3, Dr. Edgar Serfling and Dr. Stefan Klein-Hessling (University of Wuerzburg, Wuerzburg, Germany) for pRSV-NFATc1/A and pRSV-NFATc2/p encoding murine NFATc2/p, Dr. Derya Unutmaz (Vanderbilt University School, Nashville, Tennessee, United States of America) for lentiviral vector encoded human Foxp3 (H110.Foxp3-IRES-HSA). We are grateful to Eva Steinhuber for her technical assistance and DI Dr. Ulrike Resch for helpful comments. The GEN-AU program of the Austrian Federal Ministry of Science and Research and the Austrian Science Fund (FWF, grant no. P18347-B13) supported this work.
Abbreviations
- d2EGFP
destabilized enhanced green fluorescent protein
- DsRed
Discosoma red fluorescent protein
- FSC
forward scatter
- HA
hemagglutinine tag
- SSC
side scatter
References
- Allan SE, Passerini L, Bacchetta R, Crellin N, Dai M, Orban PC, Ziegler SF, Roncarolo MG, Levings MK. The role of 2 FOXP3 isoforms in the generation of human CD4+ Tregs. J Clin Invest. 2005;115:3276–3284. doi: 10.1172/JCI24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antov A, Yang L, Vig M, Baltimore D, Van Parijs L. Essential role for STAT5 signaling in CD25+CD4+ regulatory T cell homeostasis and the maintenance of self-tolerance. J Immunol. 2003;171:3435–3441. doi: 10.4049/jimmunol.171.7.3435. [DOI] [PubMed] [Google Scholar]
- Bardel E, Larousserie F, Charlot-Rabiega P, Coulomb-L’Hermine A, Devergne O. Human CD4+ CD25+ Foxp3+ regulatory T cells do not constitutively express IL-35. J Immunol. 2008;181:6898–6905. doi: 10.4049/jimmunol.181.10.6898. [DOI] [PubMed] [Google Scholar]
- Barnes MJ, Krebs P, Harris N, Eidenschenk C, Gonzalez-Quintial R, Arnold CN, Crozat K, Sovath S, Moresco EM, Theofilopoulos AN, Beutler B, et al. Commitment to the regulatory T cell lineage requires CARMA1 in the thymus but not in the periphery. PLoS Biol. 2009;7:e51. doi: 10.1371/journal.pbio.1000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassuny WM, Ihara K, Sasaki Y, Kuromaru R, Kohno H, Matsuura N, Hara T. A functional polymorphism in the promoter/enhancer region of the FOXP3/Scurfin gene associated with type 1 diabetes. Immunogenetics. 2003;55:149–156. doi: 10.1007/s00251-003-0559-8. [DOI] [PubMed] [Google Scholar]
- Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Dastrange M, Oukka M. Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc Natl Acad Sci USA. 2005;102:5138–5143. doi: 10.1073/pnas.0501675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp T, Palmetshofer A, Serfling E, Heib V, Schmitt S, Richter C, Klein M, Schild H, Schmitt E, Stassen M. NFATc2 and NFATc3 transcription factors play a crucial role in suppression of CD4+ T lymphocytes by CD4+ CD25+ regulatory T cells. J Exp Med. 2005;201:181–187. doi: 10.1084/jem.20041538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- Chan H, Bartos DP, Owen-Schaub LB. Activation-dependent transcriptional regulation of the human Fas promoter requires NF-kappaB p50-p65 recruitment. Mol Cell Biol. 1999;19:2098–2108. doi: 10.1128/mcb.19.3.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C, Bowcock AM. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106:R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Manicassamy S, Vasu C, Kumar A, Shang W, Sun Z. Differential requirement of PKC-theta in the development and function of natural regulatory T cells. Mol Immunol. 2008;46:213–224. doi: 10.1016/j.molimm.2008.08.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. Embo J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigae H. GATA transcription factors and hematological diseases. Tohoku J Exp Med. 2006;210:1–9. doi: 10.1620/tjem.210.1. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- Isomura I, Palmer S, Grumont RJ, Bunting K, Hoyne G, Wilkinson N, Banerjee A, Proietto A, Gugasyan R, Li W, McNally A, et al. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J Exp Med. 2009;206:3001–3014. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S, Jailwala P, Haribhai D, Waukau J, Glisic S, Grossman W, Mishra M, Wen R, Wang D, Williams CB, Ghosh S. The role of NF-kappaB and Smad3 in TGF-beta-mediated Foxp3 expression. Eur J Immunol. 2009;39:2571–2583. doi: 10.1002/eji.200939201. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kim HJ, Hurt EM, Chen X, Howard OM, Farrar WL. Functional and genomic analyses of FOXP3-transduced Jurkat-T cells as regulatory T (Treg)-like cells. Biochem Biophys Res Commun. 2007;362:44–50. doi: 10.1016/j.bbrc.2007.07.187. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Li Y, Sedwick CE, Hu J, Altman A. Role for protein kinase Ctheta (PKC-theta) in TCR/CD28-mediated signaling through the canonical but not the non-canonical pathway for NF-kappaB activation. J Biol Chem. 2005;280:1217–1223. doi: 10.1074/jbc.M409492200. [DOI] [PubMed] [Google Scholar]
- Long M, Park SG, Strickland I, Hayden MS, Ghosh S. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921–931. doi: 10.1016/j.immuni.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Mantel PY, Ouaked N, Ruckert B, Karagiannidis C, Welz R, Blaser K, Schmidt-Weber CB. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176:3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- Marienfeld R, Neumann M, Chuvpilo S, Escher C, Kneitz B, Avots A, Schimpl A, Serfiing E. Cyclosporin A interferes with the inducible degradation of NF-kappa B inhibitors, but not with the processing of p105/NF-kappa B1 in T cells. Eur J Immunol. 1997;27:1601–1609. doi: 10.1002/eji.1830270703. [DOI] [PubMed] [Google Scholar]
- Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medoff BD, Sandall BP, Landry A, Nagahama K, Mizoguchi A, Luster AD, Xavier RJ. Differential requirement for CARMA1 in agonist-selected T-cell development. Eur J Immunol. 2009;39:78–84. doi: 10.1002/eji.200838734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero LL, Yang J, Gajewski T, Abraham C, Farrar MA, Alegre ML. CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J Immunol. 2009;182:6736–6743. doi: 10.4049/jimmunol.0900498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan ME, van Bilsen JH, Bakker AM, Heemskerk B, Schilham MW, Hartgers FC, Elferink BG, van der Zanden L, de Vries RR, Huizinga TW, Ottenhoff TH, Toes RE. Expression of FOXP3 mRNA is not confined to CD4+CD25+ T regulatory cells in humans. Hum Immunol. 2005;66:13–20. doi: 10.1016/j.humimm.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Novy M, Pohn R, Andorfer P, Novy-Weiland T, Galos B, Schwarzmayr L, Rotheneder H. EAPP, a Novel E2F Binding Protein that Modulates E2F Dependent Transcription. Mol Biol Cell. 2005;16:2181–2190. doi: 10.1091/mbc.E04-11-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, Sakaguchi S. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- Ouaked N, Mantel PY, Bassin C, Burgler S, Siegmund K, Akdis CA, Schmidt-Weber CB. Regulation of the foxp3 gene by the Th1 cytokines: the role of IL-27-induced STAT1. J Immunol. 2009;182:1041–1049. doi: 10.4049/jimmunol.182.2.1041. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan D, Chen C, Johnston TD, Jeon H, Nagabhushan M. Curcumin inhibits mitogen stimulated lymphocyte proliferation, NFkappaB activation, and IL-2 signaling. J Surg Res. 2004;121:171–177. doi: 10.1016/j.jss.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Ruan Q, Kameswaran V, Tone Y, Li L, Liou HC, Greene MI, Tone M, Chen YH. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Courtois G, Tian J, Coyle AJ, Israel A, Rajewsky K, Pasparakis M. Mature T cells depend on signaling through the IKK complex. Immunity. 2003;19:377–389. doi: 10.1016/s1074-7613(03)00237-1. [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Tian J, Grant EP, Pasparakis M, Maehr R, Ovaa H, Ploegh HL, Coyle AJ, Rajewsky K. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proc Natl Acad Sci USA. 2004;101:4566–4571. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert LA, Jeffery E, Zhang Y, Ramsdell F, Ziegler SF. Scurfin (FOXP3) acts as a repressor of transcription and regulates T cell activation. J Biol Chem. 2001;276:37672–37679. doi: 10.1074/jbc.M104521200. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Shevach EM. Fatal attraction: tumors beckon regulatory T cells. Nat Med. 2004;10:900–901. doi: 10.1038/nm0904-900. [DOI] [PubMed] [Google Scholar]
- Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- Tran DQ, Ramsey H, Shevach EM. Induction of FOXP3 expression in naive human CD4+FOXP3 T cells by T-cell receptor stimulation is transforming growth factor-beta dependent but does not confer a regulatory phenotype. Blood. 2007;110:2983–2990. doi: 10.1182/blood-2007-06-094656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- van den Hoff MJ, Moorman AF, Lamers WH. Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res. 1992;20:2902. doi: 10.1093/nar/20.11.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- Wildin RS, Smyk-Pearson S, Filipovich AH. Clinical and molecular features of the immunodysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J Med Genet. 2002;39:537–545. doi: 10.1136/jmg.39.8.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, Bellucci R, Raderschall E, Canning C, Soiffer RJ, Frank DA, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108:1571–1579. doi: 10.1182/blood-2006-02-004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.