Abstract
Mutant p53 exerts gain-of-function effects that drive metastatic progression and therapeutic resistance, but the basis for these effects remain obscure. The RNA binding protein RBM38 limits translation of mutant p53 and is often ablated in tumors harboring it. Here we show how loss of Rbm38 significantly alters cancer susceptibility in mutant p53 knock-in mice, by shortening lifespan, altering tumor incidence and promoting T cell lymphomagenesis. Loss of Rbm38 enhanced mutant p53 expression and decreased expression of the tumor suppressor Pten, a key regulator of T cell development. Furthermore, Rbm38 was required for Pten expression via stabilization of Pten mRNA through an AU-rich element in its 3’UTR. Our results suggest that Rbm38 controls T cell lymphomagenesis by jointly modulating mutant p53 and Pten, with possible therapeutic implications for treating T cell malignancies.
Significance
An RNA-binding protein controls T cell lymphomagenesis by jointly modulating mutant p53 and PTEN, with possible therapeutic implications for treating T cell malignancies.
Keywords: Rbm38, Pten, mutant p53, mRNA stability, T-lymphoblastic lymphoma
Introduction
The tumor suppressor p53 is often referred to as the “guardian of the genome” (1, 2). p53 is the most commonly mutated gene in human cancer and loss of p53 is known to play a central role in tumor development (3). The importance of p53 in tumor suppression is underscored by its ability as a transcription factor to regulate a series of downstream target genes (4). For example, p21(WAF1/CIP1) (5) and GADD45 (6) are induced by p53 to regulate the cell cycle; PUMA (7) and Killer/DR5 (8) are induced by p53 to mediate apoptosis.
Unlike most tumor suppressor genes, which are mainly inactivated as a result of deletion or truncation, the p53 gene is frequently altered as missense mutations, which are clustered within the central DNA binding domain (9). There are several hotspot mutation sites in p53, such as R175H, R248W, and R273H, which occur frequently in cancers(10). Consequently, these mutations make p53 protein defective to induce its target genes for tumor suppression (11). Notably, some mutants acquire new and distinct oncogenic properties, generally referred to as “gain-of-function”, such as the ability to promote tumor progression and metastasis (12, 13). In support of this notion, mutant p53 knock-in mice exhibit significantly different tumor spectra and high incidence of tumor metastasis when compared with p53-null mice (14, 15). Additionally, clinical studies have shown that a high level of mutant p53 is correlated with more aggressive tumors, poorer outcomes, and enhanced resistance to chemotherapeutic drugs (16). Thus, targeting the mutant p53 pathway will lay a foundation to develop new therapeutic approaches for a broad range of cancers.
Rbm38, an RNA-binding protein, has been implicated in cell cycle regulation and differentiation (17–19). Consistent with its cellular activities, Rbm38 is necessary for normal hematopoiesis and loss of Rbm38 leads to accelerated aging and spontaneous tumors in mice (20). The diverse biological functions of Rbm38 are probably due to its role as an RNA-binding protein to regulate multiple targets. For example, Rbm38 regulates mRNA stability of Mdm2 (21), estrogen receptor (22), and p21 (17). In addition, Rbm38 can regulate both wild-type and mutant p53 translation by modulating the binding of eIF4E, a translation initial factor, to the p53 mRNA (23, 24). Interestingly, Rbm38 is a target of the p53 family proteins, including p53, p63, and p73 (17, 25) Thus, Rbm38 and the p53 family form a feedback regulatory loop. Notably, based on the TCGA database and other studies (26–28), Rbm38 expression is altered together with mutant p53 in several types of human cancers, including breast cancer and acute myeloid leukemia. Thus, it is possible that the Rbm38-mutant p53 axis plays a role in tumorigenesis.
In the current study, by using Rbm38-deficient and mutant p53-KI compound mouse models, we showed that Rbm38 deficiency alters the cancer susceptibility of mutant p53-KI mice. We also showed that loss of Rbm38 promotes the development of T-lymphoblastic lymphomas (T-LBLs) via modulating both mutant p53 and Pten. Additionally, we identified a novel mechanism by which Pten is regulated by Rbm38 via mRNA stability. Together, these data suggest that an exquisite regulation among Rbm38, p53, and Pten is critical for preventing T-cell lymphomagenesis.
Materials and Methods
Mutant mice
Rbm38-knockout mice (on a pure C57BL/6 background) were previously generated (20). The p53+/− (29), p53R270H/+ (14) and p53R172H/+ (14) mice (on a C57BL/6 background) were purchased from The Jackson Laboratory. Rbm38+/− mice were crossed with p53+/−, p53R270H/+ and p53R172H/+ mice to generate Rbm38+/−;p53+/−, Rbm38+/−;p53R270H/+, Rbm38+/−;p53R172H/+ mice. Next, Rbm38+/−;p53+/− mice were crossed with Rbm38+/−;p53R270H/+ and Rbm38+/−;p53R172H/+ mice to generate Rbm38−/−; p53R270H/− and Rbm38−/−; p53R172H/− mutant mice, respectively. The p53+/− mice were also crossed with p53R270H/+ and p53R172H/+ mice to generate p53R270H/− and p53R172H/− mice, respectively. All animals and use protocols were approved by the University of California at Davis Institutional Animal Care and Use Committee.
Cell culture, cell line generation, and primary mouse embryonic fibroblasts (MEFs) isolation
U2OS, HCT116, and MiaPaCa2 cells were cultured in DMEM (Dulbecco’s Modified Eagle’s medium, Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA). Raji and Romas cells were cultured in RPMI-1640 supplement with 10% fetal bovine serum. U2OS, HCT116, and MiaPaCa2 were purchased from the American Type Culture Collection (ATCC). Raji and Romas cells were obtained from Dr. Tuscano at UC Davis and were originally purchased from ATCC (30). Cell lines were obtained from ATCC between 2007 and 2016 and used at below passage 20 or within 2 months for this study after reception or thawing. Cells were tested negative for mycoplasma and after thawing and used within two months. Since all cell lines from ATCC have been thoroughly tested and authenticated, we did not authenticate the cell lines used in this study. To generate Rbm38-KO cell lines by CRISPR-Cas9, pSpCas9(BB)-2A-Puro vector expressing Rbm38 guide RNA#1 (5’- GTC GGT ATC TGT CGG GTG GA 3’) was transfected into U2OS whereas vector expressing Rbm38 guide RNA#2 (5’-TTT TCT CCA TCA GGG CAC GT 3’) was transfected into HCT116 and MiaPaCa2 cells. The cells were selected with puromycin and each individual clone was confirmed by western blot analysis. To isolate p53R270H/- and Rbm38−/−;p53R270H/- MEFs, Rbm38+/−;p53+/− mice were bred with Rbm38+/−;p53R270H/+ mice. At 13.5-day, embryos were isolated as described previously (31). The MEFs were cultured in DMEM supplemented with 10% FBS (HyClone), 55 μM β-mercaptoethanol, and 1× non-essential amino acids (NEAA) solution (Cellgro). To isolate p53R172H/- and Rbm38−/−;p53R172H/- MEFs, Rbm38+/−;p53+/− mice were bred with Rbm38+/−;p53R172H/+mice and the MEFs were isolated from 13.5-day embryos.
Western Blot Analysis
Western blot analysis was performed as previously described (32). The antibodies used in this study were as follows: anti-p53(1C12) and anti-Pten (Cell signaling), anti-actin (Sigma), anti-HA (Covance), and anti-Rbm38 (customized).
RNA Interference
Scrambled siRNA (GGC CGA UUG UCA AAU AAU U) and siRNAs against Rbm38 (5′-CAC CUU GAU CCA GCG GAC UUA-3′) were purchased from Dharmacon (Chicago, IL). For siRNA transfection, RNAiMax (Life Technology) was used according to the user’s manual. Both Raji and Romas cells were transfected with scrambled or Rbm38 siRNA at 100 nM for 3 days.
RNA Isolation, RT-PCR, and Quantitative PCR (qPCR)
Total RNA was isolated with Trizol reagent as described (32). cDNA was synthesized with Reverse Transcriptase (promega) and used for RT-PCR. The PCR program used for amplification was (i) 94 °C for 5 min, (ii) 94 °C for 45 s, (iii) 58 °C for 45 s, (iv) 72 °C for 30 s and (v) 72 °C for 10 min. From steps 2–4, the cycle was repeated 22 times for actin or 28 times for Rbm38 and Pten. For qPCR analysis, 20-μl reactions were set up using 2X qPCR SYBR Green Mix (ABgene, Epsom, UK) along with 5 μM primers. The reactions were run on a StepOne plus (Invitrogen) using a two-step cycling program: 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 68 °C for 30 s. A melt curve (57–95 °C) was generated at the end of each run to verify the specificity. The primers for human Pten were forward primer, 5’- ACC CAC CAC AGC TAG AAC TT-3’, and reverse primer, 5’- GGG AAT AGT TAC TCC CTT TTT GTC-3’. The primers for mouse Pten were forward primer, 5’- TGT AAA GCT GGA AAG GGA CGG ACT G −3’ and reverse primer, 5’- AGC AGT GCC ACG GGT CTG TAA TCC −3’. The primers for mouse actin were forward primer, 5’-CCC ATC TAC GAG GGC TAT-3, and reverse primer, 5’-AGA AGG AAG GCT GGA AAA-3’. The primers for human actin were forward primer, CTG AAG TAC CCC ATC GAG CAC GGC A-3’, and reverse primer, 5’-GGA TAG CAC AGC CTG GAT AGC AAC G-3’. The primers for mouse Rbm38 were forward primer 5’-GAC GCA TCG CTC AGA AAG T-3’, and reverse primer, 5’-GAG GAG TCA GCC CGT AGG T-3’.
Plasmids
pcDNA3-Rbm38 and pcDNA3-HA-EGFP expression vector was previously generated (17, 33). To generate HA-tagged EGFP expression vector carrying various regions of Pten 3’-UTR, cDNAs from HCT116 cells were used as template to amplify various regions of Pten 3’UTR. The PCR products were then clone into pcDNA3-HA-EGFP vector via NotI and XhoI. The primers to amplify 3’UTR-A were forward primer, 5’- TTG CGG CCG CAT AGA TAT TCT GAC ACC ACT GAC TCT GAT CCA G-3’, and reverse primer, 5’- TTG CGG CCG CAT AGA TAT TCT GAC ACC ACT GAC TCT GAT CCA G-3’. The primers to amplify 3’UTR-B were forward primer, 5’- TTG CGG CCG CAG TTG GGA CTA GGG CTT CAA TTT CAC TTC-3’, and reverse primer, 5’- CCC TCG AGT TAA ACA GTA GGC TTT GAA GGA CAG CAG G-3’. The primers to amplify 3’UTR-C were forward primer, 5’- TTG CGG CCG CGA TAA TGC CTC ATC CCA ATC AGA TGT CC-3’, and reverse primer, 5’- CCC TCG AGA CAT GTA ATC TGC ATC TGT GGC ATT AGA AAG-3’. The primers to amplify 3’UTR-D were forward primer, 5’- TTG CGG CCG CAC TCA TAA TCC TAT CAC CTG GAG ACA TAG CC-3’, and reverse primer, 5’- CCC TCG AGG GAA AAA TAC TCT CCA TAG GAA GGC ATT TC-3’. The primers to amplify 3’UTR-C1 were forward primer, 5’- TTG CGG CCG CGA TAA TGC CTC ATC CCA ATC AGA TGT CC-3’, and reverse primer, 5’- CCC TCG AGC TTG AAA TAA TAA GTC CCA CTG TAA CAT TGT CTA CT-3’. The primers to amplify 3’UTR-C2 were forward primer, 5’- TTG CGG CCG CAT AAA GTA GAC AAT GTT ACA GTG GGA CT-3’, and reverse primer, 5’- CCC TCG AGG ACT GGC TAC CCA CAT TCT GAT AT-3’. The primers to amplify 3’UTR-C3 were forward primer, 5’- TTG CGG CCG CAG AAT GTG GGT AGC CAG TCA GAC AAA TTC-3’, and reverse primer, 5’- CCC TCG AGA CAT GTA ATC TGC ATC TGT GGC ATT AGA AAG-3’. To generate HA-EGFP expression vector with point mutation for ARE#1, two steps PCR were used. The first round PCR were to amplify two DNA fragments (#1 and #2) using pcDNA3-HA-EGFP-3’UTR-C as template. The forward primer for fragment#1 was the same as the forward primer of 3’UTR-C and the reverse primer was 5’-CAC CAA AAT GTC TTC CCC GGG CCC CGA ATA CTT TTA GAC T-3’. The forward primer for fragment#2 was 5’- AGT CTA AAA GTA TTC GGG GCC CGG GGA AGA CAT TTT GGT G-3’ and the reverse primer was the same as the reverse primer of 3’UTR-C. The second round PCR was performed using a mixture of Fragment #1 and #2 as template with forward and reverse primer of 3’UTR-C. The PCR products were then clone into pcDNA3-HA-EGFP via NotI and XhoI. To generate HA-EGFP expression vector with point mutation for ARE#2, similar strategy was used except that different primers were used to amplify fragment #1 and #2. The primers for fragment #1 were the forward primer of 3’UTR-C and the reverse primer, 5’-TTA AAC ACT GGT GAG CCC CGG GCC CTC AAT GGA AAT GGT-3’. The primers for fragment #2 were the forward primer, 5’- ACC ATT TCC ATT GAG GGC CCG GGG CTC ACC AGT GTT TAA-3’ and the reverse primer of 3’UTR-C. To generate HA-EGFP expression vector with point mutation for both ARE#1 and #2, two steps PCR was performed by using HA-EGFP expression vector with ARE2 point mutation as a template with the primers for generating ARE1 point mutation. To generate a vector expressing Rbm38 guide RNA, a short oligo was inserted into the pSpCas9(BB)-2A-Puro (PX459) vector (34) via BbsI site. To generate Rbm38-KO HCT116 and Mia-PaCa2 cells, the Rbm38 guide RNA sequence is 5’-TTT TCT CCA TCA GGG CAC GT 3’. To generate Rbm38-KO U2OS cells, the Rbm38 guide RNA sequence is 5’- GTC GGT ATC TGT CGG GTG GA-3’.
Histological Analysis
Mouse tissues were fixed in 10% (wt/vol) neutral buffered formalin, routinely processed, and embedded in paraffin blocks. Tissue sections (5 μm) were sectioned and stained with H&E.
Results
Loss of Rbm38 shortens the lifespan of, and alters tumor incidence in, mutant p53R270H/-knockin mice
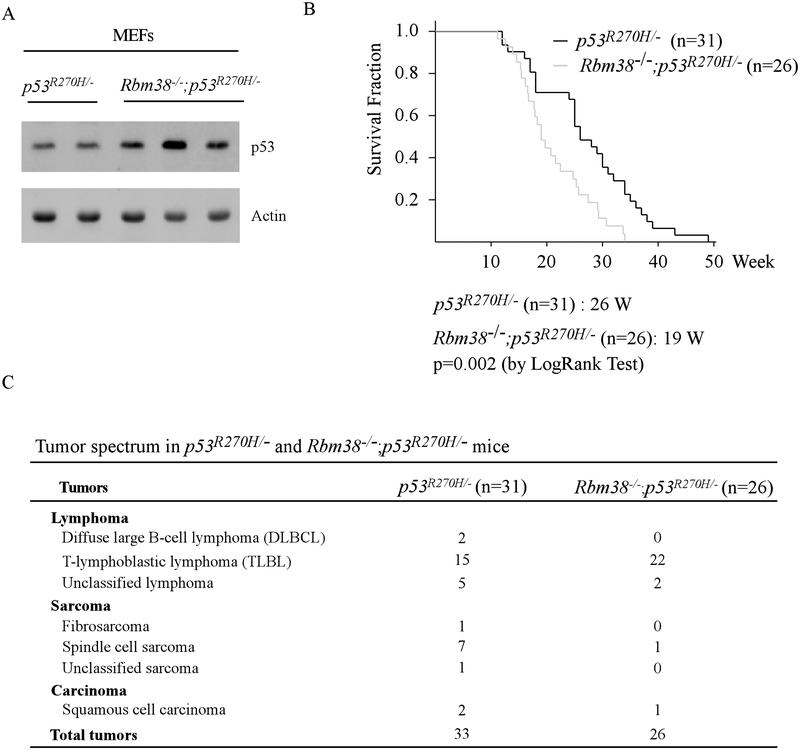
To determine the role of Rbm38-mutant p53 axis in tumorigenesis in vivo, mutant p53R270H (equivalent to human R273H, a contact mutant) knock-in mice were crossed with Rbm38-KO mice to generate a cohort of p53R270H/- and Rbm38−/−; p53R270H/- MEFs. We found that the level of mutant p53(R270H) protein was markedly increased by loss of Rbm38 (Fig. 1A), consistent with our previous report that mutant p53 expression was repressed by Rbm38 (23). Next, a cohort of p53R270H/- (n=31) and Rbm38−/−; p53R270H/- (n=26) mice was generated and monitored throughout their lifespan. We would like to note that p53R270H/- mice has been analyzed for another tumor study, which is recently published (35). We found that except one p53R270H/- mouse, all the p53R270H/- and Rbm38−/−; p53R270H/- mice succumbed to spontaneous tumors (Supplemental Table S1–2). The median lifespan for Rbm38−/−; p53R270H/- mice was 19 weeks, which was significantly shorter than that for p53R270H/- mice (26 weeks) (p=0.002 by Log Rank Test) (Fig. 1B). Moreover, loss of Rbm38 significantly altered the tumor incidence in Rbm38−/−;p53R270H/- mice (Fig. 1C). Rbm38−/−; p53R270H/- mice were prone to lymphomas (p=0.0262 by Fisher’s exact test) whereas p53R270H/- mice were prone to sarcomas (p=0.0323 by Fisher’s exact test). We also noticed that the lymphomas developed in Rbm38−/−; p53R270H/- mice were mostly T-lymphoblastic lymphomas (TLBL) (Supplemental Fig. S1A), which was significantly increased in Rbm38−/−; p53R270H/- mice as compared to p53R270H/- mice (p=0.0027 by Fisher’s exact test). The affected mutant mice usually suffered with short breath due to enlarged thymus in the thoracic cage (Supplemental Fig. S1B). We would like to mention that while this manuscript is in preparation,
Figure 1.
(A) The level of p53 and actin was examined in p53R270H/- and Rbm38−/−; p53R270H/- MEFs. (B) Kaplan Meier curves of p53R270H/- (n=31) and Rbm38−/−; p53R270H/- (n=26) mice. The median survival time was 26 weeks for p53R270H/- mice and 19 weeks for Rbm38−/−; p53R270H/- mice (p=0.002 by LogRank test). (C) The tumor spectrum of p53R270H/- (n=31) and Rbm38−/−; p53R270H/- (n=26) mice. The incidence of TLBLs is significantly higher in Rbm38−/−; p53R270H/- mice than that in p53R270H/- mice (p=0.0027 by Fisher’s exact test). The incidence of sarcomas is significantly lower in Rbm38−/−; p53R270H/- mice than that in p53R270H/- mice (p=0.0323 by Fisher’s exact test).
Loss of Rbm38 shortens the lifespan of, and alters tumor incidence in, mutant p53R172H/-knockin mice
Recent studies indicate that not all p53 mutants have the same oncogenic activities and some p53 mutants have unique activities in terms of the ability to inhibit wild-type p53 and gain-of-functions (36). In this regard, mutant p53R172H/- (equivalent to human R175H, a conformation mutant) knock-in mice were crossed with Rbm38−/− mice to generate a cohort of p53R172H/- and Rbm38−/−; p53R172H/- MEFs. We showed that the level of mutant p53(R172H) protein in Rbm38−/−; p53R172H/- MEFs was increased by loss of Rbm38 as compared to that in p53R172H/- MEFs (Fig. 2A). Next, a cohort of p53R172H/- (n=19) and Rbm38−/−; p53R172H/- (n=19) mice was generated and monitored throughout their lifespan. We found that the median survival time was 24.3 weeks for p53R172H/- mice and 21 weeks for Rbm38−/−; p53R172H/- mice (Fig. 2B). The difference in survival time between p53R172H/- and Rbm38−/−; p53R172H/- mice was statistically significant (p=0.012 by LogRank test). We also found that both p53R172H/- and Rbm38−/−; p53R172H/- mice developed similar types of tumors including TLBL, angiosarcoma, and spindle cell sarcoma (Supplemental Fig. S2). However, loss of Rbm38 altered the tumor incidence in p53R172H/- mice (Fig. 2C and Supplemental Tables S3–4). Specifically, the incidence of TLBL was significantly higher in Rbm38−/−; p53R172H/- mice than that in p53R172H/- mice (p=0.0033 by Fisher’s exact test). By contrast, the incidence of diffuse large B cell lymphoma (DLBCL) was significantly lower in Rbm38−/−; p53R172H/- mice than that in p53R172H/- mice (p=0.0182 by Fisher’s exact test).
Figure 2.
(A) The level of p53 and actin was examined in p53R172H/- and Rbm38−/−; p53R172H/- MEFs. (B) Kaplan Meier curves of p53R172H/- (n=19) and Rbm38−/−; p53R172H/- (n=19) mice. The median survival time was 24 weeks for p53R172H/- mice and 21 weeks for Rbm38−/−; p53R172H/- mice (p=0.012 by LogRank test). (C) The tumor spectrum of p53R172H/- (n=19) and Rbm38−/−; p53R172H/- (n=19) mice. The incidence of TLBLs is significantly higher in Rbm38−/−;p53R172H/- mice than that in p53R172H/- mice (p =0.0033 by Fisher’s exact test). The incidence of diffuse large B cell lymphomas is significantly lower in Rbm38−/−;p53R172H/- mice than that in p53R172H/- mice (p = 0.0182 by Fisher’s exact test).
Loss of Rbm38 increases mutant p53 expression but decreases Pten expression in TLBLs
TLBL is originated from developing thymocytes due to abnormal differentiation and proliferation in the thymus. The increased incidence of TLBL in the compound mutant mice (Fig. 1C and 2C) indicates that loss of Rbm38 may alter the regulatory mechanism for T cell differentiation and proliferation. Thus, the expression of several T-cell regulators, including NFAT1, NFAT2 and Tbet, was examined in the thymi of WT and Rbm38-KO mice. NFAT1 and NFAT2 play a critical role in T-cell development and activation (37–39) whereas Tbet is expressed in CD4+ T lymphocytes committed to Th1 T-cell development (40). We showed that loss of Rbm38 did not alter their expression (Supplemental Fig. S3A). We also examined IFN-γ, which plays a role in inducing Th1 cells (41), and IL6, which controls T cell proliferation, survival, and commitment (42). Again, these cytokines were not altered by Rbm38-KO (Supplemental Fig. S3A). Consistent with this, we showed that the level of NFAT1 and T-bet was not altered by loss of Rbm38 in the TLBLs from p53R270H/- and Rbm38−/−; p53R270H/- mice (Supplemental Fig. S3B) or from p53R172H/- and Rbm38−/−; p53R172H/- mice (Supplemental Fig. S3C).
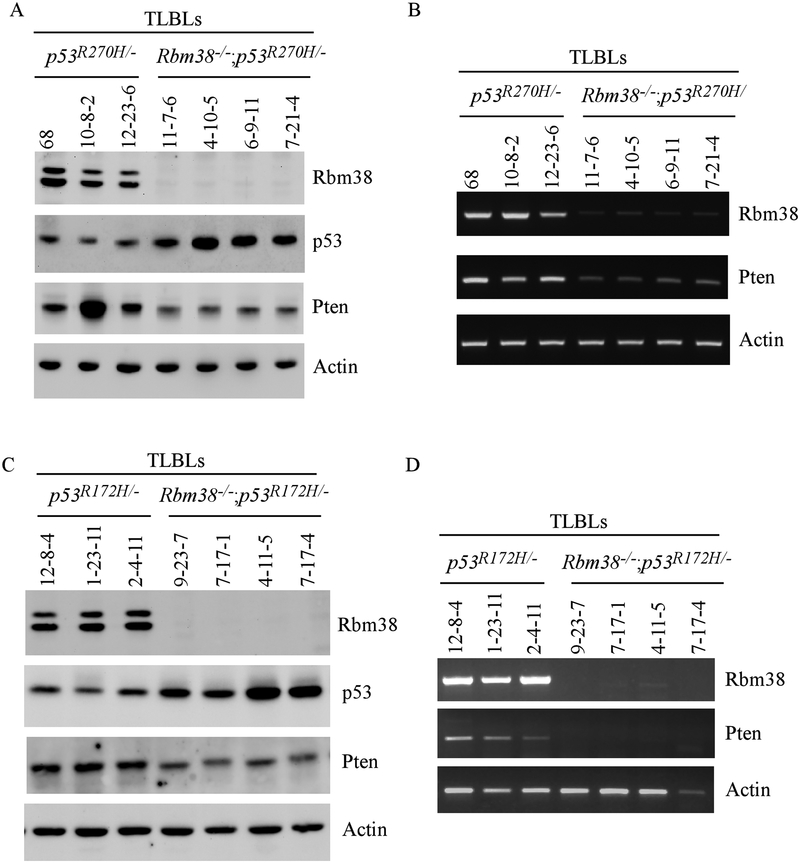
The above data let us speculate that loss of Rbm38 may affect the T-progenitor cell development. To test this, we examined expression of Pten (Phosphatase and TENsin homolog deleted on chromosome 10), a key regulator of T-progenitor cells (43). Notably, Pten+/− mice are prone to TLBLs (44). In addition, loss or reduced expression of Pten was found to be an early event in TLBLs developed in p53-null mice (45). In this regard, the levels of mutant p53(R270H) and Pten were analyzed in TLBLs from p53R270H/- and Rbm38−/−; p53R270H/- mice. We found that the level of mutant p53(R270H) protein was increased by loss of Rbm38 in TLBLs (Fig. 3A), consistent with the above observation in Rbm38−/−;p53R270H/- MEFs (Fig. 1A). Interestingly, we found that the level of Pten protein was decreased in all four TLBLs from Rbm38−/−; p53R270H/- mice as compared to the ones from p53R270H/- (Fig. 3A). Consistent with this, the level of Pten transcript was also decreased in these TLBLs from Rbm38−/−; p53R270H/- mice as compared to the ones from p53R270H/- mice (Fig. 3B). Next, we determined the effect of Rbm38-KO on the expression of mutant p53 and Pten in the TLBLs from Rbm38−/−;p53R172H/- and p53R172H/- mice. As expected, the level of mutant p53(R172H) was also increased by loss of Rbm38 (Fig. 3C). Importantly, the levels of Pten protein and transcript were decreased in the TLBLs from Rbm38−/−; p53R172H/- mice as compared to the ones from p53R172H/- (Fig. 3C–D). Together, these data suggest that loss of Rbm38 promotes the development of TLBLs in Rbm38−/−; p53R270H/- and Rbm38−/−; p53R172H/- mice by concomitantly modulating both mutant p53 and Pten.
Figure 3.
(A-B) The levels of Rbm38, p53, Pten proteins (A) and transcripts (B) were examined in T-lymphoblastic lymphomas from p53R270H/- and Rbm38−/−; p53R270H/- mice. (C-D) The levels of Rbm38, p53, Pten proteins (C) and transcripts (D) were examined in TLBLs from p53R172H/- and Rbm38−/−; p53R172H/- mice.
Loss of Rbm38 decreases Pten expression in the TLBLs from Rbm38−/−;p53−/− mice
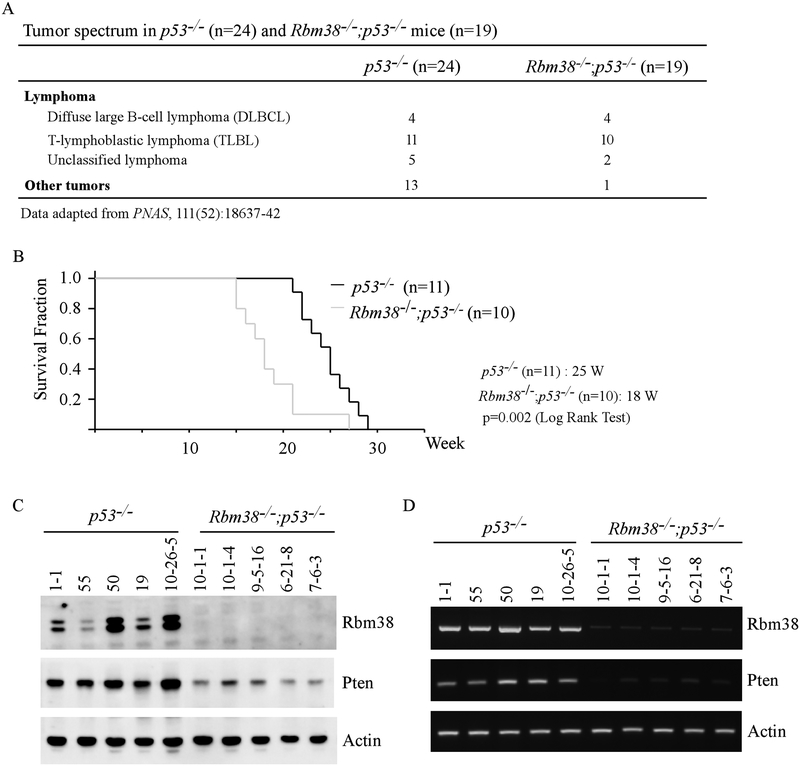
Previously, we showed that Rbm38 deficiency led to increased incidence of lymphoma in Rbm38−/−;p53−/− mice (16 lymphomas out of total 17 tumors) as compared to that in p53−/− mice (20 lymphomas out of total 33 tumors) (p=0.0183 by Fisher’s exact test) (Fig. 4A) (20). In terms of TLBLs, the incidence was not statistically higher in Rbm38−/−;p53−/− mice than that in p53−/− mice (p=0.1305 by Fisher’s exact) (Fig. 4A). However, the median survival time for mice succumbed to TLBLs was significantly shorter for Rbm38−/−;p53−/− mice (18 weeks) than that for p53−/− mice (25 weeks) (p=0.002 by LogRank Test) (Fig. 4B), suggesting that the TLBLs in Rbm38−/−;p53−/− mice are more aggressive than the ones in p53−/− mice. As Pten expression was decreased by loss of Rbm38 in Rbm38−/−;p53R270H/- and Rbm38−/−;p53R172H/- mice, we thus asked whether Pten expression is altered in the TLBLs from Rbm38−/−;p53−/− mice. We would like to note that one p53−/− (ID: 10-26-5) and one Rbm38−/−; p53−/− (ID: 10-26-5) mice were generated for current study. All other mice were generated and used in a previous study (20). Indeed, we found that the levels of both Pten proteins and transcripts were markedly decreased in the TLBLs from Rbm38−/−;p53−/− mice as compared to that from p53−/− mice (Fig. 4C–D).
Figure 4.
(A) The tumor spectrum of p53−/− (n=24) and Rbm38−/−; p53−/− (n=19) mice. These data were adapted from a previous tumor study. (B) Kaplan Meier curves of p53−/− (n=11) and Rbm38−/−; p53−/− (n=10) mice dying from T-lymphoblastic lymphoma. The median survival time was 25 weeks for p53−/− mice and 18 weeks for Rbm38−/−; p53−/− mice (p=0.002 by LogRank test). (C-D) The levels of Rbm38, p53, Pten proteins (C) and transcripts (D) were examined in TLBLs from p53−/− and Rbm38−/−; p53−/− mice. Both p53−/− (ID: 10-26-5) and Rbm38−/−; p53−/− (ID: 10-26-5) mice were generated for current study.
Rbm38 is required for Pten expression
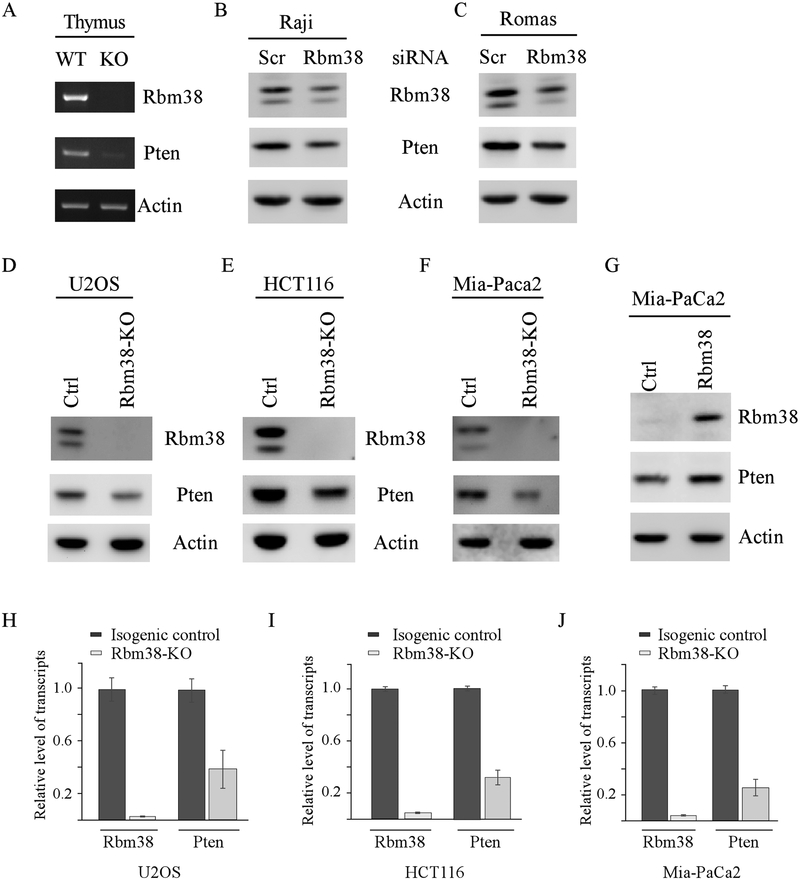
Loss of Rbm38 leads to decreased expression of Pten in the TLBLs from mutant p53 knock-in and p53-null mice (Figs. 3–4). These results let us speculate that Rbm38 directly regulates Pten expression. In this regard, we examined Pten expression in both WT and Rbm38−/− mouse thymi and found that the level of Pten transcript was reduced by loss of Rbm38 (Fig. 5A). Next, to examine whether knockdown of Rbm38 alters Pten expression in human lymphoma cells, Rbm38 siRNA along with a control siRNA were transiently transfected into Raji and Romas cells, both of which are lymphoma cell lines. We found that Rbm38 siRNA led to decreased expression of Rbm38 protein, which was accompanied with a reduction of Pten protein (Fig. 5B–C). Next, to test whether Rbm38 regulates Pten expression in non-lymphoma tissues, various cancer cell lines, including U2OS (osteosarcoma), HCT116 (colon cancer), and Mia-PaCa2 (pancreatic cancer), were used to generate Rbm38-KO cell lines by CRISPR-Cas9 technology. We would like to mention that HCT116 and U2OS cells contain wild-type p53 whereas Mia-PaCa2 cells contain mutant p53(R248W). Sequencing analysis indicated that Rbm38-KO U2OS cells had a 4-nt out of frame deletion in Exon 1 of the Rbm38 gene. Rbm38-KO HCT116 cells had a 4-nt deletion, whereas Rbm38-KO Mia-PaCa2 cells had 1-nt deletion, in exon 4 of the Rbm38 gene. As expected, the level of Rbm38 protein was undetectable in these Rbm38-KO HCT116, U2OS, and Mia-PaCa2 cells as compared to their respective isogenic control cells (Fig. 5D–F, Rbm38 panel). Importantly, the level of Pten protein was decreased by loss of Rbm38 (Fig. 5D–F, Pten panel). Conversely, we found that ectopic expression of Rbm38 increased Pten expression in Mia-PaCa2 cells (Fig. 5G, Pten panel). Next, qRT-PCR analysis was performed to examine whether loss of Rbm38 alters the level of Pten transcripts. Indeed, we found that the level of Pten transcript was reduced by loss of Rbm38 in HCT116, U2OS, and Mia-PaCa2 cells (Fig. 5H–J). Together, these data suggest that Rbm38 is required for Pten expression regardless of p53 genetic status.
Figure 5.
(A) The levels of Rbm38, Pten, and actin transcripts were examined in the thumus from WT and Rbm38−/− mice. (B-C) The levels of Rbm38, Pten, and actin proteins were examined in Raji (B) or Romas (C) cells transiently transfected with scrambled or Rbm38 siRNA at 100 nM for 3 days. (D-F) The levels of Rbm38, Pten, and actin proteins were examined in isogenic control and Rbm38-KO U2OS (D), HCT116 (E), and Mia-Paca2 (F) cells. (G) The level of Rbm38, Pten and actin prpteins were examined in Mia-PaCa2 cells transfected with a control or Rbm38 expression vector for 24 h. (H-J) The levels of Pten and actin transcripts were examined by qRT-PCR in isogenic control and Rbm38-KO U2OS (H), HCT116 (I), and Mia-Paca2 (J) cells.
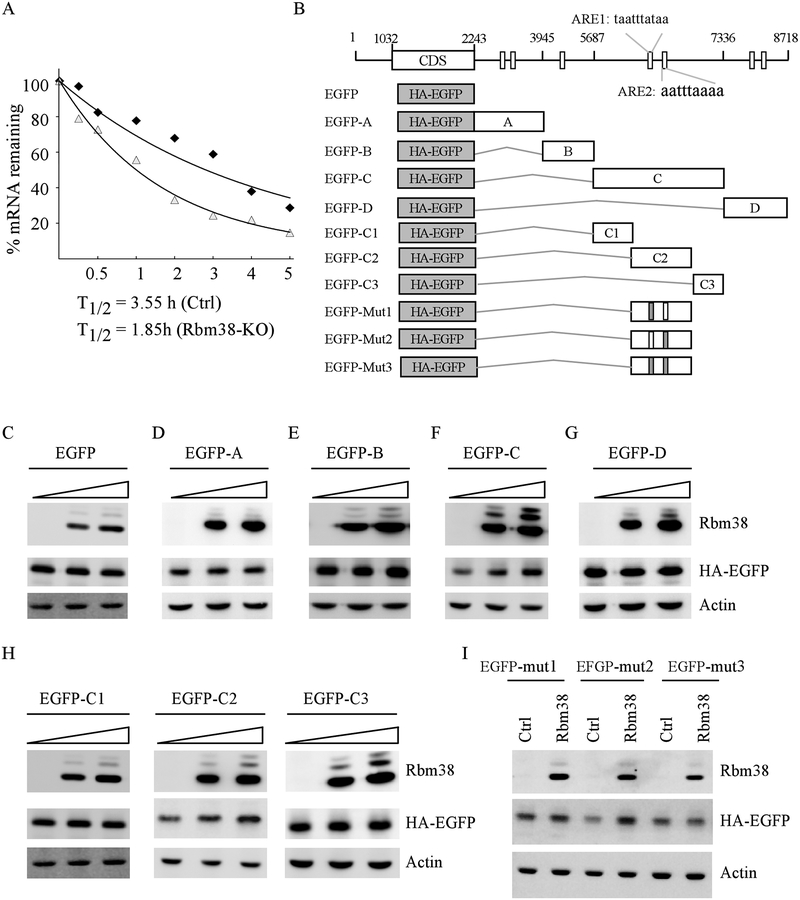
Rbm38 stabilizes Pten mRNA via an AU-rich element in its 3’UTR
As a RNA binding protein, Rbm38 is known to regulate gene expression via mRNA stability (46, 47). Thus, we examined whether Pten mRNA turnover is regulated by Rbm38. To test this, isogenic control and Rbm38-KO MiaPaCa2 cells were treated with DRB (5,6-Dichlorobenzimidazole-β-D-ribofuranoside) for various times and the half-life of Pten mRNA was determined by qRT-PCR. DRB inhibits the elongation step during RNA polymerase II transcription and subsequently, de novo mRNA synthesis (48). Thus, DRB can be used to measure the half-life of a given mRNA, which is defined as the time required to degrade 50% of the mRNA. We found that the half-life of Pten mRNA was decreased from 3.55 h in isogenic control cells to 1.85 h in Rbm38-KO cells (Fig. 6A). As Rbm38 is known to preferentially bind to AU-rich elements (AREs) (17), we searched for and found several AREs in Pten 3’UTR (Fig. 6B). To test this, we generated several EGFP reporters that contain HA-tagged EGFP alone, or together with a fragment from various regions of Pten 3’UTR (Fig. 6B). We found that Rbm38 had no effect on EGFP expression from EGFP reporter vectors that carried only EGFP (Fig. 6C) or the EGFP reporter fused with fused with A (Fig. 6D) or B (Fig.6E) fragment of Pten 3’UTR. Moreover, we found that ectopic expression of Rbm38 led to increased expression of EGFP when the EGFP reporter was fused with the C fragment of Pten 3’UTR (Fig. 6F), but not the one fused with D fragment (Fig. 6G). To further delineate Rbm38 binding site(s) in the C fragment, we generate three additional EGFP reporters that contain C1, C2, or C3 fragment from Pten 3’UTR (Fig. 6B). We found that the reporter that contained the C2 fragment, but not the C1 and C3 fragments, was responsive to Rbm38 (Fig. 6H). Upon close examination, we found two AREs (ARE1 and ARE2) located in the C2 fragment (Fig. 6B). To determine whether these AREs are responsive to Rbm38, we generated three mutant C2 fragments in that the A and U in the ARE1, ARE2, or both were substituted with G and C, respectively (Fig. 6B). We found that mutation of ARE1 nearly abolished, whereas mutation of ARE2 had no effect on, EGFP expression induced by Rbm38 (Fig. 6I). Similarly, Rbm38 was unable to induce EGFP expression when both ARE1 and ARE2 were mutated (Fig. 6I). Together, these data indicated that the ARE1 in Pten 3’UTR is likely responsible for Rbm38 to stabilize Pten mRNA.
Figure 6.
(A) Rbm38 is required for maintaining Pten mRNA stability. The level of Pten transcript was measured by qRT-PCR in isogenic control and Rbm38-KO Mia-Paca2 cells treated with DRB for various times. (B) Schematic representation of Pten transcript and the locations of 3’UTR fragments for reporter assay. AU-rich elements were shown in boxes. The grey box indicates mutation in ARE1 or ARE2. (C-G) Various amounts of Rbm38 expression vector were transfected into p53−/− HCT116 cells along with a fixed amount of EGFP (C) or EGFP expression vectors that contain Pten 3’UTR-A (D), 3’UTR-B (E), 3’UTR-C (F), or 3’UTR-D (G). The levels of Rbm38, HA-EGFP, and actin were examined by western blot analysis. (H) The levels of Rbm38, HA-EGFP, and actin proteins were examined in p53−/− HCT116 cells transfected with various amount of Rbm38 expression vector along with a fixed amount of EGFP vectors that contain Pten 3’UTR-C1, C2, or C3. (I) HA-EGFP vector that contains point mutation in ARE1, ARE2 or both was transfected into p53−/− HCT116 cells together with or without Rbm38 expression vector, followed by western blot analysis to examine the level of Rbm38, HA-EGFP, and actin.
Discussion
Recently, we showed that loss of Rbm38 promotes mutant p53 translation (23). Interestingly, loss of Rbm38 occurs frequently in tumors with mutant p53 (27, 28). Thus, we hypothesize that loss of Rbm38 is necessary for maintaining high levels of mutant p53, which then promotes tumor development. To test this, we generated compound mutant mice containing Rbm38 deficiency and knock-in mutant p53R270H or p53R172H. Indeed, we found that Rbm38 deficiency significantly enhances the cancer susceptibility of p53R270H/- or p53R172H/- mice by shortening the lifespan and altering the tumor incidence (Figs. 1–2). Interestingly, we showed previously that in a p53-heterozygous background, Rbm38 deficiency protects mice from developing tumors at least in part via enhanced wild-type p53 expression (20). These data indicate that Rbm38 deficiency may have differential effects on tumorigenesis depending on p53 status. Thus, p53 status needs to be considered when exploring Rbm38 as a target for cancer treatment.
In this study, we showed that p53R270H/- and p53R172H/- mice had a median survival of 24–26 weeks (Fig. 1B and2B) and were prone to lymphomas (Fig. 1C and2C), consistent with a previous report (14). Importantly, in an Rbm38-null background, the incidence of the TLBLs was markedly increased in both p53R270H/- and p53R172H/- mice along with increased expression of mutant p53 (Figs. 1C, 2C and Fig. 3). These data let us speculate that enhanced mutant p53 expression by loss of Rbm38 is needed to promote the formation of TLBLs, likely through its gain-of-function. In support of this notion, it was reported previously that p53R270/R270H mice developed substantially more TLBLs than p53R270H/- animals owing to increased levels of mutant p53 (42). Similarly, previous study indicated that p53R172H/R172H mice showed a high incidence of lymphomas when compared to p53R172H/+ animals (49). Together, these data indicates that Rbm38 deficiency cooperates with mutant p53 to promote proliferation of T cells with improper pre-TCR signaling and subsequently increases the incidence of TLBLs.
Pten is necessary for maintaining the hematopoietic stem cells and known to inhibit leukemia-initiating cells by suppressing the PI3K-AKT pathway (50). Additionally, both Pten and Rbm38 were found to be decreased in human leukemia patients (27, 51). Here, we showed that Pten expression was reduced by loss of Rbm38 in the TLBLs from mutant p53 knock-in mice (Fig. 3). Thus, it is possible that in a mutant p53 context, a reduced level of Pten by Rbm38 deficiency would activate the PI3K-Akt pathway, which alters T cell differentiation and proliferation, leading to TLBLs. Interestingly, Pten was reduced by loss of Rbm38 in the TLBLs from p53-null mice. Since p53-null mice were prone to TLBLs, it is not surprising that loss of Rbm38 did not significantly increase the incidence of TLBLs in p53-null mice (Fig. 4A). Nevertheless, Rbm38−/−;p53−/− mice succumbed to TLBLs had a shorter lifespan than that for p53−/− mice (Fig. 4B). Thus, we speculate that reduced Pten expression in Rbm38−/−;p53−/− mice would promote proliferation of T cells that are lymphomagenic, leading to more aggressive tumors.
We showed that Rbm38 is required for Pten expression (Fig. 5). We also showed that Rbm38 binds to an AU-rich element in Pten 3′ UTR and subsequently, stabilizes Pten mRNA (Fig. 6). To our knowledge, this is the first report that Pten is regulated by a RNA-binding protein via mRNA stability. Of note, several miRNAs were found to inhibit Pten expression, including miR-21(52), miR-214 (53) and miR-19 (54). In addition, it was found that several Pten competing endogenous RNAs (ceRNAs), such as Pten pseudogene transcripts (55), can compete for miRNAs that targeted sequences common to both Pten mRNA and its pseudo-mRNAs (56). Thus, it will be interesting to examine whether and how Rbm38 play a role in modulating Pten expression via these miRNAs or ceRNAs. Moreover, as Pten 3’UTR contains several AU-rich elements, it would be interesting to examine whether other RNA-binding proteins, such as HuR (19), known to bind an AU-rich element, can cooperate or antagonize with Rbm38 to regulate Pten expression and the biological significance of these regulations.
Our data suggest that Rbm38 controls two “hits” during the development of T-cell lymphomagenesis by concomitantly modulating mutant p53 and Pten. However, several questions remain to be addressed. First, although the TLBLs in Rbm38−/−;p53R270H/- and Rbm38−/−;p53R172H/- mice were histologically indistinguishable from the ones in p53R270H/- and p53R172H/- mice, it would be interesting to examine the pre-TCR signaling pathway in these tumors and further elucidate the role of Rbm38, Pten, and mutant p53 in T-cell development. Second, the compound mice in this study are genetically predisposed to mutant p53 throughout their lifespan. As a result, most of compound mice die early due to TLBLs, which may not fully mimic the development of T-cell malignancies in humans. Thus, it would necessary to use conditional knockout mouse models to elucidate the role of Rbm38-mutant p53 axis in tumor development. Third, as TLBLs have similar clinical presentations and cytogenetic characteristics as T-cell acute lymphoblastic leukemia (T-ALL), these compound mutant mice may be used as a model to test the efficacy of preclinical drugs for treatment of T-ALL.
In summary, we have uncovered a novel mechanism by which Pten is regulated by Rbm38 via mRNA stability. Our data indicate that loss of Rbm38 synergizes with mutant p53 to promote lymphomagenesis though down-regulation of Pten and that the Rbm38-mutant p53/Pten axes may be explored for the treatment of T cell malignancies.
Supplementary Material
Acknowledgements
This work is supported in part by the American Cancer Research Institutional Research Grant (IRG-95-125-13 to J. Zhang) and by National Institutes of Health (CA076069 to XB Chen). We would like to thank Dr. Tuscano at UC Davis to provide the Raji and Romas cell lines.
Footnotes
Competing interests: The authors declare no competing financial interests.
References
- 1.Vogelstein B, Lane D, & Levine AJ (2000) Surfing the p53 network. Nature 408(6810):307–310. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH & Prives C (2009) Blinded by the Light: The Growing Complexity of p53. Cell 137(3):413–431. [DOI] [PubMed] [Google Scholar]
- 3.Kandoth C, et al. (2013) Mutational landscape and significance across 12 major cancer types. Nature 502(7471):333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harms K, Nozell S, & Chen X (2004) The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci 61(7–8):822–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.el-Deiry WS, et al. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75(4):817–825. [DOI] [PubMed] [Google Scholar]
- 6.Kastan MB, et al. (1992) A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71(4):587–597. [DOI] [PubMed] [Google Scholar]
- 7.Nakano K & Vousden KH (2001) PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7(3):683–694. [DOI] [PubMed] [Google Scholar]
- 8.Wu GS, et al. (1997) KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet 17(2):141–143. [DOI] [PubMed] [Google Scholar]
- 9.Olivier M, et al. (2002) The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat 19(6):607–614. [DOI] [PubMed] [Google Scholar]
- 10.Freed-Pastor WA & Prives C (2012) Mutant p53: one name, many proteins. Genes Dev 26(12):1268–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko LJ & Prives C (1996) p53: puzzle and paradigm. Genes Dev 10(9):1054–1072. [DOI] [PubMed] [Google Scholar]
- 12.Oren M & Rotter V (2010) Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2(2):a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller PA & Vousden KH (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25(3):304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olive KP, et al. (2004) Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119(6):847–860. [DOI] [PubMed] [Google Scholar]
- 15.Lang GA, et al. (2004) Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119(6):861–872. [DOI] [PubMed] [Google Scholar]
- 16.He C, Li L, Guan X, Xiong L, & Miao X (2017) Mutant p53 Gain of Function and Chemoresistance: The Role of Mutant p53 in Response to Clinical Chemotherapy. Chemotherapy 62(1):43–53. [DOI] [PubMed] [Google Scholar]
- 17.Shu L, Yan W, & Chen X (2006) RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev 20(21):2961–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, & Carstens RP (2009) ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33(5):591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyamoto S, Hidaka K, Jin D, & Morisaki T (2009) RNA-binding proteins Rbm38 and Rbm24 regulate myogenic differentiation via p21-dependent and -independent regulatory pathways. Genes Cells 14(11):1241–1252. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, et al. (2014) Mice deficient in Rbm38, a target of the p53 family, are susceptible to accelerated aging and spontaneous tumors. Proc Natl Acad Sci U S A 111(52):18637–18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Xu E, & Chen X (2013) Regulation of Mdm2 mRNA stability by RNA-binding protein RNPC1. Oncotarget 4(8):1121–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi L, et al. (2015) Estrogen receptor (ER) was regulated by RNPC1 stabilizing mRNA in ER positive breast cancer. Oncotarget 6(14):12264–12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, et al. (2011) Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev 25(14):1528–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, Zhang J, Chen X, Cho SJ, & Chen X (2013) Glycogen synthase kinase 3 promotes p53 mRNA translation via phosphorylation of RNPC1. Genes Dev 27(20):2246–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, et al. (2003) Isolation and characterization of fourteen novel putative and nine known target genes of the p53 family. Cancer Biol Ther 2(1):55–62. [DOI] [PubMed] [Google Scholar]
- 26.Xue JQ, et al. (2014) RNA-binding protein RNPC1: acting as a tumor suppressor in breast cancer. BMC Cancer 14:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wampfler J, Federzoni EA, Torbett BE, Fey MF, & Tschan MP (2016) The RNA binding proteins RBM38 and DND1 are repressed in AML and have a novel function in APL differentiation. Leuk Res 41:96–102. [DOI] [PubMed] [Google Scholar]
- 28.Leveille N, et al. (2011) Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun 2:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacks T, et al. (1994) Tumor spectrum analysis in p53-mutant mice. Curr Biol 4(1):1–7. [DOI] [PubMed] [Google Scholar]
- 30.Abuhay M, et al. (2016) The HB22.7-vcMMAE antibody-drug conjugate has efficacy against non-Hodgkin lymphoma mouse xenografts with minimal systemic toxicity. Cancer Immunol Immunother 65(10):1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scoumanne A, Cho SJ, Zhang J, & Chen X (2011) The cyclin-dependent kinase inhibitor p21 is regulated by RNA-binding protein PCBP4 via mRNA stability. Nucleic Acids Res 39(1):213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dohn M, Zhang S, & Chen X (2001) p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20(25):3193–3205. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M, Xu E, Zhang J, & Chen X (2015) PPM1D phosphatase, a target of p53 and RBM38 RNA-binding protein, inhibits p53 mRNA translation via dephosphorylation of RBM38. Oncogene 34(48):5900–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ran FA, et al. (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8(11):2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang HJ, et al. (2017) Ninjurin 1 has two opposing functions in tumorigenesis in a p53-dependent manner. Proc Natl Acad Sci U S A 114(43):11500–11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mello SS & Attardi LD (2013) Not all p53 gain-of-function mutants are created equal. Cell Death Differ 20(7):855–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macian F (2005) NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol 5(6):472–484. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida H, et al. (1998) The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production. Immunity 8(1):115–124. [DOI] [PubMed] [Google Scholar]
- 39.Peng SL, Gerth AJ, Ranger AM, & Glimcher LH (2001) NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity 14(1):13–20. [DOI] [PubMed] [Google Scholar]
- 40.Szabo SJ, et al. (2000) A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100(6):655–669. [DOI] [PubMed] [Google Scholar]
- 41.Lighvani AA, et al. (2001) T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A 98(26):15137–15142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter CA & Jones SA (2015) IL-6 as a keystone cytokine in health and disease. Nat Immunol 16(5):448–457. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, et al. (2010) Distinct roles for PTEN in prevention of T cell lymphoma and autoimmunity in mice. J Clin Invest 120(7):2497–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki A, et al. (1998) High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol 8(21):1169–1178. [DOI] [PubMed] [Google Scholar]
- 45.Dudgeon C, et al. (2014) The evolution of thymic lymphomas in p53 knockout mice. Genes Dev 28(23):2613–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu E, Zhang J, & Chen X (2013) MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability. Oncogene 32(17):2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Jun Cho S, & Chen X (2010) RNPC1, an RNA-binding protein and a target of the p53 family, regulates p63 expression through mRNA stability. Proc Natl Acad Sci U S A 107(21):9614–9619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zandomeni R, Zandomeni MC, Shugar D, & Weinmann R (1986) Casein kinase type II is involved in the inhibition by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J Biol Chem 261(7):3414–3419. [PubMed] [Google Scholar]
- 49.Terzian T, et al. (2008) The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev 22(10):1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yilmaz OH, et al. (2006) Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441(7092):475–482. [DOI] [PubMed] [Google Scholar]
- 51.Gutierrez A, et al. (2009) High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood 114(3):647–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lou Y, Yang X, Wang F, Cui Z, & Huang Y (2010) MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int J Mol Med 26(6):819–827. [DOI] [PubMed] [Google Scholar]
- 53.Yang H, et al. (2008) MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res 68(2):425–433. [DOI] [PubMed] [Google Scholar]
- 54.Mavrakis KJ, et al. (2010) Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol 12(4):372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poliseno L, et al. (2010) A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465(7301):1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tay Y, et al. (2011) Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147(2):344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.