Abstract
Yellow fever (YF) was one of the most dangerous infectious diseases of the 18th and 19th centuries, resulting in mass casualties in Africa and the Americas. The etiologic agent is yellow fever virus (YFV) and its live-attenuated form, YFV-17D, remains one of the most potent vaccines ever developed. During the first half of the 20th century, vaccination combined with mosquito control eradicated YFV transmission in urban areas. However, the recent 2016–2018 outbreaks in areas with historically low or no YFV activity have raised serious concerns for an estimated 400–500 million unvaccinated people who now live in at-risk areas. Once a forgotten disease, we highlight here that YF still represents a very real threat to human health and economies. As many gaps remain in our understanding of how YFV interacts with the human host and causes disease, there is an urgent need to address these knowledge gaps and propel YFV research forward.
Keywords: yellow fever, yellow fever virus, yellow fever vaccine, flaviviruses, animal models, viral pathogenesis
Yellow fever
Yellow fever (YF), caused by yellow fever virus (YFV) which is the prototype member of the Flavivirus genus, has historically been considered one of the most dangerous infectious diseases [1]. Endemic to tropical and sub-tropical regions of South America and Africa, YFV is transmitted to humans via mosquitoes of the Haemogogus, Sabethes and Aedes genera. YFV mostly circulates in a sylvatic (or jungle) cycle between mosquitoes and non-human primates (NHPs). However, human encroachment into more rural areas can introduce YFV into urbanized areas (urban cycle), resulting in human-to-human viral circulation via anthropophilic mosquitoes (Box 1) [1].
Box 1: Yellow Fever Transmission Cycles in Africa and South America.
In Africa, YFV circulates in three main ecosystems:
The rainforest. In this homogeneous ecosystem, the incidence of YF outbreaks is low. Aedes africanus maintains a sylvatic cycle with a low transmission rate to humans.
The forest-savanna ecotone/moist savanna. Unique to Africa, this ecosystem hosts an intermediate sylvatic cycle between human and anthropophilic Aedes mosquitoes, which co-exist at a higher rate than in the rainforest. This intermediate cycle provides an easy bridge for the viral disease to reach more densely populated areas, where an urban cycle can be initiated. The existence of this intermediate cycle is one of the reasons why YF epidemics are usually of higher intensity in Africa than in South America (Fig. 1).
The dry savanna. This is the ecosystem where the urban cycle occurs. In West Africa, urban cycles are mediated primarily by the anthropophilic Aedes aegypti mosquitoes, which transmit the virus from human to human. In East Africa, another Aedes mosquito species, Aedes bromelia, is the dominant vector mediating YF urban cycles.
In South America, YFV circulates in two main ecosystems:
The rainforest. A sylvatic cycle is maintained between mosquitoes of the Haemogogus and Sabethes genera and non-human primates.
The urban areas. Although Haemogogus and Sabethes mosquitoes primarily feed on monkeys, human transmission can occur. The subsequent return of infected humans to non-endemic, more densely populated areas can then trigger an urban cycle, which is maintained by Aedes aegypti mosquitoes.
Annually, there are approximately 80,000–200,000 YFV cases worldwide. The number of fatalities annually is commonly estimated as 30,000–60,000, with a case fatality rate (CFR) ranging from 20–60% [1]. However, no exhaustive epidemiologic studies have been conducted to fully support these estimates.
YF disease is mainly characterized by a period of flu-like symptoms that can become more severe [2]. As no treatment for YF is available, mass vaccination campaigns with the highly potent, live-attenuated YFV vaccine, termed YFV-17D, are currently the only weapon to fight the disease and prevent future outbreaks. Mass vaccination campaigns during the 1940–1950s and during the 2000s had a significant impact on containing YFV outbreaks ([3, 4] and i). However, between the 1960s and the mid-2000s, vaccination coverage significantly decreased in endemic areas and was associated with a major upsurge in YFV outbreaks in South America and Africa (Fig. 1).
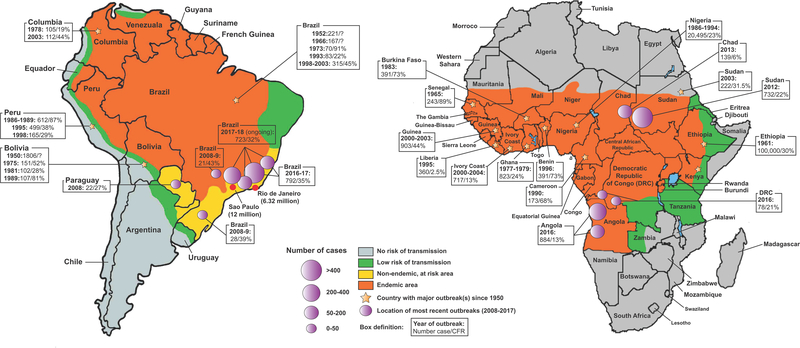
Figure 1. YFV endemic areas and epidemiological distribution of YF outbreaks since 1950.
YFV endemic areas (orange) in South America (left) and Africa (right) are shown. Non-endemic, at risk areas (yellow), areas with low risk of transmission (green) and no risk of transmission (grey) are also displayed. On each continent, countries where major YF outbreaks occurred between 1950 and 2018 are marked by a yellow star, and epidemiological information (number of cases/CFR) for some of these outbreaks are shown. Precise locations of the most recent YF outbreaks (2008–2018) are represented by purple circles, sized in proportion to the intensity of the reported outbreak. Data were extracted from the WHO Global Yellow Fever Data Base (i) and from the periodic WHO “Yellow Fever Situation Reports” (viii). Epidemiological information related to the ongoing 2017–2018 outbreak in Brazil shows the number of confirmed cases and related CFRs reported between July 1, 2017 and February 27, 2018 (vi).
However, in 2016–2018, intense YFV reemergence events have been observed in non-endemic areas and in endemic areas with historically low YFV activity, all displaying low vaccination coverage ([5–8]; ii-vi) [4]. These unusual resurgence events emphasize the urgency for reconsidering YFV as a serious threat to human health, as well as the need for better monitoring and understanding this disease. In this review, we concisely summarize the most recent knowledge on YF epidemiology, disease and YFV biology. We underscore that YF still represents a major threat and that a better understanding of the host-virus interactions and molecular mechanisms governing viral pathogenicity are critical for managing future outbreaks. Finally, we highlight several approaches to address these challenges and enhance our knowledge of YFV biology and YF disease.
1. A short history of yellow fever and vaccination efforts
Despite being endemic to tropical and sub-tropical regions of South America and Africa, numerous genetic and epidemiological lines of evidence support an African origin of YFV [1, 9]. Its introduction to the Americas, likely through the slave trade in the 16th century, triggered a dramatic chain reaction of outbreaks across the continent between the 17th and 18th centuries [10, 11]. The sub-tropical climate conditions and presence of permissive mosquito species in Central America were favorable to the introduction of YFV, which then rapidly spread toward non-endemic, more populated areas such as the coastal cities of the eastern United States. During the 18th and 19th centuries, at least 25 major outbreaks killing hundreds of thousands occurred in America, including in New York City, Philadelphia, Baltimore and New Orleans [11]. The 1793 outbreak in Philadelphia, which killed 9% of the city’s population (approximately 5,000 deaths), is a prime example of the threat YF represented for human health and economic development during this period [12]. Besides its considerable impact on human health, YF also had a profound effect on world geopolitics and economic stability. During the Spanish-American War in Cuba in 1898, YF killed more soldiers than the battlefield [13]. Thousands were also killed during the construction of the Panama Canal between 1904 and 1914, delaying the completion of this strategically important project [14].
The massive YF casualties during the Spanish-American War resulted in the U.S. Army creating a special commission, the Reed Yellow Fever Commission led by Walter Reed, to identify the mechanism behind YF transmission [15]. In 1900, consistent with the hypothesis of a Cuban physician, Carlos Finlay, the commission identified mosquitoes as the vector of YF. This was the first observation of disease transmission via an arthropod host, and it highlighted potential strategies to contain YF spread. Between 1900 and 1920, mosquito control programs led to the eradication of urban yellow fever in the U.S. and in several Central and South American countries.
In 1915, the Rockefeller Foundation Yellow Fever Commission was part of a major scientific effort to identify the cause of YF [16]. In 1927, members of this commission isolated the etiologic agent of YF, YFV, by serially passaging in rhesus macaques the serum of a mild African case of YFV. The virus isolated, subsequently designated as YFV-Asibi after the name of the originating patient, demonstrated enhanced virulence in rhesus macaques after serial passages [17]. In mice, only intracranial injection of YFV-Asibi induced significant signs of disease, and death resulted from fatal encephalitis without signs of human-like viscerotropic disease [18]. Interestingly, serial passaging of the Asibi strain in mouse brains attenuated its virulence when injected in macaques intraperitoneally [18] and conferred protection against YFV-Asibi [19]. However, when injected intracranially, the mouse-adapted virus displayed elevated neurovirulence in macaques [19], raising concerns about its use as an immunogen.
Thus, Max Theiler and colleagues adopted a different approach. After 235–240 passages of the Asibi virus in mouse embryonic tissues and medium containing minced whole chick embryo, an attenuated strain – called 17D – was isolated [20]. Following subcutaneous injection of 17D into macaques, only a mild, generalized infection was observed without evidence of significant virulence. Following subcutaneous injection of 17D in a cohort of human patients neither previously exposed or immunized to YFV, neutralizing antibodies were detectable in the serum of most individuals two weeks post vaccination [21], and no major clinical manifestations of disease were observed.
Following this preliminary evidence, larger scale human studies confirmed the potent immunogenicity of YFV-17D, and subsequent mass vaccination campaigns had a considerable impact on preventing new outbreaks worldwide [16]. In addition to YFV-17D, another YFV attenuated vaccine, the French neurotropic vaccine strain (FNV), was developed concurrently [22]. Despite successful mass vaccination campaigns in the 1950s-’60s, vaccine production was abandoned in 1982 because of an unacceptable rate of vaccine-induced encephalitis [16].
By the end of the 20th century, YF had exchanged its status as a major threat to become a neglected disease. With its high seroconversion rate, its persistent long-term immunity (for up to 30–35 years post-vaccination) [23] and the 500 million vaccinations administered since then, YFV-17D is still recognized as one of the most effective vaccines ever created.
2. Yellow fever: an actual threat
Although mass vaccination campaigns in YFV endemic areas in the 1940s-’50s and 2000s successfully contained YFV transmission and disease [4, 24], unusual reemergence events have been recently reported in areas of Africa and South America associated with low or no YFV activity (ii-vi).
In Africa, YFV has been historically endemic to the tropical and subtropical regions from Guinea to Ethiopia (Fig. 1). Outbreaks in West and East Africa account for the great majority of YF cases (around 90%) worldwide each year. In 2013, WHO reported an estimated 84,000–170,000 cases of YF and 29,000–60,000 deaths in Africa ([3]; vii). In West Africa, decreased vaccination efforts after the 1950s were associated with a surge in major YF outbreaks in the area until the mid-2000s (Fig. 1). Between 1986 and 1994, a major outbreak in Nigeria resulted in a reported 20,495 cases and a CFR up to 30% in certain localities (i). These outbreaks triggered new mass vaccination campaigns in the mid-2000s that have successfully contained transmission since then. In contrast, the vaccination coverage in endemic areas of East Africa with historically less YF activity has remained very low over the past 70 years. This low coverage likely contributed to the rise of dramatic outbreaks in the areas over the past 50 years, such as in Ethiopia in 1961 where more than 100,000 cases and 30,000 deaths were reported (i). More recently, in 2016–2017, the worst African YF outbreak in 30 years was reported in Angola, an endemic area where YFV activity is infrequent and the vaccination coverage lower than in West Africa. The outbreak subsequently spread to the Democratic Republic of Congo (and potentially Uganda) (Fig. 1). Between December 2015 and October 2016, 884 confirmed cases and 121 deaths were confirmed in these countries (ii), triggering a mass vaccination campaign of 30 million people.
In South America, YFV endemic areas extend from the tropical region of southern Paraguay to northern Colombia and Venezuela. Cases in this subcontinent usually account for a small fraction of the total annual number of cases worldwide. YF activity follows a specific weather trend in South America, with outbreaks mostly recorded during rainy season from September/October to April.
Following vaccination efforts in the middle of the 20th century, YF outbreaks significantly decreased in South America despite infrequent, but sometimes severe, reemergence events, such as in Peru in 1995 (Fig. 1). However, over the past decade, several South American countries have experienced major YF resurgence events. In 2008–2009, an outbreak in non-endemic areas of the Brazilian states of São Paulo and Rio Grande do Sul resulted in human fatalities (case fatality rate, CFR 39–43%) (Fig. 1). Despite no major outbreaks between the 2010–2015 period, an exceptional outbreak occurred in Brazil between December 2016 and May 2017, where 792 confirmed human cases (CFR 35%) were reported (Fig. 1) (iii). Although spread of the disease predominantly fell between July 2017 and early January 2018 (iv), the number of cases dramatically increased over the first three months of 2018, reaching a total of 723 cases and 274 deaths as of February 28, 2018 (v, vi). More concerning, this 2018 resurgence was associated with a significantly higher number of cases in the non-endemic areas of the states of Rio de Janeiro, São Paulo, and Minas Gerais (v, vi), suggesting that increasingly intense resurgences of YFV might be connected to an ongoing establishment of the virus in more densely populated areas with low vaccination coverage (Fig. 1).
To prevent potentially dramatic urban cycles in these areas, a mass vaccination campaign targeting more than 23 million people was started by the Brazilian authorities in January 2018 (vi). However, as of February 27, 2018, only 23% of the targeted individuals have been vaccinated. As worldwide stocks of YFV-17D are experiencing a significant shortage, such campaigns use only one-fifth of the regular vaccine dose, which is still sufficient to provide immunity for at least one year (vi; [25, 26]).
Besides low vaccination coverage, the causes of recent YF outbreaks in areas with historically low or no YFV activity remain unclear. The absence of management policies of the mosquito population, the increase of human YFV circulation locally, or the expansion of human activities into YFV endemic areas could all represent additional drivers of YFV reemergence. With increased temperatures and rainfall intensity, global warming could also favor mosquito reproduction and YFV emergence in previously unaffected areas. Research efforts aiming to elucidate the ecological connections between YFV, its vector and its environmental niche will be of critical importance to more easily anticipate and prevent future epidemics.
3. YFV replication cycle: brief overview and recent findings
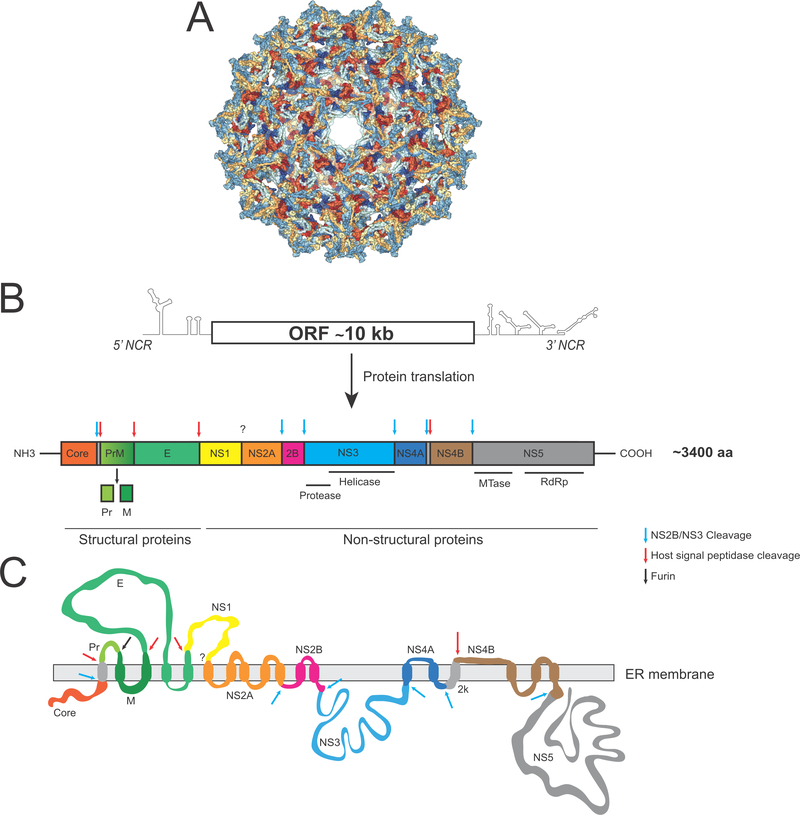
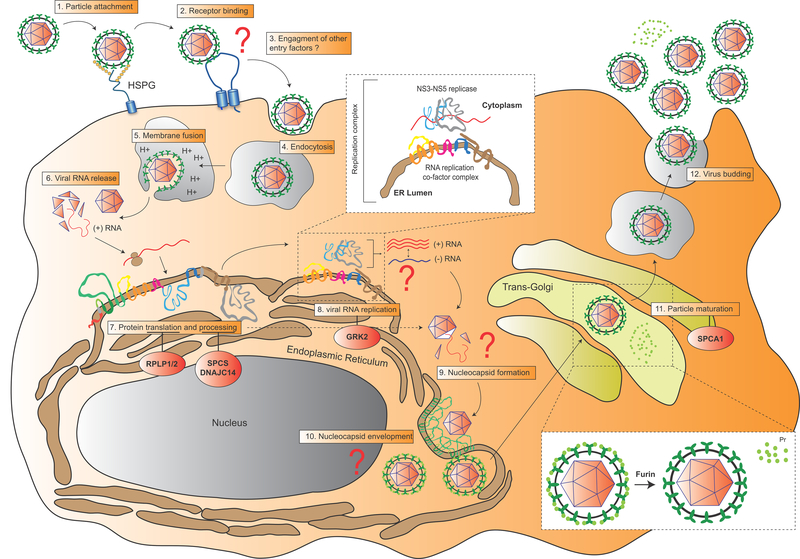
YFV is an enveloped virus that contains a single-stranded, positive-sense RNA genome of about 11,000 nucleotides (Fig. 2). A single open reading frame encodes for a large polyprotein of 3,400 amino acids that is processed into ten viral proteins: three structural proteins (Core, PrM and E) and seven non-structural proteins (NS; NS1, NS2A-2B, NS3, NS4A-B and NS5) [27] (Fig. 2). YFV binds in a non-specific manner to glycosaminoglycan heparan sulfate on the surface of host cells such as hepatocytes or dendritic cells (DCs) [28, 29] (Fig. 3). However, the host cellular receptor to which E, the major YFV envelope glycoprotein, binds before virus fusion remains unknown. Following entry via clathrin-mediated endocytosis [30] and release of the viral genome into the cytoplasm, the viral RNA genome is translated into a large polyprotein and processed by cellular signal peptidases and the NS2B/3 viral protease [27, 31–38] (Fig. 2, 3). Several host factors are involved in flavivirus protein processing or translation, such as the endoplasmic reticulum-associated signal peptidase complex (SPCS; involved in Pr-E junction processing) [39], DNAJC14 (involved in regulation of polyprotein processing) [40, 41] and the ribosomal proteins RPLP1 and RPLP2 (important for polyprotein translation) [42]. Following effective polyprotein translation and processing, the formation of the RNA replication complex − likely via recruitment of NS1 and the NS3-NS5 replicase complex by NS2A, NS4A and NS4B [43–50] − along with NS4A-induced membrane rearrangements [51, 52] promotes active viral RNA replication (Fig. 3). The G protein-coupled receptor kinase 2 (GRK2) is also suggested to play a role in YFV viral RNA replication as well as during dengue and hepatitis C virus infections [53].
Figure 2. YFV genome organization.
A. Cryo-EM representation of an immature YFV particle (PDB 1NA4). B. Schematic representation of YFV viral RNA and polyprotein. Each viral protein is represented using a distinct color. Arrows indicate cleavage sites in the polyprotein that are processed by proteases of cellular (red or black arrow) or viral (blue arrow) origin. C. Schematic representation of the YFV polyprotein anchored into the endoplasmic reticulum (ER) membrane following translation.
Figure 3. Schematic representation of YFV life cycle.
Key steps of the YFV replication life cycle are displayed from 1 to 11. The few identified host factors regulating some of these steps are shown in red circles (see section 3 for description). The viral RNA replication step and the PrM-E maturation step are enlarged in white boxes. Major gaps in our understanding of specific steps of the life cycle are highlighted by red question marks.
The assembly of YFV and other flavivirus viral particles is poorly understood. Newly assembled YFV nucleocapsids, composed of core proteins and viral RNA, are thought to be engulfed within ER membranes harboring the envelope glycoproteins E and PrM [54] (Fig. 3). These immature particles migrate to the trans-Golgi network where they mature and undergo glycosylation. During this stage, PrM is cleaved into two subunits, M and Pr, by the cellular protease furin [55]. The release of Pr from the remaining E-M complex is a critical step for subsequent exocytosis [54, 55] (Fig. 3). Besides furin, other host factors are important for YFV particle maturation in the trans-Golgi network, such as the calcium pump SPCA1 whose role in glycoprotein maturation is conserved across numerous viral families [56].
Altogether, despite the identification of several cellular factors important for infection, the life cycle of YFV remains incompletely understood. Efforts to better understand the cellular mechanisms orchestrating entry, assembly and replication are still urgently needed to unlock the development of potent and specific antiviral strategies.
4. Pathogenesis and immune responses
4.1. YFV pathogenesis and immune responses
Following the bite of an infected mosquito, individuals typically experience an incubation period of 3 to 6 days, subsequently displaying flu-like symptoms prior to a remission period of 1 to 2 days. Following remission, some patients (20–60%) progress to a more toxic phase of disease, characterized by hemorrhagic fever, jaundice, thrombocytopenia, liver and renal failure [2]. These pathological features can lead to more generalized multi-organ dysfunction, vasculopathy and even death.
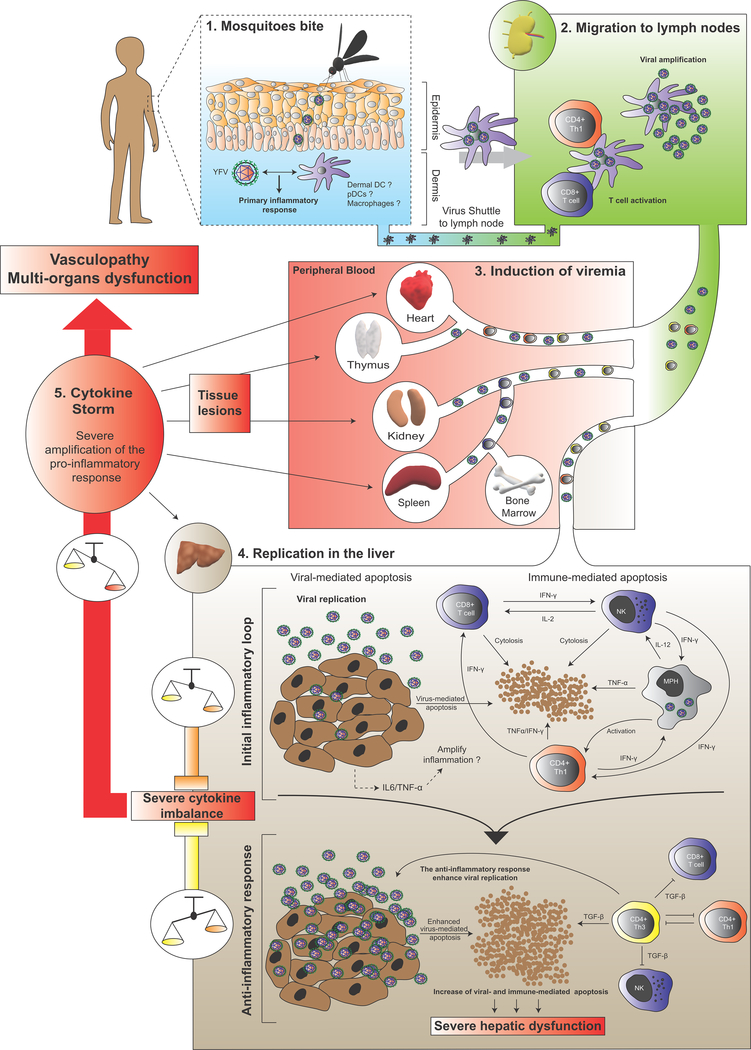
YF pathogenesis is viscerotropic in humans with viral replication in the liver central to the establishment of disease [57]. Current models suggest that after YFV transmission from the mosquito salivary glands to the host’s dermal environment, the virus infects DCs and circulates to the lymph nodes (Fig. 4). In this tissue, YFV amplifies and primes the cellular immune response before spreading through the peripheral blood, ultimately reaching the liver for active replication. In this main target organ, YFV induces hepatocyte apoptosis and lytic necrosis, which, combined with steatosis, results in most of the liver damage observed during infection (Fig. 4). Significant lesions can also occur in the heart, thymus, kidney and spleen, where evidence of replication has been reported in humans [58] and/or animal models [59, 60]. However, the extent of viral replication in these tissues and contribution to lesion formation remain unclear as viral antigens have only been detected in the liver of the YF rhesus macaque model [61]. Studies in human cells suggest virulent YFV and/or YFV-17D display(s) a very broad tissue tropism and can replicate in hepatocytes [62–64], various hematopoietically-derived cells (including DCs [65–67], monocyte-derived macrophages (MDMs) [67], T cells [68], Kupffer cells [69]) and endothelial cells [70]. Recently, an in vivo study using humanized mice, i.e. immunodeficient mouse engrafted with components of a human immune system, unveiled a broad, human-specific cellular tropism of YFV-17D for several human hematopoietic cell lineages [71]. More extensive studies are required to establish an exhaustive profile of YFV tissue tropism.
Figure 4. Model of YF pathogenesis.
Schematic model of the YF-induced pathogenesis process, starting from a mosquito bite and leading to hepatic apoptosis and cytokine storm. The pathogenesis process is divided into five distinct steps (noted from 1 to 5), and each tissue compartment into which YFV circulates is displayed in a distinctly colored box. Intense viral replication in the liver is thought to trigger a molecular chain reaction inducing severe cytokine imbalance and pro-inflammatory cytokine secretion, leading to severe vasculopathy and multi-organ dysfunction. DC, dendritic cells; pDCs, plasmacytoid dendritic cells.
The precise mechanisms of YFV-induced pathogenesis are poorly understood, especially due to the scarcity of cost-effective animal models that recapitulate human features of YFV pathogenesis. Our limited knowledge is based on human tissue biopsies from fatal YF cases [72–74] and indirect pathogenesis observations in animal models such as rhesus macaques [61, 75, 76] and hamsters [77]. Hepatocyte apoptosis is considered central in YF pathogenesis [57] (Fig. 4). In addition to the virus-induced cytopathic effect, several studies suggest that the immune response itself, via a systemic and unbalanced cytokine response (or cytokine storm), is a major driver of hepatotoxicity and YF disease [72, 78, 79] (Fig. 4). Th1 CD4+ T cells and, to a larger extent, Th3 CD4+ T cells have been detected in the livers of patients who succumbed to YF [72, 73]. Th1 CD4+ T cells of infected patients expressed the pro-inflammatory cytokines TNF-α and interferon (IFN)-γ, with TNF-α an important mediator of liver injury [80] (Fig. 4). Additionally, activated macrophages, major producers of TNF-α, were found in the livers of patients with fatal YF [74]. Excessive TNF-α concentration could also enhance CD8+ T cell cytolysis activity and liver damage. In parallel, the large number of Th3 CD4+ T cells found in the livers of patients with fatal YF expressed the pro-apoptotic cytokine TGF-β [72, 73]. TGF-β is a potent anti-inflammatory protein and a strong pro-apoptotic inducer [81], suggesting that an unbalanced pro- and anti-inflammatory cytokine response by CD4+ T cells could be central to the immunopathogenesis of YF (Fig. 4). The pro-inflammatory response of infected hepatocytes and liver endothelial cells also likely contributes to the cytokine imbalance in the liver during infection.
4.2. Innate and adaptive immune responses to yellow fever vaccination
Due to the scarcity of animal models and difficulty accessing samples from patients with fatal YF over the full course of infection, most of our knowledge of the immune response to YFV is derived from human cohorts vaccinated with YFV-17D.
The live-attenuated YFV-17D virus has been of long-standing interest for uncovering key mechanisms regulating protective immunity in humans. Multiple studies have been conducted to delineate the innate and adaptive immune responses in cohorts of human YFV-17D vaccinees at both the transcriptomic and cellular levels [82–88]. With a focus on vaccinees’ peripheral blood mononuclear cells (PBMCs), the immune response to YFV-17D appears to be a complex, multi-lineage and polyfunctional response that mobilizes all arms of the immune system. Upon vaccination, human vaccinees display robust innate immune activation involving effectors and transcription factors of interferon pathways, inflammasome and complement [82–84]. An early transcriptomic signature of vaccination in PBMCs has also been established as a correlate of vaccine immunogenicity [82], supporting the idea that a coordinated and profound activation of multiple arms of the innate immune response is critical for a potent and sustained adaptive response to YFV-17D. A strong, antigen-specific T cell response and persistent memory B cell response seem to represent the hallmarks of long-term protective immunity, as YFV-specific memory T cells and IgG antibodies can persist for decades in patient sera [23, 88–90]. Interestingly, YFV-17D can also induce long-term immunity in immunocompromised individuals at levels comparable to those of healthy individuals [91].
Several studies have described the broad and polyfunctional antigen-specific CD4+ and CD8+ T cell response upon vaccination [85–87, 92]. During the first two weeks post vaccination, peripheral YFV-specific CD8+ T cells divide extensively and then evolve rapidly toward a polyfunctional, long-lived CCR7- CD45RA+ memory phenotype [85, 87, 92]. Interestingly, YFV-specific CD8+ T cells isolated in the blood of vaccinees 10 years post vaccination possessed a similar epigenetic profile to those of early effector cells, suggesting the importance of the pool of memory CD8+ T cells in YFV-17D protective immunity [90]. During the early stages of YFV-17D infection, activation of DCs and natural killer cells (NK) is likely critical for bridging potent innate immune activation and robust priming of the T cell response by enhanced antigen presentation to T cells [65, 93–95].
Despite its high effectiveness, the safety of YFV-17D has triggered some debates over the past years. Between 2001 and 2011, 65 cases of YF vaccine-associated viscerotropic disease (YEL-AVD) have been recorded, with a relatively high CFR of 63% [1]. Despite the low prevalence of YEL-AVD (0.4/100,000), developing anti-viral therapies to manage these clinical cases during mass vaccination campaigns remains urgent. As YFV-17D is extremely genetically stable in vivo [96, 97], differences in host susceptibility and polymorphism in genes regulating anti-viral immunity are suspected to be important in the development of YEL-AVD. In addition to YEL-AVD, cases of YF vaccine-associated neurotropic disease (YEL-AND) have been reported. However, relative to YEL-AVD, the prevalence is slightly higher (0.8/100000) but rarely fatal [1]. Interestingly, a recent study using a mouse model of YFV-17D infection found that deficiency in type III IFN impairs blood brain barrier impermeability, inducing extensive YFV-17D neuroinvasion [98]. These data suggest that genetic polymorphisms affecting the type III IFN response could favor YEL-AND in some patients.
5. YFV as model to study the interface between viral pathogenicity and immunogenicity
YFV-17D and YFV-Asibi differ by only 32 amino acids and 68 nucleotides (with an extra three and six differences, respectively, depending on the Asibi clone; Fig. 5) [99]. However, infection by these two viral strains leads to opposite disease outcomes, suggesting differential host-pathogen interactions govern the potent immunogenicity of YFV-17D and pathogenicity of YFV.
Figure 5. Molecular and phenotypic differences between YFV-17D and YFV-Asibi infection.
A. Location in the polyprotein of the amino acid differences between YFV-17D and YFV-Asibi. 32 amino-acid mutations between the two strains are shown, as well as three additional mutations (either M/M,I or S/S,P) due to Asibi clonal differences. B. Major differences between YFV-Asibi (red capsid) and YFV-17D (yellow capsid) infection directly or indirectly suggested by the literature and that could play a critical role in regulating YFV virulence or attenuation. Important biological questions related to each of the listed differences are highlighted. At the bottom of the figure are three more general outstanding questions that encompass our currently limited understanding of the viral and host determinants regulating YFV course of infection.
Work in cell culture and animal models has attempted to uncover virologic and immunologic differences between infection with YFV-17D versus YFV-Asibi. In hepatoma cell lines, YFV-17D induces apoptosis and replicates more extensively than YFV-Asibi [62–64]. In primary human vascular endothelial cells, Kupffer cells, MDMs and monocyte-derived dendritic cells (MoDCs), YFV-Asibi causes a greater, more prolonged pro-inflammatory response than YFV-17D [69, 70, 100]. Comparative studies using primary human hepatocytes have yet to be performed.
The 12 amino acid differences between the E envelope glycoprotein of YFV-17D and YFV-Asibi have been suggested as a major determinant of YFV pathogenicity and attenuation. YFV-17D and virulent YFV E glycoprotein have different neutralizing epitopes [101] and use distinctive cell entry pathways to infect HeLa cells [30]. YFV-17D E glycoprotein also promotes more efficient binding to host cells [29, 30], which was associated with the induction of a more robust anti-viral innate immune response in HeLa cells [30]. Consistently, two in vivo studies identified the role of one specific domain of E (domain III) in determining YFV-17D binding enhancement, lower dissemination and attenuation [29, 102, 103]. YFV-17D replication was observed to be restricted in mature DCs, preventing their apoptosis [104, 105]. This could enable DCs to travel to the nearest lymph nodes and promote effective interactions with T cells. However, there is no direct evidence that Asibi replicates to a greater extent than YFV-17D in DCs. Interestingly, YFV-17D infected MoDCs, but not YFV-Asibi-infected MoDCs, promote IFN-γ and IL-2 production in CD4+ T cells [100], suggesting specific virus-host interactions in antigen presenting cells promotes enhanced T cell priming and adaptive immunity during YFV-17D infection.
Despite these findings, our overall understanding of the immunological mechanisms and host-pathogen interactions governing YFV attenuation and pathogenesis remains very limited (Fig. 5). Although in vitro studies have been instrumental in pinpointing key phenotypic differences between YFV-Asibi and YFV-17D infection, YFV-induced immunogenicity or pathogenicity is more likely determined by differential spatio-temporal interactions between viral, non-immune and immune components that work together as a whole. Hence, an exhaustive investigation of these interactions in vivo and in a human(-like) context is critical to elucidate the mechanisms of YFV virulence and attenuation and ultimately pave the way toward the rational design of a novel generation of vaccines. Recently, a descriptive study using rhesus macaques infected with either YFV-17D or the virulent YFV-DakH1279 strain was performed [61]. Although YFV-17D induced significant upregulation of anti-viral genes in PBMCs in comparison to YFV-DakH1279, PMBCs from DakH1279-infected animals had a transcriptomic signature of immune response dysregulation. More in vivo studies are needed to identify host-YFV interactions that differentially govern the outcome of infection during YFV-17D and wild-type YFV infection.
6. Concluding remarks
The YF outbreaks the world has experienced since 2016 are unique. Occurring at a scale not seen for decades, they have also taken place in areas with historically low or no YFV activity. These recent events suggest that mass vaccination campaigns limited only to endemic areas over the past decades [106] have left more environments, especially those densely populated, permissive to YFV due to low vaccination coverage. However, besides low vaccination coverage, the causes of such reemergence events, and why they are suddenly happening now, remains unknown.
Placing hundreds of millions of unvaccinated people at risk, the recent outbreaks have also highlighted how the threat of YF beyond its classic endemic areas has been underestimated. These outbreaks revealed a significant vaccine shortage worldwide, with current stocks insufficient to meet the standard protocol of one dose per patient. The use of a fraction of the standard vaccine dose has since been recommended, despite the lack of clear evidence for long-term protection (up to 5–10 years) (vi; [25, 26]). Additionally, these recent events suggest YFV circulation between non-human primates and mosquitoes is inadequately monitored around densely populated and under-vaccinated areas. This has raised serious concerns about the potential importation of YFV into non-endemic countries, where millions of people are unvaccinated and immunologically naïve to YFV and other flaviviruses [107]. Such risk is exemplified by the 2016 report of 10 laboratory-confirmed YF cases in China imported from Angola, the first ever reported cases in Asia.
To respond to all of these challenges and prevent potentially disastrous urban cycles, the WHO launched in 2017 the EYE (Eliminate Yellow Fever Epidemics) program, a nine-year strategy to enhance vaccination coverage in at risk-areas, prevent international spread, improve surveillance capabilities, and build up YFV vaccine stocks [108]. The effective completion of this campaign across all YFV at-risk areas is a herculean task, and its progress will be observed with particular attention.
Even after almost a century of research, our knowledge of the YFV replication cycle, YFV-induced mechanisms of disease and molecular basis of attenuation remains very limited. The availability of a potent vaccine has likely contributed to the status of YFV as a neglected disease, reducing research efforts. However, recent resurgence events demonstrate that YFV is still very much a real threat, and new research efforts are needed to better understand this virus. We believe that research efforts should primarily focus on three main axes : i) the interplay between the virus, the immune system and the liver in the development of disease and the role of the cytokine storm in this process; ii) the differential host-virus interactions that regulate wild-type YFV pathogenicity and YFV-17D immunogenicity and iii) the genetic basis regulating YF disease susceptibility in humans (see outstanding questions for additional research areas of interest).
Importantly, the recent developments in viral detection methods, genetic engineering, sequencing technologies and animal models create unprecedented opportunities to approach these questions and enhance our understanding of YFV and YF disease. The emergence of potent viral tracking methods [71, 109] provides a powerful way to better understand how the differential spatio-temporal dynamics of replication in vivo correlate with virulence or immunogenicity. Additionally, the recent development of single-cell RNA sequencing approaches and their implementation for BSL-3 pathogens [110] represent a formidable opportunity to explore differential transcriptomic regulations governing YFV virulence and attenuation at single-cell resolution in vivo and in a spatio-temporal manner.
A major need to accelerate YFV research is the development of an amenable and cost-effective animal model that can recapitulate the human-virus interplay, pathogenesis features and disease kinetics. The closest animal model to humans, rhesus macaques, have been intensively used in attempts to characterize YFV pathophysiology [61, 111]. However, rhesus macaques display distinct disease kinetics, cytokine profiles and transcriptomic regulations upon YFV infection. Recently, humanized mice were proven as permissive to YFV-17D [71]. The recent development of improved humanized mouse models with an enhanced human-like immune response [112, 113], as well as the generation of dually-engrafted humanized mouse models combining a human liver and a human immune system [114–116], thus represent unprecedented and cost-effective platforms for exploring YFV-induced pathogenesis and immune responses. In combination with single-cell RNA sequencing approaches, such platforms could be instrumental in uncovering critical human-YFV interactions that govern the mechanisms of disease and attenuation.
Highlights.
Intense YFV reemergence events have been recently reported in Africa and South America, but vaccine shortage triggered significant concerns in our ability to prevent future outbreaks.
The host-dependency of YF are poorly understood and limit the development of anti-viral therapies.
Liver disease and immune dysregulations are thought to be major driver of YF pathogenesis.
Recent insights into the human immune response to YFV-17D have helped to profile the cellular responses that define a potent vaccine.
Key differences between YFV-17D and wild-type YFV infection have been uncovered, but the molecular mechanisms governing YFV immunogenicity and pathogenicity in vivo are unknown.
Animal models have been instrumental in modeling YFV pathogenesis and immune response, but their high-cost and/or the absence of human tissues limit their use for YF research.
Recent advances in genomics and humanized animal models open a new and unprecedented path for YF research.
Outstanding questions.
What are the critical host factors required for YFV entry, replication, assembly and egress?
How does the interplay between YFV, the immune system and the liver regulate the development of YF disease?
What is the precise contribution of the cytokine storm in the process of YF disease?
What are the differential host-virus interactions and pathways that regulate wild-type YFV pathogenicity and YFV-17D immunogenicity.
What is the immunologic and genetic basis for the high immunogenicity of the YFV vaccine?
What is the host genetic basis regulating YF disease severity in humans?
What are the viral contributions to virulence?
How do environmental factors (microbiome, co-infections, etc.) impact on disease severity?
How do mosquito-derived factors influence the early phases of infection and immune responses after transmission?
What are the ecological dynamics and interactions between YFV and its natural (zoonotic) reservoirs?
How human activities and climate change impact YFV natural transmission cycles and promote reemergence events?
What governs the limited host tropism of YFV? Can immunocompetent animal models be constructed that recapitulate accurately human-like pathogenesis and immune response to YFV?
Acknowledgments:
The work was supported by grants from the National Institutes of Health (R01 AI107301, R21AI117213 to A.P.), a grant from the Grand Health Challenge program from Princeton University and an Investigator in Pathogenesis Award by the Burroughs Wellcome Fund (to A.P.). We thank members of the Ploss lab for critical discussions and comments on the manuscript, in particular Jenna Gaska for excellent editing. We apologize to all colleagues whose work could not be cited due to space constraints.
References
- 1.Monath TP and Vasconcelos PF (2015) Yellow fever. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology 64, 160–173 [DOI] [PubMed] [Google Scholar]
- 2.Quaresma JA, et al. (2013) Immunity and immune response, pathology and pathologic changes: progress and challenges in the immunopathology of yellow fever. Reviews in medical virology 23, 305–318 [DOI] [PubMed] [Google Scholar]
- 3.Garske T, et al. (2014) Yellow Fever in Africa: estimating the burden of disease and impact of mass vaccination from outbreak and serological data. PLoS medicine 11, e1001638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shearer FM, et al. (2017) Global yellow fever vaccination coverage from 1970 to 2016: an adjusted retrospective analysis. The Lancet. Infectious diseases [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paules CI and Fauci AS (2017) Yellow Fever - Once Again on the Radar Screen in the Americas. The New England journal of medicine 376, 1397–1399 [DOI] [PubMed] [Google Scholar]
- 6.Ortiz-Martinez Y, et al. (2017) Yellow fever in the Americas: the growing concern about new epidemics. F1000Research 6, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed QA and Memish ZA (2017) Yellow fever from Angola and Congo: a storm gathers. Tropical doctor 47, 92–96 [DOI] [PubMed] [Google Scholar]
- 8.(2017) Yellow fever in Africa and the Americas, 2016. Releve epidemiologique hebdomadaire 92, 442–452 [PubMed] [Google Scholar]
- 9.Bryant JE, et al. (2007) Out of Africa: a molecular perspective on the introduction of yellow fever virus into the Americas. PLoS pathogens 3, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White CR (1959) Yellow fever; history of the disease in the eighteenth and nineteenth century. The Journal of the Kansas Medical Society 60, 298–302 passim [PubMed] [Google Scholar]
- 11.Patterson KD (1992) Yellow fever epidemics and mortality in the United States, 1693–1905. Soc Sci Med 34, 855–865 [DOI] [PubMed] [Google Scholar]
- 12.Eckert J (1993) In the days of the epidemic: the 1793 yellow fever outbreak in Philadelphia as seen by physicians. Trans Stud Coll Physicians Phila 15, 31–38 [PubMed] [Google Scholar]
- 13.Bollet AJ (2005) Military medicine in the Spanish-American War. Perspectives in biology and medicine 48, 293–300 [DOI] [PubMed] [Google Scholar]
- 14.McCullough D (1978) The Path Between the Seas: The Creation of the Panama Canal, 1870–1914. New York: Simon & Schuster [Google Scholar]
- 15.Clements AN and Harbach RE (2017) History of the discovery of the mode of transmission of yellow fever virus. Journal of vector ecology : journal of the Society for Vector Ecology 42, 208–222 [DOI] [PubMed] [Google Scholar]
- 16.Frierson JG (2010) The yellow fever vaccine: a history. The Yale journal of biology and medicine 83, 77–85 [PMC free article] [PubMed] [Google Scholar]
- 17.Stokes A, et al. (1928) The transmission of yellow fever to Macacus rhesus - Preliminary note. J Amer Med Assoc 90, 253–254 [Google Scholar]
- 18.Theiler M (1930) Susceptibility of White Mice to the Virus of Yellow Fever. Science 71, 367. [DOI] [PubMed] [Google Scholar]
- 19.Sellards AW (1931) The behavior of the virus of yellow fever in monkeys and mice. Proceedings of the National Academy of Sciences of the United States of America 17, 339–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theiler M and Smith HH (1937) The Effect of Prolonged Cultivation in Vitro Upon the Pathogenicity of Yellow Fever Virus. The Journal of experimental medicine 65, 767–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theiler M and Smith HH (1937) The Use of Yellow Fever Virus Modified by in Vitro Cultivation for Human Immunization. The Journal of experimental medicine 65, 787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minor PD (2015) Live attenuated vaccines: Historical successes and current challenges. Virology 479–480, 379–392 [DOI] [PubMed] [Google Scholar]
- 23.Poland JD, et al. (1981) Persistence of neutralizing antibody 30–35 years after immunization with 17D yellow fever vaccine. Bulletin of the World Health Organization 59, 895–900 [PMC free article] [PubMed] [Google Scholar]
- 24.Barnett ED (2007) Yellow fever: epidemiology and prevention. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 44, 850–856 [DOI] [PubMed] [Google Scholar]
- 25.Calisher CH and Woodall JP (2016) Yellow Fever-More a Policy and Planning Problem than a Biological One. Emerging infectious diseases 22, 1859–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahuka-Mundeke S, et al. (2018) Immunogenicity of Fractional-Dose Vaccine during a Yellow Fever Outbreak - Preliminary Report. The New England journal of medicine [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach BD and Rice CM (2003) Molecular biology of flaviviruses. Advances in virus research 59, 23–61 [DOI] [PubMed] [Google Scholar]
- 28.Germi R, et al. (2002) Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology 292, 162–168 [DOI] [PubMed] [Google Scholar]
- 29.Lee E and Lobigs M (2008) E protein domain III determinants of yellow fever virus 17D vaccine strain enhance binding to glycosaminoglycans, impede virus spread, and attenuate virulence. Journal of virology 82, 6024–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez-Garcia MD, et al. (2016) Vaccine and Wild-Type Strains of Yellow Fever Virus Engage Distinct Entry Mechanisms and Differentially Stimulate Antiviral Immune Responses. mBio 7, e01956–01915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amberg SM and Rice CM (1999) Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. Journal of virology 73, 8083–8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chambers TJ, et al. (2005) Yellow fever virus NS2B-NS3 protease: characterization of charged-to-alanine mutant and revertant viruses and analysis of polyprotein-cleavage activities. The Journal of general virology 86, 1403–1413 [DOI] [PubMed] [Google Scholar]
- 33.Chambers TJ, et al. (1995) Mutagenesis of the yellow fever virus NS2B/3 cleavage site: determinants of cleavage site specificity and effects on polyprotein processing and viral replication. Journal of virology 69, 1600–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers TJ, et al. (1993) Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. Journal of virology 67, 6797–6807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chambers TJ, et al. (1991) Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. Journal of virology 65, 6042–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers TJ, et al. (1990) Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proceedings of the National Academy of Sciences of the United States of America 87, 8898–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Droll DA, et al. (2000) Yellow fever virus NS2B-NS3 protease: charged-to-alanine mutagenesis and deletion analysis define regions important for protease complex formation and function. Virology 275, 335–347 [DOI] [PubMed] [Google Scholar]
- 38.Lin C, et al. (1993) Cleavage at a Novel Site in the Ns4a Region by the Yellow-Fever Virus Ns2b-3 Proteinase Is a Prerequisite for Processing at the Downstream 4a/4b Signalase Site. Journal of virology 67, 2327–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang R, et al. (2016) A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535, 164-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi Z, et al. (2011) Identification and characterization of the host protein DNAJC14 as a broadly active flavivirus replication modulator. PLoS pathogens 7, e1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bozzacco L, et al. (2016) Chaperone-Assisted Protein Folding Is Critical for Yellow Fever Virus NS3/4A Cleavage and Replication. Journal of virology 90, 3212–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campos RK, et al. (2017) RPLP1 and RPLP2 Are Essential Flavivirus Host Factors That Promote Early Viral Protein Accumulation. Journal of virology 91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie XP, et al. (2015) Two Distinct Sets of NS2A Molecules Are Responsible for Dengue Virus RNA Synthesis and Virion Assembly. Journal of virology 89, 1298–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kummerer BM and Rice CM (2002) Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. Journal of virology 76, 4773–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiryaev SA, et al. (2009) NS4A regulates the ATPase activity of the NS3 helicase: a novel cofactor role of the non-structural protein NS4A from West Nile virus. The Journal of general virology 90, 2081–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umareddy I, et al. (2006) Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. The Journal of general virology 87, 2605–2614 [DOI] [PubMed] [Google Scholar]
- 47.Zou J, et al. (2015) Mapping the Interactions between the NS4B and NS3 Proteins of Dengue Virus. Journal of virology 89, 3471–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brand C, et al. (2017) Organization of the Flavivirus RNA replicase complex. Wiley interdisciplinary reviews. RNA 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindenbach BD and Rice CM (1999) Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. Journal of virology 73, 4611–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindenbach BD and Rice CM (1997) trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. Journal of virology 71, 9608–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller S, et al. (2007) The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. The Journal of biological chemistry 282, 8873–8882 [DOI] [PubMed] [Google Scholar]
- 52.Roosendaal J, et al. (2006) Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. Journal of virology 80, 4623–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le Sommer C, et al. (2012) G protein-coupled receptor kinase 2 promotes flaviviridae entry and replication. PLoS neglected tropical diseases 6, e1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Apte-Sengupta S, et al. (2014) Coupling of replication and assembly in flaviviruses. Current opinion in virology 9, 134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukhopadhyay S, et al. (2005) A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 3, 13–22 [DOI] [PubMed] [Google Scholar]
- 56.Hoffmann HH, et al. (2017) Diverse Viruses Require the Calcium Transporter SPCA1 for Maturation and Spread. Cell host & microbe 22, 460-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quaresma JAS, et al. (2013) Immunity and immune response, pathology and pathologic changes: progress and challenges in the immunopathology of yellow fever. Reviews in medical virology 23, 305–318 [DOI] [PubMed] [Google Scholar]
- 58.Debrito T, et al. (1992) Human Fatal Yellow-Fever - Immunohistochemical Detection of Viral-Antigens in the Liver, Kidney and Heart. Pathol Res Pract 188, 177–181 [DOI] [PubMed] [Google Scholar]
- 59.Meier KC, et al. (2009) A mouse model for studying viscerotropic disease caused by yellow fever virus infection. PLoS pathogens 5, e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melo-Lima BL, et al. (2015) The Attenuated Live Yellow Fever Virus 17D Infects the Thymus and Induces Thymic Transcriptional Modifications of Immunomodulatory Genes in C57BL/6 and BALB/C Mice. Autoimmune diseases 2015, 503087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Engelmann F, et al. (2014) Pathophysiologic and transcriptomic analyses of viscerotropic yellow fever in a rhesus macaque model. PLoS neglected tropical diseases 8, e3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lefeuvre A, et al. (2006) Host-cell interaction of attenuated and wild-type strains of yellow fever virus can be differentiated at early stages of hepatocyte infection. Microbes Infect 8, 1530–1538 [DOI] [PubMed] [Google Scholar]
- 63.Woodson SE and Holbrook MR (2011) Infection of hepatocytes with 17-D vaccine-strain yellow fever virus induces a strong pro-inflammatory host response. The Journal of general virology 92, 2262–2271 [DOI] [PubMed] [Google Scholar]
- 64.Woodson SE, et al. (2013) Coagulation factors, fibrinogen and plasminogen activator inhibitor-1, are differentially regulated by yellow fever virus infection of hepatocytes. Virus research 175, 155–159 [DOI] [PubMed] [Google Scholar]
- 65.Barba-Spaeth G, et al. (2005) Live attenuated yellow fever 17D infects human DCs and allows for presentation of endogenous and recombinant T cell epitopes. Journal of Experimental Medicine 202, 1179–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bruni D, et al. (2015) Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons. Science signaling 8, ra25. [DOI] [PubMed] [Google Scholar]
- 67.Cong Y, et al. (2016) Characterization of Yellow Fever Virus Infection of Human and Non-human Primate Antigen Presenting Cells and Their Interaction with CD4+ T Cells. PLoS neglected tropical diseases 10, e0004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLinden JH, et al. (2017) Yellow Fever Virus, but Not Zika Virus or Dengue Virus, Inhibits T-Cell Receptor-Mediated T-Cell Function by an RNA-Based Mechanism. The Journal of infectious diseases 216, 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woodson SE, et al. (2011) Differential cytokine responses from primary human Kupffer cells following infection with wild-type or vaccine strain yellow fever virus. Virology 412, 188–195 [DOI] [PubMed] [Google Scholar]
- 70.Khaiboullina SF, et al. (2005) Yellow fever virus strains Asibi and 17D-204 infect human umbilical cord endothelial cells and induce novel changes in gene expression. Virology 342, 167–176 [DOI] [PubMed] [Google Scholar]
- 71.Douam F, et al. (2017) Single-cell tracking of flavivirus RNA uncovers species-specific interactions with the immune system dictating disease outcome. Nature communications 8, 14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quaresma JAS, et al. (2006) Revisiting the liver in human yellow fever: Virus-induced apoptosis in hepatocytes associated with TGF-beta TNF-alpha and NK cells activity. Virology 345, 22–30 [DOI] [PubMed] [Google Scholar]
- 73.Quaresma JAS, et al. (2006) Immunohistochemical examination of the role of Fas ligand and lymphocytes in the pathogenesis of human liver yellow fever. Virus research 116, 91–97 [DOI] [PubMed] [Google Scholar]
- 74.Quaresma JAS, et al. (2007) Hepatocyte lesions and cellular immune response in yellow fever infection. Transactions of the Royal Society of Tropical Medicine and Hygiene 101, 161–168 [DOI] [PubMed] [Google Scholar]
- 75.Hudson NP (1928) The Pathology of Experimental Yellow Fever in the Macacus Rhesus: I. Gross Pathology. The American journal of pathology 4, 395–406 391 [PMC free article] [PubMed] [Google Scholar]
- 76.Monath TP, et al. (1981) Pathophysiologic correlations in a rhesus monkey model of yellow fever with special observations on the acute necrosis of B cell areas of lymphoid tissues. The American journal of tropical medicine and hygiene 30, 431–443 [DOI] [PubMed] [Google Scholar]
- 77.Xiao SY, et al. (2001) Experimental yellow fever virus infection in the golden hamster (Mesocricetus auratus). II. Pathology. Journal of Infectious Diseases 183, 1437–1444 [DOI] [PubMed] [Google Scholar]
- 78.ter Meulen J, et al. (2004) Activation of the cytokine network and unfavorable outcome in patients with yellow fever. The Journal of infectious diseases 190, 1821–1827 [DOI] [PubMed] [Google Scholar]
- 79.Quaresma JAS, et al. (2006) Midzonal lesions in yellow fever: A specific pattern of liver injury caused by direct virus action and in situ inflammatory response. Medical hypotheses 67, 618–621 [DOI] [PubMed] [Google Scholar]
- 80.Bradham CA, et al. (1998) Mechanisms of hepatic toxicity I. TNF-induced liver injury. Am J Physiol-Gastr L 275, G387–G392 [DOI] [PubMed] [Google Scholar]
- 81.Li MO, et al. (2006) Transforming growth factor-beta regulation of immune responses. Annual review of immunology 24, 99–146 [DOI] [PubMed] [Google Scholar]
- 82.Querec TD, et al. (2009) Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature immunology 10, 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gaucher D, et al. (2008) Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. The Journal of experimental medicine 205, 3119–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hou J, et al. (2017) A Systems Vaccinology Approach Reveals Temporal Transcriptomic Changes of Immune Responses to the Yellow Fever 17 D Vaccine. J Immunol 199, 1476–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akondy RS, et al. (2009) The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol 183, 7919–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Akondy RS, et al. (2015) Initial viral load determines the magnitude of the human CD8 T cell response to yellow fever vaccination. Proceedings of the National Academy of Sciences of the United States of America 112, 3050–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miller JD, et al. (2008) Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity 28, 710–722 [DOI] [PubMed] [Google Scholar]
- 88.Fuertes Marraco SA, et al. (2015) Long-lasting stem cell-like memory CD8+ T cells with a naive-like profile upon yellow fever vaccination. Science translational medicine 7, 282ra248. [DOI] [PubMed] [Google Scholar]
- 89.Niedrig M, et al. (1999) Assessment of IgG antibodies against yellow fever virus after vaccination with 17D by different assays: neutralization test, haemagglutination inhibition test, immunofluorescence assay and ELISA. Tropical medicine & international health : TM & IH 4, 867–871 [DOI] [PubMed] [Google Scholar]
- 90.Akondy RS, et al. (2017) Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wieten RW, et al. (2016) 17D yellow fever vaccine elicits comparable long-term immune responses in healthy individuals and immune-compromised patients. The Journal of infection 72, 713–722 [DOI] [PubMed] [Google Scholar]
- 92.Blom K, et al. (2013) Temporal dynamics of the primary human T cell response to yellow fever virus 17D as it matures from an effector- to a memory-type response. J Immunol 190, 2150–2158 [DOI] [PubMed] [Google Scholar]
- 93.Marquardt N, et al. (2015) The Human NK Cell Response to Yellow Fever Virus 17D Is Primarily Governed by NK Cell Differentiation Independently of NK Cell Education. J Immunol 195, 3262–3272 [DOI] [PubMed] [Google Scholar]
- 94.Ravindran R, et al. (2014) Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science 343, 313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Querec T, et al. (2006) Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. The Journal of experimental medicine 203, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beck A, et al. (2014) Comparison of the live attenuated yellow fever vaccine 17D-204 strain to its virulent parental strain Asibi by deep sequencing. The Journal of infectious diseases 209, 334–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xie H, et al. (1998) Yellow fever 17D vaccine virus isolated from healthy vaccinees accumulates very few mutations. Virus research 55, 93–99 [DOI] [PubMed] [Google Scholar]
- 98.Douam F, et al. (2017) Type III Interferon-Mediated Signaling Is Critical for Controlling Live Attenuated Yellow Fever Virus Infection In Vivo. mBio 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hahn CS, et al. (1987) Comparison of the Virulent Asibi Strain of Yellow-Fever Virus with the 17d Vaccine Strain Derived from It. Proceedings of the National Academy of Sciences of the United States of America 84, 2019–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cong Y, et al. (2016) Characterization of Yellow Fever Virus Infection of Human and Non-human Primate Antigen Presenting Cells and Their Interaction with CD4(+) T Cells. PLoS neglected tropical diseases 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sil BK, et al. (1992) Identification of Envelope Protein Epitopes That Are Important in the Attenuation Process of Wild-Type Yellow-Fever Virus. Journal of virology 66, 4265–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McElroy KL, et al. (2006) Role of the yellow fever virus structural protein genes in viral dissemination from the Aedes aegypti mosquito midgut. Journal of General Virology 87, 2993–3001 [DOI] [PubMed] [Google Scholar]
- 103.McElroy KL, et al. (2008) Characterization of the Antigen Distribution and Tissue Tropisms of Three Phenotypically Distinct Yellow Fever Virus Variants in Orally Infected Aedes aegypti Mosquitoes. Vector-Borne Zoonot 8, 675–687 [DOI] [PubMed] [Google Scholar]
- 104.Barba-Spaeth G, et al. (2005) Live attenuated yellow fever 17D infects human DCs and allows for presentation of endogenous and recombinant T cell epitopes. J Exp Med 202, 1179–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Palmer DR, et al. (2007) Restricted replication and lysosomal trafficking of yellow fever 17D vaccine virus in human dendritic cells. J Gen Virol 88, 148–156 [DOI] [PubMed] [Google Scholar]
- 106.Shearer FM, et al. (2017) Global yellow fever vaccination coverage from 1970 to 2016: an adjusted retrospective analysis. The Lancet. Infectious diseases 17, 1209–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wasserman S, et al. (2016) Yellow fever cases in Asia: primed for an epidemic. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases 48, 98–103 [DOI] [PubMed] [Google Scholar]
- 108.(2017) Eliminate Yellow fever Epidemics (EYE): a global strategy, 2017–2026. Releve epidemiologique hebdomadaire 92, 193–204 [PubMed] [Google Scholar]
- 109.Frei AP, et al. (2016) Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nature methods 13, 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gierhan TM, et al. (2017) Seq-Well: A Portable, Low-cost Platform for Single-Cell RNA-Seq of Low-Input. Nature methods In Press. [Google Scholar]
- 111.Stokes A, et al. (2001) The transmission of yellow fever to Macacus rhesus. 1928. Reviews in medical virology 11, 141–148 [DOI] [PubMed] [Google Scholar]
- 112.Theocharides AP, et al. (2016) Humanized hemato-lymphoid system mice. Haematologica 101, 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Douam F and Ploss A (2018) The use of humanized mice for studies of viral pathogenesis and immunity. Current opinion in virology 29, 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wilson EM, et al. (2014) Extensive double humanization of both liver and hematopoiesis in FRGN mice. Stem cell research 13, 404–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Strick-Marchand H, et al. (2015) A novel mouse model for stable engraftment of a human immune system and human hepatocytes. PloS one 10, e0119820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gutti TL, et al. (2014) Human hepatocytes and hematolymphoid dual reconstitution in treosulfan-conditioned uPA-NOG mice. The American journal of pathology 184, 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Resource section
- i.WHO Global Yellow Fever Data Base; http://apps.who.int/globalatlas/default.asp
- ii.WHO (2016) Situation Report Yellow Fever - 28 October 2016; http://www.who.int/emergencies/yellow-fever/situation-reports/28-october-2016/en/
- iii.WHO/PAHO (2017) Epidemiological Update Yellow Fever - 10 July 2017. http://www.paho.org/hq/index.php?option=com_docman&task=doc_view&Itemid=270&gid=40841&lang=en
- iv.WHO – Disease outbreak news of January 22nd 2018; http://www.who.int/csr/don/22-january-2018-yellow-fever-brazil/en/
- v.WHO – Disease outbreak news of February 27th 2018; http://www.who.int/csr/don/27-february-2018-yellow-fever-brazil/en/
- vi.WHO – Disease outbreak news of March 9th 2018; http://www.who.int/csr/don/09-march-2018-yellow-fever-brazil/en/
- vii.WHO (2014) Yellow fever in Africa and South America, 2013. Releve epidemiologique hebdomadaire 89, 297–306; http://www.who.int/iris/handle/10665/242235 [PubMed] [Google Scholar]
- viii.WHO - Yellow Fever Situation Reports; http://www.who.int/emergencies/yellow-fever/situation-reports/archive/en/