Abstract
IFNγ is a cytokine with important roles in tissue homeostasis, immune and inflammatory responses and tumour immunosurveillance. Signalling by the IFNγ receptor activates the Janus kinase (JAK)-signal transducer and activator of transcription 1 (STAT1) pathway to induce the expression of classical interferon-stimulated genes that have key immune effector functions. This Review focuses on recent advances in our understanding of the transcriptional, chromatin-based and metabolic mechanisms that underlie IFNγ-mediated polarization of macrophages to an ‘M1-like’ state, which is characterized by increased pro-inflammatory activity and macrophage resistance to tolerogenic and anti-inflammatory factors. In addition, I describe the newly discovered effects of IFNγ on other leukocytes, vascular cells, adipose tissue cells, neurons and tumour cells that have important implications for autoimmunity, metabolic diseases, atherosclerosis, neurological diseases and immune checkpoint blockade cancer therapy.
IFNγ is a cytokine that is primarily produced by cells of the immune system, including innate-like lymphocyte populations, such as natural killer (NK) cells and innate lymphoid cells (ILCs), and adaptive immune cells, such as T helper 1 (TH1) cells and CD8+ cytotoxic T lymphocytes (CTLs). It signals through the IFNγ receptor (IFNγR; comprising the IFNγR1 and IFNγR2 subunits), which can be expressed on most, if not all, cell types (reviewed in REF1) (FIG. 1). In innate-like lymphocytes, IFNγ production can be induced by cytokines (primarily IL-12 and IL-18) or following the activation of pattern recognition receptors (PRRs) or broadly reactive antigen receptors during microbial infection or tissue damage. As such, an early burst of IFNγ production occurs during infections before the emergence of an antigen-specific adaptive immune response. By contrast, high levels of sustained IFNγ production by TH1 cells or CTLs typically require T cell receptor (TCR)-mediated recognition of microbial (but also self or mutated self) peptides in the context of MHC class II or MHC class I molecules, respectively.
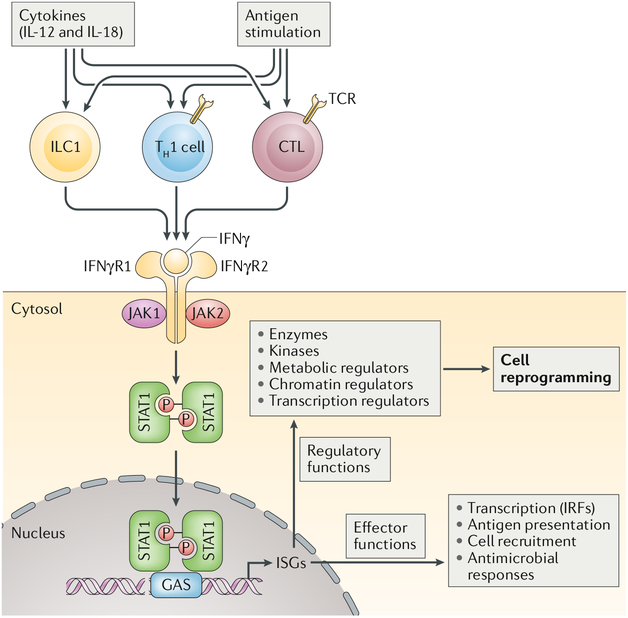
Fig. 1 |. IFNγ production and signalling.
IFNγ is produced by innate-like lymphocytes, including group 1 innate lymphoid cells (ILC1s), and by adaptive lymphocytes, including T helper 1 (TH1) cells and cytotoxic T lymphocytes (CTLs), in response to cytokine and antigen stimulation. IFNγ acts on its receptor to induce rapid and transient Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signalling and interferon-stimulated gene (ISG) induction. Over time, the cellular IFNγ response evolves by impacting the expression and function of various enzymes and regulators of metabolism, chromatin and transcription to induce a reprogrammed cellular state that is characterized not only by its gene expression profile but also by altered responsiveness to environmental challenges. GAS, IFNγ activation site; IFNγR, IFNγ receptor; IRF, interferon regulatory factor; TCR, T cell receptor.
In all cell types studied, binding of IFNγ to its receptor activates the canonical Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signalling pathway1–3 (BOX 1; FIG. 1). IFNγR ligation results in activation of the receptor-associated JAK1 and JAK2 protein-tyrosine kinases and subsequent tyrosine phosphorylation and activation of primarily STAT1, which translocates to the nucleus, binds to conserved IFNγ activation site (GAS) DNA elements and directly activates the transcription of interferon-stimulated genes (ISGs). ISGs encode many products that have direct effector immune functions, such as chemok-ines, antigen-presenting molecules (including MHC molecules), phagocytic receptors and various antiviral and antibacterial factors (FIG. 1). Compelling genetic and biochemical data support key non-redundant roles for JAK1, JAK2, STAT1 and many ISGs in mediating cellular IFNγ responses and in key IFNγ functions in vivo, such as host defence against intracellular pathogens, modulation of immune and inflammatory responses and associated tissue damage and tumour immunosurveil-lance. This core IFNγ-JAK-STAT1-ISG response, the direct immune functions of ISGs and feedback inhibition of this pathway have been extensively reviewed1–3 and are not covered here.
Box 1 |. IFnγ signalling.
IFNγ signalling has conventionally been defined as a cascade of tyrosine phosphorylation events initiated by the binding of IFNγ to IFNγ receptor (IFNγR), which results in the initiation of interferon-stimulated gene (ISG) transcription. activation of receptor-associated Janus kinases (JAKs) results in phosphorylation of tyrosine residues in the IFNγR cytoplasmic domain, creating a recognition substrate that recruits signal transducer and activator of transcription 1 (STAT1). tyrosine phosphorylation of STAT1 promotes dimerization, nuclear translocation, DNa binding to IFNγ activation site (GAS) elements and transcriptional activation by the stat1 dimers (see also Fig. 1). In addition to inducing STAT1 dimers that bind Gas elements and can cooperate with IFNγ-induced interferon regulatory factors (IRFs), IFNγ can also activate non-canonical transcriptional complexes that are similar to the interferon-stimulated gene factor 3 (ISGF3) complexes induced by type I interferons in that they contain IRF9 and bind to interferon-stimulated response elements (ISREs). The transcriptional potency of STAT1 is modulated by post-translational modifications, most notably phosphorylation of the transcription activation domain. these proximal IFNγ signalling events have recently been reviewed (REFS1–3,12). In this Review, we instead revisit IFNγ signalling and look beyond the cytoplasmic events that lead to activation of ISGs. This includes a discussion of how IFNγ signalling induces epigenetic remodelling at chromatin, the genome-wide interactions of STAT1 with IRFs and other transcriptions factors and the role of IFNγ in the modulation of metabolic pathways.
IFNγ was originally identified as ‘macrophage-activating factor’, and macrophages are a major physiological target for IFNγ action1. Thus, studies of its cellular functions and underlying mechanisms of action have been extensively performed using macrophages or cell line models. This Review focuses on the sustained and global effects of IFNγ on macrophages that cannot be readily explained by the direct functions of ISGs but are instead mediated by an IFNγ-induced transcriptional network, epigenetic mechanisms and metabolic changes in macrophages that alter their cell state and reprogramme how they respond to environmental stimuli (FIGS. 1,2a). It is increasingly appreciated that important in vivo biological and pathological effects of IFNγ are mediated at least in part by cells other than macrophages or immune cells. Therefore, this Review also covers recent insights into how IFNγ regulates various non-leukocyte cell types and the implications of this for autoimmunity, obesity and metabolic syndrome, vascular biology and atherosclerosis, neuronal function and cancer immunotherapy.
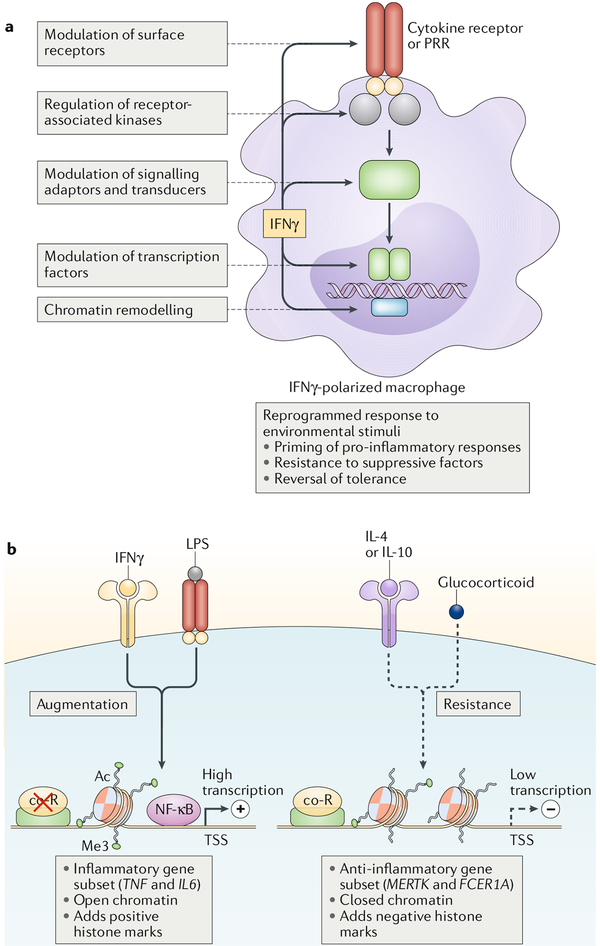
Fig. 2 |. ‘Super-activation’ of macrophages following priming by IFNγ.
Polarization of macrophages by IFNγ results in their increased responsiveness to pro-inflammatory stimuli (such as lipopolysaccharide (LPS) or type I interferons) and resistance to anti-inflammatory stimuli (such as IL-4, IL-10 and glucocorticoids). This results in ‘super-activation’ of macrophages. a | Modulation of key signalling, transcriptional and chromatin components by IFNγ mediates its cross-regulation of signalling by distinct receptors. b | IFNγ augments the transcriptional activation of a subset of pro-inflammatory genes (including TNFand IL6) by opening and priming chromatin at the gene regulatory elements while inducing resistance to anti-inflammatory signals by closing chromatin in a gene-specific manner. Ac, acetylation; co-R, co-repressor; Me3, trimethylation; NF-κB, nuclear factor-κB; PRR, pattern recognition receptor; TSS, transcription start site.
Programming of macrophages by IFNγ
Signalling by the JAK-STAT pathway, including activation of STAT1 by IFNγ, is typically transient, with a peak signal occurring at 15–60 minutes and resolution back to baseline occurring at 2–4 hours after stimulation. Accordingly, transcriptional responses of many direct STAT1 target genes peak within several hours of IFNγ stimulation. Much of the analysis of cellular responses to IFNγ has focused on these early time points, and the early IFNγ response and associated ISG induction have been extensively reviewed1–3. However, even predating the discovery of STATs, it was apparent that IFNγ induces a subset of ISGs with delayed kinetics in a manner dependent on new protein synthesis (implying indirect regulation) and also induces a pattern of sustained gene expression that persists beyond the duration of JAK-STAT signalling4,5. These delayed and sustained kinetics of gene induction could be explained in part by a feedforward loop in which IFNγ induces de novo expression of transcription factors, most notably interferon regulatory factors (IRFs) and STATs themselves, which cooperate to induce and sustain gene expression1. Another mechanism for persistence of IFNγ signalling is capture of IFNγ by cell surface phosphatidylserine on viable cells, followed by its slow release to drive long-term transcription6. Recent developments in interferon signalling (see also BOX 1) include the use of transcriptomics to fully define the set of IFNγ-induced transcription factors7, genome-wide analysis of IRF-mediated networks and their cooperation with STATs8,9, the description of alternative STAT complexes10–13 and the identification of a role for unphosphorylated STATs in mediating gene expression at later phases after initial JAK-STAT signalling has subsided14.
The IFNγ-induced transcriptional network described above can explain delayed and sustained patterns of gene expression but does not fully explain several aspects of the IFNγ-activated macrophage cell state, which has been termed M1 polarization, classical activation or priming1,15. In addition to ISG expression, there are several salient features of IFNγ-polarized macrophages. First, IFNγ-polarized macrophages (which have been referred to as M1 macrophages and more recently termed ‘M(IFNγ) cells’.16) are hyper-responsive to various inflammatory stimuli, which include cytokines such as tumour necrosis factor (TNF) and type I interferons and microbial products and ligands for Toll-like receptors (TLRs). Stimulation of IFNγ-polarized macrophages with TLR ligands results in a massive super-induction of inflammatory cytokines and canonical nuclear factor-κB (NF-κB) target genes (FIG. 2b, left panel). This phenomenon is termed priming. Second, IFNγ induces gene-specific refractoriness to anti-inflammatory factors (such as IL-10 or glucocorticoids) and IL-4 and IL-13, cytokines that promote the resolution of inflammation, tissue healing and return to homeostasis1,7,17–19 (FIG. 2b). Third, IFNγ prevents and reverses macrophage tolerance20, a cell state in which prior strong activation of macrophages by TLR ligands or TNF induces refractoriness to induction of canonical inflammatory NF-κB target genes. Refractoriness to anti-inflammatory stimuli and abrogation of tolerance enable exaggerated inflammatory responses and thus contribute to macrophage priming.
The biological importance of IFNγ-mediated polarization and priming of macrophages is supported by in vivo studies in model organisms and in human subjects (reviewed in REFS1,21). M1 macrophage polarization is regarded as a type I immune response that is promoted by IFNγ; it is important for control of infections by intracellular pathogens but can drive pathology in autoimmune diseases. A sustained IFNγ signature is seen in the inflamed tissues of patients with autoimmune diseases, such as rheumatoid arthritis, and disease-associated macrophages that express an IFNγ signature show increased sensitization to inflammatory cytokines and TLR ligands and resistance to IL-10 and glucocorticoids1,21. More recently, primed monocytes have been observed in the peripheral blood of patients with rheumatoid arthritis22, and work in a mouse model of gastrointestinal infection showed that bone marrow monocytes are primed by NK cell-derived IFNγ to exhibit increased responses to bacterial ligands before egress from the bone marrow and migration to the site of infection23. In addition, a remarkable series of experiments with human volunteers demonstrated that IFNγ prevents and reverses experimental endotoxin tolerance in vivo24. This work provides the rationale for treatment of patients with sepsis — who exhibit a tolerance-related immunoparalysis phenotype — with IFNγ to restore cellular functions25. Interferons have also been proposed to prevent tolerization of monocytes by circulating endotoxin in systemic lupus erythematosus (SLE); this would result in increased cell activation and cytokine production and thereby drive inflammatory pathogenesis26,27. The potential to therapeutically manipulate IFNγ-mediated macrophage polarization to modulate inflammatory responses for the benefit of patients provides a strong rationale for investigation of underlying mechanisms.
When studied in vitro, IFNγ-mediated priming and resistance to suppression in primary macrophages are stable for at least several days, raising the question of how these responses are sustained in the setting of diminishing IFNγR signalling and decaying expression of IFNγ-induced transcription factors. Furthermore, the gene-specific nature of IFNγ-induced resistance to tolerance and IL-10 and IL-4 (REFS7,17,20,26,28) argues against suppression of upstream signalling pathways by IFNγ. Instead, stability of gene expression that persists beyond the initiating signal suggests that epigenetic mechanisms provide short-term memory29, and gene-specific effects suggest specific regulation of individual genes at the chromatin level, as originally suggested by Medzhitov and colleagues30 (FIG. 2b).
Epigenetic regulation by IFNγ
Epigenetic mechanisms of IFNγ-mediated macrophage reprogramming.
Herein, we use the term ‘epigenetic mechanisms’ to refer to developmentally or environmentally induced chemical changes to DNA or chromatin that do not change the genetic code but instead regulate gene expression. These epigenetic changes can be moderately long-lived and persist beyond the original stimulus, thereby promoting a more stable and sustained transcriptional response. In macrophages, analysis of epigenetic regulation has focused predominantly on chromatin accessibility at gene regulatory elements (promoters and enhancers), which is determined by the balance of positive relative to negative histone marks (post-translational modifications) and nucleosome remodelling (reviewed in REFS15,31,32). A macrophage-specific pattern of stable open chromatin at promoters and enhancers (also termed the ‘epigenomic landscape’) is established during cell differentiation by the lineage-determining transcription factor PU.1 and the CCAAT-enhancer-binding protein (C/EBP) family of proteins, which often bind cooperatively with other macrophage-expressed transcription factors (FIG. 3a). This epigenomic landscape enables access of general transcriptional machinery to constitutively expressed genes and provides a poised chromatin state that enables rapid binding and function of signal-activated transcription factors, such as NF-κB and STATs, after cell stimulation. Thus, the epigenomic landscape shapes the pattern of constitutive gene expression and the nature of the early transcriptional response to environmental stimuli. It has become clear that cells partially remodel their epigenomic landscape in a gene-specific manner after cell stimulation15,32. Increases in positive histone marks and chromatin accessibility can result in increased transcription per se but can also prime genes for more rapid or augmented transcription in response to subsequent stimulation (FIG. 3a). Conversely, negative histone marks and closing of chromatin silence active genes and can make genes refractory to subsequent stimulation. Thus, remodelling of the chromatin landscape provides an attractive potential explanation for the priming and silencing effects that occur in a stable and gene-specific manner in IFNγ-polarized macrophages.
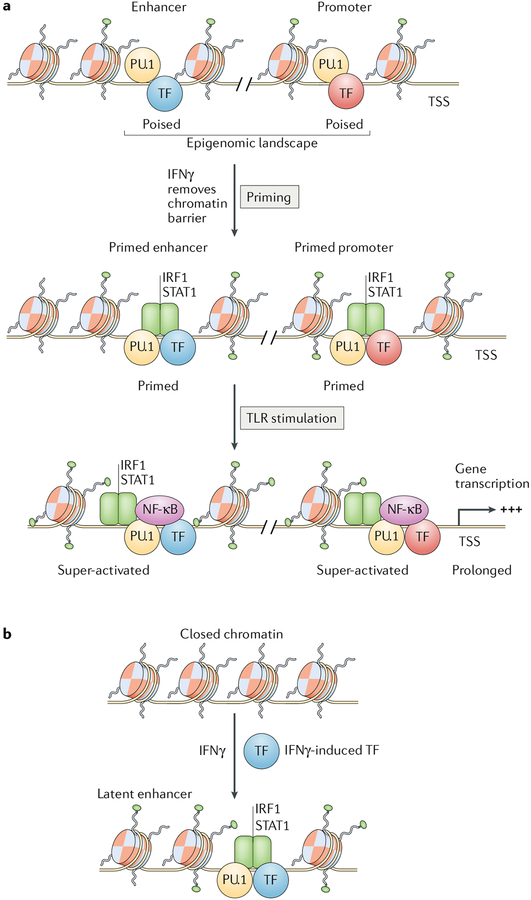
Fig. 3 |. IFNγ primes and induces de novo enhancer formation to promote activation of gene transcription.
a | IFNγ primes pre-existing enhancers and promoters via the recruitment of signal transducer and activator of transcription 1 (STAT1) and interferon regulatory factor 1 (IRF1); this is associated with increased histone acetylation and chromatin remodelling. b | IFNγ induces formation of latent enhancers by inducing transcription factors (TFs) that cooperate with transcription factor PU.1 or CCAAT-enhancer-binding protein (C/EBP) family proteins to form new enhancers. NF-κB, nuclear factor-κB; TLR, Toll-like receptor; TSS, transcriptional start site.
Epigenetic mechanisms of macrophage priming by IFNγ.
Induction of the rapid and often transient early phase of ISG transcription by IFNγ is mediated by direct binding of STAT1 to accessible GAS-containing regulatory elements2,18 (FIG. 1). STATs recruit histone acetyltransferases and chromatin-remodelling enzymes2, and as the IFNγ response evolves (at 4–24 hr), there is a shift in the genomic binding profile of STAT1 towards IRF elements, many of which are co-occupied by IRF1, and pervasive remodelling of histone acetylation at almost half of STAT1-binding regulatory elements genome wide, consistent with a primed open chromatin state18 (FIG. 3a). This re-directed binding of STAT1 towards genes that contain adjacent IRF-binding and NF-κB-binding sites allows STAT1 to access canonical NF-κB target genes such as IL6 that do not contain GAS elements and are not conventional ISGs.
This priming of regulatory elements does not necessarily activate transcription but instead ‘bookmarks’ classical inflammatory genes such as TNF, IL6 and IL12B for massive and sustained transcriptional responses to lipopoly-saccharide (LPS) (FIG. 3a). Under conditions where LPS is added simultaneously or subsequent to IFNγ for M1 polarization, LPS-induced type I interferons will induce STAT1-containing interferon-stimulated gene factor 3 (ISGF3) complexes that bind to interferon-stimulated response elements (ISREs); given the similarity between ISREs and IRF-binding sequences, this will further redirect STAT1 to IRF-binding sites.
IFNγ not only primes pre-existing enhancers but also induces de novo formation of several hundred latent enhancers33 (FIG. 3b). Although IFNγ-induced latent enhancers bind STAT1, strikingly, the DNA motif most enriched in these enhancers is not a canonical GAS but instead an IRF-binding site, suggesting indirect binding of STAT1 as part of IRF-containing complexes (FIG. 3b). Latent enhancers are formed by cooperative binding of IFNγ-induced transcription factors such as STATs and IRFs with the lineage-determining factor PU.1 to open chromatin and stably deposit the histone H3 lysine 4 monomethylation (H3K4me1) enhancer mark. Latent enhancers persist at least 48 hours after removal of IFNγ and are associated with faster and occasionally greater induction of associated genes after cytokine rechallenge, thereby conferring short-term transcriptional memory.
IFNγ induces expression of IRF1 and IRF8, potentially enabling a time-dependent increase in their interactions with STAT1 and in the binding of STAT1-IRF complexes to regulatory elements. In line with this notion, coordinate binding of STAT1-IRF1 or STAT1-IRF1-IRF8 plays a key role in basal and IFNγ-inducible expression of macrophage genes that are important in inflammatory and host defence functions, including in models of neuroinflammation and tuberculosis in vivo8,9. In the context of LPS stimulation, IRF8 contributes to formation of latent enhancers by cooperatively binding at new sites with AP1 transcription factors but plays a minimal role in STAT1 recruitment9. Overall, these studies show that IFNγ-induced STAT1 activation and the downstream transcriptional network mediated by IRFs are translated into extensive remodelling of the epigenome that alters gene transcription. Priming of chromatin with bound transcription factors and altered histone marks represents one mechanism of augmenting transcriptional responses to subsequent challenges.
Epigenetic mechanisms of resistance to antiinflammatory factors.
Major suppressors of macrophage inflammatory responses include glucocorticoids, IL-10, IL-4 and IL-13. The glucocorticoid receptor induces inhibitory genes and binds NF-κB and AP1 to inhibit their inflammatory activity, whereas IL-10 signals via STAT3 to induce genes that suppress inflammation. IL-4 and IL-13 activate STAT6 and AKT signalling to modulate inflammatory responses and promote a wound-healing reparative macrophage phenotype. Although IFNγ can transiently suppress signalling by these anti-inflammatory factors34,35, such signalling inhibition cannot explain the stable inhibition that persists after IFNγ activity is terminated or the gene-specific repression of only subsets of anti-inflammatory genes. An additional non-mutually exclusive inhibitory mechanism is gene-specific induction of stable repressive chromatin states by IFNγ.
In line with this idea, IFNγ induces histone H3 lysine 27 trimethylation (H3K27me3), a stable his-tone mark associated with gene repression, at gene promoters19. Although only a small number of genes (approximately 15) are silenced by this mechanism, these genes include functionally important genes with anti-inflammatory functions such as PPARG and MERTK. IFNγ induces recruitment of the enhancer of zeste homologue 2 (EZH2) catalytic component of polycomb repressor complex 2 (PRC2) that induces a time-dependent and stable accumulation of H3K27me3. Macrophage genes with increased promoter H3K27me3 are silenced for at least 5 days and are refractory to induction by glucocorticoids and IL-4. These results support a model whereby IFNγ induces a negative chromatin state mediated by H3K27me3 at promoters to stabilize gene silencing, thereby making them refractory to induction by anti-inflammatory signalling pathways (FIG. 2b, right panel).
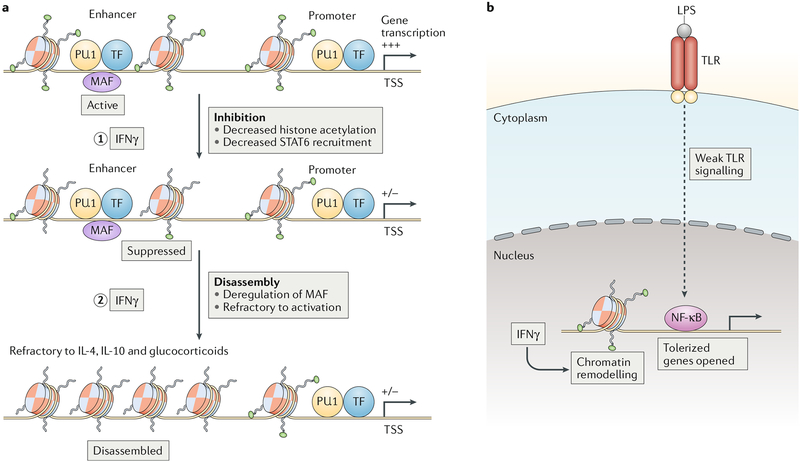
However, H3K27me3-mediated silencing of promoters does not explain the broad IFNγ-induced suppression of more than 700 macrophage genes that are induced by glucocorticoids, IL-4 and IL-10 and are associated with the M2 macrophage phenotype. Instead, this broad suppression can be explained by IFNγ-mediated downregulation of histone H3 lysine 27 acetylation (H3K27ac) and the activity of more than 5,000 enhancers and their associated genes7 (FIG. 4a). Strikingly, a subset of 12% of these enhancers loses chromatin accessibility and binding by lineage-determining transcription factors PU.1 and C/EBP family proteins, a process termed enhancer disassembly (FIG. 4a). Genes associated with disassembled enhancers remain stably repressed after IFNγ removal and are refractory to induction by glucocorticoids. The majority (77%) of disassembled enhancers is enriched for DNA-binding motifs for transcription factor MAF, and IFNγ suppresses MAF expression and binding to target enhancers. These results support a model whereby a subset of macrophage enhancers is maintained in an open chromatin state by cooperative binding by MAF and PU.1, and these enhancers are lost upon IFNγ stimulation, with downregulation of associated genes. The need to re-assemble an enhancer helps explain the stability of the refractory phenotype, whereas the specific binding of MAF to a subset of macrophage enhancers helps explain gene-specific effects. MAF-related enhancer disassembly accounts for suppression of approximately 15% of IFNγ-repressed genes (IRGs), and thus there are likely additional mechanisms of repression, possibly involving an interactive network of the more than 70 transcription factors whose expression is regulated by IFNγ7.
Fig. 4 |. Chromatin regulation by IFNγ controls gene expression.
a | IFNγ suppresses enhancer function by decreasing histone acetylation and attenuating the recruitment of signal transducer and activator of transcription 6 (STAT6) (step 1). A subset of suppressed enhancers is bound by transcription factor MAF, and these enhancers harbour STAT6-binding motifs that exhibit decreased STAT6 occupancy after IFNγ stimulation. At a subset of MAF-bound enhancers, IFNγ-mediated downregulation of MAF expression and binding results in enhancer disassembly and refractoriness to activation by IL-4, IL-10 or glucocorticoids (step 2). b | IFNγ reverses gene tolerization by enabling opening of chromatin in response to weak upstream signals. The magnitude of gene expression is determined by the combination of signalling strength and chromatin state. LPS, lipopolysaccharide; NF-κB, nuclear factor-κB; TF, transcription factor; TLR, Toll-like receptor; TSS, transcription start site.
Transcriptional and epigenomic profiling has shown that coadministration of IFNγ broadly attenuates the IL-4-induced transcriptional programme, with stronger inhibition of a subset of canonical M2 macrophage genes17. IFNγ does not have a marked effect on IL-4 signalling but substantially and broadly suppresses IL-4-induced H3K27ac at regulatory elements concomitant with a modest decrease in STAT6 binding. Accordingly, IL-4-activated enhancers that are sensitive to inhibition by IFNγ exhibit enrichment of STAT6-binding motifs (FIG. 4a); these repressed enhancers are also enriched for MAF motifs, which provides additional support for MAF as a target for inhibition by IFNγ. Interestingly, a small number (317) of IL-4-induced acetylated regions that are resistant to cross-inhibition by IFNγ show enrichment for MYC proto-oncogene protein binding motifs, and depletion studies implicate MYC in establishing resistance to suppression by IFNγ. The converse analysis of the effects of IL-4 on IFNγ responses showed that enhancers resistant to suppression by IL-4 are enriched for STAT-binding and IRF-binding motifs, while IL-4-sensitive enhancers show over-representation of motifs for binding to AP1, ATF, C/EBP and NF-κB. Thus, the core STAT1-IRF axis that is resistant to suppression by IL-4 and is important for host defence is preserved, but genes whose activation requires auxiliary factors such as JUNB and C/EBPβ are vulnerable to inhibition by IL-4. Another study showed that STAT6 can directly repress genes by recruiting histone deacetylase 3 (HDAC3) to non-canonical binding sites, with associated decreased expression of genes important for inflammasome activation36. The model emerges that IFNγ broadly suppresses gene expression by suppressing histone acetylation at gene enhancers, likely by targeting key enhancer-associated transcription factors such as MAF (FIG. 4 a). Enhancer deactivation or disassembly makes genes refractory to antagonistic anti-inflammatory factors and stabilizes an activation phenotype.
Reversal of macrophage tolerance.
Strong activation of NF-κB signalling induces a state of macrophage tolerance characterized by diminished proximal signalling (FIG. 4b) that is unable to induce the chromatin remodelling required for re-induction of inflammatory NF-κB target genes30,37. IFNγ prevents and reverses tolerance by enabling the opening of chromatin in response to weak signals20 (FIG. 4b). The underlying mechanism involves the co-activator receptor-interacting protein 140 (RIP140; also known as NRIP1)38 and most likely IRFs26 but this requires further elucidation.
Collectively, studies of the epigenetic effects of IFNγ support the idea that IFNγ polarizes macrophages by altering chromatin to reprogramme transcriptional profiles and responses to environmental stimuli. Similarities exist between the priming and tolerance-reversing effects of IFNγ and the training of innate immune cells for improved activation responses by prior exposure to microbial and tissue damage-associated products that elicit low-grade activation39,40. Such training has been shown to confer innate immune memory, including in in vivo systems, and to work via similar epigenetic and chromatin-based mechanisms. The role of IFNγ and cytokines in trained immunity has not been investigated, but it is possible that IFNγ can improve training and vice versa.
Regulation of macrophage metabolism by IFNγ
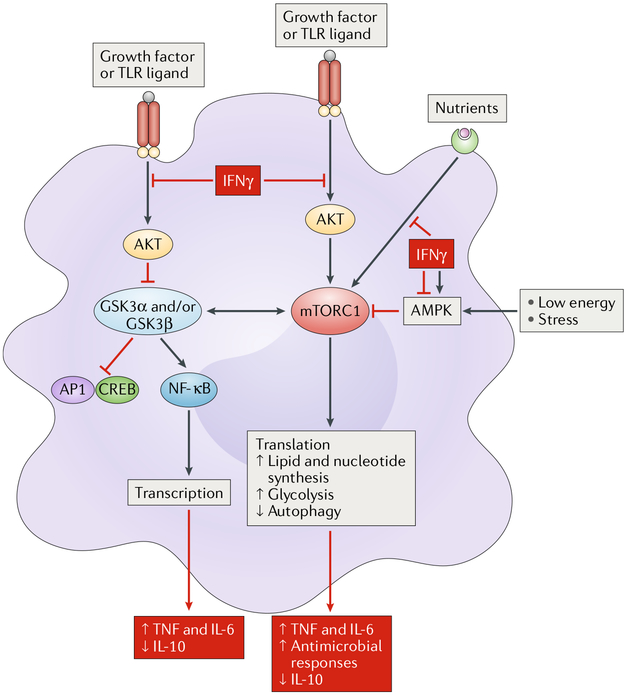
Metabolic reprogramming, defined as the altered use of metabolic pathways for the generation of energy and key metabolites, represents an important aspect of macrophage activation and polarization and has been recently reviewed41. Briefly, M1-type macrophage activation by TLR ligands induces aerobic glycolysis and disrupts the Krebs cycle, whereas M2-type macrophage polarization promotes fatty acid oxidation and oxidative phosphorylation. Detailed analyses ofthe effects of IFNγ (in the absence of co-stimulation with TLR ligands) on cell metabolites, respiration and related metabolic pathways have not been performed; instead, the effects of IFNγ on three enzymes that are major regulators of cellular metabolism — mammalian target of rapamycin complex 1 (mTORC1), 5′-AMP-activated protein kinase (AMPK) and glycogen synthase kinase 3 (GSK3) — have been reported42–44. The mTORC1 complex senses growth factors and nutrients and in a nutrient replete environment coordinates the cellular anabolic response by promoting protein, lipid and nucleotide biosynthesis (FIG. 5). IFNγ inhibits mTORC1 activity in resting human macrophages, which results in a selective decrease in translation of proteins important for tRNA charging, purine nucleotide synthesis, small molecule transport, mitochondrial function and anti-inflammatory mediators (including IL-10 and the transcription factor HES1), with increased expression of inflammatory cytokines44. Along with this shift in metabolism towards a more inflammatory phenotype, inhibition of mTORC1 is associated with increased autophagy, which promotes microbial killing and antigen presentation45. In tolerized macrophages and monocytes from patients with sepsis who exhibit severe metabolic defects, IFNγ promotes glycolytic metabolism via mTORC1, which contributes to reversal of the broadly suppressed immune state (immunoparalysis) associated with sepsis25.
Fig. 5 |. IFNγ modulates key metabolic pathways.
IFNγ suppresses growth factor and nutrient pathways to modulate activity of several central regulators of cellular metabolism, including mammalian target of rapamycin complex 1 (mTORC1), glycogen synthase kinase 3 (GSK3) and 5′-AMP-activated protein kinase (AMPK). Functionally important outcomes of metabolic regulation by IFNγ are depicted in red boxes. CREB, CCAAT-enhancer-binding protein; NF-κB, nuclear factor-κB; TLR, Toll-like receptor; TNF, tumour necrosis factor.
mTORC1 is functionally coupled with GSK3 and negatively regulated by AMPK (FIG. 5). GSK3 modulates the balance between NF-κB and AP1-CREB signalling, and its activation by IFNγ decreases IL-10 while increasing inflammatory cytokine production42. AMPK senses cellular energy deprivation and suppresses inflammation while promoting M2 polarization41. IFNγ activates AMPK under low energy conditions, which can function as a feedback loop to restrain inflammation43 but also may contribute to IFNγ-mediated suppression of mTORC1. In summary, IFNγ regulation of the upstream metabolic regulators mTORC1, GSK3 and AMPK is important for inflammatory responses. The effects of IFNγ on cellular metabolic pathways deserve further investigation, especially in light of the finding that polarization of macrophages with LPS in combination with IFNγ suppresses mitochondrial function46, the importance of mitochondrial electron transport in inflammatory responses47 and the finding that IL-10 regulates glycolysis and mammalian target of rapamycin (mTOR)-mediated mitophagy to suppress inflammation via metabolic pathways48. In addition to regulating cellular metabolism, IFNγ can affect systemic metabolism, for example, by modulating glucose tolerance via regulation of the composition of the microbiome49.
Regulation of other immune cells by IFNγ
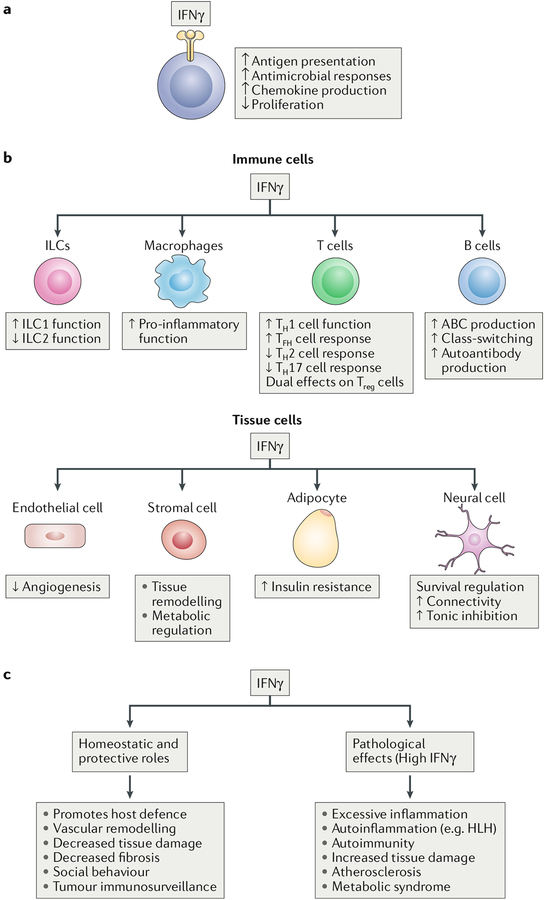
Although immune cell responses to IFNγ have been most extensively studied in macrophages, IFNγ also has important effects on T helper (TH) cells, T follicular helper (TFH) cells, regulatory T (Treg) cells, B cells and innate-like lymphocytes (Figs. 6a,b). The effects of IFNγ on promoting TH1 cell differentiation, suppressing TH2 and TH17 cells, inducing Treg cells specialized to control TH1 cell responses and promoting B cell class switching towards production of immunoglobulin G2a (IgG2a) isotypes have been previously reviewed, as have the roles of interferon in host defence, autoimmune diseases and tissue remodelling1,3. Therefore, the following sections highlight recent advances that extend our understanding of how IFNγ regulates these immune cell populations.
Fig. 6 |. Effects of IFNγ on immune and non-immune cells.
The functional outcomes of IFNγ action on tissues and organs are determined by the integration of its effects on specialized tissue cells and on resident or infiltrating immune cells. The effects of IFNγ are context-dependent and can differ under homeostatic or disease conditions; thus, IFNγ can either suppress or promote tissue damage. a | IFNγ has general effects on various cells. b | IFNγ has effects on different immune cell populations. c | IFNγ has homeostatic and pathological effects. ABC, age-associated B cell; HLH, haemophagocytic lymphohistiocytosis; ILCs, innate lymphoid cells; TFH cell, T follicular helper cell; TH cell, T helper cell; Treg cell, regulatory T cell.
Innate immunity.
As can be surmised from the above discussion of macrophages, IFNγ strongly promotes innate immune and inflammatory responses. Recent insights from infection models include several relevant findings. First, they suggest that IFNγ is important for the local differentiation of monocytes into dendritic cells (DCs) and macrophages that serve as the major sources of IL-12 at sites of infection50. Second, they suggest that the early production of IFNγ by ILC1s is important for local antiviral responses51. Third, they suggest that an important component of vaccine-induced memory is memory T cell-derived IFNγ that instructs strong expression of effector cytokines and microbiocidal pathways in monocytes, DCs, NK cells and natural killer T (NKT) cells during infectious challenge52.
In non-infectious settings, it was recently shown that excessive local production of IFNγ can impair tissue repair by increasing macrophage activation and that tissue production of IFNγ is restrained by Treg cells53. In addition, a recent study found that IFNγ can also promote type 1 immune responses by suppressing the function of tissue-resident group 2 ILCs (ILC2s)54. Type 1 immune responses have been implicated in the effector phase of multiple autoimmune diseases that are characterized by an IFNγ signature at sites of inflammation (reviewed in REF21).
Autoinflammation.
Autoinflammatory diseases are genetic disorders that typically present in childhood with severe and episodic or chronic inflammation and exuberant production of inflammatory cytokines in the absence of overt autoimmunity. Although inflammation in several autoinflammatory conditions is mediated by IL-1-related pathways, inflammation in haemophagocytic lymphohistiocytosis, and possibly in the related macrophage activation syndrome, is mediated by high levels of IFNγ55,56. A pathogenic role for IFNγ in these two conditions, which likely involves activation of macrophages, is supported by animal models, although the role of IFNγ is complex, and it may also have protective effects in certain autoinflammatory diseases56,57. Another group of autoinflammatory disorders termed proteasome disability syndromes (PDS), which includes chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature (CANDLE) and STING-associated vasculopathy with onset in infancy (SAVI), is characterized by an interferon signature in blood cells, and they appear sensitive to JAK inhibitors58. Although type I interferons have been most strongly implicated in these disorders58, aspects of the observed interferon signature and work in animal models suggest a role for IFNγ in some patients59,60.
Adaptive immunity and autoimmune responses.
Recent work in SLE-related autoimmunity models has strongly implicated IFNγ in the generation of TFH cells, germinal centres and pathogenic autoantibodies61–63. IFNγ signalling in T cells and B cells can drive this autoimmune phenotype, in both cases by promoting the expression of B cell lymphoma 6 (BCL-6). The B cell function of IFNγ can be selective for the autoimmune context and autoantibody production and not affect antibody responses against T cell-dependent foreign antigens; such selectivity supports therapeutic targeting of IFNγ. IFNγ also contributes to the formation of age-associated B cells, which are dependent on T-bet and IRF5 and accumulate prematurely and contribute to autoantibody production in SLE64. Further support for targeting IFNγ in autoimmune diseases is provided by reports documenting interferon activity in autoimmune diseases such as rheumatoid arthritis (reviewed in REFs1,21) and by more recent detailed analysis of the interferon signature in diseases such as Sjogren’s syndrome and lupus, which revealed distinct IFNγ and type I interferon signatures in patients’ cells65,66. One important issue is that many autoimmune disease states appear to involve the activity of both IFNγ and type I interferons, which have been recently reviewed67. It can be difficult to resolve IFNγ and type I interferon signatures, which involve overlapping gene sets. In theory, one could separate the activity of these cytokines on the basis of the activation of direct STAT1 targets by IFNγ and ISGF3 targets by type I interferons, but in practice, many genes are commonly induced by both type I and type II interferons by direct and indirect mechanisms. Transcriptomic comparison of gene induction by type I and type II interferons has suggested that certain gene modules or specific genes such as GBP1 and GBP2 are selectively induced by IFNγ65,66, but such conclusions are necessarily constrained by the limited cell types and time points analysed.
Collectively these findings have helped motivate early-phase studies of IFNγ blockade therapy in patients with lupus or rheumatoid arthritis. These studies have clearly demonstrated a role for IFNγ in the interferon signature of patients with SLE, although clinical efficacy remains to be determined68,69. In accord with an important pathogenic role for type I interferons, blockade of these cytokines is also promising in SLE therapy70; the pathogenic functions of type I interferons have been reviewed67 and are beyond the scope of this paper. As previously reviewed1, IFNγ can also have protective functions in restraining autoimmunity and the tissue damage associated with chronic inflammation. Recent examples of protective effects of IFNγ include its role in control of pathogenic self-reactive TH17 cell responses via IL-27 induction71, suppression of autoimmunity via nitric oxide production72,73, induction of specialized Treg cells74,75 and the IFNγ-mediated induction of prostaglandin E2 that suppresses lymphocyte function and promotes myeloid-derived suppressor cell generation in a peritonitis model76.
IFNγ in tissue-specific pathology
Regulation of non-immune cells by IFNγ contributes to tissue-specific pathology.
IFNγR is ubiquitously expressed, and thus IFNγ can act upon most cell types in various body tissues1. Previously, it was thought that the main direct effects of IFNγ on non-immune cells (FIG. 6a) mostly involved1 several cellular processes, including the induction of antiviral ISGs and a local antiviral state; the upregulation or induction of MHC class I and MHC class II molecules on non-immune cells in the tissue, which promotes immune recognition and removal of infected and malignant cells; the induction of chemokines that promote recruitment of immune cells; and the suppression of proliferation by targeting the cell cycle and regulation of cell survival. Thus, the previous paradigm posited that IFNγ acts on local tissue cells primarily, but not exclusively, by inducing expression of ISGs that mediate host defence and immune responses. More recently, evidence has been building that IFNγ has important effects on the tissue-specific functions of non-immune cells and that the combined effects of IFNγ on tissue cells and infiltrating immune cells have an important role in tissue homeostasis and pathobiology (FIG. 6b,c).
IFNγ effects on tissue remodelling, vascular cells and atherosclerosis.
The homeostatic role of IFNγ in limiting inflammation-associated tissue damage has been previously reviewed1. Important components of the protective role of IFNγ are suppression of TH17 cell differentiation, attenuation of infiltration by tissue-damaging cells such as neutrophils and suppression of expression of tissue-degrading enzymes. However, the effects of IFNγ on tissue remodelling are complex, as it antagonizes the function of the homeostatic and pro-repair cytokines IL-4, IL-13 and transforming growth factor-β (TGFβ). Such antagonism can be beneficial by suppressing fibrotic responses driven by excessive action of these cytokines but can be harmful by interfering with their homeostatic functions and the return to tissue homeostasis.
One important aspect of tissue remodelling and repair is regulation of the vasculature - an initial phase of angiogenesis is followed by maturation and regression of blood vessels to allow a return to normal tissue architecture77. IFNγ has been long known to suppress angiogenesis, in part indirectly via regulation of immune cell production of angiogenic factors, such as vascular endothelial growth factors (VEGFs), in part by direct suppression of the proliferation of vascular cells78 and also via induction of antiangiogenic chemokines78. IFNγ also regulates vascular smooth muscle cell pro-liferation, migration and apoptosis to induce loss of vascular smooth muscle cells from maternal spiral arteries during uterine arterial remodelling79. In line with a suppressive role on vascular cells, IFNγ maintains a homeostatic balance of lymph node lymphatic vessels by suppressing lymph node VEGF expression and by suppressing expression of lymphatic-specific genes (such as PROX1 and LYVE1) in lymphatic endothelial cells (LECs), suppressing sprouting and growth of lymphatic vessels and suppressing tube formation by LECs, thereby reducing lymph node lymphatic vessel formation and promoting their post-inflammatory regression80. The regulation of sprouting, tube formation and LEC-specific gene expression by IFNγ supports the idea that it regulates tissue-specific cell function in addition to its general effects on cell proliferation and survival. Effects of IFNγ on vascular cells have been linked to disease pathobiology by genetic evidence that atherosclerosis risk alleles are located in an enhancer that binds STAT1 (REF81). This enhancer is activated in human vascular endothelial cells by IFNγ and appears to regulate expression of various genes, possibly via induction of a non-coding regulatory RNA. Regulation of vascular cells is in line with an important role for IFNγ in atherosclerosis models via its effects on plaque-infiltrating immune cells, endothelial cells and smooth muscle cells78,82.
IFNγ in obesity and metabolic syndrome.
Adipose tissue homeostasis is maintained by eosinophils, ILC2s and invariant NKT (iNKT) cells that secrete type 2 cytokines such as IL-5 and IL-13 that promote M2 polarization of tissue macrophages83. In response to a high-fat diet or obesity, adipose tissue macrophages switch to an M1 phenotype and produce inflammatory mediators such as TNF and IL-1 that contribute to insulin resistance and metabolic syndrome. In line with a pathogenic role of M1 macrophages, deletion of IFNγ improves insulin resistance and metabolic parameters in these models84. Recent work has shown that a high-fat diet induces production of IFNγ by adipose tissue-associated NK cells and ILC1s, which are activated by cell surface ligands expressed by stressed adipocytes and IL-12, respectively (REFS84,85). In addition to polarizing macrophages, adipose tissue-derived IFNγ suppresses an IL-33-driven ILC2 pathway that is important for adipose tissue homeostasis86. As IL-10 regulates adipocyte function via chromatin-based mechanisms87, it is likely that IFNγ also directly regulates adipocyte function.
Effects of IFNγ on neural cell function and in Alzheimer disease.
Microglia are central nervous system (CNS)-resident myeloid cells that are derived from yolk sac progenitors. Recent work has implicated microglia in healthy brain function, such as sculpting developing neuronal circuits, synaptic pruning and guiding learning-associated plasticity, and in the pathogenesis of neurodevelopmental and neurodegenerative diseases such as autism and Alzheimer disease88–91. CNS disease states are associated with microglial cell activation, although such activation is not clearly categorized into M1 and M2 states, and it is not clear whether IFNγ action on CNS myeloid cells is predominantly pathogenic or protective. Indeed, protective functions for IFNγ signalling in myeloid cells have been suggested in the clearance of cerebral amyloid-β plaques in Alzheimer disease models92 and in the experimental autoimmune encephalomyelitis model of multiple sclerosis (reviewed in REF1).
Interestingly, IFNγ acts directly on neurons to regulate their survival and function. In the context of infection or autoimmunity and/or inflammation, IFNγ signalling has deleterious effects on neurons, either promoting cell death or dendrite and synapse loss in a viral encephalitis model (in which IFNγ blockade therapy was neuroprotective) and in human Rasmussen encephalitis93 and exacerbating spinal cord injury94. IFNγ also exerts homeostatic functions under physiological conditions by acting directly on CNS neurons to regulate neuronal connectivity and social behaviour95. Cortical neurons exhibit an IFNγ signature, likely related to IFNγ production by meningeal T cells, and IFNγ-deficient mice exhibit aberrant neuronal hyper-connectivity in fronto-cortical/insular brain regions as well as associated social behavioural deficits. STAT1 deficiency in an inhibitory subset of brain neurons also results in social behaviour deficits. One cellular mechanism of IFNγ action is the augmentation of tonic inhibitory currents, which most likely occurs through elevation of ambient concentrations of the neurotransmitter GABA95. In accord with increased tonic inhibition, IFNγ delays onset and lowers severity of seizures induced by the GABA receptor antagonist pentylenetetrazol. The specific behavioural defects of IFNγ deficiency contrast with the regulation of spatial learning behaviour by IL-4 (REF96), which also acts directly on peripheral sensory neurons to sensitize them to pruritogens and promote itching97. These neural functions of IFNγ are part of an exciting emerging area of cytokines as neuromodulators and suggest novel mechanisms by which infections that increase CNS IFNγ amounts can modify behaviour.
Pivotal role of IFNγ in cancer immunotherapy
The antitumour effects of type I and type II interferons and the effectiveness of anti-type I interferon therapies have been extensively described and previously reviewed98,99. Briefly, IFNγ can suppress tumours by acting directly on tumour cells (inhibiting their proliferation while increasing MHC expression, antigen presentation and thus antigenicity and cell death), by augmenting the function oftumour-infiltrating immune cells including TH1 cells, CTLs and macrophages, by suppressing Treg cell function and by modulating stromal cell function to alter metabolism and suppress angiogene-sis (FIG. 7). IFNγ also suppresses metastasis by altering the extracellular matrix and tumour architecture100. Extensive evidence that tumours develop resistance to the effects of interferons to escape immune eradication further supports an important role for interferons in antitumour immunity99.
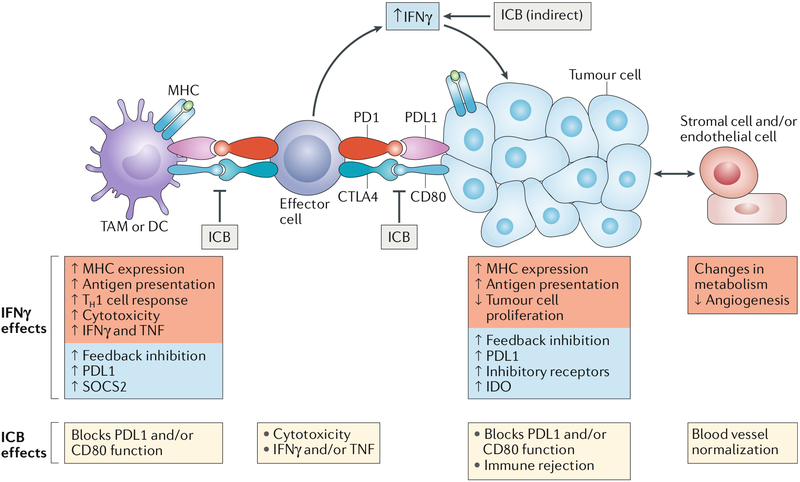
Fig. 7 |. IFnγ and cancer immunotherapy.
IFNγ plays an important role in the effectiveness of immune checkpoint blockade (ICB). ICB blocks the interaction of programmed cell death 1 ligand 1 (PDL1), CD80 and CD86 expressed on tumour cells, tumour-associated macrophages (TAMs) and dendritic cells (DCs) with their cognate inhibitory receptors programmed cell death protein 1 (PD1) and cytotoxic T lymphocyte antigen 4 (CTL A4) expressed on tumour-infiltrating effector T cells (including cytotoxic T lymphocytes (CTLs)). Two important consequences of ICB are increased T cell function (because of diminished inhibitory signalling that reverses their exhausted state) and increased intratumoural production of IFNγ, likely at least in part by T cells. Important IFNγ functions (red boxes) include direct effects on tumour cells to suppress proliferation and increase antigen presentation. The effects of IFNγ and ICB on the depicted cell types are listed under each cell type. The combination of increased CTL function and increased antigen presentation promotes immune-mediated tumour eradication. IFNγ also has feedback inhibitory effects (blue boxes) that can attenuate antitumour immunity; overcoming these inhibitory effects is an important goal for improving the efficacy of ICB. IDO, indoleamine 2,3-dioxygenase; TH1 cell, T helper 1 cell; TNF, tumour necrosis factor; SOCS2, suppressor of cytokine signalling 2.
A recent breakthrough in cancer therapy is immune checkpoint blockade (ICB), which involves blocking inhibitory receptors that are expressed on intratumoural effector T cells101–103. Most notably, ligation of cytotoxic T lymphocyte antigen 4 (CTLA4) and programmed cell death protein 1 (PD1) by their ligands (CD80 and CD86, and programmed cell death 1 ligand 1 (PDL1), respectively), which are expressed on tumour cells and tumour-associated macrophages (TAMs), suppresses T cell effector function and cytotoxicity, promotes T cell exhaustion and allows the tumour to escape immune responses (FIG. 7). ICB using blocking antibodies against CTLA4 (ipilimumab) or PD1 (pembrolizumab) strongly activates antitumour immunity and has generated striking clinical responses, but certain patients are resistant and some tumours do not respond to ICB. Thus, mechanisms of resistance and how ICB mobilizes antitumour immunity are under intense study.
Induction of intratumoural IFNγ production by ICB in patients and mouse models, and dependence of tumour infiltration by immune cells on IFNγR104–107, suggests a function for IFNγ in mediating tumour rejection (FIG. 7). A role for ICB-induced IFNγ action directly on tumour cells received strong support from studies that analysed tumour cells from patients with melanoma who were resistant to anti-CTLA4 or anti-PD1 therapy108–110. Strikingly, resistance to checkpoint blockade was found to be associated with genomic defects in the IFNγ pathway in tumour cells, including mutations in both components of IFNγR (IFNγR1 and IFNγR2), JAK2 and the downstream protein IRF1 (REFs108–110). Mutations were also found in β2-microglobulin, which is required for cell surface expression of IFNγ-inducible MHC class I molecules and presentation of intracellular antigens to T cells. These results support a model whereby ICB-induced IFNγ works in part by increasing presentation of tumour antigens to CTLs, which themselves have been directly sensitized by blockade of inhibitory receptors. Accordingly, knockdown of IFNγRl in B16 melanoma tumours results in increased in vivo tumour growth and decreased mouse survival after ICB108. This model received further support from genome-wide CRISPR-mediated screens aimed at identifying molecules important for immunotherapy and CTL function111,112. Strikingly, both screens found that defects in IFNγ signalling contribute to resistance to immunotherapy and suggested that IFNγ confers sensitivity to immunotherapy by suppressing tumour cell growth and increasing MHC class I-mediated antigen presentation, thereby increasing sensitivity of tumour cells to CTLs. Accordingly, genes important for antigen presentation were found to be mutated in more than 100 patient tumours in The Cancer Genome Atlas database. The additional finding that several molecules newly implicated in regulating responses to immunotherapy in these CRISPR screens, for example, tyrosine-protein phosphatase non-receptor type 2 (PTPN2) and apelin receptor (APLNR)111,112, actually work by modulating cellular responsiveness to IFNγ further supports the pivotal role of this cytokine in the efficacy of antitumour immunotherapy.
However, like most cytokines, IFNγ induces feedback inhibitory mechanisms to restrain the magnitude of immune responses1. In tumours, IFNγ induces expression of inhibitory receptors, including PDL1, on tumour cells and TAMs (FIG. 7) and upregulates suppressor of cytokine signalling 2 (SOCS2) in DCs102,113–116. Thus, IFNγ can also have suppressive effects on antitumour immunity. As is often the case with feedback pathways, the relative balance of activating and suppressive mechanisms induced by IFNγ determines the overall functional outcome. In ICB, anti-PD1 therapy blocks a suppressive mechanism of IFNγ, namely, the function of IFNγ-induced PDL1, and this likely potentiates its antitumour activity and therapeutic efficacy. It is plausible that resistance to ICB is mediated by distinct inhibitory receptors and molecules that are induced by IFNγ but not targeted by the ICB therapy102–113,115.
In addition to its effects on tumour cells, IFNγ contributes to immunotherapy and the efficacy of checkpoint blockade by acting on endothelial cells to promote blood vessel normalization (increased pericyte coverage, decreased leakiness and decreased hypoxia) and regression117,118 and by inducing Treg cell fragility119; additional mechanisms of action are likely to be discovered. Furthermore, IFNγ improves the efficacy of chemotherapy with cisplatin, doxorubicin, antibodies against receptor tyrosine-protein kinase ERBB2 and kinase inhibitors by targeting stromal cell functions and by as yet undiscovered mechanisms and may play a role in responses to radiation combined with ICB120–123. Thus, IFNγ is an integral component of various antitumour therapies.
Concluding remarks
Over the past decade, our understanding of cellular responses to IFNγ has been extended beyond its induction of the core JAK-STAT signalling pathway and ISGs. We now appreciate that IFNγ induces complex reprogramming of cell state and responsiveness to environmental cues, which is mediated by epigenetic and metabolic mechanisms. In parallel, our understanding of the cellular targets of IFNγ has been extended beyond immune cells, and we are now aware of the various effects of IFNγ on stromal and specialized tissue cells. One important future direction is to gain a deeper understanding of the associated epigenetic and metabolic mechanisms, especially in non-immune cells and in vivo (including in disease states), coupled with investigation of the effects of IFNγ on 3D chromatin conformation and DNA methylation. A challenge will be to link specific ISGs with the epigenetic and metabolic mechanisms described herein. Another interesting area of study is the relationship of IFNγ-mediated priming with training of innate immunity by microbial exposure that promotes more effective recall responses39. Additionally, future studies should address the polarization of tissue-resident macrophages, as their different transcriptional starting point relative to the bone marrow-derived or blood-derived macrophages typically used for polarization studies may result in distinct polarization outcomes. It will be important to understand the role of IFNγ in regulating the functions of specialized tissue cells, its effects on progenitor and stem cells and the implications for tissue and organ function under homeostatic, immune and pathological conditions. It is perhaps surprising that IFNγ plays important homeostatic roles, and determining the mechanisms underlying context-dependent IFNγ functions will be important for developing therapeutic strategies to manipulate IFNγ activity to promote health and suppress disease. Advancement of our knowledge of IFNγ functions and mechanisms of action, which have been summarized in this Review, can be harnessed to develop new therapeutic strategies to improve host defence, suppress autoimmunity and augment responses to various cancer therapies, including in patients with tumours resistant to currently available therapeutics.
Acknowledgements
This work was supported by grants from the National Institutes of Health.
Glossary
- IFNγ signature
A pattern of elevated expression of canonical IFNγ target genes in inflamed tissues; it is often detected in samples from patients with autoimmune disease.
- Endotoxin tolerance
Classically, a macrophage cell state in which prior exposure to lipopolysaccharide (LPS; an endotoxin) renders inflammatory nuclear factor-κB (NF-κB) target genes refractory to induction by subsequent LPS challenge. Tolerance can be induced by various inflammatory factors such as tumour necrosis factor (TNF), IL-1 and Toll-like receptor (TLR) ligands, and tolerized cells are resistant to multiple cell activators.
- Interferon-stimulated gene factor 3
(ISGF3). A transcription factor complex comprising signal transducer and activator of transcription 1 (STAT1), STAT2 and interferon regulatory factor 9 (IRF9) that binds to interferon-stimulated response elements and regulates the expression of interferon-stimulated genes. ISGF3 is predominantly activated by type I interferons.
- Latent enhancers
Enhancers that are inactive and associated with closed chromatin in resting myeloid cells. During cell activation, chromatin at latent enhancers becomes accessible, and they bind to transcription factors and drive expression of associated genes.
- M2 macrophage
A type of macrophage that has been polarized by IL-4, IL-13, IL-10, glucocorticoids or various anti-inflammatory factors. M2 macrophages exhibit a range of phenotypes related to resolution of inflammation, wound healing and tissue remodelling.
- Mitophagy
The selective degradation of mitochondria by autophagy.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Immunology thanks T. Decker, G. Trinchieri and P. Hertzog for their assistance with the peer review of this manuscript.
References
- 1.Hu X & Ivashkiv LB Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity 31, 539–550 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark GR & Darnell JE Jr. The JAK-STAT pathway at twenty. Immunity 36, 503–514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villarino AV, Kanno Y & O’Shea JJ Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat. Immunol 18, 374–384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy DE, Kessler DS, Pine R, Reich N & Darnell JE Jr. Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 2, 383–393 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Levy DE, Lew DJ, Decker T, Kessler DS & Darnell JE Jr. Synergistic interaction between interferon-alpha and interferon-gamma through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 9, 1105–1111 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oyler-Yaniv J et al. Catch and release of cytokines mediated by tumor phosphatidylserine converts transient exposure into long-lived inflammation. Mol. Cell 66, 635–647.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang K et al. Interferon-gamma represses M2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity 47, 235–250.e4 (2017).This study reveals mechanisms by which IFNγ represses gene transcription and identifies the functional importance of IFNΓ-repressed genes.
- 8.Langlais D, Barreiro LB & Gros P The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J. Exp. Med 213, 585–603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancino A et al. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev. 29, 394–408 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bancerek J et al. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38, 250–262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Begitt A et al. STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat. Immunol 15, 168–176 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Majoros A et al. Canonical and non-canonical aspects of JAK-STAT signaling: lessons from interferons for cytokine responses. Front. Immunol 8, 29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wienerroither S et al. Cooperative transcriptional activation of antimicrobial genes by STAT and NF-kappaB pathways by concerted recruitment of the mediator complex. Cell Rep. 12, 300–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheon H, Yang J & Stark GR The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J. Interferon Cytokine Res. 31, 33–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glass CK & Natoli G Molecular control of activation and priming in macrophages. Nat. Immunol 17, 26–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray PJ et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piccolo V et al. Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat. Immunol 18, 530–540 (2017).This study delineates mechanisms by which IFNγ and IL-4 oppose each other’s actions during macrophage polarization and shows the extent of plasticity of polarization phenotypes genome wide.
- 18.Qiao Y et al. Synergistic activation of inflammatory cytokine genes by interferon-gamma-induced chromatin remodeling and toll-like receptor signaling. Immunity 39, 454–469 (2013).This study identifies the epigenetic basis for synergistic transcriptional activation of inflammatory cytokine genes by IFNγ and TLR signalling.
- 19.Qiao Y, Kang K, Giannopoulou E, Fang C & Ivashkiv LB IFN-gamma induces histone 3 lysine 27 trimethylation in a small subset of promoters to stably silence gene expression in human macrophages. Cell Rep. 16, 3121–3129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J & Ivashkiv LB IFN-gamma abrogates endotoxin tolerance by facilitating Toll-like receptor-induced chromatin remodeling. Proc. Natl Acad. Sci. USA 107, 19438–19443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, Chakravarty SD & Ivashkiv LB Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol. Rev 226, 41–56 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karonitsch T et al. Interferon signals and monocytic sensitization of the interferon-gamma signaling pathway in the peripheral blood of patients with rheumatoid arthritis. Arthritis Rheumatism 64, 400–408 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Askenase MH et al. Bone-marrow-resident NK cells prime monocytes for regulatory function during infection. Immunity 42, 1130–1142 (2015).This study clearly establishes that IFNΓ-mediated priming occurs in vivo and is important for host defence against pathogens.
- 24.Leentjens J et al. Reversal of immunoparalysis in humans in vivo: a double-blind, placebo-controlled, randomized pilot study. Am. J. Respiratory Crit. Care Med 186, 838–845 (2012).This study demonstrates that IFNγ reverses endotoxin tolerance in vivo in human subjects.
- 25.Cheng SC et al. Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat. Immunol 17, 406–413 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Park SH et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat. Immunol 18, 1104–1116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PloS one 9, e93846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrero C et al. Reprogramming of IL-10 activity and signaling by IFN-gamma. J. Immunol 171, 5034–5041 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Monticelli S & Natoli G Short-term memory of danger signals and environmental stimuli in immune cells. Nat. Immunol 14, 777–784 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Foster SL, Hargreaves DC & Medzhitov R Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 (2007).This study introduces the concept of epigenetic regulation into the field of endotoxin tolerance.
- 31.Ivashkiv LB Epigenetic regulation of macrophage polarization and function. Trends Immunol. 34, 216–223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smale ST, Tarakhovsky A & Natoli G Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol 32, 489–511 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Ostuni R et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).This study demonstrates stable remodelling of chromatin at de novo enhancers induced by immune stimuli, including IFNγ, and helps pioneer the concept of short-term memory in macrophage responses.
- 34.Li JJ et al. IL-27/IFN-gamma induce MyD88-dependent steroid-resistant airway hyperresponsiveness by inhibiting glucocorticoid signaling in macrophages. J. Immunol 185, 4401–4409 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Yoshimura A, Ito M, Chikuma S, Akanuma T & Nakatsukasa H Negative regulation of cytokine signaling in immunity. Cold Spring Harb. Perspect. Biol 10.1101/cshperspect.a028571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czimmerer Z et al. The transcription factor STAT6 mediates direct repression of inflammatory enhancers and limits activation of alternatively polarized macrophages. Immunity 48, 75–90.e76 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biswas SK & Lopez-Collazo E Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30, 475–487 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Ho PC, Tsui YC, Feng X, Greaves DR & Wei LN NF-kappaB-mediated degradation of the coactivator RIP140 regulates inflammatory responses and contributes to endotoxin tolerance. Nat. Immunol 13, 379–386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Netea MG et al. Trained immunity: a program of innate immune memory in health and disease. Science 352, aaf1098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novakovic B et al. β-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167, 1354–1368.e14 (2016).This study extends the understanding of epigenetic mechanisms in innate immune training and establishes their importance in reversal of tolerance by training stimuli.
- 41.O’Neill LA, Kishton RJ & Rathmell J A guide to immunometabolism for immunologists. Nat. Rev. Immunol 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu X et al. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity 24, 563–574 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Meares GP, Qin H, Liu Y, Holdbrooks AT & Benveniste EN AMP-activated protein kinase restricts IFN-gamma signaling. J. Immunol 190, 372–380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su X et al. Interferon-gamma regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol 16, 838–849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deretic V, Saitoh T & Akira S Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol 13, 722–737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van den Bossche J et al. Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Rep. 17, 684–696 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Mills EL et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470.e13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ip WKE, Hoshi N, Shouval DS, Snapper S & Medzhitov R Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greer RL et al. Akkermansia muciniphila mediates negative effects of IFNgamma on glucose metabolism. Nat. Commun 7, 13329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldszmid RS et al. NK cell-derived interferon-gamma orchestrates cellular dynamics and the differentiation of monocytes into dendritic cells at the site of infection. Immunity 36, 1047–1059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weizman OE et al. ILC1 confer early host protection at initial sites of viral infection. Cell 171, 795–808. e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soudja SM, Ruiz AL, Marie JC & Lauvau G Inflammatory monocytes activate memory CD8(+) T and innate NK lymphocytes independent of cognate antigen during microbial pathogen invasion. Immunity 37, 549–562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panduro M, Benoist C & Mathis D Treg cells limit IFN-γ production to control macrophage accrual and phenotype during skeletal muscle regeneration. Proc. Natl Acad. Sci. USA 115, E2585–E2593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moro K et al. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat. Immunol 17, 76–86 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Pachlopnik Schmid J et al. Inherited defects in lymphocyte cytotoxic activity. Immunol. Rev 235, 10–23 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Canna SW Editorial: interferon-gamma: friend or foe in systemic juvenile idiopathic arthritis and adult-onset Still’s Disease? Arthritis Rheumatol. 66, 1072–1076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avau A et al. Systemic juvenile idiopathic arthritis-like syndrome in mice following stimulation of the immune system with Freund’s complete adjuvant: regulation by interferon-gamma. Arthritis Rheumatol. 66, 1340–1351 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Canna SW & Goldbach-Mansky R New monogenic autoinflammatory diseases—a clinical overview. SeminarsImmunopathol. 37, 387–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y et al. Mutations in proteasome subunit beta type 8 cause chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature with evidence of genetic and phenotypic heterogeneity. Arthritis Rheumatism 64, 895–907 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reinhardt RL et al. A novel model for IFN-gamma-mediated autoinflammatory syndromes. J. Immunol 194, 2358–2368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Domeier PP et al. IFN-gamma receptor and STAT1 signaling in B cells are central to spontaneous germinal center formation and autoimmunity. J. Exp. Med 213, 715–732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson SW et al. B cell IFN-gamma receptor signaling promotes autoimmune germinal centers via cell-intrinsic induction of BCL-6. J. Exp. Med 213, 733–750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SK et al. Interferon-gamma excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity 37, 880–892 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Manni M et al. Regulation of age-associated B cells by IRF5 in systemic autoimmunity. Nat. Immunol 19, 407–419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiche L et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis Rheumatol. 66, 1583–1595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall JC et al. Precise probes of type II interferon activity define the origin of interferon signatures in target tissues in rheumatic diseases. Proc. Natl Acad. Sci. USA 109, 17609–17614 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ivashkiv LB & Donlin LT Regulation of type I interferon responses. Nat. Rev. Immunol 14, 36–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Welcher AA et al. Blockade of interferon-gamma normalizes interferon-regulated gene expression and serum CXCL10 levels in patients with systemic lupus erythematosus. Arthritis Rheumatol. 67, 2713–2722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Werth VP et al. Brief report: pharmacodynamics, safety, and clinical efficacy of AMG 811, a human anti-interferon-gamma antibody, in patients with discoid lupus erythematosus. Arthritis Rheumatol. 69, 1028–1034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Touma Z & Gladman DD Current and future therapies for SLE: obstacles and recommendations for the development of novel treatments. Lupus Sci. Med 4, e000239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chong WP et al. NK-DC crosstalk controls the autopathogenic Th17 response through an innate IFN-gamma-IL-27 axis. J. Exp. Med 21 2, 1739–1752 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koblish HK, Hunter CA, Wysocka M, Trinchieri G & Lee WM Immune suppression by recombinant interleukin (rIL)-12 involves interferon gamma induction of nitric oxide synthase 2 (iNOS) activity: inhibitors of NO generation reveal the extent of rIL-12 vaccine adjuvant effect. J. Exp. Med 188, 1603–1610 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tarrant TK et al. Interleukin 12 protects from a T helper type 1-mediated autoimmune disease, experimental autoimmune uveitis, through a mechanism involving interferon gamma, nitric oxide, and apoptosis. J. Exp. Med 189, 219–230 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall AO et al. The cytokines interleukin 27 and interferon-gamma promote distinct Treg cell populations required to limit infection-induced pathology. Immunity 37, 511–523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levine AG et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 546, 421–425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Newson J et al. Inflammatory resolution triggers a prolonged phase of immune suppression through COX-1/mPGES-1-derived prostaglandin E2. Cell Rep. 20, 3162–3175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vannella KM & Wynn TA Mechanisms of organ injury and repair by macrophages. Annu. Rev. Physiol 79, 593–617 (2017). [DOI] [PubMed] [Google Scholar]
- 78.McLaren JE & Ramji DP Interferon gamma: a master regulator of atherosclerosis. Cytokine Growth Factor Rev. 20, 125–135 (2009). [DOI] [PubMed] [Google Scholar]
- 79.Liu W et al. dNK derived IFN-gamma mediates VSMC migration and apoptosis via the induction of LncRNA MEG3: a role in uterovascular transformation. Placenta 50, 32–39 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Kataru RP et al. T lymphocytes negatively regulate lymph node lymphatic vessel formation. Immunity 34, 96–107 (2011).This study establishes a new function for IFNγ in the regulation of lymphatic vessels and lymph node function by acting directly on tissue cells.
- 81.Harismendy O et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature 470, 264–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tabas I & Lichtman AH Monocyte-macrophages and T cells in atherosclerosis. Immunity 47, 621–634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Odegaard JI & Chawla A Type 2 responses at the interface between immunity and fat metabolism. Curr. Opin. Immunol 36, 67–72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wensveen FM et al. NK cells link obesity-induced adipose stress to inflammation and insulin resistance. Nat. Immunol 16, 376–385 (2015). [DOI] [PubMed] [Google Scholar]
- 85.O’Sullivan TE et al. Adipose-resident group 1 innate lymphoid cells promote obesity-associated insulin resistance. Immunity 45, 428–441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Molofsky AB et al. Interleukin-33 and interferon-gamma counter-regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity 43, 161–174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rajbhandari P et al. IL-10 signaling remodels adipose chromatin architecture to limit thermogenesis and energy expenditure. Cell 172, 218–233.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Herz J, Filiano AJ, Smith A, Yogev N & Kipnis J Myeloid cells in the central nervous system. Immunity 46, 943–956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ransohoff RM How neuroinflammation contributes to neurodegeneration. Science 353, 777–783 (2016). [DOI] [PubMed] [Google Scholar]
- 90.Salter MW & Stevens B Microglia emerge as central players in brain disease. Nat. Med 23, 1018–1027 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Ulland TK et al. TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell 170, 649–663.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baruch K et al. PD-1 immune checkpoint blockade reduces pathology and improves memory in mouse models of Alzheimer’s disease. Nat. Med 22, 135–137 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Kreutzfeldt M et al. Neuroprotective intervention by interferon-gamma blockade prevents CD8+ T cell-mediated dendrite and synapse loss. J. Exp. Med 210, 2087–2103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun G et al. gammadelta T cells provide the early source of IFN-gamma to aggravate lesions in spinal cord injury. J. Exp. Med 215, 521–535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Filiano AJ et al. Unexpected role of interferon-gamma in regulating neuronal connectivity and social behaviour. Nature 535, 425–429 (2016).This study introduces the importance of the direct action of IFNγ on neurons under physiologic conditions and delineates the importance of IFNγ for neuronal function and social behaviours.
- 96.Derecki NC et al. Regulation of learning and memory by meningeal immunity: a key role for IL-4. J. Exp. Med 207, 1067–1080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oetjen LK et al. Sensory neurons co-opt classical immune signaling pathways to mediate chronic itch. Cell 171, 217–228.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dunn GP, Koebel CM & Schreiber RD Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol 6, 836–848 (2006). [DOI] [PubMed] [Google Scholar]
- 99.Parker BS, Rautela J & Hertzog PJ Antitumour actions of interferons: implications for cancer therapy. Nat. Rev. Cancer 16, 131–144 (2016). [DOI] [PubMed] [Google Scholar]
- 100.Glasner A et al. NKp46 receptor-mediated interferon-gamma production by natural killer cells increases fibronectin 1 to alter tumor architecture and control metastasis. Immunity 48, 396–398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Callahan MK, Postow MA & Wolchok JD Targeting T cell co-receptors for cancer therapy. Immunity 44, 1069–1078 (2016). [DOI] [PubMed] [Google Scholar]
- 102.Minn AJ & Wherry EJ Combination cancer therapies with immune checkpoint blockade: convergence on interferon signaling. Cell 165, 272–275 (2016). [DOI] [PubMed] [Google Scholar]
- 103.Sharma P & Allison JP The future of immune checkpoint therapy. Science 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 104.Fu T, He Q & Sharma P The ICOS/ICOSL pathway is required for optimal antitumor responses mediated by anti-CTLA-4 therapy. Cancer Res. 71, 5445–5454 (2011). [DOI] [PubMed] [Google Scholar]
- 105.Liakou CI et al. CTLA-4 blockade increases IFNgamma-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc. Natl Acad. Sci. USA 105, 14987–14992 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peng W et al. PD-1 blockade enhances T cell migration to tumors by elevating IFN-gamma inducible chemokines. Cancer Res. 72, 5209–5218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shi LZ et al. Interdependent IL-7 and IFN-gamma signalling in T cell controls tumour eradication by combined alpha-CTLA-4+alpha-PD-1 therapy. Nat. Commun 7, 12335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao J et al. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404.e9 (2016).Together with references 109–112, this study establishes the pivotal role of IFNγ pathways in the efficacy of immune checkpoint blockade therapy for cancers.
- 109.Shin DS et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. CancerDiscov. 7, 188–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zaretsky JM et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Manguso RT et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Patel SJ et al. Identification of essential genes for cancer immunotherapy. Nature 548, 537–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Benci JL et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 167, 1540–1554.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garcia-Diaz A et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 19, 1189–1201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nirschl CJ et al. IFNgamma-dependent tissue-immune homeostasis is co-opted in the tumor microenvironment. Cell 170, 127–141.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zaidi MR et al. Interferon-gamma links ultraviolet radiation to melanomagenesis in mice. Nature 469, 548–553 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kammertoens T et al. Tumour ischaemia by interferon-gamma resembles physiological blood vessel regression. Nature 545, 98–102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tian L et al. Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature 544, 250–254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Overacre-Delgoffe AE et al. Interferon-gamma drives Treg fragility to promote anti-tumor immunity. Cell 169, 1130–1141.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hannesdottir L et al. Lapatinib and doxorubicin enhance the Stat1-dependent antitumor immune response. Eur. J. Immunol 43, 2718–2729 (2013). [DOI] [PubMed] [Google Scholar]
- 121.Stagg J et al. Anti-ErbB-2 mAb therapy requires type I and II interferons and synergizes with anti-PD-1 or anti-CD137 mAb therapy. Proc. Natl Acad. Sci. USA 108, 7142–7147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Twyman-Saint Victor C et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 520, 373–377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang W et al. Effector T cells abrogate stroma-mediated chemoresistance in ovarian cancer. Cell 165, 1092–1105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]