Abstract
The pathogenesis of hepatocellular carcinoma (HCC) is poorly understood, but recent advances in genomics have increased our understanding of the mechanisms by which HBV, HCV, alcohol, fatty liver disease, and other environmental factors, such as aflatoxin, cause liver cancer. Genetic analyses of liver tissues from patients have provided important information about tumor initiation and progression. Findings from these studies can potentially be used to individualize the management of HCC. In addition to sorafenib, other multikinase inhibitors have recently been approved for treatment of HCC and the preliminary success of immunotherapy has raised hopes. Continued progress in genomic medicine could improve classification of HCCs based on their molecular features and lead to new treatments for patients with liver cancer.
Keywords: HCC, epigenetics, tert, hepatitis, immunotherapy
Hepatocellular carcinoma (HCC) is the second-most common cause of cancer mortality worldwide 1. HCC is most commonly caused by chronic hepatitis or cirrhosis, resulting from infection with hepatitis B virus (HBV) and C virus (HCV), as well as alcoholic or fatty liver diseases. However, the attributable risks from different etiologies vary significantly among regions. HBV is the most common risk factor for HCC in Southeast Asia and sub-Saharan Africa 2, 3, 4, whereas HCV infection is the most common risk factor in Egypt 5, Europe 6, North America 7, and Japan 8. Despite the magnitude of the global burden of HCC, it is one of the least-understood cancers and has limited therapeutic options. Advances in genomic research have increased our understanding of HCC development, and could lead to new strategies for prevention and therapy. We review the genomic features of HCC, correlations between genotypes and phenotypes, progression of viral hepatitis-related HCC, and strategies to individualize treatment.
Genetics
Over the past decade the study of cancer has shifted from evaluation of variants of individual genes and pathways to analyses of gene expression patterns and epigenetic profiles of tumor tissues and cells. Advances in next-generation sequencing and computational data analyses can be credited for this shift. The genetic events that contribute to HCC initiation and progression can be classified as genomic (somatic mutations and genome structure changes such as gene fusions or copy number variations), epigenetic (changes in methylation, chromatin remodeling, microRNAs, and long non-coding RNAs [lnc RNAs]), and transcriptional (changes in gene expression) (Figure 1).
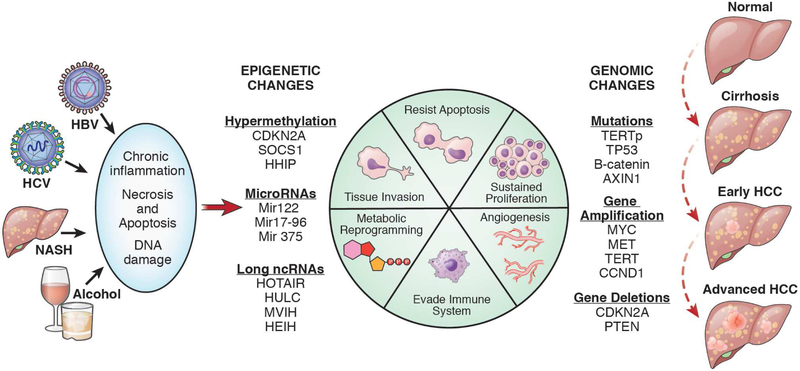
Figure 1. Pathogenesis of HCC.
Liver cirrhosis is caused by chronic infection with HBV or HCV, alcohol abuse, or fatty liver disease. The chronic inflammation, necrosis, and regeneration that occur during development of cirrhosis cause genetic and epigenetic changes in liver tissue that lead to tumor formation.
NASH, non-alcoholic steatohepatitis
Somatic Genomic Events
Somatic mutations occur in somatic (non-germ) cells and are therefore not heritable. When these mutations occur in proto-oncogenes or tumor suppressor genes or in genes involved in regulatory pathways, they can lead to cell transformation and tumorigenesis. Whole-exome and whole-genome sequencing studies have identified mutations that contribute to development of HCC 9–19. The well-characterized mutations in HCCs are in CTNNB1 (which encodes beta-catenin), TP53, AXIN1, RB1, ARID1A, ARID2, and NFE2L2. Mutations in the catalytic telomerase reverse transcriptase (TERT) have been more recently recognized as frequent driver events detected in 40%–65% of HCC samples 20–24. The first case of germline mutation in TERT was initially discovered in an analysis of data from the Cancer Genome Atlas (TCGA) of HCC, implying germline mutations in TERT might cause inherited forms of HCC 19. TERT promoter mutations cause overexpression of telomerase, which allows cells to become immortal. Mutations in the TERT promoter that increase its expression appear to be early events in hepatocarcinogenesis 20, 21. Furthermore, the TERT gene appears to be altered by HBV and HCV infection, via different mechanisms. Mutations in the TERT promoter have been more frequently associated with HCC resulting from chronic HCV infection and alcohol intake 20, 25 than with HBV-associated HCC. However, in Hep B related HCC, telomerase expression can be activated by recurrent integration of HBV into the TERT promoter26. TERT alterations promote cell immortality and transformation also via interactions with transcriptions factors such as MYC 27, beta-catenin 28 and NF-KB 29, to alter expression of their target genes.
Mutations that disrupt the function of TP53 are detected in 12%–48% of HCCs, and with high frequency in advanced tumors, but no therapeutic strategies have been developed to restore TP53 function to cells. An analysis of HCCs in TCGA identified a TP53-regulated gene expression signature that can be used to identify HCC tumors with loss of TP53 function—even when the TP53 gene is not mutated. The TP53-regulated gene expression signature was associated with clinical outcome and might be used as a biomarker to select treatment. HCCs have developed methods to reduce TP53 activity without mutating the TP53 gene. For example, TP53 levels are reduced in liver tissues from patients with chronic HBV infection via direct repression of the TP53 gene promoter by HBx 30.
Activating mutations of in CTNNB1 have been found in 11%–37% of HCC samples, and inactivating mutations in AXIN1 have been found in 5%–15% of HCCs. These mutations activate Wnt signaling, which promotes cell motility, de-differentiation, and proliferation 31. Mutations in proteins that regulate chromatin remodeling, such as ARID1A, are detected in 4%–17% of HCCs; ARID2 mutations are found in 3%–18% of HCCs 9, 14, 19. These mutations lead to transcriptional repression of genes regulated by the transcription factor E2F. In normal cells, these genes block cell proliferation by upregulating CDKN1A, which encodes the cyclin-dependent kinase inhibitor P21 32.
Many HCC cells contain copy number alterations that result in either gains or losses of segments of genomic DNA. Genes with increased copy numbers amplifications in HCC include FGF19 and CCND1. Amplification of FGF19 results in increased expression of its product and FGF pathway activation 33, 17. Brivanib, an inhibitor of VEGF and FGF, did not provide clinical benefit to patients with HCC. However, lenvatinib, another inhibitor of multiple tyrosine kinase receptors, including FGF receptors, increased survival times in patients with HCC in a phase 3 trial 34, 35. Other highly potent or irreversible FGFR inhibitors are being evaluated in patients and these might be more effective and have better safety profiles36. Other oncogenes that are frequently amplified in HCCs include TERT, VEGFA, MYC, CCND1, and MET 10, 14, 19, whereas tumor suppressor genes such as PTEN37,38 and CDKN2A (encoding P16INK4A) are frequently deleted in HCC samples 39, 40. Loss of these genes leads to cell cycle progression and proliferation.
Epigenetic Changes
Epigenetic alterations also alter gene expression to affect cell and tissue phenotypes 41. Epigenetic modifications occur via processes such as DNA methylation, covalent modifications to chromatin, alterations in nucleosome position, and changes in levels of micro-RNAs (miRNAs) and long noncoding RNAs (lncRNAs). Epigenetic and genetic events can co-operate to promote tumorigenesis or progression and metastasis. For example, TERT promoter mutations frequently co-occur with silencing of CDKN2A by promoter hypermethylation 19. The combination of telomerase overexpression and silencing of a cell cycle checkpoint inhibitor contribute to cell immortalization 42.
Some genes that are silenced by promoter hypermethylation during hepatocarcinogenesis include the suppressor of cytokine signaling 1 (SOCS1)43,44, hedgehog interacting protein (HHIP)19, 45, CDKN2A, CDKN1A, CDKN2B46, APC 47, carbamoyl-phosphate synthase 1 (CPS1, a urea cycle gene)48, TIMP metallopeptidase inhibitor 3 (TIMP3)49, and glutathione S-transferase pi 1 (GSTP1)50. HCV and HBV can induce epigenetic modifications that promote liver tumorigenesis. HCV induces overexpression of protein phosphatase 2 catalytic subunit alpha (PPP2CA), leading to deregulation of histone modifications, altered gene expression, and anchorage-independent growth51. In vivo and in vitro studies have shown that HCV can induce promoter hypermethylation and silencing of GADD45B, leading to aberrant cell cycle arrest and diminished DNA excision repair52. HBV infection also appears to lead to unique DNA methylation patterns that suppress genes including MDM2, FGF4, FGF19, and HSP90AA153. HBV alters the epigenome via HBx protein54,55. HBx increases total DNA methyltransferase (DNMT) activity and promotes regional hypermethylation of specific tumor suppressor genes54,56. HBx also promotes recruitment and transactivation of co-activators of the CREB-binding protein CBP–P300 complex, leading to acetylation and thereby activation of cellular genes57.
MicroRNAs are short (20–22 nucleotide) non-coding RNAs that pair with complementary 3’-untranslated regions mRNAs, inhibiting their translation or leading to their degradation58. A single microRNA can control levels of several mRNAs to regulate biological processes such apoptosis, differentiation, and metastasis. One of the most abundant microRNAs in the liver is microRNA 122 (MIR122), which is involved in regulating several genes in the cholesterol metabolism pathway and is also required for HCV replication59. Levels of MIR122 are significantly reduced in HCCs19, 60, which is associated with metastasis and poor outcomes. MIR122-knockout mice develop spontaneous liver tumors resembling HCCs61 and re-expression of MIR122 reduced tumor incidence and development in Mir122a−/− mice61,62. MIR375 is also downregulated in HCCs and appears to function as a tumor suppressor. Delivery of MIR375 into HCC cells, via MIR375 mimics on the surface of gold nanoparticles, reduced proliferation and induced apoptosis 63.
Several microRNAs appear to promote tumorigenesis, called oncomirs. Their levels are increased expression HCCs. MIR221 is one of the most highly expressed microRNAs in HCCs; transgenic expression in mice leads to liver tumor development 64. Inhibition of MIR221 with an anti-sense oligonucleotide delayed tumor growth in Mir221 transgenic mice64. The MIR17-92 cluster encodes at least 6 microRNAs that regulate cell survival, proliferation, differentiation, and angiogenesis. MIR17-92 is significantly overexpressed in HCCs, and its liver-specific overexpression promoted tumor development in transgenic mice 65. Delivery of anti-MIR17 oligonucleotide via lipid nanoparticles was able to delay MYC-induced tumorigenesis in mice 66. MicroRNAs might therefore serve as therapeutic targets and also as serum biomarkers. In a nested case-control study performed in China, expression patterns of 7 microRNAs (MIR29a, MIR29c, MIR133a, MIR143, MIR145, MIR192, and MIR505) could be used to identify patients with early-stage HCC 67. So far, no serum microRNA-based tests have made it to the bedside, but results are promising.
LncRNAs are made of 200–300 nucleotides and regulate gene expression by various mechanisms, including recruitment of chromatin modifying enzymes or interaction with proteins to direct their binding to DNA68,69. Aberrant overexpression of lncRNAs like HOTAIR70, HULC71, HEIH72, DREH73, and MVIH74 have been associated with HCC initiation and progression. Lau et al showed that integration of HBV DNA into the genome led to transcription of viral–human gene fusions that encode lncRNAs. These authors showed that the hybrid RNA HBx–LINE1 activated Wnt signaling to beta-catenin to promote tumor progression in transgenic mice expressing the viral-human chimeric fusion transcript75. Yang et al performed a comprehensive analysis of lncRNA expression levels in HCCs and found 917 recurrently deregulated lncRNAs whose levels correlated with clinical features 76. Many of these lncRNAs were enriched in co-expressed clusters of genes related to cell adhesion, immune responses, and metabolic processes.
A different epigenetic mechanism of gene regulation in cancer cells is histone modification. Histones regulate gene expression by regulating access to DNA based on the open or closed state of chromatin 77. Post-translational histone modifications such as methylation or acetylation can influence this process. Acetylation of specific lysine residues in histone tails reduces the affinity between histones and DNA, making DNA more accessible to transcription factors and polymerases, so acetylation generally promotes gene transcription. Transcription changes associated with histone methylation are more complex. Depending on the specific lysine residue, methylation can lead to activation or repression of transcription. Some examples of methylation changes in HCC which influence outcomes include trimethylated histone H3 lysine 4 (H3K4me3) and trimethylated lysine 27 (H3K27me3), whose overexpression correlates with reduced overall survival and poor outcomes of patients with HCC78,79. Another mechanism of epigenetic gene regulation is chromatin remodeling which involves dynamic changes in chromatin structure that regulate gene expression, apoptosis, and DNA repair80. ARID1A, ARID1B, and ARID2 encode proteins that are part of chromatin-remodeling complexes and function as tumor suppressors which explains why they frequently undergo inactivating mutations in HCC9, 19,81.
Many genomic and epigenetic events contribute to hepatocarcinogenesis, and viruses are directly or indirectly involved in several of these. What is the stepwise acquisition of genomic events during hepatic tumorigenesis?
Mechanisms of Hepatocyte Transformation and Genetic Alterations
Telomerase activation
Hepatocytes become transformed and form malignancies via a series of genetic and epigenetic alterations leading to genome diversification 82 (Figure 2). The specific mechanisms of tumorigenesis vary among patients with vs without cirrhosis, among patients with different liver diseases, and in patients exposed to different carcinogens. In patients with chronic hepatitis, non-alcoholic steatohepatitis, or alcoholic liver disease, persistent liver injury leads to cell proliferation in response to necrosis and telomere shortening due to the absence of telomerase activity in the adult liver cells83. Telomere attrition in senescent hepatocytes is characteristic of cirrhosis and could account for the reduced ability of cirrhotic liver to regenerate after liver resection84. Studies of mice have shown that telomere attrition promotes cirrhosis development. In humans, rare germline inactivating mutations in TERT were associated with cirrhosis 85, 86.
Figure 2. Genotype and Phenotype Classification of HCC.
HCCs can be classified based on their genetic features, molecular features (S1 to S3, ref 134 and G1 to G6, ref 120) pathology features, signaling pathways activated, and clinical features of patients. Many of these subgroups overlap in their features. Chrom, chromosome.
During hepatocarcinogenesis, telomerase reactivation is required for malignancy, and is observed in more than 90% of HCC samples87. Mutations in the TERT promoter were observed in premalignant nodules of patients with cirrhosis, with a prevalence of 6% in low-grade dysplastic nodules and 19% in high-grade dysplastic nodules21. TERT mutations were detected in 60% of early-stage and progressing HCCs from patients with cirrhosis20. Mutations in the TERT promoter therefore associate with tumor initiation, whereas mutations in other genes, such as TP53, CTNNB1 and ARID1A appear during later stages of HCC progression, to cause additional changes in the genome and transcription20,88.
From hepatocellular adenoma to carcinoma
In rare instances, patients without liver cirrhosis can develop hepatocellular adenomas (HCAs), which are benign but can become malignant 89 (Figure 3). Development of HCAs has been associated with exposure to estrogen (such as in oral contraceptives), so they are most commonly detected in women90. Subgroups of HCA include: HNF1A-mutated; inflammatory; HCA with mutations in exons 3, 7, or 8 of CTNNB1; and sonic hedgehog HCA90. Genetic analysis of HCCs that developed in patients with HCAs reveal a sequence of genetic alterations that led to malignancy 91.
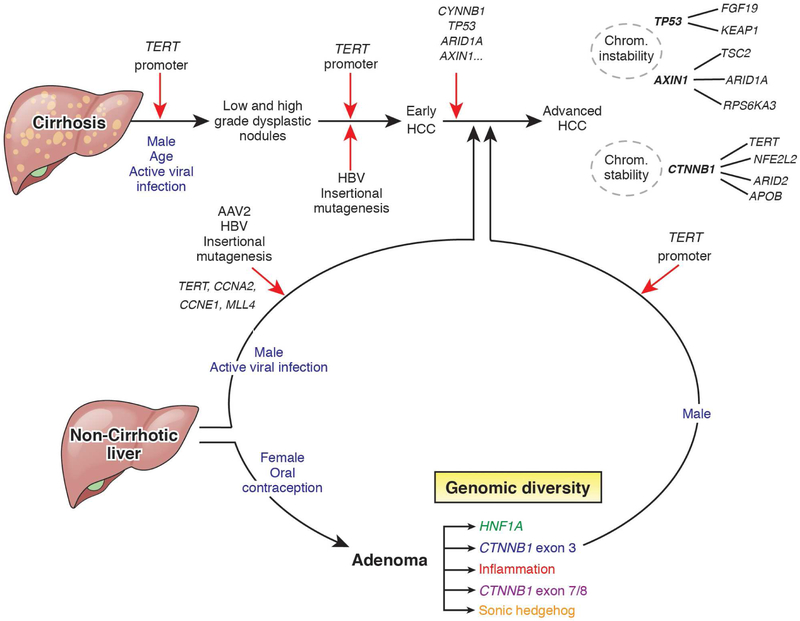
Figure 3. From Hepatocyte Transformation to Malignancy.
Genetic changes that occur during transformation of hepatocytes in patients with and without cirrhosis. Mutations in the promoter region of TERT contribute to immortality and proliferation of hepatocytes, resulting in dysplastic nodules. Additional mutations, some induced by viruses such as HBV or AAV2, lead to early-stage HCC. Mutations in genes such as TP53, CTNNB1, and AXIN1 lead to advanced HCC, with additional chromosome alterations. Patients without cirrhosis exposed to high levels of estrogen, such as through oral contraceptives, are at increased risk for hepatic adenomas (HCA), which are benign but can progress to malignant tumors if cells acquire mutations in the same genes that contribute to HCC development. Red arrows indicate genetic alterations believed to be required for heptocarcinogenesis.
Increased Wnt signaling to beta-catenin has been associated with malignancy. Mutations in exon 3 of CTNNB1 that activate its product, beta-catenin, have been associated with progression of HCAs to HCC whereas mutations in exons 7 and 8 that do not lead to beta-catenin activation have not been associated with progression to malignancy 92, 93. Mutations in the TERT promoter also appear to be required for progression of HCA to HCC91. In patients with cirrhosis, mutations in the TERT promoter allow senescent hepatocytes to bypass telomere attrition, whereas in patients without cirrhosis but with HCA, overexpression of TERT occurs after hepatocyte proliferation is induced by beta-catenin activation. These observations are important, because the sequence of accumulation of mutations during different stages of tumorigenesis might be used to select preventative or therapeutic strategies.
Mutation signatures at the start of carcinogenesis
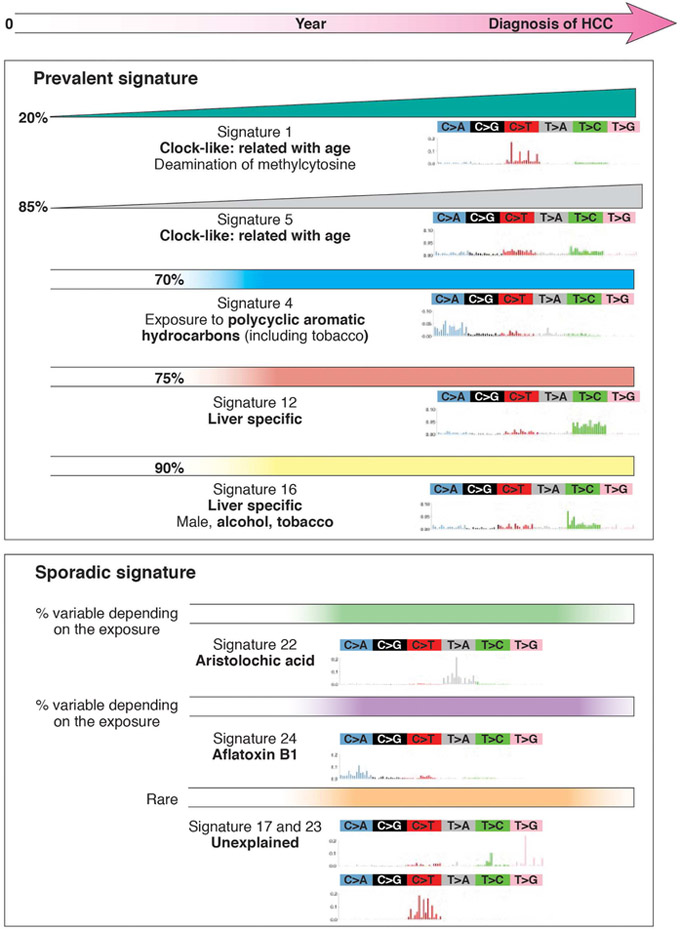
How do hepatocytes acquire DNA mutations that lead to transformation and malignancy? Researchers have categorized the types of nucleotide substitutions found in HCCs associated with different environmental factors (Figure 4). Mutation-inducing processes can occur at the same time or sequentially, during formation and development of a tumor94. Whole-exome and whole-genome studies have identified mutation signatures found in large and small proportions of HCCs 15, 17, 95. For example, COSMIC signatures 1 and 5 are related to patient age, whereas mutation signature 4 has been associated with HCC from patients with exposure to tobacco or polycyclic aromatic hydrocarbons. Mutation signatures 12 and 16 have been observed only in liver tumors, including HCCs and HCAs95, and might result from exposure to carcinogenic products of liver metabolism; this signature includes mutations in CTNNB1. Mutation signature 16 associates with HCCs from patients exposed to tobacco and alcohol. These findings support results from epidemiology studies indicating that tobacco exposure increases risk of HCC 17, 95.
Figure 4. Accumulation of Mutations During Liver Carcinogenesis.
Mutation signatures of different subgroups of HCCs. Some signatures are found in a large proportion of HCCs (percentage values given) worldwide, whereas others are found in small proportions of HCCs, related to specific carcinogen exposures or in sporadic tumors. Lines of increasing width from left to right indicate mutations that accumulate with time, whereas straight lines from left to right indicate mutations that do not increase with time.
Mutation signature 24 is found in HCCs from patients exposed to aflatoxin B1. Aflatoxin B1 is a fungal mycotoxin that contaminates crops in Asia and Africa and increases risk of HCC in these regions, in synergy with chronic HBV infection15, 17, 96. Exposure to aflatoxin causes a unique mutation profile with a strong transcriptional strand bias for C>A mutations, indicating guanine damage that is repaired by transcription- coupled nucleotide excision repair. These mutations can lead to the R249S substitution in TP53. 97, 98
Next-generation sequencing identified signature 22, characterized by sporadic mutations, in HCCs from patients exposed to aristolochic acid. This compound is derived from a medicinal plant used in Asia; 99, 100 it is used in traditional Chinese medicine and herbal supplements, and in weight-loss products in South East Asia. Exposure to aristolochic acid causes urothelial and liver cancers 98, 99, 101. Signature 22 is characterized by predominance of A–T to T–A transversions at [C|T]AG tri-nucleotide motifs, resulting in tumors that have a significant enrichment in splice-site mutations 19, 99. Aflatoxin B1 and aristolochic acid are banned in the United States by the Food and Drug Administration (FDA), and tests are available for high-risk food products such as peanuts and herbal supplements. Analyses of mutational processes have helped to identify risk factors for HCC that can be reduced by public health approaches.
Virus-induced mutagenesis
HBV can transform hepatocytes by integration of its DNA into the host genome. HBV is a 3.2 kb DNA virus found in a circular form (covalently closed circular DNA) in infected hepatocytes. Although HBV can promote HCC development indirectly, by promoting cell injury, inflammation, fibrosis, and cirrhosis, it also has direct carcinogenic effects. So, some patients with chronic HBV infection with normal liver still develop HCC. Virus oncoproteins, such as HBx or the preS2/S protein, alter cell signaling pathways to promote carcinogenesis 102. Overexpression of truncated HBx protein increases hepatocyte proliferation and prevents apoptosis103, regulating cell metabolism104 and increasing invasiveness and metastasis 105. HBV DNA sequences integrate into the human genome and can there serve as templates for viral DNA replication. 102 Insertion of HBV DNA near to or within oncogenes, or in cis, can alter expression levels to promote hepatocyte transformation 106–108. Insertion of HBV DNA near the TERT, CCNE1, CCNA2, and MLL2 genes has observed in HCC samples 108, 109. HBV DNA was detected near the TERT gene in 15% to 20% of HBV-associated HCCs, independent of mutations in the promoter region. Also, integration of HBV DNA can lead to virus–human transcript fusions with functional effects. Asian patients were reported to have integration of HBV DNA in the LINE gene, resulting an HBV–LINE1 fusion transcript. Its product can activate Wnt signaling to beta-catenin 75.
The adeno-associated virus type 2 (AAV2), a DNA virus that inserts into human DNA, considered to be non-pathogenic in the general population, also causes mutations that have been detected in HCCs110. AAV2 DNA sequences were identified in the TERT, CCNE1, MLL2, and TN6SF10 genes of HCCs from patients with normal liver, without inflammation or cirrhosis. These observations were confirmed in a Japanese study, indicating that AAV2 DNA integration can contribute to HCC development in patients with normal liver9, 110. There is controversy over whether HCV is a liver carcinogen—specific HCV proteins could have oncogenic properties. The HCV NS3, NS4B, NS5B, and HCV core protein can transform specific cell types. Mice that overexpress HCV structural proteins develop liver tumors 9, 111, 112. However, in humans, HCV infection appears to primarily promote liver cancer via inflammation and cirrhosis 113.
Genome diversity and heterogeneity
All the mechanisms of malignant transformation lead to the acquisition of additional genetic alterations that result in the development of a complex genomic architecture during tumor evolution 114. Tumor initiation, progression, and metastasis are associated with the acquisition of mutations and copy number variations in subclones, which result in considerable spatial and temporal heterogeneity in HCC. Mutations that promote HCC development, such as those in the TERT promoter or CTNNB1, have been identified in all parts of primary tumors and are therefore called core clonal alterations88. In contrast, several studies have confirmed the presence of spatial heterogeneity with mutations in subclones that are only present in specific regions of HCC tumors115, 116. There is an additional layer of complexity in liver carcinogenesis in patients with multifocal disease, who may have multifocal intra-hepatic metastases from the original tumor, along with inter-tumor heterogeneity due to the near simultaneous development of de novo tumors at multiple sites 117, 118.
Development of tumor heterogeneity is a dynamic process with complex timing. Exposure to carcinogens and acquisition of mutations by HCC clones and subclones changes with time. For example, early HCC clones can have a mutation signature associated with aflatoxin B1 among patients exposed in Africa during early life. However, if the patient develops HCC while living in a western country, HCC subclones that form later in life may no longer have the aflatoxin B1-associated mutation signature95, 118. So, tumor genomes accumulate alterations throughout life that reflect etiologic influences during the various periods of exposure. Moreover, the acquisition of chromosome or genome duplications appears to be a very late event during HCC evolution95. This spatial and temporal tumor heterogeneity of tumors is important to appreciate as it may explain the subsequent acquisition of primary or secondary resistance to targeted therapies.
Genotype and Phenotype Classifications
Interactions of genome alterations
Interactions among gene mutations, changes in transcription, alterations in epigenetic regulation, environmental factors, and histologic features should all be considered in classification of HCCs82. Whole-exome and whole-genome sequence analyses of HCCs identified 4 to 6 mutations in oncogenes per tumor; associations and exclusions among these mutations indicate redundancy and/or cooperation between factors in overlapping signaling pathways9, 10, 15, 17, 19. Mutations occur in groups of genes that are associated with specific signaling pathways. For example. tumors with mutations in CTNNB1 do not usually have mutations in TP53 or AXIN1. CTNNB1 mutations are frequently associated with mutations in the TERT promoter, APOB, NFE2L2, ARID2, and MLL219. Mutations in TP53 are frequently associated with mutations in KEAP1, CCND1, and TSC2. Mutations in AXIN1 are frequently detected with mutations in ARID1A and RPS6KA3 15, 17.
Risk factors associated with molecular profiles
HCC risk factors associate with their genetic features. For example, HBV-related HCCs have a specific pattern of mutations that result from insertion of HBV DNA into the genome, as well as mutations in TP53 and AXIN1 and acquisition of stem cell features 15, 17, 119–121. Some of these associations could result from the geographic risk factors, such as the coincidence of regions of high HBV prevalence with regions of high dietary aflatoxin exposure. Similarly, HCCs in patients with high alcohol intake frequently contain mutations in ARID1A and CTNNB1, whereas HCC of unknown etiology have fewer TERT promoter mutations and more frequent IL6ST-activating mutations 15, 17. However, no HCC mutation pattern has been confirmed to be associated with HCV infection or metabolic syndrome.
Molecular alterations related to outcome
Tumor features identified from genetic, epigenetic, and transcriptome analyses have been associated with poor outcomes of patients treated for HCC. RB1 and TP53 mutations and FGF19 amplification increase risk of tumor relapse and death 122–124 Interestingly, TP53 mutations are a risk factor for poor survival and tumor recurrence in patients with HBV-related HCC but not in patients with HCC related to other etiologies 121, 125. Transcriptome signatures from tumor tissues have been associated with tumor aggressiveness and tumor recurrence 2–3 years after surgery (early recurrence) 126–130, whereas transcriptomes of non-tumor liver tissues have been associated with carcinogenesis de novo, usually in patients with cirrhosis, and tumor recurrence after 3 years (late recurrence) 131. Moreover, signatures from non-tumor cirrhotic liver have also been associated with severity of the liver disease and are consequently linked with HCC occurrence and decompensation of liver disease 132. Expression levels of 5 genes (5-gene score) in tumor tissues, combined with an expression pattern of 186 genes in non-tumor liver tissue, were associated with early tumor recurrence and late recurrence, as well as overall survival 126. However, most prognostic transcriptome signatures were derived from specimens obtained during resection of very early- or early-stage HCCs. These findings therefore require validation in studies of biopsies from patients with intermediate- and advanced-stage tumors who received different types of treatment. Prognostic transcriptome signatures are currently not used in clinical practice 82.
Molecular features and correlations with phenotypes
Classification systems developed to assess genome diversity identified different subgroups of HCC. One group is called proliferative HCC, characterized by chromosome instability (G1 to G3 subgroups, proliferative subgroup, cluster A, S1 and S2) and a second is considered to have less proliferative HCC cells, with chromosomal stability (G4 to G6, S3, cluster B) 120, 128, 133, 134. Among the HCCs with less-proliferative cells, a subgroup was defined by somatic mutations in CTNNB1, leading to activation of genes regulated by Wnt signaling to beta-catenin (G5, G6)120, 135. Another subgroup, well-differentiated HCC, had a gene expression pattern close to that of mature hepatocytes (G4 subgroup, hepatocyte like, S3). Acquisition of progenitor cell characteristics and re-expression of fetal genes defined a group of HCC with stem cells features (G1 subgroup, progenitor like, hepatoblast like, S2). Finally, a subtype of HCC with inactivation of CDKN2A and mutations in TP53, leading to dysregulation of cell cycle genes, was associated with poor outcome (G3 subgroup)120.
HCC transcriptomes and mutation patterns were linked with specific histologic features136, 137, 138. The G1 to G3 subgroups of HCC are often poorly differentiated and have mutations in TP53. The G1 subgroup and tumors with RP6SKA3 mutations were linked with the progenitor phenotype, with expression of stem cell markers such as CK19 or EPCAM, based on immunohistochemical analyses. The scirrhous histologic subtype of HCC was linked with mutations in TSC1 and TSC2 and expression of genes of the epithelial to mesenchymal transition136, 137. HCCs from patients with steatohepatitis are frequently classified in the G4 subgroup, characterized by immune cell infiltration and activation of the JAK-STAT signaling pathway 136. In contrast, HCCs with steatosis and infiltration by inflammatory cells were associated, in a separate study, with the S1 subgroup, so additional studies are needed to classify these tumors 137. Well-differentiated HCCs of the G5 to G6 subgroups are enriched in activating mutations of CTNNB1 and are characterized by cholestasis, increased levels of glutamine synthase (determined by immunohistochemistry), and nuclear translocation of beta-catenin. A histological subtype called macrotrabecular massive is characterized by the G3 and S2 transcription profile, TP53 mutations, and FGF19 amplification136,137. This subgroup was associated with a higher rate of tumor recurrence in a large cohort of patients who underwent resection or radiofrequency ablation, so its identification in surgical samples or tumor biopsies can be helpful in clinical practice 139. Another subtype, known as chromophobe HCC with abrupt anaplasia, characterized by nuclear atypia on a background of cells with bland nuclei, has been correlated with the presence of alternative lengthening of telomeres 140.
Features of mixed hepatocholangiocarcinoma tumors
The genetic features of cholangiocarcinomas differ from those of HCCs in that cholangiocarcinomas have frequent mutations in KRAS, BRAF, BAP1, SMAD4, IDH1, and IDH2; as well as fusion of FGFR2, ROS1, and PRKACA genes, but few TERT promoter mutations 141–143. However, a continuum seems to exist among cholangiocarcinoma, mixed hepato-cholangiocarcinoma, and HCCs with stem cell features, indicating that similar early genetic alterations in different cell types results in different histologic and genetic subtypes of tumors143. Interestingly, next-generation sequencing analyses of specific areas of HCCs, cholangiocarcinomas, and hepatocholangiocarcinomas found common somatic mutations among tumor areas, indicating clonal origins for each part of these tumors144. Moreover, the proportions of tumors with TERT promoter mutations ranges from 59% in HCC, to 20% in hepatocholangiocarcinomas, to few in cholangiocarcinomas. Similar to HCC, amplifications in CCND1 and FGF19 were identified in some hepatocholangiocarcinomas. Some studies have reported gene dysregulation typical of cholangiocarcinoma in HCCs with stem cell features 145–147. More studies are needed to determine how similar and different genetic alterations contribute to development of CCA, HCC and mixed tumors.
Personalized Medicine
The goal of personalized medicine is select specific treatments for each individual tumor based on its genotype or other features. This idea is not novel but is becoming a practical reality. Success stories in precision medicine include the use of imatinib mesylate for treatment of chronic myelogenous leukemia 148, BRAF inhibitors for treatment of melanoma with the BRAF V600E mutation149, tyrosine kinase inhibitors such as erlotinib for lung adenocarcinomas with alterations in the epidermal growth factor receptor (EGFR), 150 and ALK inhibitors for lung cancer with ALK rearrangements 151.
HCC is relatively resistant to traditional chemotherapeutics such as 5-fluorouracil, cisplatin, doxorubicin, or gemcitabine. Until 2007, patients with advanced, unresectable HCC could receive only best supportive care. In 2007, sorafenib, a multi-kinase inhibitor that blocks signaling via vascular endothelial growth factor receptor (VEGFR), platelet derived growth factor receptor beta (PDGFRB), BRAF, and KIT, was the first systemic agent to increase survival times of patients with advanced HCC 152–154. Although the drug increased patient survival time by only 3–4 months, it provided hope that additional targeted therapies could be developed for HCC.
Unfortunately, the approval of sorafenib was followed by a long period of failure of agents tested in phase 3 trials of patients with HCC, including sunitinib 155, brivanib 34, linifanib, 156 and erlotinib 157. Nevertheless, there have been promising results for other kinase inhibitors, such as regorafenib, lenvatinib, and cabozantinib (Table 1). Regorafenib has been approved as a second-line therapy based on results from the RESORCE trial, which showed that this drug significantly increased survival times of patients with advanced HCC that progressed during treatment with sorafenib, compared to placebo (10.6 months vs 7.8 months)158. Lenvatinib, another multi-kinase inhibitor, was found to be non-inferior to sorafenib as first-line therapy for untreated advanced HCC in the REFLECT trial35 and has recently been approved by the FDA. Also, Cabozantinib was reported to have met clinical endpoints, compared with placebo, in a phase 3 trial (CELESTIAL), as a second-line agent159, 160. Although studies of these multi-kinase inhibitors have produced encouraging results, there is much to be desired in terms of their efficacy and safety—most of these drugs only prolong overall survival by a few months and fewer than 10% of patients achieve the objective response.
Table 1. Phase 3 Trials of Targeted Chemotherapy for HCC.
BCLC, Barcelona Clinic Liver Cancer classification; ECOG, Eastern Cooperative Oncology Group; PS, Performance Status. mAb, monoclonal antibody. VEGFR2 -Vascular endothelial growth factor receptor 2; CDK- cyclin-dependent kinase; EGFR- Epidermal growth factor receptor. TGF- Transforming growth factor.
| TRIAL NAME. DRUG | CLINICAL TRIAL ID | MECHANISM OF ACTION | TITLE OF STUDY | LINE OF THERAPY |
INCLUSION CRITERIA | STATUS | |
|---|---|---|---|---|---|---|---|
| PHASE 3 COMPLETED. POSITIVE TRIALS | Reference | ||||||
| SHARP. Sorafenib | NCT00105443 | Multiple tyrosine kinase receptor inhibitor | A Phase 3 Randomized, Placebo-controlled Study of Sorafenib in Patients With Advanced Hepatocellular Carcinoma | First | Child pugh A. BCLC B-C, ECOG PS 0-2. | Completed. Sorafenib improves overall survival over placebo. | 152 |
| ASIA-PACIFIC. Sorafenib | NCT00492752 | Multiple tyrosine kinase receptor inhibitor | A Randomized, Double-blinded, Placebo-controlled Study of Sorafenib in Patients With Advanced Hepatocellular Carcinoma | First | Child pugh A. BCLC B-C, ECOG PS 0-2. | Completed. Sorafenib improves overall survival over placebo. | 153 |
| RESOURCE. Regorafenib | NCT02042144 | Multiple tyrosine kinase receptor inhibitor | Randomized, Double Blind, Placebo Controlled, Multicenter Phase 3 Study of Regorafenib in Patients With Hepatocellular Carcinoma (HCC) After Sorafenib | Second | Child Pugh A, BCLC B-C, ECOG PS 0-1. Failed Sorafenib | Completed. Regorafenib improves overall survival over placebo in patients who progressed on Sorafenib. | 158 |
| CELESTIAL. Cabozantinib | NCT01908426 | Multiple tyrosine kinase receptor inhibitor | A Phase 3, Randomized, Double-blind, Controlled Study of Cabozantinib (XL184) vs Placebo in Subjects With Hepatocellular Carcinoma Who Have Received Prior Sorafenib | Second | Child Pugh A, BCLC B-C, ECOG PS 0-1. Failed Sorafenib | Completed. Cabozantinib improves overall survival over placebo in patients who progressed on Sorafenib. | 160 |
| REFLECT. Levatinib | NCT01761266 | Multiple tyrosine kinase receptor inhibitor | A Multicenter, Randomized, Open-Label, Phase 3 Trial to Compare the Efficacy and Safety of Lenvatinib (E7080) Versus Sorafenib in First-Line Treatment of Subjects With Unresectable Hepatocellular Carcinoma | First | Child Pugh A, BCLC B-C, ECOG PS 0-1 | Completed. Lenvatinib is non-inferior to Sorafenib in improving overall survival as first line therapy for advanced HCC. | 35 |
| PHASE 3, ACTIVE | Date of Completion | ||||||
| REACH-2. Ramucirumab | NCT02435433 | VEGFR2 inhibitor | Randomized, Double-Blind, Placebo-Controlled, Phase 3 Study of Ramucirumab and Best Supportive Care (BSC) Versus Placebo and BSC as Second-Line T reatment in Patients With Hepatocellular Carcinoma and Elevated Baseline Alpha-Fetoprotein (AFP) Following First-Line Therapy With Sorafenib | Second | Child Pugh A, BCLC B-C, ECOG PS 0-1 AFP>400ng/ml |
Completed. Ramucirumab improves overall survival over placebo inpatients with advanced HCC with AFP>400 (164). | June 2020 |
| Everolimus | NCT02081755 | mTOR inhibitor | A 36 Month Multi-center, Open Label, Randomized, Comparator Study to Evaluate the Efficacy and Safety of Everolimus Immunosuppression Treatment in Liver Transplantation for Hepatocellular Carcinoma Exceeding Milan Criteria | First | Liver transplant for HCC and high risk for recurrence | Recruiting | August 2019 |
| SATURNE. Sunitinib | NCT01164202 | Multiple tyrosine kinase receptor inhibitor | A Double-Blind, Randomized, Phase 2/3 Study Comparing the Use of Chemoembolization Combined With Sunitinib Against Chemoembolization Combined With a Placebo in Patients With Hepatocellular Carcinoma (SATURNE) | First | Child pugh A. BCLC B-C, ECOG PS 0-2. | Not Recruiting | July 2018 |
| Apatinib | NCT02329860 | VEGFR2 inhibitor | Study of Apatinib After Systemic Therapy in Patients With Hepatocellular | CSecond | Child pugh A-B. BCLC B-C, ECOG PS 0-1. | Not Recruiting | June 2018 |
| Donafenib | NCT02645981 | Multiple tyrosine kinase receptor inhibitor | Efficacy and Safety of Donafenib in Patients With Advanced Hepatocellular Carcinoma | First | Child pugh A-B. BCLC B-C, ECOG PS 0-1. | Not Recruiting | Aug 2019 |
One of the main reasons for failure of multiple targeted therapies in phase 3 trials was felt to be interpatient tumor heterogeneity and many solutions have been proposed to overcome this, including testing drugs in biomarker-stratified subpopulations. Tivantinib, a MET inhibitor, showed promising results in a phase 2 trial, especially for patients whose tumors had high MET expression161. This was followed by a biomarker-stratified phase 3 study, which included only patients with tumors that had high levels of MET, determined by immunohistochemistry. In this study, patients were randomly assigned to groups that were given tivantinib (n=226) or placebo (n=114). At a follow-up time of 18.1 months, median overall survival times were 8.4 months in the tivantinib group and 9.1 months in the placebo group (hazard ratio, 0.97). So, unfortunately, the encouraging results from the phase 2 study did not continue into the phase 3 trial 162. In analyses of the disappointing results of this phase 3 trial, researchers found that tivantinib did not act as a MET inhibitor after all, but instead as an anti-mitotic agent. So, MET overexpression is likely not a good biomarker of tumors likely to respond to tivantinib 163.
In the phase 3 REACH2 trial, ramucirumab, an inhibitor of VEGFR2, increased survival times, when given as a second-line agent, in patients with HCC and Child Pugh scores of 5 or 6 and serum levels of alpha-fetoprotein (AFP) above 400 ng/ml 164. In a phase 3 trial of only patients with high baseline serum levels of AFP (NCT02435433), ramucirumab increased survival times compared to placebo. Apatinib, another inhibitor of VEGFR2, is being tested as a first-line therapy in a phase 2 trial (NCT03046979), based on promising results from smaller studies 165, 166.
Another challenge to development of therapies for HCC is that the somatic mutations associated with tumor development lie in genes whose products are not easily or safely targeted. Mutated forms of TERT, TP53, CTNNB1, and MYC are believed to be undruggable. Although our understanding of TERT promoter mutations has rapidly expanded, we do not have small molecule inhibitor of telomerase. A synthetic hTERT DNA vaccine, INO-1400, is being tested in a phase 1 trial of patients with solid tumors (NCT02960594) and some trials are using TERT promoter mutation as a biomarker for study enrollment (NCT02766270). New strategies might be developed to target these driver genes or their pathways, such as microRNA-based therapeutics. Advances in genome research should help identify events that can be targeted or used as biomarkers to select patients for specific therapies. Table 1 and Supp table 1 presents targeted therapies for HCC that are in phase 3 and phase 2 trials respectively. Most of the agents are being explored as second-line therapies for patients with advanced HCC who were failed by sorafenib, but this may change soon.
Immunotherapy
The combination of the immune-tolerant microenvironment of the liver, ability of HCV and HBV to evade the immune response, and the immune-modulatory effects of the tumor allow for growth and progression of HCC. Hence strategies to reactivate anti-tumor immunity can be used to prevent or treat HCC. Nivolumab was recently given accelerated approval for treatment of advanced liver cancer, based on promising results from a phase 2 trial (Checkmate-040)167. Approximately 20% of the patients had complete or partial responses to nivolumab and 40% achieved stable disease; the 12-month overall rate of survival was 59.9%. Although these responses are modest, they are more promising than previous systemic agents. Multiple clinical trials of immunotherapy agents are underway in patients with HCC and there is hope the treatment paradigm will improve. It is important to continue to investigate the effects of HCC on the immune system— especially in patients with viral hepatitis, to identify patients most likely to respond to specific therapies. This is being extensively discussed in another article in this issue 168.
Future Directions
Recent advances in genetic, genomic, and proteomic analyses have increased our understanding of HCC pathogenesis and our ability to classify tumors based on genetic and histologic features. We are learning more about the specific oncogenic effects of HBV, HCV, alcohol, fatty liver disease, and environmental factors such as aflatoxin and aristolochic acid. We have been identifying genetic alterations that contribute to liver carcinogenesis, learning the sequence of acquisition of these mutations, and discovering the chromosomal and epigenetic changes required for tumor development and progression.
There is continued progress in identifying multi-kinase inhibitors of angiogenesis and other receptor tyrosine kinase signaling pathways in tumor cells and the tumor microenvironment that might slow tumor growth yet have an acceptable safety profile in persons with liver disease. Although we have not been able to use HCC subclasses to select the optimal therapy for patients, some trials have used biomarkers to identify the subsets of patients with highest rates of response to specific targeted therapies. As the key molecular drivers of HCC are identified, strategies are being developed to reduce levels of TERT, Wnt signaling to beta-catenin, MYC activation, P53 inactivation, and expression of chromatin modifying genes.
Studies are needed to determine the potential effectiveness of immunotherapies, to identify subgroups of HCCs that are most sensitive to checkpoint inhibitors or other agents, and to determine the potential of neo-adjuvant, adjuvant, and combination strategies to improve patient outcomes. It is important to continue to acquire and analyze intermediate- to advanced-stage HCC samples from participants in clinical trials of systemic targeted or immune therapies. Integrated molecular analyses of these samples will potentially identify the subsets of patients most likely to benefit from specific therapeutic agents.
Supplementary Material
Acknowledgements
Grant Funding:
1. Renumathy Dhanasekaran: NIH Grant CA222676 from the National Cancer Institute (NCI), ACG Junior Faculty Career Development Grant.
2. Lewis R Roberts: Grant Numbers CA165076 and CA186566 from the National Cancer Institute (NCI), the Mayo Clinic Hepatobiliary SPORE (NCI CA210964), the Mayo Clinic Center for Cell Signaling in Gastroenterology (NIDDK P30DK084567), the Mayo Clinic Cancer Center (CA15083), and the Mayo Clinic Center for Translational Science Activities (NIH/NCRR CTSA Grant Number UL1 TR000135)
3. Jean-Charles Nault and Jessica Zucman-Rossi- Financial supports: This work was supported by Institut National du Cancer (INCa) with the International Cancer Genome Consortium (ICGC LICA-FR project) and NoFLIC projects (PAIR HCC, INCa and ARC), INSERM with the « Cancer et Environnement » (plan Cancer), MUTHEC projects (INCa) and the HECAM project (BPI). The group is supported by the Ligue Nationale contre le Cancer (Equipe Labellisée), Labex OncoImmunology (investissement d’avenir),grant IREB, Coup d’Elan de la Fondation Bettencourt-Shueller, the SIRIC CARPEM and Fondation Mérieux.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- Lewis R Roberts: Grant Funding:
- Ariad Pharmaceuticals, Bayer, BTG International, Exact Sciences, Gilead Sciences, RedHill Biopharma, TARGET PharmaSolutions, Wako Diagnostics
- Advisory Board: Bayer, Exact Sciences, Gilead Sciences, RedHill Biopharma, TAVEC, Wako Diagnostics
- No Conflict of Interest for other authors.
REFERENCES
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer 2014;136:E359–E386. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–132. [DOI] [PubMed] [Google Scholar]
- 3.Acharya SK. Epidemiology of Hepatocellular Carcinoma in India. J Clin Exp Hepatol 2014;4:S27–S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yi S-W, Choi J-S, Yi J-J, et al. Risk factors for hepatocellular carcinoma by age, sex, and liver disorder status: A prospective cohort study in Korea. Cancer 2018. Available at: 10.1002/cncr.31406. [DOI] [PubMed] [Google Scholar]
- 5.Gomaa A, Allam N, Elsharkawy A, et al. Hepatitis C infection in Egypt: prevalence, impact and management strategies. Hepat Med 2017;9:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blachier M, Leleu H, Peck-Radosavljevic M, et al. The burden of liver disease in Europe: a review of available epidemiological data. J Hepatol 2013;58:593–608. [DOI] [PubMed] [Google Scholar]
- 7.Ryerson AB, Eheman CR, Altekruse SF, et al. Annual Report to the Nation on the Status of Cancer, 1975-2012, featuring the increasing incidence of liver cancer. Cancer 2016;122:1312–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umemura T, Ichijo T, Yoshizawa K, et al. Epidemiology of hepatocellular carcinoma in Japan. J Gastroenterol 2009;44 Suppl 19:102–107. [DOI] [PubMed] [Google Scholar]
- 9.Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012;44:760–764. [DOI] [PubMed] [Google Scholar]
- 10.Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy- number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleary SP, Jeck WR, Zhao X, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013;58:1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kan Z, Zheng H, Liu X, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res 2013;23:1422–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn S-M, Jang SJ, Shim JH, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 2014;60:1972–1982. [DOI] [PubMed] [Google Scholar]
- 14.Jhunjhunwala S, Jiang Z, Stawiski EW, et al. Diverse modes of genomic alteration in hepatocellular carcinoma. Genome Biol 2014;15:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267–1273. [DOI] [PubMed] [Google Scholar]
- 16.Shiraishi Y, Fujimoto A, Furuta M, et al. Integrated analysis of whole genome and transcriptome sequencing reveals diverse transcriptomic aberrations driven by somatic genomic changes in liver cancers. PLoS One 2014;9:e114263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimoto A, Furuta M, Totoki Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet 2016;48:500–509. [DOI] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu, Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327–1341.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nault JC, Mallet M, Pilati C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun 2013;4:2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nault JC, Calderaro J, Di Tommaso L, et al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology 2014;60:1983–1992. [DOI] [PubMed] [Google Scholar]
- 22.Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267–1273. [DOI] [PubMed] [Google Scholar]
- 23.Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu, Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327–1341.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pezzuto F, Izzo F, Buonaguro L, et al. Tumor specific mutations in TERT promoter and CTNNB1 gene in hepatitis B and hepatitis C related hepatocellular carcinoma. Oncotarget 2016;7:54253–54262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao L-H, Liu X, Yan H-X, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun 2016;7:12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang B, Xie R, Qin Y, et al. Human telomerase reverse transcriptase (hTERT) promotes gastric cancer invasion through cooperating with c-Myc to upregulate heparanase expression. Oncotarget 2015;7 Available at: 10.18632/oncotarget.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J-I, Venteicher AS, Hong JY, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 2009;460:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh A, Saginc G, Leow SC, et al. Telomerase directly regulates NF-KB-dependent transcription. Nat Cell Biol 2012;14:1270–1281. [DOI] [PubMed] [Google Scholar]
- 30.Lee SG, Rho HM. Transcriptional repression of the human p53 gene by hepatitis B viral X protein. Oncogene 2000;19:468–471. [DOI] [PubMed] [Google Scholar]
- 31.Wong CM, Fan ST, Ng IO. beta-Catenin mutation and overexpression in hepatocellular carcinoma: clinicopathologic and prognostic significance. Cancer 2001;92:136–145. [DOI] [PubMed] [Google Scholar]
- 32.Guo X-Q, Zhang Q-X, Huang W-R, et al. [Tumor suppressor role of chromatin-remodeling factor ARID1A]. Yi Chuan 2013;35:255–261. [DOI] [PubMed] [Google Scholar]
- 33.Sawey ET, Chanrion M, Cai C, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell 2011;19:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson PJ, Qin S, Park J-W, et al. Brivanib versus sorafenib as first-line therapy in patients with unresectable, advanced hepatocellular carcinoma: results from the randomized phase III BRISK-FL study. J Clin Oncol 2013;31:3517–3524. [DOI] [PubMed] [Google Scholar]
- 35.Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018;391:1163–1173. [DOI] [PubMed] [Google Scholar]
- 36.Kim R, Sharma S, Meyer T, et al. First-in-human study of BLU-554, a potent, highly-selective FGFR4 inhibitor designed for hepatocellular carcinoma (HCC) with FGFR4 pathway activation. Eur J Cancer 2016;69:S41. [Google Scholar]
- 37.Xu Z, Hu J, Cao H, et al. Loss of Pten synergizes with c-Met to promote hepatocellular carcinoma development via mTORC2 pathway. Exp Mol Med 2018;50:e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Wang W-L, Zhang Y, et al. Epigenetic and genetic alterations of PTEN in hepatocellular carcinoma. Hepatol Res 2007;37:389–396. [DOI] [PubMed] [Google Scholar]
- 39.Biden K, Young J, Buttenshaw R, et al. Frequency of mutation and deletion of the tumor suppressor gene CDKN2A (MTS1/p16) in hepatocellular carcinoma from an Australian population. Hepatology 1997;25:593–597. [DOI] [PubMed] [Google Scholar]
- 40.Jin M, Piao Z, Kim NG, et al. p16 is a major inactivation target in hepatocellular carcinoma. Cancer 2000;89:60–68. [DOI] [PubMed] [Google Scholar]
- 41.Jones PA, Baylin SB. The Epigenomics of Cancer. Cell 2007;128:683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiyono T, Foster SA, Koop JI, et al. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 1998;396:84–88. [DOI] [PubMed] [Google Scholar]
- 43.Zhang C, Guo X, Jiang G, et al. CpG island methylator phenotype association with upregulated telomerase activity in hepatocellular carcinoma. International Journal of Cancer 2008;123:998–1004. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Wang J, Cheng J, et al. An integrated analysis of SOCS1 down-regulation in HBV infection- related hepatocellular carcinoma. J Viral Hepat 2014;21:264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tada M, Kanai F, Tanaka Y, et al. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clin Cancer Res 2008;14:3768–3776. [DOI] [PubMed] [Google Scholar]
- 46.Wahid B, Ali A, Rafique S, et al. New Insights into the Epigenetics of Hepatocellular Carcinoma. Biomed Res Int 2017;2017:1609575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Csepregi A, Röcken C, Hoffmann J, et al. APC promoter methylation and protein expression in hepatocellular carcinoma. J Cancer Res Clin Oncol 2008;134:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, Dong H, Robertson K, et al. DNA methylation suppresses expression of the urea cycle enzyme carbamoyl phosphate synthetase 1 (CPS1) in human hepatocellular carcinoma. Am J Pathol 2011;178:652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Defamie V, Sanchez O, Murthy A, et al. TIMP3 controls cell fate to confer hepatocellular carcinoma resistance. Oncogene 2015;34:4098–4108. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y-J, Chen Y, Ahsan H, et al. Silencing of glutathione S-transferase P1 by promoter hypermethylation and its relationship to environmental chemical carcinogens in hepatocellular carcinoma. Cancer Lett 2005;221:135–143. [DOI] [PubMed] [Google Scholar]
- 51.Duong FHT, Christen V, Lin S, et al. Hepatitis C virus-induced up-regulation of protein phosphatase 2A inhibits histone modification and DNA damage repair. Hepatology 2010;51:741–751. [DOI] [PubMed] [Google Scholar]
- 52.Higgs MR, Lerat H, Pawlotsky J-M. Downregulation of Gadd45beta expression by hepatitis C virus leads to defective cell cycle arrest. Cancer Res 2010;70:4901–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye C, Tao R, Cao Q, et al. Whole-genome DNA methylation and hydroxymethylation profiling for HBV- related hepatocellular carcinoma. Int J Oncol 2016;49:589–602. [DOI] [PubMed] [Google Scholar]
- 54.Park IY, Sohn BH, Yu E, et al. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology 2007;132:1476–1494. [DOI] [PubMed] [Google Scholar]
- 55.Tian Y, Yang W, Song J, et al. Hepatitis B Virus X Protein-Induced Aberrant Epigenetic Modifications Contributing to Human Hepatocellular Carcinoma Pathogenesis. Mol Cell Biol 2013;33:2810–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng D-L, Zhang L, Cheng N, et al. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol 2009;50:377–387. [DOI] [PubMed] [Google Scholar]
- 57.Cougot D, Wu Y, Cairo S, et al. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J Biol Chem 2007;282:4277–4287. [DOI] [PubMed] [Google Scholar]
- 58.Macfarlane L-A, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 2010;11:537–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bandiera S, Pfeffer S, Baumert TF, et al. miR-122 - A key factor and therapeutic target in liver disease. J Hepatol 2015;62:448–457. [DOI] [PubMed] [Google Scholar]
- 60.Kishikawa T, Otsuka M, Tan PS, et al. Decreased miR122 in hepatocellular carcinoma leads to chemoresistance with increased arginine. Oncotarget 2015;6:8339–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu S-H, Wang B, Kota J, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest 2012;122:2871–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsai W-C, Hsu S-D, Hsu C-S, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 2012;122:2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xue H-Y, Liu Y, Liao J-Z, et al. Gold nanoparticles delivered miR-375 for treatment of hepatocellular carcinoma. Oncotarget 2016;7:86675–86686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Callegari E, Elamin BK, Giannone F, et al. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology 2012;56:1025–1033. [DOI] [PubMed] [Google Scholar]
- 65.Zhu H, Han C, Wu T. MiR-17–92 cluster promotes hepatocarcinogenesis. Carcinogenesis 2015;36:1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dhanasekaran R, Gabay-Ryan M, Baylot V, et al. Anti-miR-17 therapy delays tumorigenesis in MYC- driven hepatocellular carcinoma (HCC). Oncotarget 2018;9:5517–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin X-J, Chong Y, Guo Z-W, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case- control study. Lancet Oncol 2015;16:804–815. [DOI] [PubMed] [Google Scholar]
- 68.Lai F, Orom UA, Cesaroni M, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 2013;494:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marchese FP, Grossi E, Marin-Bejar O, et al. A Long Noncoding RNA Regulates Sister Chromatid Cohesion. Mol Cell 2016;63:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z, Zhou L, Wu L-M, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011;18:1243–1250. [DOI] [PubMed] [Google Scholar]
- 71.Du Y, Kong G, You X, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem 2012;287:26302–26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang F, Zhang L, Huo X-S, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011;54:1679–1689. [DOI] [PubMed] [Google Scholar]
- 73.Huang J-F, Guo Y-J, Zhao C-X, et al. Hepatitis B virus X protein (HBx)-related long noncoding RNA (IncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology 2013;57:1882–1892. [DOI] [PubMed] [Google Scholar]
- 74.Yuan S-X, Yang F, Yang Y, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology 2012;56:2231–2241. [DOI] [PubMed] [Google Scholar]
- 75.Lau C-C, Sun T, Ching AKK, et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell 2014;25:335–349. [DOI] [PubMed] [Google Scholar]
- 76.Yang Y, Chen L, Gu J, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun 2017;8:14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lawrence M, Daujat S, Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet 2016;32:42–56. [DOI] [PubMed] [Google Scholar]
- 78.He C, Xu J, Zhang J, et al. High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Hum Pathol 2012;43:1425–1435. [DOI] [PubMed] [Google Scholar]
- 79.Cai M-Y, Hou J-H, Rao H-L, et al. High expression of H3K27me3 in human hepatocellular carcinomas correlates closely with vascular invasion and predicts worse prognosis in patients. Mol Med 2011;17:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dhanasekaran R, Bandoh S, Roberts LR. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000Res 2016;5 Available at: 10.12688/f1000research.6946.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li M, Zhao H, Zhang X, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet 2011. ;43:828–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zucman-Rossi J, Villanueva A, Nault J-C, et al. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015;149:1226–1239.e4. [DOI] [PubMed] [Google Scholar]
- 83.Günes C, Lenhard Rudolph K. The Role of Telomeres in Stem Cells and Cancer. Cell 2013;152:390–393. [DOI] [PubMed] [Google Scholar]
- 84.Satyanarayana A, Manns MP, Rudolph KL. Telomeres and telomerase: a dual role in hepatocarcinogenesis. Hepatology 2004;40:276–283. [DOI] [PubMed] [Google Scholar]
- 85.Calado RT, Brudno J, Mehta P, et al. Constitutional telomerase mutations are genetic risk factors for cirrhosis. Hepatology 2011;53:1600–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hartmann D, Srivastava U, Thaler M, et al. Telomerase gene mutations are associated with cirrhosis formation. Hepatology 2011;53:1608–1617. [DOI] [PubMed] [Google Scholar]
- 87.Kolquist KA, Ellisen LW, Counter CM, et al. Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nat Genet 1998; 19:182–186. [DOI] [PubMed] [Google Scholar]
- 88.Torrecilla S, Sia D, Harrington AN, et al. Trunk mutational events present minimal intra- and inter-tumoral heterogeneity in hepatocellular carcinoma. J Hepatol 2017;67:1222–1231. [DOI] [PubMed] [Google Scholar]
- 89.Nault J-C, Bioulac-Sage P, Zucman-Rossi J. Hepatocellular benign tumors-from molecular classification to personalized clinical care. Gastroenterology 2013;144:888–902. [DOI] [PubMed] [Google Scholar]
- 90.Nault J-C, Couchy G, Balabaud C, et al. Molecular Classification of Hepatocellular Adenoma Associates With Risk Factors, Bleeding, and Malignant Transformation. Gastroenterology 2017;152:880–894.e6. [DOI] [PubMed] [Google Scholar]
- 91.Pilati C, Letouzé E, Nault J-C, et al. Genomic profiling of hepatocellular adenomas reveals recurrent FRK- activating mutations and the mechanisms of malignant transformation. Cancer Cell 2014;25:428–441. [DOI] [PubMed] [Google Scholar]
- 92.Zucman-Rossi J, Jeannot E, Nhieu JTV, et al. Genotype-phenotype correlation in hepatocellular adenoma: new classification and relationship with HCC. Hepatology 2006;43:515–524. [DOI] [PubMed] [Google Scholar]
- 93.Rebouissou S, Franconi A, Calderaro J, et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different β-catenin activity associated with liver tumor progression. Hepatology 2016;64:2047–2061. [DOI] [PubMed] [Google Scholar]
- 94.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Letouzé E, Shinde J, Renault V, et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun 2017;8:1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsia CC, Kleiner DE Jr, Axiotis CA, et al. Mutations of p53 gene in hepatocellular carcinoma: roles of hepatitis B virus and aflatoxin contamination in the diet. J Natl Cancer Inst 1992;84:1638–1641. [DOI] [PubMed] [Google Scholar]
- 97.Hsu IC, Metcalf RA, Sun T, et al. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature 1991;350:427–428. [DOI] [PubMed] [Google Scholar]
- 98.Ozturk M p53 mutation in hepatocellular carcinoma after aflatoxin exposure. Lancet 1991;338:1356–1359. [DOI] [PubMed] [Google Scholar]
- 99.Poon SL, Pang S-T, McPherson JR, et al. Genome-Wide Mutational Signatures of Aristolochic Acid and Its Application as a Screening Tool. Sci Transl Med 2013;5:197ra101–197ra101. [DOI] [PubMed] [Google Scholar]
- 100.Hoang ML, Chen C-H, Sidorenko VS, et al. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci Transl Med 2013;5:197ra102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qian GS, Ross RK, Yu MC, et al. A follow-up study of urinary markers of aflatoxin exposure and liver cancer risk in Shanghai, People’s Republic of China. Cancer Epidemiol Biomarkers Prev 1994;3:3–10. [PubMed] [Google Scholar]
- 102.Neuveut C, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol 2010;52:594–604. [DOI] [PubMed] [Google Scholar]
- 103.Ma N-F, Lau SH, Hu L, et al. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res 2008;14:5061–5068. [DOI] [PubMed] [Google Scholar]
- 104.Wang Q, Zhang W, Liu Q, et al. A Mutant of Hepatitis B Virus X Protein (HBxΔ127) Promotes Cell Growth through A Positive Feedback Loop Involving 5-Lipoxygenase and Fatty Acid Synthase. Neoplasia 2010;12:103-IN3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sze KMF, Chu GKY, Lee JMF, et al. C-terminal truncated hepatitis B virus x protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatology 2013;57:131–139. [DOI] [PubMed] [Google Scholar]
- 106.Brechot C, Pourcel C, Louise A, et al. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature 1980;286:533–535. [DOI] [PubMed] [Google Scholar]
- 107.Wang J, Chenivesse X, Henglein B, et al. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature 1990;343:555–557. [DOI] [PubMed] [Google Scholar]
- 108.Paterlini-Bréchot P, Saigo K, Murakami Y, et al. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 2003;22:3911–3916. [DOI] [PubMed] [Google Scholar]
- 109.Sung W-K, Zheng H, Li S, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012;44:765–769. [DOI] [PubMed] [Google Scholar]
- 110.Nault J-C, Datta S, Imbeaud S, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet 2015;47:1187–1193. [DOI] [PubMed] [Google Scholar]
- 111.Moriya K, Fujie H, Shintani Y, et al. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med 1998;4:1065–1067. [DOI] [PubMed] [Google Scholar]
- 112.Bartosch B, Thimme R, Blum HE, et al. Hepatitis C virus-induced hepatocarcinogenesis. J Hepatol 2009;51:810–820. [DOI] [PubMed] [Google Scholar]
- 113.Hoshida Y, Fuchs BC, Bardeesy N, et al. Pathogenesis and prevention of hepatitis C virus-induced hepatocellular carcinoma. J Hepatol 2014;61:S79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alizadeh AA, Aranda V, Bardelli A, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med 2015;21:846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Friemel J, Rechsteiner M, Frick L, et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res 2015;21:1951–1961. [DOI] [PubMed] [Google Scholar]
- 116.Xue R, Li R, Guo H, et al. Variable Intra-Tumor Genomic Heterogeneity of Multiple Lesions in Patients With Hepatocellular Carcinoma. Gastroenterology 2016;150:998–1008. [DOI] [PubMed] [Google Scholar]
- 117.Finkelstein SD, Marsh W, Demetris AJ, et al. Microdissection-based allelotyping discriminates de novo tumor from intrahepatic spread in hepatocellular carcinoma. Hepatology 2003;37:871–879. [DOI] [PubMed] [Google Scholar]
- 118.Wilkens L, Bredt M, Flemming P, et al. Differentiation of multicentric origin from intra-organ metastatic spread of hepatocellular carcinomas by comparative genomic hybridization. J Pathol 2000;192:43–51. [DOI] [PubMed] [Google Scholar]
- 119.Laurent-Puig P, Legoix P, Bluteau O, et al. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology 2001;120:1763–1773. [DOI] [PubMed] [Google Scholar]
- 120.Boyault S, Rickman DS, Reynies A de, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007;45:42–52. [DOI] [PubMed] [Google Scholar]
- 121.Woo HG, Wang XW, Budhu A, et al. Association of TP53 mutations with stem cell-like gene expression and survival of patients with hepatocellular carcinoma. Gastroenterology 2011;140:1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schulze K, Imbeaud S, Letouze E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ahn S-M, Jang SJ, Shim JH, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 2014;60:1972–1982. [DOI] [PubMed] [Google Scholar]
- 124.Woo HG, Wang XW, Budhu A, et al. Association of TP53 mutations with stem cell-like gene expression and survival of patients with hepatocellular carcinoma. Gastroenterology 2011;140:1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Amaddeo G, Cao Q, Ladeiro Y, et al. Integration of tumour and viral genomic characterisations in HBV- related hepatocellular carcinomas. Gut 2014;64:820–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nault J-C, De Reynies A, Villanueva A, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology 2013;145:176–187. [DOI] [PubMed] [Google Scholar]
- 127.Villanueva A, Hoshida Y, Battiston C, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology 2011;140:1501–12.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee J-S, Chu I-S, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 2004;40:667–676. [DOI] [PubMed] [Google Scholar]
- 129.Lee J-S, Heo J, Libbrecht L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med 2006;12:410–416. [DOI] [PubMed] [Google Scholar]
- 130.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med 2008;359:1995–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pinyol R, Nault JC, Quetglas IM, et al. Molecular profiling of liver tumors: classification and clinical translation for decision making. Semin Liver Dis 2014;34:363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hoshida Y, Villanueva A, Sangiovanni A, et al. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology 2013;144:1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chiang DY, Villanueva A, Hoshida Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res 2008;68:6779–6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hoshida Y, Nijman SMB, Kobayashi M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 2009;69:7385–7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lachenmayer A, Alsinet C, Savic R, et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res 2012;18:4997–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Calderaro J, Couchy G, Imbeaud S, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol 2017;67:727–738. [DOI] [PubMed] [Google Scholar]
- 137.Tan PS, Nakagawa S, Goossens N, et al. Clinicopathological indices to predict hepatocellular carcinoma molecular classification. Liver Int 2016;36:108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018. Available at: 10.1002/hep.29904. [DOI] [PubMed] [Google Scholar]
- 139.Ziol M, Poté N, Amaddeo G, et al. Macrotrabecular-massive hepatocellular carcinoma: A distinctive histological subtype with clinical relevance. Hepatology 2017. Available at: 10.1002/hep.29762. [DOI] [PubMed] [Google Scholar]
- 140.Wood LD, Heaphy CM, Daniel HD-J, et al. Chromophobe hepatocellular carcinoma with abrupt anaplasia: a proposal for a new subtype of hepatocellular carcinoma with unique morphological and molecular features. Mod Pathol 2013;26:1586–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jusakul A, Cutcutache I, Yong CH, et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov 2017;7:1116–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fujimoto A, Furuta M, Shiraishi Y, et al. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat Commun 2015;6:6120. [DOI] [PubMed] [Google Scholar]
- 143.Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003–1010. [DOI] [PubMed] [Google Scholar]
- 144.Wang A, Wu L, Lin J, et al. Whole-exome sequencing reveals the origin and evolution of hepato- cholangiocarcinoma. Nat Commun 2018;9:894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chaisaingmongkol J, Budhu A, Dang H, et al. Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell 2017;32:57–70.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Woo HG, Lee J-H, Yoon J-H, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res 2010;70:3034–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Brunt E, Aishima S, Clavien P-A, et al. cHCC-CCA: Consensus terminology for primary liver carcinomas with both hepatocytic and cholangiocytic differentation. Hepatology 2018. Available at: 10.1002/hep.29789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Druker BJ, Guilhot F, O’Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med 2006;355:2408–2417. [DOI] [PubMed] [Google Scholar]
- 149.Anon. Vemurafenib in Melanoma with BRAF V600E Mutation. N Engl J Med 2011;365:1448–1450. [DOI] [PubMed] [Google Scholar]
- 150.Zhou C, Wu YL, Chen G, et al. Final overall survival results from a randomised, phase III study of erlotinib versus chemotherapy as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer (OPTIMAL, CTONG-0802). Ann Oncol 2015;26:1877–1883. [DOI] [PubMed] [Google Scholar]
- 151.Anon. First-Line Crizotinib versus Chemotherapy in ALK-Positive Lung Cancer. N Engl J Med 2015;373:1582. [DOI] [PubMed] [Google Scholar]
- 152.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378–390. [DOI] [PubMed] [Google Scholar]
- 153.Cheng A, Kang Y, Chen Z, et al. Randomized phase III trial of sorafenib versus placebo in Asian patients with advanced hepatocellular carcinoma. J Clin Oncol 2008;26:4509–4509.18802164 [Google Scholar]
- 154.Cheng A-L, Kang Y-K, Chen Z, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol 2009;10:25–34. [DOI] [PubMed] [Google Scholar]
- 155.Cheng A, Kang Y, Lin D, et al. Phase III trial of sunitinib (Su) versus sorafenib (So) in advanced hepatocellular carcinoma (HCC). J Clin Oncol 2011;29:4000–4000. [Google Scholar]
- 156.Cainap C, Qin S, Huang W-T, et al. Linifanib versus Sorafenib in patients with advanced hepatocellular carcinoma: results of a randomized phase III trial. J Clin Oncol 2015;33:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhu AX, Rosmorduc O, Evans TRJ, et al. SEARCH: a phase III, randomized, double-blind, placebo- controlled trial of sorafenib plus erlotinib in patients with advanced hepatocellular carcinoma. J Clin Oncol 2015;33:559–566. [DOI] [PubMed] [Google Scholar]
- 158.Bruix J, Qin S, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;389:56–66. [DOI] [PubMed] [Google Scholar]
- 159.Anon. Cabozantinib Improves Survival in Phase III HCC Trial. OncLive. Available at: https://www.onclive.com/web-exclusives/cabozantinib-improves-survival-in-phase-iii-hcc-trial [Accessed April 22, 2018]. [Google Scholar]
- 160.Abou-Alfa GK, Meyer T, Cheng A-L, et al. Cabozantinib in Patients with Advanced and Progressing Hepatocellular Carcinoma. N Engl J Med 2018;379:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Santoro A, Rimassa L, Borbath I, et al. Tivantinib for second-line treatment of advanced hepatocellular carcinoma: a randomised, placebo-controlled phase 2 study. Lancet Oncol 2013;14:55–63. [DOI] [PubMed] [Google Scholar]
- 162.Rimassa L, Assenat E, Peck-Radosavljevic M, et al. Tivantinib for second-line treatment of MET-high, advanced hepatocellular carcinoma (METIV-HCC): a final analysis of a phase 3, randomised, placebo- controlled study. Lancet Oncol 2018. Available at: 10.1016/S1470-2045(18)30146–3. [DOI] [PubMed] [Google Scholar]
- 163.Rebouissou S, La Bella T, Rekik S, et al. Proliferation Markers Are Associated with MET Expression in Hepatocellular Carcinoma and Predict Tivantinib Sensitivity In Vitro. Clin Cancer Res 2017;23:4364–4375. [DOI] [PubMed] [Google Scholar]
- 164.Zhu AX, Baron AD, Malfertheiner P, et al. Ramucirumab as Second-Line Treatment in Patients With Advanced Hepatocellular Carcinoma: Analysis of REACH Trial Results by Child-Pugh Score. JAMA Oncol 2017;3:235–243. [DOI] [PubMed] [Google Scholar]
- 165.Kong Y, Sun L, Hou Z, et al. Apatinib is effective for treatment of advanced hepatocellular carcinoma. Oncotarget 2017;8:105596–105605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yu W-C, Zhang K-Z, Chen S-G, et al. Efficacy and Safety of apatinib in patients with intermediate/advanced hepatocellular carcinoma: A prospective observation study. Medicine 2018;97:e9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017;389:2492–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Greten LAO. Molecular- and immune-based therapies for HCC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.