Abstract
In a multicellular organism, somatic mutations represent a permanent record of the past chemical and biochemical perturbations experienced by a cell in its local microenvironment. Akin to a perpetual recording device, with every replication, genomic DNA accumulates mutations in patterns that reflect: i) the sequence context-dependent formation of DNA damage, due to environmental or endogenous reactive species, including spontaneous processes; ii) the activity of DNA repair pathways, which, depending on the type of lesion, can erase, ignore or exacerbate the mutagenic consequences of that DNA damage; and iii) the choice of replication machinery that synthesizes the nascent genomic copy. These three factors result in a richly contoured sequence context-dependent mutational spectrum that, from appearances, is distinct for most individual forms of DNA damage. Such a mutagenic legacy, if appropriately decoded can reveal the local history of genome-altering events such as chemical or pathogen exposures, metabolic stress, and inflammation, which in turn can provide an indication of the underlying causes and mechanisms of genetic disease. Modern tools have positioned us to develop a deep mechanistic understanding of the cellular factors and pathways that modulate a mutational process and, in turn, provide opportunities for better diagnostic and prognostic biomarkers, better exposure risk assessment and even actionable therapeutic targets. The goal of this Perspective is to present a bottom-up, lesion-centric framework of mutagenesis that integrates the contributions of lesion replication, lesion repair and lesion formation to explain the complex mutational spectra that emerge in the genome following exposure to mutagens. The mutational spectra of the well-studied hepatocarcinogen aflatoxin B1 are showcased here as specific examples, but the implications are meant to be generalizable.
Keywords: Mutagenesis, DNA adduct, mutational signature, mycotoxin, HCC, cancer
1. Introduction
Mutagenesis is a fundamental biological process in which the heritable information encoded in genomic DNA is irrevocably altered. A double-edged sword, mutagenesis enables beneficial events, such as evolution of species[1], diversification of antibody repertoires in immune cells [2] and rapid clearance of viral pathogens [3], while at the same time being deleterious by driving carcinogenesis [4], (premature) aging [5], and neurodegenerative [6] and autoimmune diseases [7].
Mutagenesis-induced genomic changes, which can range from single nucleotide (point mutations) to large scale, complex rearrangements of genetic material (chromosomal rearrangements) are driven by a myriad of extrinsic and intrinsic factors that in most cases chemically modify DNA. Examples of exogenous mutagenic agents include radiation (UV light and ionizing radiation), alkylating agents from either naturally occurring (N-nitroso compounds) or man-made (chemotherapeutics) sources, mycotoxins (e.g., aflatoxin B1) and polyaromatic hydrocarbons (e.g., benzo[a]pyrene). Examples of endogenous mutagenic agents include reactive oxygen, nitrogen and halogen species and their secondary damage products (e.g., reactive aldehydes formed by lipid peroxidation), and dysregulated enzymatic processes (e.g., APOBEC deaminases). Additionally, deficiencies in DNA repair pathways due to genetic defects (loss of function), epigenetic inactivation (hypermethylation of a promoter) or metabolic disruption such as lack of necessary enzymatic co-factors (e.g., lack of α-ketoglutarate for AlkB-class demethylases [8]) or building blocks (e.g., imbalanced [9] or contaminated [10] nucleotide pool) are also strong contributors to mutagenesis by potentiating the effects of DNA damage.
The diversity of factors that contribute to mutagenesis creates a challenge toward interpreting complex end-stage mutational patterns associated with disease. This Perspective lays out a unifying mechanistic framework that could explain the biochemical mechanisms underlying mutagenesis in almost all cases. The deep mechanistic understanding of a mutational process in turn can provide translational opportunities for predicting and modulating mutagenic outcomes.
2. A mechanistic analysis of mutational processes
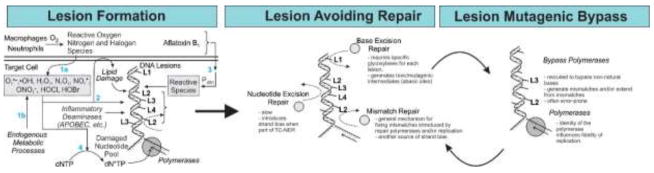
The sequence of biochemical events that introduces a specific mutational pattern in DNA is defined as a mutational process [11,12]. In terms of mechanism, the most general description of a mutational process involves three distinct steps: 1) DNA lesion formation; 2) DNA repair avoidance; 3) DNA lesion replication (Fig. 1).
Fig. 1. The mechanistic components behind a mutagenic process.
Almost all mutations can be traced back to a sequence of events in which first, the DNA is chemically altered by reactive species generating a DNA lesion (L1, L2, L3, etc.; left panel). Several sources of reactive species are depicted: exogenous chemicals, such as aflatoxin B1 and inflammatory mediators (paths 1a and 3); inflammation and associated processes that generate endogenous reactive species (reactive oxygen, nitrogen and halogen species; path 1b), as well as secondary reactive products (path 2; lipid oxidation-derived aldehydes); endogenous enzymes that directly modify DNA bases (APOBEC family deaminases). One additional pathway that leads to DNA lesions is the incorporation of a damaged nucleotide, generated by some of the same reactive species above (path 4). Most lesions are typically detected and repaired by the DNA repair pathways, before they get a chance to be replicated (middle panel). Finally, during replication, the DNA lesion may miscode, leading to a mismatch. In a subsequent replication, or during an attempt at mismatch repair, this mismatch will lead to a point mutation (right panel).
In the majority of cases, the process of mutagenesis begins with the formation of a DNA lesion. Such a process includes the chemical modification of DNA by extrinsic and intrinsic factors, as enumerated in the introduction, but also spontaneous reactions (e.g., non-enzymatic deamination, oxidation, depurination) that modify the chemical structure of DNA or DNA bases, as well as aberrant enzymatic reactions on DNA (e.g., enzymatic deaminations, overactive glycosylases, etc.). Even the electronic fluctuations intrinsic to the chemical structures of the DNA bases such as tautomerism can be included in this analysis; by virtue of their ability to alter base-pairing preferences, minor tautomers of the natural bases, for example, can be considered as very-transient DNA lesions [13–15]. Lastly, another way by which DNA accumulates mutagenic lesions is the incorporation of chemically modified nucleotides during DNA replication or via DNA repair pathways that involve DNA synthesis (Fig. 1, left panel).
Most DNA lesions, however, do not lead to mutations, owing to the successful activity of DNA repair pathways. In many ways, DNA repair acts as the lynchpin protecting the integrity of genetic information, because it controls the type and amount of DNA lesions that still remain at the moment of DNA replication. Accordingly, a mutational process may reflect either the absence of a lesion-specific DNA repair pathway (e.g., a genetic defect), or the ability of a lesion to evade repair. Repair evasion, in turn, can occur in several different ways:
Lesion overload. When the lesion formation rate exceeds the rate of repair, a number of lesions will remain unrepaired at the time replication happens [16]. This case typically occurs in the wake of a massive, acute exposure to a DNA damaging agent as might happen after DNA-damaging chemotherapy. Stochastic fluctuations in the levels of repair enzymes can also lead to situations where slowly repaired lesions might be missed (e.g., repair of deaminated 5-methylcytosine (5mC) by the TDG or MBD4 glycosylases [17]).
Repair-resistant/Stealthy lesions. Certain types of DNA modifications do not significantly disrupt the tertiary DNA structure, rendering them essentially invisible to DNA repair surveillance [18], and therefore they are repaired very slowly if at all. Such modifications typically persist until they are encountered by transcriptional or replication complexes. Owing to their planar structure, which allows DNA intercalation, certain adducts formed by benzo[a]pyrene (B[a]P) [18,19], aflatoxin B1 (AFB1) [20,21] or aristolochic acid (AA) [22] are examples of lesions that evade repair and persist in the genome, as evidenced by their unusually long half-lives in tissues [23–25].
Toxic lesions. Unlike stealthy lesions, these adducts disrupt the structure of DNA in a manner that leads to toxicity and replication stress. Such lesions include those formed by certain bulky alkylating agents and crosslinking agents (e.g., UV light, cisplatin, and reactive aldehydes).
The final step in a mutational process is DNA replication, which converts a DNA lesion that has escaped repair into a heritable base change. Here, the identity of the replication machinery is responsible for shaping the mutational outcome. Many lesions are too bulky to be accommodated by the native replicative polymerases; instead, translesion synthesis polymerases (e.g., Y-family bypass polymerases) are recruited to the replication fork, and these polymerases are sometimes intrinsically error-prone. In many cases, an extension polymerase (e.g., pol zeta) is required to extend from a mispaired DNA lesion, before the normal DNA replication can resume.
When examining the distribution of point mutations that accumulate in the genome, it has long been appreciated that not all bases mutate with the same frequency [26–28]. Rather, all known genomes, sequenced after mutagenic insults, feature a collection of hot-spots (bases that are more prone to mutate than average) and cold-spots (bases that are less prone to mutate than average) [27–33]. Many factors contribute to the uneven distribution of mutations; chromatin accessibility (open vs closed chromatin) [34], transcriptional status (transcribed vs non-transcribed strands) [35], replication status (early vs late replicating genes) [35], coding status (regulatory elements vs gene bodies) [36], all shape the distribution of mutations at a genomic scale. But at a local scale, when all other factors are the same, the differential mutational frequencies are primarily driven by sequence context [37–40]. Owing to their slightly different physico-chemical and biochemical properties (e.g., size, charge, stacking propensity and hydrogen bonding ability), the DNA bases immediately preceding (at the 5′ position) and immediately following (at the 3′) of a point mutation influence all the three steps of a mutational process: sequence context influences the reactivity of a given nucleobase with a chemical agent; it modulates the efficiency with which the lesion is recognized and repaired; and it shapes the fidelity with which a lesion is replicated by a polymerase.
a. Sequence-dependent lesion formation
One of the major contributing factors to sequence-dependent mutagenesis is the propensity of a lesion to form in a given sequence context. When enzymatic processes are involved, such reactions typically are sequence specific. To give two examples, APOBEC enzymes responsible for generating certain deaminated cytosines have strong sequence specificity for the TpCpN sequence context [41,42] and hydrolytic deamination of m5C follows the sequence specificity of m5C formation by DNA methyl transferases, the NpCpG sequence context [43].
DNA lesions formed by large and planar reactive species (such as those derived from environmental mutagens like AFB1, AA, B[a]P, etc.) usually involve a DNA intercalation step before covalent attachment. This mechanism of interaction prior to chemical reaction could therefore explain the preference of these agents to mutate certain sequence contexts over others [20,21,44].
b. Sequence-dependent lesion repair
Every DNA repair pathway has a certain sequence-dependent bias. For example, the rate with which DNA glycosylases can splay a damaged base outside the DNA helix is likely dependent on the adjacent bases and their ability to base-stack with the lesion and/or to stabilize the repair complex [45]. A similar argument can be made for mismatch repair (MMR) [46,47] and for nucleotide excision repair (NER) [48–50], where the lesion identification step may be influenced by the sequence context around the lesion. The activity of direct reversal DNA repair enzymes is also affected by sequence context. A study done in living cells by Delaney et al., (2001) showed that the activities of bacterial O6-methyl-guanine demethylases Ogt and Ada are both sequence-context dependent and distinct from one another [37].
Additionally, lesion repair is generally responsible for the mutational asymmetry between DNA strands (strand bias); the template strand in the actively transcribed regions benefits from transcription coupled repair (TC-NER) and therefore contains fewer lesions and consequently fewer mutations than the non-transcribed strand [51].
c. Polymerase- and sequence-dependent lesion replication
The identity of the polymerase replicating across a DNA lesion is a key determinant of the fidelity-outcome of the replication event. Many lesions require specialized polymerases in order to be replicated, such as Y-family bypass polymerases. While as a class, bypass polymerases are more error prone than the replicative polymerases, they can help bypass certain lesions in an error-free manner. For example, the bulky benzo[a]pyrene diolepoxide-derived guanine adduct is faithfully bypassed by pol kappa [52]; most of the mutations introduced by this adduct seem to occur when the bypass is performed by pol eta [53]. By contrast, UV-light induced pyrimidine dimers are bypassed error-free by pol eta [54], while other Y-family polymerases, pol kappa and pol iota, are more likely to introduce errors when bypassing the same lesions [55]. Additionally, auxiliary polymerases (e.g., pol zeta) are required to extend from the mismatch introduced by a DNA lesion, as is the case with aristolochic acid-adenine adducts [56]; in this case, the Y-family polymerases do not seem to contribute to the formation of the mismatch [56].
Regardless of the polymerase involved, the fidelity of replication is also sequence-context dependent; the bases immediately adjacent to the lesion [57,58] as well as more distant bases [40] influence the outcome of the replication.
d. Reconstruction of a mutational spectrum from sequence-dependent mechanistic factors
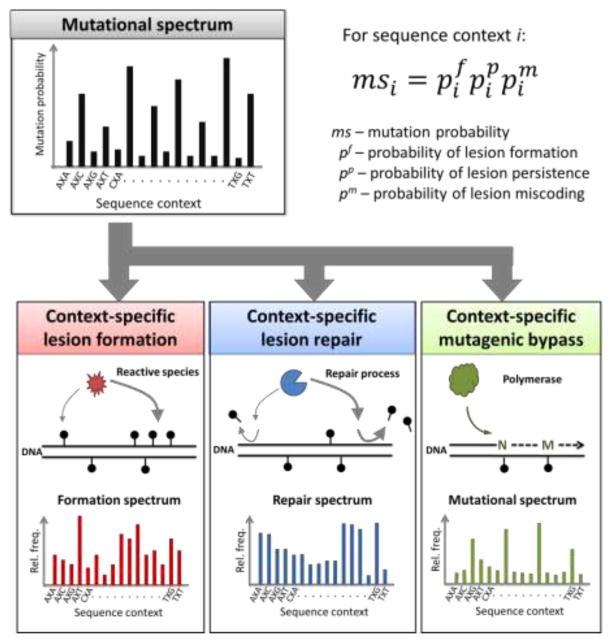
Mathematically, one could envision each of the aforementioned three factors as an independent contributor to the mutational spectrum characteristic of a mutational process; therefore, the sequence-context frequencies of a given type of point mutation can be viewed as the direct product (appropriately normalized) of the sequence-dependent frequencies of lesion formation, lesion repair avoidance and lesion mutagenic bypass (Fig. 2). As detailed in the next section, the classification of point mutations as a function of trinucleotide sequence context enabled the construction of mutational spectra characteristic of a single mutagen/carcinogen or characteristic of a defined mutational process. Therefore, in practice, the model outlined in Figure 2 may be most useful in teasing out the sequence-dependent contributions of lesion formation, lesion repair or lesion mutagenic bypass—mechanistic aspects difficult to measure directly—to an already known mutational spectrum or one easily accessed experimentally.
Fig. 2. A mathematical model of the fundamental elements underlying a mutational spectrum.
The context-specific mutational probability associated with a mutational process (i.e., mutational spectrum) can be reconstructed from context-specific lesion formation, lesion repair and lesion mutagenic bypass probabilities, in a given genetic background. For each sequence context i, the mutational probability msi reflects the product of the probabilities that the lesion forms in that sequence context (ppi), the lesion persists (i.e., avoids repair) in that sequence context (p i), and the lesion miscodes (i.e., introduces a mutation) in that sequence context (pmi).
While complex, understanding the molecular mechanisms underpinning mutational processes is key to measure accurately the mutagenic impact of known environmental agents and establish the genetic and metabolic risk factors that shape the mutagenic properties of those agents. Such knowledge can then translate into biomarkers and diagnostic tools, based on mutagenesis measurements, as well as intervention points that can modulate the risk and progression of mutational diseases (e.g., cancer, autoimmune diseases, neurodegeneration and aging).
3. Mutational signatures of human cancer
As the poster disease of mutagenesis gone awry, cancer and the carcinogenic process have remained for decades largely intractable from the mutational mechanisms point of view due to the complexity of the risk factors, and the sheer size of genomic space under analysis. The analysis of inherited or acquired driver mutations in oncogenes and tumor suppressor genes was generally insufficient to identify operative mutational processes and define causative factors of mutagenesis.
The advent of deep sequencing enabled a significant leap in understanding the distribution and diversity of mutations that accumulate in mammalian genomes, and in particular in the cancer genome. Tumor sequencing efforts quickly led to the construction of massive databases (e.g., COSMIC, TCGA) that contained the complete list of point mutations accumulated in a cancer genome. Recently, data mining algorithms (e.g., non-negative matrix factorization), pouring over these data, were able, for the first time to extract distinct mathematical patterns of mutational distributions that ostensibly corresponded to biochemically distinct mutational processes [11,12]. When evaluating primarily substitution mutations, the three-nucleotide sequence context emerged as a variable that allowed workers to start distinguishing among the many mutational processes that generate the same type of point mutation. Introduction of additional biological insights, such as the strand bias of certain mutations in transcribed regions, or the correlation of a particular spectrum with a type of cancer led to further refinements of these patterns of mutations, which are called mutational signatures [11,12,59,60].
There are currently at least 30 mathematically-derived mutational signatures that have been extracted from data encompassing tens of thousands of individual tumor exomes and genomes, from over 40 cancer types. Linear combinations of these patterns can be used, in principle, to reconstruct the mutational spectrum of any sequenced tumor (or tissue with a genetic disease), and thus provide insight on the explicit mutational processes (i.e., biochemical events) that have shaped the development of that malignancy [11,12].
For the purposes of this Perspective, we are making a distinction between mutational signatures, which are at origin mathematical constructs deconvoluted from complex samples using statistical models and simulations, and mutational spectra characteristic of a mutagen, which denote the experimentally observed collections of point mutations in a specific sample exposed to the mutagen. A mutational spectrum recorded under comparable experimental conditions is highly reproducible (see the analysis of the AFB1 mutational spectra below (Fig. 3); also Fig. S3 in Chawanthayatham et al. [61]). By contrast, the output of the mathematical deconvolution of large data sets is highly dependent not only on the choice of algorithm but also on the size, diversity and quality of the data. To illustrate this point, the initial large scale study to identify recurring patterns of mutations in human cancers (i.e., mutational signatures) found only 5 signatures [60]. The next iteration, operating on a much larger dataset, found about 21 [59]. The collection of signatures increased again to 30 when more data were considered and a more targeted analysis was performed on a larger cohort of sequenced liver tumors [62]. Importantly, with each iteration, some early signatures resolved into multiple, separate ones or became more refined in the process. Yet another even larger dataset is in the works that will expand the number of mutational signatures even further (see the Perspective by David Phillips).
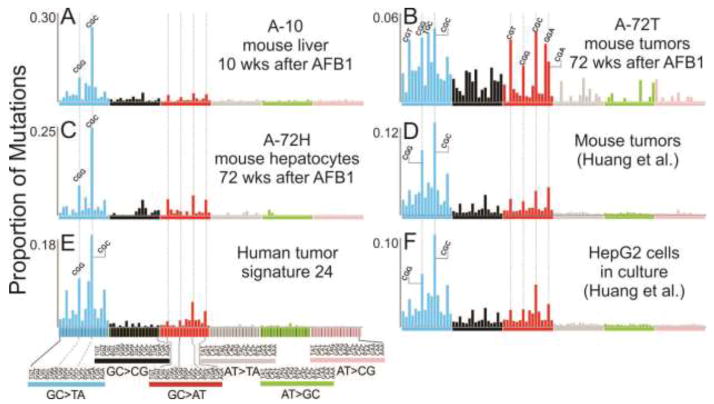
Fig. 3. Mutational spectra of aflatoxin B1.
Spectra shown in panels A, B and C are from Chawanthayatham et al., 2017, where a single dose of AFB1 was given to male mice in the first week of life. At 10 weeks, the mutations in their livers were enumerated using duplex consensus sequencing (spectrum A-10). At 72 weeks, the tumor cells were separated from the surrounding liver hepatocytes and analyzed separately, yielding spectra A-72T (tumor) and A-72H (hepatocyte fraction). Spectra shown in panels D and F were derived from Huang et al. 2017. Male mice exposed early in life to a single AFB1 dose were raised until tumors developed, which were then analyzed using whole genome sequencing (Panel D). HepG2 cells were exposed to AFB1 in cell culture, and then subjected to Whole Genome Sequencing (Panel F). Panel E shows human signature 24, as reported by Schulze et al. 2015, and on the COSMIC website. The x axis denotes triplet contexts for point mutations from the standpoint of purine-originated mutations; the order of triplets, however, is the same as the one shown in other published studies reporting mutational signatures. All spectra were normalized to reflect point mutation frequencies when trinucleotide oligomers occur with equal probabilities.
Nevertheless, the top-down approach, relying primarily on mathematical deconvolution of large datasets, has yielded a number of impressive results. The mutational signatures of several prevalent environmental carcinogens were identified: UV light (signature 7) [59], B[a]P (signature 4) [59], AA (signature 22) [11,63,64] and AFB1 (signature 24) [62]. These signatures were subsequently reproduced in simpler cell culture systems (signatures 4, 7, and 22) [65] and in both cell culture and animal models (signature 24) [61,66,67], engendering confidence in the causal link between mutagen exposure and the resulting mutational spectrum. Additionally, for each of these agents, the biochemical steps that lead to mutations, from the types of DNA lesions induced to the modulatory effects of DNA repair and replication, are generally well understood, making them excellent candidates to showcase the general molecular mechanism of mutagenesis outlined in the previous section. One other mutational signature (signature 11) is strongly associated with the brain cancer drug temozolomide treatment [11,59], and the observed dominant mutations (GC>AT) reflect the properties of the most likely mutagenic DNA adduct generated by the agent (O6mG). However, efforts to reproduce this signature in simpler, cell culture systems have not been successful yet (see the Perspective by David Philips).
Another subset of the identified mutational signatures strongly correlates with deficiencies in various aspects of DNA repair. Signature 3 reflects deficient DNA double-strand break repair by homologous recombination, usually due to loss of function of the BRCA1 or BRCA2 proteins [11,59]. Signatures 6, 15, 20, 26 all correlate with defective MMR [11]. Signature 10 reflects the activity of an exonuclease-deficient (error-prone) pol epsilon, one of the major replicative polymerases [59,68]. Signature 18 seems to reflect the absence of the MUTYH glycosylase, the key enzyme responsible for preventing the mutagenesis of 8-oxoguanine, a prevalent DNA oxidation damage [69,70]; however, a subsequent study suggested that signature 18 may be specific only to certain cancers, and proposed an additional signature (albeit very reminiscent of signature 18) to reflect MUTYH deficiency and 8-oxoguanine mutagenesis [71]. The new pattern was referred as signature 36 [71], and it is yet to be added to the curated list of signatures on the COSMIC site (https://cancer.sanger.ac.uk/cosmic/signatures). Finally, signature 30 has been recently associated with a defective NTHL1 glycosylase [72].
Each of the signatures above likely reflects a collection of mutational processes, initiated by several different DNA lesions that are the primary substrates of each of the repair pathways indicated. Related to this subset of signatures is signature 1, commonly thought to reflect the deamination of 5mC in CpG islands. As the deamination product—a T~G mismatch—is a substrate for both BER (via the TDG or MBD4 glycosylases) and MMR, the appearance of these mutations can be interpreted as a deficient or incomplete repair process. The other highly prevalent signature (signature 5) has been suggested to reflect, in part, a deficiency in the NER pathway, and in particular, a defect in the core protein ERCC2 [73,74].
The remaining mutational signatures have poorly characterized etiologies, reflecting potentially complex mechanisms. One signature worth mentioning here is signature 16, which has been strongly associated with chronic alcohol exposure [62,75,76]; however, neither the operative DNA lesions nor the modulating repair processes are currently known for this signature.
4. Aflatoxin B1 – a potent hepatocarcinogen
The discussion above lays out the intellectually appealing view that the mutational spectra of mutagens/carcinogens, either experimentally-determined or mathematically derived from large datasets, feature a finer substructure that reflects the explicit biochemical contributions of lesion formation, repair and bypass. A further deconvolution of a mutational spectrum into its mechanistic components (Fig. 2) would allow molecular explanations for the correlative metadata and other biological insights that often accompany mutational spectra/signatures (e.g., genetic background, strand bias, characteristics of the cellular microenvironment, etc.), and provide deeper insight into the molecular origins of carcinogenesis. This Perspective closes using a recent example from the literature – AFB1, which provides a richly detailed mutational spectrum ripe for deconvolution.
The mycotoxin AFB1 is a human liver carcinogen associated with more than 700,000 primary hepatocellular carcinoma (HCC) deaths worldwide each year [77–81]. Aflatoxin is carcinogenic alone, but its potency is amplified at least 60-fold by concurrent hepatitis B viral infection [82,83]. Both fungal contamination of crops and viral hepatitis are endemic factors in the developing world (specifically, sub-Saharan Africa, Central and South America, Southeast Asia), making HCC one of the leading cause of cancer death in the high risk areas [80]. Additionally, recent studies have shown that early life exposures to the toxin, including in utero exposures, come with an amplified risk [84,85], while exposure during pregnancy can magnify the deleterious consequences of the toxin for the mother [86].
Despite decades of research on AFB1, there is still an unmet medical need for timely detection of aflatoxin-induced HCC and effective therapeutic strategies. To address these challenges, most recent work in this area has focused on characterizing and detecting early the mutational processes that underlie AFB1-induced carcinogenesis [61,66,67].
a. Mutational spectra of AFB1 in vivo
The association between aflatoxin exposure and HCC has been long established, but the mutational signature of aflatoxin in human HCC emerged only a few years ago. The initial large scale study that enumerated a majority of mutational signatures [59] found many signatures operating in liver cancer, suggesting that HCC in humans has diverse etiologies. In an attempt to stratify the disease, an enlarged dataset was developed that led to a mutational signature (signature 24) that likely reflects, based on tumor sample metadata, exposure to AFB1 [62]. Figure 3E shows a version of signature 24 replotted such that the triplet sequence contexts depict the mutations at purines (rather than pyrimidines); additionally, the data are normalized to the triplet frequencies characteristic of the human genome (calculated using the SomaticSignatures package [87] applied on the GRCh38 human reference genome), thus reflecting the mutational distribution when all triplets occur with equal probability. Such plotting captures better the underlying mechanistic elements of the mutational spectra, such as the propensity to generate more DNA damage, avoid repair and/or lead to misreplication in a particular sequence context. Signature 24 is dominated by G→T transversions, which are characteristic of the AFB1 adducts [88–92]. A follow-up study further examined the presence of signature 24 in more samples, as well as some time-dependent changes of the proportion of these mutations during the carcinogenic process [75].
Efforts to demonstrate a direct causal link between AFB1 exposure and emergence of mutational signature 24 in tissues followed. In one study [67], the group of Steven Rozen exposed a human liver cell line (HepG2) to AFB1 in cell culture. Several surviving cells were expanded clonally and subjected to whole-genome sequencing [67]. In a parallel experiment, male mice were exposed to a single large dose of AFB1 in the first week of life; a year and a half later, all mice developed liver tumors, which were collected and subjected to whole-genome sequencing [67]. Typical mutational spectra from these two experiments are reproduced in Figure 3, panels D and F, once again, replotted from the perspective of the mutations at purines, and normalized for triplet frequency occurrence.
Our laboratory has been interested in evaluating the mutagenic imprint of AFB1 in vivo at a time point close to the exposure period, and long before the carcinogenic process is set in motion [61,66]. Accordingly, a transgenic mouse model (similar to the one used in the study above) was exposed to a single dose of AFB1 in the first week of life. Previous work had shown that this exposure protocol induces liver cancer in 100% of the male mice by 72 weeks of life. We, however, sacrificed a cohort of treated mice at 10 weeks, a time point at which the treated livers are indistinguishable from controls, and extracted the DNA to look for a possible early-onset mutagenic imprint of AFB1. Given the heterogeneity of the mutations and their relative rarity in the exposed tissues, standard next-generation sequencing techniques would have not been sensitive enough to detect the AFB1-induced mutations at this early time point. Thus, we turned to an ultra-high fidelity sequencing technique, denoted duplex consensus sequencing (DupSeq), developed by our collaborator, Lawrence A. Loeb [93–95]. This technique is at least three orders of magnitude more sensitive than traditional next-gen sequencing, affording accurate detection of mutations as infrequent as 1 in 107–108 bases. The resulting mutational spectrum of AFB1 in 10 week old livers, denoted A-10, is shown in Figure 3. In a separate experimental arm, the mice were raised until 72 weeks, at which point they had all developed liver tumors. We isolated the normal hepatocytes surrounding the tumors, as well as the tumors themselves, and analyzed both samples by DupSeq [61,66], yielding the spectra A-72H (hepatocytes at 72 weeks post AFB1 exposure) and A-72T (tumors) (Fig. 3).
Of note is that the tumor spectrum is visibly more complex than the tumor spectrum reported by Huang and co-workers [67]. The difference is likely due to the sequencing technology used. The whole-genome sequencing performed by Huang et al. primarily captures clonal mutations that are present in a large number of tumor cells; accordingly, such mutations must have occurred early in tumorigenesis, and thus they closely reflect AFB1-induced mutations (Fig. 3). By contrast, owing to the high fidelity of DupSeq and the ability to analyze a heterogenous population of cells directly, A-72T captures the mutations that have occurred during the most recent replication cycles. Such mutations contribute to the tumor heterogeneity and are likely contributed by mutational processes other than those provoked by mutagenic replication of the initial AFB1 adducts. Accordingly, the contribution of the AFB1 mutational process (i.e., the A-10 spectrum) is diminished, accounting for only ~30% of mutations in A-72T [61]. This phenomenon of dilution of the mutations generated by older mutational processes by more recent mutations characteristic of other mutational processes has also been observed in human tumors. A careful analysis of tumor cell lineage and clonality revealed that in certain human HCCs, an initiating mutational spectrum induced by AFB1 is essentially drowned out by subsequent mutational processes that drive the later stages of carcinogenesis [75].
There are remarkable similarities among all the mutational spectra (with the exception of A-72T) directly recorded following exposure to AFB1 and human mutational signature 24 (cosine similarity 0.9 or greater)(Fig. 3). The salient features—the dominant G→T transversions concentrated in the CGC and CGG sequence contexts, and a small proportion of G→A transitions primarily in the CGN contexts—are evident across all spectra. Additionally, the different experimental conditions used to generate the different the mutational spectra in Figure 3 provide additional insight into the mutational process characteristic of AFB1 exposure. The spectrum obtained from liver cell culture suggests that AFB1 mutagenic imprint does not require extrinsic factors, such as inputs/stimuli from immune cells; the metabolic activation of the toxin by P450 cytochromes is sufficient to induce the characteristic DNA damage and subsequently mutations. The A-10 mouse spectrum teaches us that the AFB1 mutational spectrum takes hold in the mouse liver within the first 10 weeks of life. Furthermore, the AFB1 mutational spectrum is persistent, as evidenced by both A-72H hepatocytes spectrum at 72 weeks and the mouse tumor spectrum in the Huang et al study, which largely reflects the early, clonal mutational events. Finally, the similarity between mouse spectra and human Signature 24 indicates that the mouse model is an excellent approximation of the consequences of human exposure to AFB1, at least from the point of view of the exposure spectrum, which presumably recapitulates AFB1 metabolic activation, formation of DNA lesions, and the contributions of repair pathways and mutagenic replicative bypass acting on the AFB1-induced lesions.
b. Contribution of lesion formation to the AFB1 mutational spectrum
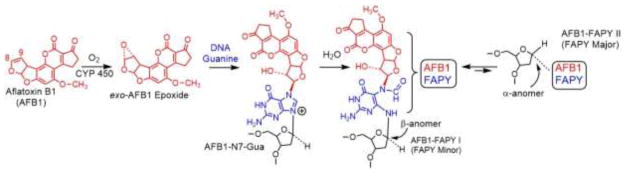
The mutational process of AFB1 is summarized in Figure 4. The mycotoxin is readily absorbed in the gut and reaches the liver through the portal circulation. Inside hepatocytes, AFB1 is bio-activated by several Phase I metabolic enzymes (cytochrome P450s) to the highly reactive species, the exo-AFB1-8,9-epoxide. The epoxide is stable enough to diffuse into the nucleus and, owing to its planar structure, intercalates in genomic DNA [96,97]. This interaction lines up the epoxide to react by an SN2 mechanism with N7 atom of guanine, forming the initial DNA damage product, the AFB1-N7-guanine (AFB1-N7-Gua) adduct [98]. Over time, the adduct may lead to an abasic site through depurination; or it can react with water to form two very chemically and biologically stable formamidopyrimidine (FAPY) adducts: FAPY I (or FAPY minor), which corresponds to the β-anomer of the deoxyribose, and FAPY II (or FAPY major), which corresponds to the α-anomer. The AFB1-N7-Gua and FAPY I DNA adducts are mutagenic; when replicated, they mispair primarily with adenine, leading to G→T transversions [88–92], but they also generate a small amount of G→A transitions [88–92]. The FAPY II adduct is strong block to replication [90]. The AFB1 epoxide can also react, to a small extent, with adenine, generating the N7 adduct.
Fig. 4.
The structure of aflatoxin B1 and the DNA lesions induced by the metabolically-generated AFB1-exo-8,9-epoxide: AFB1-N7-G, AFB1-FAPY-I (FAPY minor), and AFB1-FAPY-II (FAPY major).
All AFB1-epoxide interactions with DNA, from intercalation through DNA adduct formation, are known to be strongly influenced by the sequence context [20,96,97,99–107]. A careful, systematic study by Edward Loechler investigated the propensity of the AFB1-epoxide to form adducts in vitro when the target guanine is present in a large variety of pentanucleotide sequence contexts [106]. The data highlighted the vastly different reactivities of the AFB1-epoxide in various sequence contexts, indicating that the formation spectrum of AFB1 adducts is rugged. Several patterns emerged, such as the reaction preference for contexts with at least two adjacent guanines [106].
c. Contribution of lesion repair to the AFB1 mutational spectrum
DNA repair pathways considerably shape the mutagenic outcomes of the DNA adducts induced by AFB1. Of particular note is the observation that the most mutagenically consequential AFB1 adduct, AFB1-FAPY minor, is only partially repaired, perhaps owing to its property to cause minimal disruption of DNA structure. Supportive of this conclusion are studies that have measured the persistence of this adduct in the livers of animals acutely exposed to a high dose of AFB1 [24,108,109]. The primary repair pathway that can remove the AFB1-FAPY adducts is NER [109,110]. Also in support of this repair modality is the observed strong mutational strand bias present in transcribed genomic regions (in signature 24 [11,62] and in the AFB1-induced spectra in cells and animals [67]), which is characteristic of a lesion processed by TC-NER [51]. Base excision repair (BER) likely complements NER for repair of AFB1 lesions, as evidenced in a recent study showing the involvement of NEIL1 endonuclease in modulating the toxic and mutagenic effects of AFB1 exposure; these recent studies suggest that NEIL1 and BER play a role in the repair of AFB1-FAPY adducts [111].
d. Contribution of lesion mutagenic bypass to AFB1 mutational spectrum
The identity of the polymerase that introduces the adenine opposite the AFB1 adducts, and thus responsible for the mutagenic effects of the toxin varies with the type of adduct. The AFB1-N7-Gua (Fig. 4) adduct is replicated in an error-free manner by both the mammalian replicative polymerase pol delta and the mismatch extension polymerase pol zeta [91]. The characteristic mutations induced by the AFB1-N7-Gua adduct (~86% G→T, ~12% G→A) manifest primarily when replicated by TLS polymerases, such as pol kappa and to a lesser extent pol eta [91]. This result is consistent with earlier work in which the site specific AFB1 adduct was replicated in SOS-induced bacteria [88]; however, the lower mutation frequency observed in bacteria suggests that bacterial TLS polymerases are more efficient at error-free bypass of the adduct than mammalian ones. The involvement of bypass polymerases is also consistent with the geometry of the mispairing, which requires the adducted guanine to rotate to a syn conformation [112]. Naturally, Y-family polymerases featuring larger active sites can better accommodate such molecular gymnastics.
The other, much longer-lived AFB1 adduct, AFB1-FAPY I, is treated quite differently by the replication machinery. Although both adducts intercalate in the DNA on the 5′ face of the modified guanine [96,97], and they both induce predominantly G→T mutations (AFB1-FAPY I generates ~90% G→T, ~8% G→A), the responsible polymerases seem to be different. Unlike the AFB1-N7-Gua adduct, the AFB1-FAPY I is a replication block for replicative polymerases (such as pol delta) [92], and thus it requires TLS. However, Y-family polymerases, pol kappa, eta and iota, were shown to be inefficient at replicating past the FAPY adduct [92], although in vitro, pol kappa was able to catalyze the mutagenic mispairing with A. Instead, the main polymerase responsible for the mutagenic bypass of the AFB1-FAPY I was found to be the B family polymerase pol zeta [92]. Pol zeta introduced both the incorrect A opposite the lesion and extended several bases from the mismatch [92]. Such involvement of pol zeta in TLS has been observed for other lesions. The bulky adenine adduct generate by AA also requires pol zeta for mutagenic bypass, while the Y-family polymerases have only a very minor contribution [56].
6. Perspective and Conclusion
Mutational spectra of mutagens are mechanistically informative biomarkers of exposure, and accordingly have powerful medical applications. Understanding the biochemical and genetic mechanisms underlying a mutagenic event can lead to disease prevention and early detection. In this vein, the appearance of an early onset biomarker mutation associated with AFB1 carcinogenesis, one that appears long before the tumor becomes clinically relevant [61,66], could give an opportunity for surgical intervention at a time at which tumors are treatable. Hence, mechanistic studies on the factors that mold mutational spectra uncover fingerprints that can eventually be used for early detection of tumors caused by specific genotoxic agents. Such medically relevant early warning systems will arise from further study of the sequence-dependent contributions of formation, repair and replication of DNA damage.
Acknowledgments
The authors would like to thank Lina Kim for help with plotting the normalized mutational spectra in Figure 3. This work was supported by the following National Institutes of Health grants: P01-CA26731, R01-CA080024, R01-ES016313, and P30-ES002109.
ABBREVIATIONS
- 5mC
5-methylcytosine
- AA
aristolochic acid
- AFB1
aflatoxin B1
- AFB1-N7-Gua
AFB1 N7-guanine adduct
- A-10
mouse mutational spectrum at 10 weeks following exposure to AFB1
- A-72H
mouse mutational spectrum in the hepatocytes at 72 weeks following exposure to AFB1
- A-72T
mouse mutational spectrum in the liver tumor at 72 weeks following exposure to AFB1
- APOBEC
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like
- B[a]P
benzo[a]pyrene
- BER
base excision repair
- BRCA1
breast cancer 1 gene
- BRCA2
breast cancer 2 gene
- COSMIC
Catalogue of somatic mutations in cancer
- DupSeq
duplex sequencing
- ERCC2
excision repair cross-complementing protein 2
- FAPY
formamidepyrimidine DNA adduct
- HCC
hepatocellular carcinoma
- MBD4
methyl-CpG binding domain 4 DNA glycosylase
- MMR
mismatch repair
- MUTYH
MutY homolog glycosylase
- NEIL1
Nei-like endonuclease 1
- NER
nucleotide excision repair
- NTHL
Nth-like endonuclease 1
- O6mG
O6-methylguanine
- TC-NER
transcription-coupled NER
- TCGA
The Cancer Gene Atlas
- TDG
thymine DNA glycosylase
- TLS
trans-lesion synthesis
- UV
ultraviolet
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Padian K. Darwin’s enduring legacy. Nature. 2008;451:632–634. doi: 10.1038/451632a. [DOI] [PubMed] [Google Scholar]
- 2.Steele EJ. Somatic hypermutation in immunity and cancer: Critical analysis of strand-biased and codon-context mutation signatures. DNA Repair. 2016;45:1–24. doi: 10.1016/j.dnarep.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Petit V, Vartanian J-P, Wain-Hobson S. Powerful mutators lurking in the genome. Philos Trans R Soc Lond B Biol Sci. 2009;364:705–715. doi: 10.1098/rstb.2008.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Lenart P, Krejci L. DNA, the central molecule of aging. Mutat Res. 2016;786:1–7. doi: 10.1016/j.mrfmmm.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Pan L, Penney J, Tsai L-H. Chromatin regulation of DNA damage repair and genome integrity in the central nervous system. J Mol Biol. 2014;426:3376–3388. doi: 10.1016/j.jmb.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Detanico T, St Clair JB, Aviszus K, Kirchenbaum G, Guo W, Wysocki LJ. Somatic mutagenesis in autoimmunity. Autoimmunity. 2013;46:102–114. doi: 10.3109/08916934.2012.757597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen F, Bian K, Tang Q, Fedeles BI, Singh V, Humulock ZT, Essigmann JM, Li D. Oncometabolites d- and l-2-Hydroxyglutarate Inhibit the AlkB Family DNA Repair Enzymes under Physiological Conditions. Chem Res Toxicol. 2017;30:1102–1110. doi: 10.1021/acs.chemrestox.7b00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 10.Rudd SG, Valerie NCK, Helleday T. Pathways controlling dNTP pools to maintain genome stability. DNA Repair. 2016;44:193–204. doi: 10.1016/j.dnarep.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 11.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topal MD, Fresco JR. Complementary base pairing and the origin of substitution mutations. Nature. 1976;263:285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- 14.Wang W, Hellinga HW, Beese LS. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc Natl Acad Sci U S A. 2011;108:17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimsey IJ, Szymanski ES, Zahurancik WJ, Shakya A, Xue Y, Chu C-C, Sathyamoorthy B, Suo Z, Al-Hashimi HM. Dynamic basis for dG dT misincorporation via tautomerization and ionization. Nature. 2018;554:195–201. doi: 10.1038/nature25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otteneder M, Lutz WK. Correlation of DNA adduct levels with tumor incidence: carcinogenic potency of DNA adducts. Mutat Res. 1999;424:237–247. doi: 10.1016/s0027-5107(99)00022-6. [DOI] [PubMed] [Google Scholar]
- 17.Sjolund AB, Senejani AG, Sweasy JB. MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat Res. 2013;743–744:12–25. doi: 10.1016/j.mrfmmm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geacintov NE, Broyde S. Repair-Resistant DNA Lesions. Chem Res Toxicol. 2017;30:1517–1548. doi: 10.1021/acs.chemrestox.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Y, Geacintov NE, Broyde S. Nucleotide excision repair efficiencies of bulky carcinogen-DNA adducts are governed by a balance between stabilizing and destabilizing interactions. Biochemistry (Mosc) 2012;51:1486–1499. doi: 10.1021/bi201794x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopalakrishnan S, Byrd S, Stone MP, Harris TM. Carcinogen-nucleic acid interactions: equilibrium binding studies of aflatoxin B1 with the oligodeoxynucleotide d(ATGCAT)2 and with plasmid pBR322 support intercalative association with the B-DNA helix. Biochemistry (Mosc) 1989;28:726–734. doi: 10.1021/bi00428a047. [DOI] [PubMed] [Google Scholar]
- 21.Johnston DS, Stone MP. Refined solution structure of 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 opposite CpA in the complementary strand of an oligodeoxynucleotide duplex as determined by 1H NMR. Biochemistry (Mosc) 1995;34:14037–14050. doi: 10.1021/bi00043a009. [DOI] [PubMed] [Google Scholar]
- 22.Kathuria P, Sharma P, Abendong MN, Wetmore SD. Conformational preferences of DNA following damage by aristolochic acids: Structural and energetic insights into the different mutagenic potential of the ALI and ALII-N(6)-dA adducts. Biochemistry (Mosc) 2015;54:2414–2428. doi: 10.1021/bi501484m. [DOI] [PubMed] [Google Scholar]
- 23.Qu SX, Stacey NH. Formation and persistence of DNA adducts in different target tissues of rats after multiple administration of benzo[a]pyrene. Carcinogenesis. 1996;17:53–59. doi: 10.1093/carcin/17.1.53. [DOI] [PubMed] [Google Scholar]
- 24.Croy RG, Wogan GN. Temporal patterns of covalent DNA adducts in rat liver after single and multiple doses of aflatoxin B1. Cancer Res. 1981;41:197–203. [PubMed] [Google Scholar]
- 25.Schmeiser HH, Nortier JL, Singh R, Gamboa da Costa G, Sennesael J, Cassuto-Viguier E, Ambrosetti D, Rorive S, Pozdzik A, Phillips DH, Stiborova M, Arlt VM. Exceptionally long-term persistence of DNA adducts formed by carcinogenic aristolochic acid I in renal tissue from patients with aristolochic acid nephropathy. Int J Cancer. 2014;135:502–507. doi: 10.1002/ijc.28681. [DOI] [PubMed] [Google Scholar]
- 26.Benzer S. ON THE TOPOGRAPHY OF THE GENETIC FINE STRUCTURE. Proc Natl Acad Sci U S A. 1961;47:403–415. doi: 10.1073/pnas.47.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller JH. Mutational specificity in bacteria. Annu Rev Genet. 1983;17:215–238. doi: 10.1146/annurev.ge.17.120183.001243. [DOI] [PubMed] [Google Scholar]
- 28.Miller JH. Perspective on mutagenesis and repair: the standard model and alternate modes of mutagenesis. Crit Rev Biochem Mol Biol. 2005;40:155–179. doi: 10.1080/10409230590954153. [DOI] [PubMed] [Google Scholar]
- 29.Miller JH. Mutagenic specificity of ultraviolet light. J Mol Biol. 1985;182:45–65. doi: 10.1016/0022-2836(85)90026-9. [DOI] [PubMed] [Google Scholar]
- 30.Coulondre C, Miller JH. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977;117:577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- 31.Sargentini NJ, Smith KC. DNA sequence analysis of gamma-radiation (anoxic)-induced and spontaneous lacId mutations in Escherichia coli K-12. Mutat Res. 1994;309:147–163. doi: 10.1016/0027-5107(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 32.Cariello NF, Cui L, Skopek TR. In vitro mutational spectrum of aflatoxin B1 in the human hypoxanthine guanine phosphoribosyltransferase gene. Cancer Res. 1994;54:4436–4441. [PubMed] [Google Scholar]
- 33.Richardson KK, Richardson FC, Crosby RM, Swenberg JA, Skopek TR. DNA base changes and alkylation following in vivo exposure of Escherichia coli to N-methyl-N-nitrosourea or N-ethyl-N-nitrosourea. Proc Natl Acad Sci U S A. 1987;84:344–348. doi: 10.1073/pnas.84.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polak P, Karlić R, Koren A, Thurman R, Sandstrom R, Lawrence M, Reynolds A, Rynes E, Vlahoviček K, Stamatoyannopoulos JA, Sunyaev SR. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Sougnez C, Ambrogio L, Nickerson E, Shefler E, Cortés ML, Auclair D, Saksena G, Voet D, Noble M, DiCara D, Lin P, Lichtenstein L, Heiman DI, Fennell T, Imielinski M, Hernandez B, Hodis E, Baca S, Dulak AM, Lohr J, Landau D-A, Wu CJ, Melendez-Zajgla J, Hidalgo-Miranda A, Koren A, McCarroll SA, Mora J, Crompton B, Onofrio R, Parkin M, Winckler W, Ardlie K, Gabriel SB, Roberts CWM, Biegel JA, Stegmaier K, Bass AJ, Garraway LA, Meyerson M, Golub TR, Gordenin DA, Sunyaev S, Lander ES, Getz G. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perera D, Poulos RC, Shah A, Beck D, Pimanda JE, Wong JWH. Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature. 2016;532:259–263. doi: 10.1038/nature17437. [DOI] [PubMed] [Google Scholar]
- 37.Delaney JC, Essigmann JM. Effect of sequence context on O(6)-methylguanine repair and replication in vivo. Biochemistry (Mosc) 2001;40:14968–14975. doi: 10.1021/bi015578f. [DOI] [PubMed] [Google Scholar]
- 38.Singer B, Chavez F, Goodman MF, Essigmann JM, Dosanjh MK. Effect of 3′ flanking neighbors on kinetics of pairing of dCTP or dTTP opposite O6-methylguanine in a defined primed oligonucleotide when Escherichia coli DNA polymerase I is used. Proc Natl Acad Sci U S A. 1989;86:8271–8274. doi: 10.1073/pnas.86.21.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatahet Z, Zhou M, Reha-Krantz LJ, Morrical SW, Wallace SS. In search of a mutational hotspot. Proc Natl Acad Sci U S A. 1998;95:8556–8561. doi: 10.1073/pnas.95.15.8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibutani S, Suzuki N, Tan X, Johnson F, Grollman AP. Influence of flanking sequence context on the mutagenicity of acetylaminofluorene-derived DNA adducts in mammalian cells. Biochemistry (Mosc) 2001;40:3717–3722. doi: 10.1021/bi0027581. [DOI] [PubMed] [Google Scholar]
- 41.Suspène R, Aynaud M-M, Guétard D, Henry M, Eckhoff G, Marchio A, Pineau P, Dejean A, Vartanian J-P, Wain-Hobson S. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc Natl Acad Sci U S A. 2011;108:4858–4863. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. ELife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutsenko E, Bhagwat AS. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat Res. 1999;437:11–20. doi: 10.1016/s1383-5742(99)00065-4. [DOI] [PubMed] [Google Scholar]
- 44.Benasutti M, Ezzedine ZD, Loechler EL. Construction of an Escherichia coli vector containing the major DNA adduct of activated benzo[a]pyrene at a defined site. Chem Res Toxicol. 1988;1:160–168. doi: 10.1021/tx00003a006. [DOI] [PubMed] [Google Scholar]
- 45.Donigan KA, Sweasy JB. Sequence context-specific mutagenesis and base excision repair. Mol Carcinog. 2009;48:362–368. doi: 10.1002/mc.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.St Charles JA, Liberti SE, Williams JS, Lujan SA, Kunkel TA. Quantifying the contributions of base selectivity, proofreading and mismatch repair to nuclear DNA replication in Saccharomyces cerevisiae. DNA Repair. 2015;31:41–51. doi: 10.1016/j.dnarep.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunkel TA, Erie DA. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu Rev Genet. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adar S, Hu J, Lieb JD, Sancar A. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc Natl Acad Sci U S A. 2016;113:E2124–2133. doi: 10.1073/pnas.1603388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai Y, Patel DJ, Broyde S, Geacintov NE. Base sequence context effects on nucleotide excision repair. J Nucleic Acids. 2010;2010 doi: 10.4061/2010/174252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai Y, Kropachev K, Xu R, Tang Y, Kolbanovskii M, Kolbanovskii A, Amin S, Patel DJ, Broyde S, Geacintov NE. Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion. J Mol Biol. 2010;399:397–409. doi: 10.1016/j.jmb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jha V, Ling H. Structural basis of accurate replication beyond a bulky major benzo[a]pyrene adduct by human DNA polymerase kappa. DNA Repair. 2017;49:43–50. doi: 10.1016/j.dnarep.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 53.Klarer AC, Stallons LJ, Burke TJ, Skaggs RL, McGregor WG. DNA polymerase eta participates in the mutagenic bypass of adducts induced by benzo[a]pyrene diol epoxide in mammalian cells. PloS One. 2012;7:e39596. doi: 10.1371/journal.pone.0039596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cruet-Hennequart S, Gallagher K, Sokòl AM, Villalan S, Prendergast AM, Carty MP. DNA polymerase eta, a key protein in translesion synthesis in human cells. Subcell Biochem. 2010;50:189–209. doi: 10.1007/978-90-481-3471-7_10. [DOI] [PubMed] [Google Scholar]
- 55.Jansen JG, Temviriyanukul P, Wit N, Delbos F, Reynaud C-A, Jacobs H, de Wind N. Redundancy of mammalian Y family DNA polymerases in cellular responses to genomic DNA lesions induced by ultraviolet light. Nucleic Acids Res. 2014;42:11071–11082. doi: 10.1093/nar/gku779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hashimoto K, Bonala R, Johnson F, Grollman AP, Moriya M. Y-family DNA polymerase-independent gap-filling translesion synthesis across aristolochic acid-derived adenine adducts in mouse cells. DNA Repair. 2016;46:55–60. doi: 10.1016/j.dnarep.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganai RA, Johansson E. DNA Replication-A Matter of Fidelity. Mol Cell. 2016;62:745–755. doi: 10.1016/j.molcel.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Arana ME, Kunkel TA. Mutator phenotypes due to DNA replication infidelity. Semin Cancer Biol. 2010;20:304–311. doi: 10.1016/j.semcancer.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Imielinsk M, Jäger N, Jones DTW, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt ANJ, Valdés-Mas R, van Buuren MM, van’t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR Australian Pancreatic Cancer Genome Initiative, ICGC Breast Cancer Consortium, ICGC MMML-Seq Consortium, ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jönsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerød A, Tutt A, Martens JWM, Aparicio SAJR, Borg Å, Salomon AV, Thomas G, Børresen-Dale A-L, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chawanthayatham S, Valentine CC, Fedeles BI, Fox EJ, Loeb LA, Levine SS, Slocum SL, Wogan GN, Croy RG, Essigmann JM. Mutational spectra of aflatoxin B1in vivo establish biomarkers of exposure for human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2017;114:E3101–E3109. doi: 10.1073/pnas.1700759114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud A-L, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc J-F, Mazzaferro V, Calvo F, Villanueva A, Nault J-C, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feldmeyer N, Schmeiser HH, Muehlbauer K-R, Belharazem D, Knyazev Y, Nedelko T, Hollstein M. Further studies with a cell immortalization assay to investigate the mutation signature of aristolochic acid in human p53 sequences. Mutat Res. 2006;608:163–168. doi: 10.1016/j.mrgentox.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 64.Rosenquist TA, Grollman AP. Mutational signature of aristolochic acid: Clue to the recognition of a global disease. DNA Repair. 2016;44:205–211. doi: 10.1016/j.dnarep.2016.05.027. [DOI] [PubMed] [Google Scholar]
- 65.Nik-Zainal S, Kucab JE, Morganella S, Glodzik D, Alexandrov LB, Arlt VM, Weninger A, Hollstein M, Stratton MR, Phillips DH. The genome as a record of environmental exposure. Mutagenesis. 2015 doi: 10.1093/mutage/gev073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fedeles BI, Chawanthayatham S, Croy RG, Wogan GN, Essigmann JM. Early detection of the aflatoxin B1mutational fingerprint: A diagnostic tool for liver cancer. Mol Cell Oncol. 2017;4:e1329693. doi: 10.1080/23723556.2017.1329693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang MN, Yu W, Teoh WW, Ardin M, Jusakul A, Ng AWT, Boot A, Abedi-Ardekani B, Villar S, Myint SS, Othman R, Poon SL, Heguy A, Olivier M, Hollstein M, Tan P, Teh BT, Sabapathy K, Zavadil J, Rozen SG. Genome-scale mutational signatures of aflatoxin in cells, mice, and human tumors. Genome Res. 2017;27:1475–1486. doi: 10.1101/gr.220038.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shinbrot E, Henninger EE, Weinhold N, Covington KR, Göksenin AY, Schultz N, Chao H, Doddapaneni H, Muzny DM, Gibbs RA, Sander C, Pursell ZF, Wheeler DA. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24:1740–1750. doi: 10.1101/gr.174789.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pilati C, Shinde J, Alexandrov LB, Assié G, André T, Hélias-Rodzewicz Z, Ducoudray R, Le Corre D, Zucman-Rossi J, Emile J-F, Bertherat J, Letouzé E, Laurent-Puig P. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. J Pathol. 2017;242:10–15. doi: 10.1002/path.4880. [DOI] [PubMed] [Google Scholar]
- 70.Ohno M, Sakumi K, Fukumura R, Furuichi M, Iwasaki Y, Hokama M, Ikemura T, Tsuzuki T, Gondo Y, Nakabeppu Y. 8-oxoguanine causes spontaneous de novo germline mutations in mice. Sci Rep. 2014;4:4689. doi: 10.1038/srep04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Viel A, Bruselles A, Meccia E, Fornasarig M, Quaia M, Canzonieri V, Policicchio E, Urso ED, Agostini M, Genuardi M, Lucci-Cordisco E, Venesio T, Martayan A, Diodoro MG, Sanchez-Mete L, Stigliano V, Mazzei F, Grasso F, Giuliani A, Baiocchi M, Maestro R, Giannini G, Tartaglia M, Alexandrov LB, Bignami M. A Specific Mutational Signature Associated with DNA 8-Oxoguanine Persistence in MUTYH-defective Colorectal Cancer. EBioMedicine. 2017;20:39–49. doi: 10.1016/j.ebiom.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman JE, van Wezel T, Nik-Zainal S, Kuiper RP, Cuppen E, Clevers H. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017;358:234–238. doi: 10.1126/science.aao3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Kwiatkowski DJ, Rosenberg JE, Van Allen EM, D’Andrea A, Getz G. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48:600–606. doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu D, Abbosh P, Keliher D, Reardon B, Miao D, Mouw K, Weiner-Taylor A, Wankowicz S, Han G, Teo MY, Cipolla C, Kim J, Iyer G, Al-Ahmadie H, Dulaimi E, Chen DYT, Alpaugh RK, Hoffman-Censits J, Garraway LA, Getz G, Carter SL, Bellmunt J, Plimack ER, Rosenberg JE, Van Allen EM. Mutational patterns in chemotherapy resistant muscle-invasive bladder cancer. Nat Commun. 2017;8:2193. doi: 10.1038/s41467-017-02320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Letouzé E, Shinde J, Renault V, Couchy G, Blanc J-F, Tubacher E, Bayard Q, Bacq D, Meyer V, Semhoun J, Bioulac-Sage P, Prévôt S, Azoulay D, Paradis V, Imbeaud S, Deleuze J-F, Zucman-Rossi J. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun. 2017;8:1315. doi: 10.1038/s41467-017-01358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garaycoechea JI, Crossan GP, Langevin F, Mulderrig L, Louzada S, Yang F, Guilbaud G, Park N, Roerink S, Nik-Zainal S, Stratton MR, Patel KJ. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature. 2018;553:171–177. doi: 10.1038/nature25154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J-G, Egner PA, Ng D, Jacobson LP, Muñoz A, Zhu Y-R, Qian G-S, Wu F, Yuan J-M, Groopman JD, Kensler TW. Reduced aflatoxin exposure presages decline in liver cancer mortality in an endemic region of China. Cancer Prev Res Phila Pa. 2013;6:1038–1045. doi: 10.1158/1940-6207.CAPR-13-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kensler TW, Roebuck BD, Wogan GN, Groopman JD. Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicol Sci Off J Soc Toxicol. 2011;120(Suppl 1):S28–48. doi: 10.1093/toxsci/kfq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.El-Serag HB, Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: where are we? Where do we go? Hepatol Baltim Md. 2014;60:1767–1775. doi: 10.1002/hep.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 81.Kew MC. Aflatoxins as a cause of hepatocellular carcinoma. J Gastrointest Liver Dis JGLD. 2013;22:305–310. [PubMed] [Google Scholar]
- 82.Groopman JD, Kensler TW. Role of metabolism and viruses in aflatoxin-induced liver cancer. Toxicol Appl Pharmacol. 2005;206:131–137. doi: 10.1016/j.taap.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 83.Kew MC. Synergistic interaction between aflatoxin B1 and hepatitis B virus in hepatocarcinogenesis. Liver Int Off J Int Assoc Study Liver. 2003;23:405–409. doi: 10.1111/j.1478-3231.2003.00869.x. [DOI] [PubMed] [Google Scholar]
- 84.Woo LL, Egner PA, Belanger CL, Wattanawaraporn R, Trudel LJ, Croy RG, Groopman JD, Essigmann JM, Wogan GN, Bouhenguel JT. Aflatoxin B1-DNA adduct formation and mutagenicity in livers of neonatal male and female B6C3F1 mice. Toxicol Sci Off J Soc Toxicol. 2011;122:38–44. doi: 10.1093/toxsci/kfr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chawanthayatham S, Thiantanawat A, Egner PA, Groopman JD, Wogan GN, Croy RG, Essigmann JM. Prenatal exposure of mice to the human liver carcinogen aflatoxin B1 reveals a critical window of susceptibility to genetic change. Int J Cancer. 2015;136:1254–1262. doi: 10.1002/ijc.29102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sriwattanapong K, Slocum SL, Chawanthayatham S, Fedeles BI, Egner PA, Groopman JD, Satayavivad J, Croy RG, Essigmann JM. Pregnancy alters aflatoxin B1 metabolism and increases DNA damage in mouse liver. Toxicol Sci Off J Soc Toxicol. 2017 doi: 10.1093/toxsci/kfx171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015;31:3673–3675. doi: 10.1093/bioinformatics/btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bailey EA, Iyer RS, Stone MP, Harris TM, Essigmann JM. Mutational properties of the primary aflatoxin B1-DNA adduct. Proc Natl Acad Sci U S A. 1996;93:1535–1539. doi: 10.1073/pnas.93.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smela ME, Currier SS, Bailey EA, Essigmann JM. The chemistry and biology of aflatoxin B(1): from mutational spectrometry to carcinogenesis. Carcinogenesis. 2001;22:535–545. doi: 10.1093/carcin/22.4.535. [DOI] [PubMed] [Google Scholar]
- 90.Smela ME, Hamm ML, Henderson PT, Harris CM, Harris TM, Essigmann JM. The aflatoxin B(1) formamidopyrimidine adduct plays a major role in causing the types of mutations observed in human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2002;99:6655–6660. doi: 10.1073/pnas.102167699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin Y-C, Li L, Makarova AV, Burgers PM, Stone MP, Lloyd RS. Error-prone replication bypass of the primary aflatoxin B1 DNA adduct, AFB1-N7-Gua. J Biol Chem. 2014;289:18497–18506. doi: 10.1074/jbc.M114.561563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin Y-C, Li L, Makarova AV, Burgers PM, Stone MP, Lloyd RS. Molecular basis of aflatoxin-induced mutagenesis-role of the aflatoxin B1-formamidopyrimidine adduct. Carcinogenesis. 2014;35:1461–1468. doi: 10.1093/carcin/bgu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, Prindle MJ, Kuong KJ, Shen J-C, Risques R-A, Loeb LA. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat Protoc. 2014;9:2586–2606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018 doi: 10.1038/nrg.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gopalakrishnan S, Harris TM, Stone MP. Intercalation of aflatoxin B1 in two oligodeoxynucleotide adducts: comparative 1H NMR analysis of d(ATCAFBGAT).d(ATCGAT) and d(ATAFBGCAT)2. Biochemistry (Mosc) 1990;29:10438–10448. doi: 10.1021/bi00498a002. [DOI] [PubMed] [Google Scholar]
- 97.Mao H, Deng Z, Wang F, Harris TM, Stone MP. An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR. Biochemistry (Mosc) 1998;37:4374–4387. doi: 10.1021/bi9718292. [DOI] [PubMed] [Google Scholar]
- 98.Essigmann JM, Croy RG, Nadzan AM, Busby WF, Reinhold VN, Büchi G, Wogan GN. Structural identification of the major DNA adduct formed by aflatoxin B1 in vitro. Proc Natl Acad Sci U S A. 1977;74:1870–1874. doi: 10.1073/pnas.74.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Giri I, Jenkins MD, Schnetz-Boutaud NC, Stone MP. Structural refinement of the 8,9-dihydro-8-(N7-guanyl)-9-hydroxy-aflatoxin B(1) adduct in a 5′-Cp(AFB)G-3′ sequence. Chem Res Toxicol. 2002;15:638–647. doi: 10.1021/tx010187n. [DOI] [PubMed] [Google Scholar]
- 100.Giri I, Johnston DS, Stone MP. Mispairing of the 8,9-dihydro-8-(N7-guanyl)-9-hydroxy-aflatoxin B1 adduct with deoxyadenosine results in extrusion of the mismatched dA toward the major groove. Biochemistry (Mosc) 2002;41:5462–5472. doi: 10.1021/bi012116t. [DOI] [PubMed] [Google Scholar]
- 101.Muench KF, Misra RP, Humayun MZ. Sequence specificity in aflatoxin B1--DNA interactions. Proc Natl Acad Sci U S A. 1983;80:6–10. doi: 10.1073/pnas.80.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Refolo LM, Conley MP, Sambamurti K, Jacobsen JS, Humayun MZ. Sequence context effects in DNA replication blocks induced by aflatoxin B1. Proc Natl Acad Sci U S A. 1985;82:3096–3100. doi: 10.1073/pnas.82.10.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Denissenko MF, Cahill J, Koudriakova TB, Gerber N, Pfeifer GP. Quantitation and mapping of aflatoxin B1-induced DNA damage in genomic DNA using aflatoxin B1-8,9-epoxide and microsomal activation systems. Mutat Res. 1999;425:205–211. doi: 10.1016/s0027-5107(99)00038-x. [DOI] [PubMed] [Google Scholar]
- 104.Misra RP, Muench KF, Humayun MZ. Covalent and noncovalent interactions of aflatoxin with defined deoxyribonucleic acid sequences. Biochemistry (Mosc) 1983;22:3351–3359. doi: 10.1021/bi00283a008. [DOI] [PubMed] [Google Scholar]
- 105.Li L, Brown KL, Ma R, Stone MP. DNA Sequence Modulates Geometrical Isomerism of the trans-8,9- Dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)- 9-hydroxy Aflatoxin B1 Adduct. Chem Res Toxicol. 2015;28:225–237. doi: 10.1021/tx5003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benasutti M, Ejadi S, Whitlow MD, Loechler EL. Mapping the binding site of aflatoxin B1 in DNA: systematic analysis of the reactivity of aflatoxin B1 with guanines in different DNA sequences. Biochemistry (Mosc) 1988;27:472–481. doi: 10.1021/bi00401a068. [DOI] [PubMed] [Google Scholar]
- 107.Loechler EL, Teeter MM, Whitlow MD. Mapping the binding site of aflatoxin B1 in DNA: molecular modeling of the binding sites for the N(7)-guanine adduct of aflatoxin B1 in different DNA sequences. J Biomol Struct Dyn. 1988;5:1237–1257. doi: 10.1080/07391102.1988.10506467. [DOI] [PubMed] [Google Scholar]
- 108.Croy RG, Wogan GN. Quantitative comparison of covalent aflatoxin-DNA adducts formed in rat and mouse livers and kidneys. J Natl Cancer Inst. 1981;66:761–768. [PubMed] [Google Scholar]
- 109.Bedard LL, Massey TE. Aflatoxin B1-induced DNA damage and its repair. Cancer Lett. 2006;241:174–183. doi: 10.1016/j.canlet.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 110.Alekseyev YO, Hamm ML, Essigmann JM. Aflatoxin B1 formamidopyrimidine adducts are preferentially repaired by the nucleotide excision repair pathway in vivo. Carcinogenesis. 2004;25:1045–1051. doi: 10.1093/carcin/bgh098. [DOI] [PubMed] [Google Scholar]
- 111.Vartanian V, Minko IG, Chawanthayatham S, Egner PA, Lin Y-C, Earley LF, Makar R, Eng JR, Camp MT, Li L, Stone MP, Lasarev MR, Groopman JD, Croy RG, Essigmann JM, McCullough AK, Lloyd RS. NEIL1 protects against aflatoxin-induced hepatocellular carcinoma in mice. Proc Natl Acad Sci U S A. 2017;114:4207–4212. doi: 10.1073/pnas.1620932114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Banerjee S, Brown KL, Egli M, Stone MP. Bypass of aflatoxin B1 adducts by the Sulfolobus solfataricus DNA polymerase IV. J Am Chem Soc. 2011;133:12556–12568. doi: 10.1021/ja2015668. [DOI] [PMC free article] [PubMed] [Google Scholar]