Abstract
Background:
Circulating hematopoietic progenitors (HPCs) have been implicated in inflammatory diseases such as atherosclerosis, and in repair and regeneration of damaged tissue after injury. Recently published studies suggest the levels of blood borne HPCs in human subjects may represent a useful biomarker to predict future cardiovascular disease (CVD) events. These studies have indicated an age and CVD risk factor dependent relationship between HPC levels and future risk of CVD events. CD133 expression within the circulating CD34+ HPC pool has been used to identify a subpopulation of CD34+ cells enriched for progenitor cells. Our goal was to determine the distribution of CD133 expression within HPC sub-types amongst circulating CD34+ cells.
Methods:
The quantity of different HPC populations within the CD34+ CD133+ and CD34+ CD133− fractions of venous blood obtained from healthy human subjects was measured using multi-color flow cytometry.
Results:
The majority of circulating CD34+ cells are CD133−. CD133+ CD34+ cells express low levels of CD38, contain cell populations bearing cell surface markers of hematopoietic stem cells, multi potent progenitors, and multi lymphoid progenitors, and are largely devoid of CD38 expressing lineage specified progenitors. These findings clarify the composition of circulating CD133+ CD34+ cell types cited in epidemiological studies of CVD and other diseases.
Keywords: Hematopoietic stem cells, Flow cytometry
Introduction:
Cells expressing CD34 in human subjects contain multiple cell populations from hematopoietic stem cells (HSCs) to lineage specified progenitors of specific immune and blood cell families1. CD34+ cells have also been the focus of many studies for cell based therapy approaches to disease2, 3. The levels of circulating CD34+ cells in the blood stream correlate with the risk of mortality in patients with coronary artery disease4. In these patients, lower levels of circulating CD34+ cells are associated with as much as a three-fold increased risk of death from myocardial infarction. Measurement of CD133+ CD34+ cells improved risk prediction metrics slightly over simply measuring total circulating CD34+ cells4. However, CD34+ CD133− cells did not identify patients at risk for death. The identity of CD133+ CD34+ cells in the context of the hierarchy of hematopoietic progenitors is not well defined. We recently established a method to measure the levels of previously defined circulating hematopoietic progenitors based on the cell surface immunophenotype from less differentiated hematopoietic cell types including HSC (hematopoietic stem cells), MPP (multi potent progenitors), MLP (multi lymphoid progenitors) to the lineage specified progenitors including CMP (common myeloid progenitors), GMP (granulocyte macrophage progenitors), and MEP (megakaryocyte erythroid progenitors) using a multi-color flow cytometry based approach5. The goal of our study was to determine the contribution of CD133+ cells to the previously defined set of hematopoietic progenitors defined by standard cell surface markers5.
Methods:
Patient Consent for Participation:
Our research protocol was conducted at the University at Buffalo Clinical and Translation Research Center (Protocol). This protocol was reviewed and approved by the University at Buffalo Intramural Review Board.
Informed consent was obtained from study participants prior to entry, and conducted according to the principles expressed in the Declaration of Helsinki. Subjects were recruited as healthy control subjects. All samples were anonymized. The investigators were blinded to the clinical and demographic background of the donors.
Study Protocol:
Venous blood samples (8 mL) were collected in sodium heparin at the time of recruitment for measurement.
Preparation of mononuclear cells and antibody staining:
Cell preparation and staining was performed as previously described with minor modifications5. Mononuclear cells (MNCs) were purified by Histopaque 1077 (Sigma-Aldrich) gradient centrifugation. The MNC fraction was washed by centrifugation and resuspension twice in HBSS at room temperature to remove residual platelets. The cell concentration in each sample was then enumerated using an automated cell counter (Beckman Coulter AcT diff hematology analyzer). Five million MNCs were stained for 20 minutes with a cocktail containing saturating amounts of the following mAbs: CD45RA-FITC (Clone ALB11; Beckman Coulter), CD123-PE-Cy7 (Clone 9F5; BD Biosciences), CD38-PerCP-Cy5.5 (Clone HIT2; BD Biosciences), CD34-APC (Clone BIRMA-K3; Dako), CD90-BV421 (Clone 5E10; BD Biosciences), and CD133−PE (Clone AC133; Miltenyi Biotec). Live Dead Aqua (Thermo Fisher) was used to enumerate and exclude dead cells from the analysis. After incubation with mAbs the cells were washed once with PBS containing 0.5% bovine serum albumin, 0.1% sodium azide and 0.0004% tetrasodium EDTA.
Flow Cytometry Analysis:
Analysis was performed as previously described with minor modifications5. Flow cytometric acquisition was performed on an LSR Fortessa (BD Biosciences) equipped with three laser excitation sources (405 nm 50 mw; 488 nm 50 mw; 640 nm 40 mw) that was quality-controlled on a daily basis using CS&T beads and FACS DiVA software (BD Biosciences). The filter configurations for the PMTs measuring fluorescence emission of the applied fluorochromes were 450/50 nm (BV421); 525/50 nm (Live Dead Aqua); 530/30 nm (FITC); 582/15 nm (PE); 780/60 nm (PE-Cy7); 660/20 nm (APC); 694/50 nm (PerCP-Cy-5.5). Autofluorescence and single-color controls were acquired to perform spectral overlap compensation using the automated compensation matrix feature in FACS DiVA software. Fluorescence minus one controls were used to establish gates and separating positive from negative population. Flow cytometry data was plotted using bi-exponential plots that include axes less than zero to assure all data was visible and properly compensated. Data analysis was performed with FlowJo, software Version vX.0.7 (FlowJo LLC, Ashland, OR).
Flow Cytometry Data Analysis Details:
Gates defining specific populations of HSC and HPSCs were established as described previously6. More recently defined cell populations were included in this analysis for dendritic precursor cells7, and multi lymphoid progenitors8.
Analysis was performed using FlowJo Software using the following steps. 1) Dead cells were excluded using LD Aqua. 2) Doublets were excluded by gating outliers on SSC-A vs. SSC-H and FSC-A vs. FSC-H plots. 3) Total CD34+ cells were identified from the CD34 CD38 dot plot (Figure 1A), and segregated into CD133+ and negative populations for sub population analysis (Figure 1B). 4) CD38 PerCP-Cy5.5 versus CD34 APC plotted as a density dot plot. Total CD34+ cells were divided into CD34+ CD38+/dim and CD34dim CD38+ populations (Figure 3B and 3C). 4) Common myeloid progenitors (CMP), granulocyte macrophage progenitors (GMP), macrophage erythrocyte progenitors (MEP), and dendritic cell progenitors (DCP) populations were defined using a CD45RA versus CD123 contour plot gated CD34dim CD38+ population (Figure 3D and 3E). The CD123+ and CD45RA+ regions were set using fluorescence minus one plots with a 2% contour levels. The CD34dim CD38+ CD123− CD45RA-population is defined as megakaryocyte erythroid progenitor (MEPs); CD34dim CD38+ CD123+ CD45RA-cells are defined as common myeloid progenitors (CMPs); CD34dim CD38+ CD123+ CD45RA+ cells are granulocyte monocyte precursors (GMPs). CD34dim CD38+ CD123 bright+ CD45RA+ cells are dendritic cell precursors (DCPs)7. 5) MPP and HSC were defined using a CD90 BV421 versus CD45RA FITC dot plot gated on the CD34+ CD38 dim (Figure 4A and 4B). The CD34+ CD38dim CD90+ CD45RA− and CD34+ CD38− CD90− CD45RA-populations were selected for measurement of HSCs and MPPs, respectively. 6) MLPs were defined as the CD90 positive and CD45RA positive population based on previously reported criteria (Figure 3)8.
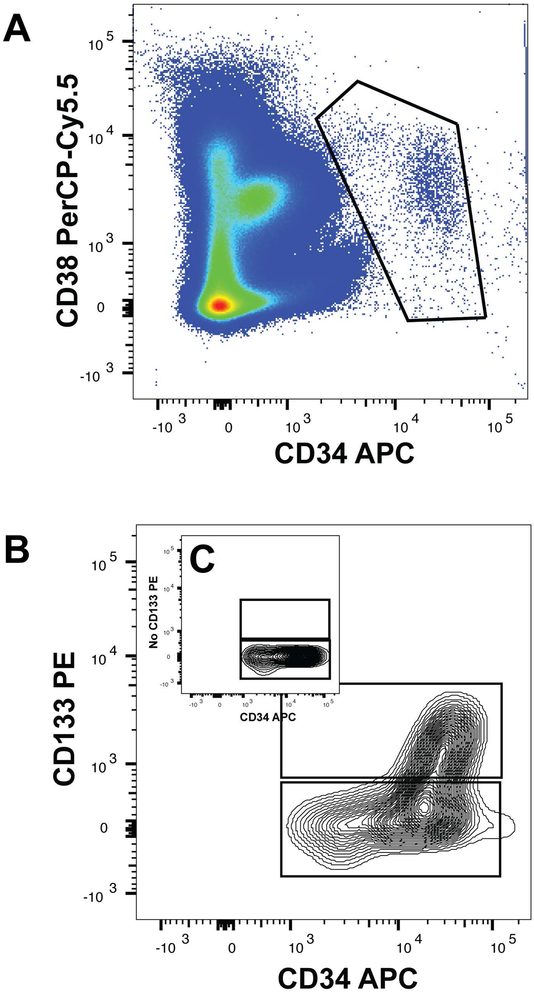
Figure 1: Heterogeneous Expression of CD133 in Circulating CD34+ Cells.
Panel A is a representative flow cytometry dot plot of CD34 and CD38 co-expressing cells. The total population of CD34+ CD38+/dim cells (identified in the figure by the octagonal box) were selected for analysis of CD133 expression. Panel B is a representative subplot using the contour presentation of CD133 and CD34 expression within the total circulating pool of CD34 and CD38+ cells. Panel C shown as an inset is a representative example of fluorescence minus one staining excluding CD133 to define the CD133+ threshold for population selection.
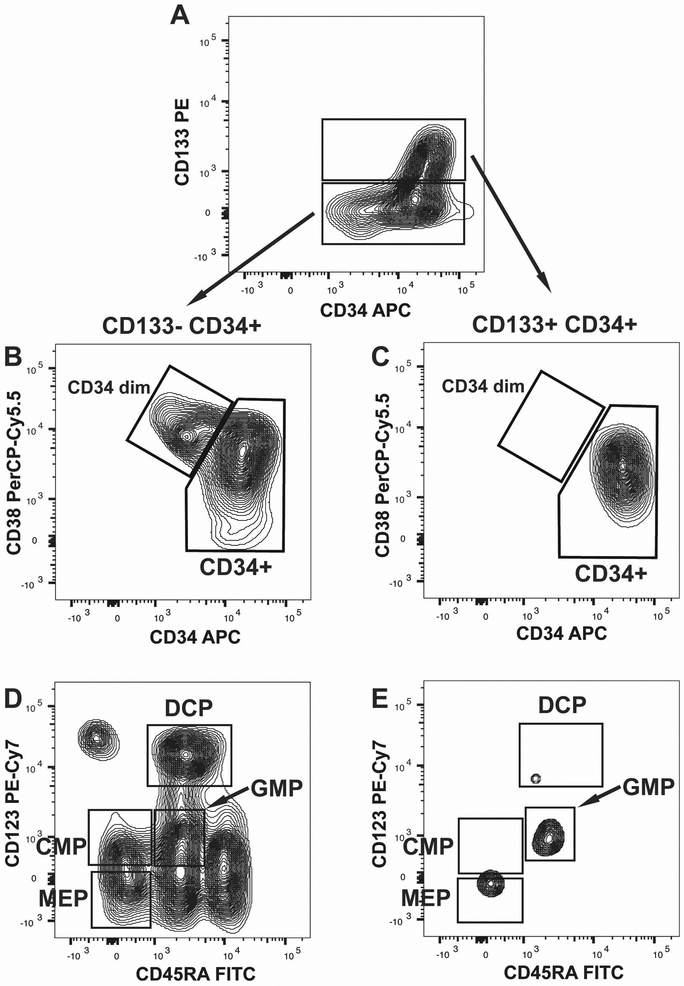
Figure 3: Circulating CD133+ CD34+ Cells Lack Significant Levels of Lineage Specified Hematopoietic Progenitor Populations:
Panel A is a representative flow cytometry contour plot of circulating CD133 and CD34+ cells. The populations were selected using the fluorescence minus CD133 staining. Panels B and C are representative flow cytometry contour plots of the CD133− CD34+ population (Panel B), and CD133+ CD34+ population (Panel C) shown in Panel A. The boxes delineate CD34 dim CD38+ and CD34+ CD38+/dim cell types observed. Panels D and E are representative flow cytometry contour plots of the CD34dim CD38+ populations from CD133− CD34+ (Panel D) and CD133+ CD34+ (Panel E) cell populations. The progenitor cell populations defined by CD123 and CD45RA expression are indicated by the boxes shown including: MEP-megakaryocyte erythroid progenitor, CMP-common myeloid progenitor, GMP-granulocyte macrophage progenitor, DCP-dendritic cell progenitor.
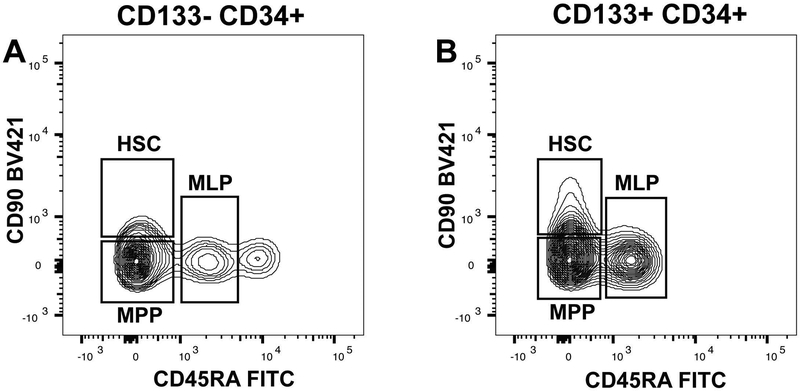
Figure 4: Circulating Hematopoietic Stem Cells, Multipotent Progenitors and Multi Lymphoid Progenitors Are Present in Both CD133− and CD133+ CD34+ Populations:
Panels A and B are representative flow cytometry contour sub-plots of circulating progenitor cell populations. CD133− CD34+ and CD133+ CD34+ cell populations were selected to exclude the CD34dim CD38+ populations (Figure 3 Panels B and C) were sub-plotted to examine for CD90 and CD45RA co-expression. Populations of hematopoietic stem cells (HSC), multi-potent progenitors (MPP), and multi lymphoid progenitors (MLP) are indicated based on their established criteria.
Statistical Analysis:
The distribution of the experimental data was determined using the Shapiro-Wilk normality test. Significant differences in the quantity of each hematopoietic progenitor cell population was determined between CD133+ CD34+ and CD133− CD34+ groups using the Mann-Whitney U test for non-normally distributed data. Statistical analysis and box-plots were created using Stata 14.2 software (Stata Corp, College Station, Texas USA). The mean values, standard error of the mean (SEM) and 95% confidence intervals for each cell population are provided in Table 1.
Table 1.
Quantity of CD133 Negative and positive cells as a percentage of total CD34+ cells
| CD133− | CD133+ | ||||||
|---|---|---|---|---|---|---|---|
| Cell Population |
Mean | SEM | 95% CI | Mean | SEM | 95% CI | p-value |
| Total CD34+ | 68.8 | 2.9 | 61 to 76 | 28.4 | 2.4 | 22 to 35 | 0.0005* |
| CD34+ CD38+ | 13.5 | 1.8 | 5 to 9 | 0.49 | 0.1 | 0.2 to 0.8 | 0.004* |
| CD34+ CD38− | 58.5 | 4.4 | 47 to 69 | 27.8 | 2.4 | 22 to 34 | 0.0001* |
| CMP | 2.7 | 0.5 | 1.3 to 4 | 0.15 | 0.05 | 0.01 to 0.29 | 0.004* |
| MEP | 2 | 0.3 | 1.3 to 2.7 | 0.11 | 0.02 | 0.06 to 0.16 | 0.004* |
| GMP | 0.9 | 0.4 | −0.13 to 1.8 | 0.07 | 0.018 | 0.03 to 0.12 | 0.007* |
| DCP | 0.83 | 0.6 | −0.24 to 1.1 | 0 | 0 | 0 | 0.003* |
| HSC | 10.7 | 2 | 5.4 to 16 | 10.1 | 2 | 4.9 to 15.4 | 0.48* |
| MPP | 35.8 | 2.2 | 30 to 41 | 13.6 | 1.7 | 9.3 to 18 | 0.0001* |
| MLP | 6.3 | 1.3 | 3 to 9.5 | 2.1 | 0.8 | 0 to 4.2 | 0.01* |
Mann-Whitney U Test
Results:
Our goal was to determine the distribution of CD133 expression within HPC sub-types amongst circulating CD34+ cells. We tested the blood of six healthy human subjects. After exclusion of dead cells and doublets, the total CD34+ CD38 pool was selected from the CD34-APC and CD38 PerCP-Cy 5.5 plot as shown in Figure 1A. This total CD34+ population was then separated into CD133+ CD34+ and CD133− CD34+ cell populations (Figure 1B) for analysis of the content of hematopoietic progenitor populations as previously defined5. Fluorescence minus CD133−PE was used to define the threshold for selection of the CD133+ cell population (Figure 1C inset within Figure 1B). We found that 0.04 ± 0.01 percent of total mononuclear (MNCs) cells were CD133+ and 0.09 ± 0.01 percent of total MNCs were CD133−. As a percentage of the total CD34+ fraction, CD133+ cells represent 28.4 ± 2.4 %, while CD133− cells were 68.8 ± 1.3 % of the total circulating CD34+ cells in the blood of healthy human subjects (p-value < 0.0005-Figure 2 Panel A). We conclude that CD133+ CD34+ cells are in the minority within the total CD34+ pool, and the difference in CD133 expression is statistically significant (Tables 1 and 2).
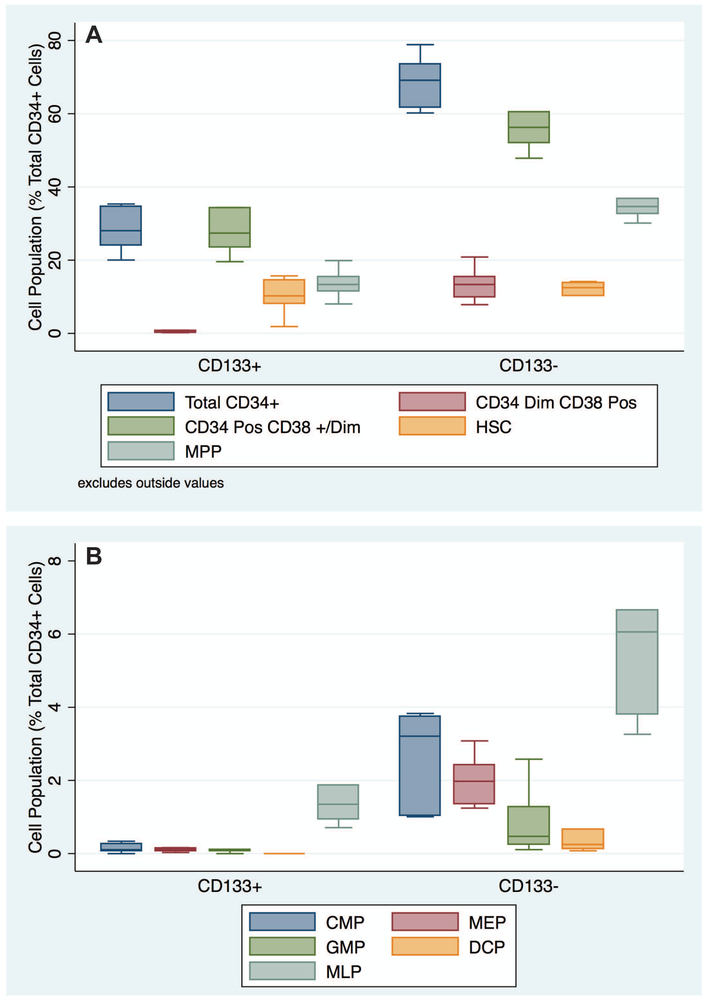
Figure 2: Quantity of Circulating Hematopoietic Progenitors in CD133− and CD133+ Cell Populations:
Panels A and B are box plot illustrations of hematopoietic progenitor populations presented as a percentage of the total circulating CD34+ cells. The populations are grouped based on the presence or absence of CD133 expression to compare their relative abundance in the total CD34+ pool. The horizontal bars in the boxes indicate the median values, and the boundaries of the boxes indicate 25th and 75th percentile ranges. Data from six healthy adult human subjects are presented.
Prior studies have indicated that CD133 expression within CD34+ cells enrich hematopoietic stem cells within the total CD34+ compartment. In the context of the hierarchy of stem cells to lineage specified progenitors, we tested to what extent the CD133+ CD34+ versus CD133− CD34+ cell fractions contributed to each previously described cell type. In our cohort of healthy human subjects, the CD133+ CD34+ cell population that co-expressed CD38 represents 0.49 ± 0.1 % of the total CD34+ cells. In contrast, the CD133− CD34 dim cell population that co-expresses CD38 represents a larger proportion 13.5 ± 1.8 % (p < 0.005) of the total CD34+ cell pool (Figure 2 Panel A). When visualized in three dimensions (CD34, CD133 and CD38) CD133 expressing cells are clearly positive for CD34 expression and are heterogeneously dim positive for CD38 (Supplemental Movie 1). While CD133 expression appears to exclude the lineage specified progenitor population co-expressing CD38, CD133 expression does not appear to enrich for the CD34+ CD38 +/dim population of hematopoietic progenitor cells. The CD133+ population of CD34+ CD38dim cells represents 27.8 ± 0.8 % of the total CD34+ cell pool while the CD133− population of CD34+ CD38dim cells accounts for 58.5 ± 4.4 % (p-value < 0.0002) of the total CD34+ pool (Figure 2 Panel A).
CD133+ CD34+ and CD133− CD34+ cell fractions were then analyzed to identify the quantity of CD34+ CD38+ cells making up the lineage specified progenitor fraction (Figure 3 Panels B and C). We observed that CD133+ CD34+ cells were virtually devoid of CD38 expression (Figure 3 Panel B), with insignificant levels of lineage specified progenitors (Figure 3 Panel E). In contrast, CD133− CD34+ cells contained the vast majority of CD34+ CD38+ lineage specified progenitors in circulating blood (Figure 3 Panel B). Based on these findings the addition of CD133 to the hematopoietic progenitor panel effectively distinguishes CD38 co-expressing lineage specified progenitor cells.
We next determined how CD133 expression within the total CD34+ cell pool partitions the quantity of cells bearing a cell surface phenotype consistent with hematopoietic progenitors including hematopoietic stem cells (HSC), multi potent progenitors (MPP) and multi lymphoid progenitors (MLP). Representative flow cytometry plots of CD133+ CD34+ and CD133− CD34+ cell populations are shown in Figure 4 Panels A and B. The populations shown are the CD34+ CD38− cell population further analyzed for CD90 and CD45RA expression which distinguishes these three cell types. The quantity of HSCs was not significantly different between CD133+ CD34+ and CD133− CD34+ cell populations (10.1 ± 2 % versus 10.7 ± 2 % of total CD34+ cells, p-value 0.48). The lack of CD133 expression amongst CD34+ cells did enrich for MPPs (13.6 ± 1.7 % of total CD34+ cells for CD133+ versus 35.8 ± 2.2 % of total CD34+ cells for the CD133− component (p-value < 0.0001). Multi lymphoid progenitors (MLP) were found in both CD133+ and CD133− cells, but were present in greater quantity in CD133− cells (6.3 ± 1.3 % of total CD34+ cells for CD133− versus 2.1 ± 0.8 % of total CD34+ cells for CD133+, p-value < 0.01).
We noted higher fluorescence intensity in CD90 within the HSC cell population when compared between CD133+ and CD133− cells (Figure 4 Panels A and B). The mean fluorescence intensity of CD90 in the HSC population from CD133− cells was 494.8 ± 27.2 while mean fluorescence intensity of CD90 from CD133+ cells was 708.2 ± 37.3, indicating significantly brighter CD90 expression in the HSC fraction from CD133+ cells (p-value < 0.0005, t-test). In conclusion, circulating CD133 expressing CD34+ cells represent roughly one-third of the total circulating CD34+ cell pool, carry equivalent levels of HSCs, significantly fewer MPPs and MLPs than CD133− CD34+ cells, but are depleted of the lineage specified progenitors, CMPs, MEPs, GMPs and DCPs.
Discussion:
We performed this study to understand the contribution of CD133 expressing cells to the pool of circulating hematopoietic progenitor cells amongst CD34+ cells, and in the hierarchy of hematopoietic progenitors commonly used by hematopoietic stem cell biologists. Prior reports indicate that CD133 expression amongst CD34+ hematopoietic cells enrich for the stem cell fraction of circulating cells. We found that minor levels of lineage specified progenitors co-express CD133. However, HSCs appear to be roughly split between CD133+ and CD133− expressing cells, and the majority of cells closely related to the HSC, MPPs and MLPs are also split between CD133+ and CD133− cells, but are found predominantly amongst the CD133− population. Therefore, the concept that CD133+ cells being more enriched for more primitive forms of hematopoietic progenitors is partially correct. Our study is limited in that we have only quantitatively determined differences between CD133+ and CD133− pools within circulating CD34+ hematopoietic cells. Clearly a test of the functional significance of CD133 expression amongst these cells is needed to understand the biological role CD133 expression plays. A head to head comparison of CD133+ and CD133− human hematopoietic cells in bone marrow transplant has not been performed to our knowledge.
In the context of cardiovascular disease, CD133 expressing cells have been associated with regeneration of vascular tissue9 and are therefore believed to have beneficial effects in the context of atherosclerotic vascular disease. Large observational studies have shown increasing age alone does not appear to have significant effects on the levels of either total CD34+ cells or CD34+ CD133+ cell types in the absence of heart disease risk factors. However, with each additional heart disease risk factor (including hypertension, diabetes mellitus, or high serum cholesterol levels), or diagnosed heart vessel disease, there is a reduction in the quantity of total CD34+ cells, and CD34+ CD133+ cells10. Other studies evaluated the impact of quantitative changes in circulating CD133+ CD34+ cells in the blood stream of patients with heart disease undergoing study for heart artery disease. In subjects with coronary artery disease, lower levels of either total circulating CD34+ cells, or CD133+ CD34+ cells have a higher risk of death from heart disease, and higher risk of future MI11. Interestingly, the population of circulating CD133−CD34+ cells was predictive of poor outcomes, however details regarding how this cell population is defined was not well documented11. Circulating cell levels are clearly influenced by glucose levels, as the presence of glucose intolerance in obese patients, but not frank diabetes mellitus was also associated with reduced levels of circulating total CD34+ cells, and CD133+ CD34+ cell types12. The levels of circulating total CD34+ cells, and CD133+ CD34+ cells also predict future atherosclerotic disease events (cardiovascular death, non-fatal myocardial infarction, non-fatal stroke), in patients with diabetes mellitus13. Collectively, measures of circulating total CD34+ and CD133+ CD34+ cell types appear to have clear predictive power to identify subjects at risk for future heart disease events. Several factors are at play to cause reduced levels of circulating CD34+, however in the early phase of disease, stimulation of the hematopoietic system by high levels of cholesterol clearly results in elevated levels of hematopoietic cells14,15. Over time, stem cell proliferation becomes limited, resulting in the decreased cell levels observed.
The expression of CD133 is limited to a subset of circulating hematopoietic cells, and the functional significance of CD133 expression has remained unclear. Recent mechanistic experiments have illustrated that CD133 is actually released from hematopoietic cells upon differentiation in the form of microvesicles that are transferred to neighboring cells16. The contents of the CD133 containing microvesicles remains to be determined. In the context of epidemiologic cardiovascular disease studies, the loss of CD133 expressing cells with progression of disease again appears to be tied to proliferation and differentiation of CD133+ hematopoietic progenitors, where inflammatory factors such as elevated cholesterol levels promote proliferation.
In conclusion, our study determined the contribution of CD133+ to the total CD34+ cell pool in the context of the hierarchy of human hematopoietic progenitors. Overall, CD133 expression is largely excluded from lineage specified progenitors, and observed predominantly in cells bearing the cell surface markers of HSCs and MPPs. Future studies may focus on how CD133 expression within HSCs and MPPs alters their function, and how they contribute to the pathogenesis of heart disease or its repair.
Supplementary Material
Acknowledgements:
Study funding provided by the University at Buffalo Clinical and Translational Science Institute, Translational Pilot Studies Fund (NIH UL1 TR001412), and The Mae Stone Goode Trust awarded to TC. Flow cytometry was performed at Roswell Park Cancer
Institute’s Flow & Image Cytometry Resource, which was established in part by equipment grants from the NIH Shared Instrument Program, and receives support from a Cancer Center Support Grant (5 P30 CA016056–29) from the National Cancer Institute to the Roswell Park Cancer Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have no conflicts of interest with the material presented.
Literature Cited:
- 1.Chao MP, Seita J, Weissman IL. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol 2008; 73: 439–449. [DOI] [PubMed] [Google Scholar]
- 2.Quyyumi AA, Vasquez A, Kerjakes DJ, Klapolz M, Schaer GL, Abdel-Latif A, Frohwein S, Henry TD, Schatz RA, Dib N, Toma C, Davidson CJ, Barsness GW, Shavelle DM, Cohen M, Poole J, Moss T, Hyde P, Kanakaraj AM, Druker V, Chung A, Junge C, Preti RA, Smith RL, Mazzo DJ, Pecora A, Losordo DW. PreSERVE-AMI: A randomized, double-blind, placebo-controlled clinical trial of intracoronary administration of autologous CD34+ cells in patients with left ventricular dysfunction post STEMI. Circ Res 2017; 120: 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu M, Li S, Menon S, Liu B, Hu MS, Longaker MT, Lorenz HP. Expansion and hepatic differentiation of adult blood-derived CD34+ progenitor cells and promotion of liver regeneration after acute injury. Stem Cells Transl Med 2016; 5: 723–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel RS, Li Q, Ghasemzadeh N, Eapen DJ, Moss LD, Janjua AU, Manocha P, Al Kassem H, Veledar E, Samady H, Taylor WR, Zafari AM, Sperling L, Vaccarino V, Waller EK, Quyyumi AA. Circulating CD34+ progenitor cells and risk of mortality in a population with coronary artery disease. Circ Res 2015; 116: 289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimato TR, Furlage RL. Conway A, Wallace PK. Simultaneous measurement of human hematopoietic stem and progenitor cells in blood using multicolor flow cytometry. Cytometry B Clin Cytom 2016; 90: 415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pang WW, Price EA, Sahoo D, Beerman I, Maloney WJ, Rossi DJ, Schrier SL, Weissman IL. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci USA 2011; 108: 20012–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breton G, Lee J, Zhou YJ, Schreiber JJ, Keler T, Puhr S, Anandasabapathy N, Schesinger S, Caskey M, Liu K, Nussenzweig MC. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J Exp Med 2015; 212: 401–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol 2010; 11: 585–93. [DOI] [PubMed] [Google Scholar]
- 9.Gehling UM, Ergun S, Schumacher U, Wagener C, Pantel K, Otte M, Schuch G, Schafhausen P, Mende T, Kilic N, Kluge K, Schafer B, Hossfeld DK, Fiedler W. In vitro differentiation of endothelial cells from AC133-positive progenitor cells. Blood 2000; 95: 3106–12. [PubMed] [Google Scholar]
- 10.Al Mheid I, Hayek SS, Ko Y-A, Akbik F, Li Q, Ghasemzadeh N, Martin GS, Long Q, Hammadah M, Zafari AM, Vaccarino V, Waller EK, Quyyumi AA. Age and human regenerative capacity: impact of cardiovascular risk factors. Circ Res 2016; 119: 801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel RS, Li Q, Ghasemzadeh N, Eapen DJ, Moss LD, Janjua AU, Manocha P, Al Kassem H, Veledar E, Samady H, Taylor WR, Zafari AM, Sperling L, Vaccarino V, Waller EK, Quyyumi AA. Circulating CD34+ progenitor cells and risk of mortality in a population with coronary artery disease. Circ Res 2015; 116: 289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Povsic TJ, Sloane R, Green J, Zhou J, Peiper CF, Pearson MP, Peterson ED, Cohen HJ, Morey MC. Depletion of circulating progenitor cells precedes overt diabetes: a substudy from the VA enhanced fitness trial. J Diabetes Complications 2013; 27: 633–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fadini GP, Rigato M, Cappellari R, Bonora BM, Avogaro A. Long-term prediction of cardiovascular outcomes by circulating CD34+ and CD34+ CD133+ stem cells in patients with type 2 diabetes. Diabetes Care 2017; 40: 125–31. [DOI] [PubMed] [Google Scholar]
- 14.Lang JK, Cimato TR. Cholesterol and hematopoietic stem cells: inflammatory mediators of atherosclerosis. Stem Cells Transl Med 2014; 3: 549–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cimato TR. Persistent stem cell-driven inflammation in patients with prior MI and stroke. Eur Heart J 2017; 38: 433–5. [DOI] [PubMed] [Google Scholar]
- 16.Bauer N, Wilsch-Brauninger M, Karbanova J, Fonseca A-V, Strauss D, Freund D, Thiele C, Huttner WB, Bornhauser M, Corbeil D. Haematopoietic stem cell differentiation promotes the release of prominin-1/CD133−containing membrane vesicles-a role of the endocytic-exocytic pathway. EMBO Mol Med 2011; 3: 398–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.