Abstract
Background
Cardiovascular (CV) diseases are major causes of mortality in uremic patients. Conventional risk factors fail to identify uremic patients with increased propensity for adverse CV outcomes. We aimed to test the hypothesis that circulating long noncoding RNAs (lncRNAs) could be a prognostic marker to predict adverse CV outcomes in uremic patients.
Methods and Results
Plasma lncRNAs were profiled in patients with end-stage renal disease (ESRD, n=28) or chronic kidney disease (CKD, n=8) and in healthy (n=12) subjects by RNA sequencing. A total of 179 lncRNAs were significantly dysregulated with ESRD; the expression signature of plasma lncRNAs distinguished ESRD from both CKD and control samples. Analysis on a microarray dataset obtained from renal biopsy samples of patients with advanced kidney disease (GSE66494) revealed that a significant proportion of plasma lncRNAs (30.7%) and mRNAs (49.5%) dysregulated with uremia were similarly dysregulated in diseased kidneys, suggesting that plasma RNA profiles mirror the transcriptomal changes in diseased kidney tissues. Further analyses identified eight plasma lncRNAs as potential predictors of adverse CV outcomes in uremic patients. Validation study in an independent cohort of ESRD patients (n=111) confirmed that elevated plasma lncRNA DKFZP434I0714 is a significant independent predictor of adverse CV outcomes in uremic patients. Additional experiments demonstrated the functional involvement of DKFZP434I0714 in the pathogenesis of endothelial dysfunction.
Conclusions
In summary, plasma lncRNA expression signature reflects the disease states of uremia. Elevated plasma levels of lncRNA DKFZP434I0714 in uremic patients portend a worse CV outcome and warrant closer monitoring and more aggressive management.
Keywords: long noncoding RNA, plasma, uremia, adverse cardiovascular events, biomarker, endothelial dysfunction
1. Introduction
Cardiovascular (CV) diseases are the major cause of morbidity and mortality in patients with end-stage renal disease (ESRD), accounting for nearly 50 percent of deaths in this population [1, 2]. It is estimated that 80% of ESRD patients with maintenance hemodialysis (HD) have one or more types of CV diseases including coronary artery disease (CAD), arrhythmia, heart failure (HF) and peripheral artery disease (PAD) [3]. In particular, endothelial dysfunction, the precursor to CAD and PAD, are highly prevalent in uremic patients [4]. Traditional risk factors, however, are much less predictive of adverse CV complications in ESRD patients [5], and biochemical markers for cardiac ischemia/injury are often unreliable with uremia [6]. In order to develop practical surveillance tools, prognostic indicators and therapeutic strategies, there is a clear need to identify novel mediators and biomarkers of CV complications in uremic patients.
Long noncoding RNAs (lncRNAs) are a heterogeneous group of non-coding transcripts longer than 200 nucleotides, which have been shown to be functionally involved in the pathogenesis of cancer [7, 8] neurodegenerative [9–11] and CV diseases [12–14] through diverse mechanisms including epigenetic modification, transcriptional, post-transcriptional and translational regulation. The wide-range functions of lncRNAs appear to depend both on sequence homology/complementarity with other nucleic acids and their secondary/tertiary structures that form frameworks and scaffolds for the assembly of macromolecular complexes [15]. While the roles of lncRNAs in various biological processes are beginning to emerge, research exploring the functional roles of lncRNAs in uremia and associated CV complications remains sparse. LncRNAs have been shown to be dysregulated in mesangial proliferative glomerulonephritis [16] and acutely rejected renal allograft [17]. In addition, lncRNA Pvt1 has been demonstrated to mediate the glomerular extracellular matrix accumulation in diabetic nephropathy [18]. A subset of lncRNAs was also shown to be associated with transforming growth factor β/Smad3-mediated renal inflammation and fibrosis [19]. Recently, a circulating lncRNA TapSaki was found to be an independent predictor of mortality in critically ill patients with acute kidney injury [20]. In addition, lncRNAs ZAP70 and BC133674 were found to be aberrantly expressed in the peripheral blood mononuclear cells of uremic patients [21]. Taken together, current evidence suggests that lncRNAs are involved in renal and cardiovascular health and diseases, adding another layer of complexity on top of the already complicated genome and proteome regulation.
In the present study, we sought to test the hypothesis that circulating lncRNA expression profiles may reflect the disease status of uremia and could serve as a reliable biomarker to predict adverse CV outcomes in uremic patients. Exploiting RNA sequencing (RNASeq)-based plasma transcriptome profiling, we identified a circulating lncRNA expression signature that mirrored the underlying kidney pathology as well as distinguished ESRD patients from healthy subjects and patients with chronic kidney diseases (CKD). Further analyses revealed that multiple circulating lncRNAs were linked to adverse CV outcomes in ESRD patients. Of these, elevated lncRNA DKFZP434I0714 level [22] was validated to be an independent predictor of adverse CV outcomes in patients with uremia. In addition, DKFZP434I0714 was strongly induced in human aortic endothelial cells (HAEC) stressed with hypoxia; knocking down DKFZP434I0714 repressed the transcript expression of intracellular adhesion molecule 1 (ICAM-1) and vascular cells adhesion molecule 1 (VCAM-1) while enhancing the expression of endothelial nitric oxide synthase (eNOS) in HAEC. In addition, DKFZP434I0714 knockdown reduced hypoxic stress-induced endothelial cell apoptosis and monocyte adhesion. These experimental results suggest a potential role of DKFZP434I0714 in the pathogenesis of endothelial dysfunction, vascular inflammation and atherosclerosis, the hallmarks of vascular complications associated with uremia.
2. Material and Methods
This section is described in the Online Data Supplement. All studies were conducted in accordance with the Declaration of Helsinki and protocols approved by the institutional review boards of National Taiwan University and Washington University in St Louis. A written informed consent was obtained from each of the study subjects. All high-throughput sequencing data have been submitted to the National Center for Biotechnology Information gene and hybridization array data repository (GEO ID: GSE97709).
3. Results
Deep sequencing of plasma RNA identified a uremia-specific RNA expression signature that mirrors pathological changes in diseased kidneys
The workflow of circulating lncRNA profiling and validation in uremic patients is provided in Figure 1A. During the exploratory study phase, plasma RNA samples were prepared from a cohort of 48 subjects (28 ESRD, 8 CKD and 12 control) followed by RNASeq to profile circulating lncRNAs and mRNAs. The characteristics of the exploratory study cohort were comparable between groups (Supplemental Table S1).
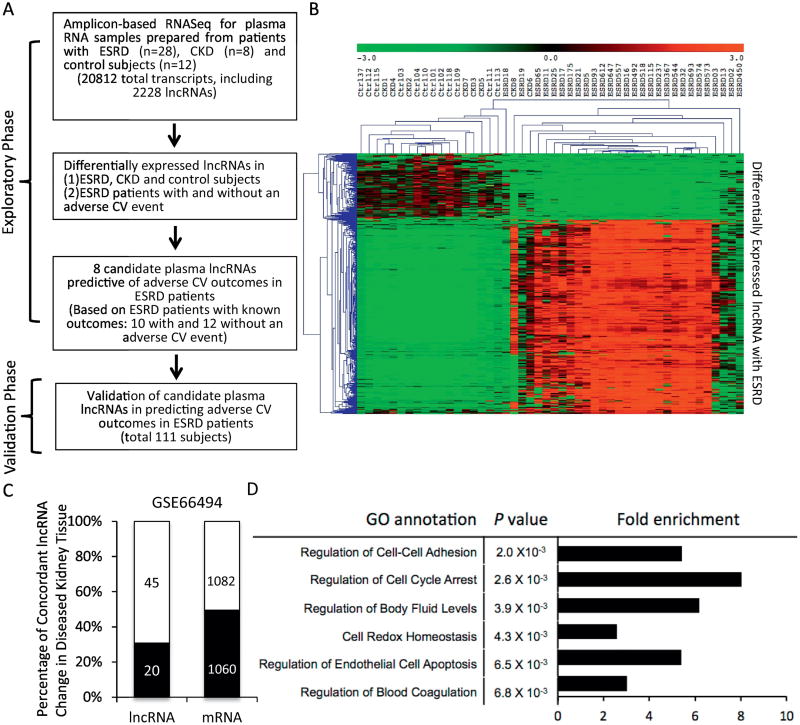
Figure 1. Study design and workflow of plasma long noncoding RNA profiling in uremic patients.
(A) During the initial exploratory phase, plasma total RNA was isolated from 48 study subjects (28 ESRD, 8 CKD and 12 controls), followed by amplicon-based RNA sequencing (AmpliSeq Transcriptome™, Thermo Fisher), which allowed simultaneous quantification of 20812 human transcripts, including 2228 lncRNAs. Differential expression analyses following RNASeq identified lncRNAs that were altered with ESRD, compared to control/CKD, as well as 8 lncRNAs that were correlated with adverse CV outcomes in uremic patients. The prognostic role of the 8 lncRNAs in patients with ESRD was then validated in an independent cohort of 111 subjects. (B) Heat map and hierarchical clustering of the plasma lncRNAs that were differentially expressed in ESRD and control subjects. Note that the expression profiles of these plasma lncRNAs distinguished ESRD from CKD/control subjects. (C) Analysis of the microarray data derived from kidney biopsy samples of patients with advance chronic kidney diseases (GSE66494) showed that a significant proportion of the dysregulated lncRNAs (30.7%, 20 out of the 65 lncRNAs from Figure 1B that were detectable in GSE66494) and mRNAs (49.5%, 1060 out of 2142 mRNAs that were detectable in GSE66494) found in the plasma from ESRD patients were also found concordantly dysregulated in diseased human kidneys. (D) Gene ontology analysis of the neighboring mRNA (cis-mRNA) of the dysregulated plasma lncRNAs with ESRD showing strong enrichment in genes that are involved in the pathogenesis of uremia.
A total of 422 million sequencing reads were obtained from 48 sequencing libraries, and more than 410 million reads were mapped to the human genome (>98%). The expression levels of individual transcript were presented as reads per million mapped reads (RPMR, see Methods in the Online Data Supplement). Using the criterion that a transcript must be expressed at ≥ 0.1 RPMR in ≥ 2 sequencing libraries, RNASeq detected 19312 transcripts in human plasma, including 1897 lncRNAs and 17415 mRNAs (Supplemental Table S2). Among the circulating lncRNAs detected in human plasma, 179 (118 up and 61 down) were significantly (absolute fold-change ≥ 2, adjusted P<0.001) dysregulated in ESRD, compared with control, samples (Supplemental Table S3); however, less than one-fifth (33 out of 179, 18.4%) of these lncRNAs showed similar dysregulation in CKD subjects (Supplemental Table S3). Unsupervised hierarchical clustering analysis demonstrated that the expression profile of these 179 lncRNAs distinguished ESRD from CKD/control samples (Figure 1B). However, the plasma lncRNA expression profiles failed to discriminate control subjects from CKD patients.
As disease-associated circulating, cell-free RNAs often derive from exosomes or microvesicles released by cells/tissues in diseased organs [23], we hypothesized that a significant proportion of uremia-associated plasma RNAs might come from malfunctioning kidney tissues and reflect the transcriptomal changes in diseased kidneys. To test this hypothesis, we analyzed the microarray data from an independent, historical cohort of human kidney samples with advanced renal dysfunction (53 advanced CKD and 8 control samples, GSE 66494) [24]. Among the 179 plasma lncRNAs dysregulated with ESRD, 65 could be detected by the probes on the arrays used in GSE66494, among which 30.7% (20 out of 65) were found to be concordantly dysregulated in diseased kidney tissues (Figure 1C and Supplemental Table S3). Similarly, 49.5% of the 2142 (all were detectable in GSE66494) plasma mRNAs dysregulated in ESRD patients were found to be congruently dysregulated in the 53 diseased human kidney tissues (Figure 1C). Taken together, these data suggest that plasma RNA expression signature reflects the disease status of advanced renal dysfunction and mirrors, at least partially, the changes in coding and noncoding transcriptome in diseased kidney tissues.
We [14] and others [25, 26] have previously shown that a significant proportion of lncRNAs function through modulating transcriptional expression of neighboring (or cis-) mRNAs (cis-regulation). Consistent with this notion, close to half (88 out of 179, 49%) of the neighboring mRNAs of these ESRD-linked lncRNAs were also significantly dysregulated in ESRD, compared to control, plasma samples (Supplemental Table S4); 46 out of the 88 lncRNA:cis-mRNA pairs showed concordant changes, whereas the other 42 pairs showed discordant changes. Gene ontology (GO) analysis of these cis-mRNAs showed significant enrichment in genes involved in the regulation of cell-cell adhesion, cell cycle arrest, regulation of body fluid levels, cell redox homeostasis, endothelial cell apoptosis and the regulation of blood coagulation, many of which have been implicated in the pathogenesis of uremia (Figure 1D) [27–30]. The strong co-expression pattern observed between the circulating lncRNA: cis-mRNA pairs, as well as the high enrichment of cis-mRNAs linked to the pathophysiology of uremia suggest the functional involvement of lncRNAs in the pathogenesis of uremia. An example of such lncRNA:cis-mRNA pair was CATSPER2P1 and its neighboring mRNA PDIA3 (ERP57). Both CATSPER2P1 and PDIA3 were significantly downregulated in the plasma from uremic patients (Supplemental Table S4), and PDIA3 has been reported to be critically involved in the pathogenesis of renal fibrosis and the progression of CKD [31].
Circulating lncRNA expression profiles discriminate between ESRD patients with and without an adverse CV event
To test the hypothesis that circulating lncRNAs could be a useful biomarker to predict adverse CV events and death in uremic patients, comparative analyses of the plasma lncRNA expression profiles were conducted in 10 ESRD patients who experienced at least one adverse CV event (defined as the need for percutaneous coronary intervention or coronary artery bypass grafting, acute coronary syndrome, heart failure, stroke and CV death. Details are described in Online Data Supplement.) during the 50-month follow-up period, as well as 12 ESRD patients who completed the 50-month follow-up without an adverse CV event. A total of 49 lncRNAs were significantly (absolute fold-change ≥ 2, adjusted P<0.05) differentially expressed (25 up- and 24 down-regulated) in ESRD patients with an adverse CV event, compared to those without one (Supplemental Table S5). Among these 49 lncRNAs, only 6 (LOC285540, CDRT15P2, LOC644656, KCNJ2-AS1, MKNK1-AS1, ZNF582-AS1) overlapped with the 179 lncRNAs that were found dysregulated with ESRD. Heat map and hierarchical clustering analyses showed that the expression signature of the 49 lncRNAs correctly clustered all but one of the ESRD patients with an CV event (labeled as eESRD) together (Figure 2A). A volcano plot illustrating the fold-change and significance levels of the 49 lncRNAs is provided in Figure 2B.
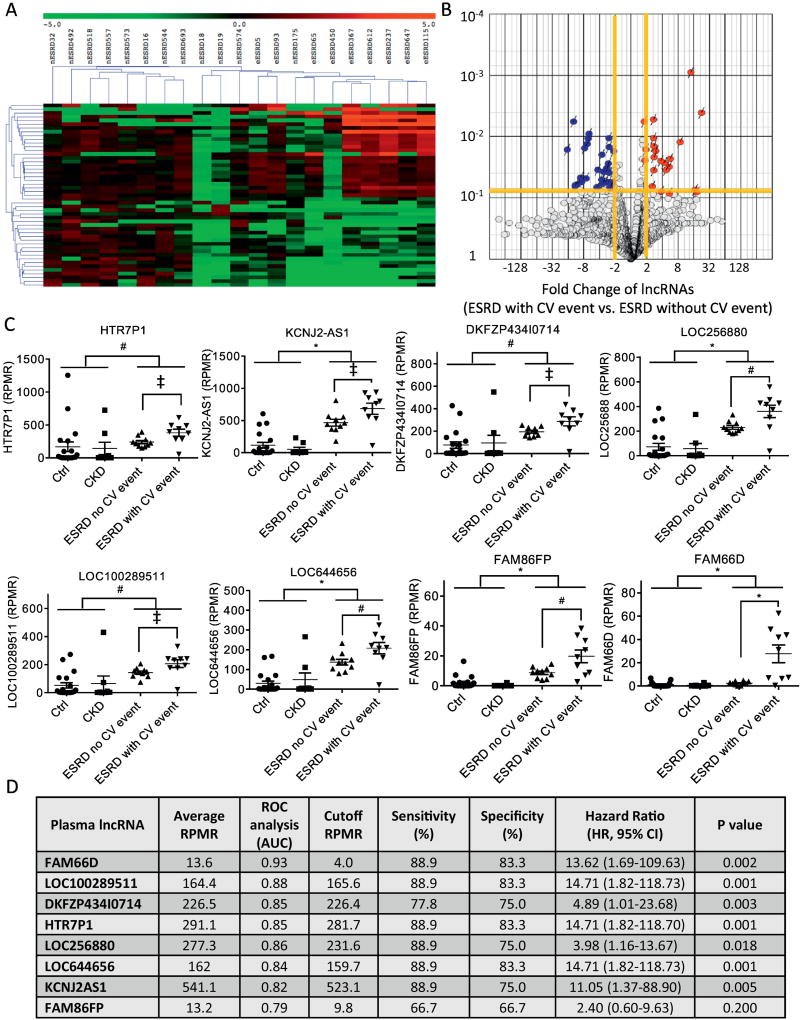
Figure 2. Identification of plasma lncRNAs linked to adverse CV outcomes in uremic patients.
(A) Heat map/hierarchical clustering analysis and (B) volcano plot of the plasma lncRNAs that were differentially expressed in ESRD patients with (eESRD) vs. without (nESRD) adverse CV events. (C) Dot plots that illustrate the distribution of the expression levels of the 8 candidate lncRNAs linked to adverse CV outcomes in uremic patients across all four groups (control, CKD, ESRD without CV event, ESRD with CV event) of study subjects. (‡, # and * denotes P <0.05, 0.01 and <0.001, respectively) (D) Table summarizing the average expression levels (of all 48 plasma samples, expressed in RPMR), results of ROC [area-under-curve (AUC)], optimal cutoff values for CV outcome prediction (in RPMR), the sensitivity/specificity of the cutoffs, and Kaplan-Meier analysis (hazard ratio and P values) of individual candidate lncRNAs in the exploratory study cohort.
To identify candidate plasma lncRNAs that might be applied clinically to determine the risk of adverse CV events in ESRD patients, the following criteria were used: (1) the expression levels of the candidate lncRNA(s) should remain unchanged in subjects without ESRD, and (2) the extent of aberrant expression of a candidate lncRNA must be greater in ESRD patients with, comparing to those without, an adverse CV event. Eight lncRNAs were found to fulfill the aforementioned criteria and the expression values of each of these lncRNAs across all four groups of study subjects were plotted in Figure 2C. Receiver operating characteristic (ROC) curve analysis was conducted to determine the overall performance of each of the 8 lncRNAs in predicting adverse CV outcomes in uremic patients. Using Cutoff Finder [32], the optimal cutoff expression levels of individual lncRNAs for CV outcome prediction were determined. Figure 2D provides the average expression levels of each lncRNA (in RPMR), the area under curve (AUC) of individual lncRNAs determined by ROC curve analysis, the optimal cutoffs of each lncRNA, as well as the sensitivity and specificity of using these cutoffs in discriminating between uremic patients with and without an adverse CV event. In addition, Kaplan-Meier analysis revealed that elevated plasma levels of all but one of these 8 lncRNAs were associated with significantly increased risks of adverse CV events in uremic patients (Figure 2D).
Validation study confirmed plasma lncRNA DKFZP434I0714 as an independent predictor of adverse CV outcomes in patients with ESRD
To further validate the prognostic value of circulating lncRNAs in predicting adverse CV outcomes in uremic patients, amplicon-based targeted RNASeq was performed to quantify the expression levels of the 8 candidate lncRNAs in the plasma samples from an independent cohort of study subjects, including 43 ESRD patients without CV events, 36 ESRD patients with at least one adverse CV event, 19 CKD patients and 13 control subjects with normal renal function. The characteristics of the 111 study subjects in this validation cohort were comparable across all groups (Supplemental Table S6).
Consistent with the lncRNA dysregulation observed in the exploratory uremic study cohort, targeted RNASeq revealed significant (P<0.001) upregulation in 6 of the 8 candidate lncRNAs (except FAM66D and FAM86FP) in CKD (P<0.05) and ESRD (P<0.001), compared with control, samples (Figure 3A, B). The expression profiles of the 8 lncRNAs also distinguished ESRD from control/CKD samples (Figure 3A). Kaplan-Meier analysis revealed that lncRNA DKFZP434I0714 (Hazard ratio [95% confidence interval], 2.38 [1.17–4.86], P=0.016, Figure 3C, D) and KCNJ2-AS1 (HR 2.13 [1.05–4.33], P=0.037, Figure 3C, Supplemental Figure S8) were significant predictors of adverse CV outcomes. Multivariate Cox regression analysis, however, showed that DKFZP434I0714 (HR 3.02 [1.09–8.32], P=0.033) and dialysis vintage (HR 1.11 [1.00–1.23], P=0.044) were the only two independent predictors of adverse CV outcomes in uremic patients after correcting for age, gender and underlying medical conditions (Figure 3C). To determine if the combination of lncRNAs and dialysis vintage could improve the predictive ability of adverse CV outcomes in uremic patients, a “combined index” was generated by combining the expression levels of lncRNAs DKFZP434I0714 and KCNJ2-AS1, as well as dialysis vintage (see Methods). This combined index showed improved sensitivity and specificity (72.2% and 62.8%, respectively, Figure 3C) as a predictor of adverse CV outcomes. Consistent with this result, both univariate (Supplemental Figures S9) and multivariate Cox regression (Figure 3C, HR 2.82 [1.36–5.84], P=0.005) analyses revealed that this combined index was a strong independent predictor of adverse CV outcomes in uremic patients.
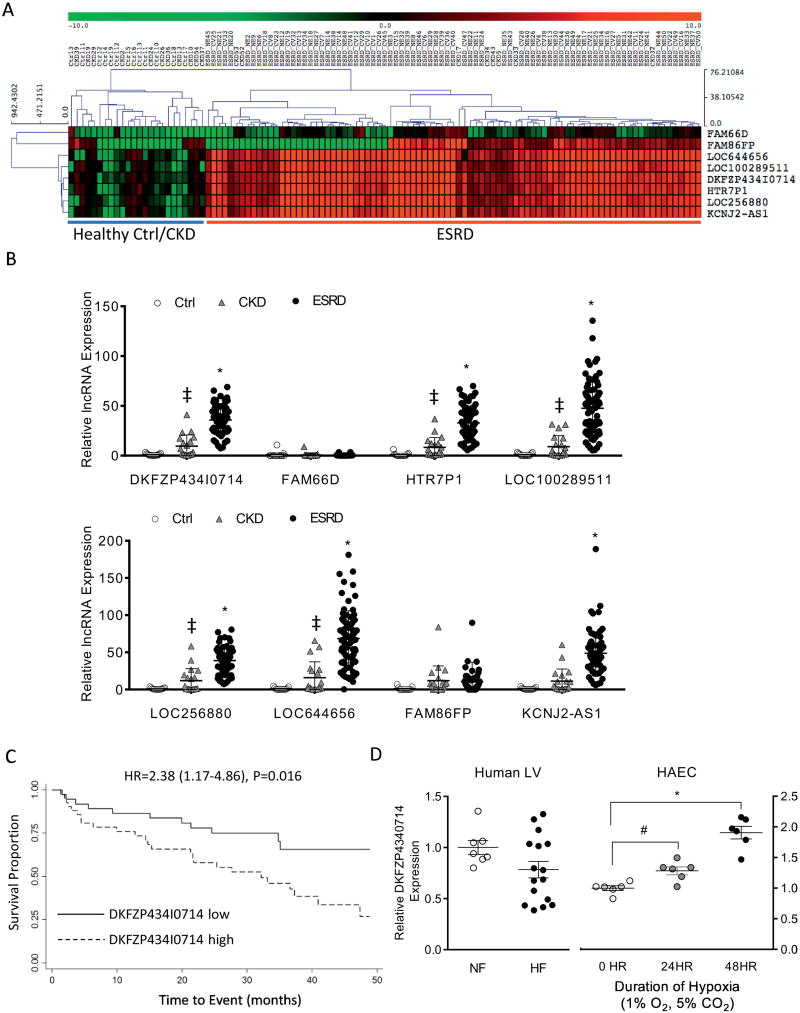
Figure 3. Validation of the prognostic role of plasma lncRNAs in patients with ESRD.
(A) Heat map and hierarchical clustering analysis of the expression levels of the 8 candidate plasma lncRNAs in an independent cohort of 111 study subjects (control 13, CKD 19, ESRD without CV event 43, ESRD with CV event 36). (B) Relative transcript expression levels of the 8 candidate lncRNAs in control, CKD and ESRD patients in the validation study cohort. (C) Univariate and multivariate analyses of candidate plasma lncRNAs linked to adverse CV outcomes in ESRD patients in the validation study cohort. #Combined index was derived from the combination of lncRNA DKFZP434I0714, KCNJ2AS1 and Dialysis Vintage (see methods). (D) Kaplan-Meier curve analysis showing significantly reduced survival in uremic patients with high plasma levels of lncRNA DKFZP434I0714 (>1.562 after normalized to the geometric mean of 4 reference genes). (D) Relative transcript expression levels of lncRNA DKFZP434I0714 in failing (HF, n=16) and non-failing (NF, n=8) human left ventricles (Human LV) and in human aortic endothelial cells (HAEC) exposed to hypoxic (1% O2, 5% CO2 for 24 and 48 hr, biological replicates, n=6 in each group) stress.
DKFZP434I0714 is induced in human aortic endothelial cells upon hypoxic stress and contributes to the regulation of atherorelevant genes in the endothelium
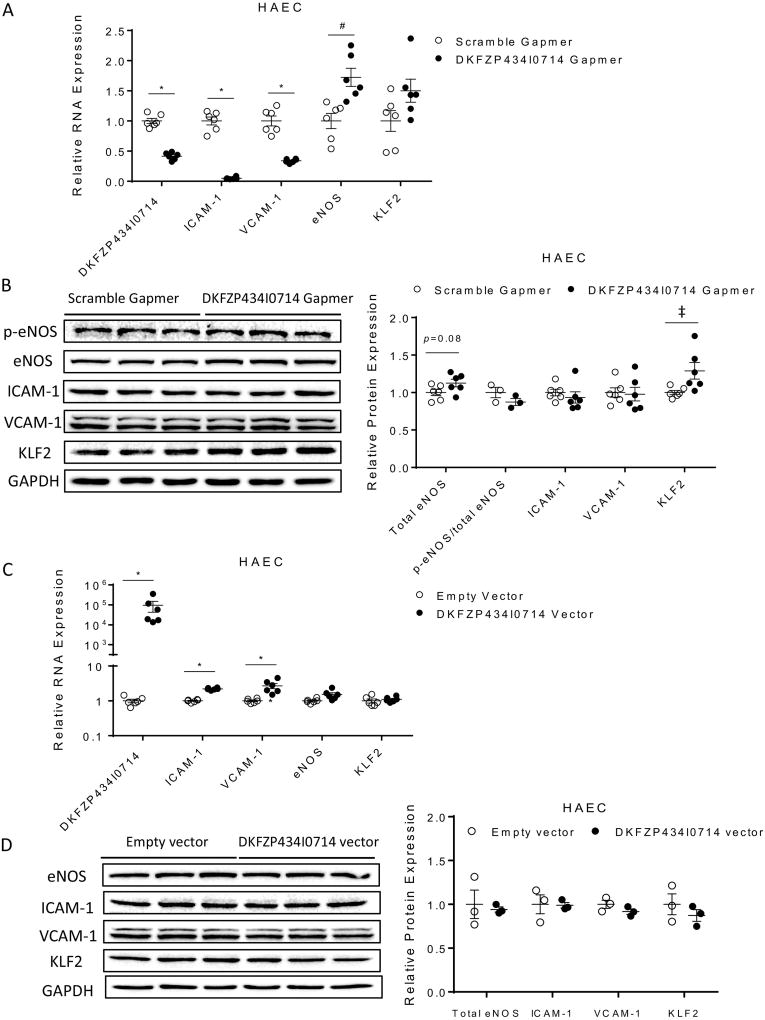
Because an elevated plasma level of lncRNA DKFZP434I0714 predicts adverse CV events in uremic patients, we hypothesized that increased DKFZP434I0714 levels could be observed in diseased cardiovascular tissues/cells. The expression levels of DKFZP434I0714 in human failing left ventricular (LV) tissues (HF, n=16), however, were not significantly different from non-failing control LV samples (NF, n=7 Figure 3E) [14]. In contrast, human aortic endothelial cells (HAEC) stressed with hypoxia (1% O2 for 24 and 48 hours) showed strong upregulation of DKFZP434I0714 (Figure 3E), as well as ICAM-1 and VCAM-1, two adhesion molecules critical for endothelial dysfunction and atherosclerosis (Supplemental Figure S1). Cell fractionation studies revealed that DKFZP434I0714 was highly enriched in the nucleus of HAECs (Supplemental Figure S2). In addition, knocking down DKFZP434I0714 in HAEC using antisense locked nucleic acid Gapmers led to significant (P<0.001) down regulation of ICAM-1 and VCAM-1, compared to control HAEC treated with scramble Gapmers (Figure 4A). In contrast, DKFZP434I0714 knockdown induced a strong transcriptional upregulation of eNOS (P<0.01) and a trend toward increased Kruppel-like factor 2 (KLF2), two atheroprotective factors, in HAEC (Figure 4A). Immunoblots were performed to assess the protein expression levels of p-eNOS, total eNOS, ICAM-1, VCAM-1 and KLF2 in HAEC with DKFZP434I0714 knockdown. As shown in Figure 4B, DKFZP434I0714 knockdown led to significant upregulation of KLF2 and a trend toward increased eNOS in HAEC. The protein expression levels of p-eNOS, ICAM-1 and VCAM-1, however, were not significantly altered with DKFZP434I0714 depletion. Consistent with the observed upregulation of eNOS in HAECs silenced for lncRNA DKFZP434I0714, NO production was also significantly increased in HAECs with DKFZP434I0714 knockdown, compared with scramble control (Supplemental Figure S6A). Moreover, knocking down lncRNA DKFZP434I0714 significantly attenuated hypoxic stress-induced apoptosis in HAEC (Supplemental Figure S3) and reduced monocyte adhesion to HAEC (Supplemental Figure S4A, B), suggesting that DKFZP434I0714 could modulate stress-induced HAEC apoptosis and plays a critical role in modulating endothelial capacity to attract and bind inflammatory cells, hence contributing to endothelial dysfunction and vascular inflammation. Overexpression of DKFZP434I0714, on the other hand, significantly increased the transcript expression levels of ICAM-1 and VCAM-1, without affecting eNOS or KLF2, in HAEC (Figure 4C), although immunoblot experiments did not reveal discernible changes in any of these proteins with increased DKFZP434I0714 expression in HAEC. Increased DKFZP434I0714 in HAEC did not result in significant changes in NO production, either (Supplemental Figure S6B). Collectively, these data strongly suggest that dysregulation of DKFZP434I0714 could be involved in the pathogenesis of endothelial dysfunction, vascular inflammation and atherosclerosis, hallmarks of vascular complications associated with uremia.
Figure 4. DKFZP434I0714 modulates atherorelevant protein genes in human aortic endothelial cells.
(A) Knocking down DKFZP434I0714 in HAEC led to significant transcriptional downregulation of VCAM-1/ICAM-1 and upregulation of eNOS, compared with scramble control. (biological replicates, n=6 in each group; ‡, # and * denotes P <0.05, 0.01 and <0.001, respectively). (B) Immunoblots showed that DKFZP434I0714 knockdown led to significant upregulation of KLF2 and a trend toward increased eNOS proteins in HAEC. The protein expression levels of ICAM-1, VCAM-1 and p-eNOS, however, were not altered by DKFZP434I0714 knockdown (biological replicates, n=3–6 in each group; ‡ denotes P <0.05). (C) Overexpression of DKFZP434I0714 significantly increased transcript expression levels of ICAM-1 and VCAM-1, without affecting eNOS or KLF2, in HAEC (biological replicates, n=3–6 in each group; * denotes P <0.001). (D) Forced expression of DKFZP434I0714, however, did not lead to discernible changes in the protein expression levels of eNOS, ICAM-1, VCAM-1 or KLF2.
4. Discussion
In the present study, RNASeq-based transcriptome profiling revealed a plasma lncRNA expression signature that distinguishes uremic patients from CKD and control subjects with normal renal function. Further analyses identified multiple lncRNAs that were associated with adverse CV outcomes in uremic patients. Validation studies in an independent cohort of uremic patients confirmed that elevated lncRNA DKFZP434I0714 was a significant predictor of adverse CV events and death in patients with ESRD. Expression analyses demonstrated that lncRNA DKFZP434I0714 was increased in human endothelial (HAEC) cells stressed with hypoxia. Knocking down DKFZP434I0714 in HAEC repressed the transcript expression levels of pro-atherogenic adhesion molecules including ICAM-1 and VCAM-1 while promoting the transcript expression of atheroprotective protein eNOS (and likely also KLF2). DKFZP434I0714 knockdown also reduced hypoxic stress-induced endothelial cell apoptosis and monocyte adhesion. These data collectively suggest that lncRNA DKFZP434I0714 plays an important role in the transcriptional regulation of atherorelevant proteins and could contribute to the pathogenesis of endothelial dysfunction, vascular inflammation and atherosclerosis.
To date, only a few studies have investigated the diagnostic or prognostic roles of circulating lncRNAs in patients with cardiovascular diseases, mostly using PCR or microarray-based experiments. Kumarswamy et al reported that plasma lncRNA LIPCAR predicts post-MI cardiac remodeling and CV death in patients with HF [33]. LncRNAs ANRIL and KCNQ1OT1 in peripheral blood cells, on the other hand, were found to be dysregulated after MI and to be predictors of post-MI LV dysfunction [34]. A plasma lncRNA CoroMarker has been shown to be a useful biomarker to facilitate the diagnosis of CAD [35]. In addition, the reciprocal changes of lncRNAs ZFAS1 and CDR1AS in whole blood were reported to be a predictor of myocardial infarction [36]. To the best of our knowledge, the present study is the first report to profile human plasma lncRNA and mRNA expression pattern using next-generation sequencing technology. Our study is also the first to demonstrate that plasma lncRNA expression signature can be a sensitive and specific biomarker to reflect the status of advanced renal dysfunction and to predict adverse CV outcomes and death in patients with ESRD.
Traditional risk factors for CV diseases including age, male gender, diabetes, hypertension and hyperlipidemia are known to be much less predictive for CV complications in uremic patients [5]. Several nontraditional risk factors such as impaired mineral metabolism (hyperphosphatemia, hypercalcemia and hyperparathyroidism) [37] and increased homocysteine levels [38] have been shown to be associated with increased mortality in ESRD patients receiving hemodialysis. The contribution of these nontraditional risk factors to increased mortality associated with uremia, however, remains controversial as interventional trials aimed at modifying these factors have failed to improve clinical outcomes in uremic patients [39–41]. Identifying lncRNAs linked to poor CV outcomes in ESRD patients could lead to the discovery of potential novel mechanisms that mediate CV complications predisposed by uremia. Indeed, the present study revealed a novel regulatory role of lncRNA DKFZP434I0714 on the transcript expression of atherorelevant molecules including ICAM-1, VCAM-1, eNOS and KLF2 in human endothelial cells, suggesting the potential involvement of DKFZP434I0714 in the pathogenesis of endothelial dysfunction and atherosclerosis. Interestingly, although knocking down DKFZP434I0714 led to significant downregulation of ICAM-1 and VCAM-1 transcripts (Figure 4A), immunoblot experiments did not show a significant reduction in the protein levels of ICAM-1 and VCAM-1 with DKFZP434I0714 depletion in HAEC (Figure 4B). It is possible that ICAM-1 and VCAM-1 proteins are relatively stable and it could take much longer than 48 hours (the course of DKFZP434I0714 knockdown experiment) to detect discernible reduction in their protein levels. Additional experiments revealed no significant differences in the plasma levels of DKFZP434I0714 in healthy control (n=20) and patients with coronary artery disease (CAD, n=35) without known renal dysfunction (Supplemental Figure S7), suggesting that DKFZP434I0714 elevation is specific to patients with uremia. Further studies are required to determine the molecular mechanisms via which lncRNA DKFZP434I0714 regulates these atherorelevant genes and to what extent it contributes to endothelial dysfunction and atherosclerosis in the context of uremia. Although it would be of great interest to study the functional role of DKFZP434I0714 in important animal models of atherosclerosis, neither DKFZP434I0714 nor the other 7 lncRNA candidates identified from RNASeq analysis were conserved between human and mouse, precluding the possibility to study their functional roles in mice.
A potential limitation of this study is that the number of study subjects in the validation cohort (n=111) was relatively small. The prognostic value of several candidate plasma lncRNAs (such as KCNJ2AS1 and LOC256880) identified from the exploratory study was borderline and could become significant if the size of the validation cohort were larger. Nevertheless, most (6 of 8) of the candidate lncRNAs showed strong upregulation in ESRD, compared to both CKD and control samples, suggesting a strong association with, and potential contribution to, the pathophysiology of uremia. Finally, although the exact cellular/tissue origin of the circulating lncRNAs linked to uremia is unclear, our data suggest that many of them could come from diseased kidney tissues and also possibly from endothelial cells.
In conclusion, using an unbiased RNASeq approach, we have demonstrated that circulating lncRNA expression signature distinguishes ESRD from CKD/health controls and that an elevated plasma level of lncRNA DKFZP434I0714 predicts adverse CV outcomes in ESRD patients. Uremic patients with increased plasma lncRNA DKFZP434I0714 levels may need closer monitoring and more aggressive management to reduce the risk of CV events. Additional experimental evidence presented suggests that DKFZP434I0714 could be involved in the pathogenesis of endothelial dysfunction and atherosclerosis. These findings strongly suggest a role for circulating lncRNAs as a new class of prognostic biomarker of adverse CV events in ESRD patients, as well as a functional role(s) for lncRNAs in the pathobiology of uremia.
Supplementary Material
Acknowledgments
This work was supported by Taiwan Ministry of Science Technology Grants 103-2320-B-002-068-MY2 (KCY), 105-2628-B-002-042-MY4 (KCY), a Taiwan National Health Research Institute Career Development Grant NHRI-EX104-10418SC (KCY), as well as grants from National Taiwan University Hospital 104-N2864 (CFL), NTUH.106-P02 (KCY, CFL) and 105-CGN01, UN106-026, 106-N3740, VN106-12 (KCY). We would also like to thank Dr. Kathryn Yamada for her critical review and comments on the article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors take responsibility for all aspects of the reliability and freedom from bias of the data presented and their discussed interpretation.
Conflict of interests
None
References
- 1.Foley RN, Parfrey PS, Sarnak MJ. Clinical Epidemiology of Cardiovascular Disease in Chronic Renal Disease. Am J Kidney Dis. 1998;32:S112–119. doi: 10.1053/ajkd.1998.v32.pm9820470. [DOI] [PubMed] [Google Scholar]
- 2.Go AS, Chertow GM, Fan D, McCulloch CE, Hsu CY. Chronic Kidney Disease and the Risks of Death, Cardiovascular Events, and Hospitalization. N Engl J Med. 2004;351:1296–1305. doi: 10.1056/NEJMoa041031. [DOI] [PubMed] [Google Scholar]
- 3.Cheung AK, Sarnak MJ, Yan G, Berkoben M, Heyka R, Kaufman A, et al. Cardiac Diseases in Maintenance Hemodialysis Patients: Results of the Hemo Study. Kidney Int. 2004;65:2380–2389. doi: 10.1111/j.1523-1755.2004.00657.x. [DOI] [PubMed] [Google Scholar]
- 4.Miyazaki H, Matsuoka H, Itabe H, Usui M, Ueda S, Okuda S, et al. Hemodialysis Impairs Endothelial Function Via Oxidative Stress: Effects of Vitamin E-Coated Dialyzer. Circulation. 2000;101:1002–1006. doi: 10.1161/01.cir.101.9.1002. [DOI] [PubMed] [Google Scholar]
- 5.Allon M. Evidence-Based Cardiology in Hemodialysis Patients. J Am Soc Nephrol. 2013;24:1934–1943. doi: 10.1681/ASN.2013060632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George SK, Singh AK. Current Markers of Myocardial Ischemia and Their Validity in End-Stage Renal Disease. Curr Opin Nephrol Hypertens. 1999;8:719–722. doi: 10.1097/00041552-199911000-00012. [DOI] [PubMed] [Google Scholar]
- 7.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a Noncoding Rna Is Elevated in Alzheimer's Disease and Drives Rapid Feed-Forward Regulation of Beta-Secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson R. Long Non-Coding Rnas in Huntington's Disease Neurodegeneration. Neurobiol Dis. 2012;46:245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved Regions Encoding Ncrnas Are Altered in Human Leukemias and Carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 10.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, et al. Xist Rna Is a Potent Suppressor of Hematologic Cancer in Mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K, et al. Epigenetic Upregulation of Lncrnas at 13q14.3 in Leukemia Is Linked to the in Cis Downregulation of a Gene Cluster That Targets Nf-Kb. PLoS Genet. 2013;9:e1003373. doi: 10.1371/journal.pgen.1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, et al. Identification of a Novel Non-Coding Rna, Miat, That Confers Risk of Myocardial Infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 13.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A Common Variant on Chromosome 9p21 Affects the Risk of Myocardial Infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 14.Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, et al. Deep Rna Sequencing Reveals Dynamic Regulation of Myocardial Noncoding Rnas in Failing Human Heart and Remodeling with Mechanical Circulatory Support. Circulation. 2014;129:1009–1021. doi: 10.1161/CIRCULATIONAHA.113.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding Rnas. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sui W, Li H, Ou M, Tang D, Dai Y. Altered Long Non-Coding Rna Expression Profile in Patients with Iga-Negative Mesangial Proliferative Glomerulonephritis. Int J Mol Med. 2012;30:173–178. doi: 10.3892/ijmm.2012.975. [DOI] [PubMed] [Google Scholar]
- 17.Sui W, Lin H, Peng W, Huang Y, Chen J, Zhang Y, et al. Molecular Dysfunctions in Acute Rejection after Renal Transplantation Revealed by Integrated Analysis of Transcription Factor, Microrna and Long Noncoding Rna. Genomics. 2013;102:310–322. doi: 10.1016/j.ygeno.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Alvarez ML, Khosroheidari M, Eddy E, Kiefer J. Role of Microrna 1207-5p and Its Host Gene, the Long Non-Coding Rna Pvt1, as Mediators of Extracellular Matrix Accumulation in the Kidney: Implications for Diabetic Nephropathy. PLoS One. 2013;8:e77468. doi: 10.1371/journal.pone.0077468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Q, Chung AC, Huang XR, Dong Y, Yu X, Lan HY. Identification of Novel Long Noncoding Rnas Associated with Tgf-Beta/Smad3-Mediated Renal Inflammation and Fibrosis by Rna Sequencing. Am J Pathol. 2014;184:409–417. doi: 10.1016/j.ajpath.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Lorenzen JM, Schauerte C, Kielstein JT, Hubner A, Martino F, Fiedler J, et al. Circulating Long Noncoding Rnatapsaki Is a Predictor of Mortality in Critically Ill Patients with Acute Kidney Injury. Clin Chem. 2015;61:191–201. doi: 10.1373/clinchem.2014.230359. [DOI] [PubMed] [Google Scholar]
- 21.Sui W, Yan Q, Li H, Liu J, Chen J, Li L, et al. Genome-Wide Analysis of Long Noncoding Rna Expression in Peripheral Blood Mononuclear Cells of Uremia Patients. J Nephrol. 2013;26:731–738. doi: 10.5301/jn.5000201. [DOI] [PubMed] [Google Scholar]
- 22.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, et al. Generation and Initial Analysis of More Than 15,000 Full-Length Human and Mouse Cdna Sequences. Proc Natl Acad Sci U S A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orozco AF, Lewis DE. Flow Cytometric Analysis of Circulating Microparticles in Plasma. Cytometry A. 2010;77:502–514. doi: 10.1002/cyto.a.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakagawa S, Nishihara K, Miyata H, Shinke H, Tomita E, Kajiwara M, et al. Molecular Markers of Tubulointerstitial Fibrosis and Tubular Cell Damage in Patients with Chronic Kidney Disease. PloS one. 2015;10:e0136994. doi: 10.1371/journal.pone.0136994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative Annotation of Human Large Intergenic Noncoding Rnas Reveals Global Properties and Specific Subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb Proteins Targeted by a Short Repeat Rna to the Mouse X Chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bro S, Moeller F, Andersen CB, Olgaard K, Nielsen LB. Increased Expression of Adhesion Molecules in Uremic Atherosclerosis in Apolipoprotein-E-Deficient Mice. J Am Soc Nephrol. 2004;15:1495–1503. doi: 10.1097/01.asn.0000128371.33195.7b. [DOI] [PubMed] [Google Scholar]
- 28.Edamatsu T, Fujieda A, Ezawa A, Itoh Y. Classification of Five Uremic Solutes According to Their Effects on Renal Tubular Cells. Int J Nephrol. 2014;2014:512178. doi: 10.1155/2014/512178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Himmelfarb J, Stenvinkel P, Ikizler TA, Hakim RM. The Elephant in Uremia: Oxidant Stress as a Unifying Concept of Cardiovascular Disease in Uremia. Kidney Int. 2002;62:1524–1538. doi: 10.1046/j.1523-1755.2002.00600.x. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Jerez A, Luengo A, Carracedo J, Ramirez-Chamond R, Rodriguez-Puyol D, Rodriguez-Puyol M, et al. Effect of Uraemia on Endothelial Cell Damage Is Mediated by the Integrin Linked Kinase Pathway. J Physiol. 2015;593:601–618. doi: 10.1113/jphysiol.2014.283887. discussion 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dihazi H, Dihazi GH, Bibi A, Eltoweissy M, Mueller CA, Asif AR, et al. Secretion of Erp57 Is Important for Extracellular Matrix Accumulation and Progression of Renal Fibrosis, and Is an Early Sign of Disease Onset. J Cell Sci. 2013;126:3649–3663. doi: 10.1242/jcs.125088. [DOI] [PubMed] [Google Scholar]
- 32.Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff Finder: A Comprehensive and Straightforward Web Application Enabling Rapid Biomarker Cutoff Optimization. PloS one. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, et al. Circulating Long Noncoding Rna, Lipcar, Predicts Survival in Patients with Heart Failure. Circ Res. 2014;114:1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 34.Vausort M, Wagner DR, Devaux Y. Long Noncoding Rnas in Patients with Acute Myocardial Infarction. Circ Res. 2014;115:668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Cai Y, Wu G, Chen X, Liu Y, Wang X, et al. Plasma Long Non-Coding Rna, Coromarker, a Novel Biomarker for Diagnosis of Coronary Artery Disease. Clin Sci (Lond) 2015;129:675–685. doi: 10.1042/CS20150121. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S, et al. Reciprocal Changes of Circulating Long Non-Coding Rnas Zfas1 and Cdr1as Predict Acute Myocardial Infarction. Sci Rep. 2016;6:22384. doi: 10.1038/srep22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Block GA, Klassen PS, Lazarus JM, Ofsthun N, Lowrie EG, Chertow GM. Mineral Metabolism, Mortality, and Morbidity in Maintenance Hemodialysis. J Am Soc Nephrol. 2004;15:2208–2218. doi: 10.1097/01.ASN.0000133041.27682.A2. [DOI] [PubMed] [Google Scholar]
- 38.Shemin D, Lapane KL, Bausserman L, Kanaan E, Kahn S, Dworkin L, et al. Plasma Total Homocysteine and Hemodialysis Access Thrombosis: A Prospective Study. J Am Soc Nephrol. 1999;10:1095–1099. doi: 10.1681/ASN.V1051095. [DOI] [PubMed] [Google Scholar]
- 39.Chertow GM, Correa-Rotter R, Block GA, Drueke TB, Floege J, Goodman WG, et al. Baseline Characteristics of Subjects Enrolled in the Evaluation of Cinacalcet Hcl Therapy to Lower Cardiovascular Events (Evolve) Trial. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2012;27:2872–2879. doi: 10.1093/ndt/gfr777. [DOI] [PubMed] [Google Scholar]
- 40.Suki WN, Zabaneh R, Cangiano JL, Reed J, Fischer D, Garrett L, et al. Effects of Sevelamer and Calcium-Based Phosphate Binders on Mortality in Hemodialysis Patients. Kidney Int. 2007;72:1130–1137. doi: 10.1038/sj.ki.5002466. [DOI] [PubMed] [Google Scholar]
- 41.Wrone EM, Hornberger JM, Zehnder JL, McCann LM, Coplon NS, Fortmann SP. Randomized Trial of Folic Acid for Prevention of Cardiovascular Events in End-Stage Renal Disease. J Am Soc Nephrol. 2004;15:420–426. doi: 10.1097/01.asn.0000110181.64655.6c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.