Abstract
R-loops are structures consisting of an RNA-DNA duplex and an unpaired DNA strand. They can form during transcription upon nascent RNA “threadback” invasion into the DNA duplex to displace the non-template strand. Although R-loops occur naturally in all kingdoms of life and serve regulatory roles, they are often deleterious and can cause genomic instability. Of particular importance are the disastrous consequences when replication forks or transcription complexes collide with R-loops. The appropriate processing of R-loops is essential to avoid a number of human neurodegenerative and other clinical disorders. We provide a perspective on mechanistic aspects of R-loop formation and their resolution learned from studies in model systems. This should contribute to improved understanding of R-loop biological functions and enable their practical applications. We propose the novel employment of artificially-generated stable R-loops to selectively inactivate tumor cells.
Keywords: R-loops, Transcription-replication collisions, RNA-DNA hybrids, DNA repair, non-canonical DNA structures
1. Introduction
During transcription, RNA polymerase (RNAP) synthesizes an RNA strand complementary to the DNA template strand. The nascent RNA must be released from that template to be available for further transactions. After generation of a short (~ 8–10 bp) RNA-DNA duplex within the “transcription complex”, the RNA product is peeled from the template strand by a special “wedge-like” protein moiety within the RNAP, and the nascent RNA is guided through the “exit channel” to prevent its re-hybridization with the template (1–7). The nascent RNA is then processed by factors involved in splicing and export from the nucleus (or in bacteria, sequestering in ribosomes), in addition to other factors that further antagonize re-hybridization (reviewed in (8,9)). However, despite these delivery and processing events, the nascent RNA sometimes hybridizes with the template DNA strand to form an R-loop, which includes the RNA-DNA duplex and the displaced non-template DNA strand (reviewed in (9,10)).
According to the most accepted model (e.g., (11)), R-loop formation occurs following extrusion of the nascent RNA from the transcription complex; in order to form an R-loop the nascent RNA must thread back into the DNA duplex upstream from the complex. An alternative model posits that the nascent RNA need not become separated from the DNA template, so that a long RNA-DNA duplex within an R-loop might form as a continuous extension from the short RNA-DNA duplex within the transcription complex. Yet, this model is not supported by crystal structures of transcription complexes (e.g., see (4)). Another pathway for R-loop formation has been proposed in which an extended RNA-DNA hybrid is generated through RNAP backtracking followed by its dislodging (reviewed in (10,12)). R-loop formation (either co-transcriptional, or post-transcriptional) also could be assisted by homologous recombination machinery ((13), reviewed in (14,15)). Post-transcriptional R-loop formation can occur in trans when the RNA invades a DNA region other than that from which it was synthesized (13)). Specialized enzymatic machinery can promote R-loop formation in trans by catalyzing invasion of short RNAs into the DNA duplex (reviewed in (16)). It has recently been suggested that posttranscriptional R-loop formation over the switch region is induced when the RNA helicase DDX1 operates upon G-quadruplex structures in long non-coding RNA, thereby facilitating the targeting of the activation-induced cytidine-deaminase (AID) to the IgH locus for class switch recombination (17).
From an historical perspective it is notable that before their discovery in biological systems, R-loops were generated artificially by hybridization of single-stranded RNA to double-stranded DNA at elevated temperatures and in the presence of formamide (18,19). Importantly, these artificial R-loops were instrumental for biological research (e.g., leading to the discovery of RNA splicing (20,21). R-loops were initially considered anomalous and generally deleterious, but now we appreciate that they have key roles in many important biological processes, e.g., immunogenesis and gene silencing (as reviewed in (9,15,22–32)). The R-loop represents an example, among many, of how non-canonical nucleic acid structures have been co-opted during evolution to provide important regulatory functions, even though they also pose formidable hazards for the essential transactions of transcription and replication (reviewed in (33–36)). Although the ubiquity of R-loops in living organisms is well established, there are certain challenges for their proper mapping and characterization (reviewed in (37)).
There has been a recent plethora of comprehensive reviews and commentaries on R-loops, so we will focus primarily upon just a few of the relevant aspects in this burgeoning field. We will consider the factors and situations that promote or suppress the formation of R-loops. These include the topological challenges for generating R-loops that arise due to the helical nature of the Watson-Crick duplex.
Although the processes involving R-loop formation and functioning in cells are complex and often involve factors beyond those of the transcription machinery (reviewed in (9,15,22–30)), comprehension may be facilitated by studies in simplified models, in which the number of components can be limited and fundamental mechanistic insights may be obtained. These insights may hopefully lead to novel applications of artificially generated R-loops for therapies.
2. Mechanistic aspects of R-loop formation
2.1. Factors affecting R-loop formation
In general, R-loop formation is facilitated by factors that provide a thermodynamic advantage to the hybrid between the nascent RNA and the DNA template strand over that of the duplex with the respective DNA strands. These factors include: Intrinsic R-loop-prone DNA sequences that form much more stable duplexes with RNA than with DNA (see refs. (38–41) and references therein), breaks in the non-template DNA strand (42,43), negative supercoiling (that facilitates DNA unwinding) (42,44), non-canonical DNA structures (e.g. G4-quadruplexes and triplexes) that encumber the non-template DNA strand (45–47) or sequestration of the non-template DNA strand by a ligand (48). In contrast, all factors that sequester the nascent RNA as it emerges from the transcription complex suppress R-loop formation (reviewed in (22)).
The superior stability of the RNA-DNA duplex versus the DNA-DNA duplex, though sufficient for R-loop maintenance, might not be sufficient for initiation of R-loop formation within intact duplex DNA in the absence of negative supercoiling: For that, an R-loop initiating sequence that contains clusters of guanines may be required, since this forms an especially stable RNA-DNA hybrid (11).
It is important to appreciate that evaluating relative stabilities of RNA-DNA and DNA-DNA duplexes for arbitrary DNA sequences is not trivial: The most stable RNA-DNA duplex (both in absolute terms and relative to the corresponding DNA-DNA duplex) is rG/dC, which is much more stable than both dG/dC and rC/dG duplexes (see (43) and references therein). This duplex could appear within an R-loop upon transcription of a DNA sequence in which the non-template DNA strand contains only guanines. As a consequence, the propensity to form an R-loop is often attributed to the GC skew (the relative abundance of guanine versus cytosine in the non-template DNA strand). However, the least stable RNA-DNA duplex (both in absolute terms, and with respect to the corresponding DNA-DNA duplex) is rU/dA, rather than rC/dG: According to our estimates (43), the former is much less stable than the corresponding DNA-DNA duplex, while the latter is slightly more stable, than the corresponding DNA-DNA duplex. Thus, detailed thermodynamic calculations might be required to estimate relative R-loop-forming propensities of various DNA sequences.
While R-loops are stabilized by negative supercoiling (i.e., DNA under-winding), positive supercoiling (i.e., DNA over-winding) tends to resolve them. However, for some DNA sequences the greater stability of DNA-RNA over the DNA-DNA duplex renders R-loops stable even with substantial positive supercoiling (e.g., see (49), and respective estimates in the Appendix of (36)).
Interestingly, the probability of R-loop formation decreases upon increasing distance between the R-loop-initiation sequence and the transcription initiation site (42). Most likely, this occurs because transcription from the region localized between the transcription initiation site and the R-loop-initiation sequence produces a long RNA “tail”, that creates a steric hindrance for RNA invasion into the DNA duplex (42). This effect is elaborated in the following Section.
Finally, note that in vivo, R-loop formation within a given sequence depends not only upon intrinsic R-loop-forming propensity, but also upon its interaction with various proteins. For example, it was reported that R-loop formation is strongly increased near polyA tracts, which are not intrinsically R-loop-prone sequences; and, it was suggested that this happens because these polyA tracts disfavor nucleosome binding (50).
2.2. Topological challenges for R-loop formation.
During R-loop formation, the nascent RNA must wind around the template DNA strand, while the non-template DNA strand unwinds from the template DNA strand. As long as the RNAP remains bound to the non-template strand, the only way for the RNA to wind around the template DNA strand (while the non-template DNA strand remains unwound) is by threading the upstream RNA “tail” between the RNA-DNA duplex and the non-template DNA strand (Fig.1, left). Thus, in order to increase the length of the RNA-DNA hybrid within an R-loop by the number of base pairs corresponding to one helical turn (i.e., roughly 10 bp), the entire RNA tail must pass through the space between the RNADNA duplex and the non-template DNA strand. The shorter the RNA tail, the easier this passage would be. So, R-loop formation is strongly facilitated if the R-loop initiation sequence is localized close to the free end of the nascent RNA, which might correspond either to the start of transcription or to a site of nascent RNA cleavage (42,51–53).
Fig.1. Pathways for R-loop propagation.
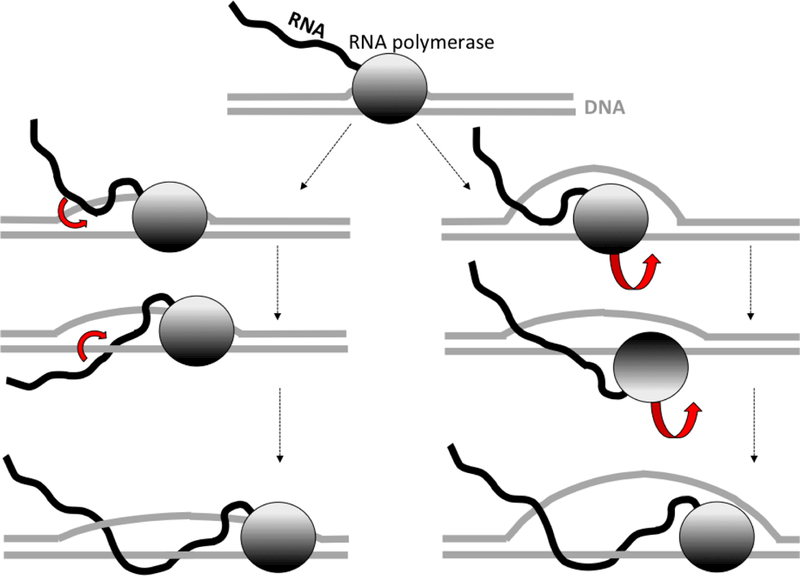
DNA is shown in gray, RNA is shown in black, RNAP is shown as a gray oval with color intensity gradient to indicate its orientation. For simplicity, the only inter-strand winding that is shown is the one between the nascent RNA and the template DNA strand. RNA passage and RNAP rotation are indicated by the red block arrows. The pathway at the left shows the intertwining between the nascent RNA and the template DNA strand mediated by the RNA “tail “ passage through the space between the RNA-DNA hybrid and the displaced non-template DNA strand. The pathway at the right shows the intertwining between the nascent RNA and the template DNA strand mediated by RNAP unbinding from the non-template DNA strand followed by rotation around the template DNA strand.
If the RNA tail is very long, its passage through the small space formed during R-loop initiation would be very difficult, especially since the nascent RNA might also form branched secondary structures that could encumber passage. In accord with that, it has been reported that R-loop formation in mammalian cells predominantly occurs near the nascent RNA free end (52,53), and it was suggested that this proximity to the nascent RNA free end is essential for R-loop formation (52).
Although it is clear that proximity of a free RNA end facilitates R-loop formation, alternative pathways for R-loop formation that are independent of a free RNA end could be possible. For example, that might happen if the RNAP could transiently unbind from the non-template DNA strand (Fig.1, right), allowing RNAP to rotate relative to the template DNA strand without involving the non-template DNA strand: Consequently, the front end of the nascent RNA would be able to wind around the template DNA strand, while the non-template DNA strand could remain unwound. This pathway for R-loop formation eliminates the need for the RNA tail to pass through the R-loop. The fact that transcription in some in vitro systems can proceed in the absence of the non-template strand (54), as well as the increased availability of the non-template strand for hybridization with a complementary probe sequence during transcription (55) argues in favor of the possibility that the non-template DNA strand can be transiently released from RNAP during transcription.
Both mechanisms for resolving topological problems require overcoming energy barriers, due either to a bulky RNA tail “squeezing” through an R-loop, or to disruption of interactions between the RNAP and the non-template DNA strand. These energy barriers are likely to make transcription in the “R-loop-mode” more prone to spontaneous pausing or termination than that in the “normal mode” (56).
As noted above, the topological challenges exist because the nascent RNA must wind around the template DNA strand while the non-template DNA strand is rendered unwound. If the non-template strand were “allowed” to wind around the growing RNADNA duplex at the same rate as the RNA winds around the template DNA strand, there would be no topological problem. In that case, R-loop formation would require neither nascent RNA “tail” threading, nor RNAP detachment from the non-template DNA strand. Therefore, R-loop initiation would not depend upon the distance from promoters. However, such tight winding of the non-template DNA strand around a duplex would be energetically costly due to loss of single-stranded DNA conformational entropy (56). This energetic cost could be (partially) compensated if the non-template DNA strand could interact with the RNA-DNA duplex to form a triplex-like structure. Such triple-stranded structures (sometimes referred to as “collapsed R-loops”) have been proposed (49,57); for example, it was hypothesized that the displaced non-template DNA strand could be localized in the minor groove of the RNA-DNA hybrid, in which the bases from this strand may form hydrogen bonds with hydroxyl groups from ribose (57). A recent report suggests that mechanically stretched single-stranded DNA can interact with the homologous DNA duplex (58). A similar interaction might occur between the displaced DNA strand and the RNA-DNA duplex within an R-loop, reducing the energetic “cost” for the displaced DNA strand to wind around the RNA-DNA duplex.
Similarly, there would be no topological problems if the RNA-DNA hybrid within an R-loop formed a structure in which the net winding is zero. For example, this could occur if the RNA-DNA hybrid contained alternating right- and left-left-handed (Z) helices; note that Z-helix formation within RNA-DNA hybrids has been demonstrated experimentally (59). Although Z-prone sequences (comprising alternating purines and pyrimidines) are not expected to be R-loop-prone, the alternation of R-loop-prone and Z-prone sequences could permit formation of an R-loop, within which the RNA-DNA duplex overall is topologically unwound.
In Fig.1, we consider the situation, in which the nascent RNA is hybridized within an open DNA region immediately upstream of the transcription complex without a double-stranded DNA region between the R-loop initiation site and the transcription complex. In principle, R-loop initiation could occur within a double-stranded DNA region sufficiently far upstream from the transcription complex, and consequently there would be a double-stranded DNA region (which we term a “spacer duplex”) between the transcription complex and the R-loop initiation site. This situation is analyzed in the Appendix. Briefly, this analysis predicts that the front of the R-loop would tend to propagate with the same speed as the transcription complex, and consequently, the length of the spacer duplex would not change.
An interesting question related to R-loop formation is whether RNAP could exit the “R-loop mode” and resume “normal” transcription accompanied by nascent RNA separation from the DNA template, while the nascent RNA “tail” remains bound (“anchored”) to the DNA via R-loop formation (Fig.2, left top).
Fig.2. Possible modes of behavior for transcription after stable R-loop formation.
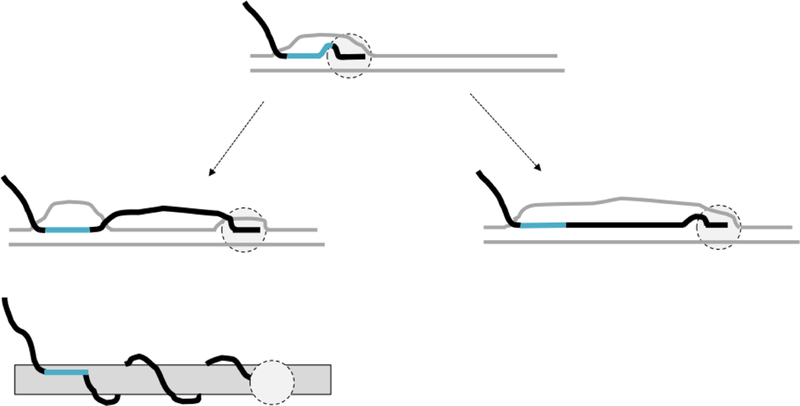
DNA is shown in gray lines, RNA is shown in black lines, except the region involved in stable R-loop formation, which is shown in turquoise. RNAP is shown as light gray oval with dashed-line borders. After stable R-loop formation (top), transcriptions could either exit “R-loop mode”(left), or continue in “R-loop mode” (right). Topological scheme below the pathway in the left shows that in this case transcription is associated with the nascent RNA wrapping around DNA.
Our analysis (56) indicated that this would be very unlikely due to the high energetic cost for nascent RNA winding around the DNA duplex (Fig.2 left bottom); and instead, transcription should continue in the R-loop mode (Fig.2, right). One of the predictions of this analysis that is that an R-loop could propagate into a non-R-loop-prone sequence (see Appendix for details). The more general prediction is that any form of stable co-transcriptional nascent RNA anchoring to the DNA duplex (e.g., by a bivalent protein like YY1(60) or triplex formation (reviewed in (61))) could cause localized negative supercoiling between the transcription complex and the anchoring site, followed by nascent RNA invasion into the duplex (36,56,62).
2.3. The actual likelihood for R-loop formation
In spite of intense study we still know little in quantitative terms about the actual probabilities of R-loop formation during transcription. The probability of R-loop formation during one round of transcription might be small, even for strongly R-loop-prone sequences, because R-loop formation within an intact DNA duplex is likely to be associated with overcoming a large energy barrier. Within the framework of the thread-back model, this barrier arises because of the requirement for transient unwinding of the DNA duplex to initiate the nascent RNA invasion, and within the framework of the continuous RNA-DNA duplex model, this barrier arises because of the requirement for disruption of interactions between the nascent RNA and the exit channel. As a result, the characteristic period for R-loop formation might be longer than the time required for the transcribing RNAP to pass through the R-loop-prone sequence; so, in most cases this passage would be completed without R-loop formation. In accord with this possibility, our recent results support the view that even for a very R-loop prone sequence localized in close vicinity of the transcription promoter, several transcriptional rounds would pass through this sequence before an R-loop would be formed (51). A notion that the probability of R-loop formation per round of transcription could be low even for R-loop-prone sequences is consistent with the observation in the yeast system that overexpression of RNase H (an enzyme that degrades RNA within RNA-DNA hybrids, and consequently, destroys transcripts sequestered within R-loops (see Section 3)) does not affect the expression of most R-loop-forming genes (63).
Important information about quantitative parameters of R-loop formation in vivo have been reported, in which the steady-state frequencies of R-loop occurrence and the kinetics of R-loop removal (i.e. half-lives of R-loops) in various genomic loci have been measured over the entire genome (64). Since the steady-state frequency of R-loop formation is defined by the ratio of the respective rates of R-loop formation and R-loop removal, these data could be used to estimate the rates of R-loop formation within a given gene.
However, the rates would depend both upon the probabilities of R-loop formation during one round of transcription and the frequencies of transcription in the respective genes.
If one could simultaneously measure both the rates of R-loop formation and the rates of transcription, one might evaluate the probabilities of R-loop formation during one round of transcription (which is reciprocal to the average number of transcriptional rounds preceding R-loop formation).
The number of transcriptional rounds preceding R-loop formation could be an important parameter for R-loop-mediated gene regulation: For example, if R-loop formation inhibits further transcription (see Section 2.4), this number can define the size of the transcriptional “burst”, which is how many transcripts are produced before the gene (temporarily) shuts down.
2.4. Interdependence between R-loop formation and transcription blockage.
R-loops have numerous stimulatory and inhibitory effects upon transcription (reviewed in (22)). The mechanisms of these effects often involve factors that are not components of the transcription machinery (e.g., RNA processing enzymes). The mechanisms can also be indirect, e.g., R-loops could affect chromatin structure and DNA modifications, and these modifications, in turn, could affect transcription (reviewed in (24)).
Examples of direct interference include RNAP collision with R-loops formed during previous round(s) of transcription. Since within an R-loop-prone sequence the RNADNA duplex is more difficult to unwind than the DNA-DNA duplex, it is likely to be more difficult for the RNAP to transcribe through it. Structural differences between RNA-DNA and DNA-DNA duplexes and switching the non-template strand from DNA to RNA upon entering the RNA-DNA duplex within the R-loop also could impede transcription through the R-loop-encumbered region. The RNAP might pause or stall when encountering an RNA-DNA hybrid within an R-loop. In accord with this expectation, partial blockage of transcription elongation by pre-formed R-loops has been observed experimentally (65). Especially strong transcription blockage by pre-formed R-loops occurs if the R-loop is localized in close vicinity to the transcription promoter (51). In this case the R-loop blocks subsequent rounds of transcription, either at initiation or during the transition from initiation to elongation.
Studies in vitro (41,43,48,66) suggest that transcription can also become blocked by R-loop formation in the wake of the RNAP (i.e., R-loop formation emanating from the transcribing RNAP is somehow able to interfere with transcription performed by that polymerase, even though the R-loop is behind it). In contrast to the collision-mediated mechanism, this “intrinsic” mechanism is more difficult to explain: Transcription in the “R-loop mode” resembles replication, in the sense that, in both cases, a long duplex comprising the nascent and the template strand is continuously produced. Thus, if replication doesn’t experience difficulties, why should transcription? As described in Section 2.2, possible reasons include the additional energy barriers for transcription in the R-loop mode due to the topological challenges, arising from the fact that the RNAP (in contrast to DNA polymerase) is normally bound to the non-template DNA strand while copying the template DNA strand. However, this cannot completely explain intrinsic R-loop-mediated transcription blockage, because a break in the non-template strand relaxes the topological problems for transcription in the R-loop mode, without abolishing R-loop mediated transcription blockage (43).
An explanation for this intrinsic R-loop mediated blockage that is consistent with all observations, is that the hybridization between the nascent RNA and the DNA template strand within an R-loop could destabilize the transcription complex; for example, by partial disruption of interactions between the nascent RNA and the RNAP, or by deformations within the transcription complex similar to those caused by hairpin formation within the nascent RNA (67–69). This destabilization could render the RNAP prone to blockage at randomly-occurring weak pausing/termination signals, which would not be significant in the normal transcription mode. This could account for the peculiar pattern of transcription blockage signals (reported in (41,43,48)) that are localized predominantly downstream from the sequence or structure that causes R-loop formation, rather than upstream from it (which would be expected for transcription blockages caused by RNAP collisions with preformed R-loops). According to this model, the role of R-loop formation in transcription blockage resembles the role of RNA hairpin formation during factor-independent transcription termination (reviewed in (70)), in which a hairpin formed by the nascent RNA destabilizes the transcription complex and causes it to terminate at sequences (e.g. oligoT/oligoA stretches), which by themselves only cause weak transcription termination. The hypothesis that an R-loop could play a role similar to that of an RNA hairpin during transcription termination was first suggested for bacterial RNAP (66) and then for T7 phage RNAP (71). The model for transcription blockage mediated by RNA-DNA hybrid formation in the wake of the RNAP is consistent with transcription blockage observed in certain experimental systems with single-stranded DNA substrates (72,73). Other steric and sequence-specific factors might also contribute to blockage and could modulate the blockage patterns for various R-loops; however, the similarity between the blockage patterns produced by R-loops from different causes (48) suggests the existence of a general mechanism that is common for all R-loops.
It is important to emphasize that transcription need not stop immediately after R-loop formation, but rather that the R-loop increases the probability of spontaneous transcription blockage. However, this probability is not necessarily very high, so transcription in the “R-loop mode” might continue for a while before blockage occurs. Important questions related to this mechanism of blockage remain to be answered, e.g., how long might RNAP transcribe in the R-loop mode before blockage, and does the RNAP remain bound or does it dissociate from DNA after blockage?
R-loop-mediated transcription blockage could be involved in transcription termination in eukaryotes. During transcription of most eukaryotic genes, an information-bearing part of the nascent RNA is cleaved and becomes involved in other transactions, while transcription can continue for a substantial distance along the DNA before termination (reviewed in (70)).The site of cleavage is usually defined by a polyadenylation (poly A) signal sequence within the nascent RNA. An R-loop-prone (G-rich) sequence immediately downstream from the poly A cleavage site causes transcription termination, which is abolished by overexpression of RNaseH (that specifically degrades RNA within RNA-DNA hybrids) implicating R-loops in termination (74).
Interestingly, this termination requires the nascent RNA cleavage to occur at a site defined by a poly A cleavage signal (74). That connection could be explained in a similar manner as that for the enhancement of R-loop formation upon reduction of the distance from the transcription start site (42). A shorter RNA tail (in this case produced by cleavage) makes it easier for the RNA to thread into DNA duplex, thus facilitating R-loop formation. R-loop accumulation is generally observed in the polyadenylation-dependent termination regions of human genes, suggesting broad involvement of R-loops in transcription termination (64). However, the mechanism(s) of R-loop formation in some of these regions could be more complex than outlined above, because the R-loops are formed not only downstream, but also upstream from the cleavage site, and in many cases the positions of terminal R-loops do not correlate with an enhanced intrinsic propensity of those sequences to form R-loops (64).
Also, R-loop-mediated transcription blockage could be involved in RNAP pausing near transcription start sites in vivo ((52,75), reviewed in (76)).
The relationship between transcription blockage and R-loop formation can go both ways: R-loop formation can cause transcription blockage, but transcription blockage, either upon encounters with DNA damage (reviewed in (77,78)) or by head-on collisions with replication machinery (79,80) appears to result in R-loop formation.
The question of whether RNAP stalling per se facilitates or opposes R-loop formation is not trivial: While stalling RNAP within an R-loop-prone sequence may increase the probability of R-loop formation, the actively transcribing RNAP creates negative supercoiling in its wake ((81,82), reviewed in (83)) that facilitates R-loop formation; but that negative supercoiling will dissipate when the RNAP is stalled.
The connection between transcription blockage and R-loop formation could be indirect, because various processes triggered by RNAP stalling, e.g., backtracking (84) or displacement of spliceosomes (85), could facilitate R-loop formation. In addition, enzymatic degradation of the RNAP may also occur during blockage (reviewed in (78)), and if that happens within an R-loop-prone sequence, the short RNA-DNA hybrid within the transcription complex might “survive” RNAP removal and initiate invasion of the rest of the nascent RNA into the DNA duplex.
Another explanation for a correlation between transcription blockage and R-loop formation in vivo could stem from the fact that R-loop detection in vivo is often based upon monitoring of RNA-DNA hybrids. RNA-DNA hybrids would be likely to form if transcription proceeds into regions, in which the non-template DNA strand is partially missing due to incomplete replication or excision repair of DNA damage. Note that within an R-loop-prone sequence, any short single-stranded DNA region might facilitate RNA invasion into the adjoining DNA duplex to generate an RNA-DNA hybrid much longer than that single-stranded DNA region. Thus, it cannot be excluded that the correlation between the transcription blockage and the accumulation of RNA/DNA hybrids in vivo might in some cases occur because the same factors that cause transcription blockage (e.g., DNA damage and transcription-replication collisions) also can result in transient stretches of single-stranded DNA. Experiments performed with in vitro systems would be required to determine whether R-loop formation can be induced simply by RNAP stalling.
2.5. Role of transcription-replication collisions in R-loop formation
When transcription and replication occur within the same genomic regions, the respective machineries may collide. However, since such collisions are generally deleterious, strategies have presumably evolved to minimize their occurrences (reviewed in (86–92)).
There are two types of transcription-replication collisions: co-directional collisions (Fig.3A), in which the replication fork and the transcription complex are moving in the same direction, and one of them (usually the replication fork) overtakes the other; and head-on collisions (Fig.3B), in which the replication fork and the transcription complex are moving towards each other (reviewed in (86–92)).
Fig. 3. R-loop formation upon transcription-replication collisions.
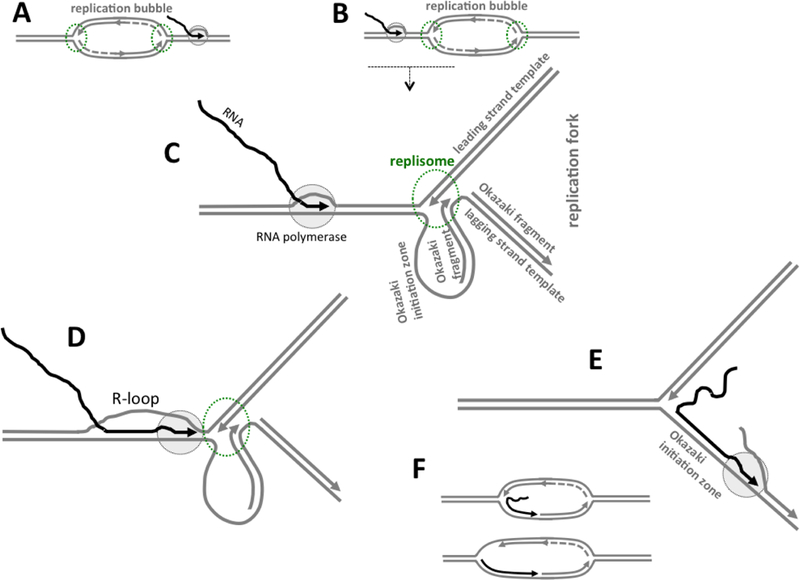
DNA is shown in gray, RNA is shown in black, growing 3’ ends of the nascent DNA or RNA strands are shown by arrows pointed in the direction of growing; RNAP is shown as light-gray oval with black dashed-line borders; replication enzymatic machinery (replisome) is symbolized as an oval area with green dashed-line borders. A: Transcription and replication in co-directional orientation. B: Transcription and replication on the head-on collision course. Replication is usually starts at some small region within the parental DNA duplex called origin, and then the replicated area is expanded forming an unwound region within the parental DNA duplex often referred to as a “replication bubble”. The tip of the replication bubble, which is moving upon the bubble growing, that contains complex multi-protein replication machinery (replisome), is called a “replication fork”. Within the replication fork, one (leading) DNA strand is replicated continuously in the direction of the fork movement, while the other (lagging) DNA strand is replicated discontinuously, forming Okazaki fragments (symbolized by a dashed line). C: More detailed depiction of the region in B underlined by a dashed line with a downward arrow, which shows the replication fork proximal to the transcription complex. The fork is shown in the moment when the synthesis of a new Okazaki fragment (in the loop together with Okazaki initiation zone) is just finished. D: RNAP is stalled upon collision with the replisome that remains intact; and R-loop formation is somehow initiated by RNAP stalling. E: Upon collision with RNAP, the replisome (or its components operating on the lagging DNA strand) dissociate, which allow RNAP to transcribe into single-stranded Okazaki initiation zone, where the likelihood of hybridization between the nascent RNA and the DNA template strand would be very high. Okazaki initiation zone is usually (partially) covered by proteins; however RNAP is probably capable to displace them. F: R-loop-like structures that are formed as a result of pathway in E. Since transcription in R-loop mode is prone to spontaneous blockage, it would probably stop somewhere within the replication bubble. In the bottom structure, the nascent RNA tail invaded into upstream DNA duplex thus extending RNA-DNA hybrid.
Note that the difference between head-on and co-directional collisions is not limited to directionality of the movement per se: In the case of co-directional collisions, the continuously replicating leading DNA strand template serves as a template for transcription; whereas in the case of head-on collision, the discontinuously replicating lagging strand template serves as a template for transcription. In general, both types of collisions could interfere with both transcription and replication; however, the deleterious consequences of head-on collisions are usually more pronounced (reviewed in (86–92)).
An interesting question is how a collision with the replication machinery affects co-transcriptional R-loop formation and its processing. Based upon topological considerations, one might expect that replication would suppress R-loop formation and facilitate preformed R-loop removal, because the extension of the newly-replicated region within the parental DNA duplex requires unwinding of the parental DNA strands within the replicating region. That induces compensatory overwinding (i.e., positive supercoiling) within the DNA outside the replicating region and, as discussed above, positive supercoiling inhibits R-loop formation and facilitates R-loop dissociation. In certain cases the replication machinery might also be able to unwind RNA-DNA hybrids within R-loops. In accord with this expectation, co-directional collisions with replication machinery reduce formation of R-loops ((79), reviewed in (93)), although especially stable R-loops (which may include the stalled RNAP) have been shown to efficiently block replication, even in a co-directional orientation ((94), reviewed in (86)).
However, unexpectedly, it was recently discovered that head-on transcription-replication collisions (schematically shown in Fig.3B and C) increase, rather than decrease R-loop formation in both human cells and bacteria (79,80). It has been suggested that R-loop formation is triggered by RNAP stalling upon collision with the replication fork (Fig.4D) (79,80); however, the broad question of whether R-loop formation could be induced by RNAP stalling per se requires further investigations.
Fig. 4. Potential strategy to induce R-loop formation for selective cell suppression.
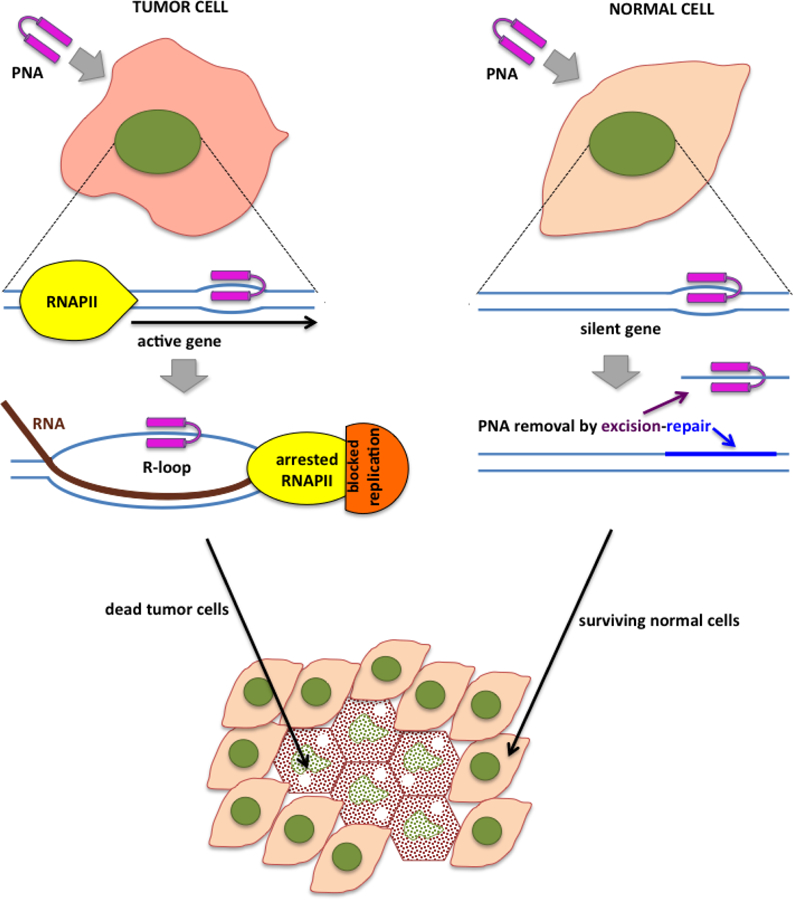
DNA is shown in blue lines, RNA is shown in brown lines, PNA is shown in magenta.
Note that the models suggested in (79,80), as well as in a more recent review (95), imply that upon collision, RNAP does not traverse the replication fork, i.e., it remains outside of the newly replicated region behind the replication fork.
If the replisome remains intact, that would prevent RNAP from traversing the replication fork. However, experimental data with bacteria suggest that upon head-on collision RNAP directly “bumps” into the replisome, rather than stalling some distance away due to the buffering effect of positive supercoiling between the respective machineries (96). Also, it was shown that in bacteria at least some components of the replisome (including DNA helicase, which in bacteria is localized on the lagging DNA strand) could dissociate from the fork upon encounter with an obstacle (97). Thus, similar (partial) disassembly of replication forks might occur upon head-on transcription-replication collisions. In addition, it was shown in yeast, deficient in the RNA-DNA helicase Sen1, that aberrant replication intermediates containing RNA-DNA hybrids were formed upon head-on collision with transcription, suggesting that the RNA-DNA hybrid containing the nascent RNA can be localized behind the replication fork (98). It was also suggested that in eukaryotes, upon head-on collisions, RNAP can displace a lagging strand DNA polymerase and continue transcription (reviewed in (99)). Thus, RNAP might be able to traverse the replication fork and transcribe into the newly-replicated region behind the fork (Fig.3E).
As mentioned above, in the case of a head-on collision, RNAP uses the lagging DNA strand template as a template for transcription. This strand is replicating discontinuously so the newly synthesized DNA strand (the non-template DNA strand for transcription) is comprised of discrete Okazaki fragments, separated by temporary gaps (reviewed in (100–102)). Moreover, due to the delay in replication of the lagging DNA strand in comparison to the leading DNA strand, the former contains a single-stranded region about the size of an Okazaki fragment, sometimes referred to as an “Okazaki initiation zone” (reviewed in (100–103)). Since a single nick in the non-template strand dramatically facilitates R-loop formation, the presence of gaps in that strand would be expected to further enhance R-loop formation. Thus, if upon head-on collision the RNAP manages to transcribe into a “freshly replicated” region behind the replication fork, (in which some gaps between Okazaki fragments and the Okazaki initiation zone are unfilled), then the likelihood of R-loop formation upon transcription through this region would be high. This might provide an additional mechanism for increased R-loop formation upon head-on collisions of transcription with replication.
The RNA-DNA structures formed through the pathway shown in Fig.3E would probably not be simple R-loops: In addition to RNA-DNA hybrids they also might also contain DNA segments hybridized to both the template and the non-template DNA strands that remain from the replication bubble (Fig.3F). However, monitoring R-loop formation by detection of RNA-DNA hybrids would not distinguish these structures from simple R-loops.
In the bottom structure in Fig.3F, the nascent RNA tail invades the DNA duplex (which would be facilitated by R-loop-prone sequences at the 5’-end of the nascent transcript). Such a structure is likely to have an increased stability against unwinding by RNA-DNA helicases (e.g., Sen1 or Senataxin) in comparison with the structure with a single-stranded 5’ RNA end, which would facilitate the helicase loading onto the nascent RNA. Traversing of the replication fork by RNAP need not be the most probable outcome of the head-on collision to increase R-loop formation: If the likelihood of R-loop formation within an intact DNA is sufficiently low, then even the relatively rare occasions in which RNAP traverses the replication fork might substantially increase the yield of R-loops. The recent revelation (79,80) of correlations between transcription-replication collisions and R-loop formation establish a new level of complexity in R-loop biology, in particular the association with DNA damage response pathways mediated by ATR and ATM (79).
3. Cellular processing of R-loops
3.1. Processing by helicases and RNase H nucleases
Multiple resolution mechanisms participate in modulating the presence of R-loops in cells. Degradation of the RNA in the RNA-DNA hybrid is ensured by two enzymes conserved from bacteria to humans, RNase H1 and H2 (104). RNase H2 is also required to remove single ribonucleotides erroneously incorporated during DNA replication and in the Okazaki primers initiated during lagging strand synthesis (105). Unwinding of the RNA from the hybrid is carried out by specialized RNA-DNA helicases (15,22,105,106). The relevance of these R-loop processing mechanisms in preserving normal cellular transactions is documented by the observation that depletion of RNase H or RNA helicases results in increased levels of DNA damage and consequent genomic instability (107,108). Furthermore, mutations in these enzymes are documented in human disease (25,26). For example, it was recently shown that mutations in RNase H1 cause adult-onset neuromuscular disease, which is characterized at the molecular level by R-loop depletion and abnormal DNA segregation of mitochondria(109). Likewise, mutations in the RNA-DNA helicase senataxin (SETX) are linked to neurodegenerative disorders characterized by altered expression of neuronal genes, by accumulation of R-loops and genomic instability (110,111). Several recent reviews detail the activities of these enzymes and their links to human disease (15,22,25,26,106).
The inolvement of R-loops in several cellular processes, ranging from regulation of gene expression and chromatin remodeling to immunoglobulin gene diversification, DNA replication and DNA repair requires that their dissolution be spatially and temporally regulated. Their removal must be avoided while they are needed during normal cellular transactions. The existence of several types of R-loop dissolution mechanisms also ensures that cells have multiple defenses against R-loops that may form erroneously as byproducts of transcription and transcription-replication collisions (discussed in Section 2.5). It will be important to learn more about the cellular functions of these enzymes and their regulation to clarify their respective roles in R-loop biology. In this regard, it was recently shown that the DEAD-box helicase DDX21, which is required in the nucleolus for RNAP I mediated-rRNA gene transcription and processing and in the nucleus for RNAP II transcription elongation (112,113), also efficiently unwinds R-loops, thus preventing accumulation of DNA damage. This activity is regulated through SIRT7 mediated deacetylation (114). A model was proposed in which both SIRT7 and DDX21 are associated with RNAP I and RNAP II, for which SIRT7 maintains DDX21 hypoacetylated. This regulation of DDX21 activity by deacetylation allows it to exert its unwinding activity on co-transcriptionally formed R-loops.
The RNA helicase, Aquarius, is part of the spliceosome, a protein complex dedicated to removing introns upon transcription of mammalian genes, and this protein is required for efficient pre-mRNA splicing (115). Knock down of Aquarius causes R-loop accumulation and genomic instability in human cells, implicating a role in R-loop prevention and/or resolution (116,117). Recently, a number of other helicases, in particular DXH9 have been implicated in R-loop resolution (118). Interestingly, RNA helicases, as well as resolving R-loops, can also facilitate their formation: For example, the DEAD-box RNA helicase DDX1 prepares RNA for R-loop formation by resolving G-quadruplex structures (17).
An interesting question is how a particular helicase may be assigned for these specialized purposes and how the other RNA-DNA helicases and, of course, RNase H can be excluded.
It will be important to assess the spatial and temporal rules that determine how these R-loop processing enzymes exercise their R-loop resolution functions. In relation to this aspect, it was recently shown that the ssDNA-binding protein Replication Protein A (RPA), which binds ssDNA at stalled replication forks and at sites of DNA damage, also interacts with RNase H1 and colocalizes with both RNase H1 and R-loops in human cells (119). Furthermore, RNase H1 mutants defective in RPA binding do not efficiently digest RNA-DNA hybrids in vivo and accumulate R-loops, resulting in genomic instability. It was proposed that the RPA-RNase H1 interaction is necessary to prevent generation of R-loop associated DNA damage and furthermore, that RPA recognizes R-loops before they are converted into double strand breaks. This effect was observed in replicating as well as in non-replicating cells, supporting the view that RPA may play a broad role as a sensor of genomic stress and suppressor of genomic instability (119).
3.2. Processing by DNA repair enzymes and other proteins
Several types of DNA alterations have been shown to cause transcription arrest, and a general mechanism implicating structural changes of the RNAP from transcription competent to incompetent has been proposed (120). These impediments are generally located on the template strand and include bulky lesions, nicks and abasic sites. The strand dependence of the blockage has been explained with respect to the different functions of the template and non-template strands during progression of the RNAP; the template strand actively participates in the nucleotide addition step of transcription whereas the non-template strand provides stability to the transcription complex (121–123). A bulky lesion in or near the nucleotide addition site of the RNAP represents an insurmountable block to polymerase translocation until the impediment is removed.
Lesion removal is mediated by one of several DNA repair mechanisms that exist in all living organisms. Of those, the nucleotide excision repair (NER) pathway is the most ubiquitous, detecting and removing a large variety of DNA adducts and structures. NER comprises two subpathways, global genomic repair (GGR) and transcription-coupled repair (TCR) ((124), reviewed in (125,126)). In TCR, lesion recognition is carried out by the RNAP that acts as a beacon to facilitate detection of lesions, including those that would otherwise be difficult to identify by GGR (e.g. those that stabilize the DNA structure). Thus, the TCR sub-pathway is independent of the XPC and DDB2 proteins involved in lesion recognition for global NER.
The requirement of transcription arrest for initiation of TCR brings up the question of whether TCR may also be initiated at sites of transcription arrest other than those caused by lesions, as in the case of R-loops. Evidence implicating TCR in resolution of R-loops was initially proposed from studies to dissect mechanisms of immunoglobulin heavy chain class switch recombination (127,128). Transcription through these highly repetitive regions generates R-loops of sizes extending to 1 kilobase in length (40). These naturally-formed R-loops were shown to be cleaved by the excision repair structure specific nucleases XPG and XPF, when these sequences were transcribed in a cell free system (128). Both nucleases efficiently cut the displaced non-template G-rich strand in the Rloop. In addition, XPF also cleaved the RNA-DNA hybrid-containing strand, suggesting that it has a less stringent single strand DNA requirement than XPG. Although it was later shown that processing of R-loops formed in these regions is mediated by activation induced cytidine deaminase rather than TCR (129), this work highlights the possibility that these enzymes from the NER pathway might cleave bubble structures formed by RNA-DNA hybrids in cells. This is consistent with previous reports showing that in cell free-systems XPF and XPG structure-specific nucleases can cleave pre-formed bubble junctions without the need for other protein factors (130,131). However, cleavage by XPG and XPF during canonical NER is a tightly regulated process requiring the coordinated action of several proteins and protein complexes (132). For example, recruitment of XPF to damaged sites is dependent upon XPA (133,134). XPG interaction with TFIIH is necessary for its localization to the damaged site as well as for correct positioning of this protein for 3’ cleavage and completion of NER (135,136). In addition, DNA cleavage by XPF and XPG is always preceded by formation of the NER complex (137), implying that if R-loops are processed by NER, other proteins may be needed in vivo. It will be important to test the activities of these enzymes in cell extracts to determine whether other NER factors normally required for DNA cleavage by XPG and XPF may be participating in the process and/or whether additional proteins may be involved in the resolution of these structures. The large size of the RNA:DNA hybrids formed in the immunoglobulin region suggests that if other NER proteins are involved, the steps for this processing may likely be altered, potentially leading to mutations. This processing would likely be independent of XPC, as shown for the repair of DNA lesions located in pre-formed DNA bubbles (138,139). However, the R-loop bubble is unique in that there is no lesion within it, and the bubble is more complicated by having an RNA/DNA hybrid on one side.
Further insights into the potential involvement of NER (and TCR in particular) into R-loop processing has come from recent studies designed to explain the accumulation of strand breaks in human cells depleted of RNA-DNA helicases Aquarius (AQR) or Senataxin (SETX), or by splicing factor ASF/SF2, or after treatment with the topoisomerase I inhibitor, camptothecin. The processing of R-loops into strand breaks was shown to require both of the structure specific nucleases XPF and XPG (116). Furthermore, it was shown that the NER factor XPA and the TFIIH helicases XPB and XPD were required for this processing. These findings suggest that R-loops generated when cells are deficient in mRNA processing factors or Topoisomerase I are converted into strand breaks by an NER-like mechanism. Based upon the requirement of the TCR specific factor CSB but not the global genome-NER factor XPC it was also proposed that this processing was mediated by a TCR-like mechanism. It will be important to determine whether the observed requirement for CSB depends upon the unique CSB role in TCR or on other functions. The use of cells from UV sensitive syndrome patients, which are exclusively deficient in TCR (e.g deficient in the UVSSA gene), might help in dissecting the TCR involvement in this processing (140). (Note that TCR is likely to be important for R-loop resolution only if enzymes that normally resolve R-loops (e.g. helicases) are insufficiently active. If the canonical TCR pathway is involved, this would be the first example in which there is no immediate blocking lesion on the template strand or structure encountered by the polymerase (unless it was another polymerase from the one that had been stalled by an R-loop). Other studies summarized in recent excellent reviews further add to this complexity by showing that crosstalk between proteins from other pathways occurs in processing unscheduled R-loops (31,141). Although further studies will be needed to reveal the details of this processing, these data suggest that unscheduled R-loops may be handled by protein factors other than the RNase H or helicases that are normally involved in resolving R-loops.
Another connection between TCR, splicing factors and R-loops was recently reported in quiescent human dermal fibroblasts, in which it was found that UV-induced transcription blocking lesions promote displacement of core components of the spliceosome, with consequent formation of R-loops. In addition it was shown that R-loop formation signaled a non-canonical ATM-induced DNA damage response (DDR), [or, more broadly, a genomic stress response (GSR) (142)] through a mechanism independent of double strand break formation or replication (143). DDR activation further increased spliceosome displacement at sites of RNAP II arrest and alternative pre-mRNA splicing genome wide. It was proposed that removal of spliceosome factors from transcribed regions is necessary for retrograde translocation of RNAP, an intermediate step in the process of TCR that renders the lesion accessible for repair (143,144). This step is mediated by TFIIS, a transcription elongation factor that promotes RNAP II reverse translocation and selective degradation of the nascent RNA product for two purposes; to reposition the transcript 3’ end to the RNAP II catalytic site to restart transcription after completion of repair, and to render the damage-containing strand accessible for cleavage by the structure specific nucleases XPG and XPF. It is not clear whether or when an R-loop might form during TCR or how resolution of this structure occurs. However, these data imply that as with the immunoglobulin genes, TCR induced R-loops may be an intermediate step in the process. These are examples of how, in addition to helicases and Rnase H, the action of NER on R-loops can have a positive outcome. Finally, it is provocative to learn that RNA-DNA hybrids, in addition to being substrates for repair, also could mediate repair of DNA lesions, as is the case for double-strand breaks (145–148).
4. Artificially targeted R-loops: harnessing “bad” R-loops for practical purposes.
Naturally occurring R-loops have been broadly labeled “good” or “bad”, depending upon their respective useful or deleterious effects (reviewed in (9,15,22–30)). One thing in common for all R-loops (and for RNA-DNA duplexes in general) is that their RNA component must eventually be removed. Even “good” R-loops, if not processed within a reasonable time frame, will interfere with DNA transactions (e.g., transcription and replication) to result in “bad” outcomes. Furthermore, if R-loops are not dissolved in the appropriate manner by specialized nucleases or helicases they can be processed into highly lethal double-strand breaks (116) (see Section 3.2).
A persistent and difficult-to-remove R-loop should cause deleterious (toxic) effects, particularly if it provokes collisions with translocating polymerases and/or attracts enzymes that cut the DNA strands in non-productive attempts at repair. In principle, such cellular toxicity might be artificially targeted to selected cells if stable R-loops can be localized to genes, which are expressed in those cells but not in the population of cells to be spared. This obviously brings up the challenge for cancer therapies, in which the cells in a tumor and its metastatic cells display unique transcriptomes in comparison to those of the normal cells from which they arose. We have proposed an approach for cancer therapy based upon this model, in which artificially targeted R-loops in particular expressed genes might selectively inactivate tumor cells, based upon their unique transcription profiles (48).
The general scheme for our proposed approach is illustrated in Fig.4.
In this example, R-loop formation is induced by sequestration of the non-template DNA strand with peptide nucleic acid (PNA), an artificial DNA mimic that can invade double-stranded DNA to bind its complementary sequence and render the complementary DNA strand unpaired ((149), reviewed in (150)). The stable R-loop is then generated upon transcription through the PNA-bound sequence. This approach for R-loop formation would be very efficient, because PNA creates a stable unwound DNA region (bubble), thus removing the energetic barrier for nascent RNA invasion into DNA duplex. PNA-induced R-loops would be difficult to eliminate, in part because their loss would be transient and would with high probability re-appear again by the next round of transcription. Therefore, this type of stable R-loop is expected to be toxic for the targeted cell.
Because co-transcriptional R-loops are formed only in transcribed regions, this approach would selectively affect the cells in which the targeted gene is transcribed. An additional factor that should enhance the selectivity of this approach is that PNA invades actively transcribed DNA regions much more efficiently, especially if the PNA-binding site is localized on the non-template DNA strand (55). Although a PNA designed to bind the template DNA strand will also block transcription, it is more likely to be removed through transcription-coupled repair than one bound to the non-transcribed strand. If excision-repair were attempted to remove the bound PNA in an R-loop, the other strand encumbered in an RNA-DNA hybrid would be unavailable as a template for repair replication. Excision-repair normally recognizes and excises lesions within the double-stranded DNA regions, rather than in long single-stranded regions (which would be the case for PNA bound to the non-template strand within an R-loop). Thus, the consequences of PNA bound within an R-loop (Fig.4, left) would be much more difficult to resolve than for PNA bound within an intact DNA duplex (Fig.4, right).
In general, an R-loop structure, in which both DNA strands are sequestered, is likely to be highly deleterious (141).
We predict that PNA-induced R-loop formation would be much more efficient than R-loop formation within an intact R-loop-prone DNA sequence, especially in the cases, in which R-loop formation occurs far from the transcription start site. This expectation is based upon a notion, that to initiate R-loop formation within an intact DNA duplex, the nascent RNA “tail” must “thread” into a transiently unpaired DNA region formed due to DNA “breathing”. Since disruption of DNA base pairing is energetically unfavorable under physiological conditions, this region is likely to be very short (probably, only a few base pairs), and it would rapidly collapse into the double helix. So it would be challenging for a RNA tail to thread into it, especially if this tail is long. In contrast to the situation for DNA “breathing”, the PNA binding to the non-template DNA strand creates a stable and relatively long unpaired region within the DNA template strand, providing an opportunity for stable “nucleation” of the RNA-DNA hybrid and much longer region to “thread through” for the RNA tail; consequently, R-loops could be efficiently initiated even far downstream from the transcription start site. In accord with this, our recent data document efficient PNA-induced R-loop formation for a PNA binding site localized over 200 bp downstream from the transcription start site (151).
An important question in relation to our proposed approach for R-loop-mediated selective cell toxicity, is to learn how many stable R-loops are required to substantially impact viability. Although we do not have a quantitative answer to this question, it has been shown that convergent transcription within a single non-essential gene containing certain microsatellite repeats can result in substantial cell death by triggering apoptosis (141,152,153); the accompanying controls implied that this deleterious situation was caused by formation of a double-R-loop (a structure in which both complementary DNA strands are bound to RNA) (141,152). The evidence that only one R-loop-like structure within a single non-essential gene could cause substantial cell death gives credibility that our proposed approach, with stable R-loop formation within only one or a few genomic locations may be sufficient to inflict substantial lethality.
It is important to emphasize that a unique feature of our proposed approach in comparison with other methods for selective gene inhibition (e.g., antisense or antigene approaches) is that by inducing R-loop formation, the very act of transcription should become toxic for the selected cells, whether the products of the targeted genes are essential or not important for cell viability. That fact dramatically increases the number of possible targets. This novel approach might be used to selectively inactivate tumor cells, in which multiple genes are intensively transcribed in comparison with those same genes in normal cells.
5. Challenges for future research on R-loops
Although the R-loop field is exploding with new information and examples, many important mechanistic aspects of R-loop formation and processing require further analysis. Among these are elucidation of the detailed mechanism of RNA invasion into DNA, quantitative estimates of the probabilities of R-loop formation under given circumstances, and the relationships between R-loop formation and transcription blockage. There could also be pathways for R-loop formation yet to be revealed (e.g., R-loop formation mediated by nascent RNA anchoring). It is provocative that enzymes involved in the canonical processes of nucleotide excision repair (and particularly transcription-coupled repair) have been implicated in the processing of R-loops. It will be important to determine whether an R-loop or the conditions under which R-loops are initiated are sufficient to induce excision-repair. The involvement of CSB is of particular interest in that it not only is required for canonical TCR, but that it has other functions in addition to that role. The possible requirement for UVSSA is also of interest, as it is not known to have any other function except that in TCR.
We have hypothesized that artificially induced stable R-loops might be employed to render the very act of transcription toxic for selected cells (such as those in a tumor) based upon their unique transcription profiles. There may also be other applications of artificially generated R-loops to be explored in future studies.
Acknowledgements:
We thank Frederic Chedin and Irina Artsimovitch for helpful discussions. We also thank Graciela Spivak and Sam Wilson for reading the manuscript and offering suggestions.
This research was supported by a grant (CA077712) from the National Cancer Institute to P.C.H., a Stanford Cancer Institute Fellowship Award (PTA 1164311-123-GHTDS) to B.P.B., and a Stanford Summer Undergraduate Research grant to A.D.D.
Abbreviations:
- RNAP
RNA polymerase
- ATR
ataxia telangiectasia and Rad3-related
- ATM
ataxia telangiectasia mutated
- NER
nucleotide-excision repair
- GGR
global genomic repair
- TCR
transcription-coupled repair
- XP(G, F, etc.)
xeroderma pigmentosum (protein G, F, etc.)
- GSR
genomic stress response
- DDR
DNA damage response
- CSB
Cokayne syndrome protein B
- UV
ultraviolet (light)
- UVSSA
UV stimulated scaffold protein A
- PNA
peptide nucleic acid
Appendix: Consequences of nascent RNA anchoring for R-loop formation.
Consider the situation in which a stable R-loop has formed co-transcriptionally within an R-loop-prone DNA region, but that the downstream DNA sequence is not R-loop-prone. Could RNAP, after passing through the R-loop-prone region, exit the “R-loop mode” and resume “normal” transcription accompanied by the nascent RNA separation from the DNA template, except for the R-loop-prone region, in which it remains bound (“anchored”) to the DNA (Fig.2, left)?
Thermodynamic considerations (56) predict that the transcription mode shown in Fig.2 (left) is very unlikely: Upon normal transcription, RNAP follows a helical path along DNA. Thus, if the nascent RNA becomes anchored to the DNA at some point, this would cause it to wrap around the DNA duplex region localized between the anchoring point and the transcription complex (Fig.2, bottom left). Our calculations show that to accommodate this wrapping, the nascent RNA (which normally adopts a loose random coil configuration) would have to be stretched almost up to its contour length. Such stretching dramatically reduces the number of possible spatial conformations that would be available for the nascent RNA, (i.e. it decreases the nascent RNA entropy.) Since a decrease in entropy is thermodynamically unfavorable, the nascent RNA wrapping around DNA would create counter-acting forces, which, according to our estimations (56), are strong enough to unwind the DNA duplex, thus promoting hybridization between the nascent RNA and the DNA template strand. (These forces could be interpreted as very high negative superhelical strain, which appears as soon as RNAP attempts to exit an R-loop mode.) Consequently, as long as an “anchoring” RNA-DNA hybrid is not disrupted, transcription is “forced” to continue in the “R-loop mode” (Fig.2, right). Thus, during transcription an R-loop could propagate into DNA regions that are not intrinsically R-loop-prone.
What is the fate of an R-loop that has extended into a non-R-loop-prone sequence, after transcription termination and RNAP dissociation?
If the RNAP dissociates from DNA while transcription proceeds in the “R-loop mode” through the non-R-loop-prone sequence, the nascent RNA could be displaced from that sequence at downstream flank of the R-loop by the non-template DNA strand.
In contrast, if transcription in the “R-loop mode” has passed through the non-R-loop-prone sequence into an R-loop-prone sequence before RNAP dissociation, the RNA/DNA hybrid within that sequence is likely to remain intact after RNAP dissociation, because the displacement would be blocked by R-loop-prone flanking sequences. These predictions are in accord with the results that on one hand, R-loops can extend through a non-R-loop-prone sequence inserted into a longer R-loop-prone region (154); but on the other hand, the locations of downstream boundaries of R-loops correlate with reduced R-loop-forming potential (155).
Importantly, the prediction that stable R-loop initiation will force transcription to continue in the “R-loop mode” regardless of sequence (Fig.2, right) is applicable for all pathways of R-loop initiation in which the DNA strands remain intact. For example, it is applicable for R-loop initiation by some agent that locally unwinds the DNA duplex and sequesters the non-template DNA strand. However, it is not applicable to R-loop initiation by a single-strand break (nick) in the non-template strand, because the nick creates a “swivel” that would allow nascent RNA unwrapping from DNA that, in principle, would allow transcription to proceed in the mode shown in Fig.2, left.
In Figs. 1, 2 we consider the situation in which R-loop formation occurs closely behind the transcription complex. What would happen if R-loop initiation occurs within a double-stranded DNA region sufficiently far upstream from the transcription complex, and consequently there would be a double-stranded DNA region (which we term a “spacer duplex”) between the transcription complex and the R-loop initiation site?
Because of the helical structure of DNA, both RNAP and an R-loop rotate relative to the spacer duplex upon their propagation along DNA. Consequently, if RNAP propagates either faster or slower than the front edge of an R-loop, then the nascent RNA would wind around the spacer duplex either in the right-handed fashion, or the left-handed fashion, respectively.
As we discussed above, the nascent RNA winding around the DNA duplex is energetically costly; thus, RNAP and the front of an R-loop would tend to propagate with the same speed to avoid this winding. Consequently, the length of the spacer duplex would tend to remain constant.
Note that the effect of the nascent RNA winding around the spacer duplex could be interpreted in terms of DNA supercoiling: when the RNA wound around the spacer duplex in either right-handed, or left-handed way would “try” to unwind itself, it would create either negative, or positive supercoiling within the spacer duplex, respectively. Consequently, the presence of Topoisomerase IB (Topo IB) activity, that can relax DNA supercoiling of either sign, could (partially) “uncouple” the rate of R-loop propagation from the rate of transcription complex movement, thus allowing the change in the length of the spacer DNA duplex.
It has been suggested that negative supercoiling generated by the nascent RNA anchoring renders RNAP-containing R-loops much more stable than “free R-loops” toward RNA-displacing factors, e.g. positive supercoiling (95). However, the negative supercoiling due to anchoring is localized between the downstream end of the R-loop and the transcription complex, and would not prevent RNA displacement from the upstream end of the R-loop. Thus, very high intrinsic stability of the anchoring RNA-DNA duplex over the DNA-DNA duplex at the upstream flank of an R-loop is required for R-loop “survival” under positive superhelical stress or other RNA-displacing factors even with the RNAP at the other end of an R-loop.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflicts of interest
References
- 1.Daube SS and von Hippel PH (1994) RNA displacement pathways during transcription from synthetic RNA-DNA bubble duplexes. Biochemistry, 33, 340347. [DOI] [PubMed] [Google Scholar]
- 2.Yin YW and Steitz TA (2004) The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell, 116, 393–404. [DOI] [PubMed] [Google Scholar]
- 3.Jiang M, Ma N, Vassylyev DG and McAllister WT (2004) RNA displacement and resolution of the transcription bubble during transcription by T7 RNA polymerase. Mol Cell, 15, 777–788. [DOI] [PubMed] [Google Scholar]
- 4.Liu X, Bushnell DA and Kornberg RD (2013) RNA polymerase II transcription: structure and mechanism. Biochim Biophys Acta, 1829, 2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korzheva N and Mustaev A (2001) Transcription elongation complex: structure and function. Curr Opin Microbiol, 4, 119–125. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Rucobo FW and Cramer P (2013) Structural basis of transcription elongation. Biochim Biophys Acta, 1829, 9–19. [DOI] [PubMed] [Google Scholar]
- 7.Ehara H, Yokoyama T, Shigematsu H, Yokoyama S, Shirouzu M and Sekine SI (2017) Structure of the complete elongation complex of RNA polymerase II with basal factors. Science, 357, 921–924. [DOI] [PubMed] [Google Scholar]
- 8.Hamperl S and Cimprich KA (2014) The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst) [DOI] [PMC free article] [PubMed]
- 9.Aguilera A and Garcia-Muse T (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell, 46, 115–124. [DOI] [PubMed] [Google Scholar]
- 10.Gowrishankar J, Leela JK and Anupama K (2013) R-loops in bacterial transcription: their causes and consequences. Transcription, 4, 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy D and Lieber MR (2009) G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol, 29, 3124–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nudler E (2012) RNA polymerase backtracking in gene regulation and genome instability. Cell, 149, 1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahba L, Gore SK and Koshland D (2013) The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife, 2, e00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahba L and Koshland D (2013) The Rs of biology: R-loops and the regulation of regulators. Mol Cell, 50, 611–612. [DOI] [PubMed] [Google Scholar]
- 15.Costantino L and Koshland D (2015) The Yin and Yang of R-loop biology. Curr Opin Cell Biol, 34, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang F and Doudna JA (2017) CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys [DOI] [PubMed]
- 17.Ribeiro de Almeida C, Dhir S, Dhir A, Moghaddam AE, Sattentau Q, Meinhart A and Proudfoot NJ (2018) RNA Helicase DDX1 Converts RNA GQuadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol Cell [DOI] [PMC free article] [PubMed]
- 18.Thomas M, White RL and Davis RW (1976) Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A, 73, 22942298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White RL and Hogness DS (1977) R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell, 10, 177–192. [DOI] [PubMed] [Google Scholar]
- 20.Berget SM, Moore C and Sharp PA (1977) Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A, 74, 3171–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chow LT, Gelinas RE, Broker TR and Roberts RJ (1977) An amazing sequence arrangement at the 5’ ends of adenovirus 2 messenger RNA. Cell, 12, 1–8. [DOI] [PubMed] [Google Scholar]
- 22.Santos-Pereira JM and Aguilera A (2015) R loops: new modulators of genome dynamics and function. Nat Rev Genet, 16, 583–597. [DOI] [PubMed] [Google Scholar]
- 23.Sollier J and Cimprich KA (2015) Breaking bad: R-loops and genome integrity. Trends Cell Biol, 25, 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chedin F (2016) Nascent Connections: R-Loops and Chromatin Patterning. Trends Genet [DOI] [PMC free article] [PubMed]
- 25.Richard P and Manley JL (2016) R Loops and Links to Human Disease. J Mol Biol [DOI] [PMC free article] [PubMed]
- 26.Groh M and Gromak N (2014) Out of balance: R-loops in human disease. PLoS Genet, 10, e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Usdin K and Kumari D (2015) Repeat-mediated epigenetic dysregulation of the FMR1 gene in the fragile X-related disorders. Front Genet, 6, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim N and Jinks-Robertson S (2012) Transcription as a source of genome instability. Nat Rev Genet, 13, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skourti-Stathaki K and Proudfoot NJ (2014) A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev, 28, 1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sebastian R and Oberdoerffer P (2017) Transcription-associated events affecting genomic integrity. Philos Trans R Soc Lond B Biol Sci, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freudenreich CH (2018) R-loops: targets for nuclease cleavage and repeat instability. Curr Genet [DOI] [PMC free article] [PubMed]
- 32.Boulianne B and Feldhahn N (2018) Transcribing malignancy: transcription-associated genomic instability in cancer. Oncogene, 37, 971–981. [DOI] [PubMed] [Google Scholar]
- 33.Mirkin SM (2008) Discovery of alternative DNA structures: a heroic decade (1979–1989). Front Biosci, 13, 1064–1071. [DOI] [PubMed] [Google Scholar]
- 34.Wang G and Vasquez KM (2014) Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair (Amst), 19, 143151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G and Vasquez KM (2017) Effects of Replication and Transcription on DNA Structure-Related Genetic Instability. Genes (Basel), 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belotserkovskii BP, Mirkin SM and Hanawalt PC (2013) DNA sequences that interfere with transcription: implications for genome function and stability. Chem Rev, 113, 8620–8637. [DOI] [PubMed] [Google Scholar]
- 37.Vanoosthuyse V (2018) Strengths and Weaknesses of the Current Strategies to Map and Characterize R-Loops. Noncoding RNA, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy D, Yu K and Lieber MR (2008) Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol, 28, 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daniels GA and Lieber MR (1995) RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res, 23, 5006–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu K, Chedin F, Hsieh CL, Wilson TE and Lieber MR (2003) R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol, 4, 442–451. [DOI] [PubMed] [Google Scholar]
- 41.Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM and Hanawalt PC (2010) Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci U S A, 107, 1281612821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy D, Zhang Z, Lu Z, Hsieh CL and Lieber MR (2010) Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol Cell Biol, 30, 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belotserkovskii BP, Neil AJ, Saleh SS, Shin JH, Mirkin SM and Hanawalt PC (2013) Transcription blockage by homopurine DNA sequences: role of sequence composition and single-strand breaks. Nucleic Acids Res, 41, 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masse E and Drolet M (1999) Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J Biol Chem, 274, 16659–16664. [DOI] [PubMed] [Google Scholar]
- 45.Grabczyk E, Mancuso M and Sammarco MC (2007) A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res, 35, 5351–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duquette ML, Handa P, Vincent JA, Taylor AF and Maizels N (2004) Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev, 18, 1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neil AJ, Liang MU, Khristich AN, Shah KA and Mirkin SM (2018) RNA-DNA hybrids promote the expansion of Friedreich’s ataxia (GAA)n repeats via break-induced replication. Nucleic Acids Res [DOI] [PMC free article] [PubMed]
- 48.Belotserkovskii BP and Hanawalt PC (2015) PNA binding to the non-template DNA strand interferes with transcription, suggesting a blockage mechanism mediated by R-loop formation. Mol Carcinog, 54, 1508–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reaban ME, Lebowitz J and Griffin JA (1994) Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J Biol Chem, 269, 21850–21857. [PubMed] [Google Scholar]
- 50.Wahba L, Costantino L, Tan FJ, Zimmer A and Koshland D (2016) S1DRIP-seq identifies high expression and polyA tracts as major contributors to Rloop formation. Genes Dev, 30, 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belotserkovskii BP, Soo Shin JH and Hanawalt PC (2017) Strong transcription blockage mediated by R-loop formation within a G-rich homopurine-homopyrimidine sequence localized in the vicinity of the promoter. Nucleic Acids Res, 45, 6589–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L, Chen JY, Zhang X, Gu Y, Xiao R, Shao C, Tang P, Qian H, Luo D, Li H et al. (2017) R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol Cell, 68, 745–757 e745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dumelie JG and Jaffrey SR (2017) Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milligan JF, Groebe DR, Witherell GW and Uhlenbeck OC (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res, 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsen HJ and Nielsen PE (1996) Transcription-mediated binding of peptide nucleic acid (PNA) to double-stranded DNA: sequence-specific suicide transcription. Nucleic Acids Res, 24, 458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belotserkovskii BP and Hanawalt PC (2011) Anchoring nascent RNA to the DNA template could interfere with transcription. Biophys J, 100, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karamychev VN, Panyutin IG, Neumann RD and Zhurkin VB (2000) DNA and RNA folds in transcription complex as evidenced by iodine-125 radioprobing. J Biomol Struct Dyn, 17 Suppl 1, 155–167. [DOI] [PubMed] [Google Scholar]
- 58.Chen J, Tang Q, Guo S, Lu C, Le S and Yan J (2017) Parallel triplex structure formed between stretched single-stranded DNA and homologous duplex DNA. Nucleic Acids Res, 45, 10032–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bae S, Kim Y, Kim D, Kim KK, Kim YG and Hohng S (2013) Energetics of Z-DNA binding protein-mediated helicity reversals in DNA, RNA, and DNA-RNA duplexes. J Phys Chem B, 117, 13866–13871. [DOI] [PubMed] [Google Scholar]
- 60.Jeon Y and Lee JT (2011) YY1 tethers Xist RNA to the inactive X nucleation center. Cell, 146, 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, Syed J and Sugiyama H (2016) RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem Biol, 23, 1325–1333. [DOI] [PubMed] [Google Scholar]
- 62.Belotserkovskii BP (2014) Relationships between the winding angle, the characteristic radius, and the torque for a long polymer chain wound around a cylinder: Implications for RNA winding around DNA during transcription. Physical Review E, 89. [DOI] [PubMed] [Google Scholar]
- 63.Hartono SR, Malapert A, Legros P, Bernard P, Chedin F and Vanoosthuyse V (2018) The Affinity of the S9.6 Antibody for Double-Stranded RNAs Impacts the Accurate Mapping of R-Loops in Fission Yeast. J Mol Biol, 430, 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, Xu X and Chedin F (2016) Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell, 63, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tous C and Aguilera A (2007) Impairment of transcription elongation by R-loops in vitro. Biochem Biophys Res Commun, 360, 428–432. [DOI] [PubMed] [Google Scholar]
- 66.Tomizawa J and Masukata H (1987) Factor-independent termination of transcription in a stretch of deoxyadenosine residues in the template DNA. Cell, 51, 623–630. [DOI] [PubMed] [Google Scholar]
- 67.Kang JY, Mishanina TV, Bellecourt MJ, Mooney RA, Darst SA and Landick R (2018) RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing. Mol Cell, 69, 802815 e801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo X, Myasnikov AG, Chen J, Crucifix C, Papai G, Takacs M, Schultz P and Weixlbaumer A (2018) Structural Basis for NusA Stabilized Transcriptional Pausing. Mol Cell, 69, 816–827 e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Artsimovitch I and Belogurov GA (2018) Uneven Braking Spins RNA Polymerase into a Pause. Mol Cell, 69, 723–725. [DOI] [PubMed] [Google Scholar]
- 70.Porrua O, Boudvillain M and Libri D (2016) Transcription Termination: Variations on Common Themes. Trends Genet, 32, 508–522. [DOI] [PubMed] [Google Scholar]
- 71.Gopal V, Brieba LG, Guajardo R, McAllister WT and Sousa R (1999) Characterization of structural features important for T7 RNAP elongation complex stability reveals competing complex conformations and a role for the non-template strand in RNA displacement. J Mol Biol, 290, 411–431. [DOI] [PubMed] [Google Scholar]
- 72.Toulokhonov I and Landick R (2006) The role of the lid element in transcription by E. coli RNA polymerase. J Mol Biol, 361, 644–658. [DOI] [PubMed] [Google Scholar]
- 73.Naryshkina T, Kuznedelov K and Severinov K (2006) The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J Mol Biol, 361, 634–643. [DOI] [PubMed] [Google Scholar]
- 74.Skourti-Stathaki K, Proudfoot NJ and Gromak N (2011) Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell, 42, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eddy J, Vallur AC, Varma S, Liu H, Reinhold WC, Pommier Y and Maizels N (2011) G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res, 39, 4975–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen FX, Smith ER and Shilatifard A (2018) Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol [DOI] [PubMed]
- 77.Mullenders L (2015) DNA damage mediated transcription arrest: Step back to go forward. DNA Repair (Amst), 36, 28–35. [DOI] [PubMed] [Google Scholar]
- 78.Steurer B and Marteijn JA (2016) Traveling Rocky Roads: The Consequences of Transcription-Blocking DNA Lesions on RNA Polymerase II. J Mol Biol [DOI] [PubMed]
- 79.Hamperl S, Bocek MJ, Saldivar JC, Swigut T and Cimprich KA (2017) Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell, 170, 774–786 e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lang KS, Hall AN, Merrikh CN, Ragheb M, Tabakh H, Pollock AJ, Woodward JJ, Dreifus JE and Merrikh H (2017) Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell, 170, 787–799 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu LF and Wang JC (1987) Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A, 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsao YP, Wu HY and Liu LF (1989) Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell, 56, 111–118. [DOI] [PubMed] [Google Scholar]
- 83.Nelson P (1999) Transport of torsional stress in DNA. Proc Natl Acad Sci U S A, 96, 14342–14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dutta D, Shatalin K, Epshtein V, Gottesman ME and Nudler E (2011) Linking RNA polymerase backtracking to genome instability in E. coli. Cell, 146, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, van IWF, Grosveld FG, Medema RH, Hoeijmakers JH et al. (2015) The core spliceosome as target and effector of non-canonical ATM signalling. Nature, 523, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mirkin EV and Mirkin SM (2007) Replication fork stalling at natural impediments. Microbiol Mol Biol Rev, 71, 13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brambati A, Colosio A, Zardoni L, Galanti L and Liberi G (2015) Replication and transcription on a collision course: eukaryotic regulation mechanisms and implications for DNA stability. Front Genet, 6, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pomerantz RT and O’Donnell M (2010) What happens when replication and transcription complexes collide? Cell Cycle, 9, 2537–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garcia-Muse T and Aguilera A (2016) Transcription-replication conflicts: how they occur and how they are resolved. Nat Rev Mol Cell Biol, 17, 553–563. [DOI] [PubMed] [Google Scholar]
- 90.Hamperl S and Cimprich KA (2016) Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell, 167, 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Merrikh H (2017) Spatial and Temporal Control of Evolution through Replication-Transcription Conflicts. Trends Microbiol, 25, 515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Helmrich A, Ballarino M, Nudler E and Tora L (2013) Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol, 20, 412–418. [DOI] [PubMed] [Google Scholar]
- 93.Lin YL and Pasero P (2017) Transcription-Replication Conflicts: Orientation Matters. Cell, 170, 603–604. [DOI] [PubMed] [Google Scholar]
- 94.Krasilnikova MM, Samadashwily GM, Krasilnikov AS and Mirkin SM (1998) Transcription through a simple DNA repeat blocks replication elongation. Embo J, 17, 5095–5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuzminov A (2017) When DNA Topology Turns Deadly - RNA Polymerases Dig in Their R-Loops to Stand Their Ground: New Positive and Negative (Super)Twists in the Replication-Transcription Conflict. Trends Genet [DOI] [PMC free article] [PubMed]
- 96.Mirkin EV and Mirkin SM (2005) Mechanisms of transcription-replication collisions in bacteria. Mol Cell Biol, 25, 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mettrick KA and Grainge I (2016) Stability of blocked replication forks in vivo. Nucleic Acids Res, 44, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M et al. (2012) Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell, 151, 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Keszthelyi A, Minchell NE and Baxter J (2016) The Causes and Consequences of Topological Stress during DNA Replication. Genes (Basel), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang D and O’Donnell M (2016) The Eukaryotic Replication Machine. Enzymes, 39, 191–229. [DOI] [PubMed] [Google Scholar]
- 101.Alberts B (2003) DNA replication and recombination. Nature, 421, 431–435. [DOI] [PubMed] [Google Scholar]
- 102.Leman AR and Noguchi E (2013) The replication fork: understanding the eukaryotic replication machinery and the challenges to genome duplication. Genes (Basel), 4, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mirkin SM (2013) DNA replication: driving past four-stranded snags. Nature, 497, 449–450. [DOI] [PubMed] [Google Scholar]
- 104.Cerritelli SM and Crouch RJ (2009) Ribonuclease H: the enzymes in eukaryotes. FEBS J, 276, 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cornelio DA, Sedam HN, Ferrarezi JA, Sampaio NM and Argueso JL (2017) Both R-loop removal and ribonucleotide excision repair activities of RNase H2 contribute substantially to chromosome stability. DNA Repair (Amst), 52, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hamperl S and Cimprich KA (2014) The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst), 19, 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Amon JD and Koshland D (2016) RNase H enables efficient repair of R-loop induced DNA damage. Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, KamieniarzGdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML et al. (2015) BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell, 57, 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Akman G, Desai R, Bailey LJ, Yasukawa T, Dalla Rosa I, Durigon R, Holmes JB, Moss CF, Mennuni M, Houlden H et al. (2016) Pathological ribonuclease H1 causes R-loop depletion and aberrant DNA segregation in mitochondria. Proc Natl Acad Sci U S A, 113, E4276–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Groh M, Albulescu LO, Cristini A and Gromak N (2017) Senataxin: Genome Guardian at the Interface of Transcription and Neurodegeneration. J Mol Biol, 429, 3181–3195. [DOI] [PubMed] [Google Scholar]
- 111.Becherel OJ, Sun J, Yeo AJ, Nayler S, Fogel BL, Gao F, Coppola G, Criscuolo C, De Michele G, Wolvetang E et al. (2015) A new model to study neurodegeneration in ataxia oculomotor apraxia type 2. Hum Mol Genet, 24, 5759–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Calo E, Flynn RA, Martin L, Spitale RC, Chang HY and Wysocka J (2015) RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature, 518, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Henning D, So RB, Jin R, Lau LF and Valdez BC (2003) Silencing of RNA helicase II/Gualpha inhibits mammalian ribosomal RNA production. J Biol Chem, 278, 52307–52314. [DOI] [PubMed] [Google Scholar]
- 114.Song C, Hotz-Wagenblatt A, Voit R and Grummt I (2017) SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev [DOI] [PMC free article] [PubMed]
- 115.De I, Bessonov S, Hofele R, dos Santos K, Will CL, Urlaub H, Lührmann R and Pena V (2015) The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes. Nat Struct Mol Biol, 22, 138–144. [DOI] [PubMed] [Google Scholar]
- 116.Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A and Cimprich KA (2014) Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell, 56, 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sakasai R, Isono M, Wakasugi M, Hashimoto M, Sunatani Y, Matsui T, Shibata A, Matsunaga T and Iwabuchi K (2017) Aquarius is required for proper CtIP expression and homologous recombination repair. Sci Rep, 7, 13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cristini A, Groh M, Kristiansen MS and Gromak N (2018) RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep, 23, 1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA and Zou L (2017) Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol Cell, 65, 832–847.e834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tornaletti S and Hanawalt PC (1999) Effect of DNA lesions on transcription elongation. Biochimie, 81, 139–146. [DOI] [PubMed] [Google Scholar]
- 121.Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR and Kornberg RD (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science, 288, 640–649. [DOI] [PubMed] [Google Scholar]
- 122.Cramer P, Bushnell DA and Kornberg RD (2001) Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science, 292, 18631876. [DOI] [PubMed] [Google Scholar]
- 123.Gnatt AL, Cramer P, Fu J, Bushnell DA and Kornberg RD (2001) Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science, 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- 124.Mellon I, Spivak G and Hanawalt PC (1987) Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell, 51, 241–249. [DOI] [PubMed] [Google Scholar]
- 125.Hanawalt PC and Spivak G (2008) Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol, 9, 958–970. [DOI] [PubMed] [Google Scholar]
- 126.Spivak G and Ganesan AK (2014) The complex choreography of transcription-coupled repair. DNA Repair (Amst), 19, 64–70. [DOI] [PubMed] [Google Scholar]
- 127.Peters A and Storb U (1996) Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity, 4, 57–65. [DOI] [PubMed] [Google Scholar]
- 128.Tian M and Alt FW (2000) Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem, 275, 24163–24172. [DOI] [PubMed] [Google Scholar]
- 129.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y and Honjo T (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell, 102, 553–563. [DOI] [PubMed] [Google Scholar]
- 130.O’Donovan A, Davies AA, Moggs JG, West SC and Wood RD (1994) XPG endonuclease makes the 3’ incision in human DNA nucleotide excision repair. Nature, 371, 432–435. [DOI] [PubMed] [Google Scholar]
- 131.de Laat WL, Appeldoorn E, Jaspers NG and Hoeijmakers JH (1998) DNA structural elements required for ERCC1-XPF endonuclease activity. J Biol Chem, 273, 7835–7842. [DOI] [PubMed] [Google Scholar]
- 132.Fagbemi AF, Orelli B and Schärer OD (2011) Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst), 10, 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tsodikov OV, Ivanov D, Orelli B, Staresincic L, Shoshani I, Oberman R, Schärer OD, Wagner G and Ellenberger T (2007) Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. EMBO J, 26, 4768–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Orelli B, McClendon TB, Tsodikov OV, Ellenberger T, Niedernhofer LJ and Schärer OD (2010) The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways. J Biol Chem, 285, 3705–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Volker M, Moné MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA and Mullenders LH (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell, 8, 213–224. [DOI] [PubMed] [Google Scholar]
- 136.Thorel F, Constantinou A, Dunand-Sauthier I, Nouspikel T, Lalle P, Raams A, Jaspers NG, Vermeulen W, Shivji MK, Wood RD et al. (2004) Definition of a short region of XPG necessary for TFIIH interaction and stable recruitment to sites of UV damage. Mol Cell Biol, 24, 10670–10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Scharer OD (2013) Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol, 5, a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Evans E, Fellows J, Coffer A and Wood RD (1997) Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J, 16, 625–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mu D and Sancar A (1997) Model for XPC-independent transcription-coupled repair of pyrimidine dimers in humans. J Biol Chem, 272, 7570–7573. [DOI] [PubMed] [Google Scholar]
- 140.Spivak G and Hanawalt PC (2015) Photosensitive human syndromes. Mutat Res, 776, 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lin Y and Wilson JH (2011) Transcription-induced DNA toxicity at trinucleotide repeats: double bubble is trouble. Cell Cycle, 10, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hanawalt PC (2015) Historical perspective on the DNA damage response. DNA Repair (Amst), 36, 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, van IJcken WF, Grosveld FG, Medema RH, Hoeijmakers JH et al. (2015) The core spliceosome as target and effector of non-canonical ATM signalling. Nature, 523, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Steurer B and Marteijn JA (2017) Traveling Rocky Roads: The Consequences of Transcription-Blocking DNA Lesions on RNA Polymerase II. J Mol Biol, 429, 3146–3155. [DOI] [PubMed] [Google Scholar]
- 145.Wei L, Nakajima S, Bohm S, Bernstein KA, Shen Z, Tsang M, Levine AS and Lan L (2015) DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc Natl Acad Sci U S A, 112, E3495–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I and Fischer T (2016) Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell, 167, 1001–1013 e1007. [DOI] [PubMed] [Google Scholar]
- 147.Welty S, Teng Y, Liang Z, Zhao W, Sanders LH, Greenamyre JT, Rubio ME, Thathiah A, Kodali R, Wetzel R et al. (2018) RAD52 is required for RNA-templated recombination repair in post-mitotic neurons. J Biol Chem, 293, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McDevitt S, Rusanov T, Kent T, Chandramouly G and Pomerantz RT (2018) How RNA transcripts coordinate DNA recombination and repair. Nat Commun, 9, 1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nielsen PE, Egholm M, Berg RH and Buchardt O (1991) Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science, 254, 1497–1500. [DOI] [PubMed] [Google Scholar]
- 150.Nielsen PE (2004) PNA Technology. Mol Biotechnol, 26, 233–248. [DOI] [PubMed] [Google Scholar]
- 151.D’Souza AD, Belotserkovskii BP and Hanawalt PC (2018) A novel mode for transcription inhibition mediated by PNA-induced R-loops with a model in vitro system. Biochim Biophys Acta, 1861, 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lin Y, Leng M, Wan M and Wilson JH (2010) Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis. Mol Cell Biol, 30, 4435–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin WY, Lin Y and Wilson JH (2014) Convergent transcription through microsatellite repeat tracts induces cell death. Mol Biol Rep, 41, 5627–5634. [DOI] [PubMed] [Google Scholar]
- 154.Zhang ZZ, Pannunzio NR, Hsieh CL, Yu K and Lieber MR (2014) The role of G-density in switch region repeats for immunoglobulin class switch recombination. Nucleic Acids Res, 42, 13186–13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Huang FT, Yu K, Hsieh CL and Lieber MR (2006) Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc Natl Acad Sci U S A, 103, 50305035. [DOI] [PMC free article] [PubMed] [Google Scholar]


