Abstract
Astrovirus VA1/HMO-C (VA1) is the representative genotype of mamastrovirus 9, a species of the single stranded, positive-sense RNA viral family, Astroviridae. Astroviruses have been traditionally considered pathogens of the gastrointestinal tract, but they have been recently associated with neurological diseases in humans, cattle, mink, sheep, and pigs. VA1 is the astrovirus genotype most commonly identified from human cases of meningoencephalitis and has been recently propagated in cell culture. VA1 can now be used as a model system to study pathogenesis of the neurological diseases associated with astrovirus infection. In this unit, we describe two fundamental assays to quantify replication and propagation of VA1, a qRT-PCR to measure viral RNA and a TCID50 assay to measure infectious viral particles.
Keywords: Astroviruses, astrovirus VA1, cell culture, replication, titration
INTRODUCTION
The application of a broad number of molecular assays and metagenomic sequencing have detected a diverse number of novel viral genomes (Chiu et al., 2008; Finkbeiner, Allred, et al., 2008; Gaynor et al., 2007; Greninger et al., 2009; Holtz et al., 2009; Janowski, Krishnamurthy, et al., 2017; Krishnamurthy, Janowski, Zhao, Barouch, & Wang, 2016; Nishizawa et al., 1997; Phan et al., 2011; Shi et al., 2016; Wang et al., 2003). However, the understanding of the fundamental biology of these novel viruses is severely limited because only a small minority have been successfully propagated in cell culture, including a novel astrovirus genotype (Chiu et al., 2010; Hao et al., 2012; Janowski, Bauer, Holtz, & Wang, 2017; Woo et al., 2005).
Since 2008, three novel astrovirus species have been identified from human specimens through molecular assays and metagenomic sequencing, including mamastrovirus 6, 8, and 9 (Finkbeiner, Allred, et al., 2008; Finkbeiner, Holtz, et al., 2009; Finkbeiner, Kirkwood, & Wang, 2008; Finkbeiner, Li, et al., 2009; Jiang et al., 2013; Kapoor et al., 2009; Meyer et al., 2015; Phan et al., 2014). Like mamastrovirus 1 (classic human astroviruses), these novel astrovirus species have been predominantly detected from human stool samples and were considered to also be primarily pathogens of the gastrointestinal tract (Bosch, Pinto, & Guix, 2014). Humans are frequently exposed to the novel human astroviruses, with >50% seropositivity in adulthood to two genotypes (Burbelo et al., 2011; Holtz et al., 2014).
However, astrovirus research has garnered further interest because of the recent discovery that some members of this viral family are neurotropic. To date, astroviruses have been described in nine cases of meningoencephalitis in humans and have been associated with neurological disease in cattle, minks, sheep, and pigs (Arruda et al., 2017; Blomstrom, Widen, Hammer, Belak, & Berg, 2010; Boros et al., 2017; Brown et al., 2015; Cordey et al., 2016; Fremond, 2015; Lum et al., 2016; Naccache et al., 2015; Pfaff et al., 2017; Quan et al., 2010; Sato et al., 2016; Selimovic-Hamza, Boujon, Hilbe, Oevermann, & Seuberlich, 2017; Wunderli et al., 2011). Astrovirus VA1/HMO-C (VA1; mamastrovirus 9), is the most frequently detected astrovirus genotype in human cases of encephalitis (Brown et al., 2015; Fremond, 2015; Lum et al., 2016; Naccache et al., 2015; Quan et al., 2010). Subsequently, the originally detected genotype of this species, VA1, has been successfully propagated in cell culture (Janowski, Bauer, et al., 2017). With this important laboratory reagent, the fundamental biology of VA1 can be studied and this virus may serve as an experimental model of astrovirus central nervous system infection.
We have described some of the biology of VA1 in cell culture, and VA1 appears to have divergent in vitro biology when compared to mamastrovirus 1 (Janowski, Bauer, et al., 2017). The addition of trypsin to cell culture media is necessary for propagation of mamastrovirus 1 (Lee & Kurtz, 1981). In addition, pretreatment of the virus with trypsin prior to inoculation also increases infectivity (Bass & Qiu, 2000; Mendez, Fernandez-Luna, Lopez, Mendez-Toss, & Arias, 2002). It is currently hypothesized that the mamastrovirus 1 coat requires further cleavage by trypsin to generate an infectious particle (Bass & Qiu, 2000; Mendez et al., 2002). In contrast, VA1 does not require trypsin to be added during any step for propagated in cell culture (Janowski, Bauer, et al., 2017). The significance of this difference remains to be determined and whether it reflects different pathogenicity of these viruses.
This unit describes the methods used to propagate VA1 in cell culture (Basic Protocol 1) and methods for quantification of viral RNA (Basic Protocol 2) and titration of infectious virus by a TCID50 assay (Basic Protocol 3). Additionally, this unit describes the maintenance of Caco-2 cells (Support Protocol 1), the primary cell line that supports astrovirus replication.
Caution:
Koch’s postulates for astroviruses causing gastroenteritis in humans have been fulfilled. The role of astroviruses in causing neurological diseases is currently under investigation; therefore, it is recommended that assays using astroviruses be performed in a Class II biological safety cabinet in a BSL-2 facility. Contaminated reusable laboratory items and surfaces should be disinfected with 10% bleach (v/v) and 70% ethanol. Other laboratory waste should be autoclaved prior to discarding.
BASIC PROTOCOL 1
Propagation of astrovirus VA1 in Caco-2 cell culture
The Caco-2 cell line is the prototypical cell line used in astrovirus research as it supports replication of all mamastrovirus 1 serotypes and mamastrovirus 9. In addition, most of the studies of the replicative cycle of the mamastrovirus 1 have been performed in Caco-2 cells. This protocol can be used to make additional stocks of astrovirus VA1. This protocol should be performed using sterilized equipment in BSL-2 conditions using a Class II biological safety cabinet.
Materials
Astrovirus VA1 stock
Caco-2 cells (ATCC HTB-37)
Caco-2 cell medium
Dulbecco’s Modified Eagle Medium with high glucose (DMEM)
T75 flasks or other sterile tissue culture plates or flasks
Serological pipettes and pipette controller
Disposable glass Pasteur pipettes
15 mL conical centrifuge tubes
1.5 mL microcentrifuge tubes
Vacuum aspirator and collection vessel for cell culture media
Tissue culture incubator (set at 37°C and 5% CO2)
Water bath set to 37°C
−80 °C freezer
Sorvall Legend RT centrifuge and rotor for conical tubes or tissue culture plates
Generation of additional astrovirus VA1 stock virus
Small scale astrovirus stocks can be created in T75 tissue culture flasks, while larger preparations may utilize several T175 or larger tissue culture flasks. The following example describes infection of a single T75 flask.
-
1
Grow a single T75 flask to 80–100% confluency with Caco-2 cells (see support protocol 1 for propagation of Caco-2 cells in cell culture).
Unlike other cell lines, Caco-2 cells remain biologically active once they reach confluency. Once confluent, they will form a monolayer with tight junctions, and the cells will differentiate with apical/basal polarity.
-
2
On the day of infection, prepare an inoculum of astrovirus VA1 suspended in warmed DMEM. For a single T75 flask with Caco-2 cells at 100% confluency, there are approximately 7.5×106 total cells. For a MOI of 0.001, 7.5×103 TCID50 units are suspended in 7.5 mL of DMEM.
For generation of viral stocks, commonly a low MOI (0.001–0.01) is used to reduce the generation of defective particles. Using approximately 50% the recommended volume for a flask enables a higher density of virus to be inoculated onto the cells, increasing the probability that infectious viral particles will bind to their target receptors.
Unlike classic human astroviruses (mamastrovirus 1), VA1 does not require preactivation of viral particles with trypsin or incubation with trypsin in the cell culture media after inoculation.
-
3
Remove the cell media by using a disposable Pasteur pipette with a vacuum aspirator and add 7.5 mL of inoculum. Incubate the flask in a tissue culture incubator set at 37°C and 5% CO2 for 1 hour.
-
4
After one hour, aspirate the inoculum and add 15 mL of warmed DMEM to the flask.
-
5
Aspirate the DMEM wash and add 15 mL of fresh Caco-2 cell medium
Cell lysis and generation of viral stock
-
6
For maximal production of infectious particles, incubate the cells for 5–7 days at 37°C with 5% CO2.
We have observed that the maximum yield of infectious particles occurs between 5–7 days. Further incubation beyond this time period does not result in further increases in the viral titer of the cell lysate.
-
7
Freeze thaw the flask three times. First, place the flask at −80°C until the contents are fully frozen (approximately 30 minutes). Next, thaw the flask in a 37°C warm room until the contents have thawed (approximately 30 minutes). Repeat this process for two further cycles.
During freezing, ice crystals will form, promoting lysis of the cells during repeated freeze-thaw cycles. Freezing the cells may also be performed in an ethanol/dry ice bath. Alternative methods that could be used for cell lysis include sonication or bead homogenization. We have not observed any differences in efficiency of recovery of infectious VA1 particles by using any alternative protocol of cell lysis.
-
8
Collect the cell lysate into a 15-mL centrifuge tube and centrifuge at 1,000 g x 5 min at 4°C.
Centrifugation removes cellular debris from the final viral stock.
-
9
Aliquot the viral stock into 1.5 mL microcentrifuge tubes and store at −80°C.
Alternatively, the viral stock can be filtered through a 0.22 µm filter to remove any potential bacterial contaminants. Further purification of the viral particles could also be performed after this step.
SUPPORT PROTOCOL 1
Propagation of Caco-2 cells in tissue culture
Caco-2 cells are derived from a human case of colorectal adenocarcinoma. The cells differentiate when grown to confluency and develop characteristics that mimic the epithelium of the gastrointestinal tract. For this reason, this cell line is commonly used in a variety of experimental protocols including permeability assays for pharmaceutical drugs. Caco-2 cells support replication and propagation of human astroviruses, including VA1, and have been often used to study host-virus interactions during infection.
The difficulties with working with this cell line include a slow doubling time, rated at 62 hours by ATCC. In addition, Caco-2 cells are sensitive to cell density as they often grow poorly at low density and at high density they will begin to differentiate. For these reasons, practicing good Caco-2 culture technique is essential for proper maintenance of this cell line in tissue culture.
Materials
Caco-2 cells (ATCC HTB-37) frozen in liquid nitrogen
Caco-2 cell medium
Dulbecco’s phosphate buffered saline (DPBS)
Trypsin-EDTA 0.25%
Dry ice
Sterile T75 flasks
Sterile 15mL conical centrifuge tubes
Serological pipettes and pipette controller
Disposable glass Pasteur pipettes
Water bath set to 37°C
Sorvall Legend RT centrifuge and rotor for conical tubes
Vacuum aspirator and collection vessel for cell culture media
Thawing an aliquot of Caco-2 cells
Steps should be carried out in a Class II biological safety hood. The following protocol utilizes T75 flasks for tissue culture, but volumes can be scaled up and down according to the number of cells required for downstream experiments.
-
1
Pre-warm Caco-2 cell medium in a 37°C water bath. Place 9 mL of prewarmed media into a 15 mL conical centrifuge tube.
We have optimized our protocols for VA1 and Caco-2 cells using the Caco-2 cell medium. Other protocols for classic human astroviruses have used the same or slightly different media compositions for Caco-2 cells including Eagle’s modified minimum essential media or different concentrations of FBS. Currently, it is unclear if these different media compositions affect astrovirus infection in Caco-2 cells.
-
2
Retrieve a cryogenic tube of Caco-2 cells frozen in liquid nitrogen. Place on dry ice.
It is important to keep the cells frozen until ready for thawing.
-
3
Dip the cryogenic tube of Caco-2 cells into the water of a 37°C water bath to thaw the aliquot. Do not submerge the tube under water as the contents could be contaminated by the water bath. Only thaw the contents partially, until a small bead of ice remains in the tube.
Because the cells are frozen in media containing dimethyl sulfoxide (DMSO), they must be rapidly thawed and diluted into fresh media. DMSO is toxic to cells at high concentrations.
-
4
Pipette the contents of the cryogenic tube containing the Caco-2 cells into the 15 mL conical centrifuge tube containing pre-warmed Caco-2 media.
-
5
Centrifuge the tube at 300 x g for five minutes to pellet the cells at room temperature.
Centrifuging the cells will enable aspiration of the media containing DMSO.
-
6
Aspirate off the media while being careful to not to disturb the cell pellet.
-
7
Resuspend the cells in 15mL of fresh warmed Caco-2 cell medium.
-
8
Pipette cell suspension into a T75 flask.
-
9
Incubate the flask at 37°C with 5% CO2.
-
10
24 hours after thawing, feed the cells fresh Caco-2 media by aspirating the media and then adding fresh media to the flask.
After thawing, dead cells will be floating in the culture media. Removing these dead cells will aid in propagation of the cells. Most Caco-2 cells will be adherent to the plastic surface 24 hours after thawing and most cells will flatten out on the surface by 48 hours.
-
11
Incubate the cells at 37°C with 5% CO2 cells until 80–90% confluency and replace the media every 3–4 days.
Caco-2 cells may take several days after thawing to return to their baseline metabolic activity. Slow growth may be observed in the first few days after thawing.
Subculturing Caco-2 cells
-
12
Once the cells reach 80–90% confluency, they can be passaged for subculturing. Aspirate off the media and add 15 mL of DPBS to the T75 flask.
If the cells are allowed to grow to confluency, the cells will begin to differentiate which may affect downstream experiments (Figure 1).
Addition of DPBS further dilutes residual FBS which contains trypsin inhibitors. In addition, the DPBS does not contain calcium and magnesium, important co-factors in cell adherence to the tissue culture flask.
-
13
Aspirate off the DPBS and add 3 mL of warmed 0.25% Trypsin-EDTA to the flask.
-
14
Incubate the cells at 37°C for approximately five minutes or until all cells are detached and floating.
As trypsin can be toxic to cells, only incubate the cells in trypsin for the minimum time needed for cell detachment. It is not recommended to tap the flask on a surface for promoting cell detachment as this often results in clumping and uneven distribution of the Caco-2 cells when passaging.
-
15
Dilute the cells suspended in trypsin with 10 mL of fresh Caco-2 cell medium.
FBS in the media contains trypsin inhibitors and other proteins that will inactivate the trypsin. Using at least a ratio of Caco-2 cell medium to trypsin of 3:1 (v:v) is recommended to inactive the trypsin and to reduce cell toxicity.
-
16
Pipette the media/cell suspension into a 15 mL conical centrifuge tube and centrifuge at 300 x g for 5 minutes.
-
17
Aspirate off the supernatant while being careful to not to disturb the pellet.
-
18
Resuspend the cells in fresh Caco-2 cell medium. A subcultivation ratio of 1:2–1:4 is often used for Caco-2 cells. For a 1:2 ratio, resuspend the cells in 30 mL of media, and pipette 15 mL into two T75 flasks.
Cells can also be quantified at this step by a hemocytometer.
-
19
Gently rock the flask to allow for even distribution over the surface.
-
20
Incubate the flask at 37°C with 5% CO2.
Figure 1:
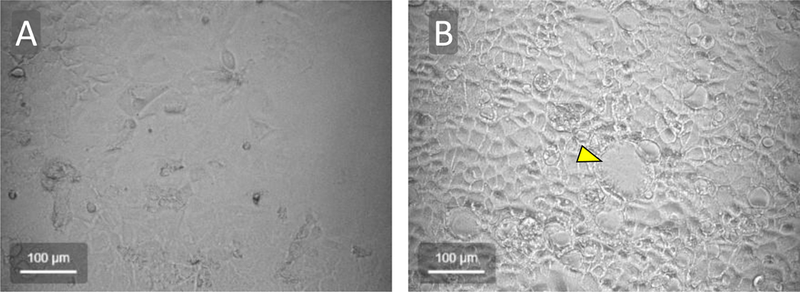
Brightfield microscopic images of Caco-2 cells grown in cell culture. In (A), cells are near 90% confluency while in (B) the cells have reached 100% confluency for two days. The cells achieve a more cobblestone appearance with distinct cell borders once the cells reach confluency and begin to differentiate. In addition, some areas of confluent Caco-2 cells develop dome-like structures (central structure denoted by yellow arrowhead in B).
BASIC PROTOCOL 2
Quantification of astrovirus VA1 replication by quantitative reverse transcription PCR (qRT-PCR)
With newer molecular assays like real time PCR, viral RNA copies can be quantified over time to demonstrate ongoing viral replication. qRT-PCR can also be used as a sensitive assay to detect virus from clinical or animal specimens.
Materials
Astrovirus VA1 stock
Caco-2 cells (ATCC HTB-37)
Caco-2 cell medium
Dulbecco’s Modified Eagle Medium (DMEM, high glucose)
Guanidinium thiocyanate phenol reagent (TRIzol: Invitrogen cat. no. 15596018 or Tri-Reagent: Sigma-Aldrich cat. no. T9424)
Sterile tissue culture plates or flasks
Serological pipettes and pipette gun
1.5 mL microcentrifuge tubes
P-1000 micropipette and tips
P-1000 multichannel micropipette
P-200 multichannel micropipette and tips
P-100 multichannel micropipette and tips
Sorvall Legend RT centrifuge and rotors for conical tubes and 96-well plates
95–100% ethanol
- Direct-zol 96-well RNA extraction kit (Zymo Research cat. no. R2056) which includes:
- 96-well column extraction plate
- 96-well collection plates (x2)
- Pre-wash buffer
- Wash buffer
- Elution buffer
PCR plate adhesive film (Thermo Fisher Scientific cat. no. 232702)
Forward primer AJ0071: 5’-CTAGTGGTGGGGAAGAAC-3’
Reverse primer AJ0072 5’-CCTTGGCTATTTGCTTTGC-3’
TaqMan probe AJ0073 5’-/56-FAM/CCATGACTT/ZEN/TGCTTTGGACCTCCC/3IABkFQ/−3’
In vitro transcribed VA1 RNA
Taqman Fast Virus 1-step master mix (Thermo Fisher Scientific cat. no. 4444434)
DEPC treated water
Sterile disposable reagent reservoirs
MicroAmp 96-well base (Thermo Fisher Scientific cat. no. N8010531)
MicroAmp Optical 96-well reaction plate with barcode (Thermo Fisher Scientific cat. no. 4306737)
MicroAmp Optical adhesive film (Applied Biosystems cat. no. 4311971)
MicroAmp Adhesive film applicator (Applied Biosystems cat. no. 4333183)
ViiA 7 Real-Time PCR system
ViiA software
Infection of Caco-2 cells
Viral multi-step growth curves are performed with a low inoculum of virus (multiplicity of infection, MOI, 0.001–0.01) to measure replication and propagation of a virus over several cycles of infection. The following example describes a multi-step growth curve of astrovirus VA1 in Caco-2 cells grown in a 24-well plate format with samples collected at 1, 12, 24, 36, 48, and 96 hours after inoculation.
-
1
After counting cells by a hemocytometer, we commonly plate 1×105 Caco-2 cells in existing media per well in a 24-well plate. Use six different 24-well plates as each plate will be harvested at a different time-point.
This protocol will use reagents containing guanidinium thiocyanate and phenol which can aerosolize and is toxic to cells in culture. Any plastic coming into contact with these reagents cannot be placed back in the cell incubator and must be discarded according to your local environmental health and safety policies.
-
2
Allow cells to incubate for at least 48 hours.
Allowing the Caco-2 cells to reach confluency and develop tight-junctions between cells results in reduced cell detachment in subsequent wash steps. Decreasing cell detachment will reduce well-to-well variability.
-
3
Aspirate the media from each well and add 500 µL warmed DMEM to each well (working volume).
-
4
Dilute astrovirus VA1 stock in DMEM to achieve a MOI of 0.001–0.01 per well. Use 50% of the working volume for the inoculum (for a 24-well plate, use 250 µL per well).
For example, the wells of a 24-well plate typically yield 2×105 Caco-2 cells per well when fully confluent. When using a MOI of 0.01, 2×103 TCID50 units will be added to each well. Each well will be inoculated using a 250 µL inoculum, so 2×103 TCID50 units will be suspended in DMEM to a final total volume of 250 µL.
-
5
Aspirate the DMEM wash and add the inoculum to each well.
-
6
Incubate the cells at 37°C with 5% CO2 for one hour. Gently rock the plates every 20 minutes to promote mixing of the media within the wells.
-
7
Aspirate off the inoculum and add 500 µL of warmed DMEM to each well.
Adding fresh DMEM will serve as a wash to remove unbound viral particles.
-
8
Aspirate off the DMEM and add 500 µL freshly warmed Caco-2 cell medium to each well.
-
9
Incubate all but one of the 24-well plates at 37°C with 5% CO2. Label each plate as the 1, 12, 24, 36, 48, and 96-hour timepoints.
Harvesting samples for multi-step growth curve
Samples will be harvested at 1, 12, 24, 36, 48, and 96 hours after inoculation. The 1-hour sample will be harvested immediately after washing the cells with DMEM and adding fresh Caco-2 cell medium.
-
10
Using a P-1000 micropipette, collect the media (supernatant fraction) from the first well and add it to a labeled 1.5 mL microcentrifuge tube.
-
11
Add 400 µL of guanidinium thiocyanate-phenol reagent to the well, using the micropipette to rinse the plastic surface to collect the cellular fraction. No cells should remain attached.
-
12
Remove the guanidinium thiocyanate-phenol reagent and pipette it into a separately labeled 1.5 mL microcentrifuge tube.
-
13
Collect all other supernatant and cell fractions from the wells for the 1-hour timepoint.
-
14
Store samples at −80°C.
-
15
At the indicated times (12, 24, 36, 48, 96 hours) repeat the collection for the labeled samples for those timepoints and store samples at −80°C.
Any plasticware or tips coming into contact with guanidinium thiocyanate-phenol reagent must be discarded according to your local environmental health and safety policies.
RNA extraction
RNA from cellular and supernatant fractions will be extracted using a 96-well column-based extraction kit. Smaller scale extractions can be performed using individual columns. Alternative protocols for using chloroform are also available.
-
16
Thaw previously collected supernatant and cellular fractions on ice. Plan plate layout of samples for future reference.
-
17
Dilute 100 µL of supernatant in 300 µL guanidinium thiocyanate-phenol reagent.
For most forms of commercially produced guanidinium thiocyanate-phenol reagent, a maximum ratio of 100 µL of liquid sample can be extracted with 300 µL of guanidinium thiocyanate-phenol reagent.
-
18
Add an equal volume of ethanol to the supernatant or cell fractions in guanidinium thiocyanate-phenol reagent.
For example, 400 µL of ethanol is added to 400 µL of supernatant mixed with guanidinium thiocyanate-phenol reagent.
-
19
Place the 96-well extraction plate on top of a collection plate and transfer samples to the columns of the 96-well extraction plate.
-
20
Centrifuge at 2,500 x g for five minutes at room temperature.
-
21
Discard the collection plate now containing guanidinium thiocyanate-phenol reagent and place the 96-well extraction plate on top of a new collection plate.
-
22
Add 800 µL of pre-wash buffer to each well and centrifuge at 2,500 x g for five minutes.
-
23
Pour out flow-through from the collection plate and add 750 µL of pre-wash buffer to each well of the extraction plate. Centrifuge at 2,500 x g for five minutes.
-
24
Pour out flow-through from the collection plate and repeat centrifugation at 2,500 x g for five minutes to dry the column.
-
25
Add 50 µL of elution buffer to each well and centrifuge at 2,500 x g for five minutes.
-
26
Resulting plate can be immediately used for qRT-PCR or frozen at −80°C.
If RNA samples are to be used immediately, the samples should be kept on ice to minimize degradation.
qRT-PCR of samples
Using the extracted RNA, the copies of VA1 can now be quantified by qRT-PCR. This protocol utilizes a master mix that contains the reagents for the reverse transcription and PCR steps. The primers and probe used in this protocol target a highly conserved region between ORF1b and ORF2 in the VA1 genome. Generation of a standard curve of VA1 genomic copies will allow conversion of the qRT-PCR cycle threshold value to RNA copies.
To minimize the potential of amplicon contamination, the reaction master mix should be setup in a dedicated PCR hood system. In a separate hood, the reaction mix should be added to the RNA samples in a PCR plate. Used pipette tips should be carefully discarded to minimize contamination of the laboratory.
-
27
Thaw reagents, RNA standards, and RNA samples on ice. Place equipment into PCR hoods and UV treat the equipment for at least 15 minutes.
-
28
Make a sufficient quantity of qRT-PCR master mix for the experimental samples. (Table 1).
-
29
Transfer 5 µL of the RNA samples to the qRT-PCR plate while being careful to not introduce air bubbles into the samples. Also transfer serial dilutions of previously quantified in vitro transcribed VA1 RNA and negative control samples containing water only.
Great care must be taken when pipetting samples with a multichannel pipette to minimize cross-contamination of samples. After transfer, the RNA plate can be resealed and stored at −80°C.
-
30
Pour qRT-PCR master mix into a sterile disposable reagent reservoir and pipette 15 µL of the master mix to each well containing RNA. Carefully pipette up and down to mix the samples while avoiding introduction of bubbles into the mixture.
-
31
Seal the PCR plate with an optical adhesive film using the film applicator to form a strong seal around each well.
-
32
Centrifuge the PCR plate at 300 x g for two minutes.
-
33
Run the qRT-PCR reaction on the real-time PCR machine.
For the astrovirus VA1 reaction using a Fast PCR system, a reverse transcription step is run for 5 minutes at 50°C then 20 seconds at 95°C to inactivate the reverse transcriptase. Then 40 cycles of amplification are performed using the following: 95°C for 3 seconds and 55°C for 30 seconds.
-
34
Use the serial dilutions of in vitro transcribed astrovirus VA1 RNA to develop a standard curve to convert cycle threshold values to RNA copies.
Table 1:
Volumes used for astrovirus VA1 qRT-PCR.
| 1x | 110x | |
|---|---|---|
| 4x Taqman Fast virus 1-step master mix |
5 µL | 550 µL |
| 10 pmol/ µL of AJ0071 (Forward primer) |
1 µL | 110 µL |
| 10 pmol/ µL of AJ0072 (Forward primer) |
1 µL | 110 µL |
| 10 pmol/ µL of AJ0073 (Probe) |
0.5 µL | 55 µL |
| DEPC H2O | 7.5 µL | 825 µL |
| Sample | 5 µL | |
| Total volume | 20 µL |
SUPPORT PROTOCOL 2
Production of astrovirus VA1 in vitro transcribed RNA
To generate a standard curve, a known quantity of astrovirus VA1 RNA is used to convert cycle threshold values to RNA copies. This protocol utilizes a plasmid already containing a region that encompasses the amplification site for qRT-PCR. This plasmid (pCR4-TOPO) also contains a T7 priming site for RNA transcription.
Alternatively, RT-PCR can be performed on RNA extracted from the viral stock to generate an amplicon containing the amplification site. This amplicon can then be cloned into a plasmid and used in the following protocol.
Materials
Plasmid DW639
Spe-I (New England Biolabs ca. no. R0133S)
Cutsmart Buffer (New England Biolabs ca. no. B7204S)
MEGAscript T7 Transcription Kit (Thermo Fisher Scientific ca. no. AM1334)
RNeasy Mini Kit (Qiagen ca. no. 74104)
1.5 mL microcentrifuge tubes
RNase-free water
Nanodrop spectrophotometer
-
Linearize 5 µg of purified DW639 plasmid dissolved in water by incubating with 10 units of Spe-I in Cutsmart Buffer overnight at 37°C.
Plasmid DW639 contains the target region of VA1 used in qRT-PCR with the amplicon downstream of a T7 promoter site. For any plasmid using this protocol, the correct orientation and insertion site of the amplicon into a plasmid must be determined prior to selection of a restriction enzyme used for digestion. In addition, the restriction enzyme selected for linearization should not have a corresponding restriction digest site within the amplicon.
The T7 polymerase is very processive. Linearization of the plasmid significantly enhances transcription efficiency of T7 polymerase as it prevents formation of multimers of RNA transcripts.
Terminate the reaction by adding 1/20th the volume of 0.5 M EDTA, 1/10th volume of 3 M sodium acetate, and 2 volumes of ethanol. Mix and incubate the mixture at −20°C for 15 minutes.
Pellet the DNA using a microcentrifuge at top speed (>8,000 x g) and resuspend in dH2O at a final concentration of 0.2–1 µg/µL.
-
Add 1 µg of linearized plasmid to the MEGAscript T7 reaction mixture.
The MEGAscript T7 reaction mixture includes 2 µL each of 75 mM ATP solution, 75 mM CTP solution, 75 mM GTP solution, and 75 mM UTP solution, 2 µL of 10x reaction buffer, 2 µL of enzyme mix, and sufficient water with linearized plasmid for a final volume of 20 µL.
Incubate the mixture at 37°C for four hours.
Place the reaction on ice and add 80 µL of RNase-free water.
Add 350 µL of Buffer RLT from the RNeasy Mini kit.
Add 250 µL of 96–100% ethanol and mix.
Transfer sample to a spin column from the RNeasy Mini kit and centrifuge for >15 seconds at >8,000 x g.
Empty the spin column and add 500 µL of Buffer RPE. Centrifuge for >15 seconds at >8,000 x g.
Empty the spin column and add 500 µL of Buffer RPE. Centrifuge for 2 minutes at >8,000 x g.
Empty the spin column and centrifuge for 1 minute at >8,000 x g.
-
Place the spin column into a new 1.5 mL microcentrifuge tube and add 50 µL of RNase-free water. Centrifuge for 1 minute at >8,000 x g.
The RNA yield may be increased by taking the eluate from this step and pipetting it on the spin column membrane for a second centrifugation step.
Quantify RNA concentration measuring the UV absorbance at 260 nm using an instrument like a Nanodrop spectrophotometer.
-
Using the expected length of the RNA transcript, convert the concentration of RNA to copies of RNA. Dilute the RNA to make 10-fold dilutions. These dilutions will be used to generate a standard curve for the qRT-PCR assay.
Commonly, we obtain yields around 1×1011 copies of RNA per µL.
BASIC PROTOCOL 3
Quantification of astrovirus VA1 by a TCID50 assay
Quantification of infectious viral particles is an essential assay used in virology. Two classic methods to quantify the infectious titer of a virus in a sample are the plaque assay and end-point dilution assays like a TCID50 assay. A plaque assay requires a virus that causes cytopathic effect in a susceptible cell line that can be visualized. By contrast, a TCID50 assay can be used to quantify infectious virus by determining at what dilution 50% of wells are infected. VA1 does not cause a distinguishable cytopathic effect in Caco-2 cells within the first few days after inoculation, so we describe a TCID50 assay to quantify infectious virus. In this instance we use qRT-PCR with a cycle threshold (CT) <30 to define wells in which infection has occurred.
Materials
Sample of astrovirus VA1 to be quantified
Astrovirus VA1 stock
Caco-2 cells (ATCC HTB-37)
Caco-2 cell medium
Dulbecco’s Modified Eagle Medium (DMEM, high glucose)
TCID50 protocol
Plate 3×104 Caco-2 cells per well in a 96-well flat-bottom tissue culture plate (see Support Protocol 1).
-
Incubate the cells for 48 hours.
Allowing the cells to reach confluency increases their adhesiveness to the plate and diminished cell detachment during subsequent wash steps.
-
If the infectious viral titer is to be measured from the cellular fraction of a sample, cell lysis must be performed prior to performing the inoculation steps of the TCID50 assay.
For tissue culture samples, we commonly quantify the infectious titer from whole cell lysate (containing both cell and media fractions). We maintain the cells in their tissue culture media and freeze-thaw the samples three times prior to usage in the TCID50 assay. Cell lysis should also be performed on tissue samples, while tissue culture supernatant may be used in the TCID50 assay without any additional sample processing.
-
Make 10-fold dilutions of each sample in DMEM, with 4 replicates for each dilution. Add dilutions and replicates to the wells of an empty 96-well plate.
We commonly use 10 µL of whole cell lysate per well as the initial inoculum.
Remove the Caco-2 media and add the serial dilutions and replicates to the wells containing Caco-2 cells.
Incubate the cells for 1 hour at 37°C with 5% CO2.
-
Aspirate the inoculum and add 100 µL of warm DMEM.
This wash steps removes free viral particles in the cell culture medium.
Aspirate the DMEM and add 100 µL of warm Caco-2 cell medium.
Incubate the cells for three days at 37°C with 5% CO2.
Remove the media and add 200 µL guanidinium thiocyanate-phenol reagent to each well.
Proceed with RNA extraction and qRT-PCR (see Basic Protocol 2)
-
Wells that have a cycle threshold <30 are considered infected while wells with a cycle threshold >30 are considered uninfected.
Based on our standard curve, a CT value of 30 corresponds to 588 RNA copies. The cutoff threshold can be adjusted based on standard curve results.
-
Calculate the TCID50 units of the sample by the Spearman-Karber or Reed-Muench method (Kärber, 1931; Reed & Muench, 1938; Spearman, 1908)
While both methods can be used to calculate the TCID50 units of a sample, only one method should be used for consistency between samples. We routinely use the Spearman-Karber method.
REAGENTS AND SOLUTIONS
Caco-2 cell medium
1 liter of DMEM (high glucose)
112.5 mL of fetal bovine serum
11.3 mL of 10,000 u/mL of penicillin/streptomycin
Store at 4°C for up to 6 months
COMMENTARY
Background Information
Astroviruses are a family of single stranded, positive sense RNA viruses originally described in 1975 (Appleton & Higgins, 1975; Madeley & Cosgrove, 1975a, 1975b). The name “astrovirus” was derived from the starry appearance of some viral particles when viewed by electron microscopy (Madeley & Cosgrove, 1975a, 1975b). Subsequently, two genera of astroviruses have been described, mamastroviruses and avastroviruses (Bosch et al., 2014). Mamastroviruses have been generally detected from mammals while avastroviruses have been detected from birds (Bosch et al., 2014). The prototype mamastrovirus species is mamastrovirus 1 (classic human astrovirus) which includes eight currently recognized serotypes. Mamastrovirus 1 causes gastrointestinal disease in humans and has been quantified as the 3rd to 5th most common cause of viral gastroenteritis/diarrhea (Buss et al., 2015; Cunliffe et al., 2010; Kirkwood, Clark, Bogdanovic-Sakran, & Bishop, 2005; Platts-Mills et al., 2015).
While traditionally considered pathogens of the gastrointestinal tract, astroviruses have now been identified from diseased central nervous system tissue of humans, minks, cattle, pigs, and sheep (Arruda et al., 2017; Blomstrom et al., 2010; Boros et al., 2017; Brown et al., 2015; Cordey et al., 2016; Fremond, 2015; Lum et al., 2016; Naccache et al., 2015; Pfaff et al., 2017; Quan et al., 2010; Sato et al., 2016; Selimovic-Hamza et al., 2017; Wunderli et al., 2011). To date, nine cases of meningoencephalitis have been attributed to human astroviruses, with astrovirus VA1 (VA1) as the most frequently astrovirus genotype (Brown et al., 2015; Cordey et al., 2016; Fremond, 2015; Lum et al., 2016; Naccache et al., 2015; Quan et al., 2010; Sato et al., 2016; Wunderli et al., 2011). In one study of cattle, astroviruses account for 34% of cases of encephalitis that were previously without an identified etiology (Selimovic-Hamza et al., 2017). Because astroviruses are not routinely tested from diseased humans or other mammals, the epidemiology of astrovirus-associated central nervous system infection is unknown. Astroviruses could be a significant cause of central nervous system infection because comprehensive studies of the causes of human encephalitis have only identified an etiology in under 50% of cases (Ambrose et al., 2011; Glaser et al., 2006; Kolski et al., 1998; Koskiniemi et al., 1997).
VA1 is the first of the recently identified human astrovirus genotypes to be propagated in cell culture (Janowski, Bauer, et al., 2017). With added interest in astroviruses, additional assays to study the biology of this virus will likely become available in the near future. Currently, there is no in vivo model in which to study any astrovirus genotype identified from humans. It is also unknown whether many of the host-virus interactions that have been identified for mamastrovirus 1 are also true for astrovirus VA1. In vitro, astrovirus VA1 does not require trypsin for propagation in cell culture, unlike mamastrovirus 1, suggesting that there is divergent biology between the astrovirus species (Janowski, Bauer, et al., 2017).
Critical Parameters and Troubleshooting
Cell culture
The most common cell line used to study astrovirus biology is Caco-2 cells. This cell line is more difficult to work with than other cell lines commonly used to propagate viruses. Caco-2 cells grow more slowly than other cell types (doubling time around 62 hours) and their phenotype will change once grown to confluency. Special attention must be given to passaging this cell line in culture to achieve replicable results.
Viral stocks
Astroviruses replicate using a RNA-dependent RNA polymerase that lacks a proofreading function; thus, they have a high mutational rate compared to DNA viruses. This highlights the importance of using a virus isolate that has not been passaged several times in cell culture as mutations can occur with subsequent passages. Generally, a low passage number virus isolate is used as the primary stock to seed subsequent viral stocks. In addition, it is important to use the same viral stock for the same experiments, as the phenotype of the virus could change with development of further mutations with each passage.
qRT-PCR
The advantage of qRT-PCR is the high sensitivity when detecting a nucleic acid target. However, the high sensitivity also places this assay at high risk for spurious results due to contamination. For example, a retrovirus xenotrophic murine leukemia virus (XMRV) was associated with various human diseases when it was detected by PCR, but further analysis suggested that detection of this virus was often due to laboratory contamination (Arias & Fan, 2014). Special steps must be taken to avoid contamination including using designated laboratory space for PCR, using reagents rated as PCR clean, and careful handling of samples and controls. Using a series of negative controls (using water for RNA extraction or PCR reactions) will aide in detecting potential sources of contamination. Additional attention to avoiding contamination is necessary when using a 96-well plate and multi-channel pipettes.
In addition, avoiding contamination with RNases is also important because RNases can degrade RNA. Usage of DEPC treated water, treatment of laboratory equipment with cleaning agents to RNases, and careful handling/storage of samples will aide in avoiding RNA degradation.
Understanding Results
Generation of astrovirus VA1 stock
Caco-2 cells have typically produced 1×105-5×107/mL infectious astrovirus VA1 TCID50 units. Further optimization and purification of viral particles could lead to a higher concentration of viral particles. Further characterization of the replication kinetics of astrovirus VA1 in other cell lines may identify cell types that support even more robust growth.
Quantification of astrovirus VA1 by qRT-PCR by multi-step growth curve
Using a MOI of 0.01, we have routinely detected a >100–1000-fold increase in astrovirus VA1 RNA 24 hours after infection with further increases at later timepoints. Monitoring the increase in viral RNA is also an important readout when quantifying the effect of antiviral interventions. We have previously demonstrated a 20-fold decrease in viral RNA 96-hours post-inoculation when pretreating Caco-2 cells with interferon-β (Janowski, Bauer, et al., 2017).
Measurement of Astrovirus VA1 infectious titer by TCID50 assay
We use qRT-PCR to differentiate between infected and uninfected wells. We have set a stringent CT value as we expect a >1000 fold increase in astrovirus VA1 RNA 72 hours post-inoculation if infectious particles are present. We routinely observe infected wells of a 96-well plate with CT values <25. Most uninfected wells have a CT that is undetectable, and only rarely do we detect wells with CT values between 30–35. Thus, use of 30 as the cutoff is conservative.
It is essential that a sufficient number of replicates and dilutions are performed in order to obtain accurate tittering of infectious virus when performing the TCID50 assay.
Time Considerations
Astrovirus VA1 stock generation
We estimate around 6–8 days are necessary to develop a viral stock as much of the time is for incubation of the virus and cells. Quantification of the stock by the TCID50 assay will take another 3 days. Once aliquoted and frozen at −80°C, the stock will be stable for usage for several years.
Quantification of astrovirus VA1 by qRT-PCR by multi-step growth curve
Performing the multi-step growth curve under our routine parameters (obtaining samples at 1, 12, 24, 36, 48, and 96 hours post-inoculation) will take 4 days to complete. Completing the RNA extraction and qRT-PCR will take about 4–5 hours. We often extract the RNA from four different growth curve experiments as this is an optimal workflow for a single researcher and a sufficient number of samples to fill two 96-well RNA extraction plates.
Measurement of Astrovirus VA1 infectious titer by TCID50 assay
Quantification of the viral titer by the TCID50 assay will take approximately 3.5 days, with the majority of the time during the 3-day incubation of the virus with the cells. Because the cells are infected in a 96-well format, the time required for transferring the samples into a 96-well plate is significantly reduced.
Significance Statement.
Over the past two decades, the recognized diversity of the virosphere has tremendously increased. Despite the significant increase in the number of viral genomes that have been identified, only a minute subset of novel viruses has been successfully isolated and propagated in cell culture. We describe the cell culture techniques involved in the infection and propagation of astrovirus VA1, a novel neurotropic human virus associated with multiple cases of encephalitis. Assays for quantification in this unit include qRT-PCR for viral RNA and a TCID50 assay to measure infectious viral particles. Together, these assays form the foundation to study the basic biology and pathogenesis of this virus.
ACKNOWLEDGEMENTS:
ABJ is supported by NIH grant K08 AI132745. DW is supported in part by NIH grant R21 NS101371. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
LITERATURE CITED
- Ambrose HE, Granerod J, Clewley JP, Davies NW, Keir G, Cunningham R, … Group, U. K. A. o. E. S. (2011). Diagnostic strategy used to establish etiologies of encephalitis in a prospective cohort of patients in England. J Clin Microbiol, 49(10), 3576–3583. doi: 10.1128/JCM.00862-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton H, & Higgins PG (1975). Letter: Viruses and gastroenteritis in infants. Lancet, 1(7919), 1297. [DOI] [PubMed] [Google Scholar]
- Arias M, & Fan H (2014). The saga of XMRV: a virus that infects human cells but is not a human virus. Emerg Microbes Infect, 3(4), e. doi: 10.1038/emi.2014.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda B, Arruda P, Hensch M, Chen Q, Zheng Y, Yang C, … Li G (2017). Porcine Astrovirus Type 3 in Central Nervous System of Swine with Polioencephalomyelitis. Emerg Infect Dis, 23(12), 2097–2100. doi: 10.3201/eid2312.170703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass DM, & Qiu S (2000). Proteolytic processing of the astrovirus capsid. J Virol, 74(4), 1810–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomstrom AL, Widen F, Hammer AS, Belak S, & Berg M (2010). Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol, 48(12), 4392–4396. doi: 10.1128/JCM.01040-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros A, Albert M, Pankovics P, Biro H, Pesavento PA, Phan TG, … Reuter G (2017). Outbreaks of Neuroinvasive Astrovirus Associated with Encephalomyelitis, Weakness, and Paralysis among Weaned Pigs, Hungary. Emerg Infect Dis, 23(12), 1982–1993. doi: 10.3201/eid2312.170804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A, Pinto RM, & Guix S (2014). Human astroviruses. Clin Microbiol Rev, 27(4), 1048–1074. doi: 10.1128/CMR.00013-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Morfopoulou S, Hubb J, Emmett WA, Ip W, Shah D, … Breuer J (2015). Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis, 60(6), 881–888. doi: 10.1093/cid/ciu940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo PD, Ching KH, Esper F, Iadarola MJ, Delwart E, Lipkin WI, & Kapoor A (2011). Serological studies confirm the novel astrovirus HMOAstV-C as a highly prevalent human infectious agent. PLoS One, 6(8), e22576. doi: 10.1371/journal.pone.0022576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, … Bourzac KM (2015). Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol, 53(3), 915–925. doi: 10.1128/JCM.02674-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CY, Greninger AL, Chen EC, Haggerty TD, Parsonnet J, Delwart E, … Ganem D (2010). Cultivation and serological characterization of a human Theiler’s-like cardiovirus associated with diarrheal disease. J Virol, 84(9), 4407–4414. doi: 10.1128/JVI.02536-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CY, Greninger AL, Kanada K, Kwok T, Fischer KF, Runckel C, … DeRisi JL (2008). Identification of cardioviruses related to Theiler’s murine encephalomyelitis virus in human infections. Proc Natl Acad Sci U S A, 105(37), 14124–14129. doi: 10.1073/pnas.0805968105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordey S, Vu DL, Schibler M, L’Huillier AG, Brito F, Docquier M, … Kaiser L (2016). Astrovirus MLB2, a New Gastroenteric Virus Associated with Meningitis and Disseminated Infection. Emerg Infect Dis, 22(5), 846–853. doi: 10.3201/eid2205.151807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe NA, Booth JA, Elliot C, Lowe SJ, Sopwith W, Kitchin N, … Regan M (2010). Healthcare-associated viral gastroenteritis among children in a large pediatric hospital, United Kingdom. Emerg Infect Dis, 16(1), 55–62. doi: 10.3201/eid1601.090401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, & Wang D (2008). Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog, 4(2), e1000011. doi: 10.1371/journal.ppat.1000011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner SR, Holtz LR, Jiang Y, Rajendran P, Franz CJ, Zhao G, … Wang D (2009). Human stool contains a previously unrecognized diversity of novel astroviruses. Virol J, 6, 161. doi: 10.1186/1743-422X-6-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner SR, Kirkwood CD, & Wang D (2008). Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol J, 5, 117. doi: 10.1186/1743-422X-5-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner SR, Li Y, Ruone S, Conrardy C, Gregoricus N, Toney D, … Tong S (2009). Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol, 83(20), 10836–10839. doi: 10.1128/JVI.00998-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fremond ML, Perot P, Muth E, Cros G, Dumarest M, Mahlaoui N, Seilhean D, Desguerre I, Hebert C, Corre-Catelin N, Neven B, Lecuit M, Blanche S, Picard C, Eloit M. (2015). Next-Generation Sequencing for Diagnosis and Tailored Therapy: A Case Report of Astrovirus-Associated Progressive Encephalitis. Journal of the Pediatric Infecctious Diseases Society, 1–5. [DOI] [PubMed]
- Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, … Wang D (2007). Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog, 3(5), e64. doi: 10.1371/journal.ppat.0030064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser CA, Honarmand S, Anderson LJ, Schnurr DP, Forghani B, Cossen CK, … Tureen JH (2006). Beyond viruses: clinical profiles and etiologies associated with encephalitis. Clin Infect Dis, 43(12), 1565–1577. doi: 10.1086/509330 [DOI] [PubMed] [Google Scholar]
- Greninger AL, Runckel C, Chiu CY, Haggerty T, Parsonnet J, Ganem D, & DeRisi JL (2009). The complete genome of klassevirus - a novel picornavirus in pediatric stool. Virol J, 6, 82. doi: 10.1186/1743-422X-6-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Bernard K, Patel N, Ulbrandt N, Feng H, Svabek C, … Zhu Q (2012). Infection and propagation of human rhinovirus C in human airway epithelial cells. J Virol, 86(24), 13524–13532. doi: 10.1128/JVI.02094-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz LR, Bauer IK, Jiang H, Belshe R, Freiden P, Schultz-Cherry SL, & Wang D (2014). Seroepidemiology of astrovirus MLB1. Clin Vaccine Immunol, 21(6), 908–911. doi: 10.1128/CVI.00100-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz LR, Finkbeiner SR, Zhao G, Kirkwood CD, Girones R, Pipas JM, & Wang D (2009). Klassevirus 1, a previously undescribed member of the family Picornaviridae, is globally widespread. Virol J, 6, 86. doi: 10.1186/1743-422X-6-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski AB, Bauer IK, Holtz LR, & Wang D (2017). Propagation of astrovirus VA1, a neurotropic human astrovirus, in cell culture. J Virol doi: 10.1128/JVI.00740-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski AB, Krishnamurthy SR, Lim ES, Zhao G, Brenchley JM, Barouch DH, … Wang D (2017). Statoviruses, A novel taxon of RNA viruses present in the gastrointestinal tracts of diverse mammals. Virology, 504, 36–44. doi: 10.1016/j.virol.2017.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Holtz LR, Bauer I, Franz CJ, Zhao G, Bodhidatta L, … Wang D (2013). Comparison of novel MLB-clade, VA-clade and classic human astroviruses highlights constrained evolution of the classic human astrovirus nonstructural genes. Virology, 436(1), 8–14. doi: 10.1016/j.virol.2012.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Li L, Victoria J, Oderinde B, Mason C, Pandey P, … Delwart E (2009). Multiple novel astrovirus species in human stool. J Gen Virol, 90(Pt 12), 2965–2972. doi: 10.1099/vir.0.014449-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärber G (1931). Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn-Schmiedebergs Archiv für experimentelle pathologie und pharmakologie, 162(4), 480–483. [Google Scholar]
- Kirkwood CD, Clark R, Bogdanovic-Sakran N, & Bishop RF (2005). A 5-year study of the prevalence and genetic diversity of human caliciviruses associated with sporadic cases of acute gastroenteritis in young children admitted to hospital in Melbourne, Australia (1998–2002). J Med Virol, 77(1), 96–101. doi: 10.1002/jmv.20419 [DOI] [PubMed] [Google Scholar]
- Kolski H, Ford-Jones EL, Richardson S, Petric M, Nelson S, Jamieson F, … MacGregor D (1998). Etiology of acute childhood encephalitis at The Hospital for Sick Children, Toronto, 1994–1995. Clin Infect Dis, 26(2), 398–409. [DOI] [PubMed] [Google Scholar]
- Koskiniemi M, Korppi M, Mustonen K, Rantala H, Muttilainen M, Herrgard E, … Vaheri A (1997). Epidemiology of encephalitis in children. A prospective multicentre study. Eur J Pediatr, 156(7), 541–545. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy SR, Janowski AB, Zhao G, Barouch D, & Wang D (2016). Hyperexpansion of RNA Bacteriophage Diversity. PLoS Biol, 14(3), e1002409. doi: 10.1371/journal.pbio.1002409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TW, & Kurtz JB (1981). Serial propagation of astrovirus in tissue culture with the aid of trypsin. J Gen Virol, 57(Pt 2), 421–424. [DOI] [PubMed] [Google Scholar]
- Lum SH, Turner A, Guiver M, Bonney D, Martland T, Davies E, … Wynn R (2016). An emerging opportunistic infection: fatal astrovirus (VA1/HMO-C) encephalitis in a pediatric stem cell transplant recipient. Transpl Infect Dis doi: 10.1111/tid.12607 [DOI] [PubMed] [Google Scholar]
- Madeley CR, & Cosgrove BP (1975a). Letter: 28 nm particles in faeces in infantile gastroenteritis. Lancet, 2(7932), 451–452. [DOI] [PubMed] [Google Scholar]
- Madeley CR, & Cosgrove BP (1975b). Letter: Viruses in infantile gastroenteritis. Lancet, 2(7925), 124. [DOI] [PubMed] [Google Scholar]
- Mendez E, Fernandez-Luna T, Lopez S, Mendez-Toss M, & Arias CF (2002). Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J Virol, 76(16), 7996–8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CT, Bauer IK, Antonio M, Adeyemi M, Saha D, Oundo JO, … Holtz (2015). Prevalence of classic, MLB-clade and VA-clade Astroviruses in Kenya and The Gambia. Virol J, 12, 78. doi: 10.1186/s12985-015-0299-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache SN, Peggs KS, Mattes FM, Phadke R, Garson JA, Grant P, … Chiu CY (2015). Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis, 60(6), 919–923. doi: 10.1093/cid/ciu912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, & Mayumi M (1997). A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun, 241(1), 92–97. doi: 10.1006/bbrc.1997.7765 [DOI] [PubMed] [Google Scholar]
- Pfaff F, Schlottau K, Scholes S, Courtenay A, Hoffmann B, Hoper D, & Beer M (2017). A novel astrovirus associated with encephalitis and ganglionitis in domestic sheep. Transbound Emerg Dis, 64(3), 677–682. doi: 10.1111/tbed.12623 [DOI] [PubMed] [Google Scholar]
- Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, & Delwart EL (2011). The fecal viral flora of wild rodents. PLoS Pathog, 7(9), e1002218. doi: 10.1371/journal.ppat.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Nordgren J, Ouermi D, Simpore J, Nitiema LW, Deng X, & Delwart E (2014). New astrovirus in human feces from Burkina Faso. J Clin Virol, 60(2), 161–164. doi: 10.1016/j.jcv.2014.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platts-Mills JA, Babji S, Bodhidatta L, Gratz J, Haque R, Havt A, … Investigators, M.-E. N. (2015). Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health, 3(9), e564–575. doi: 10.1016/S2214-109X(15)00151-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, … Lipkin WI (2010). Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis, 16(6), 918–925. doi: 10.3201/eid1606.091536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, & Muench H (1938). A simple method of estimating fifty per cent endpoints. American journal of epidemiology, 27(3), 493–497. [Google Scholar]
- Sato M, Kuroda M, Kasai M, Matsui H, Fukuyama T, Katano H, & Tanaka-Taya K (2016). Acute encephalopathy in an immunocompromised boy with astrovirus-MLB1 infection detected by next generation sequencing. J Clin Virol, 78, 66–70. doi: 10.1016/j.jcv.2016.03.010 [DOI] [PubMed] [Google Scholar]
- Selimovic-Hamza S, Boujon CL, Hilbe M, Oevermann A, & Seuberlich T (2017). Frequency and Pathological Phenotype of Bovine Astrovirus CH13/NeuroS1 Infection in Neurologically-Diseased Cattle: Towards Assessment of Causality. Viruses, 9(1). doi: 10.3390/v9010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, … Zhang YZ (2016). Redefining the invertebrate RNA virosphere. Nature doi: 10.1038/nature20167 [DOI] [PubMed] [Google Scholar]
- Spearman C (1908). The method of ‘right and wrong cases’(‘constant stimuli’) without Gauss’s formulae. British Journal of Psychology, 1904‐1920, 2(3), 227–242. [Google Scholar]
- Wang D, Urisman A, Liu YT, Springer M, Ksiazek TG, Erdman DD, … DeRisi JL (2003). Viral discovery and sequence recovery using DNA microarrays. PLoS Biol, 1(2), E2. doi: 10.1371/journal.pbio.0000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, … Yuen KY (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol, 79(2), 884–895. doi: 10.1128/JVI.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderli W, Meerbach A, Gungor T, Berger C, Greiner O, Caduff R, … Tapparel C (2011). Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS One, 6(11), e27483. doi: 10.1371/journal.pone.0027483 [DOI] [PMC free article] [PubMed] [Google Scholar]


