Abstract
The existence of eukaryotic ribosomes with distinct ribosomal protein (RP) stoichiometry and regulatory roles in protein synthesis has been speculated for over sixty years. Recent advances in mass spectrometry and high throughput analysis have begun to identify and characterize distinct ribosome stoichiometry in yeast or mammalian systems. In addition to RP stoichiometry, ribosomes host a vast array of protein modifications, effectively expanding the number of human RPs from 80 to many thousands of distinct proteoforms. Is it possible these proteoforms combine to function as a ‘ribosome code’ to tune protein synthesis? We outline the specific benefits that translational regulation by specialized ribosomes can offer and discuss the means and methodologies available to correlate and characterize RP stoichiometry with function. We highlight previous research with a focus on formulating hypotheses that can guide future experiments and crack the ‘ribosome code’.
Keywords: Ribosome, Stoichiometry, Heterogeneity, Translation, Mass Spectrometry
Ribosomes, the cellular machinery of protein synthesis, are present at up to ten million copies per cell in mammals. Despite their abundance and the wide array of known modifications to both ribosomal proteins (RPs) and rRNA, study of the direct role of the ribosome in tuning cellular translation has until recently taken a back seat to post- transcriptional regulation at the level of translation initiation. The hypothesis that ribosomes actively regulate protein synthesis as part of normal development and physiology dates back to the 1950s [1]. In the ensuing decades, numerous albeit inconclusive, observations have supported this hypothesis, and a subset of those are shown in Figure 1.
Figure 1. Timeline.
The concept of eukaryotic ribosome specialization has existed for decades, and recent methodological advances have resulted in renewed interest and the ability to explore and characterize these phenomena. In this timeline, a few key manuscripts are colored by the area of ribosome heterogeneity they have described.
For many years, the dominant ‘abundance’ model suggested a limited role for ribosomes in translation regulation [2]. In this model, if ribosomes have different initiation affinity for different transcripts, a global decrease in the availability of free ribosomes selectively decreases the initiation rates of different transcripts to varying degrees [2]. This mechanism, recently reviewed by Mills and Green [3], relies on the non-linear dependence of translation initiation on free ribosomes. Recent research suggested this mechanism could explain the failure of erythroid lineage commitment seen with Diamond-Blackfan anemia (DBA) [4]. Under this model, cell or tissue specificity of ribosomopathies such as DBA (Box 1) is explained as a cell-type specific response to ribosome-induced cell stress [3]. However, other experiments performed in yeast suggested this model alone is insufficient to fully capture the details of ribosome-mediated translational control in wild-type cells [5]. Translational regulation by the abundance model is limited in magnitude by changes in total ribosomal content and in flexibility since it provides unidirectional regulation for all proteins.
Box 1: Ribosomopathies.
Ribosomopathies are conditions resulting from abnormalities in either ribosomal proteins, ribosomal RNAs or a subset of related genes. These abnormalities may impact ribosome availability or function, or both [85]. Typically, these result from haploinsufficiency of an RP where one copy of a RP gene is knocked out or non- functional and the single remaining copy is insufficient for normal ribosome function [85]. Ribosomopathies target different tissues, each ribosomeopathy targeting only one or a few specific tissues, and show a large degree of variation even between patients with the same condition [85,86]. Some of the conditions associated with aberrant ribosomes and RP mutations include:
Diamond blackfan anaemia: a group of conditions defined by specific reduction of erythrocytes. Approximately half of patients exhibit other symptoms such as cleft palate, and cardiac defects. Typically diagnosed early in life, DBA was recently suggested to be a result of low ribosome abundance, with a number of RP mutations linked to the condition including RPS19, RPS17 and RPS24 [4,24].
Isolated congenital asplenia: Is characterized as the absence of the spleen at birth in the absence of other developmental issues. Linked to haploinsufficiency of RPSA [87,88].
Neurodevelopmental disorders: RPL10 mutations have been linked to neurodevelopmental conditions including autism spectrum disorders and microcephaly [89].
Cancer: Patients with ribosomopathies appear predisposed to certain cancers [3]. In addition, ribosomal genes are frequently dysregulated during cancer and mutations in RPs may promote cellular transformation, for example RPL5, RPL10, and RPL21 [35,90,91]
The concept of ribosome specialization
In the ‘specialized’ ribosome model (Box 2), ribosomes do not possess constant structure or composition (Figure 2A, B), and instead exhibit altered stoichiometry of what were previously thought to represent ‘core’ ribosomal proteins (Figure 1) [6–11]. In this model, different ribosomal compositions are functional and have specific roles in translation. Specialized ribosomes could co-exist within cells, or between different cells or tissues [12,13]. Ribosome heterogeneity is a related concept where ribosomes within or between cells can have altered stoichiometry or composition, but do not necessarily play a functional role.
Box 2: Specialized ribosomes and the ribosome code.
Recent data support the idea that ribosomes are not simply passive recipients in the translation control process, but in fact play a more central role in translation regulation.
Ribosome abundance: Is a major model, also termed the ribosome concentration hypothesis [3], that explains how ribosomes could exert control over host translation by the modulation of ribosome abundance within a cell. As long as ribosome association with mRNAs is a non-linear function of ribosome concentration, this function will have different slopes for different ribosome concentrations. Thus, the translation rates of mRNAs whose response functions are steeper over the range of concentration changes will be affected disproportionately [2]. This model could explain some of the translation control often attributed to specialized ribosomes. Still, it is limited in the extent to which it could provide regulatory functionality as it can only provide uni-directional translation regulation, i.e., as ribosome levels decrease, the translation of all mRNAs decreases, albeit to a different degree [2]. This model may explain some tissue-specific phenotypes if mRNAs change their ribosome association rates in a tissue specific manner, e.g., a particular transcript has high translation-initiation rate in one tissue (and is thus not affected by decreased ribosome availability in that tissue) but low translation-initiation rate in another tissue (and is thus affected by decreased ribosome availability in that tissue) [85].
Ribosome heterogeneity: Is a broad concept reflecting that ribosome composition may vary across different ribosomes. This variation can include the absence of some RPs, modifications of RPs, or modifications of rRNAs [22,92]. This variation does not necessarily imply functional differences. Recent mass spectrometry data provide strong evidence for ribosome heterogeneity by demonstrating that ribosomal proteins in purified ribosome fractions are not all present at stoichiometric levels [22]. Since the isolated ribosomes originate from many cells, it remains unclear whether this variation in RP ribosome association extends to within-cell heterogeneity. Within cell heterogeneity is suggested by single-cell protein measurements, but remain inconclusive because the ribosomes were not isolated [64]. Additionally, the observation that some populations of ribosomes exhibit altered stoichiometry does not necessarily mean that such populations are functional or have distinct functions [85]. Where heterogenous ribosomes are confirmed to have distinct functions, these are referred to as specialized ribosomes.
Specialized ribosomes: the term specialized ribosomes refers to a subset of heterogenous ribosomes, where that heterogeneity has been demonstrated to result in functionally distinct ribosomes, with specific roles [85]. Ribosome specialization may take the form of the ‘ribosome code’.
The ribosome code: refers to the concept that different modifications to the ribosome, such as altered RP stoichiometry, different post-translational modifications to the RPs, or the use of alternate rRNA transcripts or rRNA modifications all combine to combinatorically regulate ribosome function. This is analogous to the ‘histone code’ hypothesis introduced by David Allis [93], however at this time the existence of a ribosome code remains unproven.
We emphasize that the ribosome abundance and specialized ribosome models are mutually compatible. The ribosome abundance model rests on well-validated principles, but it does not necessarily exclude the existence of specialized ribosomes. Still, it must always be considered first in explaining ribosome-related phenotypes, before invoking the more complex ribosome specialization model.
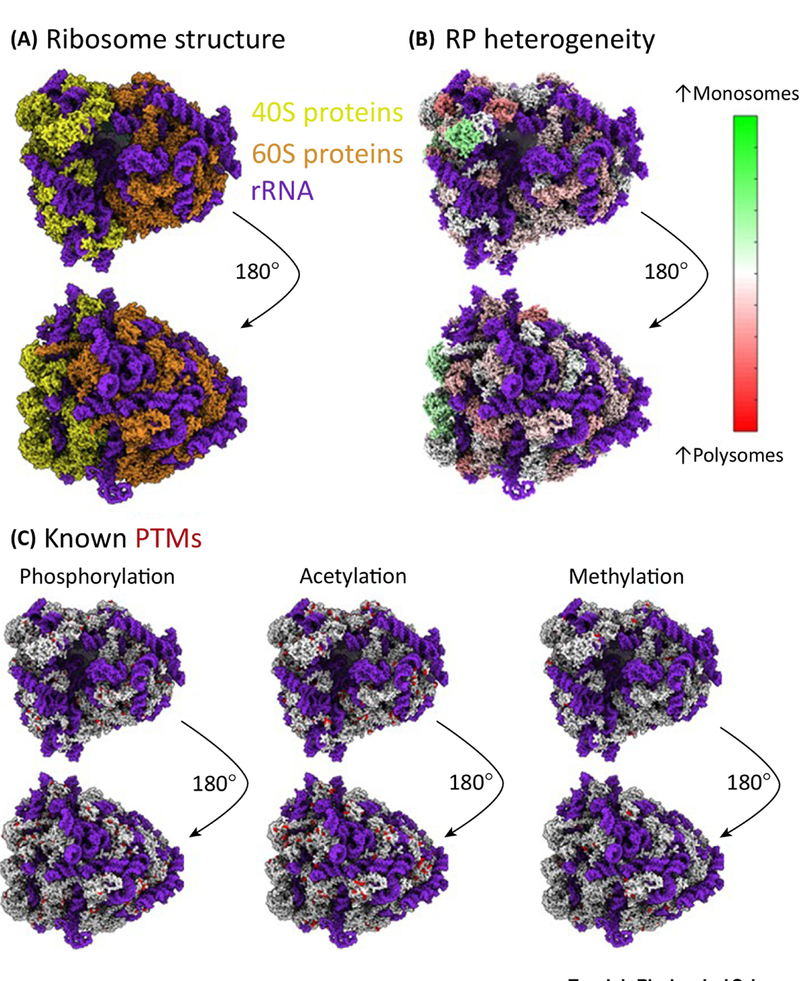
Figure 2. Heterogenous Ribosomes and their PTMs.
A) Ribosomes can be divided into the small (40S, shown in gold) and large (60S, shown in bronze) subunits, which in humans comprises 4 rRNAs (shown in blue) and 80 ribosomal proteins. B) Mass spectrometry analysis of human ribosomes reveals RPs are not all present at stoichiometric levels (Levels in monosomes compared to polysomes, unpublished data, U-937 human monocyte cells). C) RPs are highly modified with over 2500 modifications listed in Phosphositeplus [66] as of January 2018. The most abundant RP modifications currently known are phosphorylation, acetylation and methylation. Modification sites are shown in red. The human ribosome structures presented here were generated using PDB structure 5T2C [99] in the UCSF ChimeraX software.
A partial parallel for the specialization model is found with epigenetics and the ‘histone code’, where different post-translational modification (PTMs) states of the histone proteins can drive activation or repression of transcription [14–17]. Similar to histones, RPs are known to harbor a wide range of PTMs (Figure 2C). The key concept of the histone code hypothesis is that these modifications serve not only to modulate the specific interactions between histones and the DNA, but also to recruit accessory factors which can recognize the modified histones, providing further functionality and regulation. These modifications are proposed to function combinatorically with these modified proteins or proteoforms massively expanding the level of control histones can exert over transcription.
A ‘ribosome code’ could function similarly, with modifications to the ribosome- residence or PTM status of RPs, or rRNA modifications, driving either the recruitment of accessory factors [18], or modifying the mRNA-binding biases of particular RPs and therefore the host ribosome. However, there are features that would distinguish these two regulatory codes. The ribosome combines the roles of both histones and the RNA polymerase, and rather than acting in cis on the specific gene bound by the histones, the ribosome code would function in trans on its target mRNAs. By taking on the additional roles of the polymerase, the ribosome code would offer the potential to control ribosome elongation and error rates as well as subcellular localization of translation. Whilst the authors of this review have focused on the roles of RPs and RP modifications, distinct rRNA transcripts and modifications may also contribute to ribosome specialization [7,12,19,20]. Regulation by specialized ribosomes can provide unique advantages for the cell – such as direct integration between cytoplasmic metabolites, and translational regulation [21], lower gene expression noise, spatial localization, and very short timescales (Figure 3).
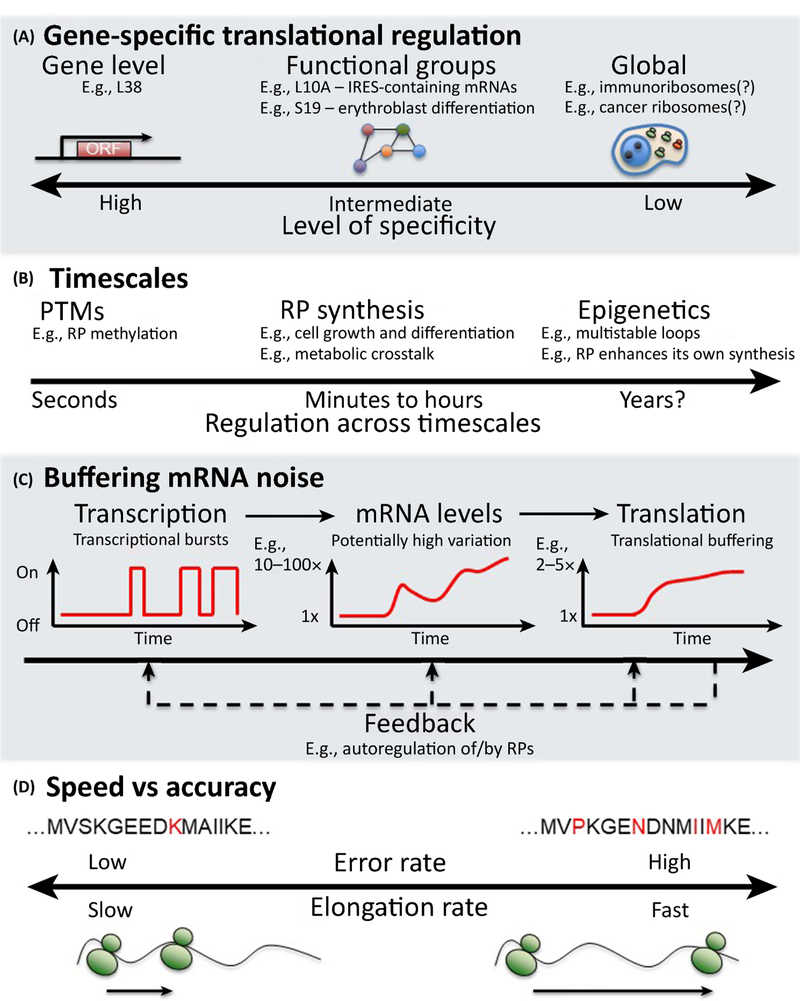
Figure 3. Ribosome specialization.
If populations of ribosomes exhibit distinct phenotypes there are multiple ways in which these functional differences could exist. A) Distinct ribosome subpopulations could have a range of specificities for their mRNAs. These could be from the individual mRNA level to global translation regulation. B) The timescale at which changes to RP stoichiometry or PTMs could exert effects on translation can potentially range from the extremely rapid/seconds (especially in the case of PTMs), to the very long term e.g. years. C) mRNA expression is noisy and buffered at the level of translation. D) The elongation rate of a ribosome represents a tradeoff between speed and accuracy. Further the elongation rate is not constant on a given mRNA with some sections of an mRNA being translated more rapidly than others.
We review the evidence for ribosome specialization and focus on experiments that can rigorously explore and discriminate between these two models. Future studies of ribosome specialization can benefit from well formulated hypotheses about the degree of mRNA-specificity, the time-scale of regulation, and the potential regulatory benefits to the host cell of ribosome specialization.
Evidence for ribosome specialization
Wildtype cells make ribosomes with altered stoichiometry [22,23] (Figure 2). Genetic perturbation of RPs have highly specific phenotypes [24–26] (Figure 1, Box 1), yet it remains possible that such specific phenotypes may be mediated by extra-ribosomal functions of RPs, or a general depletion of functional ribosomes that decreases the translation of some transcripts more than others [2,3]. The biochemical evidence for specialized ribosomes fulfilling physiological roles in wildtype cells had until recently remained indirect, mostly limited to differential RP-transcript levels. The lack of technologies to accurately identify and quantify proteins limited most early studies of RP stoichiometries in 30S and 40S fractions of bacterial ribosomes purified from sucrose gradients [27,28]. These fractions also contained immature ribosome biogenesis particles [29–31] that complicated the interpretation of measured stoichiometries. More recently, quantitative mass-spectrometry (MS) has begun to provide direct evidence for differential RP synthesis [32] and stoichiometry in isolated ribosomes [22]. In addition, advances in cryo-EM make it feasible to identify missing RPs [33].
Regulating gene expression
There is substantial evidence that modified ribosomes can specifically alter the translation of particular classes of mRNA, or even individual transcripts (Figure 3A) [4,34,35]. Conceivably, each ribosomal structure – characterized by its rRNA and protein composition, and their modifications – might be specific to a single mRNA transcript, or even transcript isoform. In favor of broader specificity, wild-type cells with ribosomes enriched in RPL10A preferentially translate a subset of mRNAs containing IRES elements [23]. Recent work in yeast identified RPS26-deficient ribosomes that preferentially bind mRNAs involved in select stress response pathways [36] and Horos et al., 2012 [37] reported that RPS19, affects the ribosomal density along hundreds of mRNAs essential for the differentiation of murine and human erythroblasts. Other studies also report that RP perturbations can affect the translation of hundreds of genes organized in coherent functional groups [23,38]. RPs are routinely dysregulated in the context of cancer [35,39], and adjusted throughout cell growth and metabolic cycles [40], for example by mTOR regulation via 5’ terminal oligopyrimidine (TOP) motifs in RP mRNAs [41].
More limited examples exist for mRNA-specific ribosomal regulation of translation. Barna and colleagues have suggested that RPL38, affects specifically the synthesis of only three proteins. However, the authors did not measure genome-wide translation so the possibility that the synthesis of other proteins is altered as well cannot be excluded [38,42]. Loss of RPS25 also resulted in inhibition of viral IRES-based translation, though not cap-dependent cellular translation [43]. The interferon-gamma regulated release of RPL13a from the ribosome is also postulated to impact the translation of around fifty genes [44,45]. These findings argue for a very high degree of specificity. This ability to preferentially translate individual or functional clusters of mRNAs could also allow the cell to help control localized translation by targeting the ribosomes responsible to specific subcellular destinations [46,47]. Targeting of mRNAs, either individually or in groups can be achieved through recognition of mRNA motifs, such as the aforementioned TOP motifs [41], or structures in the untranslated regions of mRNAs [48].
Many of these data on the mRNA-specificity of specialized ribosomes were obtained using sucrose gradient fractionation or immune enrichment, and thus these data reflect population averages over all ribosomal structures in each sucrose fraction, and likely capture only general trends that affect a large fraction of ribosomes and mRNAs, not ribosomal structures with single-gene specificity. Ribosomes vastly outnumber mRNA molecules in mammalian cells. If mRNA-specialized ribosomes play an important role, even modest changes to the ribosome population identified from sucrose gradient fractions could exert a significant effect.
A further benefit of mRNA-specific ribosomes could be in buffering mRNA noise (Figure 3C). Gene expression noise tends to be dominated by transcriptional noise due to transcriptional bursts and low copy number mRNAs. This can clearly be seen when examining transcriptomic and proteomic data from the same experimental system, with 10–100-fold changes in mRNA levels resulting in comparatively modest protein level changes. If post-transcriptional mechanisms did not actively buffer mRNA variability, these large fold changes would propagate to the protein levels. One buffering mechanism may involve microRNAs or other translational regulators such as RNA binding proteins (RBPs) [49,50]. Others may involve proteins interacting with specialized ribosomes and exerting direct feedback on the translation of their mRNAs. Good candidates for this mechanism of noise reduction are the RPs themselves.
RP levels correlate very poorly to their corresponding mRNA levels [51]. This poor correlation may reflect many posttranscriptional mechanisms such as protein degradation. Indeed, when RPL3 mRNAs is transcribed 7.5 times as much as in wildtype cells, RPL3 levels increase by less than 20% [52–55]. A particularly intriguing mechanism could be that some RPs, when incorporated into ribosomes, inhibit the translation of their own mRNAs, thus providing an efficient feedback-loop.
Speed vs. Accuracy: The elongation and error rates of the ribosome
Whilst altered RP stoichiometry may influence which mRNAs a specific ribosome may bind, it could also allow for modulation of ribosome behavior once it has bound a target mRNA. For example, the elongation rate of the ribosome. The elongation rate is usually understood as a tradeoff between the speed of translation and accuracy, with improved accuracy benefiting from a lower elongation rate via kinetic proofreading [56,57] (Figure 3D). Elongation rates have been understood to be variable for decades [58], with cellular tRNA pools impacting the relative and local elongation rates [59,60].
Lipopolysaccharide treatment of monocytes altered the translation rates of hundreds of proteins [32]. This was especially true for housekeeping proteins which are generally highly expressed and understood to be more translationally robust [61,62]. Indeed, while fold changes were dominated by altered mRNA levels, absolute protein abundance was dominated by altered translation and degradation rates. The ability to tune the elongation rate in response to changing conditions would permit cells the ability to produce certain proteins more rapidly, albeit with higher error rates.
Identifying ribosome heterogeneity
Recent advances have begun to demonstrate functional specialization of ribosomes within a species. We have demonstrated differences in RP stoichiometry in ribosomes purified from wildtype cells [22], though the functional specificity is implied by a correlation, not shown by direct measurement. Even the prominent example suggesting ribosome specialization, RPL38 regulating HOX genes [38,42], falls short of direct proof since (i) its exclusive specificity to 3 HOX is implied and not directly measured and (ii) the existence of ribosomes lacking RPL38 in wildtype cells is assumed, not measured. However, more recent data identified distinct mRNA subsets exhibiting enriched or diminished ribosome association with ribosomes enriched for RPL10a [23], and Ferretti & colleagues demonstrated a specific role for RPS26- containing ribosomes [36]. A rigorous experimental proof of specialization should demonstrate functional specialization of distinct ribosomal structures/compositions found in wildtype cells. Dynamic settings, such as a time course or differentiation protocol, offer the most straightforward means of inducing heterogeneity within a well- controlled framework, thus minimizing the potential for introducing artifacts. A first requirement is to identify what variation in RP stoichiometry exists in the system under study.
The separation of translating ribosomes on a sucrose gradient is a long-established method in the translation field. It allows the isolation of intact ribosomes and by isolating individual peaks along the gradient, comparisons can be made between the composition of the various monosome and polysome fractions. Mass spectrometry approaches (Box 3) using isobaric or metabolic labeling can be applied to these fractions to yield data on the relative abundance of core and ribosome-associated proteins. We successfully applied this approach in Slavov et al., 2015, identifying differences in monosome and polysome RP stoichiometry, as well as between fractions isolated from cells following stress such as yeast grown in ethanol or glucose (Figure 4A). However, the method has limited resolution for separating ribosome populations and the underlying populations do not represent pure ribosomal populations, and instead represent different levels of individual ribosome sub- populations.
Box 3: Mass spectrometry approaches to studying specialized ribosomes.
Mass spectrometry approaches offer a powerful tool with which to characterize functional specialization of ribosomes.
- Identifying heterogeneity
- o Bottom up and Top-down: Methods based on liquid chromatography coupled to tandem mass-spectrometry (LC-MS/MS) allow the quantification of RP’s with high throughput, specificity and accuracy [22,23,92,94]. These methods can be broadly divided into two categories. The first is bottom-up proteomics, which involves quantifying peptides resulting from digesting proteins with one or more proteases such as trypsin or Lys-C [95]. The second is top-down proteomics, which involves quantifying whole proteins or complexes such as ribosomes. Most current research on ribosomes is conducted with bottom-up approaches since these methods are currently more robust and accessible than top-down methods [75].
- o Quantification of relative RP abundance: Capturing the true heterogeneity of ribosomes poses a challenge for their LC-MS/MS analysis. Even when purified, e.g., by sucrose gradients, the ribosomes represent mixed populations. Measurements in such populations average across the heterogeneity. This averaging can substantially diminish the observed alterations to ribosomal protein (RP) stoichiometry to the limits of detection and quantification of existing methods [22]. The three major methods for mass spectrometry-based quantification are label-free, metabolic labelling approaches such as stable isotope labelling of amino acids in cell culture (SILAC), and isobaric labelling approaches such as Tandem mass tags (TMT) or iTRAQ [95,96]. Metabolic and isobaric labelling approaches permit relative comparison of multiple samples within the same run. This enables both increased multiplexing and canceling out sources of noise, e.g., variations in LC and ionization efficiency, when estimating the relative changes of a protein across samples [97]. Metabolic labelling approaches produce a shifted pattern of isotopic peaks for the peptide at the MS1 level. Relative quantification is achieved by comparing the peak areas for the unlabeled and labelled peptides. Isobaric labelling approaches including TMT result in labelled peptides that migrate identically at the MS1 level, but release unique reporter ions that can be quantified during MS/MS following peptide fragmentation [95].
- Determining functional specialization
- o Protein synthesis: Stable isotope-based pulse-labelling approaches provide the most direct means of measuring the synthesis and degradation rates of thousands of proteins when analysed by mass spectrometry [98]. The most recent iterations of this approach have combined SILAC pulse labelling with enhanced multiplexing through the use of TMT reagents [68].
- o Mis-translation rates: Current estimates for mis-translation rates are on the order of 10−3 to 10−4 , however a recent preprint has suggested that the sensitivity of modern mass spectrometers and data processing algorithms can be used to investigate the error rate of protein synthesis. [73].
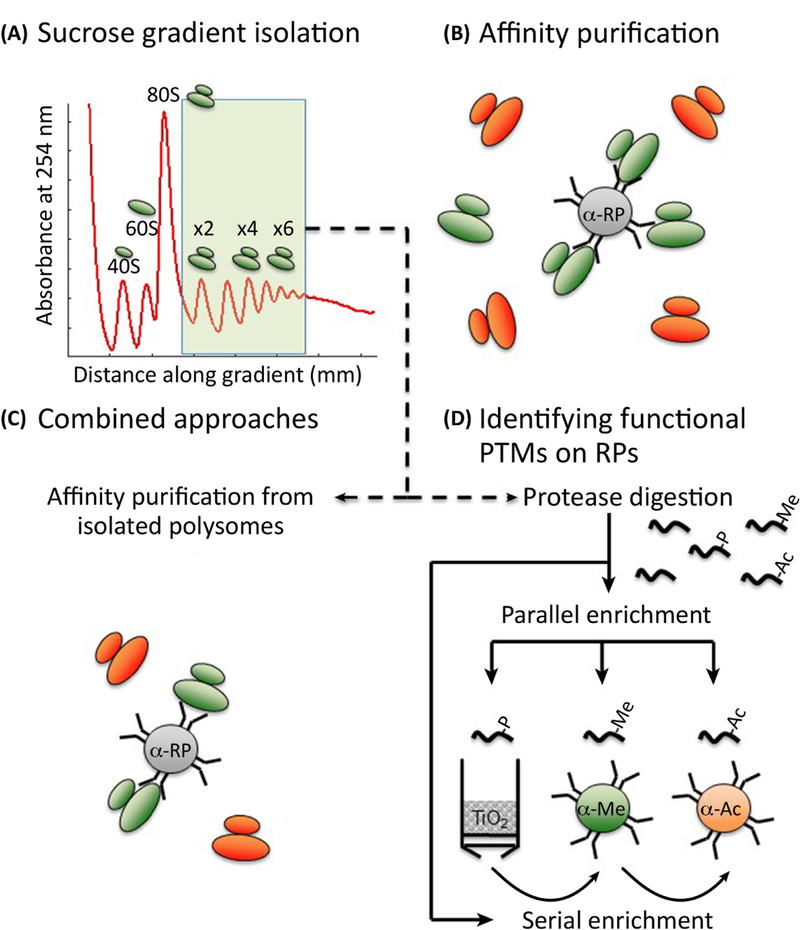
Figure 4. Identifying altered RP stoichiometry and PTMs.
A) For decades, the gold standard approach for isolating RPs in the context of intact, functional, ribosomes has been sucrose gradient centrifugation. B) Affinity purification is a powerful means of identifying differential RP association with complexes, however it cannot definitively say whether an RP is resident in a ribosome or represents an extra-ribosomal population of the RP. C) A combined approach whereby affinity purification is performed on sucrose gradient fractions allows the advantages of affinity purification to be applied to samples where the RP is known to be ribosome-resident. D) Heterogeneity amongst ribosomal protein modifications is a promising new area of research, and the methods required to explore this are an extension of those for identifying changes to RP association. Protease-digested peptides from sucrose gradient fractions or affinity purification can be enriched for a particular PTM of choice, either individually, or serially whereby the flow-through of one enrichment is applied to the next enrichment process.
Subcellular compartmentalization of specific ribosome populations is an emerging area for research. Advances in methods for detecting ongoing translation in cells have helped underline subcellular variation in translation [47]. Whilst ribosomes are generally taught as being free-cytoplasmic, or ER-associated, translation can be found localized near synapses in neuronal cells, sequestered in virus factories following infection, or even in the nucleus. Subcellular fractionation or purification methods e.g.LOPIT [63] could therefore be applied to distinguish ribosome populations of interest. A further area where heterogeneity could exist is between individual single cells. Our group recently made advances in this area with the advent of a first method for performing single cell mass spectrometry on average-sized mammalian cells [64]. Indeed, the data suggested altered RP stoichiometry between the two cell lines under study. However, in both the case of single cell, and subcellular localization-based approaches, it is uncertain whether the RPs demonstrating altered abundance are incorporated into fully assembled ribosomes or not, and therefore follow-up experiments would be required to determine whether changes determined in whole cell lysates are representative of assembled ribosomes.
Once RPs exhibiting altered stoichiometry are known, specific isolation of more homogenous ribosome populations can be attempted. Methods for this include affinity purification (Figure 4B). A caveat with this approach is that a simple affinity purification will isolate both ribosome associated, and free-RPs. Particularly in the case of epitope tagged RPs, the incorporation of the tagged RP into the ribosome may be poor relative to the endogenous RP. Several means exist to ameliorate these issues. Prior removal of nuclei followed by affinity purification will reduce the background from incomplete/assembling ribosomes but still yield a mix of ribosome-associated and free RP. An improved approach would combine sucrose gradient centrifugation and affinity purification, with the affinity purification being conducted on pooled, or individual gradient fractions thus ensuring the isolated protein was derived from intact ribosomes (Figure 4C).
RPs are host to a huge array of PTMs [65], with over 2500 modifications of core human RPs known [66] (Figure 2C). The identification of RP PTMs represents an extension of the methods required to investigate RP stoichiometry. Large scale PTM screens can be conducted by mass spectrometry by enriching for individual PTMs such as phosphorylation, methylation and acetylation which represent a majority of currently known RP PTMs. One additional consideration when investigating RP PTMs is the case where the addition of a PTM induces the loss of ribosome association of the modified RP. A known example of this is L13a where phosphorylation at Ser-77 is associated with dissociation from the 60S ribosomal subunit [44]. As such, the inclusion of either whole cell lysate or soluble cytoplasmic extracts prepared from the same cells used for sucrose gradient fractionation would allow determination of whether PTM status is affecting ribosome association of the RP. Ideally, PTM- enrichment should be performed on the same samples used for investigating RP stoichiometry, allowing the inference of PTM stoichiometry [67]. Functional validation of the impact of PTMs could be determined using inhibitors, knock-out or mutagenesis approaches, and examining their impacts on the outputs described above.
Demonstrating functional specialized ribosomes
A conclusive demonstration of altered RP stoichiometry does not prove functional specialized ribosomes. A key task is the identification of outputs which can be directly attributed to the ribosome itself, rather than noise from transcriptional or translation initiation events which may also be influenced by a perturbation of choice. Ideally, several outputs would be examined, as illustrated in Figure 5A. Pulsed time course experiments have been employed for decades in the study of protein synthesis and turnover, and non-radioactive versions are amenable to mass spectrometry-based analysis (Box 3). These approaches allow for the investigation of the turnover and degradation rates of thousands of proteins, with a recent study characterizing the dynamics of over 6000 proteins [68,69] However, protein synthesis rate per mRNA can change not only because of ribosome remodeling but also because of translation factors affecting translation initiation and elongation. It therefore may provide a potential functional readout, rather than definitive confirmation of functional specialization. A similar claim can be made for investigating the association of specific mRNAs with ribosome subsets following a perturbation. mRNA specificity represents a key area where ribosome specialization could play a role (Figure 5B). However, the degree of association of an individual mRNA with specific ribosomes can be determined not only by increased affinity of specialized ribosomes for the mRNA but also by altered mRNA abundance, and translation initiation factors.
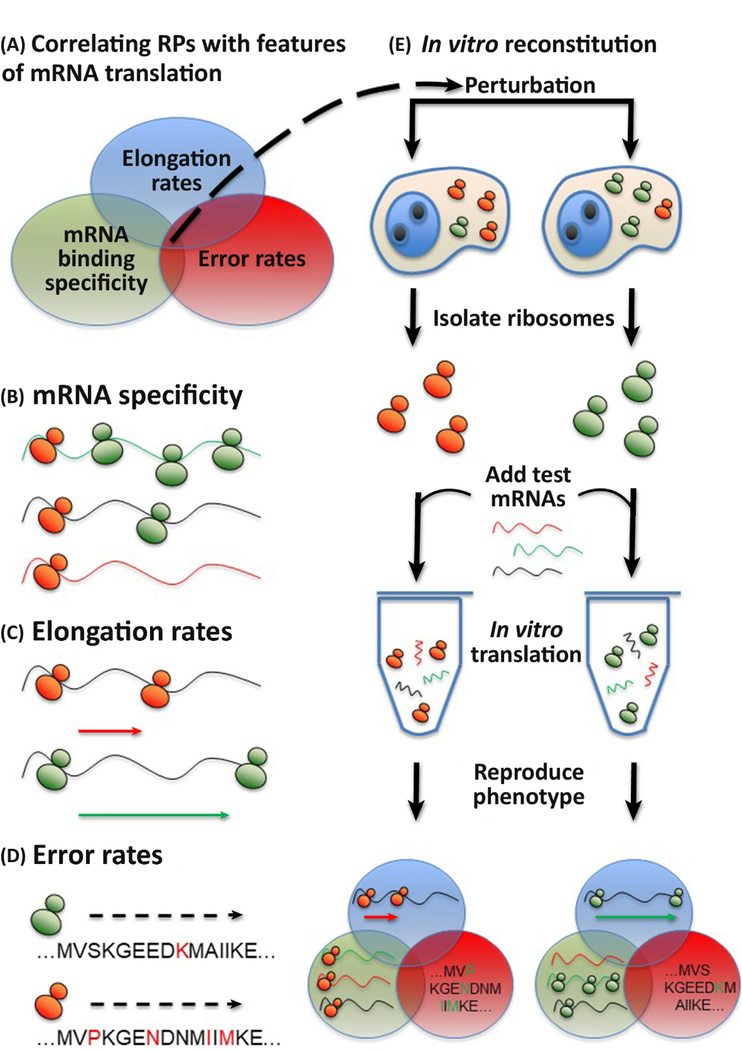
Figure 5. Testing for functional specialization.
A) To experimentally prove ribosome specialization, several outputs for measurement stand out. These are the mRNA binding specificity, elongation rates and error rates. A conclusive demonstration of functional ribosome specialization will likely employ several or all of these. B) If specific mRNAs are favored by individual ribosome conformations then this can be assessed by immunoprecipitation with tagged RPs. C) Elongation rates for the ribosome on particular mRNA substrates can be estimated from pulse-chase data. D) The error rate for individual ribosomes can be monitored using luminescent or fluorescent reporters for specific substrates, or in a higher-throughput manner by mass spectrometry. E) Functional validation of specialized ribosomes can be investigated through in vitro reconstitution of the phenotype.
For a definitive result, the elongation and error rates stand out for investigation because they relate directly to ribosome activity, though can still be influenced by trans-factors [70] (Figure 5C, 5D). One possibility makes use of the inhibitor harringtonine which stalls translation at the initiation codon. Using a modification of the widely adopted ribosome profiling method [71], reduced ribosome density on a given mRNA at extending intervals after the addition of harringtonine are used to calculate the average time it takes a ribosome to completely traverse an mRNA. When the length of the mRNA is known this can be used to calculate the elongation rate [72], though this method has yet to be widely adopted by the translation community.
The error rate of the translating ribosome also offers a promising target for investigating ribosome specialization (Box 3). Typically, such assays are low- throughput, and rely on stop codon readthrough or frameshift/coding errors to generate a detectable signal, typically by a luciferase or a fluorescent reporter. However, these methods are very context-dependent and may therefore miss trends in error rates outside their specific context. However, a recent preprint has suggested a possible mass spectrometry-based approach [73] to identify mis-translation products. While the sensitivity of the approach may limit it to study of the more abundant mis-translation products, the authors data included altered error rates following perturbations such as amino acid starvation and the addition of an antibiotic known to effect ribosomal proofreading function, suggesting the method holds promise as a high-throughput means of investigating ribosomal error rates.
Complementary approaches
Mass spectrometry represents a powerful tool for investigating RP stoichiometry, though ultimately its conclusions are drawn from mixed, albeit enriched populations of ribosomes. Single molecule methods and imaging offer a powerful means of identifying the precise composition of individual ribosomes. Recent work highlighted how cryo-EM could be used to map the proportions of yeast ribosomes containing or lacking RPL10 and RPS1A/B [33]. Alternative approaches include super-resolution microscopy which would allow imaging of ribosomes directly in cells. It does require fluorophore labeling which can be limited in throughput, by epitope occlusion, or lead to artifacts if fluorescent proteins as used. Alternatively, top-down mass spectrometry approaches, where intact proteins or complexes can be analyzed to determine structural and conformational information have also begun to identify altered ribosome compositions [74,75]. This ability to precisely define specific, individual ribosome conformations, will prove invaluable for proving true RP heterogeneity within single cells.
Finally, whilst the above methods can validate the existence of altered RP stoichiometry, and of functional ribosome specialization in cells, there remains a large degree of overlap where the impact of the ribosome and of other, linked translational events can contribute to this heterogeneity. The ability to extract specific ribosome conformations from cells and reproduce translational phenotypes in vitro is key (Figure 5E). Various methods for preparing translational components from cells are known, ranging from crude preparations [76], to methods requiring extensive fractionation [77–81]. The reproduction of specific translational phenotypes present in cells, including mRNA specificity, elongation and error rates with specific ribosomes in vitro offers the most stringent demonstration of functional ribosome specialization.
Ribosome specialization: more than just RPs
We have focused on the impact of RP stoichiometry for ribosome function. However, equally important and interesting are modifications of the rRNAs that may also confer ribosome specificity as discussed by Mauro and Matsuda, 2016 [7]. Indeed, rRNA isoforms are expressed in tissue-specific patterns [12], complementing observations of cell-specific RP transcripts [82,83]. rRNA’s exhibit extensive and pervasive variation at the level of rDNA between individuals [12], and rRNA modifications were identified at sub-stoichiometric amounts in recent studies [19,20]. Technological advances such as the ability to directly sequence full-length RNA molecules and identify modifications through the use of nanopore sequencing [84], could be combined with the above proteomic approaches to investigate rRNA heterogeneity and function in order to obtain a more complete perspective on the constellation of features that distinguish individual ribosomes and their function.
Highlights.
New and emerging methods permit the identification of ribosomes with distinct ribosomal protein stoichiometry from eukaryotic cells, and the exploration of ways in which ribosome-mediated regulation could contribute to the control of protein synthesis.
Two models for ribosome-mediated translation regulation exist: 1) The abundance model whereby the abundance of ribosomes within the cell can exert bi-directional global regulation of mRNAs. 2) The specialized ribosome model in which changes to ribosome stoichiometry or post-translational modifications permit finer control of ribosome function.
Regulation by specialized ribosomes could include mRNA-specific or functional class- specific ribosomes, and modification of ribosome elongation and error rates in response to stress or stimuli.
This review proposes a path to unambiguously identify functional ribosome specialization within and between mammalian cells.
Outstanding Questions:
To what degree do specialized ribosomes contribute to the regulation of protein synthesis in wild-type cells? Is this a common or niche regulatory mechanism?
How mRNA-specific are specialized ribosomes? Are there ribosomes for specific isoforms or splice variants of an mRNA?
Do ribosomes exist in wild-type cells with distinct elongation or error rates? Are these regulated (or dysregulated) in response to stress or stimuli?
How much of a role to tissue-specific specialized ribosomes play in differentiation and cell-specific translation?
Acknowledgements
The authors thank Annie Schide, Aleksandra Petelski, and the rest of the Slavov lab for constructive feedback. The authors also wish to apologize to their colleagues whose work was omitted due to space considerations. This work was funded by a New Innovator Award from the NIGMS from the National Institutes of Health to N.S. under Award Number DP2GM123497.
Glossary
- 40S
The small ribosomal subunit in eukaryotic ribosomes. The prokaryotic equivalent is the 30S subunit.
- 60S
The large ribosomal subunit in eukaryotic ribosomes. The prokaryotic equivalent is the 50S subunit.
- 80S
See monosome.
- Bottom up (mass spectrometry)
Bottom up proteomics is the analysis of protease digested protein samples. These methods measure peptides, not full-length proteins, and the peptide-level data is used to infer protein levels.
- Elongation rate
The rate at which the ribosome is able to extend the growing polypeptide chain as it proceeds along a mRNA.
- Error rate
The rate at which the incorrect amino acid is mis- incorporated into elongating polypeptide chain by the ribosome.
- Extra-ribosomal
Some RPs are proposed to have roles within the cell separate to their role in forming part of the ribosome. When these proteins are found outside the ribosome they are termed extra-ribosomal.
- Heterogenous ribosomes
Ribosomes possessing variation in their RP, rRNA or PTM stoichiometry. Specialized ribosomes are a subset of heterogenous ribosomes where the heterogeneity has been linked to specific functions.
- Histone code
The ‘histone code’ is the hypothesis that post-translational modifications of histone proteins function combinatorically to provide highly customizable control of transcription.
- Housekeeping protein(s)
Proteins required for the basic functioning of the cell. Constitutively expressed, often to high levels.
- IRES
Internal ribosome entry site. A RNA sequence allowing cap-independent translation of a target mRNA containing the IRES.
- Kinetic proofreading
A method for correcting errors in biochemical reactions. By separating a reaction into multiple irreversible intermediate steps, error rates far lower than would otherwise be possible with a single-step reaction can be achieved.
- Label-free
Methods of analyzing one sample at a time that does not contain isotopic labels. These methods can be DDA or DIA.
- Monosome
A single ribosome (80S) comprising both small (40S) and large (60S) ribosomal subunits. Isolated monosomes may not necessarily be associated with a translating mRNA and as such monosome populations can not be assumed to be fully translationally active.
- MS1
Following liquid chromatography and introduction into a mass spectrometer, MS1 analysis of a peptide reveals its charge state and mass. Whilst this might sometimes reveal the amino acid content of a peptide, sequencing at the MS2 level provides the amino acid order and more definitive identification.
- MS/MS (MS2)
A peptide of interest, identified at MS1 level, can be isolated within the mass spectrometer, fragmented, and the fragment m/z ratios analyzed. These fragments form a ladder of ions which can be used to determine the sequence order of amino acids within the peptide.
- Polysome
Multiple ribosomes present on a single mRNA. Polysome fractions from cells are studied as unlike monosome fractions, the presence of multiple ribosomes on a mRNA indicates active translation.
- Proteoform
A term that describes different modification states of a single protein. For example, unmodified RPS6, and RPS6 phosphorylated at Serine 148 represent different proteoforms of the same protein, and potentially possess distinct functions or behavior.
- PTM
Post-translational modification. Common post- translational modifications of proteins include phosphorylation, acetylation and methylation. The addition or removal of a PTM can cause changes to a proteins structure, binding partners or function.
- Ribosome code
The hypothesis that modifications to ribosome stoichiometry or PTM state of individual RPs can function in a combinatorial manner to generate specialized ribosomes with a high degree of customizability. A similar concept is the histone code.
- Ribosomopathy
A pathological condition resulting from a mutation or absence of a particular ribosomal protein, rRNA, or ribosome biogenesis factor.
- RP
Ribosomal protein. In humans there are approximately 80 ribosomal proteins.
- SILAC
Stable isotope labelling of amino acids in cell culture. A metabolic-labelling technique permitting relative quantification of proteins in a sample by mass spectrometry at the MS1 level.
- Top-down (mass spectrometry)
The analysis of intact proteins or complexes by mass spectrometry, in contrast to the more common ‘bottom-up’ approach where proteins are first digested to peptides and proteins are identified and quantified based on these peptides.
- TMT
Tandem Mass Tags. An isobaric labelling method allowing multiplexing and quantification of multiple samples by mass spectrometry. Unlike SILAC-quantification, quantification occurs at the MS2 level.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Crick FH (1958) On protein synthesis. Symp. Soc. Exp. Biol 12, 138–63 [PubMed] [Google Scholar]
- 2.Lodish HF (1974) Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251, 385–8 [DOI] [PubMed] [Google Scholar]
- 3.Mills EW and Green R (2017) Ribosomopathies: There’s strength in numbers. Science 358, eaan2755 [DOI] [PubMed] [Google Scholar]
- 4.Khajuria RK et al. (2018) Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell DOI: 10.1016/j.cell.2018.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzl-Raz E et al. (2017) Principles of cellular resource allocation revealed by condition-dependent proteome profiling. Elife 6, e28034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert WV (2011) Functional specialization of ribosomes? Trends Biochem. Sci 36, 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mauro VP and Matsuda D (2016) Translation regulation by ribosomes: Increased complexity and expanded scope. RNA Biol 13, 748–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue S and Barna M (2012) Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol 13, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinman JD (2016) Pathways to Specialized Ribosomes: The Brussels Lecture. J. Mol. Biol 428, 2186–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genuth NR and Barna M (2018) Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat. Rev. Genet 2018 DOI: 10.1038/s41576-018-0008-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preiss T (2016) All Ribosomes Are Created Equal. Really? Trends Biochem. Sci 41, 121–123 [DOI] [PubMed] [Google Scholar]
- 12.Parks MM et al. (2018) Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv 4, eaao0665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta V and Warner JR (2014) Ribosome-omics of the human ribosome. RNA 20, 1004–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komili S et al. (2007) Functional Specificity among Ribosomal Proteins Regulates Gene Expression. Cell 131, 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenuwein T and Allis CD (2001) Translating the histone code. Science 293, 1074–80 [DOI] [PubMed] [Google Scholar]
- 16.Prakash K and Fournier D (2018) Evidence for the implication of the histone code in building the genome structure. Biosystems 164, 49–59 [DOI] [PubMed] [Google Scholar]
- 17.Strahl BD and Allis CD (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 18.Simsek D et al. (2017) The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell 169, 1051–1065.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popova AM and Williamson JR (2014) Quantitative Analysis of rRNA Modifications Using Stable Isotope Labeling and Mass Spectrometry. J. Am. Chem. Soc 136, 2058–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogh N et al. (2016) Profiling of 2′- O -Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res 44, 7884–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seip B and Innis CA (2016) How Widespread is Metabolite Sensing by Ribosome-Arresting Nascent Peptides? J. Mol. Biol 428, 2217–2227 [DOI] [PubMed] [Google Scholar]
- 22.Slavov N et al. (2015) Differential Stoichiometry among Core Ribosomal Proteins. Cell Rep 13, 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Z et al. (2017) Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol. Cell 67, 71–83.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horos R et al. (2012) Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood 119, 262–272 [DOI] [PubMed] [Google Scholar]
- 25.Lee AS-Y et al. (2013) A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc. Natl. Acad. Sci. U. S. A 110, 324–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortier S et al. (2015) Haploinsufficiency screen highlights two distinct groups of ribosomal protein genes essential for embryonic stem cell fate. Proc. Natl. Acad. Sci 112, 2127–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber HJ (1972) Stoichiometric measurements of 30S and 50S ribosomal proteins from Escherichia coli. Mol. Gen. Genet 119, 233–48 [DOI] [PubMed] [Google Scholar]
- 28.Westermann P et al. (1976) On the stoichiometry of proteins in the small ribosomal subunit of hepatoma ascites cells. FEBS Lett 62, 132–5 [DOI] [PubMed] [Google Scholar]
- 29.Granneman S and Baserga SJ (2004) Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res 296, 43–50 [DOI] [PubMed] [Google Scholar]
- 30.Sykes MT and Williamson JR (2009) A Complex Assembly Landscape for the 30S Ribosomal Subunit. Annu. Rev. Biophys 38, 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen SS and Williamson JR (2013) Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J. Mol. Biol 425, 767–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jovanovic M et al. (2015) Dynamic profiling of the protein life cycle in response to pathogens. Science (80-. ) 347, 1259038–1259038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun M et al. (2018) Identification of changing ribosome protein compositions using cryo-EM and mass spectrometry. bioRxiv DOI: 10.1101/271833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhuri S et al. (2007) Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA 13, 2224–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bastide A and David A (2018) The ribosome, (slow) beating heart of cancer (stem) cell. Oncogenesis 7, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferretti MB et al. (2017) Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements. Nat. Struct. Mol. Biol 24, 700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horos R et al. (2012) Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood 119, 262–272 [DOI] [PubMed] [Google Scholar]
- 38.Kondrashov N et al. (2011) Ribosome-Mediated Specificity in Hox mRNA Translation and Vertebrate Tissue Patterning. Cell 145, 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guimaraes JC and Zavolan M (2016) Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 17, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slavov N and Botstein D (2011) Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol. Biol. Cell 22, 1997–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoreen CC et al. (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue S et al. (2015) RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landry DM et al. (2009) RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 23, 2753–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazumder B et al. (2003) Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115, 187–98 [DOI] [PubMed] [Google Scholar]
- 45.Vyas K et al. (2009) Genome-wide polysome profiling reveals an inflammation-responsive posttranscriptional operon in gamma interferon- activated monocytes. Mol. Cell. Biol 29, 458–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rangaraju V et al. (2017) Local translation in neuronal compartments: how local is local? EMBO Rep 18, 693–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.David A et al. (2012) Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J. Cell Biol 197, 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinnebusch AG et al. (2016) Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 352, 1413–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faure AJ et al. (2017) Systematic Analysis of the Determinants of Gene Expression Noise in Embryonic Stem Cells. Cell Syst 5, 471–484.e4 [DOI] [PubMed] [Google Scholar]
- 50.Schmiedel J et al. (2017) Noise control is a primary function of microRNAs and post-transcriptional regulation. bioRxiv DOI: 10.1101/168641 [DOI] [Google Scholar]
- 51.Franks A et al. (2017) Post-transcriptional regulation across human tissues. PLoS Comput. Biol 13, e1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yates JL et al. (1981) E. coli ribosomal protein L10 inhibits translation of L10 and L7/L12 mRNAs by acting at a single site. Nature 294, 190–2 [DOI] [PubMed] [Google Scholar]
- 53.Pearson NJ et al. (1982) Yeast use translational control to compensate for extra copies of a ribosomal protein gene. Cell 29, 347–55 [DOI] [PubMed] [Google Scholar]
- 54.Warner JR et al. (1985) Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol. Cell. Biol 5, 1512–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sung MK et al. (2016) A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. Elife 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hopfield JJ (1974) Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. U. S. A 71, 4135–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodnina MV and Wintermeyer W (2001) Fidelity of Aminoacyl-tRNA Selection on the Ribosome: Kinetic and Structural Mechanisms. Annu. Rev. Biochem 70, 415–435 [DOI] [PubMed] [Google Scholar]
- 58.Talkad V et al. (1976) Evidence for variable rates of ribosome movement in Escherichia coli. J. Mol. Biol 104, 299–303 [DOI] [PubMed] [Google Scholar]
- 59.Spencer PS et al. (2012) Silent Substitutions Predictably Alter Translation Elongation Rates and Protein Folding Efficiencies. J. Mol. Biol 422, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu C-H et al. (2015) Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding. Mol. Cell 59, 744–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drummond DA et al. (2005) Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. U. S. A 102, 14338–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drummond DA et al. (2006) A single determinant dominates the rate of yeast protein evolution. Mol. Biol. Evol 23, 327–37 [DOI] [PubMed] [Google Scholar]
- 63.Mulvey CM et al. (2017) Using hyperLOPIT to perform high-resolution mapping of the spatial proteome. Nat. Protoc 12, 1110–1135 [DOI] [PubMed] [Google Scholar]
- 64.Budnik B et al. (2018) SCoPE-MS: mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biology DOI: 10.1186/s13059-018-1547-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simsek D and Barna M (2017) An emerging role for the ribosome as a nexus for post-translational modifications. Curr. Opin. Cell Biol 45, 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hornbeck PV et al. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43, D512–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malioutov D et al. (2018) Quantifying homologous proteins and proteoforms. Molecular & Cellular Proteomics, DOI: 10.1074/mcp.TIR118.000947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zecha J et al. (2018) Peptide level turnover measurements enable the study of proteoform dynamics. Mol. Cell. Proteomics DOI: 10.1074/mcp.RA118.000583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Savitski MM et al. (2018) Multiplexed Proteome Dynamics Profiling Reveals Mechanisms Controlling Protein Homeostasis. Cell 173, 260–274.e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noel JK and Whitford PC (2016) How EF-Tu can contribute to efficient proofreading of aa-tRNA by the ribosome. Nat. Commun 7, 13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ingolia NT et al. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ingolia NT et al. (2011) Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell 147, 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mordret E et al. (2018) Systematic detection of amino acid substitutions in proteome reveals a mechanistic basis of ribosome errors. bioRxiv DOI: 10.1101/255943 [DOI] [PubMed] [Google Scholar]
- 74.van de Waterbeemd M et al. (2017) High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods 14, 283–286 [DOI] [PubMed] [Google Scholar]
- 75.Belov AM et al. (2017) Analysis of Proteins, Protein Complexes, and Organellar Proteomes Using Sheathless Capillary Zone Electrophoresis - Native Mass Spectrometry. J. Am. Soc. Mass Spectrom 28, 2614–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Favre D and Trepo C (2001) Translational extracts active biologically in vitro obtained from eukaryotic monolayer cells: a versatile method for viral RNA studies. J. Virol. Methods 92, 177–181 [DOI] [PubMed] [Google Scholar]
- 77.Lomakin IB et al. (2006) The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J 25, 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pisarev AV et al. (2007) Assembly and Analysis of Eukaryotic Translation Initiation Complexes. In Methods in enzymology 430pp. 147–177 [DOI] [PubMed] [Google Scholar]
- 79.Acker MG et al. (2007) Reconstitution of Yeast Translation Initiation. In Methods in enzymology 430pp 111–145 [DOI] [PubMed] [Google Scholar]
- 80.Benne R and Hershey JW (1978) The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem 253, 3078– 87 [PubMed] [Google Scholar]
- 81.Trachsel H et al. (1977) Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J. Mol. Biol 116, 755–67 [DOI] [PubMed] [Google Scholar]
- 82.Ramagopal S and Ennis HL (1981) Regulation of synthesis of cell-specific ribosomal proteins during differentiation of Dictyostelium discoideum. Proc. Natl. Acad. Sci. U. S. A 78, 3083–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramagopal S (1990) Induction of cell-specific ribosomal proteins in aggregation-competent nonmorphogenetic Dictyostelium discoideum. Biochem. Cell Biol 68, 1281–7 [DOI] [PubMed] [Google Scholar]
- 84.Garalde DR et al. (2018) Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 [DOI] [PubMed] [Google Scholar]
- 85.Mills EW and Green R Ribosomopathies: There’s strength in numbers. ,Science, 358 (2017) [DOI] [PubMed] [Google Scholar]
- 86.McCann KL and Baserga SJ (2013) Mysterious ribosomopathies. Science 341, 849–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bolze A et al. (2018) Incomplete penetrance for isolated congenital asplenia in humans with mutations in translated and untranslated RPSA exons. Proc. Natl. Acad. Sci. U. S. A DOI: 10.1073/pnas.1805437115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bolze A et al. (2013) Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science 340, 976–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brooks SS et al. (2014) A Novel Ribosomopathy Caused by Dysfunction of RPL10 Disrupts Neurodevelopment and Causes X-Linked Microcephaly in Humans. Genetics 198, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Keersmaecker K et al. (2013) Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet. 45, 186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rao S et al. (2012) Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 120, 3764– 73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Imami K et al. (2018) Phosphorylation of the Ribosomal Protein RPL12/uL11 Affects Translation during Mitosis. Mol. Cell 72, 84–98.e9 [DOI] [PubMed] [Google Scholar]
- 93.Jenuwein T and Allis CD (2001) Translating the histone code. Science 293, 1074–80 [DOI] [PubMed] [Google Scholar]
- 94.Samir P et al. (2018) Identification of Changing Ribosome Protein Compositions using Mass Spectrometry. Proteomics DOI: 10.1002/pmic.201800217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sinitcyn P et al. (2018) Computational Methods for Understanding Mass Spectrometry–Based Shotgun Proteomics Data. Annu. Rev. Biomed. Data Sci 1, 207–234 [Google Scholar]
- 96.Altelaar AFM et al. (2013) Benchmarking stable isotope labeling based quantitative proteomics. J. Proteomics 88, 14–26 [DOI] [PubMed] [Google Scholar]
- 97.Thompson Andrew, † et al. (2003) Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by MS/MS DOI: 10.1021/AC0262560 [DOI] [PubMed] [Google Scholar]
- 98.Selbach M et al. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 99.Zhang X et al. (2016) Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes. Nat. Commun 7, 13223. [DOI] [PMC free article] [PubMed] [Google Scholar]