Dear Editor,
Next-generation sequencing (NGS) is rapidly being adapted in clinical practice, and numerous clinical laboratories are using this technology to assess patients with acute myeloid leukemia (AML). Among recurrent somatic mutations in AML, the FLT3 internal tandem duplication (ITD) has major clinical implications and is associated with adverse outcomes [1,2,3]. Identification of the mutation is critical for risk stratification and the decision of early hematopoietic stem cell transplantation [2,3]. Furthermore, as activated kinase targets such as FLT3 and JAK are under active clinical investigation [4,5], it has become increasingly important to identify activating FLT3 mutations. During NGS, however, accurate mapping and consequent variant calling is not always possible because the duplication size could be small or large, and the inserted segment is essentially, but not necessarily, a part of the normal reference sequence.
We performed NGS for patients with AML. Bone marrow aspirates of 229 patients diagnosed as having AML between March 2017 and February 2018 in Severance Hospital, Seoul, Korea, were prospectively collected after obtaining informed consent. The study was approved by our Institutional Review Board (4-2016-0869). Thirty patients harbored the FLT3 ITD mutation, identified by PCR and fragment analysis. To determine whether FLT3 ITD mutations could be accurately identified from targeted multigene NGS data, we analyzed the NGS data with different algorithms and compared the results with those of PCR and fragment analysis, a current standard test.
NGS was performed using the NextSeq 550 instrument (Illumina, San Diego, CA, USA), with custom probes (Celemics, Seoul, Korea) for AML. The Burrows-Wheeler alignment tool (0.7.12) was used for sequence alignment [6]. GATK MuTect2 (3.8–0), and Pindel (0.2.0) algorithms were used to detect FLT3 ITD mutations [7,8], which were implemented in the DxSeq algorithm (Dxome, Seoul, Korea). PCR amplification and fragment detection analyses were performed using a 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and GeneMapper 3.2 software (Applied Biosystems), with a modified protocol based on that previously described [9].
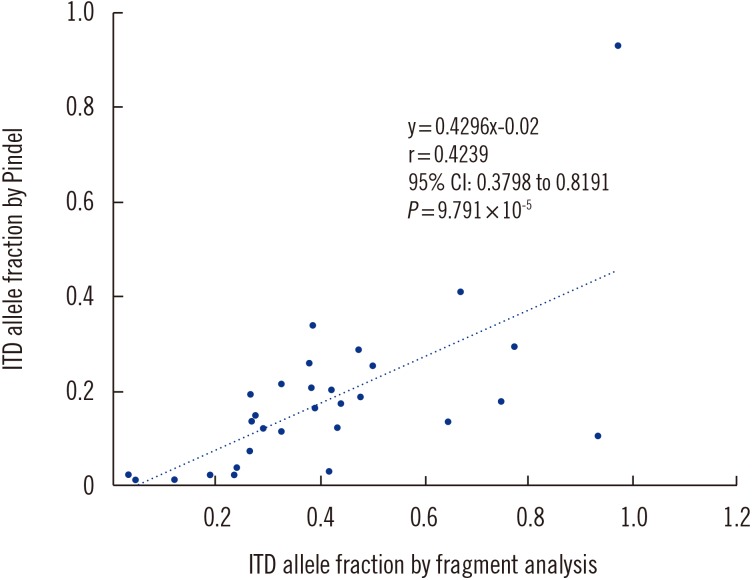
The median depth of the FLT3 gene was 4,412×, ranging from 2,626× to 7,595×. GATK MuTect2 detected FLT3 ITD mutations in 15 of 30 cases, most of which had a relatively short duplicated segment. The Pindel algorithm detected FLT3 ITD mutations in all cases. The duplication ranged from 18 to 201 bp, and the mutant allele burden was estimated to be 0.028–0.930 on PCR fragment analysis. The duplication sizes estimated using Pindel algorithm were concurrent with that by PCR fragment analysis in all cases, while the mutant allele fraction calculated using Pindel tended to be lower than that obtained by PCR fragment analysis (Fig. 1). Values from the two methods were not strongly correlated (Fig. 1).
Fig. 1. Correlation of the ITD mutant allele fraction between fragment analysis and the Pindel (0.2.0) algorithm for patients harboring FLT3 ITD mutation.
Abbreviation: ITD, internal tandem duplication.
The different results of the two variant callers stem from their algorithms. GATK MuTect2 helps detect insertions and deletions (indels), using initially identified indels during read alignment [10]. Owing to process limitations among read pairs with discordant sizes or orientations mapped with high confidence, the algorithm can detect only insertions contained entirely within individual read alignments, typically less than approximately 15–20 bp. In contrast, Pindel is a pattern-growth algorithm using read pairs, wherein one is partially or completely unaligned [8]. Inclusion of these reads is critical for identifying medium-size insertions, such as those in FLT3 ITD mutations, because this size range is too large to be detected on the basis of indels within single reads and is often too small for detection by analysis of discordant insert sizes.
However, the Pindel algorithm tends to underestimate the mutant allele fraction. Inaccurate estimation of allele burden is a major disadvantage because the FLT3 ITD mutant allele burden is one of the prognostic markers for AML [2,3]. Identification of FLT3 ITD is challenging because wild-type DNA yields shorter amplicons than mutant DNA, thereby having a competitive advantage leading to a PCR bias in NGS analyses adopting amplicon-based enrichment. NGS analyses with hybrid capture probes may be limited in capturing mutant DNAs with large insertions or deletions partly owing to large mismatches between capture probes and mutant DNAs. The capture bias tends to increase with an increase in duplication length. In one case, this difference was the largest at a 138-bp insertion. Although the true mutant allele burden could be deduced from the variant allele frequency of each tool, the existence of a larger portion of leukemic cells harboring the mutation warrants consideration.
In conclusion, the Pindel algorithm was highly effective in detecting FLT3 ITD mutations in the NGS assessment of AML patients and was superior to GATK MuTect2. Although the mutant allele fraction calculated tended to be lower than that obtained by PCR fragment analysis, Pindel has clinical utility for identifying clinically significant FLT3 ITD mutations in a single NGS analysis.
Footnotes
Authors' Disclosures of Potential Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.National Comprehensive Cancer Network. Acute Myeloid Leukemia (Version 2.2018) [Updated on Aug 2018, registration required]. https://www.nccn.org/professionals/physician_gls/pdf/aml.pdf.
- 2.Chevallier P, Labopin M, Turlure P, Prebet T, Pigneux A, Hunault M, et al. A new Leukemia Prognostic Scoring System for refractory/relapsed adult acute myelogeneous leukaemia patients: a GOELAMS study. Leukemia. 2011;25:939–944. doi: 10.1038/leu.2011.25. [DOI] [PubMed] [Google Scholar]
- 3.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degryse S, Cools J. JAK kinase inhibitors for the treatment of acute lymphoblastic leukemia. J Hematol Oncol. 2015;8:91. doi: 10.1186/s13045-015-0192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wander SA, Levis MJ, Fathi AT. The evolving role of FLT3 inhibitors in acute myeloid leukemia: quizartinib and beyond. Ther Adv Hematol. 2014;5:65–77. doi: 10.1177/2040620714532123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zwaan CM, Meshinchi S, Radich JP, Veerman AJ, Huismans DR, Munske L, et al. FLT3 internal tandem duplication in 234 children with acute myeloid leukemia: prognostic significance and relation to cellular drug resistance. Blood. 2003;102:2387–2394. doi: 10.1182/blood-2002-12-3627. [DOI] [PubMed] [Google Scholar]
- 10.Spencer DH, Abel HJ, Lockwood CM, Payton JE, Szankasi P, Kelley TW, et al. Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J Mol Diagn. 2013;15:81–93. doi: 10.1016/j.jmoldx.2012.08.001. [DOI] [PubMed] [Google Scholar]