Abstract
Purpose: Melanosis coli (MC) is a disorder of pigmentation of the wall of the colon, often identified at the time of colonoscopy. The aim of the present study is to identify candidate biomarkers for MC. Methods: The transcriptome data for MC (GSE78933) with five MC tissues and five corresponding normal tissues is obtained from the NCBI Gene Expression Omnibus (GEO) database. R/Bioconductor package limma was used to screen differently expressed genes (DEGs). ClueGO of cytoscape was applied for Gene Ontology (GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. Based on STRING V10 database, protein–protein interaction (PPI) network was constructed. The pathological tissue and normal tissue from 23 MC patients and 23 controls were collected, respectively. The relative expression of hub nodes was detected by qRT-PCR and Western blot. For regulating the expression of these genes, overexpression vector was constructed or siRNA transfection was used. Finally, apoptosis was detected by flow cytometry. Results: Total 1342 DEGs were screened, including 786 up-regulated and 556 down-regulated genes. These genes were mainly enriched in stimulatory C-type lectin receptor signaling pathway, polysaccharide biosynthetic process, intracellular, and oxidative phosphorylation. PPI network was then constructed with 426 DEGs and 895 interactions. Thereinto, G-protein subunit γ 5 (GNG5), lysophosphatidic acid receptor 3 (LPAR3), mitogen-activated protein kinase 8 (MAPK8), NHP2L1, proteasome 26S subunit, ATPase 6 (PSMC6), and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit β (PIK3CB) were hub nodes with higher degree. RT-PCR and Western blot results showed that GNG5, LPAR3, MAPK8, and PSMC6 were differently expressed with significance. The expression of these screened genes is also related with cell apoptosis. Conclusion: GNG5, LPAR3, MAPK8, and PSMC6 might be candidate biomarkers associated with apoptosis in MC.
Keywords: biomarkers, cell apoptosis, differently express, Melanosis coli, protein-protein interaction network
Introduction
Melanosis coli (MC) is a non-inflammatory bowel disease characterized by melanin deposition in the colonic mucosa [1]. The essence of this disease is that macrophages in the lamina propria of the colonic mucosa contain a large amount of lipofuscin [2]. The incidence rate in males is higher than that of females, and the age of onset is always more than 60 years [3]. The main symptoms are bloating, constipation, and difficulty in defecation, while few patients have symptoms of lower abdominal pain and poor appetite [4]. Some patients are with hypokalemia, hyponatremia, and low blood calcium. There is currently no specific drug treatment for MC. Most scholars believe that MC is a benign and reversible non-inflammatory intestinal mucosal lesion [3]. With the improvement of constipation symptoms and the withdrawal of laxatives, a large amount of lipofuscin is digested and decomposed by lysosomes, and the pigmentation of MC can be alleviated and even disappeared [5]. However, what is frustrating that more patients were with complications such as colon cancer, adenoma, and polyps [6]. Thereby, early detection and treatment of MC are very necessary to reduce the risk of disease and its complications.
In recent years, biological analysis and molecular mechanism analysis are widely used for various diseases. For instance, Liu et al. [7] conducted an analysis on amino acid and cDNA sequence, and confirmed that metallopanstimulin-1 was closely associated with tumorigenesis. Interestingly, the expression of several colorectal carcinoma genes, including Bcl-2, Ki-67, K-ras, and Cox-2, were related to cells’ mucosal apoptosis in elderly MC patients [8]. Though these mechanisms and genes were researched, complications’ prevention of MC by genetic means has not yet reached. More precise biomarkers need to be studied to promote the progress of this event.
With the implementation of the Human Genome Project, the development of bioinformatics has been greatly promoted, and the ensuing and analysis of the large amount of accounting and protein data has become the main task of biology [9]. However, the results of bioinformatics analysis are often extensive and inaccurate. In order to provide a theoretical basis for researching MC development and preventing complications in the early stages, bioinformatics analysis was used to screen candidate biomarkers of MC, and clinical data and in vitro experiments were combined to research the molecular mechanism of screened genes in MC.
Materials and methods
Acquisition and preprocessing of data
The expression profile of MC was acquired from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo) database of NCBI (GEO accession: GSE78933) with the platform of [HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version]. The profile included five colonic mucosa samples in MC groups and five colonic mucosa samples in MC groups. The raw data of expression profile were preprocessed by background correction and standardization. According to the annotation information of the chip, the probes were mapped to the corresponding genes, and the average value was calculated as expression value of each gene.
Screening of differently expressed genes and enrichment analysis
The moderated T-test of R/Bioconductor package limma was used to screen differently expressed genes (DEGs) between MC and control samples with the threshold of P<0.05.
ClueGO is a Cytoscape plug-in that visualizes the non-redundant biological terms for large clusters of genes in a functionally grouped network. It can be also combined with GOlorize. ClueGO charts showed underlying specificity and the common aspects of biological role. The significance of the terms is automatically calculated. ClueGO is easily updatable based on the newest files, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome. In the present study, it was used for GO and KEGG signal enrichment analysis. P<0.05 was chosen as the threshold. R/Bioconductor package pathview was used for visualization of significant KEGG signaling pathway.
Construction of protein–protein interaction network
The interaction between DEGs was extracted from the STRING V10 (https://string-db.org/) database for protein–protein interaction (PPI) construction. The combined score for each protein interaction pair could be obtained from STRINE database, which is distributed from 0 to 1. DEG.PPI was constructed with the threshold of P<0.7.
qRT-PCR detected the expression of hub nodes in DEG.PPI
In order to verify the clinical value of screened hub nodes in DEG.PPI, qRT-PCR was used to detect the expression of hub nodes. First, the pathological tissue and normal tissue from 23 MC patients and 23 controls were collected, respectively. All specimens were collected from colonoscopy of patients in Hangzhou Cancer Hospital from December 2016 to December 2017. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki, and was approved by Hangzhou Cancer Hospital. All patients signed an informed consent form. The obtained fresh specimens were washed with RNAase-free water, put into RNA later immediately and frozen in −80°C until use. The relative expression of hub nodes was detected by qRT-PCR. Total RNA was extracted by cDNA synthesis kit according to the manufacturer’s instructions (Agilent Technologies, CA, U.S.A.). qRT-PCR was undertaken using SYBR Green Mastermix (Applied Biosystems, CA, U.S.A.). The experiment was repeated for three times. Primers were shown as follows:
GAPDH: (F) 5′-TCG ACA GTC AGC CGC ATC TTC TTT-3′; (R) 5′-ACC AAA TCC GTT GAC TCC GAC CTT-3′;
G-protein subunit γ 5 (GNG5): (F) 5′-AGC ACA GAA CCG GAA ACT TAG-3′; (R) 5′-TCA CTT TTA CGC GGT TGA GTC-3′;
Lysophosphatidic acid receptor 3 (LPAR3): (F) 5′-AGG GCT CCC ATG AAG CTA AT-3′; (R) 5′-TTC ATG ACG GAG TTG AGC AG-3′;
Mitogen-activated protein kinase 8 (MAPK8): (F) 5′-TCT CCA GCA CCC ATA CAT CA-3′; (R) 5′-CCT CCA AAT CC ATTA CCT CC-3′;
NHP2L1: (F) 5′- CAG CTG ACC AAC CAG TTG AA-3′; (R) 5′-AAA TCG TCG CAG ATT GCT TT-3′;
Proteasome 26S subunit, ATPase 6 (PSMC6): (F) 5′-GCT GCG TCC AGG AAG ATT AG-3′; (R) 5′-TGC GAA CAT ACC TGC TTC AG -3′;
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit β ( (PIK3CB): (F) 5′-CAT TGA AGT TGT GAG CAC CTC TGA A-3′; (R) 5′-ATG GCT CGG TCC AGG TCA TC-3′.
Cell culture
Normal colon cell line (FHC) was purchased from ATCC and cultured in DMEM with 10% FBS. The cells were incubated in humidified environment with 5% CO2 at 37°C.
Overexpression or silencing of interest genes
Total RNA was extracted by TRIzol method, and cDNA was synthesized by reverse transcription. According to the mRNA sequence of GNG5, LPAR3, and MSMC6 genes, primers were designed by software Clone Manager 7, PCR was used to amplify the target gene, and product was electrophoresed on 1% agarose gel. The electrophoresis was carried out by QIA-quick’s agarose gel electrophoresis recovery kit. The PCR product and pcDNA3.1 Vector were digested with BamH1 and EcoR1, and the recombinant plasmid was extracted by plasmid extraction reagent. The recombinant plasmid was digested by enzyme and identified. The positive recombinants were identified and transfected with Lipofectamine 2000.
SiRNA (SI02757209) for MAPK8 was synthesized by the Qiagen. The FHC cells were seeded in 12-well plates, and then incubated for more than 12 h in antibiotic-free medium. The siRNA were transfected into FHC cells with the help of Lipofectamine 2000 reagent, and the transfected cells were incubated for 60 h.
Western blot
Cells were harvested and lysed. Antibodies used for Western blot analysis were GNG5, LPAR3, MAPK8, NHP2L1, PSMC6, and PIK3CB (1:1000, Santa Cruz Biotechnology) and GAPDH 1:3000 (Santa Cruz Biotechnology) were used as loading controls. Signals were visualized using ECL chemiluminescence. Changes in protein expression were quantitated by ImageJ software.
Flow cytometry
Flow cytometric analysis was processed to detect the apoptosis rate of FHC cells after overexpression or silencing of interest genes. After treatment, FHC cells were stained with 7-aminoactinomycin D and annexin V based on instructions (BD Biosciences, CA, U.S.A.). GACSCalibur flow cytometer (BD Biosciences) was used to analyze the stained cells.
Statistical analysis
Statistical analysis was performed using SPSS 19.0 software. t test was used to compare the mean between two groups. P<0.05 was regarded as significantly different.
Results
DEGs screening and enrichment analysis
Compared with controls, a total of 1342 DEGs, including 786 up-regulated and 556 down-regulated were screened. These screened DEGs were mainly enriched in several GO terms, such as polysaccharide biosynthetic process (P=0.007), mitochondrial proton-transporting ATP synthase complex, coupling factor F(o) (P=6.10E-11), and innate immune response activating cell surface receptor signaling pathway (P=0.014) (Table 1). Simultaneously, these DEGs were involved in a number of pathways, including Oxidative phosphorylation (P=0.003), Alanine, aspartate and glutamate metabolism (P=0.043) and Glycerolipid metabolism (P= 0.045) (Table 2).
Table 1. Top six functional terms enriched by DEGs.
| GO ID | GO term | Nr. genes | Term P-value |
|---|---|---|---|
| GO:0000271 | Polysaccharide biosynthetic process | 10 | 0.007 |
| GO:0002220 | Innate immune response activating cell surface receptor signaling pathway | 11 | 0.048 |
| GO:0000276 | Mitochondrial proton-transporting ATP synthase complex, coupling factor F | 4 | 6.10E-11 |
| GO:0000315 | Organellar large ribosomal subunit 5 | 5 | 0.031 |
| GO:0002220 | Innate immune response activating cell surface receptor signaling pathway | 11 | 0.014 |
| GO:0002223 | Stimulatory C-type lectin receptor signaling pathway | 11 | 0.012 |
Table 2. KEGG signaling pathway enriched by DEGs.
| GOID | Ontology source | GO term | Nr. Genes | % Associated genes | Term P-value |
|---|---|---|---|---|---|
| KEGG:00190 | KEGG_09.11.2015 | Oxidative phosphorylation | 16 | 12.03008 | 0.002893328 |
| KEGG:00250 | KEGG_09.11.2015 | Alanine, aspartate, and glutamate metabolism | 5 | 14.28571 | 0.043279967 |
| KEGG:00561 | KEGG_09.11.2015 | Glycerolipid metabolism | 7 | 11.86441 | 0.044958189 |
| KEGG:03050 | KEGG_09.11.2015 | Proteasome | 10 | 22.72727 | 1.18E-04 |
| KEGG:04910 | KEGG_09.11.2015 | Insulin signaling pathway | 14 | 10.07194 | 0.022857026 |
| KEGG:05010 | KEGG_09.11.2015 | Alzheimer’s disease | 15 | 8.928572 | 0.048119193 |
Construction of PPI network
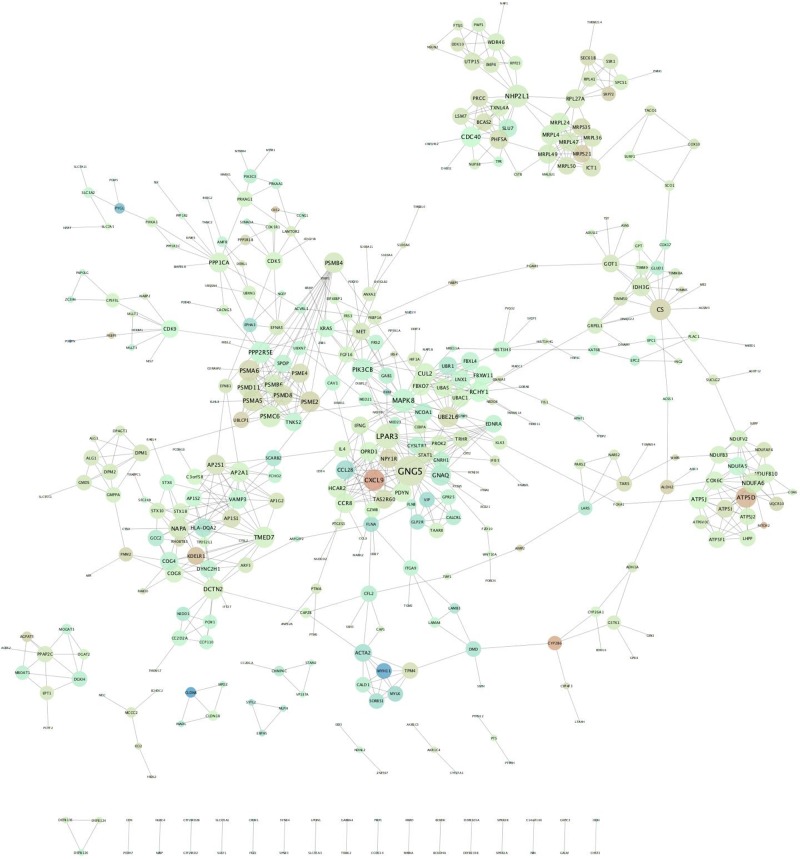
DEG.PPI network was successfully constructed with 426 DEGs and 895 edges (Figure 1). The distribution of node degrees in the network approximated the exponential distribution (correlation = 0.896, R-squared = 0.797). Therefore, the network is a scale-free network (SCN). The nodes with higher degrees were regarded as hub genes. Top six genes with the highest degree were shown, including GNG5 (logFC = 0.323), LPAR3 (logFC = 0.246), MAPK8 (logFC = −0.189), NHP2L1 (logFC = 0.213), PSMC6 (logFC = 0.202), and PIK3CB (logFC = 0.212) (Table 3).
Figure 1. PPI networks.
The nodes present screened DEGs, while the edges present their relationships. The color of Node from green to red indicates the change in the degree of differential expression logFC from negative to positive. The Node size grows from small to large, indicating a change in the node degree.
Table 3. Top six hub nodes of DEG.PPI.
| Gene | Degree | logFC |
|---|---|---|
| GNG5 | 26 | 0.323217200000002 |
| LPAR3 | 18 | 0.246190400000001 |
| MAPK8 | 16 | −0.1897492 |
| NHP2L1 | 15 | 0.2130782 |
| PSMC6 | 14 | 0.202464600000002 |
| PIK3CB | 14 | −0.211626399999998 |
The expression level of hub nodes in DEG.PPI
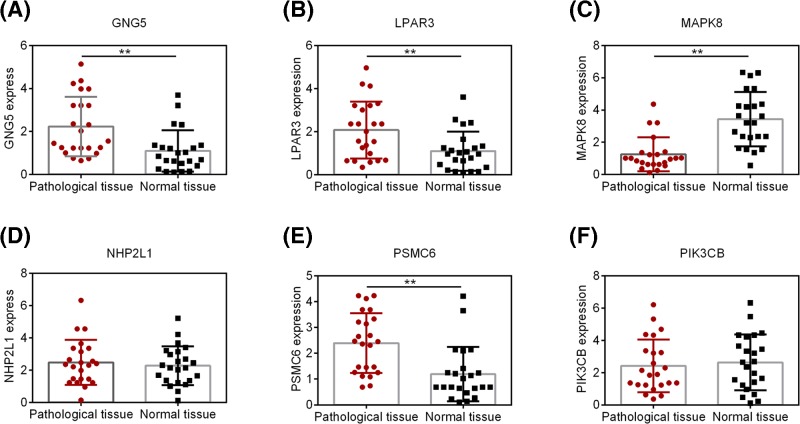
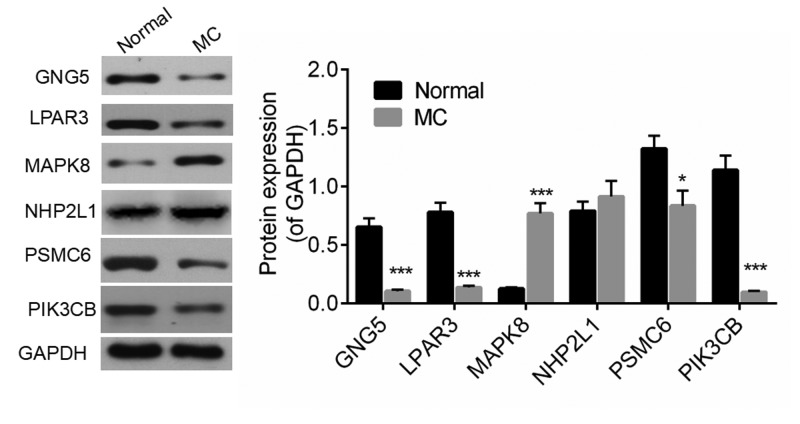
The expression levels of top six DEGs in DEG.PPI network were detected by qRT-PCR. As shown in Figure 2, the expression levels of GNG5, LPAR3, MAPK8, and PSMC6 were with significant difference between pathological and normal tissue, while no different expression of Small Nuclear Ribonucleoprotein 13 (NHP2L1) and PIK3CB were found between two groups. It is worth noting that consistent with bioinformatics analysis, the expression levels of GNG5, LPAR3, and Proteasome 26S Subunit, ATPase 6 (PSMC6) were up-regulated; the expression level of MAPK8 was down-regulated. Similarly, with the results of qRT-PCR, Western blot assay also confirmed that GNG5, LPAR3, and PSMC6 were significantly overexpressed, while MAPK8 was expressed lower in pathological tissue (Figure 3).
Figure 2. The expression level of hub nodes in pathological tissue and normal tissue.
(A) GNG5; (B) LPAR3; (C) MAPK8; (D) NHP2L1; (E) PSMC6; (F) PIK3CB.**P<0.01 compared with Normal tissue.
Figure 3. Western blot detected the expression level of hub nodes.
*P<0.05, ***P<0.001 compared with Normal group.
Cell apoptosis
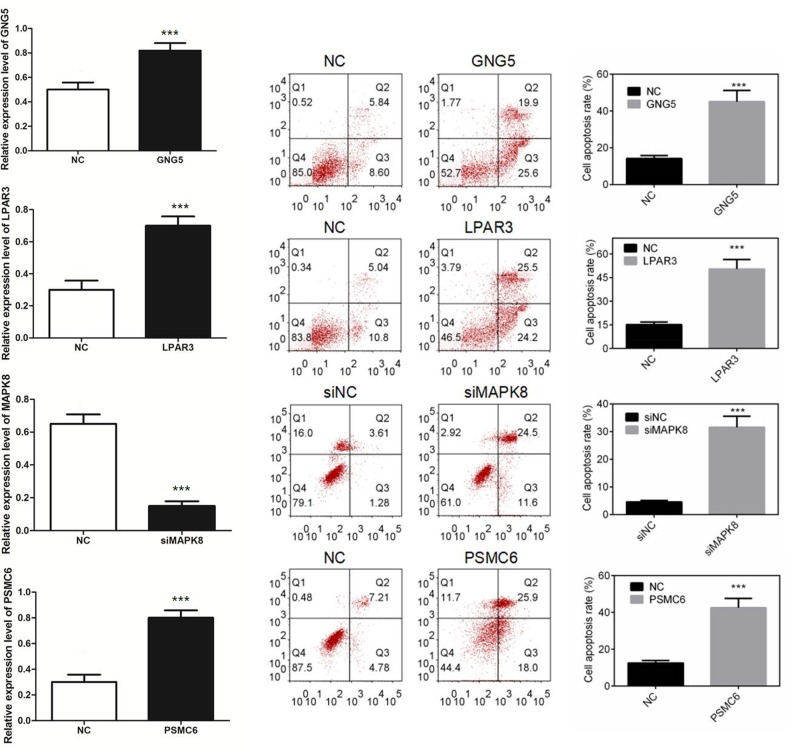
By overexpression or silencing of above four genes, Western blot was used to detect if the positive recombinants and siRNA were successfully transfected. As shown in Figure 4, the expression of GNG5, LPAR3, and PSMC6 were significantly up-regulated, while MAPK8 was down-regulated. By up-regulating the expression of GNG5, LPAR3, and PSMC6 or down-regulating the expression of MAPK8, the cell apoptosis rate of FHC cells were significantly higher. The result indicated that the expression levels of GNG5, LPAR3, PSMC6, and MAPK8 were closely related to cell apoptosis (Figure 4).
Figure 4. Cell apoptosis of colon cell line FHC by regulating the overexpression of GNG5, LPAR3, PSMC6, or silencing the expression of MAPK8.
***P<0.001 compared with NC group.
Discussion
In order to screen accurate biomarkers and provide a theoretical basis for researching MC development and preventing complications in the early stages, bioinformatics analysis was used to screen candidate biomarkers of MC, and clinical data and in vitro experiments were combined to research the molecular mechanism of screened genes in MC. And the results, total six key genes, including GNG5, LPAR3, MAPK8, NHP2L1, PSMC6, and PIK3CB were hub nodes with higher degree in PPI network of MC. RT-PCR and Western blot results confirmed that GNG5, LPAR3, MAPK8, and PSMC6 were differently expressed with significance, and flow cytometry results further showed that expression of these screened genes related with cell apoptosis.
GNG5 encodes a member of G protein, which plays a critical role in internalization, trafficking, and signaling pathways of various G-protein coupled receptors [10]. As shown in previous studies, G-protein could induce apoptotic response in various cancers, such as murine colon adenocarcinoma, human melanoma cells [11,12]. Interestingly, RGS6, a G-protein inactivator, was confirmed to mediate doxorubicin-induced myocardial cell apoptosis [13]. Based on the above information, G-protein and its’ inactivator could regulate cell apoptosis process in cancer. In the present study, GNG5 was screened and involved in functions of generation of precursor metabolites and energy, and contractile fiber. So far, fiber colonoscopy was the main and accurate detection method to diagnose the MC [14]. Thereby, GNG5 might participate in pathogenesis of MC by regulating contractile fiber and apoptotic response.
LPAR3 and PSMC6 were also up-regulated DEGs in MC patients. In the present study, LPAR3 was found to participate in regulation of protein phosphorylation. Moreover, the protein phosphorylation was required for anti-apoptotic function [16]. Importantly, Ali et al. [17] found that serine phosphorylation of vasodilator-stimulated phosphoprotein regulated cell survival and apoptosis of colon cancer. Besides, PSMC6 was involved in innate immune response activating cell surface receptor signaling pathway and pathway of proteasome in this study. In human multiple myeloma cells, lacking the PSMC6 expression might induce resistance to apoptosis [18].
It is worth noting that, MAPK8 was screened and identified as the unique down-regulated gene of MC patients in the present study. In addition, it was involved in regulation of innate immune response and pathway of insulin signaling pathway. Similarly with previous study, phosphorylation of MAPK8 was related with cell apoptosis [15]. A case–control study of Corredoira-Sanchez et al. [19] showed that the relationship between Streptococcus gallolyticus subsp. gallolyticus and colorectal neoplasia has been established, and innate immune response played an important role in this process. So far, no evidence confirmed the connection between MC and insulin signaling pathway. However, a number of evidence confirmed that the differently expressed MAPK8 or phosphorylation of MAPK8 was associated with cell apoptosis [15]. Thence, MAPK8 might regulate the process of immunity and apoptosis, and further effect the development of MC.
Based on the above evidence, we found that the screened key genes were more or less associated with apoptosis. Previous studies have shown that constipation and long-term oral laxatives are the leading causes of colonic melanosis [20]. Amongst them, oral steroids and diphenylmethane laxatives are the most important drugs. The study of Wang et al. [21] have confirmed that the apoptosis rate of colonic melanosis group was significantly higher than that of the control group. When the various laxatives enter the colon, the drug could cause transient, meter-related apoptosis of intestinal epithelial cells [22]. Consistent with the results of this study, after up-regulating the expression of GNG5, LPAR3, and PSMC6, or down-regulating the expression of MAPK8, the cell apoptosis rate of FHC cells were significantly higher. Thereby, the expression of GNG5, LPAR3, MAPK8, and PSMC6 were closely related with apoptosis of normal colonic mucosal cells.
In conclusion, GNG5, LPAR3, MAPK8, and PSMC6 might be candidate biomarkers associated with apoptosis in MC. The result might provide a theoretical basis for researching MC development and preventing complications in the early stages.
Highlights
Total 1342 DEGs were screened between MC and controls.
PPI network with 426 DEGs and 895 edges was constructed.
GNG5, LPAR3, MAPK8, and PSMC6 were differently expressed in pathological and normal tissues.
GNG5, LPAR3, MAPK8, and PSMC6 were related to cell apoptosis.
Abbreviations
- ATCC
American type culture collection ATCC
- DEG
differently expressed gene
- DMEM
Dulbecco’s Modified eagle medium
- FHC
Fetal human cells
- GEO
Gene Expression Omnibus
- GNG5
G-protein subunit γ 5
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- logFC
log Fold change
- LPAR3
lysophosphatidic acid receptor 3
- MAPK8
mitogen-activated protein kinase 8
- MC
melanosis coli
- PIK3CB
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit β
- PPI
protein–protein interaction
- PSMC6
proteasome 26S subunit, ATPase 6
- qRT-PCR
quantitative real-time quantitative polymerase chain reaction
- RT-PCR
real-time quantitative polymerase chain reaction
- SCN
scale-free network
Funding
This work was supported by the Hangzhou Municipal Science and Technology Bureau [grant number 20150633B41].
Author contribution
X.H. contributed to the conception of the study and analysis and manuscript preparation. J.C. performed the data analyses and wrote the manuscript. L.W. helped perform the analysis with constructive discussions.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.dos santos Rodrigues A.L. et al. (2009) Melanosis coli associated with laxative herbal teas ingestion in a brazilian amazon woman: case report. Rev. Para. Med. 23 (4) [Google Scholar]
- 2.Pranesh N., Haboubi N.Y. and O’Dwyer S.T. (2005) Pigmented mesenteric lymphadenopathy in familial adenomatous polyposis - an unusual cause of intraoperative abandonment of ileo-anal pouch. Ann. R. Coll. Surg. Engl. 87, 1–4 10.1308/147870805X50708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang S. et al. (2018) Gender, age, and concomitant diseases of melanosis coli in China: a multicenter study of 6,090 cases. Peer J. 6, e4483 10.7717/peerj.4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuike T. et al. (2002) Effects of polydextrose on habitual constipation with melanosis coli. J. Jpn. Assoc. Dietary Fiber Res. 6, 55–60 [Google Scholar]
- 5.Chen L. (2017) Effect of evidence-based nursing in patients with melanosis coli and the influence on improving constipation. Clin. Med. Eng. 13, 2180–2184 [Google Scholar]
- 6.Nusko G. et al. (1993) Retrospective study on laxative use and melanosis coli as risk factors for colorectal neoplasma. Pharmacology 47, 234–241 10.1159/000139863 [DOI] [PubMed] [Google Scholar]
- 7.Liu J. et al. (2014) cDNA and amino acid sequence analysis of melanosis coli associated gene metallopanstimulin-1. Chin. Rem. Clin. 14, 1182–1184 [Google Scholar]
- 8.Guang-Huia X.U. et al. (2013) Relation between melanosis coli and expression of oncogene as well as its clinical significance. Clin. Focus 28, 294–297 [Google Scholar]
- 9.van Ommen G.J. (2002) The Human Genome Project and the future of diagnostics, treatment and prevention. J. Inherit. Metab. Dis. 25, 183–188 10.1023/A:1015673727498 [DOI] [PubMed] [Google Scholar]
- 10.Brooks C. et al. (2018) Farnesylation of the Transducin G protein gamma subunit is a prerequisite for its ciliary targeting in rod photoreceptors. Front. Mol. Neurosci. 11, 16–26 10.3389/fnmol.2018.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bill M.A. et al. (2009) Curcumin induces proapoptotic effects against human melanoma cells and modulates the cellular response to immunotherapeutic cytokines. Mol. Cancer Ther. 8, 2726–2735 10.1158/1535-7163.MCT-09-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jalili A. et al. (2002) A single injection of immature dendritic cells is able to induce antitumour response against a murine colon adenocarcinoma with a low apoptotic index. Oncol. Rep. 9, 991–994 [PubMed] [Google Scholar]
- 13.Yang J. et al. (2013) G-protein inactivator RGS6 mediates myocardial cell apoptosis and cardiomyopathy caused by doxorubicin. Cancer Res. 73, 1662–1667 10.1158/0008-5472.CAN-12-3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jao C.C. et al. (2011) Endoscpic and histological studies of 7 cases of melanosis coli. Gastroenterol. Endosc. 14, 276–282_1 [Google Scholar]
- 15.Show M.D. et al. (2008) Phosphorylation of mitogen‐activated protein kinase 8 (MAPK8) is associated with germ cell apoptosis and redistribution of the Bcl2‐modifying factor (BMF). J. Androl. 29, 338 10.2164/jandrol.107.003558 [DOI] [PubMed] [Google Scholar]
- 16.Ito T. et al. (1997) Bcl-2 phosphorylation required for anti-apoptosis function. J. Biol. Chem. 272, 11671–11673 10.1074/jbc.272.18.11671 [DOI] [PubMed] [Google Scholar]
- 17.Ali M., Rogers L.K. and Pitari G.M. (2015) Serine phosphorylation of vasodilator-stimulated phosphoprotein (VASP) regulates colon cancer cell survival and apoptosis. Life Sci. 123, 1–8 10.1016/j.lfs.2014.12.018 [DOI] [PubMed] [Google Scholar]
- 18.Shi C.X. et al. (2015) Crispr sgRNAs genome-wide screen identifies the proteasome regulatory subunit PSMC6 as a bortezomib resistance gene in human multiple myeloma cells. Blood 126, 450–461 [Google Scholar]
- 19.Corredoira-Sánchez J. et al. (2012) Association between bacteremia due to Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis I) and colorectal neoplasia: a case-control study. Clin. Infect. Dis. 55, 491–496 10.1093/cid/cis434 [DOI] [PubMed] [Google Scholar]
- 20.Sharma A. and Rao S. (2017) Constipation: pathophysiology and current therapeutic approaches. Handb. Exp. Pharmacol. 239, 59 10.1007/164_2016_111 [DOI] [PubMed] [Google Scholar]
- 21.Wang X.M., Hong-Mei X.U. and Liu Y.L. (2005) Endoscopic characteristics of colonic polyps in patients with melanosis coli. China J. Endosc. 11, 1013–1015 [Google Scholar]
- 22.Steigmann F., Wozasek O. and Dyniewicz J.M. (1943) Studies on the influence of various substances on the colon: I. Phenolphthalein and other laxatives. American Journal of Digestive Diseases 10 (6), 208–216 [Google Scholar]