Abstract
Introduction: The increasing incidence of infections caused by extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in sub-Saharan Africa is of serious concern. Studies from countries with a highly industrialized poultry industry suggest the poultry production-food-consumer chain as a potential transmission route. In Africa, integrated studies at this human–animal interface are still missing.
Aim: To determine the molecular epidemiology of ESBL-producing E. coli from the intestinal tract of humans and poultry in rural Ghana.
Methods: During a 6-month period, fecal samples from all children admitted to the Agogo Hospital (Ghana) and broilers at eight poultry farms located within the hospital catchment area were collected. After screening on selective ESBL agar, whole genome sequencing (WGS) was performed on all ESBL isolates. The genomes were analyzed using multilocus sequence typing (MLST), ESBL genotyping and genome-based phylogenetic analyses.
Results: Of 140 broilers and 54 children, 41 (29%) and 33 (61%) harbored ESBL E. coli, respectively, with prevalences on farms ranging between 0 and 85%. No predominant sequence type (ST) was detected among humans. ST10 was most prevalent among broilers (n = 31, 69%). The ESBL gene blaCTX-M-15 was predominant among broilers (n = 43, 96%) and humans (n = 32, 97%). Whole-genome-based phylogenetic analysis revealed three very closely related broiler/human isolate clusters (10% of ESBL isolates) with chromosomal and plasmid-mediated ESBL genes.
Conclusion: The findings demonstrate a high frequency of intestinal ESBL-producing E. coli in rural Ghana. Considering that animal and human samples are independent specimens from the same geographic location, the number of closely related ESBL isolates circulating across these two reservoirs is substantial. Hence, poultry farms or meat products might be an important source for ESBL-producing bacteria in rural Ghana leading to difficult-to-treat infections in humans.
Keywords: Escherichia coli, Ghana, microbial drug resistance, poultry, extended spectrum β-lactamases (ESBL), transmission
Introduction
The inappropriate use of antibiotics, not only in human medicine but also in animal husbandry, has been considered a main driver leading to the increase of multidrug-resistant bacteria (Collignon et al., 2013; Chantziaras et al., 2014). Consequently, in Europe, the European Food Safety Authority (EFSA) and the European Medicines Agency (EMA) recently advocated measures to reduce use of antimicrobial agents in animal husbandry in the European Union (Murphy et al., 2017). Food-producing animals, especially poultry, have been suggested as a potential source for transmission of extended-spectrum beta-lactamase (ESBL)-producing bacteria to humans, either by direct contact or consumption of contaminated meat products, leading to the colonization of the intestinal tract and eventually to severe infections (Lazarus et al., 2014).
A number of previous studies on the inter-sectoral transmission of ESBL-producing bacteria did only use methods with low discriminatory power, which target a small number of genes (e.g., ESBL genotyping, multilocus sequence typing), and therefore may suggest, but not prove clonal transmission events (Leverstein-van Hall et al., 2011; Overdevest et al., 2011; Kluytmans et al., 2013; Lazarus et al., 2014; Valentin et al., 2014; Dahms et al., 2015). As an example, a Dutch study could not confirm previously suggested clonal transmission of ESBL-producing E. coli isolates between humans and poultry, when using the same collection of isolates but changing the typing method to a whole genome sequencing (WGS) approach. Instead, results rather demonstrated that antibiotic resistance between different reservoirs is likely to be transmitted by the spread of plasmids (de Been et al., 2014). Similarly, a Swedish study estimates that less than 0.1% of the Swedish population carry poultry-associated ESBL-producing isolates, but 5% are colonized with ESBL-encoding plasmids identical to those found in isolates from chicken meat and poultry (Börjesson et al., 2016).
Those findings may not be directly transferable to developing countries. In these regions, the inappropriate use of veterinary antibiotics, which are often readily sold in shops and markets without prescriptions, is considered to be very high, potentially selecting for antibiotic resistance (Mainda et al., 2015). Nevertheless, a recent review found a lower prevalence of ESBL/pAmpC E. coli among poultry meat products in African countries (average 16.3%) compared to reports from many European countries such as Spain (84–93%) and the Netherlands (77%) (Alonso et al., 2017). In comparison to industrialized countries, it can be assumed that inter-host transmission is more likely to happen in rural areas of sub-Saharan Africa with mainly subsistence-based agricultural communities, where people frequently live in close contact with livestock animals (Alonso et al., 2017). However, bacterial transmission among poultry and humans has not been adequately addressed in this region until now (Alonso et al., 2017).
This study aims to compare ESBL-producing E. coli found in the intestinal tract of humans and poultry using highly discriminatory WGS methods in order to assess potential transmission routes in a rural community of Central Ghana.
Materials and Methods
Study Site and Sample Collection
The study was conducted in Agogo, a town with 32,000 inhabitants situated in the Asante Akim North District within the Ashanti Region of Ghana. Between January and June 2015, all children below 15 years of age living in Agogo and nearby surroundings, who were admitted to the Agogo Presbyterian Hospital, were recruited into the study. If available, a stool sample was collected at the day of admission and transported to the hospital laboratory within 2–4 h.
A total of eight poultry farms, with around 250–1,000 chickens on each farm, are situated within and around the town of Agogo. The infrastructure with wooden shelters and open sides is comparable on all farms. Between February and April 2015, all eight farms were visited once and six farms were visited a second time between May and June 2015. On each farm visit, 10 single fecal droppings were collected with an eSwabTM (Hain Lifescience, Nehren, Germany) and transported to the laboratory within 2–4 h.
The Committee on Human Research, Publications and Ethics, School of Medical Science, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana and the Ethics Committee of the Ärztekammer Hamburg, Germany approved the study design and the informed consent procedures. All participants were informed about the study’s purpose and procedures in accordance with the Declaration of Helsinki. Written informed consent was obtained from the parents or the guardian on behalf of the study children prior to enrolment.
ESBL Detection and Antibiotic Susceptibility Testing
On arrival in the laboratory, each fecal sample was directly inoculated on two selective MacConkey agar plates containing 1 mg/L ceftazidime and 1 mg/L cefotaxime, respectively. Plates were incubated at 37°C for 24–48 h in normal atmosphere. All colonies with typical E. coli morphology were selected and confirmed biochemically as E. coli by API 20E tests (bioMérieux, Marcy L’Etoile, France). For all E. coli, ESBL-production was confirmed by the combined disk test with cefotaxime and ceftazidime alone and in combination with clavulanic acid (Becton, Dickinson and Company, Sparks, MD, United States) as described before by the EUCAST [EUCAST guideline on detection of resistance mechanisms v 1.0 (2013-12-11)]. Quality control for each batch of cephalosporin-containing MacConkey agar plates was performed with E. coli ATCC 25922 and a blaCTX-M-15 positive E. coli isolate.
Whole Genome Sequencing
E. coli isolates were stored at -80°C in cryobank tubes (Pro-lab Diagnostics, Richmond Hill, ON, Canada) and shipped on dry ice to Germany, where WGS was performed for all ESBL-producing E. coli isolates. In brief, whole genome DNA was isolated using the Purelink Genome DNA Mini kit (Invitrogen, Darmstadt, Germany) according to the manufacturer’s instruction. Sequencing was carried out using an Illumina Nextera XT library with 2 × 300 bp paired-end reads on an Illumina MiSeq instrument (Illumina, San Diego, CA, United States) achieving an average of 2,170,898 reads per isolate (61× average coverage). The raw data was assembled using SPAdes (version 3.0). Multilocus sequence type (ST) determination was performed using MLST 1.0 (Zankari et al., 2013). Antibiotic resistance genes were identified using ResFinder (Zankari et al., 2012) and the genetic location of the ESBL-genes was investigated using the ESBL-gene containing contigs by blastn (Altschul et al., 1990) using the nt database. Contigs depicting hits only to chromosomal sequences were considered as chromosomal hits, while those depicting hits to plasmids were considered as plasmid hits. Contigs depicting hits to both the chromosome and plasmids were considered as “ambiguous.”
The single nucleotide polymorphisms (SNPs) were called by SAMtools (Li, 2011). The tree was generated using FastTree 2 (Price et al., 2010) and drawn by Evolview (He et al., 2016).
To analyze possible transmission, isolates depicting an identical ST and present in both reservoirs were analyzed in more detail. SNPs were called as mentioned above, and regions of high recombination were excluded manually.
The raw sequencing data are available at the European Nucleotide Archive (ENA) under the project accession number PRJEB26592.
Results
Prevalence of ESBL-Producing Isolates in Hospitalized Children and Broilers
Between January and June 2015, 54 hospitalized children, with a median age of 20 months (IQR: 11–36 months), provided a stool sample at admission, regardless of the admission diagnosis. All children had contact to free-roaming chickens in town, but had no contact or access to the poultry farms. Chicken meat is part of the regular diet. Of all children, 33 (61%) harbored an ESBL-producing E. coli. From one child, two different E. coli were isolated, leading to a total of 34 ESBL-producing E. coli.
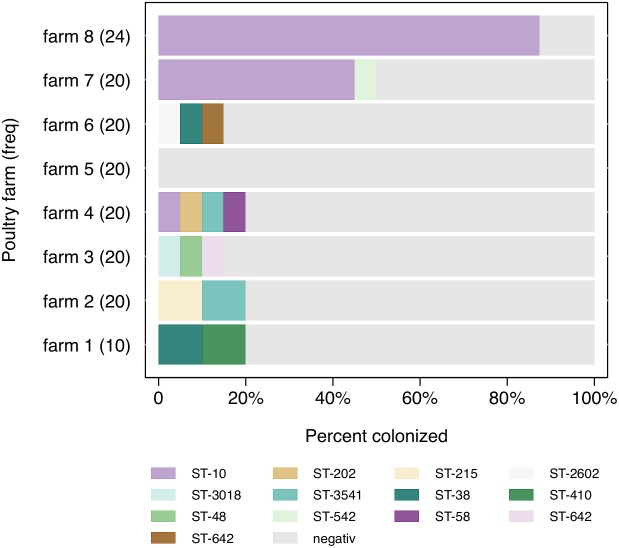
During the same time-period, fecal samples were obtained from 140 broilers, of which 41 (29%) were positive for an ESBL-producing E. coli. Prevalences differed widely between farms, ranging from 0 to 85% (Figure 1). Four broilers were detected with two different E. coli isolates, which results in 45 ESBL-producing E. coli isolates from broilers.
FIGURE 1.
Distribution of ESBL-producing Escherichia coli sequence types (ST) (n = 45) among farms.
Multilocus Sequence Determination of the ESBL-Producing Isolates
The 34 human isolates displayed 22 different sequence types (ST). ST-131 (12%; n = 4), ST-167 (9%; n = 3) and ST-617 (9%; n = 3) were most frequently detected. No clusters with more than two isolates with the same ST during a 1-month period were identified (Table 1).
Table 1.
Frequency of sequence type (ST) in human and poultry samples (sorted by ST frequency).
| STs | Human samples (n) | Poultry samples (n) | Detected in human and poultry samples |
|---|---|---|---|
| ST-10 | 0 | 31 | |
| ST-131 | 4 | 0 | |
| ST-38 | 2 | 2 | x |
| ST-6359 | 2 | 1 | x |
| ST-3541 | 1 | 2 | x |
| ST-167 | 3 | 0 | |
| ST-617 | 3 | 0 | |
| ST-3268 | 2 | 0 | |
| ST-3018 | 1 | 1 | x |
| ST-46 | 2 | 0 | |
| ST-58 | 1 | 1 | x |
| ST-443 | 2 | 0 | |
| ST-2602 | 0 | 1 | |
| ST-295 | 1 | 0 | |
| ST-542 | 0 | 1 | |
| ST-940 | 1 | 0 | |
| ST-642 | 0 | 1 | |
| ST-34 | 1 | 0 | |
| ST-44 | 1 | 0 | |
| ST-90 | 1 | 0 | |
| ST-202 | 0 | 1 | |
| ST-4450 | 1 | 0 | |
| ST-2141 | 1 | 0 | |
| ST-48 | 0 | 1 | |
| ST-155 | 1 | 0 | |
| ST-1706 | 1 | 0 | |
| ST-215 | 0 | 1 | |
| ST-410 | 0 | 1 | |
| ST-773 | 1 | 0 | |
| ST-648 | 1 | 0 | |
STs detected in both humans and poultry samples are indicted.
Among all broiler isolates (n = 45), 13 different STs were identified, with ST-10 being most prevalent (69%; n = 31), although ST-10 has been only detected on three farms. On farm 7 and 8, ST-10 comprised 90% (n = 9) and 100% (n = 21) of all ESBL-producing isolates, respectively (Figure 1). There was no reported exchange of broilers, feed or personnel between those two farms. All other STs were not detected more than twice on each farm.
The overlap of sequence types between the two different sources (human and broiler) involved five sequence types (ST-38, ST-58, ST-3018, ST-3541, ST-6359), comprising seven broiler and seven human isolates (Table 1).
Presence and Location of ESBL Genes
The most common ESBL genes in E. coli from both populations were blaCTX-M-15 (n = 76) followed by blaSHV -12 (n = 2) and blaCTX-M-14 (n = 1). The ESBL gene blaCTX-M-15 was equally distributed in both populations, whereas blaSHV -12 was exclusively found in broiler and blaCTX-M-14 in human isolates, respectively (Table 2). The location of the ESBL genes differed between human and broiler isolates. Among human isolates, 29% (n = 10) harbored plasmid and 44% (n = 15) chromosomally inserted ESBL genes, while the exact location could not be identified among the remaining 26% (n = 9). Among broiler isolates, 82% (n = 36) harbored a chromosomally inserted ESBL gene (Table 2). Of note, all but one (C105F) of the 31 ST-10 isolates, which were isolated from three different farms, harbored blaCTX-M-15 at an identical chromosomal location (Supplementary Table 1), indicating the spread of a distinct clone on these farms.
Table 2.
Characteristics of the sequenced Escherichia coli isolates.
| Isolate | Source | MLST | ESBL gene | Location of ESBL gene |
|---|---|---|---|---|
| C011F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C012F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C013F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C014F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C015F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C016_1F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C016_2F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C017F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C018F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C019F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C028F | Chicken | 3541 | blaCTX-M-15 | Chromosome |
| C029F | Chicken | 58 | blaSHV -12 | Plasmid |
| C050F | Chicken | 3018 | blaCTX-M-15 | Plasmid |
| C060F | Chicken | 38 | blaCTX-M-15 | Plasmid |
| C065F | Chicken | 542 | blaCTX-M-15 | Ambiguous |
| C068F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C071F | Chicken | 3541 | blaCTX-M-15 | Chromosome |
| C075F | Chicken | 215 | blaCTX-M-15 | Chromosome |
| C081F | Chicken | 38 | blaCTX-M-15 | Chromosome |
| C088F | Chicken | 410 | blaCTX-M-15 | Plasmid |
| C091_1F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C091_2F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C092F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C093_1F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C093_2F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C095F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C096F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C097F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C099_1F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C099_2F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C100F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C105F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C106F | Chicken | 202 | blaCTX-M-15 | Plasmid |
| C111F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C113F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C114F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C115F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C116F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C118F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C119F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C120F | Chicken | 10 | blaCTX-M-15 | Chromosome |
| C124F | Chicken | 2602 | blaCTX-M-15 | Plasmid |
| C125F | Chicken | 642 | blaCTX-M-15 | Plasmid |
| C132F | Chicken | 6359 | blaCTX-M-15 | Chromosome |
| C137F | Chicken | 48 | blaSHV -12 | Plasmid |
| 701460 | Human | 44 | blaCTX-M-15 | Plasmid |
| 701499 | Human | 131 | blaCTX-M-14 | Chromosome |
| 701518 | Human | 167 | blaCTX-M-15 | Plasmid |
| 701519 | Human | 3541 | blaCTX-M-15 | Chromosome |
| 701533 | Human | 131 | blaCTX-M-15 | Chromosome |
| 701538 | Human | 617 | blaCTX-M-15 | Plasmid |
| 701571 | Human | 131 | blaCTX-M-15 | Chromosome |
| 701592 | Human | 46 | blaCTX-M-15 | Plasmid |
| 701611 | Human | 38 | blaCTX-M-15 | Chromosome |
| 701625 | Human | 617 | blaCTX-M-15 | Plasmid |
| 701670 | Human | 295 | blaCTX-M-15 | Plasmid |
| 701674 | Human | 2141 | blaCTX-M-15 | Ambiguous |
| 701684 | Human | 46 | blaCTX-M-15 | Plasmid |
| 701698 | Human | 3018 | blaCTX-M-15 | Plasmid |
| 701744 | Human | 443 | blaCTX-M-15 | Ambiguous |
| 701769 | Human | 773 | blaCTX-M-15 | Ambiguous |
| 701790 | Human | 3268 | blaCTX-M-15 | Plasmid |
| 701805 | Human | 58 | blaCTX-M-15 | Ambiguous |
| 701846 | Human | 617 | blaCTX-M-15 | Ambiguous |
| 701853 | Human | 167 | blaCTX-M-15 | Plasmid |
| 701856 | Human | 940 | blaCTX-M-15 | Plasmid |
| 701861 | Human | 6359 | blaCTX-M-15 | Chromosome |
| 701867 | Human | 6359 | blaCTX-M-15 | Chromosome |
| 701872 | Human | 1706 | blaCTX-M-15 | Plasmid |
| 701874 | Human | 3268 | blaCTX-M-15 | Plasmid |
| 701875 | Human | 155 | blaCTX-M-15 | Ambiguous |
| 701889 | Human | 443 | blaCTX-M-15 | Ambiguous |
| 701902 | Human | 90 | blaCTX-M-15 | Plasmid |
| 701903 | Human | 131 | blaCTX-M-15 | Ambiguous |
| 701495_1 | Human | 38 | blaCTX-M-15 | Chromosome |
| 701495_2 | Human | 648 | blaCTX-M-15 | Chromosome |
| 701572_1 | Human | 34 | blaCTX-M-15 | Ambiguous |
| 701842_1 | Human | 4450 | blaCTX-M-15 | Chromosome |
| 701887_1 | Human | 167 | blaCTX-M-15 | Plasmid |
Single Nucleotide Polymorphism (SNP) Detection in Isolates With Identical ST in Humans and Broilers
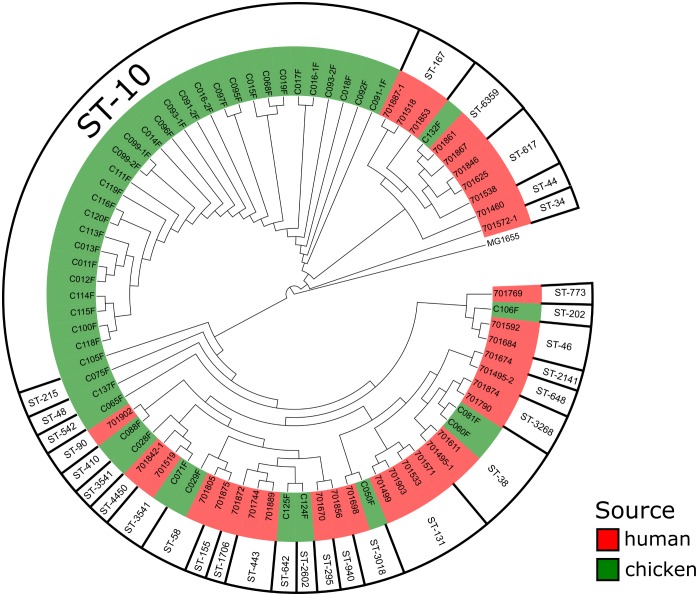
Five human/broiler isolate clusters depicting an identical ST were regarded as possible clonal spread events and therefore investigated for SNPs (Figure 2). The ST-3541 isolates (C28F/C071F/701519) revealed a difference in 49 and 24 SNPs, respectively. In all three isolates, the blaCTX-M-15 gene was located on the chromosome at an identical position, thus indicating a common origin of these isolates.
FIGURE 2.
Whole genome-based phylogenetic analysis of the ESBL-producing Escherichia coli using Escherichia coli MG1655 as a reference strain.
An almost identical situation was found in the ST-6359 isolates (C132F/701861/701867). The human isolate 701861 differed by 99 SNPs from C132F, whereas 701867 differed by 17 SNPs, implying close genetic relation to the 701867 isolate. Interestingly, all ST-6359 isolates harbored blaCTX-M-15 at an identical chromosomal location, implying a common ancestor of these isolates. Similarly, the ST-3018 pair (C50F/701698) differed by only 21 SNPs and harbored blaCTX-M-15 on a plasmid.
A complete different pattern was detected in the ST-58 (C29F/701805) and ST-38 isolates (C060/C081F/701495-1/701611). ST-58 isolates differed by 4449 SNPs, thereby indicating that these are two genetically not related isolates and no transmission occurred. ST-38 isolates differed already in the chicken isolates (2034 SNPs), while the human isolates depicted 9588 (701495-1) and 9560 (701611) SNPs, respectively. This indicates that ST-38 isolates are highly diverse.
Discussion
The study demonstrates genetic links of ESBL-producing E. coli that may indicate transmission between the poultry and human population in rural Ghana. With four human and four broiler isolates being closely related, 10% of all ESBL-producing isolates in this study may have been transmitted at a certain point in time between the two different populations.
The potential risk for the transmission of antimicrobial-resistant bacteria between animals or animal food products and humans in sub-Saharan Africa has been highlighted before, however, from this region no data on the clonal spread between human and animal reservoirs has been published (Soge et al., 2006; Alonso et al., 2017). Other studies using MLST-based typing methods or ESBL-genotyping do not allow to conclusively interpret the clonal spread of ESBL-producing E. coli (Leverstein-van Hall et al., 2011; Overdevest et al., 2011; Kluytmans et al., 2013). More recently, a Dutch study using WGS methods concluded that clonal transfer between humans and poultry of antibiotic resistant E. coli occurs less frequently than the transfer of resistance plasmids between the two reservoirs (de Been et al., 2014). Similarly, studies from Romania, Sweden and the United Kingdom found evidence for the transmission of ESBL-harboring plasmids instead of clonal transmission of isolates between poultry and humans (Stokes et al., 2012; Börjesson et al., 2013; Maciuca et al., 2015).
In contrast, the present results suggest clonal transmission between the two reservoirs in a rural Ghanaian town, where children were sampled at hospital admission, independent from any contact to the poultry farms to which children have no access. Taking into account the low level of sanitary and hygiene conditions in rural Ghana, inter-host transmission of bacteria is expected to be more predominant compared to industrialized countries and not necessarily limited to farm workers. Furthermore, transmission via consumption of meat products has been suggested as a potential source of multidrug resistant bacteria in Africa (Alonso et al., 2017; Eibach et al., 2018).
The high rate of ESBL-producing E. coli on farms is in sharp contrast to previous Ghanaian data from 2009, where no ESBL-producing E. coli isolates (n = 103) were found in broilers from Accra (Donkor et al., 2012). However, this previous study did not use any ESBL screening plates for the detection of E. coli, which might underestimate the ESBL production.
Notably, a high diversity of STs has been observed, with only 5 out of 30 STs overlapping between the two sampling groups. ST-10 clearly predominates within two farms. Indeed, ST-10 ESBL-producing E. coli have been frequently identified worldwide in the chicken production chain, including reports from Nigeria and Tanzania (Ojo et al., 2016; Seni et al., 2016). Interestingly, studies from the Netherlands, Sweden and Vietnam detected ESBL-producing ST-10 E. coli not only in chickens, but also in high numbers among humans (Huijbers et al., 2014; Ueda et al., 2015; van Hoek et al., 2015; Börjesson et al., 2016). However, in the present study ST-10 could not be detected among children.
In this study, ST-131, although not very frequent (n = 4), is the most prevalent E. coli subtype among children (12%). ST-131 E. coli have been described as an clinically relevant multidrug-resistant ST, frequently harboring blaCTX-M-15 and fluoroquinolone resistance (Mathers et al., 2015). High virulence leading to serious infections, such as invasive bloodstream, urinary tract and intra-abdominal infections have been attributed to ST131, also in Ghana (Eibach et al., 2016). Although identified in different animal reservoirs, the ST131 lineage seems to be specifically adapted to the human host and reservoirs to be rather unclear (Platell et al., 2011; Mathers et al., 2015). It has been suggested that due to acquired virulence factors ST131 is responsible for the global increase of E. coli harboring blaCTX-M-15 (Mathers et al., 2015).
The CTX-M-15 β-lactamase is the most prevalent ESBL type among chicken (96%) and human (97%) isolates in this study. While in many countries CTX-M-15 is one of the most frequent ESBL types in ESBL-producing bacteria, causing human infections (Valentin et al., 2014), it showed a relatively low prevalence among bacteria isolated from fecal poultry samples in previous studies using ESBL screening plates as an isolation method in the Netherlands (0%), Belgium (2%), and the United Kingdom (12%) (Smet et al., 2008; Randall et al., 2011; Huijbers et al., 2014). Studies from Japan and China also demonstrate low prevalences (0–2%) of CTX-M-15 in fecal poultry samples (Kameyama et al., 2013; Rao et al., 2014), in contrast to most surveys of African livestock, which show a predominance of blaCTX-M-15 positive ESBL-producing E. coli in a recent review by Alonso et al. (2017). This geographical distribution suggests that transmission between humans and poultry may occur more frequently on the African continent.
The present study has some limitations. Firstly, it does not allow any conclusions on the transmission of plasmids, but only indicates clonal bacterial transmission between poultry and humans. Therefore, the focus on clonal transfer alone will probably underestimate the transmission of resistance genes. In addition, the collection of stool samples at hospital admission may result in a higher ESBL prevalence among children compared to the general population, due to a different health care seeking behavior and potential previous antibiotic prescriptions.
Conclusion
Apart from a high intestinal detection frequency, this study shows that highly similar ESBL-producing E. coli strains circulate among the human and poultry populations within a small town in rural Ghana. Hence, poultry farms or meat products might be an important source for ESBL-producing bacteria in rural Ghana leading to difficult-to-treat infections in humans. Despite widespread agreement that integration of human and animal data is desirable for antimicrobial resistance surveillance, there is very little, if any, integration of data in most low and middle-income countries. An integrated “One Health” surveillance system would be able to monitor transmission events and detect resistant bacteria in a timely manner from both sectors.
Data Availability Statement
The raw sequencing data are available at the European Nucleotide Archive (ENA) under the project accession number PRJEB26592. All other raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
LF, CI, SP, JM, and DE designed and coordinated the study. KO, BH, VL, NS, and EO-D conducted and supervised fieldwork. KO and CA conducted laboratory work. RK and OS performed the epidemiological and bioinformatics analysis. DE and LF wrote the first draft of the paper. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express our gratitude to all patients and their caregivers for participating in this study. We thank all dedicated fieldworkers study nurses and participating farms. Further, we thank Christina Gerstmann (Institute of Medical Microbiology, JLUG) and Doris Winter (Bernhard Nocht Institute for Tropical Medicine, Hamburg) for excellent technical assistance and Anna Jaeger (Bernhard Nocht Institute for Tropical Medicine, Hamburg) for organizing the data management.
Footnotes
Funding. This study was supported by grants to the German Center of Infection Research (DZIF), through the German Federal Ministry of Education and Research [BMBF; Grant Numbers 8000 701-3 (HZI) TI06.001, 8032808811 and 8000 201-3]. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03358/full#supplementary-material
References
- Alonso C. A., Zarazaga M., Ben Sallem R., Jouini A., Ben Slama K., Torres C. (2017). Antibiotic resistance in Escherichia coli in husbandry animals. the african perspective. Lett. Appl. Microbiol. 64 318–334. 10.1111/lam.12724 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Börjesson S., Jernberg C., Brolund A., Edquist P., Finn M., Landén A., et al. (2013). Characterization of plasmid-mediated AmpC-producing E. coli from Swedish broilers and association with human clinical isolates. Clin. Microbiol. Infect. 19 E309–E311. 10.1111/1469-0691.12192 [DOI] [PubMed] [Google Scholar]
- Börjesson S., Ny S., Egervärn M., Bergström J., Rosengren Å, Englund S., et al. (2016). Limited dissemination of extended-spectrum β-lactamase– and plasmid-encoded AmpC–producing Escherichia coli from food and farm animals, Sweden. Emerg. Infect. Dis. 22 634–640. 10.3201/eid2204.151142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantziaras I., Boyen F., Callens B., Dewulf J. (2014). Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: a report on seven countries. J. Antimicrob. Chemother. 69 827–834. 10.1093/jac/dkt443 [DOI] [PubMed] [Google Scholar]
- Collignon P., Aarestrup F. M., Irwin R., McEwen S. (2013). Human deaths and third-generation cephalosporin use in Poultry, Europe. Emerg. Infect. Dis. 19 1339–1340. 10.3201/eid1908.120681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahms C., Hübner N.-O., Kossow A., Mellmann A., Dittmann K., Kramer A. (2015). Occurrence of ESBL-producing Escherichia coli in livestock and farm workers in mecklenburg-western pomerania, Germany. PLoS One 10:e0143326. 10.1371/journal.pone.0143326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Been M., Lanza V. F., de Toro M., Scharringa J., Dohmen W., Du Y., et al. (2014). Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 10:e1004776. 10.1371/journal.pgen.1004776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor E. S., Newman M. J., Yeboah-Manu D. (2012). Epidemiological aspects of non-human antibiotic usage and resistance: implications for the control of antibiotic resistance in Ghana. Trop. Med. Int. Health 17 462–468. 10.1111/j.1365-3156.2012.02955.x [DOI] [PubMed] [Google Scholar]
- Eibach D., Campos C. B., Krumkamp R., Al-Emran H. M., Dekker D., Boahen K. G., et al. (2016). Extended spectrum beta-lactamase producing Enterobacteriaceae causing bloodstream infections in rural Ghana, 2007-2012. Int. J. Med. Microbiol. 306 249–254. 10.1016/j.ijmm.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Eibach D., Dekker D., Gyau Boahen K., Wiafe Akenten C., Sarpong N., Belmar Campos C., et al. (2018). Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in local and imported poultry meat in Ghana. Vet. Microbiol. 217 7–12. 10.1016/j.vetmic.2018.02.023 [DOI] [PubMed] [Google Scholar]
- He Z., Zhang H., Gao S., Lercher M. J., Chen W.-H., Hu S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44 W236–W241. 10.1093/nar/gkw370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers P. M., Graat E. A., Haenen A. P., van Santen M. G., van Essen-Zandbergen A., Mevius D. J., et al. (2014). Extended-spectrum and AmpC -lactamase-producing Escherichia coli in broilers and people living and/or working on broiler farms: prevalence, risk factors and molecular characteristics. J. Antimicrob. Chemother. 69 2669–2675. 10.1093/jac/dku178 [DOI] [PubMed] [Google Scholar]
- Kameyama M., Chuma T., Yabata J., Tominaga K., Iwata H., Okamoto K. (2013). Prevalence and epidemiological relationship of CMY-2 AmpC beta-lactamase and CTX-M extended-spectrum beta-lactamase-producing Escherichia coli isolates from broiler farms in Japan. J. Vet. Med. Sci. 75 1009–1015. 10.1292/jvms.12-0453 [DOI] [PubMed] [Google Scholar]
- Kluytmans J. A., Overdevest I. T., Willemsen I., Kluytmans-van den Bergh M. F., van der Zwaluw K., Heck M., et al. (2013). Extended-spectrum β-lactamase-producing Escherichia coli from retail chicken meat and humans: comparison of strains, plasmids, resistance genes, and virulence factors. Clin. Infect. Dis. 56 478–487. 10.1093/cid/cis929 [DOI] [PubMed] [Google Scholar]
- Lazarus B., Paterson D. L., Mollinger J. L., Rogers B. A. (2014). Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin. Infect. Dis. 60 439–452. 10.1093/cid/ciu785 [DOI] [PubMed] [Google Scholar]
- Leverstein-van Hall M. A., Dierikx C. M., Cohen Stuart J., Voets G. M., van den Munckhof M. P., van Essen-Zandbergen A., et al. (2011). Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17 873–880. 10.1111/j.1469-0691.2011.03497.x [DOI] [PubMed] [Google Scholar]
- Li H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciuca I. E., Williams N. J., Tuchilus C., Dorneanu O., Guguianu E., Carp-Carare C., et al. (2015). High prevalence of Escherichia coli- producing CTX-M-15 extended-spectrum beta-lactamases in poultry and human clinical isolates in romania. Microb. Drug Resist. 21 651–662. 10.1089/mdr.2014.0248 [DOI] [PubMed] [Google Scholar]
- Mainda G., Bessell P. R., Muma J. B., McAteer S. P., Chase-Topping M. E., Gibbons J., et al. (2015). Prevalence and patterns of antimicrobial resistance among Escherichia coli isolated from Zambian dairy cattle across different production systems. Sci. Rep. 5:12439. 10.1038/srep12439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers A. J., Peirano G., Pitout J. D. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28 565–591. 10.1128/CMR.00116-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D., Ricci A., Auce Z., Beechinor J. G., Bergendahl H., Breathnach R., et al. (2017). EMA and EFSA joint scientific opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European union, and the resulting impacts on food safety (RONAFA). EFSA J. 15:4666 10.2903/j.efsa.2017.4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojo O. E., Schwarz S., Michael G. B. (2016). Detection and characterization of extended-spectrum β-lactamase-producing Escherichia coli from chicken production chains in Nigeria. Vet. Microbiol. 194 62–68. 10.1016/j.vetmic.2016.04.022 [DOI] [PubMed] [Google Scholar]
- Overdevest I., Willemsen I., Rijnsburger M., Eustace A., Xu L., Hawkey P., et al. (2011). Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg. Infect. Dis. 17 1216–1222. 10.3201/eid1707.110209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platell J. L., Johnson J. R., Cobbold R. N., Trott D. J. (2011). Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet. Microbiol. 153 99–108. 10.1016/j.vetmic.2011.05.007 [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. P., Clouting C., Horton R. A., Coldham N. G., Wu G., Clifton-Hadley F. A., et al. (2011). Prevalence of Escherichia coli carrying extended-spectrum-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J. Antimicrob. Chemother. 66 86–95. 10.1093/jac/dkq396 [DOI] [PubMed] [Google Scholar]
- Rao L., Lv L., Zeng Z., Chen S., He D., Chen X., et al. (2014). Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003–2012. Vet. Microbiol. 172 534–541. 10.1016/j.vetmic.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Seni J., Falgenhauer L., Simeo N., Mirambo M. M., Imirzalioglu C., Matee M., et al. (2016). Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in mwanza, tanzania, harbor commonly occurring plasmids. Front. Microbiol. 7:142. 10.3389/fmicb.2016.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smet A., Martel A., Persoons D., Dewulf J., Heyndrickx M., Catry B., et al. (2008). Diversity of extended-spectrum β-lactamases and class C β-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob. Agents Chemother. 52 1238–1243. 10.1128/AAC.01285-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soge O. O., Queenan A. M., Ojo K. K., Adeniyi B. A., Roberts M. C. (2006). CTX-M-15 extended-spectrum (beta)-lactamase from Nigerian Klebsiella pneumoniae. J. Antimicrob. Chemother. 57 24–30. 10.1093/jac/dki429 [DOI] [PubMed] [Google Scholar]
- Stokes M. O., Cottell J. L., Piddock L. J. V., Wu G., Wootton M., Mevius D. J., et al. (2012). Detection and characterization of pCT-like plasmid vectors for blaCTX-M-14 in Escherichia coli isolates from humans, turkeys and cattle in England and Wales. J. Antimicrob. Chemother. 67 1639–1644. 10.1093/jac/dks126 [DOI] [PubMed] [Google Scholar]
- Ueda S., Ngan B. T. K., Huong B. T. M., Hirai I., Tuyen L. D., Yamamoto Y. (2015). Limited transmission of bla(CTX-M-9)-type-positive Escherichia coli between humans and poultry in Vietnam. Antimicrob. Agents Chemother. 59 3574–3577. 10.1128/AAC.00517-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin L., Sharp H., Hille K., Seibt U., Fischer J., Pfeifer Y., et al. (2014). Subgrouping of ESBL-producing Escherichia coli from animal and human sources: an approach to quantify the distribution of ESBL types between different reservoirs. Int. J. Med. Microbiol. 304 805–816. 10.1016/j.ijmm.2014.07.015 [DOI] [PubMed] [Google Scholar]
- van Hoek A. H., Schouls L., van Santen M. G., Florijn A., de Greeff S. C., van Duijkeren E. (2015). Molecular characteristics of extended-spectrum cephalosporin-resistant Enterobacteriaceae from humans in the community. PLoS One 10:e0129085. 10.1371/journal.pone.0129085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Kaas R. S., Seyfarth A. M., Agersø Y., Lund O., et al. (2013). Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J. Antimicrob. Chemother. 68 771–777. 10.1093/jac/dks496 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data are available at the European Nucleotide Archive (ENA) under the project accession number PRJEB26592. All other raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.