Abstract
Both cervical and throat cancers are associated with human papillomavirus (HPV). HPV infection requires cleavage of the minor capsid protein L2 by furin. While furin is present in the vaginal epithelium, it is absent in oral epithelial basal cells where HPV infection occurs. The objective of this study was to investigate whether common oral bacteria express furin‐like peptidases. By screening strains representing 12 oral Streptococcus and Enterococcus species, we identified that eight Streptococcus strains displayed high levels of furin‐like peptidase activity, with S. gordonii V2016 the highest. We constructed null mutations for 14 genes encoding putative endopeptidases in S. gordonii V2016. Results showed that three endopeptidases, PepO, PulO, and SepM, had furin‐like activities. All three mutants showed decreased natural transformation by chromosomal DNA, while the pepO mutant also showed reduced transformation by plasmid DNA, indicating involvement of these endopeptidases in competence development. The purified S. gordonii PepO protein promoted infection of epithelial 293TT cells in vitro by HPV16 pseudovirus. In conclusion, oral bacteria might promote HPV infection and contribute to HPV tissue tropism and subsequent carcinogenesis in the oral cavity and throat by providing furin‐like endopeptidases.
Keywords: cancer, furin, HPV, peptidase, Streptococcus, transformation
1. INTRODUCTION
Oral and oropharyngeal (throat) cancers are the sixth most common cancer world‐wide. Each year, about 650,000 cases are diagnosed and nearly 40% of them are fatal (Ferlay, Pisani, & Parkin, 2004). While cervical cancer is the most common human papillomavirus (HPV)‐associated cancer in women, oral and oropharyngeal cancers are the most common HPV‐associated cancers in men (CDC, 2012). In recent decades, the incidences of HPV‐associated oropharyngeal cancers have been increasing consistently in industrialized countries (Chaturvedi et al., 2011; Shiboski, Schmidt, & Jordan, 2005). HPV‐related cervical cancer is a predominant cause of mortality among women in African countries (De Vuyst et al., 2013).
To infect human tissues, viruses often require activation by host peptidases such as furin. Furin is a serine endopeptidase expressed in many mammalian cells, mainly localized in the trans‐Golgi network (Nakayama, 1997). It belongs to the family of subtilisin‐like proprotein convertases that process latent precursor proteins into their biologically active products, and it is a calcium‐dependent protease and can efficiently cleave precursor proteins at their proteolytic cleavage consensus site (‐RXXR↓‐) (Thomas, 2002). Proteolytic activation of viral attachment factors at the site of infection is a strategy shared among many viruses (Klenk & Garten, 1994), where many viral proproteins contain consensus sites for furin. These include the minor capsid L2 protein of HPV (Richards, Lowy, Schiller, & Day, 2006) and the envelope glycoprotein gB of cytomegalovirus (CMV) (Jean et al., 2000) and Epstein–Barr virus (EBV) (Sorem & Longnecker, 2009).
Among many HPV subtypes, HPV16 is the most oncogenic. Infection by HPV16 is initiated by binding to the receptor heparan sulfate proteoglycans (HSPGs) on exposed basement membrane. This exposes the L2 cleavage site. Following cleavage by furin, HPV16 invades the tissue (Raff et al., 2013; Richards et al., 2006; Schiller, Day, & Kines, 2010). Although HSPGs exist in nearly all tissues throughout the body (Sarrazin, Lamanna, & Esko, 2011), the expression of furin varies. In all layers of the mouse vaginal epithelium, furin expression is high (Kines, Thompson, Lowy, Schiller, & Day, 2009), while in the human oral epithelium, furin expression is low, detectable only in the upper spinous and surface layers, not in the lower basal cell layer, where HPV initiates infection (López de Cicco, Bassi, Page, & Klein‐Szanto, 2002). The absence of furin in local tissues cannot explain why HPV‐induced cancers commonly occur in the oral cavity and oropharynx. Apparently, a gap exists in current understanding of HPV infection. It is largely based on in vitro cell culture studies under germ‐free conditions (Schiller et al., 2010), experiments with nonhuman animals (Kines et al., 2009), and level of infection reported in specific populations, such as young men in the United States and women in Africa (Curado & Boyle, 2013; De Vuyst et al., 2013).
The oral cavity is not sterile. It is colonized by billions of microorganisms, collectively referred to as the oral microbiome (McLean, 2014) mostly growing as biofilm attached to the mucosal and tooth surfaces (Kolenbrander, 2000). For HPV to infect the oral tissue, the lack of host furin in epithelial basal cells might be compensated by furin‐like peptidases from the oral microbiome. In our previous publications, we reported that Streptococcus promotes HPV16 entry into oral keratinocytes and oral squamous cell carcinoma cells. These suggest a possible interplay for Streptococcus and other similar bacteria to contribute to HPV16 entry (Schwartz et al., 2012; Tao, Pavlova, Gasparovich, Jin, & Schwartz, 2015). It is, therefore, reasonable to investigate how these microorganisms could play a role in determining HPV tissue tropism, and subsequently, how they might contribute to the development of tissue‐specific neoplasia.
Several anatomic sites in the oral cavity and oropharynx, including the back of the mouth, the base of the tongue, and the tonsils, are subject to HPV infection. These sites have nonkeratinized thin mucosal lining that form crypts, which house a complex microbial population. Included are various bacteria and viruses that cause periodontitis, common cold, influenza, and cancer. Tissue tropism varies among different viruses, and bacteria found at specific locations in the body might provide necessary conditions for infection by specific viruses (Doolittle & Webster‐Cyriaque, 2014). Although a furin‐like peptidase has been reported in the eukaryotic parasite Cryptosporidium parvum (Wanyiri et al., 2007), in hundreds of bacterial species comprising the human microbiome (Human Microbiome Project, 2012), furin‐like peptidases have not been reported. Here, we continue our studies of interaction between Streptococcus and HPV16 entry capability. We report finding of three furin‐like peptidases in S. gordonii, characterizing one of them, and observation of its promotion of HPV infection in vitro.
2. MATERIALS AND METHODS
2.1. Bacterial strains, growth conditions, and plasmids
Bacterial strains and plasmids used in these studies are described in Table 1. Unless sources are indicated in the table, oral bacterial strains were isolated from the saliva of healthy volunteers. Their species were identified by 16S rRNA gene sequence. The saliva sampling study was approved by the Institutional Review Board of the University of Illinois at Chicago. All Streptococcus and Entrococcus strains were grown in Todd‐Hewitt (TH) broth supplemented with 0.5% yeast extract (THY) at 37°C without agitation in a candle jar. For growth of E. coli strains, the Luria‐Bertani (LB) broth or agar was used; for growth of Lactobacillus strains, the MRS broth or agar was used. For transformation of S. gordonii, heat‐inactivated horse serum (56°C for 30 min) was added into TH broth at 5% (THS). An overnight culture of S. gordonii strain in THS was diluted 1:40 into fresh THS. After 2 hr of incubation at 37°C, DNA was added and the bacterial culture was incubated for 1 hr and then plated onto TH agar supplemented with appropriate antibiotics (kanamycin, 250 μg/ml; erythromycin, 10 μg/ml; or tetracycline, 15 μg/ml). The plates were incubated at 37°C for 24 hr in a candle jar for selection of transformants. All chemicals and reagents unless otherwise indicated were purchased from Sigma‐Aldrich (St. Louis, Missouri).
Table 1.
Bacterial strains and plasmids used in this study
| Strain/Plasmid | Characteristics | Reference |
|---|---|---|
| Streptococcus anginosus ARB136 | Wild‐type isolate | This study |
| Streptococcus gordonii V2016 | Wild‐type isolate | Pavlova et al. (2013) |
| Streptococcus infantis ARB112‐2 | Wild‐type isolate | This study |
| Streptococcus mitis 110‐5 | Wild‐type isolate | This study |
| Streptococcus mutans ARB117‐1 | Wild‐type isolate | This study |
| Streptococcus oralis 108 | Wild‐type isolate | This study |
| Streptococcus parasanguinis ARB104‐2 | Wild‐type isolate | This study |
| Streptococcus pneumoniae CP2000 | CP1250; Δcps; SmR | Weng, Piotrowski, & Morrison (2013) |
| Streptococcus pyogenes NZ131 | Wild‐type isolate | Simon & Ferretti (1991) |
| Streptococcus pyogenes NS05‐24 | Wild‐type isolate | This study |
| Streptococcus salivarius 101‐1 | Wild‐type isolate | This study |
| Streptococcus sanguinis SK36 | ATCC BAA‐1455 | Todd Kitten |
| Enterococcus faecalis IS51. | Wild‐type isolate | This study |
| Lactobacillus casei OLB1a | Wild‐type isolate | This study |
| Lactobacillus coryniformis OLB5 | Wild‐type isolates | This study |
| Lactobacillus fermentum OLB19b, 20b | Wild‐type isolates | This study |
| Lactobacillus lactis OLB12, 28, 32b, 36b, 43b | Wild‐type isolates | This study |
| Lactobacillus oris OLB9a | Wild‐type isolate | This study |
| Lactobacillus plantarum OLB14a, | Wild‐type isolate | This study |
| Lactobacillus rhamnosus OLB24a, 25a, 25b, 33 | Wild‐type isolates | This study |
| Lactobacillus salivarius OLB21, 22L, 22S, 160S | Wild‐type isolates | This study |
| S. gordonii V2016 mutant | ||
| ΔsubAB | Deletion of SGO_0316 & SGO_0317 with KmR | This study |
| Δsgc | Deletion of SGO_0566 with KmR | This study |
| ΔsecA2 | Deletion of SGO_0974 with KmR | This study |
| ΔsepA | Deletion of SGO_2150 with KmR | This study |
| pepM22 | Inactivation of SGO_0221 with pSF151, KmR | This study |
| cppA | Inactivation of SGO_0237 with pSF151, KmR | This study |
| pepM26 | Inactivation of SGO_0408 with pSF151, KmR | This study |
| sepM | Inactivation of SGO_0652 with pSF151, KmR | This study |
| pulO | Inactivation of SGO_0664 with pSF151, KmR | This study |
| pepZ1 | Inactivation of SGO_0796 with pSF151, KmR | This study |
| sepB | Inactivation of SGO_0847 with pSF151, KmR | This study |
| pepSY | Inactivation of SGO_1298 with pSF151, KmR | This study |
| pepO | Inactivation of SGO_1799 with pSF151, KmR | This study |
| pepZ2 | Inactivation of SGO_2009 with pSF151, KmR | This study |
| sepM‐pepO | Inactivations with pVA891 and pSF151, EmR, KmR, | This study |
| sepM‐pulO‐pepO | Inactivations with pVA891, pSF152 and pSF151, EmR, SpR, KmR | This study |
| S. sanguinis SK36 mutant | ||
| pepO | Transformed with the S. gordonii pepO DNA, KmR | This study |
| pulO | Transformed with the S. gordonii pulO DNA, KmR | This study |
| Escherichia coli strain JM109 | Cloning strain | Yanisch‐Perron, Vieira, & Messing, (1985) |
| E. coli DH5α | Cloning strain | Hanahan (1983) |
| E. coli BL21(DE3) | Protein overexpression strain | New England BioLabs |
| Plasmid | ||
| pSF151 | Streptococcal integration plasmid, 3.5 kb, KmR | Tao (1998) |
| pSF152 | Streptococcal integration plasmid, 3.2 kb, SpR | Tao (1998) |
| pVA891 | Streptococcal integration plasmid, 5.9 kb, EmR | Tao (1998) |
| pA13 | Lactic acid bacteria‐E. coli shuttle plasmid, 4.6 kb, EmR | Kojic et al. (2010) |
| pET21a | Cloning/expression, 5.4 kb, AmpR | Novagen |
| pET21a‐pepO | pET21a containing S gordonii pepO | This study |
2.2. Furin‐like peptidase assay
Since certain viruses require furin activation in order to infect, we analyzed oral and throat bacterial strains for furin‐like peptidase activity. Bacteria were grown to mid‐exponential phase to OD600 of ~0.7 in THY broth for Streptococcus or Enterococcus and MRS broth for Lactobacillus, and 100 μl culture was harvested, washed, and resuspended in Na/MES buffer for analysis of furin‐like peptidase activity. The fluorogenic furin substrate Boc‐RVRR‐AMC (Enzo Life Sciences) at 50 μmol/L in Na/MES buffer [pH 7.0, 20 mmol/L MES [2‐(N‐morpholino) ethanesulfonic acid], 1 mmol/L CaCl2, and 0.01% Triton X‐100] was added. The mixture was incubated for 30 min at 37°C. The cells were removed by centrifugation. The fluorescence released from the substrate in the supernatant was read with the VICTOR X5 Plate Reader of PerkinElmer (Excitation 360 nm; Emission 460 nm). For analysis of purified peptidases, the protein was added to the reaction mixture and incubated for 30 min at 37°C before fluorescence reading. The purified recombinant human furin from New England BioLabs (NEB) was used as control.
2.3. Construction of mutant strains
The V2016 sgc, subAB, sepA, and secA2 deletion mutants were obtained by allelic exchange (Pavlova, Jin, Gasparovich, & Tao, 2013), while the remaining 10 genes were inactivated by using the streptococcal integration plasmid pSF151 (Tao, 1998). Therefore, a total of 13 genetic loci were inactivated by allelic replacement or insertion duplication. Streptococcus sanguinis SK36 pepO mutant was also obtained by transforming SK36 with chromosomal DNA isolated from the S. gordonii V2016 pepO mutant. Standard recombinant DNA techniques were employed as described (Sambrook, Fritsch, & Maniatis, 1989). Multiple pairs of oligonucleotides (Integrated DNA Technologies) used in this study are shown in Table 2. Chromosomal DNA was prepared by the glass bead method (Ranhand, 1974). PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, Valencia, California). DNA restriction enzymes were used under the conditions specified by the manufacturer (New England BioLabs, Ipswich, Massachusetts). Each mutant was confirmed genotypically by PCR. Due to insertion of the antibiotic resistance cassette, the size of the PCR DNA fragment became larger in the mutant than that in the wild type with primers flanking the insertion site (not shown).
Table 2.
Oligonucleotides used in this study
| Oligonucleotide (5′→3′) | Sequence | Restriction enzyme |
|---|---|---|
| Construction of ΔsubAB (SGO_0316‐0317) | ||
| SubA‐F1 | ataaggttaactcccttcgacaagctggtg | |
| SubA‐R1 | tgatttcccggatccttgactgg | BamHI |
| SubB‐F2 | ggttctacagaattccctgatgg | EcoRI |
| SubB‐R2 | actgaccaaatgctggttactcttagcatc | |
| Construction of Δsgc (SGO_0566) | ||
| Sgc‐F1 | cgtaggatcccttcgtgaccaaggat | |
| Sgc‐R1 | tcgagaattcagtttgaggaaccttgatgat | EcoRI |
| Sgc‐F2 | caggacggatccgagtttccatg | BamHI |
| Sgc‐R2 | gccttagcttgtgcataggcctctaag | |
| Construction of ΔsecA2 (SGO_0974) | ||
| secA2‐F1 | ttgccagaagcctatgctg | |
| secA2‐R1 | gctagaattcttatggaggccatggcacg | EcoRI |
| secA2‐F2 | atgcggatccgcagagattatcctgatcgg | BamHI |
| secA2‐R2 | ttaagaatagccaggcgctgg | |
| Construction of ΔsepA (SGO_2150) | ||
| SepA‐F1 | gcgaccgttcgcttagaaggcgaatgctct | |
| SepA‐R1 | gctaggatccttcgctagctacctgatcat | BamHI |
| SepA‐F2 | gaccagcattaggaattcaaatg | EcoRI |
| SepA‐R2 | ttagagagactaatctttacttcgactccc | |
| Construction of SGO_0221 mutant | ||
| Sgo‐221F | ctgagaattgcggtggcgac | TasI |
| Sgo‐221R | gagccaattttcctcggctt | TasI |
| Construction of SGO_0237 mutant | ||
| Sgo‐237F | ttgggagatcagacaaagag | Sau3AI |
| Sgo‐237R | gcatgatcgccaaaaggttgataat | Sau3AI |
| Construction of SGO_0408 mutant | ||
| Sgo‐408F | gcagctgtggatcctgcagt | BamHI |
| Sgo‐408R | cagctgacctttgaattcac | EcoRI |
| Construction of SGO_0652 mutant | ||
| Sgo‐652F | gactggtggatcatcggacc | Sau3AI |
| Sgo‐652R | cgtaaggttggatctgcaag | Sau3AI |
| Construction of SGO_0664 mutant | ||
| Sgo‐664F | gaatgagaatttccgtcaga | TasI |
| Sgo‐664R | cacttggtacaatttctatc | TasI |
| Construction of SGO_0796 mutant | ||
| Sgo‐796F | tttagatcaacaaggtattc | Sau3AI |
| Sgo‐796R | gaaagaacctgcagactgca | PstI |
| Construction of SGO_0847 mutant | ||
| Sgo‐847F | ggaatggaaattactactat | TasI |
| Sgo‐847R | ttggcaatttactataatct | TasI |
| Construction of SGO_1298 mutant | ||
| Sgo‐1298F | gacttcccaaattggccggt | TasI |
| Sgo‐1298R | gctcgttctaattccattga | TasI |
| Construction of SGO_1799 mutant | ||
| Sgo‐1799F | gtgaagaattcctcagcaga | EcoRI |
| Sgo‐1799R | gcggctatcctgcaggcgcc | PstI |
| Construction of SGO_2009 mutant | ||
| Sgo‐2009F | gaagggggatcggctagctac | Sau3AI |
| Sgo‐2009R | cactgatcaggtacagaatagccgt | Sau3AI |
| Cloning S. gordonii pepO gene | ||
| RW112 | tttaagaaggagatatacatatgACACGACTGCAAGATGATTTTTATG | |
| RW113 | cagtggtggtggtggtggtgctcgagCCAAATAATCACACGATCTCC | |
| plasmid flanking region in lower case | ||
2.4. Purification of recombinant S. gordonii PepO
The pepO (SGO_1799) gene was amplified from S. gordonii V2016 genomic DNA using primers RW112 and RW113. The fragment was assembled into pET21a, following digest with XhoI and NdeI, upstream of a 6‐HIS tag using NEB HiFi builder (NEB). The following product was transformed into E. coli DH5α cells. Isolated plasmid bearing the insert of interest was verified by transformation into E. coli BL21(DE3), induction with 0.5 mmol/L IPTG and the demonstrated ability of protein of the correct molecular mass in the lysate to bind nickel resin. Protein was purified as described in Wilkening, Chang, and Federle (2016). In brief, cells were grown to an OD600 of ~0.7 and induced with 0.5 mmol/L IPTG. Cells were grown with agitation at 30°C for 6 hr. Harvested cells were suspended in buffer A (0.2 mol/L phosphate buffer, 0.0054 mol/L potassium chloride, 0.274 mol/L sodium chloride, 20 mmol/L imidazole, 0.07% β‐mercaptoethanol) with the addition of 1x protease inhibitor cocktail (Pierce). Cells were lysed, and the soluble fraction was separated by centrifugation. The resulting lysate was passed over a HisTrap‐HP Nickel column (GE Bioscience) and washed with buffer A. rPepO was then eluted using 10% buffer B (buffer A + 500 mmol/L imidazole). Fractions containing rPepO were assessed by SDS–PAGE, pooled, and concentrated to 2.8 mg/ml via a 50,000 MWCO Amicon column. Imidazole was removed via buffer replacement with buffer C (buffer A with no imidazole). Concentrated rPepO was stored in 20% glycerol at −80°C.
2.5. Interaction of peptidases with bacteria and mammalian cells
Because the purified rPepO from bacteria displayed low peptidase activity compared with enzyme associated with bacteria, we studied interaction between the rPepO enzyme and bacteria or mammalian cells, using furin as control. Bacterial cells were those that naturally displayed a low furin‐like peptidase activity, including S. salivarius 101‐1, S. pyogenes NS05‐24, and E. coli JM109 and the S. gordonii pepO mutant. Streptococcus strains were grown in THY and E. coli was in LB broth overnight, and washed in Na/MES before use. Mammalian cells included two cell lines: the human lung carcinoma A549 cells and the human tongue squamous carcinoma SCC9 cells. Cells were grown in the DMEM medium with 2 mmol/L glutamine and 10% fetal bovine serum until confluence. The cells were harvested and washed in Na/MES buffer. One milliliter of washed bacterial cells (about 109 per ml) or mammalian cells (about 105 per ml) were used to perform the assay. Furin‐like peptidase activities of washed cells alone, the cells with added rPepO or Furin proteins, and the purified proteins in buffer without cells as control were assayed for peptidase activity as described above.
2.6. Interaction of PepO with bacterial cell wall peptidoglycans
Two bacterial cell wall peptidoglycans were used to interact with furin and the S. gordonii PepO protein. One is from Bacillus subtilis and the other is from S. pyogenes. These two cell walls were tested because B. subtilis cell wall was available commercially (Sigma), and S. pyogenes cell was isolated in our laboratory for another project. S. pyogenes NZ131 was grown overnight in THY broth. Cells were isolated by centrifugation and suspended in 0.1 mol/L Tris–HCl pH 6.8 with 0.25% SDS. The cells in solution were placed inside a conical tube and incubated in a boiling water bath for 2 hr to kill the bacteria and inactivate wall enzymes. Bacterial debris was collected by centrifugation at 5,000 xg in a Beckman JA‐20 rotor (Beckman Coulter) for 10 min. The pellet was washed four times in 0.1 mol/L Tris pH 6.8 to remove residual SDS. The pellet was suspended in 0.1 mol/L Tris (pH 7.5), 200 μg/ml RNase A, 100 μg/ml DNase I, and 0.2 mol/L MgSO4 and subjected to mechanical disruption at 15,000 psi using an EmulsiFlex‐C5 homogenizer (Avestin). After disruption, the solution was incubated with shaking at 37°C for 2 hr, allowing for degradation of nucleic acid polymers. Trypsin (250 μg/ml) and 10 mmol/L CaCl2 were added to the solution and continue incubation and shaking overnight. Finally, the insoluble fraction was centrifuged and the pellet suspended in 1 N HCl for 4 hr with shaking at 37°C to eliminate associated teichoic acids. Finally, the S. pyogenes peptidoglycan was suspended in 1 ml PBS at about 100 mg/ml. The purified recombinant human furin and S. gordonii rPepO protein were mixed with these bacterial cell wall peptidoglycans for 60 min at room temperature and assayed for furin activity.
2.7. Activation of PepO by proteases
Since interaction with bacteria or cells boosted the furin‐like peptidase activity of the human furin and purified PepO of S. gordonii, we tested if other proteases could activate PepO. Briefly, proteases including trypsin, chymotrypsin, papain, pepsin, pronase, protease, and proteinase K (from Sigma) solutions were made at three concentrations, 1 mg/ml, 100 μg/ml, and 10 μg/ml in Na/MES buffer (pH 7.0). The rPepO protein at 2.8 μg was incubated with 100 μl of each enzyme at three concentrations for 30 min at 37°C. Buffer alone and buffer with rPepO without proteases were used as controls. Furin‐like peptidase activities of the protease‐treated rPepO and furin were assayed by the fluorogenic furin substrate Boc‐RVRR‐AMC (Enzo Life Sciences) in the VICTOR X5 Plate Reader of PerkinElmer (Excitation 360 nm; Emission 460 nm).
2.8. Bacterial competence assay
Streptococcus gordonii V2016 wild‐type and its mutant strains with defect in three peptidases, PepO, PulO and SepM, were grown overnight in THS (TH broth with 5% horse serum) broth. In the second morning, the cultures were diluted 1:40 into fresh THS broth and continued to incubate for 2 hr or at time points indicated in the chart to develop competence. The plasmid pA13 DNA (EmR) and a chromosomal DNA that carries an erythromycin resistance marker were added, and the cultures were incubated for 2 hr and plated on TH agar plates containing erythromycin 10 μg/ml. After incubation at 37°C in a candle jar for 24 hr, colonies were counted for transformants.
2.9. HPV16 entry assay
293TT cells of an adenovirus transformed human embryonic kidney cell line with a stably integrated SV40 genome with high levels of large T antigen were cultured in DMEM (Dulbecco's Modified Eagle Medium) + Enhanced GLU (GIBCO, Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (Sigma‐Aldrich, St. Louis MO) and 250 ng/ml Hygromycin B (Invitrogen). The HPV16 pseudovirion (PsV) was produced with an Optiprep purification or a maturation method using overnight incubation of crude cell lysate at 37°C. HPV16 PsV packaging plasmids (p16L1‐GFP and pfwB) and the expression vector for luciferase (pCLucf) driven by the CMV promoter were used (Buck & Thompson, 2007). This system relies upon a co‐propagation of L1/L2 expression plasmid together with a reporter plasmid [green fluorescent protein (GFP)] to generate high titers of mature PsV stocks for visualization of viral entry.
PsV particles (20 μl/well) were placed into wells in a 6‐well plate or (5 μl/well) into a 96‐well plate coated with Type I collagen containing 293TT cells at 50%–60% confluence after the furin inhibitor decanoyl‐RVKR‐chloromethylketone (CMK) (ENZO Life Sciences. Farmingdale, NY; stock: 100 mmol/L, working concentration 1.0 μmol/L) was added to suppress internal furin activity of the cell. Infection was monitored by observation under a fluorescent microscope on day 3 and by reading with a fluorometer for GFP expression due to plasmid replication in 293TT cells on day 4 and day 6, post infection.
2.10. Statistical analysis
The paired 2‐tailed Student t test with a confidence limit of p < .05 determined the level of significance between two comparative groups, which consisted of control and experimental groups. The results presented in the figures are means of triplicate counts with standard deviations unless otherwise stated.
3. RESULTS
3.1. Bacterial genes encoding furin‐like peptidase activity
We analyzed 12 oral and throat bacterial strains representing 11 Streptococcus and 1 Enterococcus species (Table 1) for furin‐like peptidase activities using a fluorogenic furin substrate Boc‐RVRR‐AMC (Enzo Life Sciences) (Cruz, Biryukov, Conway, & Meyers, 2015). Upon incubation of bacterial cells with the substrate, 8 of the 11 streptococcal strains tested were positive for furin‐like activity (73%; Figure 1a), whereas the Enterococcus strain was negative. S. gordonii V2016 displayed the highest furin‐like peptidase activity. We also tested 19 oral Lactobacillus strains for furin‐like activity (Table 1) and found that 18 were negative (no higher than the buffer control), and 1 (L. rhamnosus OLB25b) was positive. Thus, in comparison to a panel of Lactobacillus strains, the Streptococcus species were more likely to express extracellular furin‐like enzymes.
Figure 1.
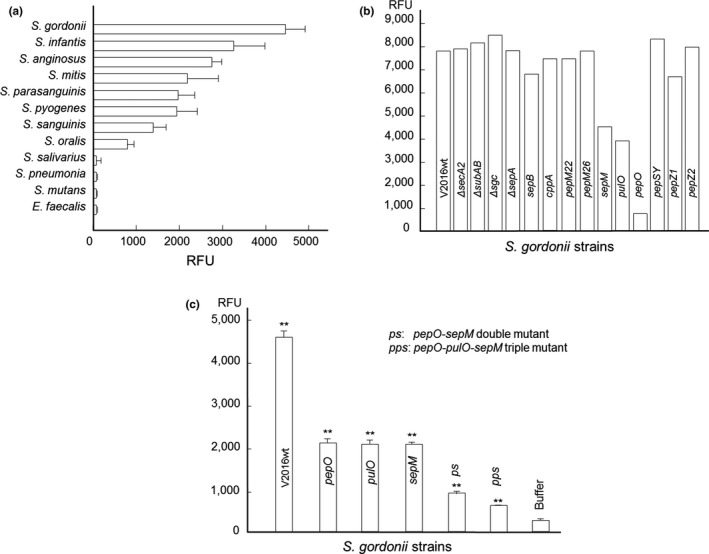
(a) Furin‐like activity of strains representing 12 oral and throat Streptococcus and Enterococcus species. (b) Furin‐like activities of S. gordonii V2016 and its 13 peptidase and the secA2 mutants (one measure per sample). (c) Furin‐like activities of S. gordonii and its furin‐like peptidase‐negative mutants. **Statistically very significant difference (p < .01) between different groups: the wild‐type, single, double and triple mutants, and the buffer control. Each bar represents the mean of triplicate values ± standard deviation
To identify genes encoding furin‐like peptidases in Streptococcus, we used the human furin protein sequence to BLAST against the genomic sequences of the Streptococcus genus. The subtilisin‐like serine protease was found as a homolog. However, blasting the S. gordonii genome (Vickerman, Iobst, Jesionowski, & Gill, 2007) with the furin sequence did not yield significant similarity. We then performed in silico analysis of S. gordonii genome for all genes encoding putative peptidases and identified 47 genes. As furin is an endopeptidase, we dismissed genes encoding exopeptidases (aminopeptidases and carboxypeptidases), leaving 14 genes to consider as candidates for furin activity. Among them, two genes, SGO_0316 and SGO_0317, are linked, and thus, a total of 13 genetic loci were subjected to mutagenesis with the kanamycin resistance marker to achieve allelic exchange (Pavlova et al., 2013) or inactivation by insertion duplication with the plasmid pSF151 (Tao, 1998) (Table 1). In addition, to identify if the Sec2 system of S. gordonii (Bensing & Sullam, 2009) is required for secretion of these furin‐like peptidases without a typical signal peptide, we constructed a secA2 deletion mutant.
Each mutant was subjected to the furin‐like activity assay with the fluorogenic furin substrate. Results showed that inactivation of 3 genetic loci, SGO_1799, SGO_0664 and SGO_0652, each had reduced furin‐like activity (Figure 1b). We used chromosomal DNA isolated from one mutant as a substrate for natural transformation by another mutant containing a different antibiotic resistance marker to generate double‐ and triple‐protease mutants (Tao, 1998). These mutants showed further reduced furin‐like activity compared with the wild‐type and single‐gene knockout mutants (Figure 1c), suggesting that these enzymes have redundant activities. Among the three proteins, none contain a classical signal peptide for secretion, and since the secA2 mutation (Bensing & Sullam, 2009) did not affect furin‐like activity in S. gordonii V2016, alternative pathways unrelated to Sec or SecA2 may allow for the observed extracellular peptidase activity.
The SGO_1799 locus encodes a 630‐amino acid protein with a calculated molecular weight of 71.5 kDa and is homologous to the conserved endopeptidase O (PepO) in other bacteria (Oetjen, Fives‐Taylor, & Froeliger, 2001). Without a signal peptide or a transmembrane domain, S. gordonii PepO is presumably a cytoplasmic protein and/or an extracellular protein secreted by an alternative pathway. Due to a zinc‐binding site, it is a presumed zinc‐dependent metalloendopeptidase. Separated by 148 and 149 base pairs from its neighboring genes, S. gordonii pepO appears to be an independent gene, not part of a multicistronic operon.
The SGO_0664 locus encodes a 220‐amino acid endopeptidase with a calculated molecular weight of 24.7 kDa. The protein does not have a signal peptide but has 4‐6 transmembrane domains. It is predicted that the protein may be an intramembrane peptidase specifically cleaving transmembrane domains of other integral membrane proteins. It is homologous to the prepilin signal peptidase PulO of other bacteria. Separated by 169 base pairs, S. gordonii pulO is divergently located upstream to the gene encoding Dps (DNA starvation/stationary phase protection protein), but overlaps convergently by 31 base pairs with the tryptophan biosynthesis (trp) operon downstream. The pulO gene also appears to be independent and not part of an operon.
The SGO_0652 locus encodes a 347‐amino acid with a calculated molecular weight of 38.1 kDa. Without a signal peptide, this locus is homologous to the conserved sepM gene in S. mutans, which encodes a membrane‐associated peptidase required for processing competence stimulating peptide (CSP) for competence development and quorum sensing (Biswas, Cao, Kim, & Biswas, 2015; Hossain & Biswas, 2012). With one transmembrane domain, S. gordonii sepM is also predicted as a membrane‐associated peptidase. The genomic location shows that S. gordonii sepM appears to be the last gene in an operon with two other genes, which encode phosphopantetheine adenylyltransferase (coaD) involved in coenzyme A biosynthesis and rRNA methyltransferase. It is unknown why the three seemingly unrelated genes co‐transcribe in an operon.
To test if these genetic loci in other Streptococcus species also encode peptidases with furin‐like activity, we transformed chromosomal DNA isolated from S. gordonii pepO, pulO, and sepM mutants into S. sanguinis SK36. We were able to obtain transformants with S. gordonii pepO and pulO DNAs, but not with sepM DNA (Table 1). Furin assay with these two S. sanguinis peptidase mutants showed reduction of furin‐like peptidase activity in comparison with the wild‐type strain SK36. Therefore, at least two furin‐like endopeptidases, PepO and PulO, may be present in multiple oral streptococcal species.
3.2. Interacting with cells and chymotrypsin activates S. gordonii rPepO
We cloned and expressed the S. gordonii pepO gene tagged with 6xHIS in E. coli and purified the recombinant protein (rPepO) using a HisTrap‐HP Nickel column (GE Bioscience), and SDS–PAGE analysis confirmed its molecular size and purity (Figure 2a). The rPepO protein was concentrated to about 2.8 mg/ml and tested for furin‐like peptidase activity, but its peptidase activity was no higher than a buffer control. We then tested the possibility that activity of rPepO, or the human furin protein (rFurin, New England BioLabs), would be enhanced in the context of bacterial or mammalian cells. The S. gordonii pepO knockout mutant and other cells, including S. salivarius 101‐1, S. pyogenes NS05‐24, and E. coli JM109 bacterial strains, and the mammalian SCC9 and A549 cells, all naturally expressing furin‐like peptidase activity, were combined with purified rPepO and added to the fluorogenic substrate. Results (Figure 2b) showed that all six cells activated rFurin, but only four (except E. coli and S. salivarius) activated rPepO. Data from 293TT could not be interpreted due to high intrinsic furin‐like activity of the cell. In addition, bacterial peptidoglycan preparations from Streptococcus pyogenes and Bacillus subtilis both activated rFurin, but not rPepO (Figure 2c), indicating that rPepO may require activation that is more specific. By interacting with various proteases, including trypsin, chymotrypsin, papain, pepsin, pronase, protease, and proteinase K, only chymotrypsin promoted furin‐like activity of rPepO (Figure 2d) and in a dose‐dependent manner (Figure 2e). As pronase (protease from Streptomyces griseus) (Calbiochem) showed strong furin‐like peptidase activities with and without added rPepO on the fluorogenic furin substrate, it could not be used for evaluating rPepO activation.
Figure 2.
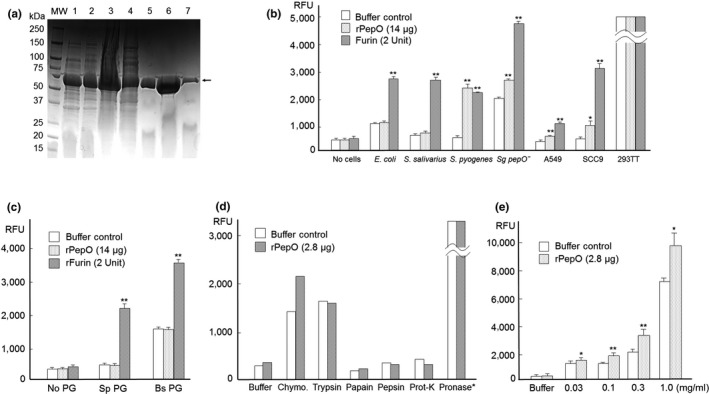
(a) Affinity purification of recombinant S. gordonii PepO from E. coli shown in SDS–PAGE. 1) Pellet without IPTG; 2) Pellet with IPTG for 6 hr; 3) Crude lysate; 4) Elution fraction 2 (flow through); 5) Fraction 8; 6) Fraction 9; and 7) Fraction 10. Arrow: rPepO. (b) rPepO activation by bacteria (E. coli JM109, S. salivarius 101‐1, S. pyogenes NS05‐24, and S. gordonii pepO mutant) and mammalian cells (A549, SCC9 and 293TT). Buffer controls are buffer with indicated cells. 293TT cells released about 19,000 RFU fluorescence from the furin substrate with or without added rPepO and furin, showing high endogenous furin level. (c) rPepO activation by bacterial peptidoglycan (PG). Sp. S. pyogenes; Bs, B. subtilis. Buffer controls are buffer with indicated PG. Note: B. subtilis PG in buffer displayed high furin‐like protease activity, indicating that these PGs may contain proteases. (d) rPepO activation by chymotrypsin, trypsin, papain, pepsin, and proteinase K. Pronase (protease from Streptomyces griseus) (Calbiochem) released 12,000 RFU fluorescence from the furin substrate with or without added rPepO (one measure per sample). Buffer controls are buffer with indicated protease. (e) rPepO activation by chymotrypsin. Both buffer control and rPepO samples contain chymotrypsin with indicated amount in the figure. *Significant difference (p < .05); **very significant difference (p < .01) compared with controls
3.3. PepO inactivation reduces competence in S. gordonii
While generating the double and triple mutants, we noticed reduction of transformation efficiency with chromosomal DNA in these S. gordonii peptidase‐defective mutants. Therefore, we studied their genetic transformation. As shown in Figure 3a, the peak number of transformants was reached at 120 min after a 1:40 dilution for both the wild‐type V2016 and the pepO mutant, but the pepO mutant displayed substantially reduced transformation rates. When compared among three different peptidase‐defective mutants against the wild type for uptake of circular plasmid and linear chromosomal DNAs, the pepO mutant showed the highest reductions in transformation by both DNA types (Figure 3b), while the pulO and sepM mutants only showed moderate reductions in transformation by chromosomal DNA (data not shown). Both pepO and sepM mutants had a same growth rate as the wild‐type strain V2016 with a doubling time of 41 min, while the pulO mutant had a slower growth rate with a doubling time of 63 min. Therefore, inactivation of pepO might suppress competence development of S. gordonii independent of growth rate. However, addition of purified PepO protein to the competent S. gordonii pepO mutant culture (at 8 μg/ml) prior to the addition of plasmid or chromosomal DNA did not restore its rate of transformation to the level of the wild‐type strain (data not shown).
Figure 3.
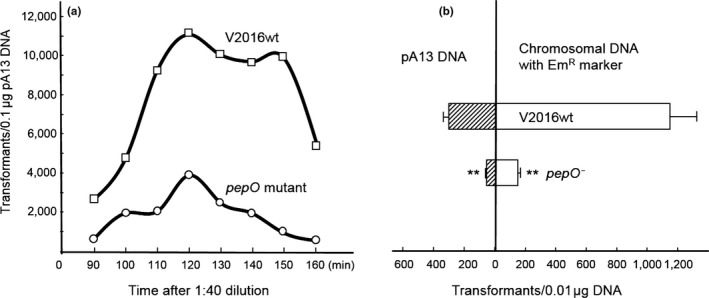
Transformation of S. gordonii V2016 and its pepO‐defective mutant. (a) Competence development as function of time. (b) Transformation rates between plasmid and chromosomal DNAs. Comparing with the wild type, the pepO mutant shows significant decrease in both plasmid and chromosomal DNA transformations. **Very significant difference (p < .01) when compared with the control
3.4. S. gordonii PepO promotes HPV infection
We studied HPV16 pseudovirus (PsV) entry into the 293TT cells to evaluate the role of furin‐like activity of S. gordonii PepO in viral activation. Since the 293TT cell has endogenous furin activity, to study the role of exogenous furin‐like peptidase, we added the furin inhibitor decanoyl‐RVKR‐chloromethylketone (CMK) to suppress its endogenous furin activity (Richards et al., 2006). As shown in Figure 4Aa and Ba, without CMK treatment, the HPV16 PsV displayed high rates of infection into 293TT cells promoted by its endogenous furin. With CMK treatment (Figures 4Ab and Bb), HPV16 PsV entry into 293TT cells was significantly reduced due to inhibition of endogenous furin. Addition of exogenous furin (Figures 4Ac and Bc) or the S. gordonii PepO protein (Figures 4Ad and Bd) to these cells, in which the endogenous furin was suppressed by CMK, HPV16 PsV entry into 293TT cells showed significant increase (p < .01) in a dose‐dependent manner in comparison with the CMK‐treated control cells (Figures 4Ab and 4Bb). The S. gordonii PepO protein‐treated group (Figure 4Ad) showed a higher HPV16 PsV entry than the human furin‐treated one (Figure 4Ac).
Figure 4.
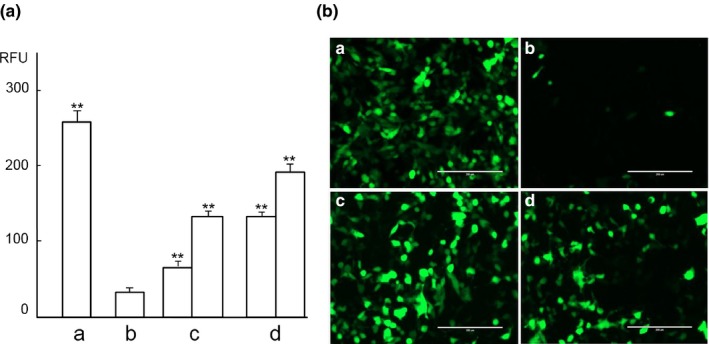
HPV16 PsV infection of 293TT cells boosted by S. gordonii rPepO. (A) HPV16 PsV infection on day 6 detected by fluorometer. a, HPV16 PsV. b, HPV‐16 PsV + CMK. c, HPV‐16 PsV + CMK + furin (2 Units, left; 10 Units, right). d, HPV‐16 PsV + CMK + S. gordonii rPepO (14 μg, left; 56 μg, right). **Very significant difference (p < .01) when compared with the control (b). (B) HPV16 PsV infection on day 3 detected by fluorescent microscope. Bar: 200 μm. a, HPV16 PsV infection of 293TT. B, HPV‐16 PsV infection of 293TT inhibited by CMK. c, HPV‐16 PsV infection of 293TT with CMK promoted with 10 units of furin. d, HPV‐16 PsV infection of 293TT with CMK promoted with 56 μg of rPepO
4. DISCUSSION
It has been reported that bacteria colonizing the human airway activate influenza viruses by cleaving the viral surface glycoprotein hemagglutinin with peptidase (Böttcher‐Friebertshäuser, Klenk, & Garten, 2013). However, there have been no reports on HPV activation by bacteria. Unlike influenza viruses, activation of HPV requires the cleavage of its L2 protein at the furin consensus site (‐RXXR↓‐) (Richards et al., 2006; Schiller et al., 2010). To our knowledge, this is the first report of a furin‐like peptidase in bacteria capable of promoting HPV16 infection in vitro.
After screening 12 representative strains of Streptococcus and Enterococcus species for furin‐like peptidases, we found that S. gordonii V2016 displayed the highest furin‐like peptidase activity among tested strains. S. gordonii not only colonizes the human oral cavity but also the throat (Frandsen, Pedrazzoli, & Kilian, 1991), which is a hot spot for HPV infection‐associated cancers. By analyzing the S. gordonii genome, we identified 14 genes encoding endoproteases and constructed null mutations in these genes. Defects in any of the three genes, pepO, pulO, and sepM, showed reduction in furin‐like peptidase activity (Figure 1B). Although the initial screening showed that the pepO mutant had the most reduction, repeated assays with triplicate samples showed comparable reductions in furin‐like peptidase among these three mutants (Figure 1C).
PepO, a member of the M13 family of zinc metalloendopeptidases, has been well studied in several different bacteria. Streptococcus parasanguinis PepO is similar to the human endothelin‐converting enzyme 1 (ECE‐1) (Oetjen et al., 2001). In Streptococcus pneumoniae (Agarwal et al., 2013, 2014), S. pyogenes (Honda‐Ogawa et al., 2017), Streptococcus mutans (Alves et al., 2017) and P. gingivalis (Ansai, Yu, Urnowey, Barik, & Takehara, 2003), PepO is a virulence protein facilitating evasion of innate immunity and invasion of host cells. In S. pyogenes, PepO degrades peptide pheromones of a quorum sensing system (Wilkening et al., 2016).
PulO is homologous to the prepilin peptidase (PilD) involved in Type IV pili production in other bacteria. These pili were found recently in the Gram‐positive bacterium Streptococcus sanguinis as a part of filamentous nano‐machine for bacterial motility (Gurung et al., 2016). In S. pneumoniae, the type IV pilus mediates DNA binding during natural transformation (Laurenceau et al., 2013). The S. gordonii CSP induces PulO expression (Vickerman et al., 2007). We found that inactivation of pulO retarded S. gordonii growth rate by about 50%.
SepM, a cell membrane‐associated peptidase, is required for the CSP maturation in S. mutans (Biswas et al., 2015; Hossain & Biswas, 2012). SepM proteins from S. mutans, S. pyogenes and S. agalactiae all activate the S. mutans CSP by cleaving CSP‐21 between the Ala18 and Leu19 residues, but SepM from S. gordonii does not (Biswas et al., 2015).
Although these peptidases exit in many bacteria, their furin‐like activity may be species specific. By transforming chromosomal DNAs from pepO and pulO‐inactivated S. gordonii strains into S. sanguinis SK36, the furin‐like activity reduced in the latter (data not shown). This suggests that PepO and PulO in S. sanguinis may have furin‐like activity, similar to that of S. gordonii. However, these peptidases may not have or have weak furin‐like activities in Streptococcus salivarius, S. pneumoniae, S. mutans, and Enterococcus faecalis, because the tested strains representing these species did not show detectable furin‐like activities (Figure 1a).
Among these three furin‐like peptidases, PulO and SepM are membrane proteins that are bound to the bacterial cell surface. This may limit their ability to interact with viruses. PepO is not membrane‐bound, so it may be released into the environment to interact with viruses in distance. Therefore, we focused our efforts on characterizing S. gordonii PepO.
After cloning and purification of S. gordonii PepO, we found it had a low activity against the fluorogenic furin‐specific substrate. Apparently, protein maturation may be required. By interacting with different bacteria, cells, bacterial peptidoglycans, and multiple different proteases, we found that chymotrypsin enhanced S. gordonii PepO in a dose‐dependent manner (Figure 2e). This suggests that maturation of S. gordonii PepO may require a chymotrypsin‐like protease. Interestingly, interaction with two bacterial peptidoglycans increased the activity of rFurin (Figure 2c). The mechanism is unknown, but it may be due to bacterial peptidoglycan‐associated protease. For example, the B. subtilis peptidoglycan with buffer alone displayed high furin‐like activity.
Without a signal peptide, PepO normally would be considered as a cytoplasmic protein. However, because inactivation of pepO substantially reduced the furin‐like peptidase activity, PepO appears to be present outside the cell. Since furin‐like peptidase activity is not reduced in the secA2 mutant, a Sec or SecA2 independent pathway (Bensing & Sullam, 2009) might secrete PepO. In Streptococcus pneumoniae (Agarwal et al., 2013), S. pyogenes (Honda‐Ogawa et al., 2017) and Porphyromonas gingivalis (Ansai et al., 2003), PepO is secreted by an unknown pathway to facilitate bacteria invasion into host cells.
Because inactivation of pepO substantially reduced competence in S. gordonii, the intended function of PepO might be associated with competence development. Although S. gordonii pepO expression is not induced by CSP (Vickerman et al., 2007), at least two peptidases in Streptococcus, ComA (Ishii et al., 2010) and SepM (Biswas et al., 2015), have been reported to promote competence by processing CSP to its mature form. The mechanism by which S. gordonii PepO facilitates genetic transformation is unknown, but it appears to play this role intracellularly, because adding purified PepO to competent pepO mutant culture did not restore its transformation to the level of the wild type. This is different from the SepM peptidase in S. mutans, which works extracellularly to process CSP (Hossain & Biswas, 2012). Nonetheless, linking PepO to bacterial competence is novel, as currently identified bacterial competence factors have not included PepO (Straume, Stamsås, & Håvarstein, 2015). Although PepO was identified to inactivate CSP in S. pneumoniae, it was not proven to regular competence in this bacterium (Bergé, Langen, Claverys, & Martin, 2002).
The proprotein convertase furin not only is essential for activation of certain viruses such as HPV (Richards et al., 2006; Schiller et al., 2010) but also promotes cancer development and metastasis (Jaaks & Bernasconi, 2017). HPV‐induced cancers mostly occur in the cervix and oropharynx. Unlike the cervix, where furin expression is high and exists in all layers of the vaginal epithelium (Kines et al., 2009), furin expression in the oral epithelium is low, only detectable in the upper spinous and surface layers, but not in the lower basal layer, where HPV initiates infection (López de Cicco et al., 2002). We observed that S. gordonii PepO promoted HPV16 PsV infection of cultured 293TT cells in vitro in a dose‐dependent manner similar to that of furin. This suggests that bacterial furin‐like peptidases might compensate the deficiency of host furin in oral epithelium basal cells to assist HPV infection. Therefore, Streptococcus PepO, perhaps also PulO and SepM, might have a moonlighting role (Henderson, 2014) in activation of viruses, such as HPV, as a furin‐like peptidase.
The human saliva contains small amounts of proteases including furin and furin inhibitors of the host origin (Sun, Salih, Oppenheim, & Helmerhorst, 2009). The trace amount of furin in the saliva may promote oral HPV infection and tumorigenesis, but due to presence of nearly equal amount of furin inhibitors, such as alpha‐1‐antitrypsin, in the saliva, the effect of salivary furin, if any, might be minimal. Therefore, furin‐like peptidases released by oral bacteria might play a role in oral HPV infection and tumorigenesis. The microbiome findings have been varied. One group reported decrease (Schmidt et al., 2014), while two other groups reported increase (Guerrero‐Preston et al., 2016; Mukherjee et al., 2017) of Streptococcus species in oral cancer lesions in comparison with controls. We think that microbial cancer‐promoting factors might not be limited to specific bacterial species. Furin‐like peptidases released by any microorganism could promote oral cancer. Identifying Streptococcus peptidases with furin‐like activity that promotes HPV infection may help us understand the linkage among Streptococcus colonization sites, HPV tissue tropism, and HPV‐associated tumorigenesis. Because only some, not all, bacterial species studied showed a high furin‐like activity, identification of specific furin‐positive bacteria and/or bacteria‐released furin‐like peptidases may provide novel drug targets for blocking viral infection and subsequent carcinogenesis.
About 18% of the global cancer burden has been attributed to infectious agents, including hepatitis B and C viruses, HPV, EBV, and Helicobacter pylori (Vedham, Divi, Starks, & Verma, 2014). However, most infected individuals do not develop cancer. Pathogenic infections are necessary but are not sufficient for cancer initiation. Additional cofactors, including secondary infections by different microbial agents, may be required. Our study, for the first time, has revealed the possibility that bacterial furin‐like peptidases might promote viral infection. Further studies will be needed to characterize such bacterium–virus interaction in vivo.
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGMENTS
This study was supported, in part, by the National Institutes of Health grant R01‐AI091779 and the Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases Award. RW was partially supported by the American Heart Association Predoctoral Fellowship 15PRE22710027.
Pavlova SI, Wilkening RV, Federle MJ, Lu Y, Schwartz J, Tao L. Streptococcus endopeptidases promote HPV infection in vitro. MicrobiologyOpen. 2019;8:e628 10.1002/mbo3.628
REFERENCES
- Agarwal, V. , Kuchipudi, A. , Fulde, M. , Riesbeck, K. , Bergmann, S. , & Blom, A. M . (2013). Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen‐ and fibronectin‐binding protein, facilitating evasion of innate immunity and invasion of host cells. Journal of Biological Chemistry, 288, 6849–6863. 10.1074/jbc.M112.405530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal, V. , Sroka, M. , Fulde, M. , Bergmann, S. , Riesbeck, K. , & Blom, A. M. (2014). Binding of Streptococcus pneumoniae endopeptidase O (PepO) to complement component C1q modulates the complement attack and promotes host cell adherence. Journal of Biological Chemistry, 289, 15833–15844. 10.1074/jbc.M113.530212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves, L. A. , Harth‐Chu, E. N. , Palma, T. H. , Stipp, R. N. , Mariano, F. S. , Höfling, J. F. , … Mattos‐Graner, R. O . (2017). The two‐component system VicRK regulates functions associated with Streptococcus mutans resistance to complement immunity. Molecular Oral Microbiology, 32, 419–431. 10.1111/omi.12183 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansai, T. , Yu, W. , Urnowey, S. , Barik, S. , & Takehara, T. (2003). Construction of a pepO gene‐deficient mutant of Porphyromonas gingivalis: Potential role of endopeptidase O in the invasion of host cells. Oral Microbiology and Immunology, 18, 398–400. 10.1046/j.0902-0055.2003.00080.x [DOI] [PubMed] [Google Scholar]
- Bensing, B. A. , & Sullam, P. M. (2009). Characterization of Streptococcus gordonii SecA2 as a paralogue of SecA. Journal of Bacteriology, 191, 3482–3491. 10.1128/JB.00365-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergé, M. , Langen, H. , Claverys, J. P. , & Martin, B. (2002). Identification of a protein that inactivates the competence‐stimulating peptide of Streptococcus pneumoniae . Journal of Bacteriology, 184, 610–613. 10.1128/JB.184.2.610-613.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, S. , Cao, L. , Kim, A. , & Biswas, I. (2015). SepM, a streptococcal protease involved in quorum sensing, displays strict substrate specificity. Journal of Bacteriology, 198, 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher‐Friebertshäuser, E. , Klenk, H. D. , & Garten, W. (2013). Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathogens and Disease, 69, 87–100. 10.1111/2049-632X.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, C. B. , & Thompson, C. D . (2007). Production of papillomavirus‐based gene transfer vectors. Current Protocols in Cell Biology 26, Unit 26.1. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) (2012). Human papillomavirus–associated cancers—United States, 2004–2008. MMWR, 61, 258–261. [PubMed] [Google Scholar]
- Chaturvedi, A. K. , Engels, E. A. , Pfeiffer, R. M. , Hernandez, B. Y. , Xiao, W. , Kim, E. , … Liu, L. (2011). Human papillomavirus and rising oropharyngeal cancer incidence in the United States. Journal of Clinical Oncology, 29, 4294–4301. 10.1200/JCO.2011.36.4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, L. , Biryukov, J. , Conway, M. J. , & Meyers, C. (2015). Cleavage of the HPV16 minor capsid protein L2 during virion morphogenesis ablates the requirement for cellular furin during De Novo infection. Viruses, 7, 5813–5830. 10.3390/v7112910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado, M. P. , & Boyle, P. (2013). Epidemiology of head and neck squamous cell carcinoma not related to tobacco or alcohol. Current Opinion in Oncology, 25, 229–234. [DOI] [PubMed] [Google Scholar]
- De Vuyst, H. , Alemany, L. , Lacey, C. , Chibwesha, C. J. , Sahasrabuddhe, V. , Banura, C. , … Parham, G. P. (2013) The burden of human papillomavirus infections and related diseases in sub‐Saharan Africa. Vaccine, 31(Suppl 5), F32–F46. 10.1016/j.vaccine.2012.07.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle, J. M. , & Webster‐Cyriaque, J. (2014). Polymicrobial infection and bacterium‐mediated epigenetic modification of DNA tumor viruses contribute to pathogenesis. MBio, 5, e01015–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay, J. , Pisani, P. , & Parkin, D. M . (2004). GLOBOCAN 2002. Cancer incidence, mortality and prevalence worldwide. IARC Cancer Base (2002 estimates). Lyon: IARC Press.
- Frandsen, E. V. , Pedrazzoli, V. , & Kilian, M. (1991). Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiology and Immunology, 6, 129–133. 10.1111/j.1399-302X.1991.tb00466.x [DOI] [PubMed] [Google Scholar]
- Guerrero‐Preston, R. , Godoy‐Vitorino, F. , Jedlicka, A. , Rodríguez‐Hilario, A. , González, H. , Bondy, J. , … Thangavel, R. . (2016). 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget, 7, 51320–51334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung, I. , Spielman, I. , Davies, M. R. , Lala, R. , Gaustad, P. , Biais, N. , & Pelicic, V. (2016). Functional analysis of an unusual type IV pilus in the Gram‐positive Streptococcus sanguinis . Molecular Microbiology, 99, 380–392. 10.1111/mmi.13237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology, 166, 557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- Henderson, B. (2014). An overview of protein moonlighting in bacterial infection. Biochemical Society Transactions, 42, 1720–1727. 10.1042/BST20140236 [DOI] [PubMed] [Google Scholar]
- Honda‐Ogawa, M. , Sumitomo, T. , Mori, Y. , Hamd, D. T. , Ogawa, T. , Yamaguchi, M. , … Kawabata, S. (2017). Streptococcus pyogenes endopeptidase O contributes to evasion from complement‐mediated bacteriolysis via binding to human complement factor C1q. Journal of Biological Chemistry, 292, 4244–4254. 10.1074/jbc.M116.749275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, M. S. , & Biswas, I. (2012). An extracelluar protease, SepM, generates functional competence‐stimulating peptide in Streptococcus mutans UA159. Journal of Bacteriology, 194, 5886–5896. 10.1128/JB.01381-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012). A framework for human microbiome research. Nature, 486, 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, S. , Yano, T. , Ebihara, A. , Okamoto, A. , Manzoku, M. , & Hayashi, H . (2010). Crystal structure of the peptidase domain of Streptococcus ComA, a bifunctional ATP‐binding cassette transporter involved in the quorum‐sensing pathway. Journal of Biological Chemistry, 285, 10777–10785. 10.1074/jbc.M109.093781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaaks, P. , & Bernasconi, M. (2017). The proprotein convertase furin in tumour progression. International Journal of Cancer, 141, 654–663. 10.1002/ijc.30714 [DOI] [PubMed] [Google Scholar]
- Jean, F. , Thomas, L. , Molloy, S. S. , Liu, G. , Jarvis, M. A. , Nelson, J. A. , & Thomas, G. (2000). A protein‐based therapeutic for human cytomegalovirus infection. Proceedings of the National Academy of Sciences of the United States of America, 97, 2864–2869. 10.1073/pnas.050504297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kines, R. C. , Thompson, C. D. , Lowy, D. R. , Schiller, J. T. , & Day, P. M. (2009). The initial steps leading to papillomavirus infection occur on the basement membrane prior to cell surface binding. Proceedings of the National Academy of Sciences of the United States of America, 106, 20458–20463. 10.1073/pnas.0908502106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk, H. D. , & Garten, W. (1994). Host cell proteases controlling virus pathogenicity. Trends in Microbiology, 2, 39–43. 10.1016/0966-842X(94)90123-6 [DOI] [PubMed] [Google Scholar]
- Kojic, M. , Lozo, J. , Jovcic, B. , Strahinic, I. , Fira, D. , & Topisirovic, L. (2010). Construction of a new shuttle vector and its use for cloning and expression of two plasmid‐encoded bacteriocins from Lactobacillus paracasei subsp. paracasei BGSJ2‐8. International Journal of Food Microbiology, 140, 117–124. 10.1016/j.ijfoodmicro.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Kolenbrander, P. E. (2000). Oral microbial communities: Biofilms, interactions, and genetic systems. Annual Review of Microbiology, 54, 413–437. 10.1146/annurev.micro.54.1.413 [DOI] [PubMed] [Google Scholar]
- Laurenceau, R. , Péhau‐Arnaudet, G. , Baconnais, S. , Gault, J. , Malosse, C. , Dujeancourt, A. , … Fronzes, R. (2013). A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae . PLoS Pathogens, 9, e1003473 10.1371/journal.ppat.1003473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Cicco, R. , Bassi, D. E. , Page, R. , & Klein‐Szanto, A. J. (2002). Furin expression in squamous cell carcinomas of the oral cavity and other sites evaluated by tissue microarray technology. Acta Odontol Latinoam, 15, 29–37. [PubMed] [Google Scholar]
- McLean, J. S. (2014). Advancements toward a systems level understanding of the human oral microbiome. Frontiers in Cellular and Infection Microbiology, 4, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, P. K. , Wang, H. , Retuerto, M. , Zhang, H. , Burkey, B. , Ghannoum, M. A. , & Eng, C. (2017). Bacteriome and mycobiome associations in oral tongue cancer. Oncotarget, 8, 97273–97289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, K. (1997). Furin: A mammalian subtilisin/Kex2p‐like endoprotease involved in processing of a wide variety of precursor proteins. Biochemical Journal, 327, 625–635. 10.1042/bj3270625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetjen, J. , Fives‐Taylor, P. , & Froeliger, E. (2001). Characterization of a streptococcal endopeptidase with homology to human endothelin‐converting enzyme. Infection and Immunity, 69, 58–64. 10.1128/IAI.69.1.58-64.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova, S. I. , Jin, L. , Gasparovich, S. R. , & Tao, L. (2013). Multiple alcohol dehydrogenases but no functional acetaldehyde dehydrogenase causing excessive acetaldehyde production from ethanol by oral streptococci. Microbiology, 159, 1437–1446. 10.1099/mic.0.066258-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff, A. B. , Woodham, A. W. , Raff, L. M. , Skeate, J. G. , Yan, L. , Da Silva, D. M. , … Kast, W. M. (2013). The evolving field of human papillomavirus receptor research: A review of binding and entry. Journal of Virology, 87, 6062–6072. 10.1128/JVI.00330-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranhand, J. M. (1974). Simple, inexpensive procedure for the disruption of bacteria. Applied Microbiology, 28, 66–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, R. M. , Lowy, D. R. , Schiller, J. T. , & Day, P. M . (2006). Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proceedings of the National Academy of Sciences of the United States of America, 103, 1522–1527. 10.1073/pnas.0508815103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E. F. , & Maniatis, T. (1989). Molecular cloning: A laboratory manual. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sarrazin, S. , Lamanna, W. C. , & Esko, J. D . (2011) Heparan sulfate proteoglycans. Cold Spring Harbor Perspectives in Biology, 3, pii: a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller, J. T. , Day, P. M. , & Kines, R. C. (2010). Current understanding of the mechanism of HPV infection. Gynecologic Oncology, 118, S12–S17. 10.1016/j.ygyno.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, B. L. , Kuczynski, J. , Bhattacharya, A. , Huey, B. , Corby, P. M. , Queiroz, E. L. , … Olshen, A. B. (2014). Changes in abundance of oral microbiota associated with oral cancer. PLoS ONE, 9, e98741 10.1371/journal.pone.0098741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, J. , Pavlova, S. , Kolokythas, A. , Lugakingira, M. , Tao, L. , & Miloro, M. (2012). Streptococci‐human papilloma virus interaction with ethanol exposure leads to keratinocyte damage. Journal of Oral and Maxillofacial Surgery, 70, 1867–1879. 10.1016/j.joms.2011.08.005 [DOI] [PubMed] [Google Scholar]
- Shiboski, C. H. , Schmidt, B. L. , & Jordan, R. C. (2005). Tongue and tonsil carcinoma: Increasing trends in the U.S. population ages 20‐44 years. Cancer, 103, 1843–1849. 10.1002/(ISSN)1097-0142 [DOI] [PubMed] [Google Scholar]
- Simon, D. , & Ferretti, J. J. (1991). Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiology Letters, 66, 219–224. 10.1111/j.1574-6968.1991.tb04868.x [DOI] [PubMed] [Google Scholar]
- Sorem, J. , & Longnecker, R. (2009). Cleavage of Epstein‐Barr virus glycoprotein B is required for full function in cell‐cell fusion with both epithelial and B cells. Journal of General Virology, 90(Pt 3), 591–595. 10.1099/vir.0.007237-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straume, D. , Stamsås, G. A. , & Håvarstein, L. S. (2015). Natural transformation and genome evolution in Streptococcus pneumoniae. Infection, Genetics and Evolution, 33, 371–380. 10.1016/j.meegid.2014.10.020 [DOI] [PubMed] [Google Scholar]
- Sun, X. , Salih, E. , Oppenheim, F. G. , & Helmerhorst, E. J. (2009). Activity‐based mass spectrometric characterization of proteases and inhibitors in human saliva. Proteomics Clinical Applications, 3, 810–820. 10.1002/prca.200800242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, L. (1998). Streptococcal integration vectors for gene inactivation and cloning. Methods in Cell Science, 20, 59–64. 10.1023/A:1009756819641 [DOI] [Google Scholar]
- Tao, L. , Pavlova, S. I. , Gasparovich, S. R. , Jin, L. , & Schwartz, J. (2015). Alcohol metabolism by oral streptococci and interaction with human papillomavirus leads to malignant transformation of oral keratinocytes. Advances in Experimental Medicine and Biology, 815, 239–264. 10.1007/978-3-319-09614-8 [DOI] [PubMed] [Google Scholar]
- Thomas, G. (2002). Furin at the cutting edge. From protein traffic to embryogenesis and disease. Nature Reviews Molecular Cell Biology, 3, 753–766. 10.1038/nrm934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedham, V. , Divi, R. L. , Starks, V. L. , & Verma, M. (2014). Multiple infections and cancer: Implications in epidemiology. Technology in Cancer Research & Treatment, 13, 177–194. 10.7785/tcrt.2012.500366 [DOI] [PubMed] [Google Scholar]
- Vickerman, M. M. , Iobst, S. , Jesionowski, A. M. , & Gill, S. R. (2007). Genome‐wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. Journal of Bacteriology, 189, 7799–7807. 10.1128/JB.01023-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanyiri, J. W. , O'Connor, R. , Allison, G. , Kim, K. , Kane, A. , Qiu, J. , … Ward, H. D (2007). Proteolytic processing of the Cryptosporidium glycoprotein gp40/15 by human furin and by a parasite‐derived furin‐like protease activity. Infection and Immunity, 75, 184–192. 10.1128/IAI.00944-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, L. , Piotrowski, A. , & Morrison, D. A. (2013). Exit from competence for genetic transformation in Streptococcus pneumoniae is regulated at multiple levels. PLoS ONE, 8, e64197 10.1371/journal.pone.0064197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkening, R. V. , Chang, J. C. , & Federle, M. J. (2016). PepO, a CovRS‐controlled endopeptidase, disrupts Streptococcus pyogenes quorum sensing. Molecular Microbiology, 99, 71–87. 10.1111/mmi.13216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch‐Perron, C. , Vieira, J. , & Messing, J. (1985). Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene, 33, 103–119. 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]


