Abstract
Chlamydospores are specific structures that are of great significance to the commercialization of fungal biopesticides. To explore the genes associated with chlamydospore formation, a biocontrol fungus Clonostachys rosea 67‐1 that is capable of producing resistant spores under particular conditions was investigated by transcriptome sequencing and analysis. A total of 549,661,174 clean reads were obtained, and a series of differentially expressed genes potentially involved in fungal chlamydospore formation were identified. At 36 hr, 67 and 117 genes were up‐ and downregulated in C. rosea during chlamydospore production, compared with the control for conidiation, and 53 and 24 genes were up‐ and downregulated at 72 hr. GO classification suggested that the differentially expressed genes were related to cellular component, biological process, and molecular function categories. A total of 188 metabolism pathways were linked to chlamydospore production by KEGG analysis. Sixteen differentially expressed genes were verified by reverse transcription quantitative PCR, and the expression profiles were consistent with the transcriptome data. To the best of our knowledge, it is the first report on the genes associated with chlamydospore formation in C. rosea. The results provide insight into the molecular mechanisms underlying C. rosea sporulation, which will assist the development of fungal biocontrol agents.
Keywords: chlamydospore, Clonostachys rosea, differentially expressed gene, reverse transcription quantitative PCR, transcriptome
1. INTRODUCTION
Filamentous fungi can produce two types of spores in their anamorphic stages; conidia and chlamydospores. Chlamydospores are specific structures with thick cell walls that are derived from the cytoplasm concentration of fungal hyphae or the conversion of conidia (Gow, 2002; Kang, Kim, Kim, Lee, & Lee, 2016; Martin, Douglas, & Konopka, 2005). Chlamydospores are usually produced in harsh environments such as low temperatures, unfavorable pH, and nutrient deficiency, since they are more resistant to adverse conditions (Eisman et al., 2006; Goh, Daida, & Vujanovic, 2009; Hughes, 1985). Various species of fungi can produce chlamydospores under particular conditions, including yeasts (Candida albicans and C. dubliniensis) (Citiulo, Moran, Coleman, & Sullivan, 2009; Staib & Morschhäuser, 2007), filamentous fungi (Fusarium oxysporum, Trichoderma harzianum, Metarhizium anisopliae, and Ceratocystis platani) (Baccelli et al., 2012; Bae & Knudsen, 2000; Iida, Kurata, Harimoto, & Tsuge, 2008; Li, Song, Li, & Chen, 2016; Ment et al., 2010), and macrofungi (Cryptococcus laurentii and Coprinus cinereus) (Kües, Walser, Klaus, & Aebi, 2002; Kurtzman, 1973).
As a specific propagule, chlamydospores make a distinct contribution to the life and survival of certain fungal species. Chlamydospores produced by the plant pathogenic fungi F. oxysporum and Cylindrocarpon destructans were found to survive for a long time under harsh environmental conditions, and to participate in the infection of plants as primary inocula (Couteaudier & Alabouvette, 1990; Kang et al., 2016). In human fungal pathogens such as in C. neoformans, chlamydospores generate infectious basidiospores (Lin & Heitman, 2005), and chlamydospores also act as diagnostic evidence in infection with C. albicans (Bottcher, Pollath, Staib, Hube, & Brunke, 2016; Cole, Seshan, Phaneuf, & Lynn, 1991). Chlamydospores can play a vital role in the activity of fungal biological control agents against pathogens. Chlamydospores of Duddingtonia flagrans can survive and germinate in animals, resulting in a reduction in parasitization by the larvae of nematodes and trematodes in host animals (Ojeda‐Robertos et al., 2009; Paz‐Silva et al., 2011). The resistant spores of T. harzianum and Clonostachys rosea have also been associated with the biocontrol of numerous plant fungal pathogens (Li, Qu, Tian, & Zhang, 2005). In some fungal species such as Candida and Fusarium, chlamydospores can be used as a taxonomic criterion (Li et al., 2012; Sancak, Colakoglu, Acikgoz, & Arikan, 2005).
Conditions for the induction of fungal chlamydospores, such as the temperature, lipopeptide inducers, and fermentation parameters, are receiving increasing attention in the field of biocontrol fungi research on species such as Trichoderma spp., C. rosea and D. flagrans (Federica, Alberto, Emilia, Carina, & Alfredo, 2013; Li et al., 2005). The genes associated with chlamydospore formation have been investigated, and genes encoding dolichol phosphate mannose synthase (Juchimiuk, Kruszewska, & Palamarczyk, 2015), hog1 mitogen‐activated protein kinase (Alonso‐Monge et al., 2003), transcription factors (Ghosh, Wangsanut, Fonzi, & Rolfes, 2015), the chromatin remodeling complex (Navarathna, Pathirana, Lionakis, Nickerson, & Roberts, 2016), and mitochondrial ATP‐dependent RNA helicase (Nobile, Bruno, Richard, Davis, & Mitchell, 2003) were linked to chlamydospore formation in C. albicans. Deletion of these genes inhibited or blocked the generation of chlamydospores. Furthermore, disruption of the sterile alpha motif domain‐encoding gene FVS1 in F. oxysporum resulted in the defective development of conidiophores, but a dramatically increased chlamydospore yield, implying negative regulation of chlamydospore formation (Iida, Fujiwara, Yoshioka, & Tsuge, 2014). However, the knowledge of the genes involved in chlamydospore formation, especially in biocontrol fungi, remains limited, and the molecular mechanisms underlying chlamydospore formation are elusive.
Clonostachys rosea (syn Gliocladium roseum) is a mycoparasite and effective biocontrol agent against various plant fungal pathogens such as Sclerotinia sclerotiorum, Rhizoctonia solani, and Botrytis cinerea (Cota, Maffia, Mizubuti, Macedo, & Antunes, 2008; Ma, Wu, Yang, & Lu, 2004; Morandi, Maffia, Mizubuti, Alfenas, & Barbosa, 2003; Rodríguez, Cabrera, Gozzo, Eberlin, & Godeas, 2011). Previous studies revealed that C. rosea produced chlamydospores under specific conditions (Li et al., 2005; Sun, Chen, Liu, Li, & Ma, 2014). Compared with conidia, chlamydospores exhibit higher stress resistance, longer shelf‐life and similar control ability (Dong, Sun, Li, Peng, & Luo, 2014). However, the genes related to chlamydospore formation in C. rosea and their regulatory mechanisms are poorly understood. Herein, we investigated the differentially expressed genes (DEGs) in this biocontrol fungal species during chlamydospore formation and conidiation by transcriptome sequencing and analysis. The results provide a platform for further understanding the molecular mechanisms underlying chlamydospore formation, and may promote the development of highly efficient and stable chlamydospore‐producing biocontrol fungi.
2. MATERIALS AND METHODS
2.1. Strain
Clonostachys rosea 67‐1 was originally obtained from a vegetable garden in Hainan Province, China (Zhang, Gao, Ma, & Li, 2004) and preserved in the Agricultural Culture Collection of China (strain number: ACCC 39160).
2.2. Sample collection
Strain 67‐1 was incubated on potato dextrose agar (PDA) plate at 26°C for 10 days for sporulation. Spores were then eluted with 10 ml sterile distilled water and inoculated in PD broth with a concentration of 1 × 107 spores·ml−1 on a rotary shaker at a speed of 180 r·min−1. After incubation at 27°C for 36 hr, the seed liquid was inoculated at a rate of 2% to produce chlamydospores in medium containing 2.5% glucose, 0.7% soybean cake powder, 0.035% urea, 0.1% K2HPO4·3H2O, 0.05% MgSO4, and 0.005% ZnSO4·7H2O in 1 L distilled water with the pH value of 6.5. The fungus was cultured at 27°C on a shaking table at a speed of 180 r·min−1, and the thallus was collected at 36 and 72 hr after inoculation by centrifuging at 8,000 g for 15 min. Samples following conidiation (control) prepared in the same culture condition, but in medium containing 0.9% glucose, 1.6% corn flour, 0.7% soybean cake powder, 0.035% urea, 0.1% K2HPO4·3H2O, 0.05% MgSO4, and 0.005% of ZnSO4·7H2O, which did not yield chlamydospores, were taken as controls. Three completely independent replicates were performed.
2.3. Construction of cDNA libraries and transcriptome sequencing
Total RNAs from each sample were extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instruction, and residual genomic DNA was removed using DNase I (TaKaRa, Dalian, China). The concentration of RNA, and OD260/280 and OD260/230 values, were determined using a micro‐spectrophotometer (SimpLiNano, Cambridge, UK), and the RNA integrity number and 28S/18S value were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
mRNAs from fungal samples were enriched using magnetic beads harboring oligo (dT) and broken into short fragments using fragmentation buffer in a Thermomixer (Eppendorf, Hamburg, Germany). Reverse transcription was conducted and cDNA fragments were synthesized after purification, end reparation, addition of adenine to the 3’ end, and adapter connection. Suitable fragments were amplified to construct cDNA libraries. After quantity and quality monitoring, 12 cDNA libraries were sequenced using the Illumina HiSeq 4000 platform (Illumina Inc., CA, USA) at the Beijing Genome Institute (BGI).
2.4. Bioinformatic analysis of the 67‐1 transcriptome during sporulation
Raw transcriptome sequence data were filtered by removing disqualified reads containing adapters, unknown bases larger than 10%, and low‐quality sequences in which the number of bases with a quality value ≤10 was larger than 50% of the reads. Clean reads were submitted to the NCBI sequence read archive (SRA), and mapped onto the C. rosea 67‐1 genome (Sun, Sun, & Li, 2015) using Burrows‐Wheeler Aligner (BWA) software (Li & Durbin, 2009). These genes were further analyzed for changes in expression levels.
2.5. Analysis of DEGs
Transcript levels of 67‐1 during generation of the two types of spores were quantified using RNASeq by Expectation Maximization (RSEM) and calculated using the fragments per kb per million fragments (FPKM) method (Audic & Claverie, 1997). The NOISeq software package was used to screen DEGs, and genes for which expression varied more than twofold and the probability divergence was higher than 0.8 during the formation of chlamydospore were considered significantly differentially expressed compared with the control (Tarazona, Garcia‐Alcalde, Dopazo, Ferrer, & Conesa, 2011). The functions of identified DEGs were then investigated by Genome Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis.
2.6. Validation of transcriptome data by reverse transcription quantitative PCR (RT‐qPCR)
A total of 16 DEGs were selected from the transcriptome of C. rosea 67‐1 for quantification under different cultural conditions using RT‐qPCR to confirm the reliability of the transcriptome data (Table 1). Primer pairs for each gene were designed using Primer Premier 6.0 software, and their specificity was verified using conventional PCR with the following program: 94°C for 3 min, 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 20 s, and a final step at 72°C for 10 min.
Table 1.
Primers used for RT‐qPCR in this study
| Transcript ID | Predicted function | Primer (5’‐3’) | Product length (bp) |
|---|---|---|---|
| Cch5729 | Catalase/peroxidase HPI | F: ATCTGGTCTGGAGGTTATCTGG | 126 |
| R: TTTGCGACGAACTGTGGG | |||
| Cch43218 | Trypsin‐like protease | F: ACTTCTGCGGTGGTGTCTT | 137 |
| R: AACGCTGACCTGAGTTCCT | |||
| Cch379 | Hypothetical protein | F: TCAGCCTTGGCGTAGAAG | 129 |
| R: GGAACTCCCTCAGTAAATCG | |||
| Cch1803 | Glucose transporter | F: AGATGCTGGCTGACAACG | 131 |
| R: GATGGGCAGAGGGAAGAT | |||
| Cch9554 | Hypothetical protein | F: ACTACCAAATCTGTGGCTCT | 198 |
| R: CAGTCGTACTCTGGCTCAA | |||
| Cch21025 | Translation initiation factor | F: TACCTCCATCTCAGTCAGTTCA | 177 |
| R: ACAGCAAAGGGACCAAGC | |||
| Cch295103 | Endochitinase‐like protein | F: ATGCGTTCCTCTATGGTTTCC | 166 |
| R: CGAACAGATCGGAGGTTGC | |||
| Cch30431 | Ketosteroid isomerase | F: CCCTCTGCACCTTGTAATT | 147 |
| R: ATCTTGCCGTTGTCGTTG | |||
| Cch30429 | Polyketide hydroxylase | F: AGATGCCCATAACCTTGC | 120 |
| R: GCCTGATAAACGGTAAACTG | |||
| Cch56332 | Glycosyltransferase family 4 protein | F: TCTTGAGAATGCCGTTTG | 188 |
| R: TGTAGAATCCCGTGTCCC | |||
| Cch45345 | Nitrosoguanidine resistance protein | F: TGGCTCTTGGGTCACATT | 102 |
| R: ATCCGTATCCTCCGCATT | |||
| Cch30433 | Aldehyde dehydrogenase | F: TTGGTCCCGTCACAAACA | 147 |
| R: AGCAGCAGCACCGAAGTA | |||
| Cch30437 | Glutathione S‐transferase | F: GCAGCATCAGAGTTCCGAATA | 150 |
| R: TCAAACGCAGGGACAAGC | |||
| Cch47820 | Dihydrodipicolinate synthase | F: GCCCGAGCCCTTGTATGA | 198 |
| R: AGGAGTGGTGAGCGGACAG | |||
| Cch30435 | Acyl esterases | F: GTCACATTGTCGCCCACT | 179 |
| R: TGAGACTCGCTGCCATCC | |||
| Cch34648 | Hypothetical protein | F: AATACACGCCTCCCAATC | 142 |
| R: GGTAATGTGCCGCTAATG |
Total RNAs extracted from thallus samples were reverse transcribed into cDNA using a cDNA FastQuant RT Kit (Tiangen, Beijing, China). The expression levels of the 16 DEGs were assayed using SYBR Premix Ex Taq II (TaKaRa, Dalian, China) with actin and succinate‐semialdehyde dehydrogenase (SSD) serving as internal reference genes (Zhang, Sun, Li, & Sun, 2017) on a IQ 5 multicolor real‐time PCR detection system (Bio‐Rad, CA, USA) in 25 μl reactions containing 1 μl of 10‐fold diluted cDNA, 1 μl of forward and reverse primer, 12.5 μl of SYBR Premix, and 9.5 μl of RNase‐free water. qRT‐PCR was performed in a 96‐well plate with the following program; 95°C for 30 s, followed by 40 cycles of 95°C 5 s and 55°C for 30 s. Fluorescence values were measured from 55°C to 95°C at 0.5°C intervals for 81 cycles to check for non‐specific amplification. The 2−ΔΔCt method was used to calculate relative expression levels for all DEGs (Livak, & Schmittgen, 2001). Three replicates were performed for each reaction.
2.7. Statistical analysis
The expression levels of DEGs were analyzed using SAS 9.1.3 statistical software (SAS Institute Inc., Cary, NC, USA). t tests were used for comparisons of the means of each treatment and p‐values lower than 0.01 were regarded as significant.
3. RESULTS
3.1. Sporulation of C. rosea 67‐1
In the appropriate medium, a large amount of chlamydospores were detected after incubation for 3 days, and the generation of conidia was inhibited. Using microscopy (Olympus BX61, Tokyo, Japan), we could see that chlamydospores were derived from mycelia, either in the middle or at the tip, and most of the mycelia were transformed into chlamydospores by the end of the incubation. While in the medium for conidiation that was used as the control for transcriptome analysis of sporulation patterns in C. rosea 67‐1, chlamydospores were not detected (Figure 1). Compared with conidia, the resistant spores were ellipsoidal to spheroidal, 5.8 ± 0.4 × 5.0 ± 0.4 μm in size, compared to conidia (2.9 ± 0.3 × 1.6 ± 0.1 μm).
Figure 1.
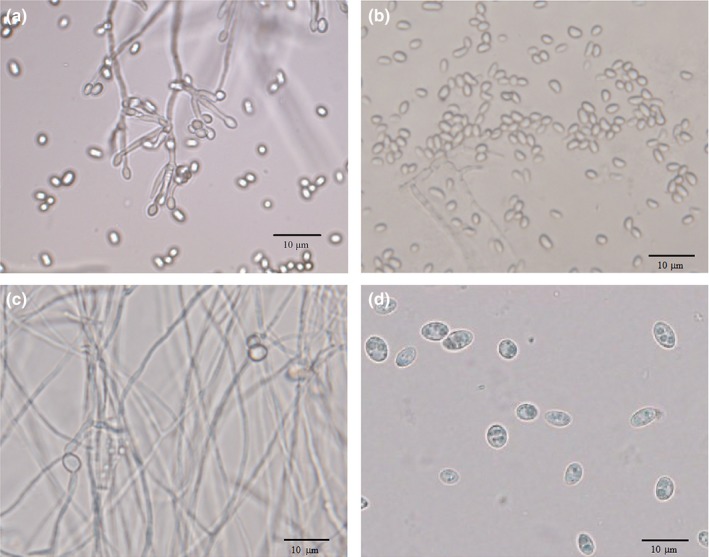
Microstructures of two types of spores produced by C. rosea 67‐1 growing in different media. (a) Conidia and hyphae in 36 hr. (b) Conidia in 72 hr. (c) Hyphae and newly formed chlamydospores in 36 hr. (d) Chlamydospores in 72 hr. Photos were taken at 400 × magnification. Scale bars = 10 μm
3.2. Bioinformatic analysis of the transcriptome
The RNA Q20 and Q30 values were above 96.1% and 90.6%, respectively, suggesting the quality and quantity of the samples met the requirements for transcriptome sequencing. All clean data were uploaded to the NCBI SRA repository with the accession numbers SRR4017358, 4017394−4017397, 4017399−4017403, 4017405, and 4017407. The average number of clean reads of the samples was 45,805,098, and each sample was mapped to the genome of C. rosea 67‐1 (Sun et al., 2015) with a map rate above 92.42%. The average number of expressed genes during all sampling time points was 14,755 (Table 2). Among them, unknown and metabolism‐related genes were the most highly expressed.
Table 2.
Analysis of the transcriptome of C. rosea 67‐1 during sporulation
| Samplea | Number of clean reads | Genome map rate (%) | Number of expressed genes |
|---|---|---|---|
| A36‐1 | 45,977,522 | 93.26 | 14,439 |
| A36‐2 | 46,070,230 | 93.28 | 15,014 |
| A36‐3 | 45,683,824 | 92.80 | 15,445 |
| A72‐1 | 45,840,164 | 93.22 | 14,491 |
| A72‐2 | 45,812,684 | 92.90 | 14,574 |
| A72‐3 | 45,674,486 | 92.66 | 15,059 |
| B36‐1 | 46,021,872 | 92.98 | 14,279 |
| B36‐2 | 45,767,546 | 92.67 | 14,576 |
| B36‐3 | 45,764,932 | 92.42 | 14,744 |
| B72‐1 | 45,711,280 | 92.92 | 15,039 |
| B72‐2 | 45,645,686 | 92.67 | 14,514 |
| B72‐3 | 45,690,948 | 92.75 | 14,889 |
Letters “A” and “B” represent the processes of conidiation (control) and chlamydospore generation in C. rosea 67‐1, respectively. Numbers after letters indicate the sampling time (hours), and the last numbers in sample lines refer to three completely independent replicates.
3.3. Analysis of DEGs
At 36 hr, a small amount of chlamydospores were detected in the medium. Analysis of the transcriptome showed that most of genes were not differentially expressed relative to the control during chlamydospore formation, however, a total of 184 genes were differentially expressed, among which 67 were upregulated, and 117 were downregulated (Figure 2). Non‐redundant (NR) annotation indicated that 31 of the upregulated DEGs were hypothetical or predicted genes, 11 were related to metabolic processes, seven were involved in transfer functions, seven were linked to hydrolysis, and others without any annotation information were regarded as new genes. For the down‐regulated DEGs, hypothetical genes were dominant (48%), followed by new genes without known functions (24%), genes encoding metabolic process‐related enzymes (12%), genes related to transfer functions (10%), and genes linked to hydrolysis (6%). Among them, MFS transporter, glucose transporter, cytochrome P450 protein, dihydrodipicolinate synthase and salicylate hydroxylase protein had a large variation.
Figure 2.
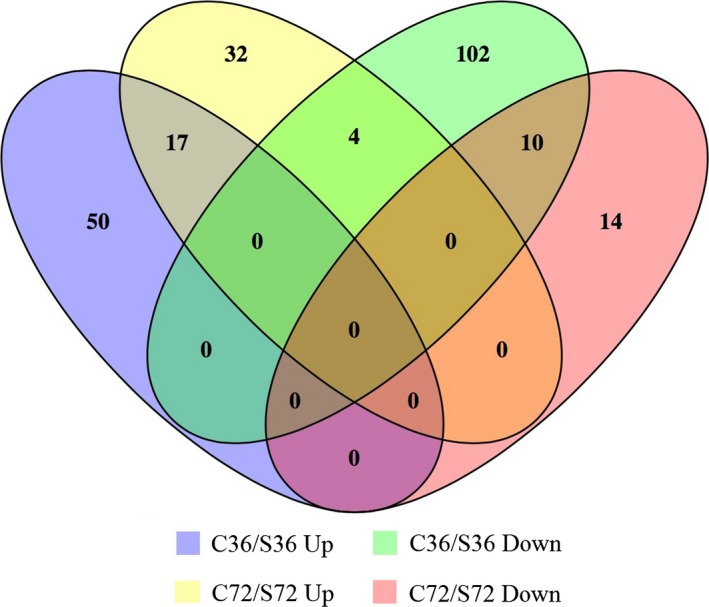
Analysis of DEGs during chlamydospore formation in C. rosea 67‐1. Letters “C” and “S” represent conidiation (control) and chlamydospore generation in C. rosea 67‐1, respectively. Numbers after letters indicate the sampling time (hrs). Up and Down refer to genes that are up‐ and downregulated during chlamydospore formation, respectively
At 72 hr, the number of DEGs related to chlamydospore formation was lower than observed at 36 hr; 53 genes were upregulated, while only 24 were downregulated (Table S1). Among these, hypothetical genes accounted for the biggest proportion, of which 30 were upregulated and 17 were downregulated. Genes related to metabolic processes were also well represented, of which 10 were upregulated and three were downregulated. Six hydrolases encoding genes were found upregulated. At this time point, no new genes were detected.
GO functional classification of DEGs during chlamydospore formation in 67‐1 was performed, and 19 functional terms in three categories were identified at 36 hr. Metabolic process, membrane and catalytic activity were the most significant in the biological process, cellular component, and molecular function categories (Figure 3a). The dominant terms in these three categories were unaltered at 72 hr, however, biological regulation and regulation of biological process terms in the biological process category that were present during the initial stage of chlamydospore formation were not evident at this later timepoint (Figure 3b), suggesting that some signals regulating chlamydospore generation might be initiated at the early stage.
Figure 3.
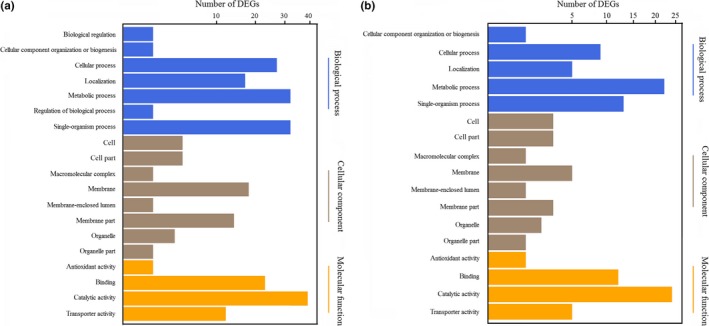
Gene ontology (GO) functional classification of differentially expressed genes during chlamydospore formation in C. rosea 67‐1. (a) Gene functions were divided into three categories (biological process, cellular component, and molecular function) to analyze DEGs during the early stage (36 hr) of chlamydospore generation and conidiation, during which the categories of biological regulation and regulation of biological process are specific. (b) Functional analysis of DEGs during the spore mass production period (72 hr)
A total of 188 metabolism pathways potentially involved in chlamydospore production in 67‐1 were identified using KEGG analysis. At 36 hr, 148 pathways belonging to 36 terms were found, while at 72 hr, the number of pathways was reduced to 120, covering 31 terms. Although differences existed at different timepoints, the main pathways were consistent throughout the production of chlamydospores. The term of global and overview maps in the metabolism category was dominant. Pathways involved in environmental information processing and genetic information processing were the next most abundant during the initial and stable phases of sporulation, respectively (Figure 4). The pathways associated with signaling molecules and interactions appeared at 36 hr but not at 72 hr, which also indicated that signal regulation‐related genes were essential to form chlamydospores. Most of the pathways were linked to metabolic pathways; biosynthesis of secondary metabolites and microbial metabolism in diverse environments were also detected frequently.
Figure 4.
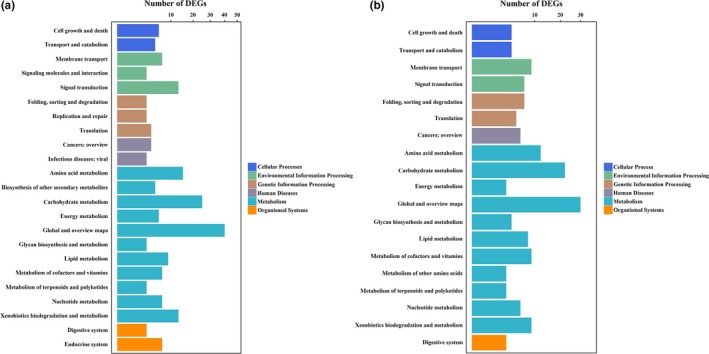
KEGG pathway analysis of DEGs during chlamydospore formation in C. rosea 67‐1. (a) Pathways were divided into six categories to analyze DEGs during the early stage (36 hr) of chlamydospore generation and conidiation. (b) Pathway analysis during the spore mass production period (72 hr). Different colours represent different categories of KEGG pathways; DEGs with numbers lower than 3 are not shown
3.4. Validation of transcriptome data
Based on the results of RT‐qPCR, the expression levels of the 16 selected DEGs were divided into three groups (Figure 5). Group 1 included a large number of DEGs encoding polyketide hydroxylase, ketosteroid isomerase, glutathione S‐transferase, aldehyde dehydrogenase, nitrosoguanidine resistance protein, glycosyltransferase family 4 protein, acyl esterases, endochitinase‐like protein, trypsin‐like protease, and a hypothetical protein, the expression of which was continuously upregulated during the production of chlamydospores. In group 2, DEGs encoding a hypothetical protein, dihydrodipicolinate synthase, translation initiation factor, and catalase/peroxidase HPI were continuously downregulated. A glucose transporter‐encoding gene was placed in a third group, and was downregulated initially, but upregulated during the later stages. The expression levels of these genes were consistent with the results of transcriptome sequencing, indicating that the DEGs derived from the transcriptome during chlamydospore formation were accurate and reliable, and hence suitable for further investigation of the genes associated with chlamydospore formation in C. rosea 67‐1.
Figure 5.
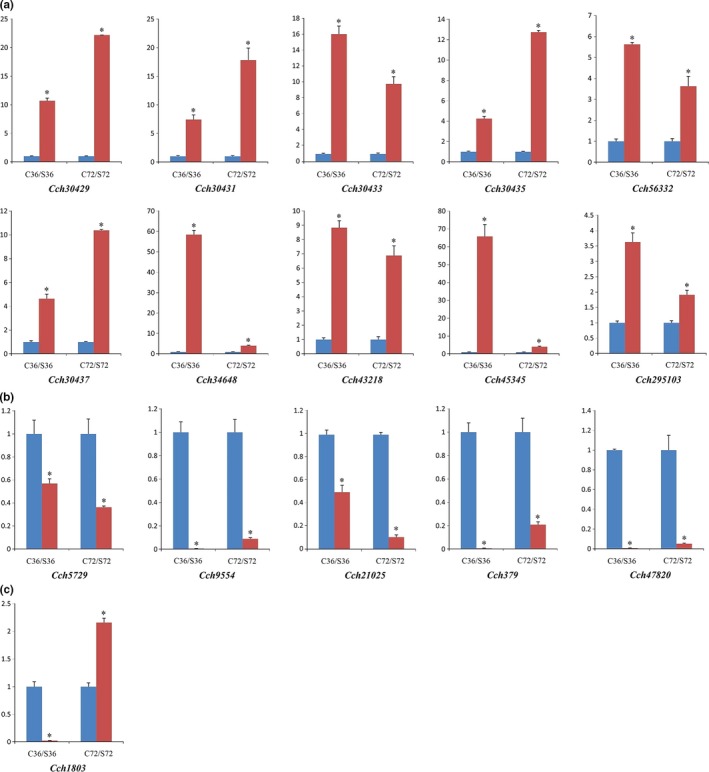
RT‐qPCR detection of 16 selected DEGs during chlamydospore formation in C. rosea 67‐1. (a) DEGs whose expression levels are continuously upregulated. (b) DEGs whose expression levels are continuously downregulated. (c) DEGs whose expression levels are downregulated initially, but upregulated during the later period. “C” represents controls (conidiation) and “S” represents chlamydospore generation. Numbers after letters indicate the sampling time (hours). Vertical axis represents the relative expression levels of DEGs in both sporulation processes. Error bars represent the standard deviation of three independent replicates for each tested gene, and stars on the columns represent significantly different (p < .01) according to the t test
4. DISCUSSION
Chlamydospores are propagules in various fungal species that are of great significance to fungal growth and survival. Chlamydospores are highly resistant to adverse environmental conditions, and display excellent biocontrol activity for fungal biopesticides (Braga, Silva, Carvalho, Araújo, & Pinto, 2011; Maciel, Freitas, Campos, Lopes, & Araújo, 2010; Sanyal, Sarkar, Patel, Mandal, & Pal, 2008). To understand the molecular mechanisms underlying chlamydospore formation and regulation with the aim of developing biocontrol chlamydospore agents, we investigated the transcriptome of C. rosea 67‐1 during chlamydospore formation and compared it with the transcriptome during conidiation. Genes potentially related to the formation and development of resistant spores were subsequently identified.
In this study, chlamydospores and conidia of C. rosea 67‐1 were cultured in different media. In the chlamydospore‐forming medium, conidiation was greatly inhibited, while in the control medium, chlamydospores were not detected during the sampling time, and initiation of conidiation was accompanied with the development of initial hyphae during the very beginning of incubation (less than 20 hr, data not shown), which suggested that the different fungal structures could not be completely separated. Unlike in previous research on gene expression under different stress conditions and developmental stages in which conidia and mycelia were collected separately (Chen, Xie, Ye, Jensen, & Eilenberg, 2016; Llanos, François, & Parrou, 2015; Xu et al., 2015), we aimed to investigate DEGs in the biocontrol fungus C. rosea during chlamydospore formation by comparing with the typical developmental processes that generate conidia from mycelia. Therefore, it was not necessary to distinguish between spores and hyphae during cultivation in each condition. We harvested the thalli of 67‐1 in both media to construct C. rosea sporulation transcriptome therefrom.
In the transcriptome of 67‐1 during sporulation, we selected two sampling timepoints (36 hr and 72 hr). At the first sampling timepoint, a few chlamydospores were found in the medium, indicating that sporulation had switched from conidiation to chlamydospore formation. Therefore, we considered 36 hr as the initial stage of chlamydospore formation. During this stage, 67 and 117 genes were up‐ and downregulated, respectively, suggesting genes related to chlamydospore formation were activated, while those involved in conidiation were inhibited. At 72 hr, a much larger amount of chlamydospores were detected, and this timepoint was therefore considered to represent chlamydospore mass production, during which 53 genes were upregulated and 24 were downregulated. Compared to the early stage, downregulated genes were decreased more sharply. We deduced that this might be due to the marked reduction in genes expressed during conidiation (Figure 2), but this hypothesis requires further experimental verification.
Through transcriptome analysis, we identified a series of DEGs (184 at 36 hr and 77 at 72 hr) that were potentially involved in chlamydospore formation in C. rosea. Some of the identified DEGs encoded transporters and had a transfer function, as reported previously in C. albicans. Franke et al. (2006) found that deletion of the putative vesicle transport protein‐encoding gene vac1p affected the formation of chlamydospores. Additionally, the putative transporter gene QDR1/CD36_29520 was confirmed to be distinctly downregulated during chlamydospore development (Palige et al., 2013). Therefore, we deduced that these genes might play roles in transporting substances needed for fungal sporulation. To date, transcription factors have attracted the most attention among the relatively few reported factors regulating chlamydospore formation. A series of genes encoding transcription factors were isolated including MAPK pathway transcription factor cph1, yeast‐hyphal transition transcription factor efg1, homeobox transcription factor grf10 and GATA transcription factor Gln3, all of which were reported as key factors in chlamydospore formation and development in C. albicans (Bottcher et al., 2016; Ghosh et al., 2015; Maiti et al., 2015; Sonneborn, Bockmühl, & Ernst, 1999). In our present study on C. rosea, we also identified genes encoding transcription factors that were differentially expressed, indicating an involvement in the regulation of transcription during chlamydospore formation and development.
Among the DEGs, we also identified a number of cell wall degrading enzymes including two chitinases and a serine protease that had been previously reported in C. rosea. The expression of chitinase encoding gene cr‐ech42 and chiB2 in C. rosea IK726 was found to be triggered by chitin, and cr‐ech42 was also induced by F. culmorum cell walls (Mamarabadi, Jensen, & Lübeck, 2008; Tzelepis, Dubey, Jensen, & Karlsson, 2015). A serine protease gene prC from C. rosea was found upregulated under some environmental stresses, for example, oxidant and heat shock, and its expression also highly increased when encountering nematode cuticle (Zou, Tao, et al., 2010; Zou, Xu, et al., 2010) . We speculate that these hydrolases might take part in the dissolve of C. rosea hyphae when the resistant spores release. Glycosyltransferase is an important kind of transferase and has been proved to be involved in cell wall remodeling in Neurospora crassa (Martínez‐Núñez & Riquelme, 2015). In our study, a glycosyltransferase encoding gene was also identified during the formation of chlamydospores. We deduced that this gene might involve in chlamydospore cell wall formation. Some genes, for example, aldehyde dehydrogenase, glucose transporter and polyketide hydroxylase that have not been reported to link with chlamydospore formation in other species were also detected, and their specific functions need further explored. A number of hypothetical genes were significantly differentially expressed as well, implying that they might be linked with interesting functions during sporulation. We determined 16 DEGs related to metabolic process, cell wall degrading enzyme and transporter using RT‐qPCR and found that their expression sharply changed in the specific condition, indicating that these genes might involve in the formation of chlamydospores in C. rosea. However, the functions of these DEGs in C. rosea during chlamydospore formation need to be further verified by gene knockout and complementation and biological assay, and the pathways involved need to be explored.
Clonostachys rosea is a promising biocontrol fungus active against various fungal plant pathogens, and agents from C. rosea, with the components of conidia and hyphae, have been registered in Europe and North America in recent decades (Gwynn, 2014). However, problems such as instability of control activity and short shelf‐life have proved a bottleneck for the industrial production and field application of C. rosea and other biocontrol fungi. The development of chlamydospore agents can provide effective fungal biopesticides of more consistent quality that are better suited to large‐scale applications owing to higher resistance, more stable control efficiency and longer shelf‐life. Investigation of the genes associated with chlamydospore production and determination of the molecular mechanisms underlying chlamydospore formation and regulation will greatly facilitate the development and application of C. rosea.
5. CONCLUSION
In this study, the transcriptomes of C. rosea during chlamydospore formation and conidiation were sequenced and analyzed, and a series of differentially expressed genes potentially involved in chlamydospore formation were identified, which were related to cellular component, biological process, and molecular function. A total of 188 metabolism pathways were suggested to be linked to chlamydospore production. To the best of our knowledge, this is the first report on genes related to chlamydospore formation in C. rosea. This study provides insight into the molecular mechanisms underlying C. rosea sporulation, and will facilitate the development of fungal biocontrol agents.
CONFLICT OF INTEREST
None declared.
Supporting information
ACKNOWLEDGMENTS
We thank Professor Enrique Monte in University of Salamanca, Spain, for advising on paper writing. This work was supported by the National Natural Science Foundation of China (grant number 31471815), the Program of Modern Agricultural Industry Technology System (grant number CARS‐25‐B‐02), and the Special Project of Ministry of Agriculture (grant number 201503112‐2).
Sun Z‐B, Zhang J, Sun M‐H, Li S‐D. Identification of genes related to chlamydospore formation in Clonostachys rosea 67‐1. MicrobiologyOpen. 2019;8:e624 10.1002/mbo3.624
Funding information
This work was supported by the National Natural Science Foundation of China (grant number 31471815), the Program of Modern Agricultural Industry Technology System (grant number CARS‐25‐B‐02), and the Special Project of Ministry of Agriculture (grant number 201503112‐2).
Contributor Information
Man‐Hong Sun, Email: sunmanhong2013@163.com.
Shi‐Dong Li, Email: sdli@ippcaas.cn.
REFERENCES
- Alonso‐Monge, R. , Navarro‐García, F. , Román, E. , Negredo, A. I. , Eisman, B. , Nombela, C. , & Pla, J. (2003). The Hog1 mitogen‐activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans . Eukaryotic Cell, 2, 351–361. 10.1128/EC.2.2.351-361.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic, S. , & Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Research, 7, 986–995. 10.1101/gr.7.10.986 [DOI] [PubMed] [Google Scholar]
- Baccelli, I. , Comparini, C. , Bettini, P. P. , Martellini, F. , Ruocco, M. , Pazzagli, L. , … Scala, A. (2012). The expression of the cerato‐platanin gene is related to hyphal growth and chlamydospores formation in Ceratocystis platani . FEMS Microbiology Letters, 327, 155–163. 10.1111/j.1574-6968.2011.02475.x [DOI] [PubMed] [Google Scholar]
- Bae, Y. S. , & Knudsen, G. R. (2000). Cotransformation of Trichoderma harzianum with beta‐glucuronidase and green fluorescent protein genes provides a useful tool for monitoring fungal growth and activity in natural soils. Applied and Environmental Microbiology, 66, 810–815. 10.1128/AEM.66.2.810-815.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher, B. , Pollath, C. , Staib, P. , Hube, B. , & Brunke, S. (2016). Candida species rewired hyphae developmental programs for chlamydospore formation. Frontiers in Microbiology, 7, 1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga, F. R. , Silva, A. R. , Carvalho, R. O. , Araújo, J. V. , & Pinto, P. S. (2011). Ovicidal activity of different concentrations of Pochonia chlamydosporia chlamydospores on Taenia taeniaeformis eggs. Journal of Helminthology, 85, 7–11. 10.1017/S0022149X10000179 [DOI] [PubMed] [Google Scholar]
- Chen, C. , Xie, T. , Ye, S. , Jensen, A. B. , & Eilenberg, J. (2016). Selection of reference genes for expression analysis in the entomophthoralean fungus Pandora neoaphidis . Brazlilan Journal of Microbiology, 47, 259–265. 10.1016/j.bjm.2015.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citiulo, F. , Moran, G. P. , Coleman, D. C. , & Sullivan, D. J. (2009). Purification and germination of Candida albicans and Candida dubliniensis chlamydospores cultured in liquid media. FEMS Yeast Research, 9, 1051–1060. 10.1111/j.1567-1364.2009.00533.x [DOI] [PubMed] [Google Scholar]
- Cole, G. T. , Seshan, K. R. , Phaneuf, M. , & Lynn, K. T. (1991). Chlamydospore‐like cells of Candida albicans in the gastrointestinal tract of infected, immunocompromised mice. Canadian Journal of Microbiology, 37, 637–646. 10.1139/m91-108 [DOI] [PubMed] [Google Scholar]
- Cota, L. V. , Maffia, L. A. , Mizubuti, E. S. G. , Macedo, P. E. F. , & Antunes, R. F. (2008). Biological control of strawberry gray mold by Clonostachys rosea under field conditions. Biological Control, 46, 515–522. 10.1016/j.biocontrol.2008.04.023 [DOI] [Google Scholar]
- Couteaudier, Y. , & Alabouvette, C. (1990). Survival and inoculum potential of conidia and chlamydospores of Fusarium oxysporum f. sp. lini in soil. Canadian Journal of Microbiology, 36, 551–556. 10.1139/m90-096 [DOI] [PubMed] [Google Scholar]
- Dong, P. P. , Sun, M. H. , Li, S. D. , Peng, Y. , & Luo, M. (2014). Biological characteristics of chlamydospores of Clonostachys rosea 67‐1. Mycosystema, 33, 1242–1252. [Google Scholar]
- Eisman, B. , Alonso‐Monge, R. , Roman, E. , Arana, D. , Nombela, C. , & Pla, J. (2006). The Cek1 and Hog1 mitogen‐activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans . Eukaryotic Cell, 5, 347–358. 10.1128/EC.5.2.347-358.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federica, S. M. , Alberto, F. L. , Emilia, I. L. , Carina, M. F. , & Alfredo, S. C. (2013). Optimization of production of chlamydospores of the nematode‐trapping fungus Duddingtonia flagrans in solid culture media. Parasitology Research, 112, 1047–1051. 10.1007/s00436-012-3231-0 [DOI] [PubMed] [Google Scholar]
- Franke, K. , Nguyen, M. , Härtl, A. , Dahse, H. M. , Vogl, G. , Würzner, R. , … Eck, R. (2006). The vesicle transport protein Vac1p is required for virulence of Candida albicans . Microbiology, 152, 3111–3121. 10.1099/mic.0.29115-0 [DOI] [PubMed] [Google Scholar]
- Ghosh, A. K. , Wangsanut, T. , Fonzi, W. A. , & Rolfes, R. J . (2015). The GRF10 homeobox gene regulates filamentous growth in the human fungal pathogen Candida albicans . FEMS Yeast Research, 15, ov093 10.1093/femsyr/fov093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, Y. K. , Daida, P. , & Vujanovic, V. (2009). Effects of abiotic factors and biocontrol agents on chlamydospore formation in Fusarium graminearum and Fusarium sporotrichioides . Biocontrol Science and Technology, 19, 151–167. 10.1080/09583150802627033 [DOI] [Google Scholar]
- Gow, N. A. R. (2002). Cell biology and cell cycle of Candida In Calderone R. A. (Ed.), Candida and candidiasis (pp. 145–158). Washington, D.C.: ASM Press. [Google Scholar]
- Gwynn, R . (2014). The manual of biocontrol agents. Hampshire: BCPC. [Google Scholar]
- Hughes, S. J. (1985). The term chlamydospore In Arai T. (Ed.), Filamentous microorganisms (pp. 1–20). Tokyo: Japan Scientific Societies Press. [Google Scholar]
- Iida, Y. , Fujiwara, K. , Yoshioka, Y. , & Tsuge, T. (2014). Mutation of FVS1, encoding a protein with a sterile alpha motif domain, affects asexual reproduction in the fungal plant pathogen Fusarium oxysporum . FEMS Microbiology Letters, 351, 104–112. 10.1111/1574-6968.12356 [DOI] [PubMed] [Google Scholar]
- Iida, Y. , Kurata, T. , Harimoto, Y. , & Tsuge, T. (2008). Nitrite reductase gene upregulated during conidiation is involved in macroconidium formation in Fusarium oxysporum . Phytopathology, 98, 1099–1106. 10.1094/PHYTO-98-10-1099 [DOI] [PubMed] [Google Scholar]
- Juchimiuk, M. , Kruszewska, J. , & Palamarczyk, G. (2015). Dolichol phosphate mannose synthase from the pathogenic yeast Candida albicans is a multimeric enzyme. Biochimica et Biophysica Acta, 1850, 2265–2275. 10.1016/j.bbagen.2015.08.012 [DOI] [PubMed] [Google Scholar]
- Kang, Y. , Kim, M. R. , Kim, K. H. , Lee, J. , & Lee, S. H. (2016). Chlamydospore induction from conidia of Cylindrocarpon destructans isolated from Ginseng in Korea. Mycobiology, 44, 63–65. 10.5941/MYCO.2016.44.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües, U. , Walser, P. J. , Klaus, M. J. , & Aebi, M. (2002). Influence of activated A and B mating‐type pathways on developmental processes in the basidiomycete Coprinus cinereus . Molecular Genetics and Genomics, 268, 262–271. [DOI] [PubMed] [Google Scholar]
- Kurtzman, C. P. (1973). Formation of hyphae and chlamydospores by Cryptococcus laurentii . Mycologia, 65, 388–395. 10.2307/3758110 [DOI] [PubMed] [Google Scholar]
- Li, H. , & Durbin, R. (2009). Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics, 25, 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Ma, M. C. , Huang, R. , Qu, Q. , Li, G. H. , Zhou, J. W. , … Luo, J. (2012). Induction of chlamydospore formation in Fusarium by cyclic lipopeptide antibiotics from Bacillus subtilis C2. Journal of Chemical Ecology, 38, 966–974. 10.1007/s10886-012-0171-1 [DOI] [PubMed] [Google Scholar]
- Li, L. , Qu, Q. , Tian, B. , & Zhang, K. Q. (2005). Induction of chlamydospores in Trichoderma harzianum and Gliocladium roseum by antifungal compounds produced by Bacillus subtilis C2. Journal of Phytopathology, 153, 686–693. 10.1111/j.1439-0434.2005.01038.x [DOI] [Google Scholar]
- Li, Y. Q. , Song, K. , Li, Y. C. , & Chen, J. (2016). Statistical culture‐based strategies to enhance chlamydospore production by Trichoderma harzianum SH2303 in liquid fermentation. Journal of Zhejiang University Science B, 17, 619–627. 10.1631/jzus.B1500226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X. , & Heitman, J. (2005). Chlamydospore formation during hyphal growth in Cryptococcus neoformans . Eukaryotic Cell, 4, 1746–1754. 10.1128/EC.4.10.1746-1754.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2−∆∆Ct method. Methods, 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Llanos, A. , François, J. M. , & Parrou, J. L. (2015). Tracking the best reference genes for RT‐qPCR data normalization in filamentous fungi. BMC Genomics, 16, 71 10.1186/s12864-015-1224-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, G. Z. , Wu, X. R. , Yang, W. L. , & Lu, G. Z. (2004). Inhibition of zymotic liquid from different isolates of Gliocladium spp. to three pathogenic fungi. Journal of Huazhong Agricultural University, 23, 96–99. [Google Scholar]
- Maciel, A. S. , Freitas, L. G. , Campos, A. K. , Lopes, E. A. , & Araújo, J. V. (2010). The biological control of Ancylostoma spp. dog infective larvae by Duddingtonia flagrans in a soil microcosm. Veterinary Parasitology, 173, 262–270. 10.1016/j.vetpar.2010.06.027 [DOI] [PubMed] [Google Scholar]
- Maiti, P. , Ghorai, P. , Ghosh, S. , Kamthan, M. , Tyagi, R. K. , & Datta, A. (2015). Mapping of functional domains and characterization of the transcription factor Cph1 that mediate morphogenesis in Candida albicans . Fungal Genetics and Biology, 83, 45–57. 10.1016/j.fgb.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Mamarabadi, M. , Jensen, B. , & Lübeck, M. (2008). Three endochitinase‐encoding genes identified in the biocontrol fungus Clonostachys rosea are differentially expressed. Current Genetics, 54, 57–70. 10.1007/s00294-008-0199-5 [DOI] [PubMed] [Google Scholar]
- Martin, S. W. , Douglas, L. M. , & Konopka, J. B. (2005). Cell cycle dynamics and quorum sensing in Candida albicans chlamydospores are distinct from budding and hyphal growth. Eukaryotic Cell, 4, 1191–1202. 10.1128/EC.4.7.1191-1202.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Núñez, L. , & Riquelme, M. (2015). Role of BGT‐1 and BGT‐2, two predicted GPI‐anchored glycoside hydrolases/glycosyltransferases, in cell wall remodeling in Neurospora crassa . Fungal Genetics and Biology, 85, 58–70. 10.1016/j.fgb.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Ment, D. , Gindin, G. , Glazer, I. , Perl, S. , Elad, D. , & Samish, M. (2010). The effect of temperature and relative humidity on the formation of Metarhizium anisopliae chlamydospores in tick eggs. Fungal Biology, 114, 49–56. 10.1016/j.mycres.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Morandi, M. A. B. , Maffia, L. A. , Mizubuti, E. S. G. , Alfenas, A. C. , & Barbosa, J. G. (2003). Suppression of Botrytis cinerea sporulation by Clonostachys rosea on rose debris: A valuable component in Botrytis blight management in commercial greenhouses. Biological Control, 26, 311–317. 10.1016/S1049-9644(02)00134-2 [DOI] [Google Scholar]
- Navarathna, D. H. , Pathirana, R. U. , Lionakis, M. S. , Nickerson, K. W. , & Roberts, D. D. (2016). Candida albicans ISW2 regulates chlamydospore suspensor cell formation and virulence in vivo in a mouse model of disseminated candidiasis. PLoS ONE, 11, e0164449 10.1371/journal.pone.0164449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile, C. J. , Bruno, V. M. , Richard, M. L. , Davis, D. A. , & Mitchell, A. P. (2003). Genetic control of chlamydospore formation in Candida albicans . Microbiology, 149, 3629–3637. 10.1099/mic.0.26640-0 [DOI] [PubMed] [Google Scholar]
- Ojeda‐Robertos, N. F. , Torres‐Acosta, J. F. , Ayala‐Burgos, A. J. , Sandoval‐Castro, C. A. , Valero‐Coss, R. O. , & Mendoza‐de‐Gives, P. (2009). Digestibility of Duddingtonia flagrans chlamydospores in ruminants: In vitro and in vivo studies. BMC Veterinary Research, 5, 46 10.1186/1746-6148-5-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palige, K. , Linde, J. , Martin, R. , Böttcher, B. , Citiulo, F. , Sullivan, D. J. , … Staib, P . (2013). Global transcriptome sequencing identifies chlamydospore specific markers in Candida albicans and Candida dubliniensis . PLoS ONE, 8, e61940 10.1371/journal.pone.0061940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz‐Silva, A. , Francisco, I. , Valero‐Coss, R. O. , Cortiñas, F. J. , Sánchez, J. A. , Francisco, R. , & deGives, P. M . (2011). Ability of the fungus Duddingtonia flagrans to adapt to the cyathostomin egg‐output by spreading chlamydospores. Veterinary Parasitology, 179, 277–282. 10.1016/j.vetpar.2011.02.014 [DOI] [PubMed] [Google Scholar]
- Rodríguez, M. A. , Cabrera, G. , Gozzo, F. C. , Eberlin, M. N. , & Godeas, A. (2011). Clonostachys rosea BAFC3874 as a Sclerotinia sclerotiorum antagonist: Mechanisms involved and potential as a biocontrol agent. Journal of Applied Microbiology, 110, 1177–1186. 10.1111/j.1365-2672.2011.04970.x [DOI] [PubMed] [Google Scholar]
- Sancak, B. , Colakoglu, S. , Acikgoz, Z. C. , & Arikan, S. (2005). Incubation at room temperature may be an independent factor that induces chlamydospore production in Candida dubliniensis . Diagnositc Microbiology and Infectious Disease, 52, 305–309. 10.1016/j.diagmicrobio.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Sanyal, P. K. , Sarkar, A. K. , Patel, N. K. , Mandal, S. C. , & Pal, S. (2008). Formulation of a strategy for the application of Duddingtonia flagrans to control caprine parasitic gastroenteritis. Journal of Helminthology, 82, 169–174. [DOI] [PubMed] [Google Scholar]
- Sonneborn, A. , Bockmühl, D. P. , & Ernst, J. F. (1999). Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect and Immunity, 67, 5514–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib, P. , & Morschhäuser, J. (2007). Chlamydospore formation in Candida albicans and Candida dubliniensis‐an enigmatic developmental programme. Mycoses, 50, 1–12. 10.1111/j.1439-0507.2006.01308.x [DOI] [PubMed] [Google Scholar]
- Sun, M. H. , Chen, Y. M. , Liu, J. F. , Li, S. D. , & Ma, G. Z. (2014). Effects of culture conditions on spore types of Clonostachys rosea 67‐1 in submerged fermentation. Letters in Applied Microbiology, 58, 318–324. 10.1111/lam.12193 [DOI] [PubMed] [Google Scholar]
- Sun, Z. B. , Sun, M. H. , & Li, S. D. (2015). Draft genome sequence of mycoparasite Clonostachys rosea strain 67‐1. Genome Announcements, 3, e00546–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona, S. , Garcia‐Alcalde, F. , Dopazo, J. , Ferrer, A. , & Conesa, A. (2011). Differential expression in RNA‐seq: A matter of depth. Genome Research, 21, 2213–2223. 10.1101/gr.124321.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzelepis, G. , Dubey, M. , Jensen, D. F. , & Karlsson, M. (2015). Identifying glycoside hydrolase family 18 genes in the mycoparasitic fungal species Clonostachys rosea . Microbiology, 161, 1407–1419. 10.1099/mic.0.000096 [DOI] [PubMed] [Google Scholar]
- Xu, Z. , Xu, J. , Ji, A. , Zhu, Y. , Zhang, X. , Hu, Y. , … Chen, S. (2015). Genome‐wide selection of superior reference genes for expression studies in Ganoderma lucidum . Gene, 574, 352–358. 10.1016/j.gene.2015.08.025 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. H. , Gao, H. L. , Ma, G. Z. , & Li, S. D. (2004). Mycoparasitism of Gliocladium roseum 67–1 on Sclerotinia sclerotiorum . Acta Phytopathological Sinica, 34, 211–214. [Google Scholar]
- Zhang, J. , Sun, Z. B. , Li, S. D. , & Sun, M. H . (2017). Identification of suitable reference genes during the formation of chlamydospores in Clonostachys rosea 67‐1. MicrobiologyOpen, 6, e505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, C. G. , Tao, N. , Liu, W. J. , Yang, J. K. , Huang, X. W. , Liu, X. Y. , … Zhang, K. Q. (2010). Regulation of subtilisin‐like protease prC expression by nematode cuticle in the nematophagous fungus Clonostachys rosea . Environmental Microbiology, 12, 3243–3252. 10.1111/j.1462-2920.2010.02296.x [DOI] [PubMed] [Google Scholar]
- Zou, C. G. , Xu, Y. F. , Liu, W. J. , Zhou, W. , Tao, N. , Tu, H. H. , … Zhang, K. Q. (2010). Expression of a serine protease gene prC is up‐regulated by oxidative stress in the fungus Clonostachys rosea: Implications for fungal survival. PLoS One, 5, e13386 10.1371/journal.pone.0013386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


