Abstract
Germline variants of PIP4K2A impact susceptibility of acute lymphoblastic leukemia (ALL) through inducing its overexpression. Although limited reports suggested the oncogenic role of PIP4K2A in cancers, regulatory network and prognostic effect of this gene remains poorly understood in tumorigenesis and leukemogenesis. In this study, we conducted genome-wide gene expression association analyses in pediatric B-ALL cohorts to discover expression associated genes and pathways, which is followed by the bioinformatics analyses to investigate the prognostic role of PIP4K2A and its related genes in multiple cancer types. 214 candidates were identified to be significantly associated with PIP4K2A expression in ALL patients, with known cancer-related genes rankings the top (e.g., RAC2, RBL2, and TFDP1). These candidates do not only tend to be clustered in the same types of leukemia, but can also separate the patients into novel molecular subtypes. PIP4K2A is noticed to be frequently overexpressed in multiple other types of leukemia and solid cancers from cancer cohorts including TCGA, and associated with its candidates in subtype-specific and cancer-specific manners. Interestingly, the association status varied in tumors compared to their matched normal tissues. Moreover, PIP4K2A and its related candidates exhibit stage-independent prognostic effects in multiple cancers, mostly with its lower expression significantly associated with longer overall survival (p < 0.05). Our findings reveal the transcriptional regulatory network of PIP4K2A in leukemia, and suggest its potentially important role on molecular subtypes of multiple cancers and subsequent treatment outcomes.
Keywords: acute lymphoblastic leukemia, cancer, TCGA, TFDP1, PIP4K2A
Introduction
Acute lymphoblastic leukemia (ALL) is the most common pediatric malignancy (Greaves, 2006; Pui and Evans, 2006), induced through multifactorial mechanisms involving both environmental and genetic factors (Buffler et al., 2005). A series of genome-wide association studies (GWASs) have identified several common risk loci for genetic predisposition basis of ALL in several genes (e.g., ARID5B, IKZF1, CEBPE, CDKN2A, PIP4K2A-BMI1, and GATA3) (Papaemmanuil et al., 2009; Trevino et al., 2009; Sherborne et al., 2010; Migliorini et al., 2013; Perez-Andreu et al., 2013; Xu et al., 2013; Perez-Andreu et al., 2015; Xu et al., 2015). As more and more causal variants are considered to impact on gene expression rather than inducing amino acid changes, analyses for expression quantitative trait loci (eQTL) in ALL have also been conducted. For instance, PIP4K2A expression is strongly impacted by the genotypes of its adjacent single nucleotide polymorphisms (SNPs), which have been validated in several independent patient cohorts for ALL susceptibility (Walsh et al., 2013; Liao et al., 2016), suggesting PIP4K2A SNPs may play role on leukemogenesis through inducing its overexpression. Meanwhile, current evidence has also shown a ethnicity-specific and subtype-specific correlation between PIP4K2A SNPs and ALL susceptibility (Migliorini et al., 2013; Walsh et al., 2013; Xu et al., 2013), which is similar as ARID5B SNPs (Xu et al., 2012, 2013). However, how PIP4K2A contributes to leukemogenesis of ALL and its subtypes remains to be poorly understood.
PIP4K2A encodes a major component of phosphatidylinositol 5-phosphate 4-kinase type-II (Rameh et al., 1997). As actively expressed in peripheral blood cells (Divecha et al., 1995), PIP4K2A may facilitate the conversion of phosphorylation of phosphatidylinositol 5-phosphate (PtdIns5P) to generate various phosphoinositide species, which regulate crucial cellular functions including proliferation, survival, glucose uptake and migration (Fiume et al., 2015). A few reports have demonstrated that PIP4K2A may play a critical role on development and cellular activities maintaining. For instance, PIP4K2A expression can influence cell response to oxidative stress by regulating the level of PtdIns5P (Wilcox and Hinchliffe, 2008; Jones et al., 2013). Depletion of PIP4K2A is associated with decreased mTOR signaling, and induces developmental defects in Drosophila and zebrafish (Elouarrat et al., 2013; Gupta et al., 2013). Besides, PIP4K2A is considered to control neuronal KCNQ channels, disorders of which are implicated in schizophrenia (Fedorenko et al., 2008, 2009), and may also involve in insulin signaling by regulating PI3/AKT pathway. Knocking out of PIP4K2B, another component of phosphatidylinositol 5-phosphate 4-kinase type-II, can increase insulin sensitivity and reduce adiposity (Carricaburu et al., 2003; Lamia et al., 2004). However, the role of PIP4K2A on tumorigenesis (including leukemogenesis) has been mentioned in limited reports. Recently, a cell-line based experiment indicates that knockdown of PIP4K2A in acute myeloid leukemia (AML) results in accumulation of the cyclin-dependent kinase inhibitors (i.e., CDKN1A and CDKN1B), G1 cell cycle arrest and apoptosis (Jude et al., 2015). Nevertheless, PIP4K2A knockdown in normal hematopoietic stem cells and progenitor cells doesn’t impair cell growth. Since knockdown of PIP4K2A only exerts inhibitory effects on tumor cells, this selective effect implies that PIP4K2A might be a candidate target for cancer treatment. In fact, PIP4K2A is noted to be up-regulated in breast tumor tissue and high expression of PIP4K2A is correlated with loss of p53. Knockdown of both PIP4K2A and PIP4K2B in a p53-deficient breast cell line dramatically abrogates cell proliferation (Emerling et al., 2013). This could be explained by impaired glucose metabolism and consequently increased reactive oxygen species (ROS), which leads to senescence. Also, PIP4K2A is considered to impact on sensitivity of some cancer drugs (e.g., paclitaxel), thus may be associated with overall survival of the treated cancer patients (Niu et al., 2012).
Single nucleotide polymorphisms around PIP4K2A are associated with ALL susceptibility through impacting expression level of PIP4K2A, and accumulating evidence indicates the potential role of PIP4K2A on cancer cells, however, it is poorly understood that how PIP4K2A impact on tumorigenesis (including leukemogenesis). In this study, we conducted array based genome-wide gene expression association analyses to investigate the potential regulatory network of PIP4K2A in leukemia, which is proved to be efficient in our previous work (Hou et al., 2017). Besides, we will also systematically examine the PIP4K2A status in multiple cancers and evaluate its prognostic effect.
Results
Screening of PIP4K2A-Related Genes in B-ALL and Their Association Status in B-ALL Subtypes
As the expression of PIP4K2A was considered to impact on leukemogenesis, we thus downloaded array-based expression data of eight independent pediatric B-ALL patient cohorts from public resource (Supplementary Table S1), and evaluated the expression correlation of PIP4K2A with the rest of other genes using a linear regression model. A series of filter criteria (e.g., p-value cutoff, r2, and correlation direction) were applied to determine more reliable PIP4K2A-related candidates for regulation network construction (Supplementary Figure S1). Altogether, 234 probes in 214 genes were significantly correlated to PIP4K2A expression with p ≤ 0.05 in all patient cohorts in the same direction (Supplementary Figure S1 and Supplementary Table S2). Through a stricter filter (i.e., p ≤ 2 × 10-6 and association r2 ≥ 0.1 in at least five cohorts), 23 genes were eventually identified as the strongest candidates (Supplementary Table S3).
Since PIP4K2A is a major component phosphate kinase, we sought to reveal its regulatory network by focusing on 10 transcription factors we screened out (Table 1). For instance, RBL2 is a member of retinoblastoma-susceptibility (RB) anti-oncogene family, which regulates cell cycle through inhibiting E2F-mediated transcription. Dysfunction of this gene is commonly involved in evolution of ALL (Ahuja et al., 1991; Stock et al., 2000; Schmitz et al., 2005), thus indicating a potentially significant role of RBL2 on B-ALL development. Pathway analysis was further conducted using DAVID Functional Annotation Tools, and three annotation clusters of gene sets exhibited significance with at least one term having Bonferroni p-value < 0.05 (Supplementary Table S4), including leukocyte migration pathway. Additionally, we illustrated the interactions between candidates by constructing a protein–protein interaction network based on STRING (Figure 1B), and identified multiple PIP4K2A related proteins, including the top gene of RAC2, which is highly expressed in immune organs. Since RAC2 belongs to the Rho small GTPase subfamily, which can regulate cell movement, proliferation, and survival, involvement of PIP4K2A in leukemogenesis may be partially explained by their interactions. In this part, we firstly built the regulatory network of PIP4K2A in B-ALL.
Table 1.
Candidates of PIP4K2A related DNA-dependent transcriptional regulatory genes in different dataset of B-ALL.
| Probe ID | 202098_s_at | 202449_s_at | 204529_s_at | 205038_at | 209107_x_at | 209930_s_at | 210249_s_at | 212330_at | 212331_at | 216241_s_at | 43544_at | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | PRMT2 | RXRA | TOX | IKZF1 | NCOA1 | NFE2 | NCOA1 | TFDP1 | RBL2 | TCEA1 | MED16 | |
| GSE7440 (n = 99) | p-Value | 7.49 × 10-10 | 7.10 × 10-13 | 8.58 × 10-7 | 4.89 × 10-5 | 1.69 × 10-10 | 2.64 × 10-9 | 2.93 × 10-13 | 2.94 × 10-12 | 8.99 × 10-10 | 8.56 × 10-8 | 3.64 × 10-11 |
| Coefficient | 0.64 | 0.61 | 0.33 | 0.38 | 0.84 | 0.38 | 0.92 | 0.51 | 0.58 | 0.65 | 0.74 | |
| se | 0.09 | 0.07 | 0.06 | 0.09 | 0.12 | 0.06 | 0.11 | 0.06 | 0.09 | 0.11 | 0.10 | |
| Adjusted r2 | 0.32 | 0.41 | 0.21 | 0.15 | 0.34 | 0.30 | 0.42 | 0.39 | 0.32 | 0.25 | 0.36 | |
| GSE10255 (n = 161) | p-Value | 0.01 | 0.01 | 7.56 × 10-4 | 0.03 | 4.10 × 10-3 | 4.80 × 10-15 | 1.84 × 10-3 | 1.35 × 10-3 | 4.50 × 10-3 | 3.65 × 10-3 | 9.86 × 10-9 |
| Coefficient | 0.40 | 0.20 | 0.16 | 0.26 | 0.57 | 0.26 | 0.51 | 0.15 | 0.37 | 0.49 | 0.72 | |
| se | 0.15 | 0.08 | 0.05 | 0.12 | 0.20 | 0.03 | 0.16 | 0.05 | 0.13 | 0.17 | 0.12 | |
| Adjusted r2 | 0.04 | 0.03 | 0.06 | 0.02 | 0.04 | 0.32 | 0.05 | 0.06 | 0.04 | 0.05 | 0.18 | |
| GSE10792 (n = 160) | p-Value | 5.74 × 10-7 | 0.01 | 0.02 | 0.03 | 0.02 | 1.67 × 10-3 | 0.04 | 8.94 × 10-7 | 6.12 × 10-4 | 2.96 × 10-3 | 5.79 × 10-4 |
| Coefficient | 0.53 | 0.22 | 0.16 | 0.33 | 0.48 | 0.24 | 0.44 | 0.29 | 0.64 | 0.84 | 0.51 | |
| se | 0.10 | 0.08 | 0.07 | 0.14 | 0.21 | 0.07 | 0.21 | 0.06 | 0.18 | 0.28 | 0.14 | |
| Adjusted r2 | 0.26 | 0.07 | 0.05 | 0.05 | 0.05 | 0.11 | 0.04 | 0.26 | 0.13 | 0.10 | 0.13 | |
| GSE11877 (n = 207) | p-Value | 4.93 × 10-9 | 1.65 × 10-3 | 2.19 × 10-3 | 1.03 × 10-6 | 1.02 × 10-4 | 1.60 × 10-5 | 1.21 × 10-4 | 2.24 × 10-3 | 8.65 × 10-13 | 1.96 × 10-4 | 2.71 × 10-7 |
| Coefficient | 0.39 | 0.17 | 0.17 | 0.33 | 0.49 | 0.15 | 0.46 | 0.20 | 0.59 | 0.43 | 0.34 | |
| se | 0.06 | 0.05 | 0.05 | 0.07 | 0.12 | 0.03 | 0.12 | 0.06 | 0.08 | 0.11 | 0.06 | |
| Adjusted r2 | 0.15 | 0.04 | 0.04 | 0.11 | 0.07 | 0.08 | 0.07 | 0.04 | 0.22 | 0.06 | 0.12 | |
| GSE13351 (n = 107) | p-Value | 4.53 × 10-9 | 3.81 × 10-4 | 3.94 × 10-3 | 1.02 × 10-3 | 1.80 × 10-5 | 1.89 × 10-6 | 3.54 × 10-5 | 1.72 × 10-3 | 3.46 × 10-12 | 4.43 × 10-5 | 9.53 × 10-6 |
| Coefficient | 0.58 | 0.24 | 0.19 | 0.31 | 0.91 | 0.28 | 0.82 | 0.20 | 0.67 | 0.58 | 0.45 | |
| se | 0.09 | 0.07 | 0.07 | 0.09 | 0.20 | 0.06 | 0.19 | 0.06 | 0.08 | 0.14 | 0.10 | |
| Adjusted r2 | 0.31 | 0.12 | 0.08 | 0.10 | 0.18 | 0.22 | 0.16 | 0.09 | 0.41 | 0.16 | 0.19 | |
| GSE13425 (n = 190) | p-Value | 9.14 × 10-5 | 1.01 × 10-6 | 5.30 × 10-3 | 1.18 × 10-3 | 2.07 × 10-4 | 2.99 × 10-3 | 3.79 × 10-3 | 5.78 × 10-3 | 1.32 × 10-6 | 3.31 × 10-4 | 1.47 × 10-3 |
| Coefficient | 0.36 | 0.39 | 0.25 | 0.31 | 0.66 | 0.18 | 0.52 | 0.20 | 0.54 | 0.54 | 0.33 | |
| se | 0.09 | 0.08 | 0.09 | 0.09 | 0.17 | 0.06 | 0.18 | 0.07 | 0.11 | 0.15 | 0.10 | |
| Adjusted r2 | 0.09 | 0.14 | 0.04 | 0.06 | 0.08 | 0.05 | 0.05 | 0.04 | 0.14 | 0.08 | 0.06 | |
| GSE33315 (n = 575) | p-Value | 4.32 × 10-17 | 9.08 × 10-5 | 3.21 × 10-3 | 1.88 × 10-9 | 1.38 × 10-12 | 1.32 × 10-26 | 8.79 × 10-14 | 3.64 × 10-16 | 1.81 × 10-23 | 8.81 × 10-11 | 5.83 × 10-21 |
| Coefficient | 0.56 | 0.16 | 0.11 | 0.34 | 0.75 | 0.22 | 0.67 | 0.24 | 0.68 | 0.57 | 0.60 | |
| se | 0.06 | 0.04 | 0.04 | 0.05 | 0.10 | 0.02 | 0.09 | 0.03 | 0.06 | 0.09 | 0.06 | |
| Adjusted r2 | 0.13 | 0.03 | 0.02 | 0.07 | 0.10 | 0.21 | 0.11 | 0.13 | 0.19 | 0.08 | 0.17 | |
| GSE635 (n = 173) | p-Value | 2.01 × 10-6 | 1.38 × 10-4 | 5.41 × 10-7 | 0.02 | 3.65 × 10-3 | 1.23 × 10-3 | 2.97 × 10-3 | 2.86 × 10-3 | 3.44 × 10-5 | 6.07 × 10-5 | 1.01 × 10-3 |
| Coefficient | 0.41 | 0.28 | 0.29 | 0.20 | 0.51 | 0.14 | 0.49 | 0.18 | 0.44 | 0.64 | 0.30 | |
| se | 0.08 | 0.07 | 0.05 | 0.09 | 0.17 | 0.04 | 0.16 | 0.06 | 0.10 | 0.16 | 0.09 | |
| Adjusted r2 | 0.12 | 0.08 | 0.13 | 0.02 | 0.04 | 0.05 | 0.04 | 0.05 | 0.09 | 0.08 | 0.06 | |
FIGURE 1.
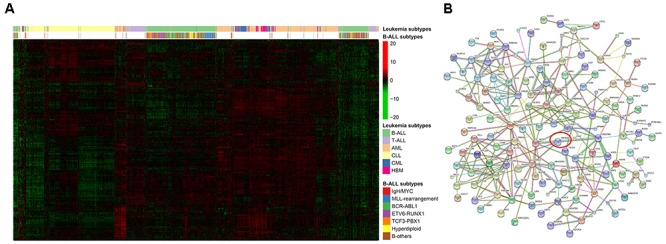
Regulatory statement of PIP4K2A and its related candidates in leukemia. (A) Expression clustering of PIP4K2A-related genes, with leukemia types and B-ALL subtypes were labeled. (B) Protein–protein interaction network of PIP4K2A-related genes. Line thickness indicates the strength of data support, and nodes disconnected with the main network were hidden.
Next, the association between PIP4K2A and its candidates were investigated in different molecular subtypes of the largest pediatric B-ALL cohort with available subtype information (GSE33315) (Supplementary Table S5). To exclude the impact of small sample size of some subtypes, such as BCR-ABL and MLL rearrangement, we mainly focused on four large patient groups, including hyperdiploid, ETV6-RUNX1, TCF3-PBX1 molecular subtypes as well as B-others. Totally 85 out of 214 genes are significantly associated with PIP4K2A expression in all four patient groups with the same direction, including seven transcription factors (Table 1 and Figure 2). Although similar association status was observed in different subtypes in terms of r2 and coefficients of these transcription factors (e.g., TFDP1, RBL2, and NFE2), subtype-specificity was also noticed. For instance, NCOA1 is significantly associated with PIP4K2A in all four patient groups with varied coefficients in different subtypes, while TCEA1 is even unrelated to PIP4K2A in TCF3-RUNX1 subtypes, indicating regulation network of PIP4K2A may vary in subtype-specific manner.
FIGURE 2.
Expression association status of PIP4K2A with some of important candidates in different subtypes of B-ALL (i.e., B-others, ETV6-RUNX1, hyperdiploid, and TCF3-PBX1) in the largest available pediatric B-ALL cohort (i.e., GSE33315).
Varied Association Status of PIP4K2A With Candidate Genes in Different Leukemia Types
Next, dataset GSE13204 (3248), which contains a large number of patients with B-ALL, T-ALL, AML, chronic lymphoblastic leukemia (CLL), chronic myeloid leukemia (CML) as well as healthy bone marrow (HBM), was selected for validation and further analyses. Expression of all PIP4K2A-related genes in B-ALL tends to be clustered in terms of leukemia types, illustrating distinct expression patterns of these candidates in different leukemia types and their different roles on leukemogenesis (Figure 1A). Not surprisingly, 99.07% (212/214) of the candidates were significantly associated with PIP4K2A expression in B-ALL of this cohort with the same direction, proving the reliability of their correlations. Importantly, the consistent rate for significant association between PIP4K2A and its candidates declined to 96.26% (206/214), 91.1% (195/214), 87.85% (188/214), 66.36% (142/214) and 49.53% (106/214) in CLL, AML, T-ALL, CML and HBM, respectively, even with opposite association directions to that in B-ALL for some genes, which account for 0.97% (2/206), 3.08% (6/195), 0% (0/188), 2.11% (3/142) and 4.72% (5/106) of in CLL, AML, T-ALL, CML and HBM, respectively (Supplementary Table S6). While the transcription factors are more stable, with higher consistent rates of significant association and no opposite direction (Figure 3). Above results implies that the association of PIP4K2A with its candidates discovered in B-ALL may also exists in other leukemia types at different levels. Notably, association status of PIP4K2A and its candidates exhibit the lowest consistent rate in health bone marrow in terms of p-value as well as r2 (Supplementary Table S6), indicating the dysfunction of PIP4K2A regulation network in leukemia compared to normal tissues.
FIGURE 3.
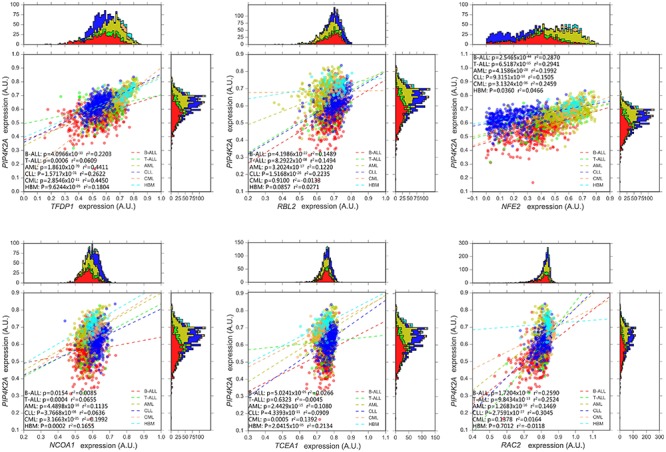
Expression association status of PIP4K2A with some of important candidates in different types of leukemia, including B-ALL (N = 576), T-ALL (N = 174), AML (N = 542), CLL (N = 448), CML (N = 76), and healthy bone marrow (HBM, N = 74) based on GSE13204.
PIP4K2A Expression Profile and Potential Genes Associated With PIP4K2A Expression in Multiple Cancer Types
Although higher PIP4K2A expression may contribute to higher risk of leukemogenesis, normal B cells are unavailable as matched control for further analyses. Therefore, we further investigated the expression profile of PIP4K2A and its associated genes in other cancer types with paired tumor/normal tissues from The Cancer Genome Atlas (TCGA) (Supplementary Figure S6). With implicated expression information in multiple types of cancers, up to 8% of patients exhibited up-regulated PIP4K2A in tumors compared with their matched normal tissues in all available cancer types except a few down-regulations (e.g., KIRC) (Supplementary Figure S2), suggesting the potential widespread oncogenic role of PIP4K2A in tumorigenesis apart from leukemia.
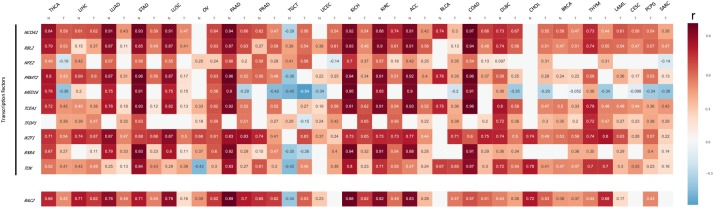
Next, we downloaded expression data from TCGA to investigate the association of PIP4K2A with its associated transcription factors in tumors and normal separately in each cancer type (Supplementary Tables S7, S8), Among the 23 types of cancers each with at least 100 patients having available follow-up information, nearly 70% of these transcription factors exhibit consistent association in both tumor and normal tissues, yet with varied coefficients and r2, even opposite directions (Figure 4). Interestingly, tumor-specific or control-specific associations were noticed. The candidates exhibited stronger association in normal tissues (e.g., normal tissues of KIRP, KICH, KIRC, ACC, and COAD) than tumors, whereas at least half of genes lost their correlation, or even turn to be negatively associated with PIP4K2A preferentially in normal tissues of OV, TGCT, UCEC, BLCA, and CHOL. For instance, NCOA1 is more correlated to PIP4K2A in normal tissues than that in tumors in terms of r2. The correlations of PIP4K2A with TFDP1 tend to exhibit tumor-specific (i.e., significant in 21/23 tumors but 9/20 normal tissues) (Figure 4 and Supplementary Figure S3), while that with MED16 is tend to be in control-specific manner (i.e., significant in 14/23 tumors but 15/20 normal tissues) (Figure 4). Additionally, opposite association directions of tumor vs. normal were also observed, such as MED16 in THCA (Figure 4), indicating the varied regulatory status and role of PIP4K2A on tumor vs. normal tissues in different types of cancers.
FIGURE 4.
Expression association status of PIP4K2A with 10 transcriptional regulators in different cancer types of TCGA. Heatmap of p-value and r2 of correlation of PIP4K2A-related transcriptional regulators with normal and tumor tissues of 23 cancer types. White box indicates non-significant (p-value > 0.05).
Impact of PIP4K2A and Its Correlated Gene on Overall Survival Rate
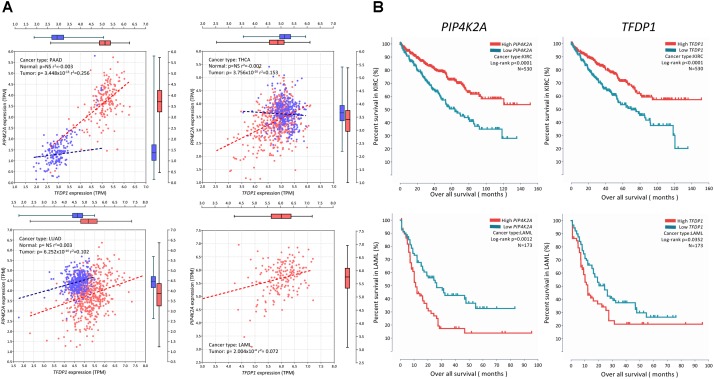
Due to the potentially important role of PIP4K2A in tumors, we further explored the possible role of PIP4K2A expression on survival rate in different cancer types. We focused on the transcription factors because of their important roles on PIP4K2A network described above. Totally, significant correlations were observed in 6 out of 23 cancer types (i.e., BLCA, KIRC, LAML, LIHC, SARC, and STAD) (Supplementary Figure S4). In particular, lower expression of PIP4K2A in tumors is associated with longer overall survival in all these cancer types except KIRC (Figure 5), suggesting the potential prognostic role of PIP4K2A overexpression in multiple cancer types, which is frequently observed as described above. Moreover, we also evaluated the correlation of survival status in these six cancer types with the expression levels of PIP4K2A related transcription factors (Supplementary Table S9). Strikingly, TFDP1 exhibits the strongest consistency with PIP4K2A in the same direction in five out of six cancer types (i.e., KIRC, LAML, SARC, BLCA, and LIHC) but not STAD (Supplementary Figure S4 and Supplementary Table S9). On the contrary, such consistency is lower for other transcription factors. For non-transcription factors, an opposite direction was even identified, such as RAC2 (Supplementary Figure S4 and Supplementary Table S9). Hence, TFDP1 may play an important role on the transcriptional regulatory network of PIP4K2A to impact on the survival rate in multiple cancer types.
FIGURE 5.
Correlation of PIP4K2A and its related candidates with survival status in different cancer types. (A) Expression association status of TFDP1 with PIP4K2A in LAML and KIRC; (B) association status of TFDP1 and PIP4K2A with overall survival in LAML and KIRC.
Discussion
PIP4K2A is largely implicated in basal cellular activities such as proliferation, survival, glucose uptake and migration, and plays an essential role for tumor growth. Due to extensive expression and pleiotropic effect of PIP4K2A in multiple tissues, it is important to unravel the transcriptional regulatory network of PIP4K2A in different cancer types, which is yet time/effect-consuming. Based on the assumption that genes participating in the same regulatory network are associated in terms of expression level, and closer transcriptional factors exhibit more significant correlation, it will be much easier to screen potential PIP4K2A-related genes by genome-wide gene expression association analyses by using available public microarray datasets. A series of GWASs and subsequent validations have revealed that SNPs in PIP4K2A locus are significantly associated with ALL susceptibility, possibly through impact on PIP4K2A expression, indicating the potential role of PIP4K2A over-expression in leukemogenesis. Therefore, 214 genes were identified to be associated with PIP4K2A expression with the similar pipeline we used previously (Hou et al., 2017). Although association strength varied in different types of leukemia, or even subtypes of B-ALLs, almost all of these candidates can be validated in B-ALL of another big patient cohort, while a large proportion of them can also be validated in other types of leukemia in terms of expression association. These findings strongly indicate the consistency and stability of PIP4K2A network. Since PIP4K2A encodes a kinase, it is desirable to focus on characteristic regulatory network of PIP4K2A in different B-ALL subtypes, respectively, by analyzing larger samples. Additionally, the over-expression of PIP4K2A could be influenced by the chemotherapeutic treatment applied, so we have searched all the datasets to see if the influence of chemotherapeutic treatment existed in these cohorts. Among eight patient cohorts analyzed in Table 1, the patients from GSE7440 and GSE11877 received chemotherapy, while the patients from GSE10255 and GSE13425 were newly diagnosed with ALL without any initial therapy. Other cohorts didn’t tell what treatment the patients received. Therefore, we compared the association status of transcription factors with PIP4K2A in abovementioned four cohorts. It turns out that no significant difference in correlation exists between the cohorts who received chemotherapy and those who did not, indicating that the chemotherapeutic treatment does not significantly influence the association between PIP4K2A and candidates, at least in the analyzed cohorts.
Noteworthily, our findings identified some genes that have been reported to be related to leukemogenesis or ALL development. As one of the strongest PIP4K2A-related candidates, the transcription factor RBL2 belongs to retinoblastoma-susceptibility (RB) anti-oncogene family. The RB gene is an anti-oncogene regulating cell cycle through inhibiting E2F-mediated transcription, which is frequently altered and thus quite commonly involved in evolution of ALL (Ahuja et al., 1991; Stock et al., 2000; Schmitz et al., 2005). RBL2 demonstrates statistically significant and positive correlation with PIP4K2A expression in four relatively larger samples of B-ALL subtypes (i.e., hyperdiploid, ETV6-RUNX1, B-other and TCF3-PBX1 subtype). RBL2 also exhibits significant correlation exclusively in tumor tissues of OV, BLCA, and CHOL, indicating a cancer type-specific correlation between RBL2 and PIP4K2A. Another example is TFDP1, this gene exhibits extraordinarily strong association with PIP4K2A. Same as RBL2, TFDP1 also shows evident association with PIP4K2A in four large samples of B-ALL subtypes as well as all types of leukemia. And in 23 other cancer types, TFDP1 remains the significance in 21 types of cancer. Specifically, in KIRC and PAAD, PIP4K2A expression is prominently affected by TFDP1. Besides, the impact of PIP4K2A and TFDP1 expression on survival status in various cancers demonstrates great consistency, and these two genes are also significantly correlated in these cancers. Above findings reveal quite close interactions between PIP4K2A and TFDP1 as well as the important role of TFDP1 in regulatory network of PIP4K2A. However, no clear binding motif information is available due to the limited studies on TFDP1. Some other candidates are linked to PIP4K2A according to PPI prediction, and we considered it is appropriate to start with these leukemia-related or cancer-related genes in order to reveal the transcriptional regulatory network of PIP4K2A in B-ALL. With the limited database, some of the candidate transcription factors have the predicted binding sites in PIP4K2A promoter or enhancer region, including RXRA (Supplementary Figure S5 and Supplementary Table S10). However, further experimental analyses are largely needed to test whether PIP4K2A is regulated by these transcription factors through direct binding. Unfortunately, we didn’t screen out the well-known genes in mTOR pathway or KCNQ channel, probably because PIP4K2A may be involved in these pathways through post-transcriptional regulation or protein interactions, which can’t be detected through expression association approaches.
Importantly, the candidate transcription factors are also significant associated with PIP4K2A expression in multiple cancer types apart from leukemia. Generally, candidate genes demonstrate much stronger association in normal tissues than tumors. Among 10 transcription factors, some genes exhibit significant associations in all 23 cancer types such as RBL2 and IKZF1, indicating their important role on PIP4K2A and ubiquitous interactions with PIP4K2A in the regulatory network among different cancers, as well as the reliability of our screening process. Interestingly, IKZF1 is one of most important transcription factors involved in hematopoietic development, mutations and deletions of this gene are frequently observed in leukemia cells of B-ALL patients. Meanwhile, SNPs in IKZF1 are also significantly associated with ALL susceptibility across multiple ethnicities, probably through impacting on the expression level of IKZF1. Therefore, the expression correlation of IKZF1 with PIP4K2A indicates that these two genes may interact with each other to facilitate leukemogenesis as well as tumorigenesis. Although insignificant in some cancer types, more than 70% of associations between PIP4K2A and the rest of the transcription factors exhibit statically significant, including cancer-specific or normal-specific manner. Moreover, loss or even opposite direction of correlations were observed in quite a few genes between tumor and normal tissues, suggesting tumor-specific dysfunction of PIP4K2A regulatory network may contribute to tumorigenesis.
Due to the lack of survival information of the ALL patients, we can’t estimate the impact of SNP genotypes and subsequent SNP-induced expression changes on survival rate. However, we noticed that PIP4K2A overexpression is related to worse treatment outcomes of multiple cancer types of TCGA, except higher PIP4K2A expression related to better survival status in KIRC. Consistently, down-regulation of PIP4K2A in tumors is much more frequently observed in KIRC than any other types of cancer in TCGA (Supplementary Figure S2). Not surprisingly, some of the PIP4K2A-related genes are also related to survival rate in the same direction, among which TFDP1 exhibits the highest consistency, suggesting that TFDP1 may interact with PIP4K2A to impact on both tumorigenesis as well as treatment outcomes. However, the prognostic value of PIP4K2A and its regulatory partners should be evaluated in independent cancer cohorts.
To sum up, we have utilized available public microarray datasets and developed a systematic screening to find expression-based regulatory network of PIP4K2A. Because very limited studies have reported to explore the functional role of PIP4K2A on leukemogenesis despite of its significant association with ALL susceptibility as well as overexpression in multiple types of cancers, our findings may shed a new light on epigenetic regulation of PIP4K2A, and provide a clue for the further functional studies on this gene in cancer.
Materials and Methods
Construction of Regulatory Network of PIP4K2A
Expression level of all genes in B-ALL patients were downloaded from Gene Expression Omnibus [GSE7440 (Bhojwani et al., 2008), GSE11877 (Kang et al., 2010), GSE10792 (Bungaro et al., 2009), GSE13351 (Den Boer et al., 2009), GSE13425 (Holleman et al., 2004), GSE635 (Sorich et al., 2008), GSE10255 (Kirschner-Schwabe et al., 2006), GSE4698 (Zhang et al., 2012), GSE33315 (Haferlach et al., 2010), and GSE13204 (Curtis et al., 2012)]. Associations of PIP4K2A expression with all the rest genes were calculated using linear regression model. Afterward, we used multiple steps to conduct candidate screening and determine the strong candidates (Supplementary Figure S1).
All the PIP4K2A-related candidates were imported into the STRING1 to show the known protein–protein interaction network (Szklarczyk et al., 2015). We used line thickness to indicate the strength of data support, and the nodes disconnected with the main network were hidden.
Functional enrichment was determined using the DAVID functional annotation tool2 (Huang da et al., 2009a,b). The functional categories used were GO term related to biological process (BP), cellular component (CC), and molecular function (MF)3. Multiple criteria were applied for pathway analyses of candidates, including P-value, FDR, etc. (Huang da et al., 2009b). Various top-related pathways were selected (i.e., lysosome, porphyrin biosynthesis and adhesion, and diapedesis of granulocytes).
With limited information of transcription factors binding motif, we predicted some candidate transcription factors by importing the specific sequence of PIP4K2A in to JASPAR4. Some of these transcription factors got high relative scores and had predicted binding sites in PIP4K2A promoter or enhancer regions.
Exploration of Differences Between PIP4K2A Regulatory Networks in Different Subtypes
We divided the patients into multiple groups according to the subtypes of B-ALL and the subtypes of leukemia from the large two cohorts, GSE33315 and GSE13204, respectively. To study the difference of PIP4K2A regulatory networks in various subtypes, we calculated the association of PIP4K2A with all the rest genes by linear regression model in diverse subtypes. The proportions of PIP4K2A-related candidates in all significant related genes were computed.
We selected 133 PIP4K2A-related probes by various criteria (i.e., p ≤ 2 × 10-6 and association r2 ≥ 0.1 in at least five cohorts, and r2 ≥ 0.3 in at least one cohort). Then, the expression levels of these candidates in GSE13204 were used to generate the heatmap, clustering by the subtypes of leukemia and the subtypes of B-ALL, respectively.
Discovery of Association of PIP4K2A With Its Related Genes in Multiple Cancers and Corresponding Normal Tissues
We compared the expression level of PIP4K2A in various cancer types in COSMIC dataset5, and computed the frequency of high-regulated and low-regulated PIP4K2A samples in different status.
Some functional PIP4K2A-related transcription factors were picked out using the Animal TFDB database (42)6. Gene expression RNAseq [log2(tpm+0.001)] of the candidates in 23 TCGA cancers (THCA, LIHC, LUAD, STAD, LUSC, OV, PAAD, PRAD, TGCT, UCEC, KICH, KIRC, ACC, BLCA, COAD, DLBC, CHOL, BRCA, THYM, LAML, CESC, PCPG, SARC) (Supplementary Table S7) and corresponding normal tissues in both TCGA and GTEX were acquired from UCSC Xena (Vivian et al., 2017)7. The association of the expression level of the selected transcription factors with PIP4K2A was subsequently calculated using linear regression models as well.
To further detect the association of PIP4K2A with these genes, the r2 of PIP4K2A and its related transcription factors as well as RAC2, that we calculated by linear model as we described above, were imported to generate the heatmap, in which white indicates non-significance.
Comparison of PIP4K2A and Its Correlated Gene on Overall Survival Rate
The clinical information for survival analyses was also acquired from UCSC Xena, including patient ID, overall survival information, etc. We divided the expression levels of PIP4K2A-related transcription factors into two groups, high-expression level and low-expression level, by the significant low point of their histogram, and used this standard to conduct survival analysis in different cancer types.
Data Availability Statement
Datasets analyzed for this study are available in GEO repository (http://www.ncbi.nlm.nih.gov/gds): GSE635, GSE10792, GSE13351, GSE13425, GSE10255, GSE4698, GSE33315, GSE7440, GSE11877, and GSE13204, and TCGA database (https://cancergenome.nih.gov/).
Author Contributions
SZ and ZL designed the study. XiY, YD, FZ, PW, JZ, DY, FL, XZ, and DZ wrote the manuscript. LB, XX, HW, XuY, WaZ, HG, and WeZ performed the data analyses. LY, QH, HX, YS, YZ, and YW contributed to the conception of the study and drafted the manuscript. All authors contributed significantly in writing the manuscript and read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported byNational Key Research Development Program [Grant No. 2016YFC0905000 (2016YFC0905002)], and the National Natural Science Foundation of China (Grant Nos. 81522028 and 81673452).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00721/full#supplementary-material
References
- Ahuja H. G., Jat P. S., Foti A., Bar-Eli M., Cline M. J. (1991). Abnormalities of the retinoblastoma gene in the pathogenesis of acute leukemia. Blood 78 3259–3268. [PubMed] [Google Scholar]
- Bhojwani D., Kang H., Menezes R. X., Yang W., Sather H., Moskowitz N. P., et al. (2008). Gene expression signatures predictive of early response and outcome in high-risk childhood acute lymphoblastic leukemia: a Children’s Oncology Group Study [corrected]. J. Clin. Oncol. 26 4376–4384. 10.1200/JCO.2007.14.4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffler P. A., Kwan M. L., Reynolds P., Urayama K. Y. (2005). Environmental and genetic risk factors for childhood leukemia: appraising the evidence. Cancer Invest 23 60–75. 10.1081/CNV-46402 [DOI] [PubMed] [Google Scholar]
- Bungaro S., Dell’Orto M. C., Zangrando A., Basso D., Gorletta T., Lo Nigro L., et al. (2009). Integration of genomic and gene expression data of childhood ALL without known aberrations identifies subgroups with specific genetic hallmarks. Genes Chromos. Cancer 48 22–38. 10.1002/gcc.20616 [DOI] [PubMed] [Google Scholar]
- Carricaburu V., Lamia K. A., Lo E., Favereaux L., Payrastre B., Cantley L. C., et al. (2003). The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc. Natl. Acad. Sci. U.S.A. 100 9867–9872. 10.1073/pnas.1734038100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C., Shah S. P., Chin S. F., Turashvili G., Rueda O. M., Dunning M. J., et al. (2012). The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486 346–352. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Boer M. L., van Slegtenhorst M., De Menezes R. X., Cheok M. H., Buijs-Gladdines J. G., Peters S. T., et al. (2009). A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 10 125–134. 10.1016/S1470-2045(08)70339-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N., Truong O., Hsuan J. J., Hinchliffe K. A., Irvine R. F. (1995). The cloning and sequence of the C isoform of PtdIns4P 5-kinase. Biochem. J. 309(Pt 3) 715–719. 10.1042/bj3090715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elouarrat D., van der Velden Y. U., Jones D. R., Moolenaar W. H., Divecha N., Haramis A. P. (2013). Role of phosphatidylinositol 5-phosphate 4-kinase alpha in zebrafish development. Int. J. Biochem. Cell Biol. 45 1293–1301. 10.1016/j.biocel.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Emerling B. M., Hurov J. B., Poulogiannis G., Tsukazawa K. S., Choo-Wing R., Wulf G. M., et al. (2013). Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 155 844–857. 10.1016/j.cell.2013.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko O., Strutz-Seebohm N., Henrion U., Ureche O. N., Lang F., Seebohm G., et al. (2008). A schizophrenia-linked mutation in PIP5K2A fails to activate neuronal M channels. Psychopharmacology (Berl) 199 47–54. 10.1007/s00213-008-1095-x [DOI] [PubMed] [Google Scholar]
- Fedorenko O., Tang C., Sopjani M., Foller M., Gehring E. M., Strutz-Seebohm N., et al. (2009). PIP5K2A-dependent regulation of excitatory amino acid transporter EAAT3. Psychopharmacology (Berl) 206 429–435. 10.1007/s00213-009-1621-5 [DOI] [PubMed] [Google Scholar]
- Fiume R., Stijf-Bultsma Y., Shah Z. H., Keune W. J., Jones D. R., Jude J. G., et al. (2015). PIP4K and the role of nuclear phosphoinositides in tumour suppression. Biochim. Biophys. Acta 1851 898–910. 10.1016/j.bbalip.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Greaves M. (2006). Infection, immune responses and the aetiology of childhood leukaemia. Nat. Rev. Cancer 6 193–203. 10.1038/nrc1816 [DOI] [PubMed] [Google Scholar]
- Gupta A., Toscano S., Trivedi D., Jones D. R., Mathre S., Clarke J. H., et al. (2013). Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates TOR signaling and cell growth during Drosophila development. Proc. Natl. Acad. Sci. U.S.A. 110 5963–5968. 10.1073/pnas.1219333110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach T., Kohlmann A., Wieczorek L., Basso G., Kronnie G. T., Bene M. C., et al. (2010). Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the international microarray innovations in leukemia study group. J. Clin. Oncol. 28 2529–2537. 10.1200/JCO.2009.23.4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleman A., Cheok M. H., den Boer M. L., Yang W., Veerman A. J., Kazemier K. M., et al. (2004). Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N. Engl. J. Med. 351 533–542. 10.1056/NEJMoa033513 [DOI] [PubMed] [Google Scholar]
- Hou Q., Liao F., Zhang S., Zhang D., Zhang Y., Zhou X., et al. (2017). Regulatory network of GATA3 in pediatric acute lymphoblastic leukemia. Oncotarget 8 36040–36053. 10.18632/oncotarget.16424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jones D. R., Foulger R., Keune W. J., Bultsma Y., Divecha N. (2013). PtdIns5P is an oxidative stress-induced second messenger that regulates PKB activation. FASEB J. 27 1644–1656. 10.1096/fj.12-218842 [DOI] [PubMed] [Google Scholar]
- Jude J. G., Spencer G. J., Huang X., Somerville T. D., Jones D. R., Divecha N., et al. (2015). A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene 34 1253–1262. 10.1038/onc.2014.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Chen I. M., Wilson C. S., Bedrick E. J., Harvey R. C., Atlas S. R., et al. (2010). Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood 115 1394–1405. 10.1182/blood-2009-05-218560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner-Schwabe R., Lottaz C., Todling J., Rhein P., Karawajew L., Eckert C., et al. (2006). Expression of late cell cycle genes and an increased proliferative capacity characterize very early relapse of childhood acute lymphoblastic leukemia. Clin. Cancer Res. 12 4553–4561. 10.1158/1078-0432.CCR-06-0235 [DOI] [PubMed] [Google Scholar]
- Lamia K. A., Peroni O. D., Kim Y. B., Rameh L. E., Kahn B. B., Cantley L. C. (2004). Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta-/- mice. Mol. Cell. Biol. 24 5080–5087. 10.1128/MCB.24.11.5080-5087.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F., Yin D., Zhang Y., Hou Q., Zheng Z., Yang L., et al. (2016). Association between PIP4K2A polymorphisms and acute lymphoblastic leukemia susceptibility. Medicine (Baltimore) 95:e3542. 10.1097/MD.0000000000003542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorini G., Fiege B., Hosking F. J., Ma Y., Kumar R., Sherborne A. L., et al. (2013). Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood 122 3298–3307. 10.1182/blood-2013-03-491316 [DOI] [PubMed] [Google Scholar]
- Niu N., Schaid D. J., Abo R. P., Kalari K., Fridley B. L., Feng Q., et al. (2012). Genetic association with overall survival of taxane-treated lung cancer patients – A genome-wide association study in human lymphoblastoid cell lines followed by a clinical association study. BMC Cancer 12:422. 10.1186/1471-2407-12-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E., Hosking F. J., Vijayakrishnan J., Price A., Olver B., Sheridan E., et al. (2009). Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 41 1006–1010. 10.1038/ng.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Andreu V., Roberts K. G., Harvey R. C., Yang W., Cheng C., Pei D., et al. (2013). Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat. Genet. 45 1494–1498. 10.1038/ng.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Andreu V., Roberts K. G., Xu H., Smith C., Zhang H., Yang W., et al. (2015). A genome-wide association study of susceptibility to acute lymphoblastic leukemia in adolescents and young adults. Blood 125 680–686. 10.1182/blood-2014-09-595744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui C. H., Evans W. E. (2006). Treatment of acute lymphoblastic leukemia. N. Engl. J. Med. 354 166–178. 10.1056/NEJMra052603 [DOI] [PubMed] [Google Scholar]
- Rameh L. E., Tolias K. F., Duckworth B. C., Cantley L. C. (1997). A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature 390 192–196. 10.1038/36621 [DOI] [PubMed] [Google Scholar]
- Schmitz N. M., Leibundgut K., Hirt A. (2005). CDK2 catalytic activity and loss of nuclear tethering of retinoblastoma protein in childhood acute lymphoblastic leukemia. Leukemia 19 1783–1787. 10.1038/sj.leu.2403900 [DOI] [PubMed] [Google Scholar]
- Sherborne A. L., Hosking F. J., Prasad R. B., Kumar R., Koehler R., Vijayakrishnan J., et al. (2010). Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat. Genet. 42 492–494. 10.1038/ng.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorich M. J., Pottier N., Pei D., Yang W., Kager L., Stocco G., et al. (2008). In vivo response to methotrexate forecasts outcome of acute lymphoblastic leukemia and has a distinct gene expression profile. PLoS Med. 5:e83. 10.1371/journal.pmed.0050083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock W., Tsai T., Golden C., Rankin C., Sher D., Slovak M. L., et al. (2000). Cell cycle regulatory gene abnormalities are important determinants of leukemogenesis and disease biology in adult acute lymphoblastic leukemia. Blood 95 2364–2371. [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43(Database issue) D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino L. R., Yang W., French D., Hunger S. P., Carroll W. L., Devidas M., et al. (2009). Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 41 1001–1005. 10.1038/ng.432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian J., Rao A. A., Nothaft F. A., Ketchum C., Armstrong J., Novak A., et al. (2017). Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 35 314–316. 10.1038/nbt.3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K. M., de Smith A. J., Chokkalingam A. P., Metayer C., Dahl G. V., Hsu L. I., et al. (2013). Novel childhood ALL susceptibility locus BMI1-PIP4K2A is specifically associated with the hyperdiploid subtype. Blood 121 4808–4809. 10.1182/blood-2013-04-495390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox A., Hinchliffe K. A. (2008). Regulation of extranuclear PtdIns5P production by phosphatidylinositol phosphate 4-kinase 2alpha. FEBS Lett. 582 1391–1394. 10.1016/j.febslet.2008.03.022 [DOI] [PubMed] [Google Scholar]
- Xu H., Cheng C., Devidas M., Pei D., Fan Y., Yang W., et al. (2012). ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J. Clin. Oncol. 30 751–757. 10.1200/JCO.2011.38.0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Yang W., Perez-Andreu V., Devidas M., Fan Y., Cheng C., et al. (2013). Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J. Natl. Cancer Inst. 105 733–742. 10.1093/jnci/djt042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhang H., Yang W., Yadav R., Morrison A. C., Qian M., et al. (2015). Inherited coding variants at the CDKN2A locus influence susceptibility to acute lymphoblastic leukaemia in children. Nat. Commun. 6:7553. 10.1038/ncomms8553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S. L., Payne-Turner D., et al. (2012). The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481 157–163. 10.1038/nature10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets analyzed for this study are available in GEO repository (http://www.ncbi.nlm.nih.gov/gds): GSE635, GSE10792, GSE13351, GSE13425, GSE10255, GSE4698, GSE33315, GSE7440, GSE11877, and GSE13204, and TCGA database (https://cancergenome.nih.gov/).