Abstract
OBJECTIVE
Previously generated genetic risk scores (GRSs) for type 1 diabetes (T1D) have not captured all known information at non-HLA loci or, particularly, at HLA risk loci. We aimed to more completely incorporate HLA alleles, their interactions, and recently discovered non-HLA loci into an improved T1D GRS (termed the “T1D GRS2”) to better discriminate diabetes subtypes and to predict T1D in newborn screening studies.
RESEARCH DESIGN AND METHODS
In 6,481 case and 9,247 control subjects from the Type 1 Diabetes Genetics Consortium, we analyzed variants associated with T1D both in the HLA region and across the genome. We modeled interactions between variants marking strongly associated HLA haplotypes and generated odds ratios to create the improved GRS, the T1D GRS2. We validated our findings in UK Biobank. We assessed the impact of the T1D GRS2 in newborn screening and diabetes classification and sought to provide a framework for comparison with previous scores.
RESULTS
The T1D GRS2 used 67 single nucleotide polymorphisms (SNPs) and accounted for interactions between 18 HLA DR-DQ haplotype combinations. The T1D GRS2 was highly discriminative for all T1D (area under the curve [AUC] 0.92; P < 0.0001 vs. older scores) and even more discriminative for early-onset T1D (AUC 0.96). In simulated newborn screening, the T1D GRS2 was nearly twice as efficient as HLA genotyping alone and 50% better than current genetic scores in general population T1D prediction.
CONCLUSIONS
An improved T1D GRS, the T1D GRS2, is highly useful for classifying adult incident diabetes type and improving newborn screening. Given the cost-effectiveness of SNP genotyping, this approach has great clinical and research potential in T1D.
Introduction
Type 1 diabetes (T1D) involves autoimmune destruction of insulin-producing pancreatic β-cells. While prominent in childhood, it may present at any age (1). Measurement of islet autoantibodies (AAb) in venous blood can reveal active disease years before the clinical diagnosis (2). Early, preclinical identification of T1D can minimize morbidity and medical costs at clinical onset and might facilitate prevention therapy (3). However, AAb surveillance is expensive and difficult in young children. Better selection of infants at high risk will improve the cost-effectiveness of any prediction program (3–5). Further, AAb measurement in incident cases of adult diabetes is not sufficiently sensitive (6) or specific (7) for accurate classification of diabetes subtype, which is important for appropriate treatment (8–10).
T1D has a largely heritable component, evidenced by a twin concordance rate of up to 70% (11) and a sibling risk of ∼8% (12). The majority of risk is explained by variation at a few very strongly associated loci including the HLA class 2 DR-DQ loci, the HLA class 1 region, and >50 associated non-HLA single nucleotide polymorphisms (SNPs) including INS and PTNP22 (12). This high genetic heritability creates the potential for powerful diagnostic discrimination if the majority of genetic risk for T1D can be captured (13,14). We and others have shown that incorporation of multiple T1D risk loci into an integrated genetic risk score (GRS) is more powerful than HLA DR-DQ genotyping alone in the discrimination of T1D, with scores incorporating 10–40 SNPs demonstrating a discriminative receiver operating characteristic (ROC) area under the curve (AUC) of 0.84–0.87 (4,15–17). Additionally, the simplification of T1D genetic risk as a continuous variable allows non-HLA scientists and clinicians much easier access to T1D genetic information for a variety of clinical and research uses.
To date, published GRSs have not fully captured all information on risk of T1D. HLA class 2 DR-DQ haplotypes (at the IDDM1 locus) are by far the most important factor in heritable T1D risk (18,19). Full assessment of HLA DR-DQ risk is complex owing to the association of many common haplotypes with T1D and the nonadditive interactions between them (19–21). Our original T1D GRS (16), herein referred to as T1D GRS1, included the susceptible HLA haplotypes DR3-DQ2 and DR4-DQ8 and the resistant haplotype DR15-DQ6 but missed many other DR-DQ haplotypes important in T1D genetic risk or protection. Advances in the density of SNP arrays combined with ever larger sets of reference data (such as the 1000 Genomes Project) have led to improvements in the accuracy of both genome-wide and HLA imputation, thereby allowing for more precise measurement of HLA-associated T1D risk. Better tagging of HLA loci using SNPs that can be cheaply and rapidly genotyped offers the possibility of capturing T1D genetic risk associated with HLA DR-DQ more comprehensively into an improved T1D GRS.
In this study, we aimed to improve SNP capture of HLA DR-DQ risk, including haplotype interactions. We also aimed to improve the GRS by capture of additional non–DR-DQ loci via additional SNPs found to be associated with T1D risk. We generate a new T1D GRS, which we term the “T1D GRS2,” with this information. We assess the additional discriminative power from this approach, demonstrate how the improvement might enhance clinical applicability, and create a framework within which different GRSs may be compared.
Research Design and Methods
We used HLA and genome-wide imputation of a large T1D case-control data set (22) to assess and improve capture of HLA and non-HLA T1D-associated loci into an improved GRS, the T1D GRS2. We first focused on IDDM1, the HLA DR-DQ locus characterized by strong linkage disequilibrium and unusually frequent polymorphisms. The extensive HLA T1D literature with published odds ratios were used to select, among common DR-DQ haplotypes, those that conferred significant risk for or protection against T1D. This list comprised many more haplotypes than those included in previously published T1D GRS. To determine HLA haplotypes in our samples, we imputed HLA alleles and used correlation statistics to identify tagging SNP variants. We further analyzed the relationship of specific combinations of two DR-DQ haplotypes (haplogenotypes). We next analyzed the impact of HLA alleles outside of DR-DQ and of additional published T1D risk loci across the rest of the genome. Finally, we combined variants into a GRS and validated our results using the approach shown in Supplementary Fig. 1. To differentiate from our original T1D GRS (16), we refer to the score based on the new combination of loci and haplotypes as T1D GRS2.
Research Subjects: Type 1 Diabetes Genetic Consortium
To investigate genetic associations, we used the Type 1 Diabetes Genetics Consortium (T1DGC) ImmunoChip case-control genetic data with ∼164,000 genotyped variants (22). The classification of T1D was made based on clinical features previously described (22). The T1DGC data consisted of a collection of white Caucasian case and control subjects (6,670 T1D case and 9,416 control subjects) combined from previous studies such as the U.K. Wellcome Trust Case Control Consortium (WTCCC). Age at clinical diagnosis among case subjects ranged from <1 year to a maximum age of 16 years. The mean age of diagnosis was 7.75 years.
Research Subjects: UK Biobank
To validate our results, we used the UK Biobank Affymetrix Axiom Array data with ∼821,000 variants genotyped in a subset of ∼374,000 individuals identified as European Caucasian by genetic clustering methods (23). Case subjects were defined by a strict set of criteria favoring specificity among selected individuals for T1D rather than sensitivity to include all T1D case subjects:
Clinical diagnosis of diabetes at ≤20 years of age
On insulin within 1 year from the time of diagnosis
Still on insulin at the time of recruitment
Not using oral antihyperglycemic agents
Did not ever self-report as having type 2 diabetes (T2D)
The total number of T1D case subjects fitting these criteria was 387. We also identified 11,885 cases of T2D by a similarly strict definition:
Clinical diagnosis of diabetes at age ≥35 years
Not on insulin within 1 year from the point of diagnosis
Did not ever self-report as having T1D
Excluded if diagnosed within last 12 months as unable to confirm insulin-free for minimum of 12 months
Imputation
We used the 1000 Genomes Project reference panel to impute a total of 80.6 million variants in the T1DGC discovery data. We used data that were centrally imputed by UK Biobank to a combination of the Haplotype Reference Consortium (HRC) panel and the UK10K plus 1000 Genomes Project panels (23). To impute HLA alleles in the T1DGC data set, we used SNP2HLA (24) with the T1DGC reference panel consisting of 5,225 SNP-genotyped and HLA-genotyped North Americans. We imputed a total of 424 alleles at the following loci: HLA-A/B/C/DQA1/DRB1/DQB1/DPB1. Alleles were imputed to four-digit resolution, and average imputation accuracy was high (mean imputation r2 = 0.977).
HLA DRB1-DQA1-DQB1 Haplotypes
Using imputed T1DGC data, we identified every possible combination of DRB1, DQA1, and DQB1 alleles occurring on the same haplotype. Each haplotype present in ≥0.5% of case and control subjects was evaluated as a predictor of T1D in a logistic regression model adjusted for every possible haplotype. Common haplotypes not observed to be independently associated (P > 0.05) but with suspected interaction effects were also included in further analysis. We used HLA imputation data from a subset of ∼128,000 UK Biobank samples to identify variants that would best mark these haplotypes from their correlation r2 and d′ statistics.
Interaction Modeling
To assess whether interactions existed between 136 possible DR-DQ haplotype pairings (haplogenotypes), we generated multiplicative interaction terms from genotype dosage values. Logistic regression, with the interaction term as the independent variable and each haplotype of the pairing included as a covariate, was used to test the strength of interaction. A Bonferroni-corrected cutoff (P < 3.7 × 10−4) was applied. Interaction terms below the P value threshold identified haplotype combinations with evidence of interaction.
Additional Loci
A total of 323 imputed alleles from HLA loci outside DR-DQ were tested in a logistic regression adjusting for the DR-DQ haplotypes previously identified—this was necessary, as long-range linkage is well-known to occur in the HLA region between class I, class II, and class III loci. A Bonferroni-corrected threshold was applied (P < 1.6 × 10−4). HLA alleles that remained significant after adjustments for both DR-DQ and for multiple testing were added to the model. Outside of the HLA region, we included variants from our genome-wide association studies (GWAS) of the T1DGC data in addition to variants identified in previously published GWAS.
Generating the GRS
We took the natural log of each odds ratio identified in the discovery data to generate a β-statistic for each pair of haplotype combinations and each allele. For haplogenotypes with an identified interaction, each person was assigned a single score at the DR-DQ locus. For haplogenotypes with no interaction identified, the score was generated from the sum of each allele weighted by its beta. The remaining loci were then added to the total score by multiplying the number of risk alleles by the β for each variant. Odds ratios and βs were coded to correspond with the minor allele for HLA loci, as this tagged the presence of an allele subtype. Positive β-values therefore imply a risk-increasing effect from an allele and negative values imply a risk-decreasing effect. An equation summarizing generation of the score is presented in Supplementary Fig. 2.
Statistical Methods
Logistic regression, with T1D as the outcome and variants of interest as independent variables, was used in discovery data to identify and validate associations and generate odds ratios. Covariates were included as described above.
A linear mixed model, as implemented in BOLT-LMM (25), was used to perform GWAS of all imputed variants in the discovery data with T1D status as the outcome. Covariates included were principal components and sex. Variants from T1D GRS1 were included in further conditional analysis. Simulations of population screening performance in both GRS and HLA typing were based on an assumed T1D prevalence of 0.3% and sensitivity and specificity derived from the UK Biobank data set.
We tested the ability of the GRS to discriminate case from control subjects in T1DGC and replicated our findings in UK Biobank by using the AUC of the ROC. ROC statistics were generated with the DeLong algorithm. The Youden index was calculated as j = sensitivity + specificity − 1. To compare the discriminative power of different GRSs, we used a χ2 test on a subset containing all T1D case subjects and 24,000 randomly selected control subjects. Bonferroni correction was used where appropriate to account for multiple testing.
Results
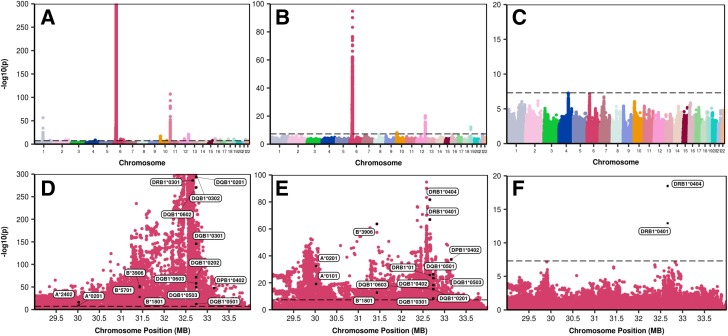
Genome-wide association analysis of T1DGC case and control subjects identified additional significant T1D genetic associations not captured by T1D GRS1 both within and outside of HLA (Fig. 1B and E). We assembled genetic components into a new risk score, the T1D GRS2, grouping them as 1) HLA DRB1-DQA1-DQB1 alleles, 2) genes in the HLA region but distinct from DR-DQ, and 3) genes outside the HLA region. Our approach to generation of the T1D GRS2 is summarized in Supplementary Figs. 1 and 2.
Figure 1.
Manhattan plots of GWAS before and after adjustment for GRSs. Top row, GWAS: unadjusted (A), after adjustment for GRS1 (B), and after adjustment for GRS2 (C). Bottom row, high-resolution plot of association scores for the full HLA region on chromosome 6q: unadjusted (D), after adjustment for GRS1 (E), and after adjustment for GRS2 (F). MB, megabases.
Improved Capture of HLA Risk
We identified tag SNPs for 14 HLA DQA1-DQB1 haplotypes (11 more than for T1D GRS1) associated with T1D in the T1DGC case-control cohort (Supplementary Table 1). We chose to study SNP variants linked to DQ haplotypes rather than DRB1 for three reasons. First, DQ has a higher impact on T1D risk than DR. Second, for most DQ haplotypes each was linked to only a single DRB1 allele in T1DGC anyway. Third, owing to low frequency in the imputation panel, we were unable to discriminate all DRB1 subtypes known to impact T1D risk, such as DRB1*04:0X. Once DQ haplotypes were identified, we sought to detect and use their interactions to refine our estimates of T1D risk. Supplementary Table 2 illustrates a heat map of DQ haplotype combination (haplogenotypes) odds ratios based on imputed DQ haplotypes in T1DGC. From DQ tag SNPs, we identified 18 DQ haplotypes with significant interaction terms and generated interaction-specific odds ratios (Supplementary Table 3). For less common haplogenotypes with insufficient data to estimate interaction odds ratios (n < 50 case subjects in T1DGC) or where no significant interaction was identified, we generated odds ratios for the individual haplotypes in remaining case-control data (Supplementary Table 1). This approach resulted in an HLA class II (IDDM1) component of the GRS derived from 14 HLA SNPs covering most major T1D-associated class II haplotypes and specific odds ratio terms for 18 haplogenotype combinations.
We identified 21 further HLA region SNPs not representing DR-DQ haplotypes that each associated independently with T1D (Supplementary Table 4). Some mark known T1D-associated alleles at class I A, B or C, or at class II DP, while others were not correlated with any specific HLA gene but, rather, were located near the XL9 regulatory region between DRB1 and DQA1 or in presumed regulatory regions near DRA1 or BTNL2 (26,27).
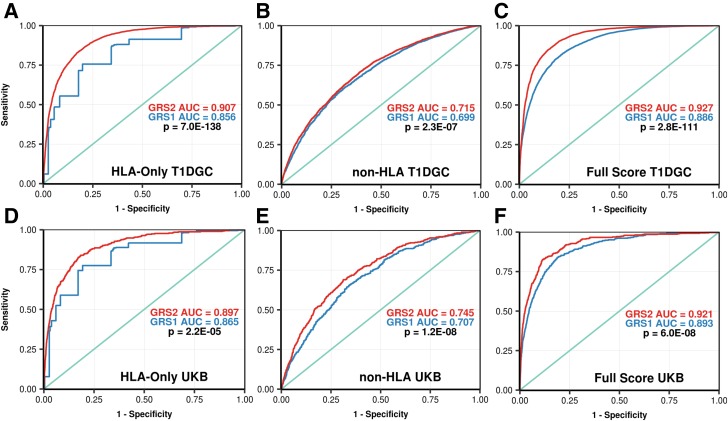
The combined model of 35 total HLA-region SNPs was much more discriminative of T1D than the 5-SNP model in T1D GRS1 in both the T1DGC discovery data set (ROC AUC 0.907 vs. 0.856; P < 0.0001) and UK Biobank validation set (ROC AUC 0.897 vs. 0.865; P < 0.0001) (Fig. 2A and D).
Figure 2.
ROC curves comparing the ability of GRS1 and GRS2, and their components, to discriminate T1D. For each pair of curves in a panel, ROC AUCs are shown as well as the P value for their comparison by the DeLong algorithm. A: HLA DR-DQ haplotypes and interaction terms for GRS1 vs. GRS2 in the T1DGC. B: Both non–DR-DQ HLA and non-HLA loci for GRS1 vs. GRS2 in the T1DGC. C: Full GRS1 vs. full GRS2 in the T1DGC discovery data set. D: HLA DR-DQ haplotypes and interaction terms, for GRS1 vs. GRS2, in the UK Biobank (UKB). E: Both non–DR-DQ HLA and non-HLA loci, for GRS1 vs. GRS2, in UK Biobank. F: Full GRS1 vs. full GRS2 in UK Biobank.
Improved Capture of Non-HLA Genetic Risk
We then identified 32 non-HLA loci (summarized in 28) that each, again, associated independently with T1D (Supplementary Table 5). Taken together, these non-HLA SNPs improved discrimination of T1D compared with GRS1 in the T1DGC discovery set (ROC AUC 0.715 vs. 0.699; P < 0.001) and in the validation data set (ROC AUC 0.75 vs. 0.707; P < 0.001) (Fig. 2E). The improved discrimination was most clearly observed in the validation data owing to the denser SNP array and better imputation of non-HLA variants.
T1D GRS2, Using 67 SNPs, Offers Significantly Improved Prediction of T1D
The final combined T1D GRS2 used a total of 67 SNPs (14 DR-DQ, 21 other HLA, and 32 non-HLA SNPs) and showed markedly improved discrimination of T1D versus T1D GRS1. The ROC AUC increased from 0.886 to 0.927, P < 0.0001, in the T1DGC discovery data set and from 0.893 to 0.921 in the UK Biobank validation P < 0.0001 (Fig. 2C and F). Genome-wide association analysis of the T1DGC case and control subjects using T1D GRS2 as a conditional variable demonstrated little remaining genome-wide significant SNP associations, indicating vastly improved capture of T1D-associated information (Fig. 1).
Assessment of T1D GRS Against Diabetes Type
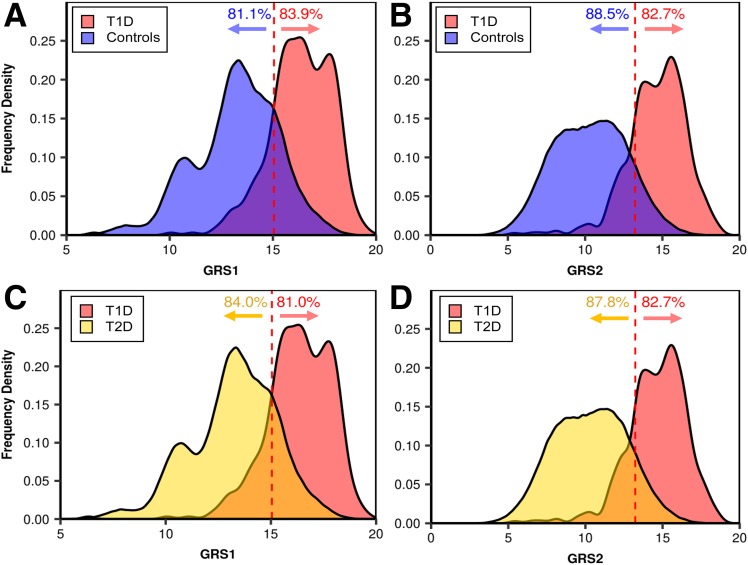
We then compared how well GRS1 and the new T1D GRS2 separated those with known T1D from the background population in UK Biobank. For example, using the optimal T1D GRS2 cutoff (at maximum Youden index) allowed inclusion of 82.7% of patients with T1D patients with 88.5% of the background population properly excluded (Fig. 3B). In a similar exercise comparing patients with T1D versus patients with T2D, T1D GRS2 alone excluded 87.8% of patients with T2D but included 82.7% of patients with T1D (Fig. 3D). Both represent substantially increased specificity at similar sensitivity versus T1D GRS1 (Fig. 3A and C).
Figure 3.
A and B: Comparison of how well GRS1 (A) and GRS2 (B) discriminate those with T1D from control subjects. C and D: Comparison of how well GRS1 (C) and GRS2 (D) discriminate those with T1D from those with T2D. Note the similar distribution of T1D GRS2 scores in the background population and in those with T2D.
Using the T1D GRS2 to Predict Future T1D Among Infants
Simulations show that the T1D GRS2 could be used for newborn screening to select babies requiring follow-up for AAb surveillance (Table 1). Individuals with a T1D GRS2 >90th centile in a general population represent >77% of cases of future T1D, with a T1D risk of 2.4%; those with a score >99.9th centile have a T1D risk >20%, but this cutoff would only identify 7% of future T1D cases. Comparative simulations of general population screening demonstrated T1D GRS2 prediction performance to be much better than that of T1D GRS1 (Table 1) but, most importantly, more than twice as good (less than half as many children requiring AAb surveillance) compared with prior methods using only HLA DR-DQ (Table 1).
Table 1.
Simulated population-based prediction of T1D using HLA screening, the original T1D GRS, and the T1D GRS2
| T1D centile* | Population centile** | GRS2 | Specificity (%) | Sensitivity (%) | 1-Specificity (%) | Youden index (j) | T1D risk (%)*** |
|---|---|---|---|---|---|---|---|
| 5 | 70.2 | 11.68 | 69.5 | 94.8 | 30.5 | 0.643 | 0.9 |
| 10 | 79.4 | 12.36 | 78.9 | 89.4 | 21.1 | 0.683 | 1.3 |
| 25 | 90.6 | 13.45 | 90.4 | 77.5 | 9.6 | 0.679 | 2.4 |
| 50 | 96.8 | 14.60 | 96.7 | 53.7 | 3.3 | 0.505 | 4.7 |
| 75 | 99.1 | 15.65 | 99.1 | 30.2 | 0.9 | 0.293 | 9.1 |
| 90 | 99.8 | 16.54 | 99.8 | 13.2 | 0.2 | 0.130 | 15.7 |
| 95 |
99.9 |
17.06 |
99.9 |
7.2 |
0.1 |
0.072 |
22.8 |
| T1D centile* |
Population centile** |
GRS1 |
Specificity (%) |
Sensitivity (%) |
1-Specificity (%) |
Youden index (j) |
T1D risk (%)*** |
| 5 | 53.3 | 13.48 | 51.9 | 95.9 | 48.1 | 0.478 | 0.6 |
| 10 | 60.7 | 14.06 | 63.9 | 92.5 | 36.1 | 0.564 | 0.8 |
| 25 | 73.8 | 15.07 | 81.7 | 82.9 | 18.3 | 0.646 | 1.3 |
| 50 | 86.1 | 16.16 | 94.0 | 56.3 | 6.0 | 0.503 | 2.7 |
| 75 | 93.3 | 17.17 | 98.3 | 32.6 | 1.7 | 0.308 | 5.4 |
| 90 | 96.5 | 17.83 | 99.5 | 18.3 | 0.5 | 0.178 | 9.9 |
| 95 |
97.8 |
18.19 |
99.8 |
9.0 |
0.2 |
0.088 |
11.8 |
| T1D centile* |
Risk category |
HLA type |
Specificity (%) |
Sensitivity (%) |
1-Specificity (%) |
Youden index (j) |
T1D risk (%)*** |
| — | Background | Other | 0.0 | 100.0 | 0.0 | 0.000 | 0.3 |
| 57.0 | Moderate | DR3/3, DR4/X | 79.1 | 77.0 | 23.0 | 0.561 | 0.6 |
| 81.1 | High | DR4/4 | 96.3 | 41.3 | 58.7 | 0.376 | 2.5 |
| 84.5 | Very High | DR3/4 | 97.2 | 37.0 | 63.1 | 0.342 | 3.8 |
Risk of T1D is calculated assuming a 0.3% population prevalence of T1D.
*T1D cases in T1DGC.
**Centile in UK Biobank European population.
***Risk of T1D is calculated assuming a 0.3% population prevalence of T1D.
Assessment of T1D GRS2 Against Age of Diagnosis
Stronger genetic risk of T1D is known to associate with younger age of diagnosis (17,29). We correlated the T1D GRS2 with age of diabetes diagnosis in the T1DGC data (Supplementary Fig. 3) and demonstrated improved performance at very young onset ages (P = 7 × 10−44, Pearson correlation). The effect appeared to be explained by greater HLA risk in those children (HLA only, P = 6 × 10−39; non-HLA, P = 1 × 10−4).
Performance in Comparison With Other Genetic Scores
To date, a variety of genetic scores have been described and used in T1D. Genetic scores vary according to SNPs available from array data, cost and sample size limitations within a study, and the methods of score calculation. Through analysis of different scores against a reference population (either a background population or a cohort with T1D) it is possible to compare different scores based on centiles in the reference set to compare the sensitivity and specificity of each score. In Supplementary Table 6, the Youden index is used to compare performance of several recently published GRSs used in T1D. A Youden index of 1 describes perfect discrimination at a particular threshold. The T1D GRS2 had the highest Youden index (0.698) across a range of centiles and the highest ROC AUC (P < 0.0001 vs. all scores). The Youden index value as UK Biobank centile varies is plotted for multiple GRSs in Supplementary Fig. 2.
Conclusions
We combined 67 SNPs into an improved T1D GRS termed T1D GRS2. With improved capture of HLA DR-DQ risk, HLA interactions, and additional non-HLA SNPs, the T1D GRS2 included all independent genome-wide significant T1D risk from T1DGC ImmunoChip data. When directly compared with other genetic scores used to date, T1D GRS2 significantly improved discrimination of those with T1D from those with T2D and control subjects. Indeed, while all published T1D GRSs, derived by a variety of methods, have had ROC AUCs of ≥0.86 (4,15–17), GRS2 yielded the greatest T1D discrimination, with an ROC AUC of 0.93 in the T1DGC data set. This in large part resulted from inclusion of many more HLA features in GRS2 compared with prior scores (4,15,16).
We used the T1D GRS2 to compare the proportion of screened newborn infants in a population who must be followed after screening to capture differing proportions of future T1D cases. For example, the detection of 77% of future cases would require following 20.9% of screened infants using only HLA DR-DQ selection criteria and 14.3% of infants using our T1D GRS1 but only 9.5% of infants using T1D GRS2 (Table 1 and Supplementary Table 6), representing a major cost savings. Each iteration represents a critical improvement in cost-effectiveness for general population pediatric T1D prediction strategies. More stringent cutoff points can identify pediatric populations at very high risk who are suitable for trials of primary prevention therapies, such as the 0.1% of a population with >10% absolute disease risk. This approach is already being tested (4), and improved selection of individuals at high risk using scores like T1D GRS2 will only enhance these efforts.
Discrimination of T1D from T2D cases can help classify incident diabetes cases in adulthood, where incorrect classification is common and can lead to incorrect treatment, greater medical costs, and greater morbidity (8,10). T1D GRS2 improved discrimination between incident T1D and T2D case subjects to a degree similar to that between T1D and healthy control subjects (Fig. 3). This is expected owing to the very minimal genetic overlap between T1D and T2D. Our previous findings (16) suggest that a combined model using onset age, AAb status, and BMI will improve discrimination of incident cases much further. A similar approach of combining genetic and nongenetic risk successfully predicted progression rate to T1D in at-risk relatives in the TrialNet Pathway to Prevention Study (combining AAb data, the T1D GRS1, age, and a metabolic score) (30). A logical next step is to use the T1D GRS2 in similar settings to assess discriminative power in combination with these additional predictors of T1D.
The T1D GRS2 was most predictive of T1D in very early life, consistent with existing genetic data (17,29). For very young ages, there are less additional risk data available (such as AAb and BMI) to combine with genetic risk; yet, unexpected T1D onset can carry particularly high morbidity and even mortality (31). A powerful GRS may be clinically useful in this setting to identify people at risk for mortality and morbidity from T1D in early childhood. Our estimate of cases captured assumes equivalent genetic risk across all childhood, and T1D GRS2 may be even more sensitive and specific to identify patients at risk of very young T1D onset.
Our T1D GRS2 development approach captured multiple HLA effects. Previous T1D genetic scores captured the commonest and best described HLA class 2 DR3-DQ2 and DR4-DQ8 risk alleles (4,15,16), a single protective allele (DR15-DQ6) (16), and two class 1 alleles (HLA A*24 and HLA B*57) (4,16). We now capture many more of the DR-DQ haplotypes, nearby regulatory regions, more class I alleles, and HLA DP alleles that confer susceptibility or resistance to T1D (20,32). We worked to ensure that GRS2 captured interaction effects between these haplotypes. Complex interactions in the class 2 HLA region have previously been described (21) but have not been integrated into methods for T1D prediction. We also discovered T1D-associated HLA region SNPs not tracking classic HLA alleles but in intergenic regulatory regions between DRB1 and DQA1 that regulate class II gene expression, e.g., near XL9 and BTNL2. These have been described in the context of systemic lupus erythematosus (26) and sarcoidosis (27) but are not well characterized in T1D. Conditional approaches similar to ours may yield further mechanistic insights; however, investigating these further is outside the scope of this work. Future more dense arrays, sequencing, and analyses of gene expression are likely to better characterize these regions, their interactions, and potential mechanisms. Finally, T1D GRS2 covered many more HLA class I and HLA DP alleles known to affect T1D risk (32). One limitation was that we struggled to capture DRB1*04 subtypes. These are currently not well imputed, and we were unable to find SNP tags that distinguished the high-risk DR4 alleles from protective DRB1*04:03 and DRB1*04:07 alleles on the DQA1*03:01–DQB1*03:02 background. Use of more dense SNP arrays and better imputation reference sets to achieve this goal may improve future GRS performance.
The amount of genetic information incorporated into a T1D GRS that can be easily used in the clinic and for research must necessarily represent a balance between effective cost of implementation and maximization of genetic information. The most genetic information could theoretically be obtained from using whole-genome sequencing, which is currently not feasible, or from genomic risk scores that use all genetic association information, unfiltered for genome-wide association significance. Genome-wide risk scores have recently been very effective at improving prediction of non–HLA-linked disease (33), but the extremely strong HLA bias of T1D heritability, and the inability of these models to currently analyze the HLA region, means that these methods are less suitable for T1D prediction. At the other end of the spectrum, SNP-based GRSs can vary from containing all associated T1D SNPs to containing smaller numbers of SNPs, such as the previously described 10-SNP version of GRS1 (16). We believe that while it is difficult to envisage a whole-genome sequencing approach to population-wide screening, the falling cost of SNP typing and the very strong association of T1D with SNPs in the HLA region make a more comprehensive GRS a practical choice for use in such population-based research and public health settings.
A limitation of T1D GRS2 is its use of genetic information discovered and validated in European Caucasian cohorts. We currently lack large, well-described, case-control cohorts of other ethnicities. Initial data from Perry et al. (17) and others suggest that the T1D GRS2 will be discriminative in Hispanics, but possibly less so in Africans, although our own unpublished work (R.A.O., M.N.W., and C.S. Yajnik, personal communication) suggests that the score works well in South Asian populations. Larger data sets examining genetic associations in these populations are required to fully define the utility of GRSs in prediction and classification of T1D. Additionally, it is possible that even in populations of similar ethnic background, different environments might mediate different genetic associations, requiring score adjustment. However, there is currently little evidence for strong gene-environment effects in T1D. These limitations of stratification, and the assumptions made when summing genetic risk into a score, must be weighed against ease of use and translatability. A single T1D GRS2, not fully adjusting for ethnicity or environmental interactions, may capture most genetic risk while avoiding the increased complexity necessary in more customized GRSs. We and others are working to develop and improve GRSs for use in diverse populations customized for ethnicity and race (17,34).
A further limitation of our T1D GRS2 is its origin in T1DGC ImmunoChip data that is itself limited in SNP coverage and density. As increasingly powerful and up-to-date SNP arrays become more widely used, and broader reference data sets such as TOPMed (Trans-Omics for Precision Medicine) (35) allow higher-quality imputation, the sensitivity to more directly identify causal variants, rare T1D risk alleles, and complex interactions will likely improve GRSs even further. However, the step change in improved prediction in our study is using new information from the most important region in T1D genetics, and the current predictive power of the T1D GRS2 suggests that there is relatively less predictive information remaining to be captured by future efforts. If we assume that conservative estimates of T1D heritability (e.g., sibling risk ratio of ∼6) are correct (14), then we will have explained the vast majority of T1D heritability with current knowledge and there will be little scope for improvement with future genetic discovery. However, the only way to know this is with future sequencing studies and studies of lower-frequency variants that are not well covered in current GWAS.
As genotyping costs fall and our knowledge of T1D genetic risk increases, additional T1D GRSs will be developed and tested. These might vary in methods of generation, for example, where SNPs available for score generation are different, where costs prohibit a full T1D GRS2 from being calculated, or where different populations are used for discovery. It is important to be able to directly compare scores to plan future use and research. A simple but robust approach for this is to compare the variety of published genetic scores for T1D that can be directly compared using score distributions in a reference population. We believe that such direct comparisons will aid the future development and application of T1D GRSs.
Supplementary Material
Article Information
Funding. S.A.S. is supported by a Diabetes UK PhD studentship (17/0005757). W.A.H. is supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant U01-DK-063829. S.S.R. is supported by the National Institutes of Health (NIH) (R01-DK-096926) and a grant from the University of Virginia Strategic Investment Fund. D.A.S. is supported by NIH training grant 2T32-DK-00724. M.N.W. is supported by the Wellcome Trust Institutional Support Fund (WT097835MF). R.A.O. is supported by a Diabetes UK Harry Keen Fellowship (16/0005529). This research was performed under the auspices of the T1DGC, a collaborative clinical study sponsored by the NIDDK, National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, National Institute of Child Health and Human Development, and JDRF International.
The views expressed are those of the authors.
Duality of Interest. R.A.O. holds a U.K. Medical Research Council institutional Confidence in Concept grant to develop a 10-SNP biochip T1D genetic test in collaboration with Randox. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. S.A.S., M.N.W., W.A.H., and R.A.O. designed the study, contributed to analysis, and wrote the manuscript. A.R.W., S.E.J., R.N.B., J.W.H., D.A.S., J.M.L., and J.T. contributed to analysis and reviewed the manuscript. S.S.R. contributed to study design, provided access to ImmunoChip data from T1DGC, contributed to analysis, and reviewed the manuscript. All authors contributed to the discussion and reviewed or edited the manuscript. R.A.O. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the Diabetes UK Professional Conference, London, U.K., 14–16 March 2018, and the American Society of Human Genetics 2018 Annual Meeting, San Diego, CA, 16–20 October 2018.
Footnotes
References
- 1.Thomas NJ, Jones SE, Weedon MN, Shields BM, Oram RA, Hattersley AT. Frequency and phenotype of type 1 diabetes in the first six decades of life: a cross-sectional, genetically stratified survival analysis from UK Biobank. Lancet Diabetes Endocrinol 2018;6:122–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziegler AG, Rewers M, Simell O, et al. . Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 2013;309:2473–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziegler AG, Danne T, Dunger DB, et al. . Primary prevention of beta-cell autoimmunity and type 1 diabetes - The Global Platform for the Prevention of Autoimmune Diabetes (GPPAD) perspectives. Mol Metab 2016;5:255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifacio E, Beyerlein A, Hippich M, et al.; TEDDY Study Group . Genetic scores to stratify risk of developing multiple islet autoantibodies and type 1 diabetes: A prospective study in children. PLoS Med 2018;15:e1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Florez JC. Found in translation: a type 1 diabetes genetic risk score applied to clinical diagnosis. Diabetes Care 2016;39:330–332 [DOI] [PubMed] [Google Scholar]
- 6.Bingley PJ. Clinical applications of diabetes antibody testing. J Clin Endocrinol Metab 2010;95:25–33 [DOI] [PubMed] [Google Scholar]
- 7.Landin-Olsson M, Arnqvist HJ, Blohmé G, et al. . Appearance of islet cell autoantibodies after clinical diagnosis of diabetes mellitus. Autoimmunity 1999;29:57–63 [DOI] [PubMed] [Google Scholar]
- 8.de Lusignan S, Sadek N, Mulnier H, Tahir A, Russell-Jones D, Khunti K. Miscoding, misclassification and misdiagnosis of diabetes in primary care. Diabet Med 2012;29:181–189 [DOI] [PubMed] [Google Scholar]
- 9.Lasserson D, Fox R, Farmer A. Late onset type 1 diabetes. BMJ 2012;344:e2827. [DOI] [PubMed] [Google Scholar]
- 10.Hope SV, Wienand-Barnett S, Shepherd M, et al. . Practical classification guidelines for diabetes in patients treated with insulin: a cross-sectional study of the accuracy of diabetes diagnosis. Br J Gen Pract 2016;66:e315–e322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redondo MJ, Jeffrey J, Fain PR, Eisenbarth GS, Orban T. Concordance for islet autoimmunity among monozygotic twins. N Engl J Med 2008;359:2849–2850 [DOI] [PubMed] [Google Scholar]
- 12.Pociot F, Lernmark Å. Genetic risk factors for type 1 diabetes. Lancet 2016;387:2331–2339 [DOI] [PubMed] [Google Scholar]
- 13.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet 2010;6:e1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clayton DG. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genet 2009;5:e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler C, Krumsiek J, Buettner F, et al. . Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia 2014;57:2521–2529 [DOI] [PubMed] [Google Scholar]
- 16.Oram RA, Patel K, Hill A, et al. . A type 1 diabetes genetic risk score can aid discrimination between type 1 and type 2 diabetes in young adults. Diabetes Care 2016;39:337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry DJ, Wasserfall CH, Oram RA, et al. . Application of a genetic risk score to racially diverse type 1 diabetes populations demonstrates the need for diversity in risk-modeling. Sci Rep 2018;8:4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet 1996;59:1134–1148 [PMC free article] [PubMed] [Google Scholar]
- 19.Noble JA. Immunogenetics of type 1 diabetes: a comprehensive review. J Autoimmun 2015;64:101–112 [DOI] [PubMed] [Google Scholar]
- 20.Erlich H, Valdes AM, Noble J, et al.; Type 1 Diabetes Genetics Consortium . HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008;57:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, Deutsch AJ, Lenz TL, et al. . Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat Genet 2015;47:898–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rich SS, Akolkar B, Concannon P, et al. . Overview of the Type I Diabetes Genetics Consortium. Genes Immun 2009;10(Suppl. 1):S1–S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bycroft C, Freeman C, Petkova D, et al. Genome-wide genetic data on ∼500,000 UK Biobank participants. bioRxiv. 20 July 2017 [preprint]. DOI: 10.1101/166298 [DOI]
- 24.Jia X, Han B, Onengut-Gumuscu S, et al. . Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 2013;8:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh PR, Tucker G, Bulik-Sullivan BK, et al. . Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet 2015;47:284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raj P, Rai E, Song R, et al. . Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. eLife 2016;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann S, Fischer A, Nothnagel M, et al. . Genome-wide association analysis reveals 12q13.3-q14.1 as new risk locus for sarcoidosis. Eur Respir J 2013;41:888–900 [DOI] [PubMed] [Google Scholar]
- 28.Onengut-Gumuscu S, Chen WM, Burren O, et al.; Type 1 Diabetes Genetics Consortium . Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 2015;47:381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howson JM, Rosinger S, Smyth DJ, Boehm BO, Todd JA; ADBW-END Study Group . Genetic analysis of adult-onset autoimmune diabetes. Diabetes 2011;60:2645–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redondo MJ, Geyer S, Steck AK, et al.; Type 1 Diabetes TrialNet Study Group . A type 1 diabetes genetic risk score predicts progression of islet autoimmunity and development of type 1 diabetes in individuals at risk. Diabetes Care 2018;41:1887–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Usher-Smith JA, Thompson MJ, Sharp SJ, Walter FM. Factors associated with the presence of diabetic ketoacidosis at diagnosis of diabetes in children and young adults: a systematic review. BMJ 2011;343:d4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noble JA, Erlich HA. Genetics of type 1 diabetes. Cold Spring Harb Perspect Med 2012;2:a007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khera AV, Chaffin M, Aragam KG, et al. . Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redondo MJ, Oram RA, Steck AK. Genetic Risk Scores for Type 1 Diabetes Prediction and Diagnosis. Curr Diab Rep 2017;17:129. [DOI] [PubMed] [Google Scholar]
- 35.Brody JA, Morrison AC, Bis JC, et al.; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium; TOPMed Hematology and Hemostasis Working Group; CHARGE Analysis and Bioinformatics Working Group . Analysis commons, a team approach to discovery in a big-data environment for genetic epidemiology. Nat Genet 2017;49:1560–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.