Abstract
Diabetic retinopathy remains the leading cause of acquired blindness in working-age adults. While the cutting-edge research in the field has identified many molecular, functional, and structural abnormalities, the exact molecular mechanism of this devastating disease remains obscure. Diabetic environment drives dysfunction of the power generator of the cell and disturbs the homeostasis of mitochondrial dynamic. Mitochondrial DNA (mtDNA) is damaged, the transcription of mtDNA-encoded genes is impaired, and the electron transport chain is compromised, fueling into a vicious cycle of free radicals. The hyperglycemic milieu also alters the epigenetic machinery, and mtDNA and other genes associated with mitochondrial homeostasis are epigenetically modified, further contributing to the mitochondrial damage. Thus, mitochondria appear to have a significant role in the development of diabetic retinopathy, and unraveling the mechanism responsible for their damage as well as the role of epigenetic modifications in mitochondrial homeostasis should identify novel therapeutic targets. This will have a major impact on inhibiting/halting diabetic retinopathy and preventing the loss of vision.
Introduction
Diabetic retinopathy is the leading cause of vision loss among working-age adults in the industrialized nations. One-third of patients with diabetes have some form of retinopathy, and among those 10% develop a vision-threatening form of the disease. Worldwide, ∼127 million people were diagnosed with diabetic retinopathy in 2010, and with the incidence of diabetes increasing at a very fast pace, over 190 million people are expected to be affected by this blinding disease in 2030 (1).
Diabetic retinopathy is a progressive disease; microaneurysms and deposits may appear in the retina, but the disease remains largely asymptomatic in the initial stages. With time, preretinal hemorrhage and neovascularization begin to appear, and if not treated, the retina detaches, leading to blindness. The pathogenesis of the disease is strongly associated with the duration of diabetes, and uncontrolled blood glucose is considered as the primary driving factor; however, blood pressure and hyperlipidemia also play significant roles in its development. Although the clinical pathology of diabetic retinopathy is mainly observed in the retinal microvasculature, recent evidence has clearly demonstrated neurodegeneration as an early event in the pathogenesis of this blinding disease (2,3).
High glucose initiates a whole array of molecular, biochemical, and functional abnormalities affecting many metabolic pathways including activation of protein kinase C, increased production of advanced glycation end products, and polyol and hexosamine pathways. Auto-oxidation of glucose, and/or initiation of many metabolic abnormalities, in the hyperglycemic milieu increases the levels of reactive oxygen species (ROS) in the retina, creating an environment with excessive free radicals (4,5). Mitochondria are the sites of cellular respiration, a process that ultimately generates fuel for the cell. During oxidative phosphorylation, the respiratory chain utilizes ∼95% of the oxygen in aerobic animals to produce ATP. However, 1–5% of all oxygen used in complexes I and III of the electron transport chain (ETC) escapes as superoxide radicals, making mitochondria the organelle with high ROS and also a target for the damaging free radicals. The majority of superoxide radicals are quickly converted to diatomic oxygen and hydrogen peroxide by mitochondrial manganese superoxide dismutase (Sod2). Under normal physiological conditions, free radicals are important for many cellular processes including cell signaling, apoptosis, and phagocytosis, but increased accumulation of these dangerous radicals induces oxidative stress. In many diseases, including diabetic retinopathy, oxidative stress, resulting from an imbalance between the production of ROS and their compensation, is considered one of the key drivers for cellular dysfunction and loss (6).
Mitochondrial Quality Control
Mitochondria have elongated structure, bounded by a double membrane, and while the outer mitochondrial membrane has integral proteins, porins, that allow small molecules to pass into the mitochondria, the inner membrane has cristae. Mitochondria are highly dynamic organelles, and to maintain proper functioning, intracellular quality control of mitochondria is maintained by fission, fusion, mitophagy, and biogenesis. Mitochondrial efficiency reflects the net balance between two opposing processes, fission and fusion. While fission is characterized by the division of one mitochondrion into two daughter mitochondria, fusion unites two mitochondria into one mitochondrion and dilutes injured mitochondrial proteins and DNA by mixing their contents. Fusion promotes the formation of elongated mitochondria with capacity to produce more ATP (7). Outer membrane fusion is mediated by fusion proteins, mitofusin 1 and 2 (Mfn1 and Mfn2), and inner membrane fusion is mediated by optic atrophy protein 1 (OPA1). Both Mfn1 and Mfn2 share ∼80% similarity and have the same structural motifs. Mfn2, however, has a proline-rich region that tethers to the mitochondria and is sufficient and essential to modulate mitochondrial metabolism. The fission protein, dynamin-related protein 1 (Drp1), constricts mitochondrial membranes in a GTP-dependent manner (8). Fusion and fission occur almost simultaneously, and unhealthy mitochondria are segregated for turnover by the autophagosomal pathway mitophagy (9). Microtubule-associated protein light chain 3 (LC3), a common autophagy marker, is expressed at a basal level, but in pathological conditions, it is proteolytically cleaved to LC3II during autophagy. Accumulation of PTEN-induced putative kinase 1 (PINK1) at the outer membrane of depolarized mitochondria allows recruitment of the E3 ubiquitin ligase Parkin. This polyubiquitinates proteins on the outer mitochondrial membrane and degrades the damaged mitochondria, and biogenesis replenishes mitochondrial mass (10). Defective mitochondrial dynamic has been implicated in many chronic diseases, including cancer and diabetes (11).
Mitochondrial Genome
The nucleus is the main site of gene transcription, and mitochondria are the only other subcellular component with their own circular DNA. Compared with nuclear DNA, mitochondrial DNA (mtDNA) is small with ∼16,500 base pairs. Each mitochondria contains 10,000–100,000 copies compared with >50 copies/nucleus. mtDNA encodes 22 tRNAs, 2 rRNAs, and genes for only 13 proteins, and all of these 13 proteins are critical in the proper functioning of the ETC (5,12). mtDNA has a noncoding region, the displacement-loop (d-loop), which is the site for transcription initiation and replication. Biogenesis of mtDNA requires a coordinated synthesis of nuclear and mitochondrial encoded proteins and mtDNA replication and is dependent mainly on mtDNA polymerase γ (POLG), master transcription factor A (TFAM), and transcription factor B2 (13).
Mitochondria and Diabetic Retinopathy
As mentioned above, mitochondria have an important function to preserve cell integrity, and any defect in this powerhouse is detrimental to cell functioning and survival. Diabetic retinopathy is a slow-progressing multifactorial complication, and increased oxidative stress has been shown to play a major role in its development (2,4,5). The retina is a highly metabolic tissue in the body, and due to high circulating glucose, electron flux through ETC is increased and the activity of complex III is reduced, culminating in increased accumulation of ROS (14). Retinal mitochondria are damaged and their ultrastructure shows partial cristolysis, their membrane integrity is impaired and cytochrome c leaks out into the cytosol, and mtDNA copy numbers are decreased. To make the bad situation worse, cytosolic ROS-producing enzymes NADPH oxidases are also activated, and constant hammering by the cytosolic ROS damages mitochondrial integrity, fueling into a vicious cycle of free radicals (5,15,16). Increased ROS also activate gelatinase matrix metalloproteinase 9 (MMP-9), and activated MMP-9, with the help of heat shock proteins, is translocated inside the mitochondria. This results in the breakdown of mitochondrial membranes and initiation of the apoptotic process (17). In a normal situation, ROS generation is kept under control by endogenous antioxidants like glutathione (GSH), Sod, and catalase, but the suboptimal cytosolic and mitochondrial antioxidant redox system in diabetes further exacerbates the damage (5,15,16).
As mentioned above, mitochondria contain several copies of the circular DNA, and their close proximity to ROS makes this histone-free “naked” DNA vulnerable to free radical damage. In diabetes, the levels of oxidized nucleoside 8-hydroxy-2-deoxy-guanosine (8-OHdG) are increased in the mitochondria, and mtDNA shows increased sequence variants. The d-loop region shows more damage and sequence variants than other coding regions of the mtDNA. The transcription of mtDNA is impaired, and the ETC system is further compromised (5,16). Mitochondria have an efficient system to repair DNA damage consisting of base excision repair enzyme 8-oxoguanine glycosylase 1 (OGG1) to remove 8-oxo-deoxyguanine moiety, and mismatch repair enzymes to repair/remove uncomplimentary base pairs and the insertion and/or deletion loops formed during DNA replication (18). However, in diabetes, these DNA repair systems are also compromised, and the damage to the mtDNA is sustained with increased sequence variants (5,15,16).
Mitochondrial fusion-fission machinery is also damaged in diabetes; the swollen retinal mitochondria have decreased expression of Mfn2, but the expression of Drp1 is increased (19). Although our knowledge about the role of mitophagy in diabetic retinopathy is still in its early stages, decreased mitophagy and inflammasome activation is seen in the retina in diabetes (20), which further contributes to the worsening of mitochondrial homeostasis.
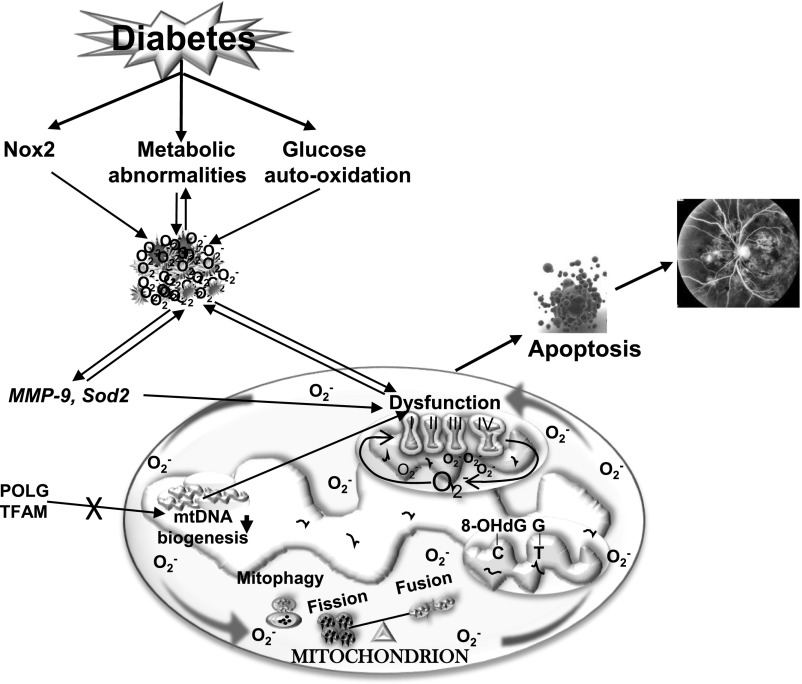
The coding capacity of mtDNA is only 13 proteins, and the other hundreds of gene products, essential for mitochondrial biogenesis, are derived from nuclear DNA (13). In diabetes, although the transcripts of PGC1 and NRF1 are increased, mtDNA biogenesis is impaired. One of the possible reasons for decreased biogenesis appears to be the posttranslational modification of TFAM, which hinders its transport in the mitochondria. Furthermore, mtDNA replication, catalyzed by POLG, is also impaired, and the translocation of POLG and Twinkle (mitochondrial 5′–3′ DNA helicase) inside the mitochondria is decreased (21). Thus, overall retinal mitochondrial stability is severely compromised in diabetes (Fig. 1).
Figure 1.
Mitochondrial dysfunction and diabetic retinopathy. Hyperglycemia induces many metabolic abnormalities, including activation of the polyol pathway and protein kinase C and advanced glycation formation, and also activates cytosolic Nox2. These abnormalities lead to increased ROS, and increased ROS, in turn, fuel in the metabolic abnormalities. High glucose also auto-oxidizes, further increasing ROS levels. ROS activate MMP-9 and inhibit antioxidant enzyme Sod2, and while activated MMP-9 damages the mitochondria, inactivated Sod2 impairs scavenging of mitochondrial free radicals produced. The activity of complex III (of ETC) is decreased, further increasing ROS levels. mtDNA is oxidatively modified, mismatches are increased and mismatch repair enzyme Mlh1 is compromised, and the transcription of mtDNA is impaired, further contributing to free radical accumulation. The vicious cycle of free radicals continues to self-perpetuate. The damaged mitochondria increase cytochrome c leakage in the cytosol and activate the apoptotic machinery, resulting in the formation of acellular capillaries and pericyte ghosts, the early histopathological lesions of diabetic retinopathy.
Diabetic retinopathy is now being appreciated as a low-grade chronic inflammatory disease (22), and the role of dysfunctional mitochondria in inflammation, along with mitochondria-associated endoplasmic reticulum stress in regulating cellular homeostatic processes, is now gaining interest (23). However, the relationship between mitochondrial dysfunction and inflammation in diabetic retinopathy remains poorly understood.
Epigenetics
Diabetes results in many structural, functional, metabolic, and molecular abnormalities, and expression of genes associated with these abnormalities is altered. Gene expression is also regulated by epigenetic modifications, the heritable modifications that do not change the primary DNA sequence. These modifications are influenced by external factors including lifestyle and disease state; they could be imprinted within the genome or could also be erased (24). Some of the major epigenetic modifications include DNA methylation, histone modifications, and noncoding RNAs. While DNA and histone modifications can either close or open the chromatin structure to regulate binding of the transcription factor, noncoding RNAs control gene expression at the RNA level (24).
Transcription of >50% of the genes in vertebrate genomes is associated with cytosine-phosphate-guanine (CpG) islands (25); addition of a methyl group at the fifth carbon atom of the cytosine changes the activity of a DNA segment without changing the sequence, and methylated cytosine (5mC) physically restricts the binding of transcription factors. DNA methylation is catalyzed by a family of enzymes, the DNA methyltransferases (Dnmts), and among this family, Dnmt1 is the maintenance enzyme and Dnmt3a and 3b are de novo enzymes (26). 5mC can also undergo hydroxymethylation by deoxygenases ten-eleven translocation (Tets), and while formation of 5mC represses gene transcription, that of 5-hydroxymethylcytosine (5hmC) activates it. 5hmC can further be converted to 5-formylcytosine and 5-carboxycytosine, and they can be converted back to unmodified cytosine by a thymine DNA glycosylase and base excision repair enzymes. In addition to active demethylation of 5mC by Tets, 5mC can also be passively demethylated during replication process (27).
Modification of histones can organize the genome into active (euchromatin) or inactive (heterochromatin) regions, making DNA respectively more or less accessible for transcription. Lysine (and arginine) on histones can be acetylated, methylated, phosphorylated, ubiquitinated, or sumoylated, and the level of any such posttranslational modification is maintained by specific enzymes with opposing functions (28). For example, while histone acetylase adds an acetyl group, deacetylase removes it from the acetylated histone. These modifications influence the binding of histones to the DNA and their ability to recruit other proteins. Acetylation of lysine on histones is an active transcription mark; however, methylation of lysine is complex, and the position of the lysine and the degree of methylation (addition of one, two, or three methyl groups) dictates the fate of histone methylation (29). These histones can have an active cross talk with other histone modifications and also with DNA methylation.
A cell also has many functional noncoding RNAs (ncRNA) that are transcribed from DNA but not translated into proteins. Epigenetic-related ncRNAs include microRNA (miRNA), long noncoding RNA (lncRNA), and piwi-interacting RNA (pi-RNA). miRNA is a small and highly conserved noncoding RNA molecule that binds the target mRNA to prevent protein production. These miRNAs are considered the new fine tuners of signaling pathways and cellular processes to posttranscriptionally repress gene expression in a targeted manner. Precursors of miRNA are commonly found in clusters through many different regions of the genome, most frequently within intergenic regions and introns of protein-coding genes. pi-RNAs are very complex and do not have much specificity; lncRNAs are also a group of heterogeneous regulatory ncRNAs, but their transcript lengths are >200 nucleotides (30).
Epigenetics and Mitochondrial Dysfunction in Diabetic Retinopathy
Epigenetic modifications are dynamic, reflecting a complex interplay between an organism and its environment, and are now being recognized as some of the key contributors in many chronic diseases including cancer, diabetes, and autoimmune diseases (31). A strong association between DNA methylation and metabolic memory has been observed in patients enrolled in the Diabetes Control and Complications Trial (DCCT)/Epidemiology of Diabetes Interventions and Complications (EDIC) study (32). The role of epigenetics in diabetes complications including retinopathy is now becoming an emerging area; recent work has shown that retinal epigenetic machinery is altered in diabetes, affecting regulation (up or down) of many genes implicated in the pathogenesis of diabetic retinopathy (5,16).
In recent years, mtDNA has received attention also as being a target of epigenetic modifications; human mtDNA, though histone free, has 435 CpG sites and 4,747 cytosine residues at non-CpG sites. In addition, Dnmts and Tets are also found in the mitochondria. As with nuclear DNA methylation, abnormal mtDNA methylation is also affected by external factors including drugs and diseases (33); for example, with aging, 5hmC levels in mtDNA are decreased (34). In diabetic retinopathy, retinal mtDNA is hypermethylated with increased 5mC levels, and, consistent with mtDNA damage, 5mC levels are higher at the d-loop region compared with the other regions. Due to increased DNA methylation, transcription of mtDNA-encoded genes, important in the functioning of the ETC system, is attenuated and the ETC continues to be compromised (5,15,16,35). Cytosine is also prone to undergo spontaneous deamination to form uracil, and 5mC to thymine, generating base mismatches or mutations in the DNA (36). The mutation rate of 5mC is several fold higher than that of cytosine, and the mutation rate of mtDNA is 10–17 times higher than that of nuclear DNA (37). To make the bad situation worse, the frequency of mutations is significantly higher in the d-loop region compared with other regions of mtDNA (38). Our recent study has shown that inhibition of DNA methylation ameliorates diabetes-induced mismatches in the d-loop region and has suggested a positive correlation between DNA methylation and base mismatches (35). In addition, DNA in the promoter regions of mismatch repair enzymes Mlh1 and POLG in the retina is hypermethylated and the binding of Dnmt1 is increased, contributing to impaired mtDNA mismatch repair and decreased mtDNA copy numbers (39 and data not shown). Dynamic DNA methylation of the promoter of Mfn2 is also associated with its decreased expression and increased mitochondrial fission (40). Thus, epigenetic modifications are important in maintaining mitochondrial dynamics and quality control.
Epigenetic modifications are also implicated in the regulation of genes responsible for protecting mitochondria from damage; e.g., various histone modifications of retinal Sod2 promoter/enhancer have been shown to play important roles in its transcriptional expression in diabetes. While the levels of mono- and dimethylated H3K4 are decreased, trimethylated H4K20 and acetylated H3K9 levels are increased, facilitating the binding of the transcription factor NF-κB and increasing its expression (5,15,16). The transcriptional activity of Nrf2, a major regulator of cytoprotective responses to endogenous and exogenous stresses caused by ROS and electrophiles, is decreased in diabetic retinopathy. Epigenetic modification of the intracellular inhibitor of Nrf2, Kelch-like ECH-associated protein 1 (Keap1) is implicated in inhibition of transcriptional activity of Nrf2. The levels of H3K4me1 are increased at the Keap1 promoter, facilitating the binding of the transcription factor, stimulating protein 1, and increasing Keap1 expression. Due to increased interactions of Keap1 with Nrf2, the movement of Nrf2 into the nucleus in impeded, and this impairs the transcriptional activity of Nrf2 and decreases expressions of antioxidant response genes including Sod2. Nrf2 is also intimately associated with GSH biosynthesis by binding at the ARE4 region (a region with strong transcription activation function) of the GSH biosynthesis enzyme glutamate-cysteine ligase (Gclc). Alterations in H3K4 methylation at Gclc-ARE4 have been shown to be one of the major factors in the impaired Nrf2 binding at Gclc-ARE4; in diabetic retinopathy, total and mitochondrial GSH levels are decreased in the retina and its vasculature (5,15,16).
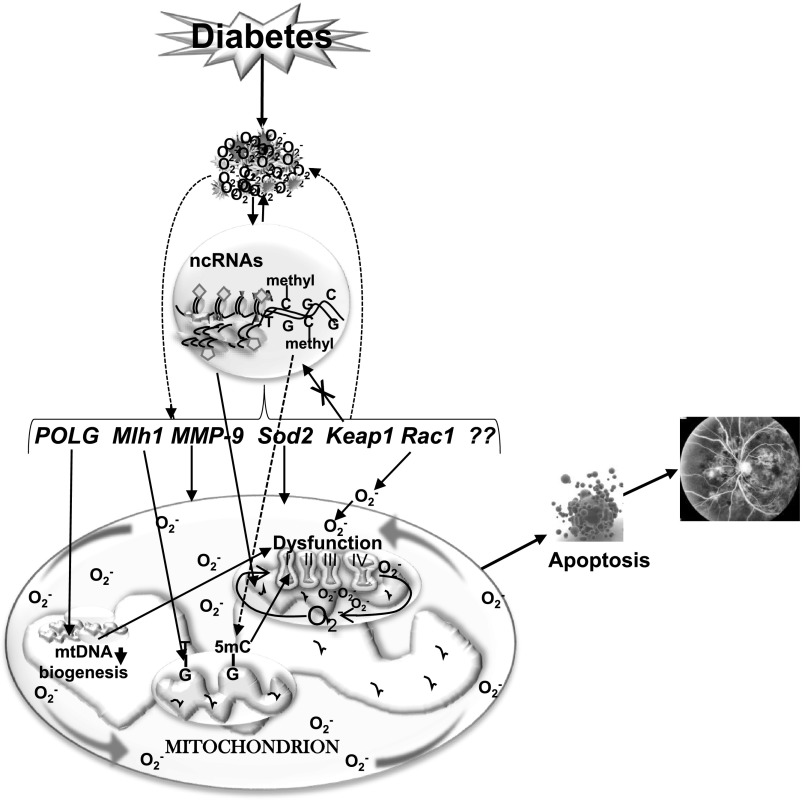
As detailed above, Rac1-Nox2–mediated increase in cytosolic ROS and MMP-9 activation damage mitochondrial membranes, and mitochondrial damage precedes the appearance of retinal histopathology (5,16). Recent work from our laboratory has shown that the promoter of Rac1 undergoes dynamic DNA methylation; while the binding of Dnmt1 is increased in the hyperglycemic milieu, a parallel increase in Tet2 hydroxymethylates the promoter, resulting in Rac1 transcriptional activation (41). Activation of HDACs is also implicated in decreased interleukin-10 (IL-10) levels in the peripheral blood of patients with diabetic retinopathy (42), and recent studies have shown involvement of HDAC6 in glucagon-like peptide 1–mediated alleviation of retinal oxidative stress and autophagy (43). Moreover, the retinal MMP-9 promoter also has increased acetylated H3K9 levels in diabetes, which, via increasing the recruitment of transcriptional factors, upregulates MMP-9 transcription. H3K27me3 levels and recruitment of the dimethylation/trimethylation enzyme Enhancer of Zeste homolog 2 (Ezh2) are also elevated. In addition, MMP-9 promoter also undergoes dynamic DNA methylation-hydroxymethylation, with increased bindings of both Dnmt1 and Tet2 and elevated levels of 5hmC (17). Increased recruitment of Ezh2 at the MMP-9 promoter facilitates Dnmt1-Tet2 recruitment, modulating transcriptional activation of MMP-9 (44). Thus, many epigenetic modifications are implicated in maintaining mitochondrial homeostasis in diabetic retinopathy (Fig. 2).
Figure 2.
Epigenetics and diabetic retinopathy. Increased ROS affect the epigenetic machinery, and the activities of enzymes responsible for maintaining DNA and histone methylation and histone acetylation are altered. Due to dynamic DNA methylation and histone modifications, gene expressions of MMP-9 and Rac1 are increased and that of Mlh1 and Sod2 are decreased. Furthermore, due to epigenetic modifications at the promoter of Keap1, a negative regulator of master transcription factor Nrf2, its expression is increased, and this impedes the translocation of Nrf2 inside the nucleus, compromising the antioxidant defense system. Hypermethylation of the POLG promoter further decreases its activity, and mtDNA replication is attenuated. mtDNA itself is also hypermethylated, and this decreases the transcription of mtDNA-encoded genes that are critical for functioning of the ETC complex. The compromised ETC system continues to fuel into a vicious cycle of free radicals, and release of cytochrome c in the cytosol activates apoptosis, ultimately resulting in the development of diabetic retinopathy.
In addition to histone and DNA modifications, many miRNAs are also now being implicated in the regulation of mitochondrial homeostasis; e.g., miR-663 has been identified as a regulator of nuclear-encoded respiratory chain subunits involved in complexes of ETC (45). Furthermore, miR-19a and HDAC11 are implicated in IL-17–mediated suppression of IL-10 expression in peripheral blood of patients with diabetic retinopathy (42). Using mouse model of diabetic retinopathy, 303 aberrantly expressed lncRNAs have been identified in the retina with 214 being downregulated and 89 upregulated, and a conserved lncRNA, MALAT1, has been shown to be significantly upregulated (46), suggesting MALAT1 as a therapeutic target for diabetic retinopathy. Epigenome-wide association studies have identified several DNA methylation markers associated with type 2 diabetes including significant methylation of an oxidative stress mediator, thioredoxin-interacting protein (47), and have revealed an association between persistant DNA methylation and metabolic memory in the type 1 diabetes cohort enrolled in the DCCT/EDIC study (32), but identification of DNA methylation markers associated with the development of diabetic retinopathy remains to be established. Thus, although epigenetics is still an evolving area of research, it is clear that these modifications have a significant role in the development of diabetic retinopathy by regulating both the damaging and protective genes associated with its development.
Since epigenetic modifications are influenced by external factors, they provide a link between the environment and disease development. These modifications can be inherited or reversed, making them attractive therapeutic targets. There is clearly a growing interest in the therapeutic use of regulators of DNA methylation and histone modifications in the cancer field (48,49), but their application in diabetes and its complications remains to be initiated. In addition, although miRNAs are gaining importance in serving as potential diagnostic biomarkers for a disease, with miRNA mimics and anti-mRNA antisense oligodeoxyribonucleotide targeting specific miRNA (50), the challenge posed by the blood-retinal barrier for these mimics/antisense oligodeoxyribonucleotide to reach to the retina needs to be recognized. As detailed above, epigenetic modifications play an important role in maintaining mitochondrial homeostasis in diabetes, and the possibility of these therapeutic modalities (chemical or natural) for the treatment of retinopathy will be a welcoming sign for patients with diabetes.
Conclusions
In summary, although mitochondria research began in 19th century, many issues concerning their relevance to health and diseases remain obscure. While the role of mitochondria in cell survival has been very well established, dysfunction of this hardest working cell component in chronic diseases is now getting appreciation. Moreover, how external factors affect the gene transcription is also gaining steam, and many epigenetic modifications are implicated in chronic diseases. Thus, the role of mitochondria in diabetic retinopathy as well as epigenetics in regulating mitochondrial dysfunction, although still in early stages, is opening up many opportunities for therapeutically targeting mitochondrial dysfunction/epigenetic modifications to halt/slow down the progression of diabetic retinopathy.
Article Information
Funding. Research discussed is this article was supported in part by grants from the National Institutes of Health National Eye Institute (EY014370, EY017313, and EY022230) and from the Thomas Foundation to R.A.K. as well as an unrestricted grant from Research to Prevent Blindness to the Ophthalmology Department.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
References
- 1.Zheng Y, He M, Congdon N. The worldwide epidemic of diabetic retinopathy. Indian J Ophthalmol 2012;60:428–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank RN. Diabetic retinopathy. N Engl J Med 2004;350:48–58 [DOI] [PubMed] [Google Scholar]
- 3.Antonetti DA, Klein R, Gardner TW. Diabetic retinopathy. N Engl J Med 2012;366:1227–1239 [DOI] [PubMed] [Google Scholar]
- 4.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes 2005;54:1615–1625 [DOI] [PubMed] [Google Scholar]
- 5.Kowluru RA, Kowluru A, Mishra M, Kumar B. Oxidative stress and epigenetic modifications in the pathogenesis of diabetic retinopathy. Prog Retin Eye Res 2015;48:40–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowaltowski AJ, de Souza-Pinto NC, Castilho RF, Vercesi AE. Mitochondria and reactive oxygen species. Free Radic Biol Med 2009;47:333–343 [DOI] [PubMed] [Google Scholar]
- 7.Liang Q, Kobayashi S. Mitochondrial quality control in the diabetic heart. J Mol Cell Cardiol 2016;95:57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pernas L, Scorrano L. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol 2016;78:505–531 [DOI] [PubMed] [Google Scholar]
- 9.Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal 2011;14:1939–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melser S, Lavie J, Bénard G. Mitochondrial degradation and energy metabolism. Biochim Biophys Acta 2015;1853:2812–2821 [DOI] [PubMed] [Google Scholar]
- 11.Williams M, Caino MC. Mitochondrial dynamics in type 2 diabetes and cancer. Front Endocrinol (Lausanne) 2018;9:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenzie M, Liolitsa D, Hanna MG. Mitochondrial disease: mutations and mechanisms. Neurochem Res 2004;29:589–600 [DOI] [PubMed] [Google Scholar]
- 13.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 2008;88:611–638 [DOI] [PubMed] [Google Scholar]
- 14.Madsen-Bouterse SA, Mohammad G, Kanwar M, Kowluru RA. Role of mitochondrial DNA damage in the development of diabetic retinopathy, and the metabolic memory phenomenon associated with its progression. Antioxid Redox Signal 2010;13:797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowluru RA, Mishra M. Oxidative stress, mitochondrial damage and diabetic retinopathy. Biochim Biophys Acta 2015;1852:2474–2483 [DOI] [PubMed] [Google Scholar]
- 16.Kowluru RA, Mishra M. Therapeutic targets for altering mitochondrial dysfunction associated with diabetic retinopathy. Expert Opin Ther Targets 2018;22:233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowluru RA, Mishra M. Regulation of matrix metalloproteinase in the pathogenesis of diabetic retinopathy. Prog Mol Biol Transl Sci 2017;148:67–85 [DOI] [PubMed] [Google Scholar]
- 18.Zinovkina LA. Mechanisms of mitochondrial DNA repair in mammals. Biochemistry (Mosc) 2018;83:233–249 [DOI] [PubMed] [Google Scholar]
- 19.Zhong Q, Kowluru RA. Diabetic retinopathy and damage to mitochondrial structure and transport machinery. Invest Ophthalmol Vis Sci 2011;52:8739–8746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh LP, Devi TS, Yumnamcha T. The role of Txnip in mitophagy dysregulation and inflammasome activation in diabetic retinopathy: a new perspective. JOJ Ophthalmol 2017;4:4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tewari S, Santos JM, Kowluru RA. Damaged mitochondrial DNA replication system and the development of diabetic retinopathy. Antioxid Redox Signal 2012;17:492–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kern TS. Contributions of inflammatory processes to the development of the early stages of diabetic retinopathy. Exp Diabetes Res 2007;2007:95103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grazioli S, Pugin J. Mitochondrial damage-associated molecular patterns: from inflammatory signaling to human diseases. Front Immunol 2018;9:832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation 2011;123:2145–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA 1993;90:11995–11999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999;99:247–257 [DOI] [PubMed] [Google Scholar]
- 27.An J, Rao A, Ko M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp Mol Med 2017;49:e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen HP, Zhao YT, Zhao TC. Histone deacetylases and mechanisms of regulation of gene expression. Crit Rev Oncog 2015;20:35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012;13:343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang N, Jiang S, Yang Y, et al. . Noncoding RNAs as therapeutic targets in atherosclerosis with diabetes mellitus. Cardiovasc Ther 2018;36:e12436. [DOI] [PubMed] [Google Scholar]
- 31.Moosavi A, Motevalizadeh Ardekani A. Role of epigenetics in biology and human diseases. Iran Biomed J 2016;20:246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Miao F, Paterson AD, et al.; DCCT/EDIC Research Group . Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci U S A 2016;113:E3002–E3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iacobazzi V, Castegna A, Infantino V, Andria G. Mitochondrial DNA methylation as a next-generation biomarker and diagnostic tool. Mol Genet Metab 2013;110:25–34 [DOI] [PubMed] [Google Scholar]
- 34.Dzitoyeva S, Chen H, Manev H. Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol Aging 2012;33:2881–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra M, Kowluru RA. DNA methylation—a potential source of mitochondria dna base mismatch in the development of diabetic retinopathy. Mol Neurobiol 21 April 2018 [Epub ahead of print]. DOI: 10.1007/s12035-018-1086-9 [DOI] [PubMed] [Google Scholar]
- 36.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology 2013;38:23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holliday R, Grigg GW. DNA methylation and mutation. Mutat Res 1993;285:61–67 [DOI] [PubMed] [Google Scholar]
- 38.Yuan RT, Sun Y, Bu LX, Jia MY. Gene mutations in the D-loop region of mitochondrial DNA in oral squamous cell carcinoma. Mol Med Rep 2015;11:4496–4500 [DOI] [PubMed] [Google Scholar]
- 39.Tewari S, Zhong Q, Santos JM, Kowluru RA. Mitochondria DNA replication and DNA methylation in the metabolic memory associated with continued progression of diabetic retinopathy. Invest Ophthalmol Vis Sci 2012;53:4881–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kowluru RA, Mishra M. Epigenetics and mitochondrial stability in diabetic retinopathy (Abstract). Diabetes 2018;67(Suppl. 1):A239
- 41.Duraisamy AJ, Mishra M, Kowluru A, Kowluru RA. Epigenetics and regulation of oxidative stress in diabetic retinopathy. Invest Ophthalmol Vis Sci 2018;59:4831–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, You Q, Cao X, Guo H, Gao X, Peng X. Micro RNA-19a suppresses interleukin-10 in peripheral B cells of patients with diabetic retinopathy. Am J Transl Res 2017;9:1410–1417 [PMC free article] [PubMed] [Google Scholar]
- 43.Cai X, Li J, Wang M, et al. . GLP-1 treatment improves diabetic retinopathy by alleviating autophagy through GLP-1R-ERK1/2-HDAC6 signaling pathway. Int J Med Sci 2017;14:1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duraisamy AJ, Mishra M, Kowluru RA. Crosstalk between histone and DNA methylation in regulation of retinal matrix metalloproteinase-9 in diabetes. Invest Ophthalmol Vis Sci 2017;58:6440–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carden T, Singh B, Mooga V, Bajpai P, Singh KK. Epigenetic modification of miR-663 controls mitochondria-to-nucleus retrograde signaling and tumor progression. J Biol Chem 2017;292:20694–20706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci 2014;55:941–951 [DOI] [PubMed] [Google Scholar]
- 47.Walaszczyk E, Luijten M, Spijkerman AMW, et al. . DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case-control sample of the Lifelines study. Diabetologia 2018;61:354–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zwergel C, Valente S, Mai A. DNA methyltransferases inhibitors from natural sources. Curr Top Med Chem 2016;16:680–696 [DOI] [PubMed] [Google Scholar]
- 49.Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet 2016;17:630–641 [DOI] [PubMed] [Google Scholar]
- 50.Joglekar MV, Januszewski AS, Jenkins AJ, Hardikar AA. Circulating microRNA biomarkers of diabetic retinopathy. Diabetes 2016;65:22–24 [DOI] [PubMed] [Google Scholar]