Abstract
Type I interferons (IFNs) induce expression of multiple genes that control innate immune responses to invoke both antiviral and antineoplastic activities. Transcription of these interferon-stimulated genes (ISGs) occurs upon activation of the canonical Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signaling pathways. Phosphorylation and acetylation are both events crucial to tightly regulate expression of ISGs. Here, using mouse embryonic fibroblasts and an array of biochemical methods including immunoblotting and kinase assays, we show that sirtuin 2 (SIRT2), a member of the NAD-dependent protein deacetylase family, is involved in type I IFN signaling. We found that SIRT2 deacetylates cyclin-dependent kinase 9 (CDK9) in a type I IFN–dependent manner and that the CDK9 deacetylation is essential for STAT1 phosphorylation at Ser-727. We also found that SIRT2 is subsequently required for the transcription of ISGs and for IFN-driven antiproliferative responses in both normal and malignant cells. These findings establish the existence of a previously unreported signaling pathway whose function is essential for the control of JAK–STAT signaling and the regulation of IFN responses. Our findings suggest that targeting sirtuin activities may offer an avenue in the development of therapies for managing immune-related diseases and cancer.
Keywords: signal transduction, post-translational modification (PTM), sirtuin, interferon, signal transducers and activators of transcription 1 (STAT1), innate immunity, histone deacetylase (HDAC), cytokine, deacetylation, immune regulation, SIRT2, STAT1, type I IFN
Introduction
The interferons (IFNs)7 are multifunctional cytokines that regulate diverse cellular biological responses, including important antitumor, antiviral, and immunomodulatory effects (1). IFNs have been used widely in the treatment of many human diseases, including viral infections, autoimmune disorders, and malignancies (2, 3). Type I IFNs engage their specific receptor complex, IFNAR, and initiate a sequence of events that lead to Janus-activated kinase–signal transducer and activator of transcription (JAK–STAT) signaling engagement and transcriptional activation of IFN-stimulated genes (ISGs) (1, 4). Activation of JAKs results in tyrosine phosphorylation of STAT1 and STAT2. Once phosphorylated, these STATs heterodimerize and bind to interferon regulatory factor 9 to assemble the transcription complex ISGF3 (interferon-stimulated gene factor 3), which binds to IFN-stimulated response elements to initiate gene expression in the nucleus. IFNs also invoke formation of STAT1 homodimers that translocate to the nucleus and bind to distinct IFNγ-activated site (GAS) elements (1, 2, 5). IFN–dependent nuclear translocation of STAT proteins requires STAT tyrosine phosphorylation, although the serine phosphorylation of STAT1 is essential for maximum transcriptional activation of ISGs (1, 6, 7). Beyond JAK–STAT activation, several other signaling cascades are activated during engagement of IFNAR and these are essential for IFN responses, associated with both transcriptional regulation and mRNA translation of ISGs (1, 2, 8–10).
Beyond phosphorylation, acetylation plays a crucial role in post-translational regulation of STAT1, affecting its activity (11, 12). Moreover, histone deacetylases (HDACs) are essential for transcription of IFN-responsive genes (12–14). Sirtuin 2 (SIRT2) is a class III HDAC, which requires NAD+ as a cofactor. SIRT2 is predominantly localized in the cytoplasm, where it deacetylates lysine (Lys) residues on several targets including tubulin. SIRT2 shuttles between the cytosol and nucleus, where, during mitosis, it regulates key components of the mitotic machinery (15–20). SIRT2 participates in the modulation of numerous biological processes, including cell cycle control, metabolism, differentiation, and has been described to regulate important oncogenes, such as Myc and KRAS (21–27). Mice deficient in Sirt2 develop various malignancies, including breast and liver cancer, suggesting that SIRT2 functions as a tumor suppressor (18, 21, 28). The tumor suppressor function of SIRT2 can be mechanistically explained in part through deacetylation of specific targets including Cdc20/Cdh1 (cell-division cycle protein 20/CDC20 homologue 1) of the APC/C (anaphase-promoting/cyclosome) complex, H4K16 (histone H4 lysine 16), and the cyclin–dependent kinase 9 (CDK9) (21, 22, 29).
In the present study, we examined the potential involvement of SIRT2 in type I IFN signaling and its role in the induction of IFN responses. We report that SIRT2 controls IFN-driven gene transcription via a novel mechanism involving control of phosphorylation of STAT1 on serine 727. Furthermore, we provide evidence that SIRT2-dependent STAT1 phosphorylation is important for transcriptional activation of ISGs.
Results
SIRT2 is required for type I IFN-induced STAT1 serine 727 phosphorylation
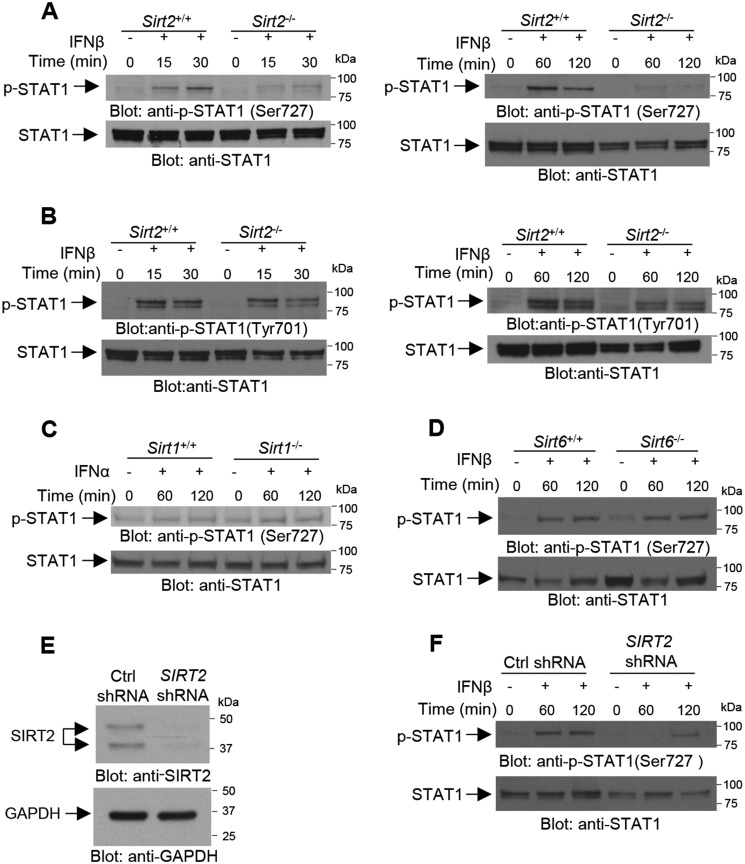
At the outset we examined whether IFNβ treatment of Sirt2−/− mouse embryonic fibroblasts (MEFs) leads to phosphorylation of STAT1. Whereas IFNβ treatment of Sirt2+/+ MEFs resulted in phosphorylation of STAT1 on Ser-727, IFNβ-induced phosphorylation of STAT1 on Ser-727 was defective in Sirt2−/− MEFs (Fig. 1A). By contrast, there was no SIRT2-dependent requirement for tyrosine phosphorylation of STAT1, as both Sirt2+/+ and Sirt2−/− MEFs exhibited IFNβ-inducible phosphorylation of STAT1 on Tyr-701 (Fig. 1B). To evaluate the potential involvement of other sirtuins in the control of IFNα/β–dependent STAT1 serine phosphorylation, we employed Sirt1−/− and Sirt6−/− MEFs. As shown in Fig. 1, C and D, IFNα/β–dependent serine phosphorylation of STAT1 occurs in both Sirt1−/− and Sirt6−/− MEFs, to similar levels observed in their WT counterparts, Sirt1+/+ and Sirt6+/+ MEFs.
Figure 1.
SIRT2 is required for type I IFN-induced STAT1 Ser-727 phosphorylation. A and B, effects of IFNβ treatment on phosphorylation of STAT1 on Ser-727 (A) and Tyr-701 (B) in Sirt2+/+ versus Sirt2−/− MEFs. Cells were either left untreated or were treated with IFNβ as indicated. Cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. C–F, effects of type I IFN treatment on phosphorylation of STAT1 on Ser-727 in, Sirt1+/+ versus Sirt1−/− MEFs (C), Sirt6+/+ versus Sirt6−/− MEFs (D), and U937 cells stably expressing control (Ctrl) shRNA versus SIRT2 shRNA (E and F). Cells were either left untreated or were treated with type I IFN as indicated. Cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
To further establish the role of SIRT2 in the regulation of type I IFN–dependent STAT1 serine phosphorylation, we conducted studies in U937 hematopoietic cells, stably expressing control shRNA or SIRT2 shRNA (Fig. 1E). As show in Fig. 1F, IFNβ–dependent phosphorylation of STAT1 on Ser-727 was substantially impaired in cells expressing SIRT2 shRNA.
SIRT2 regulates CDK9-mediated phosphorylation of STAT1 in a type I IFN–dependent manner
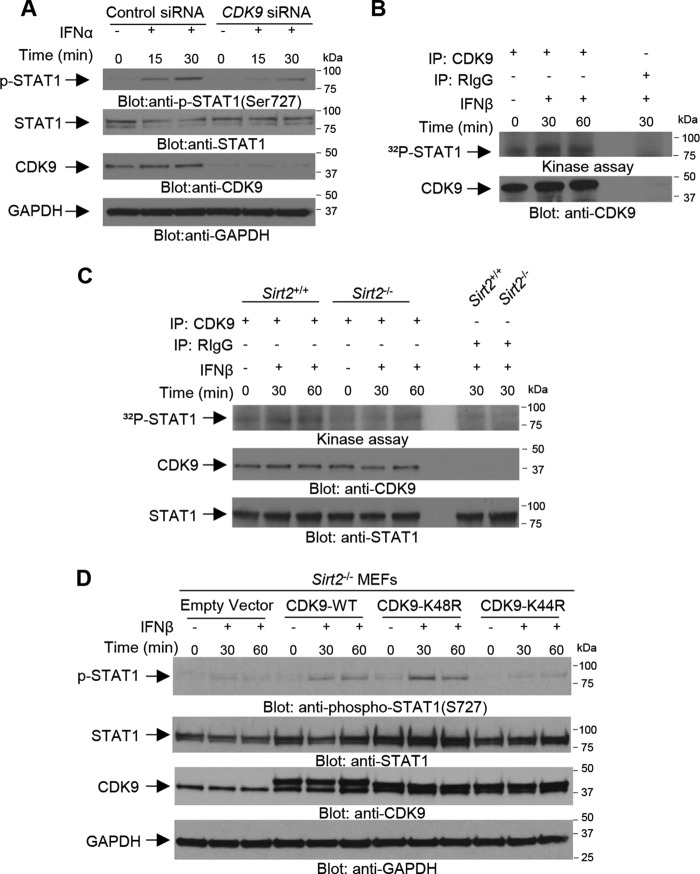
To define the mechanisms by which SIRT2 activity may regulate type I IFN–dependent phosphorylation of STAT1 on Ser-727, we examined whether this event requires deacetylation of CDK9 on Lys residue 48. Previous studies have established that Lys-48 in CDK9 is a major SIRT2 downstream target and its deacetylation is required for activation of CDK9 (29). Notably, there is evidence that silencing another member of the CDK family, CDK8, affects IFNγ–induced phosphorylation of STAT1 on Ser-727 (30). Accordingly, we examined whether CDK9 controls phosphorylation of STAT1 at Ser-727 in the type I IFN system. Specific siRNA–mediated knockdown of CDK9 in SET-2 cells resulted in a reduction of type I IFN-induced STAT1 Ser-727 phosphorylation (Fig. 2A). In vitro kinase assays were performed to directly determine whether STAT1 is phosphorylated by the CDK9 kinase after IFNβ treatment. These studies demonstrated that STAT1 is a substrate for the kinase activity of CDK9 (Fig. 2B). Importantly, such CDK9-mediated phosphorylation of STAT1 was defective in Sirt2−/− MEFs compared with Sirt2+/+ MEFs (Fig. 2C), suggesting that SIRT2 could mediate the activation of CDK9 via deacetylation in an IFNβ–dependent manner. In parallel studies in which we ectopically expressed CDK9 wildtype (WT) plasmid in Sirt2−/− MEFs, we observed an increase in the phosphorylation of STAT1 on Ser-727. In contrast, expression of a mutant CDK9 in which Lys-44 was mutated to arginine (K44R) and therefore cannot be deacetylated, showed no significant increase in the IFN–dependent phosphorylation of STAT1 at Ser-727 as determined by immunoblotting (Fig. 2D). On the other hand, mutation of CDK9 on Lys-48 to arginine (K48R), thereby blocking acetylation of the SIRT2-specific deacetylation target residue, induced potent phosphorylation of STAT1 on Ser-727 in Sirt2−/− MEFs (Fig. 2D), suggesting that SIRT2-dependent deacetylation of CDK9 on Lys-48 controls phosphorylation of STAT1 on Ser-727 after IFNβ treatment.
Figure 2.
SIRT2 activity is required for CDK9-mediated phosphorylation of STAT1 in an IFNβ-dependent manner. A, SET-2 cells were transfected with either control siRNA or CDK9 siRNA and 48 h after transfection cells were left untreated or were treated with IFNα for 15 and 30 min, as indicated. Cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. B and C, KT-1 cells (B) and Sirt2+/+ and Sirt2−/− MEFs (C) were either left untreated or were treated with IFNβ for 30 and 60 min, as indicated. After cell lysis, equal amounts of proteins were immunoprecipitated (IP) with either CDK9-specific antibody or control nonimmune rabbit IgG (RIgG). In vitro kinase assays to detect CDK9 activity were performed on the immunoprecipitates, using STAT1 recombinant inactive protein as an exogenous substrate. Immunoblot demonstrating total immunoprecipitated CDK9 expression used in each condition for the in vitro kinase assay is shown. Autoradiography film demonstrating CDK9-induced phosphorylation of STAT1 after IFNβ treatment is shown. Note: a lane between CDK9 and RIgG immunoprecipitates was loaded with 1× loading dye for best separation between the wells. D, Sirt2−/− MEFs were transfected with: empty vector, CDK9-WT, CDK9-K48R (deacetylated mutant on lysine 48), or CDK9-K44R (deacetylated mutant on lysine 44), as indicated. Forty-eight hours after transfection, the cells were treated with mouse IFNβ for the indicated times. Equal amounts of total cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
Regulation of transcriptional activation of ISGs by SIRT2 activity
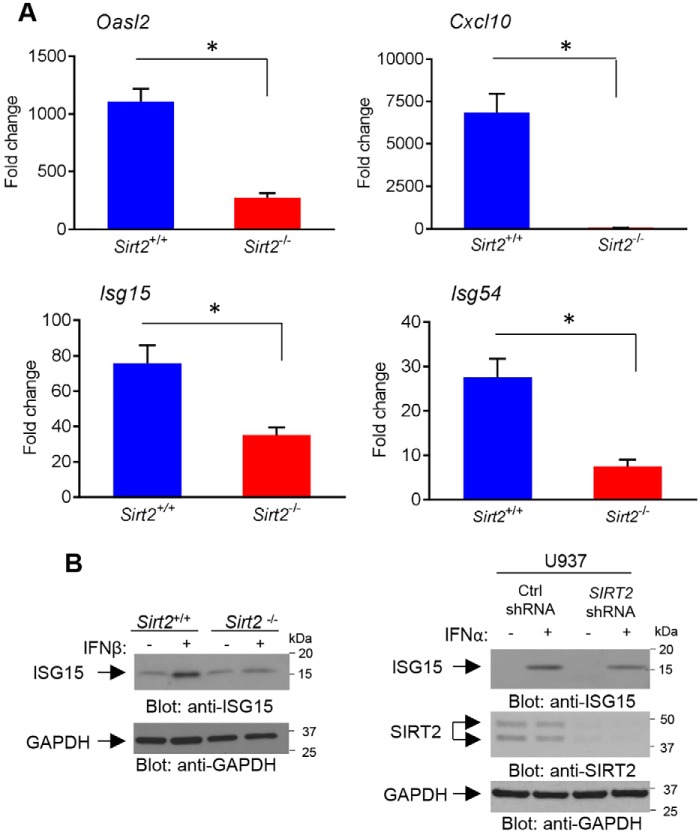
Having established that SIRT2 is required for type I IFN-induced STAT1 Ser-727 phosphorylation and cognizant that STAT1 is essential for transcription of ISGs, we next examined whether SIRT2 activity is required for IFN–dependent transcriptional activation of ISGs, using Illumina gene expression microarrays. Analysis of microarray data revealed IFN-inducible differential expression of 370 genes in Sirt2+/+ MEFs (Fig. 3, A and C) and 240 genes in Sirt2−/− MEFs (Fig. 3, B and C). Although 198 genes were differentially expressed upon IFNβ treatment in both Sirt2+/+ and Sirt2−/− MEFs (Fig. 3C), 44 of those genes showed higher expression in Sirt2+/+ MEFs compared with Sirt2−/− MEFs (Table S1, genes expressed higher are highlighted in red). Moreover, 172 genes were found to be differentially expressed only in IFNβ-treated Sirt2+/+ MEFs (Fig. 3, C and D, and Table S2), whereas 42 genes were differentially expressed only in IFNβ-treated Sirt2−/− MEFs (Fig. 3, C and E, and Table S3). To further characterize the differentially expressed genes found only in IFNβ-treated Sirt2+/+ MEFs, gene set pathways and process enrichment analyses were performed. The group of genes whose transcriptional induction by IFNβ is Sirt2-dependent (Table S2) were associated with pathways involved in the defense response to virus, lymphocyte migration, inflammatory responses, regulation of innate immune responses, regulation of erythrocyte differentiation, regulation of growth, and positive regulation of the extrinsic apoptotic signaling pathway (Fig. 3F and Table S4), suggesting that SIRT2 might play an important role in type I IFN–dependent biological functions. In further studies, we confirmed the requirement for SIRT2 activity in the expression of several key ISGs using quantitative RT-PCR (Fig. 4A). As anticipated, expression of several ISGs was defective in the absence of Sirt2. Specifically, induction of Oasl2, Cxcl10, Isg15, and Isg54 by IFNβ was significantly defective in Sirt2−/− MEFs compared with Sirt2+/+ MEFs (Fig. 4A). We also determined whether SIRT2 activity is required for expression of ISG protein products. IFNβ treatment of Sirt2+/+ MEFs resulted in induction of ISG15 protein, however, this induction was defective in Sirt2−/− MEFs (Fig. 4B, left panel). Similar results were obtained with human U937 stably expressing control shRNA or SIRT2 shRNA (Fig. 4B, right panel).
Figure 3.
SIRT2 activity is essential for ISG transcription. A and B, volcano plots of differentially expressed genes after IFNβ treatment are shown for (A) Sirt2+/+ MEFs and (B) Sirt2−/− MEFs. C, Venn diagram showing the gene expression overlap between differentially expressed genes in Sirt2+/+ MEFs (blue ellipse) and Sirt2−/− MEFs (red ellipse) after treatment with IFNβ. D, hierarchical clustering of differentially expressed genes identified only in IFNβ-treated Sirt2+/+ MEFs. E, hierarchical clustering of differentially expressed genes identified only in IFNβ-treated Sirt2−/− MEFs. F, bar graph showing enriched ontology clusters of the uniquely differentially expressed genes in IFNβ-treated Sirt2+/+ MEFs (see also Table S4).
Figure 4.
Requirement of SIRT2 activity for type I IFN–dependent gene transcription and protein expression. A, quantitative RT-PCR analyses of the relative mRNA expression of ISGs after IFNβ treatment in Sirt2+/+ and Sirt2−/− MEFs are shown. Expression levels of the indicated genes were determined using Gapdh for normalization. Data are expressed as fold-change over control untreated cells, and bar graphs represent mean ± S.E. of three independent experiments. B, protein levels of ISG15 in Sirt2+/+ and Sirt2−/− MEFs (left panel) and U937 cells stably expressing Control (Ctrl) shRNA or SIRT2 shRNA (right panel) after 24 h of treatment with or without type I IFNs, as indicated. Equal amounts of total cell lysates were resolved by SDS-PAGE and consecutively immunoblotted with the indicated antibodies. A, statistical analyses were performed using unpaired two-tailed t test with Welch's correction between treated groups (*, p < 0.05).
Role of sirtuins in the generation of type I IFN–dependent biological responses
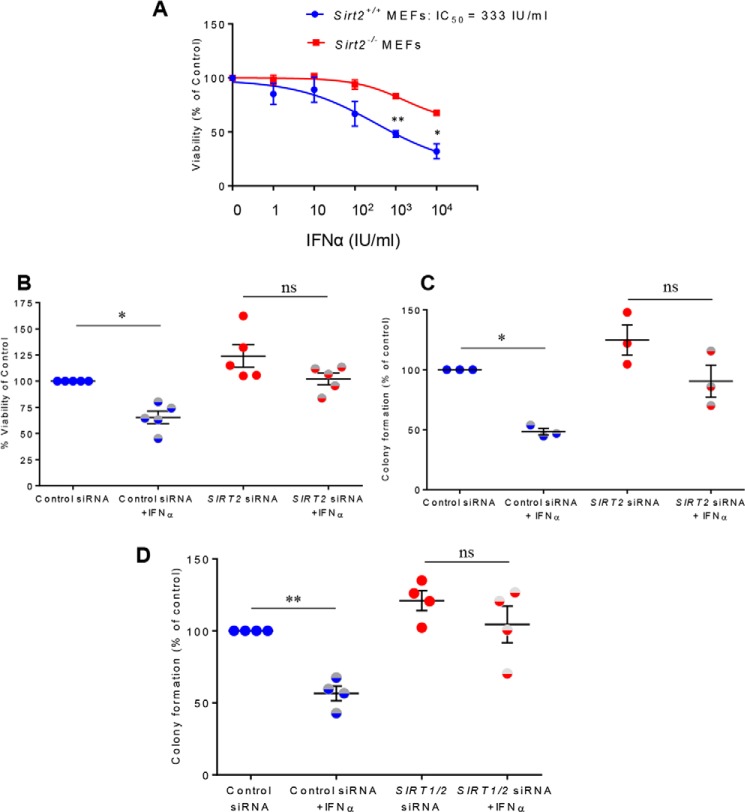
To determine the potential effects of SIRT2 on IFNα–mediated antiproliferative responses, we assessed cellular proliferation of Sirt2+/+ and Sirt2−/− MEFs treated with increasing doses of IFNα, using the viability WST-1 assay. Depletion of Sirt2 in MEFs resulted in decreased cellular sensitivity to type I IFN-induced antiproliferative responses (Fig. 5A). Next, we examined the role of SIRT2 in the generation of type I IFN–dependent anti-leukemic activity in cells expressing the JAK2(V617F) mutation, which is a critical pathogenic mutation in myeloproliferative neoplasms (31). For this purpose, we performed studies involving siRNA-mediated knockdown of SIRT2 in human erythroleukemia HEL cells and in SET-2 cells derived from an essential thrombocythemia patient at the megakaryoblastic leukemic transformation phase that express the JAK2(V617F) mutation (32) and are sensitive to type I IFN–mediated growth inhibition (Fig. S1). As shown in Fig. 5B, IFNα treatment significantly reduced proliferation of SET-2 cells, however, siRNA-mediated knockdown of SIRT2 (Fig. S2A) decreased the sensitivity of these cells to the antiproliferative effects of IFNα. Similarly, IFNα treatment suppressed the growth of primitive malignant hematopoietic precursors from HEL cells transfected with control siRNA, but this inhibition was suppressed by SIRT2 knockdown (Fig. 5C and Fig. S2B), indicating a requirement for SIRT2 in the generation of the inhibitory effects of type I IFNs on JAK2(V617F)-transformed cells. We also determined the potential involvement of SIRT2 in the generation of type I IFN anti-leukemic responses in the BCR-ABL expressing KT-1 leukemic cell line (33). IFNα treatment suppressed growth of KT1-derived primitive leukemic progenitors (CFU-L) in clonogenic assays in methylcellulose (Fig. 5D). However, these suppressive effects were blocked by siRNA-mediated SIRT1/2 knockdown (Fig. S2C), indicating that SIRT1/2 activity is required for IFNα–dependent anti-leukemic effects. In further studies, we examined whether engagement of SIRT activity is necessary for the generation of the suppressive effects of IFNα on normal hematopoiesis. Normal human bone marrow-derived CD34+ cells were transfected with siRNA to knockdown the expression of SIRT1/2 and the inhibitory effects of IFNα were determined. As anticipated, treatment with IFNα resulted in significant suppression of normal myeloid (CFU-GM) and early erythroid (BFU-E) hematopoietic progenitors growth in colony formation assays (Fig. S3). Targeted inhibition of SIRT1/2 reversed these effects (Fig. S3). Together, these data indicate that SIRT2 activity is required for induction of type I IFN-driven antiproliferative effects in both normal and malignant cells.
Figure 5.
Role of sirtuins in the generation of the biological effects of type I IFNs. A, Sirt2+/+ and Sirt2−/− MEFs were incubated with the indicated doses of mouse IFNα, and cell viability was assessed after 5 days using WST-1 reagent. Data are expressed as percentage of viability over control untreated cells. Shown are mean ± S.E. of three independent experiments. B, SET-2 cells were transfected with either control siRNA or SIRT2 siRNA and 24 h after transfection cells were left untreated or were treated with IFNα for 72 h. Viability was assessed using WST-1 reagent. Data are expressed as percentages of viability over control siRNA-transfected untreated cells, and the scatter plot represents mean ± S.E. of five independent experiments. C, HEL cells were transfected with either control siRNA or SIRT2 siRNA, and leukemic colony formation was assessed in clonogenic assays in methylcellulose in the presence or absence of human IFNα, as indicated. Data are expressed as percentage of colony formation over control siRNA-transfected untreated cells, and the scatter plot represent mean ± S.E. of three independent experiments. B and C, one-way ANOVA followed by Tukey's test was used to evaluate statistically significant differences (*, p < 0.05; ns, not significant). Note: there are no statistical significant differences seen between untreated control siRNA and untreated SIRT2 siRNA conditions. D, KT-1 cells were transfected with either control siRNA or SIRT1/2 siRNA and leukemic colony formation was assessed in clonogenic assays in methylcellulose in the presence or absence of human IFNα, as indicated. Data are expressed as percentage of colony formation over control siRNA-transfected untreated cells, and the scatter plot represents mean ± S.E. of four independent experiments. One-way ANOVA followed by Tukey's test was used to evaluate statistically significant differences (**, p < 0.01). Note: there are no statistical significant differences seen between untreated control siRNA and untreated SIRT1/2 siRNA conditions.
Discussion
Control of innate and adaptive immunity by IFN-activated signaling effectors involves JAK–STAT signaling (34) and the subsequent regulation of ISGs with antiviral and antineoplastic properties (1, 2, 35, 36). Optimal transcription of ISGs requires STAT1 phosphorylation on Ser-727, an event dependent on prior IFN-induced phosphorylation of STAT1 on tyrosine 701 (1, 6, 37). There is also evidence that acetylation/deacetylation events are important for induction of expression of ISGs (11, 12, 38). In a previous study, siRNA-mediated knockdown of HDAC3, a member of class I HDACs, was shown to reduce STAT1 phosphorylation on tyrosine 701 upon IFN treatment (12). Similarly, the class I HDAC inhibitors, valproic acid and entinostat (MS-275), were shown to impair IFN-induced phosphorylation of STAT1 (11). Additionally the class I, IIa, and IIb, inhibitor, trichostatin A, was shown to block IFN-induced gene expression (5, 12–14). These studies support a potential role for deacetylases in type I IFN signaling. However, the IFN–dependent effectors that control STAT1 function via deacetylation are not well understood.
In the present study, we provide the first evidence implicating SIRT2, a class III HDAC (16), in the control of IFN-inducible transcriptional and biological responses. Employing cells with targeted disruption of the Sirt2 gene we found that IFNβ-induced phosphorylation of STAT1 on Ser-727, but not tyrosine 701, depends on the presence of Sirt2, implicating SIRT2 in the control of IFN responses.
HDAC activity is critical for chromatin remodeling to allow for transcriptional induction of ISGs (13, 14, 39, 40). HDACs are required to recruit RNA polymerase II (RNA pol II) to the promoter regions of ISGs (5). CDK9 is a component of the positive transcription elongation factor b (P-TEFb), which phosphorylates the C-terminal domain of the large subunit of RNA pol II (29, 30). SIRT2 promotes deacetylation of lysine 48 of CDK9, which is required for induction of its kinase activity (29). Treatment with flavopiridol, a CDK inhibitor with increased specificity for CDK8 and CDK9, blocks type I and type II IFN-induced phosphorylation of STAT1 on Ser-727 (30). In the present study, we demonstrate that CDK9 is required for phosphorylation of STAT1 on Ser-727 in a type I IFN–dependent manner. Notably, our work suggests that SIRT2 may regulate type I IFN–dependent STAT1 phosphorylation on Ser-727 via deacetylation of CDK9 on lysine 48. Additionally, we provide evidence that SIRT2 activity is required for transcription of ISGs and the generation of type I IFN–dependent antiproliferative responses. In summary, our study supports a model in which SIRT2 regulates acetylation/deacetylation of CDK9 on Lys-48, which in turn is required for full STAT1 phosphorylation on Ser-727 and consequent optimal transcription of type I IFN-induced genes and biological responses. It remains to be determined whether SIRT2 regulates activation of other type I IFN–dependent signaling pathways.
Viewed together with previous studies, our results provide further support for the concept that not only phosphorylation, but other post-translational modifications, such as acetylation, glycosylation, and ubiquitination, are required for optimal type I IFN signaling (6, 12) and we now provide evidence for the involvement of SIRT2. By establishing the first association between sirtuins and the IFN system, our data also raise the possibility that targeting sirtuins might provide a unique approach for the development of therapies for immune-related diseases.
Experimental procedures
Cells and reagents
KT-1 (33), U937 (CRL-1593.2; ATCC), and HEL (TIB-180; ATCC) cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), and SET-2 (ACC 608; DSMZ) were grown in RPMI 1640 medium supplemented with 20% FBS. Sirt2+/+ and Sirt2−/− MEFs have been previously described (25). Sirt2+/+ and Sirt2−/− MEFs were cultured in Dulbecco's modified Eagle's medium supplemented with 15% FBS, 1 mm sodium pyruvate, 1 mm nonessential amino acids, 1× β-mercaptoethanol, and antibiotics. The Sirt1+/+, Sirt1−/−, Sirt6+/+, and Sirt6−/− MEFs were kindly provided by Dr. Chu-Xia Deng (NIDDKD, National Institutes of Health) (41, 42) and cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS and antibiotics. U937 cells were transduced with lentiviral control shRNA or lentiviral SIRT2 shRNA (43). Transduction was carried out by spinoculation and stably transduced cells were selected using puromycin (2 μg/ml). The empty vector, CDK9 WT, CDK9-K44R, and CDK9-K48R plasmids have been previously described (29). Control siRNA-B and human SIRT2 and SIRT1 siRNA were from Santa Cruz Biotechnology. Control siRNA and human CDK9 siRNA were purchased from Dharmacon. Recombinant human IFNα was from Hoffman-La Roche Inc. and recombinant human and mouse IFNβ were from Biogen ldec. Antibody against SIRT2 was from Proteintech. Antibodies against phospho-STAT1 (Ser-727), phospho-STAT1 (Tyr-701), and CDK9 were from Cell Signaling. A mAb recognizing human ISG15 was kindly provided by Ernest Borden (Taussing Cancer Center, Cleveland, OH). A rabbit polyclonal antibody against mouse ISG15 has been previously described (44). The anti-STAT1 and nonimmune rabbit IgG antibodies were from Santa Cruz Biotechnology. The antibody against GAPDH and SIRT1 were purchased from Millipore.
Immunoblotting, immunoprecipitation, and in vitro kinase assays
Cells were either left untreated or were treated with the indicated type I IFN (104 IU/ml for short term treatments and 103 IU/ml for 24-h treatments, as indicated). For immunoblotting analyses, cells were lysed in phosphorylation lysis buffer (1 m HEPES, pH 7.3, 5 m NaCl, 1 m MgCl2, 0.5 m EDTA, pH 8.0, 100 μm sodium fluoride, 100 μm sodium pyrophosphate, 0.5% Triton X-100, and 10% glycerol) supplemented with protease and phosphatase inhibitors. Equal amounts of total cell lysates were resolved by SDS-PAGE and transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore). Membranes were then probed with primary antibodies, followed by horseradish peroxidase-conjugated secondary antibodies, and antibody binding was detected by enhanced chemiluminescence using Amersham Biosciences ECL Prime Western blotting detection reagent (GE Healthcare Life Sciences). For kinase assays, KT-1 and Sirt2+/+ and Sirt2−/− MEFs were treated with human IFNβ and mouse IFNβ, respectively, for 30 and 60 min, and lysed in Nonidet P-40 buffer (20 mm HEPES, pH 7.4, 180 mm KCl, 0.2 mm EGTA, 10% glycerol, 0.1% Nonidet P-40) supplemented with protease and phosphatase inhibitors. 300 μg of protein (total cells lysates) from each sample were used for immunoprecipitation of CDK9 using anti-CDK9 rabbit polyclonal antibody (1:100), followed by incubation with protein G-Sepharose 4 Fast Flow beads (GE Healthcare Life Sciences). As control, the same procedure was followed, but using nonimmune rabbit IgG antibody, instead of anti-CDK9 antibody. The beads were washed three time with Nonidet P-40 buffer and twice with kinase buffer (20 mm HEPES, pH 7.4, 10 mm magnesium acetate, and 1 mm DTT) prior to the kinase reaction. In vitro kinase reactions to detect CDK9 kinase activity were performed for 30 min at 30 °C as in previous studies (45). STAT1 recombinant human inactive protein (5 μg) (Thermo Fisher Scientific) was used as an exogenous substrate. [γ-32P]ATP was from PerkinElmer Life Science.
Transfection, hematopoietic progenitor assays, and cell viability assays
Sirt2−/− MEFs were transfected with empty vector, CDK9 WT, CDK9-K44R, CDK9-K48R (29) plasmids using Amaxa Biosystems MEF2 Nucleofector Kit (Lonza) per manufacturer's instructions. The next day, cells were left untreated or were treated with IFNβ as indicated and processed for immunoblotting analysis. SET-2 megakaryoblastic cells were transfected with either control siRNA, CDK9, or SIRT2 siRNA and HEL cells were transfected with either control siRNA or SIRT2 siRNA using Amaxa Biosystems Nucleofector Kit V (Lonza) per the manufacturer's instructions. Cellular proliferation of transfected SET-2 cells in the absence or presence of human IFNα (102 IU/ml) was assessed using WST-1 assay (Hoffman-La Roche Inc.). Leukemic colony formation of transfected HEL cells was assessed in clonogenic assays in methylcellulose in the presence or absence of human IFNα (103 IU/ml). KT-1 cells were transfected with control siRNA or siRNA targeting SIRT1 and SIRT2 using TKO transfection reagent (Mirus) per the manufacturer's instructions. Clonogenic assays in methylcellulose (StemCell Technologies) in the absence or presence of human IFNα (103 IU/ml) were then performed. Human normal bone marrow CD34+ cells (Stem Cell Technologies) were transfected with either control or SIRT1 and SIRT2 siRNA using TKO transfection reagent (Mirus) per the manufacturer's instructions. Clonogenic assays in methylcellulose (StemCell Technologies) in the absence or presence of human IFNα (103 IU/ml) were then performed and erythroid (BFU-E) or myeloid (CFU-GM) colonies were scored. Cellular viability was assessed by plating Sirt2+/+, Sirt2−/− MEFs, SET-2, and HEL cells in 96-well plates in triplicates. Sirt2+/+ and Sirt2−/− MEFs were incubated with the indicated doses of mouse IFNα for 5 days. SET-2 and HEL were treated with human IFNα for 7 days at the indicated doses. Cell viability and proliferation were quantified using WST-1 Reagent (Hoffman-La Roche Inc.) according to the manufacturer's protocol. IC50 values were calculated using GraphPad Prism 6.0 for PC.
Microarray analysis
Sirt2+/+ and Sirt2−/− MEFs were plated and were then left untreated or were treated for 4 h with 2.5 × 103 IU/ml of mouse IFNβ (3 independent replicates). Total RNA was isolated using the RNeasy Mini Kit (Qiagen) per the manufacturer's instructions. The expression analysis was performed at the Genomics Core Facility at Northwestern University. From the total RNA, cRNA was synthesized and labeled using Illumina TotalPrep RNA Amplification Kit (Thermo Fisher). Mouse WG-6 v2.0 Expression BeadChips were hybridized with the labeled cRNA. After staining, the chips were scanned with Illumina iScan (Illumina) according to the manufacturer's protocol. Raw data were imported to GenomeStudio (Illumina), quantile normalized, and the average signal intensities were analyzed in Partek Genomic Suite v.6.6 (Partek, Inc.) after log2 transformation. Qualitative principal component analysis (PCA) did not reveal any outlier samples or artifacts on the microarray. Differential expression between the experimental groups, Sirt2−/− versus Sirt2−/− + IFNβ, and Sirt2+/+ versus Sirt2+/+ + IFNβ, was assessed by using two-way analysis of variance (ANOVA) and Method of Moments together with a false-discovery rate correction of the p value below 0.05 (and with −1.5 > fold-change > 1.5) (46). Fisher's least significant difference was used as the contrast method to compare the experimental groups.
Gene set enrichment analysis
The list of differentially expressed genes identified only in IFNβ-treated Sirt2+/+ cells was submitted to the Metascape database, a gene annotation and analysis resource (http://metascape.org/) (48),8 for pathway and process enrichment analysis. For each given gene list, Metascape carries pathway and process enrichment analysis using the following ontology sources: GO Biological Processes, KEGG Pathway, and Reactome Gene Sets. All genes in the genome were used as the enrichment background. Terms with p value <0.01, minimum count 3, and enrichment factor >1.5 are collected and grouped into clusters based on their membership similarities. More specifically, p values are calculated based on accumulative hypergeometric distribution, q-values are calculated using the Banjamini-Hochberg procedure to account for multiple testing. Kappa scores were used as the similarity metric when performing hierarchical clustering on the enriched terms and then subtrees with similarity >0.3 are considered a cluster. The most statistically significant term within a cluster is chosen as the one representing the cluster.
Quantitative RT-PCR analysis
Sirt2+/+ and Sirt2−/− MEFs were plated and then either left untreated or treated for 6 h with 5 × 103 IU/ml of mouse IFNβ. Total RNA was isolated using the RNeasy Mini Kit (Qiagen) per the manufacturer's instructions. 2 μg of total cellular mRNA was reverse-transcribed into cDNA using the OmniScript RT kit (Qiagen) and oligo(dT)12–18 primers (Life Technologies). Quantitative RT-PCR was carried out using an ABI7500 sequence detection system (Applied Biosystems) using commercially available FAM-labeled probes and primers (Applied Biosystems) to determine mouse Oasl2, Cxcl10, Isg15, and Isg54 mRNA expression. Mouse Gapdh was used for normalization. The mRNA amplification was calculated as described previously (47).
Statistical analyses
All statistical analyses were performed using Prism GraphPad 6.0. Unpaired two-tailed t test with Welch's correction was used for comparison of one observation between two groups and one-way ANOVA was used to compare more than two groups followed by Tukey's multiple comparisons test. Differences were considered statistically significant when p values were less than 0.05.
Author contributions
E. M. K. formal analysis; E. M. K., S. M., D. S., B. K., B. M.-K., D. G., E. N. F., A. V., and L. C. P. validation; E. M. K., S. M., C. D., A. R., A. T., T. L., and L. C. P. investigation; E. M. K., S. M., D. S., B. K., B. M.-K., P. L., C. D., A. R., A. T., T. L., D. G., E. N. F., A. V., and L. C. P. methodology; E. M. K., E. N. F., and L. C. P. writing-original draft; E. M. K., S. M., and L. C. P. project administration; E. M. K., D. S., E. N. F., A. V., and L. C. P. writing-review and editing; P. L. visualization; L. C. P. supervision.
Supplementary Material
Acknowledgment
We thank Dr. Chu-Xia Deng (NIDDK, National Institutes of Health) for kindly providing the Sirt1+/+, Sirt1−/−, Sirt6+/+, and Sirt6−/− MEFs.
This work was supported in part by National Institutes of Health Grants CA77816 and CA155566 and Grant I01CX000916 from the Department of Veterans Affairs. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Tables S1–S4.
The NCBI GEO accession number for the microarray data reported in this paper is GSE66033.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- IFN
- interferon
- JAK
- Janus kinase
- STAT
- signal transducer and activator of transcription
- ISG
- interferon-stimulated gene
- HDAC
- histone deacetylase
- CDK9
- cyclin–dependent kinase 9
- MEF
- mouse embryonic fibroblast
- shRNA
- short hairpin RNA
- FBS
- fetal bovine serum
- ANOVA
- analysis of variance
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Platanias L. C. (2005) Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 2. Hervas-Stubbs S., Perez-Gracia J. L., Rouzaut A., Sanmamed M. F., Le Bon A., and Melero I. (2011) Direct effects of type I interferons on cells of the immune system. Clin. Cancer Res. 17, 2619–2627 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 3. Fuertes M. B., Woo S. R., Burnett B., Fu Y. X., and Gajewski T. F. (2013) Type I interferon response and innate immune sensing of cancer. Trends Immunol. 34, 67–73 , [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ivashkiv L. B., and Donlin L. T. (2015) Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sakamoto S., Potla R., and Larner A. C. (2004) Histone deacetylase activity is required to recruit RNA polymerase II to the promoters of selected interferon-stimulated early response genes. J. Biol. Chem. 279, 40362–40367 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 6. Fish E. N., and Platanias L. C. (2014) Interferon receptor signaling in malignancy: a network of cellular pathways defining biological outcomes. Mol. Cancer Res. 12, 1691–1703 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stark G. R., and Darnell J. E. Jr. (2012) The JAK-STAT pathway at twenty. Immunity 36, 503–514 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saleiro D., Mehrotra S., Kroczynska B., Beauchamp E. M., Lisowski P., Majchrzak-Kita B., Bhagat T. D., Stein B. L., McMahon B., Altman J. K., Kosciuczuk E. M., Baker D. P., Jie C., Jafari N., Thompson C. B., et al. (2015) Central role of ULK1 in type I interferon signaling. Cell Rep. 11, 605–617 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y., Sassano A., Majchrzak B., Deb D. K., Levy D. E., Gaestel M., Nebreda A. R., Fish E. N., and Platanias L. C. (2004) Role of p38α Map kinase in type I interferon signaling. J. Biol. Chem. 279, 970–979 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 10. Kaur S., Kroczynska B., Sharma B., Sassano A., Arslan A. D., Majchrzak-Kita B., Stein B. L., McMahon B., Altman J. K., Su B., Calogero R. A., Fish E. N., and Platanias L. C. (2014) Critical roles for Rictor/Sin1 complexes in interferon-dependent gene transcription and generation of antiproliferative responses. J. Biol. Chem. 289, 6581–6591 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ginter T., Bier C., Knauer S. K., Sughra K., Hildebrand D., Münz T., Liebe T., Heller R., Henke A., Stauber R. H., Reichardt W., Schmid J. A., Kubatzky K. F., Heinzel T., and Krämer O. H. (2012) Histone deacetylase inhibitors block IFNγ-induced STAT1 phosphorylation. Cell Signal. 24, 1453–1460 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 12. Krämer O. H, Knauer S. K., Greiner G., Jandt E., Reichardt S., Gührs K. H., Stauber R. H., Böhmer F. D., and Heinzel T. (2009) A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 23, 223–235 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nusinzon I., and Horvath C. M. (2003) Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc. Natl. Acad. Sci. U.S.A. 100, 14742–14747 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang H. M., Paulson M., Holko M., Rice C. M., Williams B. R., Marié I., and Levy D. E. (2004) Induction of interferon stimulated gene expression and antiviral responses require protein deacetylase activity. Proc. Natl. Acad. Sci. U.S.A. 101, 9578–9583 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glozak M. A., Sengupta N., Zhang X., and Seto E. (2005) Acetylation and deacetylation of non-histone proteins. Gene 363, 15–23 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 16. North B. J, Marshall B. L, Borra M. T, Denu J. M., and Verdin E. (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 17. Rothgiesser K. M., Erener S., Waibel S., Lüscher B., and Hottiger M. O. (2010) SIRT2 regulates NF-κB dependent gene expression through deacetylation of p65 Lys310. J. Cell Sci. 123, 4251–4258 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 18. Hoffmann G., Breitenbücher F., Schuler M., and Ehrenhofer-Murray A. E. (2014) A novel sirtuin 2 (SIRT2) inhibitor with p53-dependent pro-apoptotic activity in non-small cell lung cancer. J. Biol. Chem. 289, 5208–5216 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inoue T., Hiratsuka M., Osaki M., Yamada H., Kishimoto I., Yamaguchi S., Nakano S., Katoh M., Ito H., and Oshimura M. (2007) SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene 26, 945–957 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 20. Dryden S. C., Nahhas F. A., Nowak J. E., Goustin A. S., and Tainsky M. A. (2003) Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol. Cell Biol. 23, 3173–3185 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim H. S., Vassilopoulos A., Wang R. H., Lahusen T., Xiao Z., Xu X., Li C., Veenstra T. D., Li B., Yu H., Ji J., Wang X. W., Park S. H., Cha Y. I., Gius D., and Deng C. X. (2011) SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20, 487–499 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi J. E., and Mostoslavsky R. (2014) Sirtuins, metabolism, and DNA repair. Curr. Opin. Genet. Dev. 26, 24–32 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haigis M. C., and Guarente L. P. (2006) Mammalian sirtuins: emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20, 2913–2921 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 24. O'Callaghan C., and Vassilopoulos A. (2017) Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell 16, 1208–1218 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song H. Y., Biancucci M., Kang H. J., O'Callaghan C., Park S. H., Principe D. R., Jiang H., Yan Y., Satchell K. F., Raparia K., Gius D., and Vassilopoulos A. (2016) SIRT2 deletion enhances KRAS-induced tumorigenesis in vivo by regulating K147 acetylation status. Oncotarget 7, 80336–80349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu P. Y., Xu N., Malyukova A., Scarlett C. J., Sun Y. T., Zhang X. D., Ling D., Su S. P., Nelson C., Chang D. K., Koach J., Tee A. E., Haber M., Norris M. D., Toon C., et al. (2013) The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 20, 503–514 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jing H., Hu J., He B., Negrón, Abril Y. L., Stupinski J. Weiser K., Carbonaro M., Chiang Y. L., Southard T., Giannakakou P., Weiss R. S., and Lin H. (2016) A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and exhibits broad anticancer activity. Cancer Cell 29, 297–310 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Serrano L., Martínez-Redondo P., Marazuela-Duque A., Vazquez B. N., Dooley S. J., Voigt P., Beck D. B., Kane-Goldsmith N., Tong Q., Rabanal R. M., et al. (2013) The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 27, 639–653 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H., Park S. H., Pantazides B. G., Karpiuk O., Warren M. D., Hardy C. W., Duong D. M., Park S. J., Kim H. S., Vassilopoulos A., Seyfried N. T., Johnsen S. A., Gius D., and Yu D. S. (2013) SIRT2 directs the replication stress response through CDK9 deacetylation. Proc. Natl. Acad. Sci. U.S.A. 110, 13546–13551 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bancerek J., Poss Z. C., Steinparzer I., Sedlyarov V., Pfaffenwimmer T., Mikulic I., Dölken L., Strobl B., Müller M., Taatjes D. J., and Kovarik P. (2013) CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38, 250–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kiladjian J. J., Mesa R. A., and Hoffman R. (2011) The renaissance of interferon therapy for the treatment of myeloid malignancies. Blood 117, 4706–4715 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 32. Quentmeier H., MacLeod R. A., Zaborski M., and Drexler H. G. (2006) JAK2 V617F tyrosine kinase mutation in cell lines derived from myeloproliferative disorders. Leukemia 20, 471–476 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 33. Yanagisawa K., Yamauchi H., Kaneko M., Kohno H., Hasegawa H., and Fujita S. (1998) Suppression of cell proliferation and the expression of a bcr-abl fusion gene and apoptotic cell death in a new human chronic myelogenous leukemia cell line, KT-1, by interferon-α. Blood 91, 641–648 [PubMed] [Google Scholar]
- 34. González-Navajas J. M., Lee J., David M., and Raz E. (2012) Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 12, 125–135 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levy D. E., and Darnell J. E. Jr. (2002) Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 36. Au-Yeung N., Mandhana R., and Horvath C. M. (2013) Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT 2, e23931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sadzak I., Schiff M., Gattermeier I., Glinitzer R., Sauer I., Saalmüller A., Yang E., Schaljo B., and Kovarik P. (2008) Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 105, 8944–8949 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang X., Gao J. S., Guan Y. J, McLane K. E., Yuan Z. L., Ramratnam B., and Chin Y. E. (2007) Acetylation-dependent signal transduction for type I interferon receptor. Cell 131, 93–105 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 39. Bell O., Tiwari V. K., Thomä N. H., and Schübeler D. (2011) Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 12, 554–564 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 40. Shakespear M. R., Halili M. A., Irvine K. M., Fairlie D. P., and Sweet M. J. (2011) Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 32, 335–343 [DOI] [PubMed] [Google Scholar]
- 41. Wang R. H., Sengupta K., Li C., Kim H. S., Cao L., Xiao C., Kim S., Xu X., Zheng Y., Chilton B., Jia R., Zheng Z. M., Appella E., Wang X. W., Ried T., and Deng C. X. (2008) Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14, 312–323 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim H. S., Xiao C., Wang R. H., Lahusen T., Xu X., Vassilopoulos A., Vazquez-Ortiz G., Jeong W. I., Park O., Ki S. H., Gao B., and Deng C. X. (2010) Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 12, 224–236 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park S. H., Ozden O., Liu G., Song H. Y., Zhu Y., Yan Y., Zou X., Kang H. J., Jiang H., Principe D. R., Cha Y. I., Roh M., Vassilopoulos A., and Gius D. (2016) SIRT2-mediated deacetylation and tetramerization of pyruvate kinase directs glycolysis and tumor growth. Cancer Res. 76, 3802–3812 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kaur S., Sassano A., Dolniak B., Joshi S., Majchrzak-Kita B., Baker D. P., Hay N., Fish E. N., and Platanias L. C. (2008) Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc. Natl. Acad. Sci. U.S.A. 105, 4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kroczynska B., Kaur S., Katsoulidis E., Majchrzak-Kita B., Sassano A., Kozma S. C., Fish E. N., and Platanias L. (2009) C. Interferon-dependent engagement of eukaryotic initiation factor 4B via S6 kinase (S6K)- and ribosomal protein S6K-mediated signals. Mol. Cell Biol. 29, 2865–2875 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reiner A., Yekutieli D., and Benjamini Y. (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375 10.1074/jbc.RA117.001068 , [DOI] [PubMed] [Google Scholar]
- 47. Kosciuczuk E. M., Saleiro D., Kroczynska B., Beauchamp E. M., Eckerdt F., Blyth G. T., Abedin S. M., Giles F. J., Altman J. K., and Platanias L. C. (2016) Merestinib blocks Mnk kinase activity in acute myeloid leukemia progenitors and exhibits antileukemic effects in vitro and in vivo. Blood 128, 410–414 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tripathi S., Pohl M. O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D. A., Moulton H. M., DeJesus P., Che J., Mulder L. C., Yángüez E., Andenmatten D., Pache L., Manicassamy B., Albrecht R. A., et al. (2015) Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18, 723–735 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.