Abstract
The centromere is an evolutionarily conserved eukaryotic protein machinery essential for precision segregation of the parental genome into two daughter cells during mitosis. Centromere protein A (CENP-A) organizes the functional centromere via a constitutive centromere-associated network composing the CENP-T complex. However, how CENP-T assembles onto the centromere remains elusive. Here we show that CENP-T binds directly to Holliday junction recognition protein (HJURP), an evolutionarily conserved chaperone involved in loading CENP-A. The binding interface of HJURP was mapped to the C terminus of CENP-T. Depletion of HJURP by CRISPR-elicited knockout minimized recruitment of CENP-T to the centromere, indicating the importance of HJURP in CEPN-T loading. Our immunofluorescence analyses indicate that HJURP recruits CENP-T to the centromere in S/G2 phase during the cell division cycle. Significantly, the HJURP binding–deficient mutant CENP-T6L failed to locate to the centromere. Importantly, CENP-T insufficiency resulted in chromosome misalignment, in particular chromosomes 15 and 18. Taken together, these data define a novel molecular mechanism underlying the assembly of CENP-T onto the centromere by a temporally regulated HJURP–CENP-T interaction.
Keywords: mitosis, centromere, chromosomes, chaperone, kinetochore, CENP-A, CENP-E, CENP-T, HJURP, Synthelin
Introduction
Chromosome movements during mitosis are orchestrated by dynamic interactions between spindle microtubules and the kinetochore, a multiprotein complex assembled onto centromeric DNA of the chromosome (1–2). Numerous conserved proteins are recruited to the locus for robust and flexible kinetochore assembly before mitotic entry (3–4). In vertebrates, every single kinetochore contains more than a hundred proteins, which can be divided into couples of subcomplexes. Among them, two important structural subunits are the constitutive centromere-associated network (CCAN)3 and the KNL1, Mis12, and Ndc80 complex (5).
The CCAN contains 16 structural proteins that assemble into five functional complexes: CENP-C, CENP-L/N, CENP-H/I/K/M, CENP-O/P/Q/R/U, and CENP-T/W/S/X (6–15). However, the spatiotemporal assembly pattern of these complexes and the underlying regulatory mechanisms are largely uncharacterized.
CENP-T as a basic CCAN element was first observed in the CENP-A nucleosome-associated complex, which also includes CENP-M, CENP-N, CENP-U, CENP-C, and CENP-H (16). Importantly, previous results have demonstrated that CENP-M, CENP-N, and CENP-T are required for CENP-A nucleosome-associated complex assembly at the centromere (17–20). In addition, several lines of evidence demonstrated that the CENP-T/W/S/X complex forms a nucleosome-like structure and may coordinate with the CENP-A nucleosome in building chromosome architecture and maintaining the stability of the genome (21–23). Recent biochemical and structural characterization indicated that the CENP-T/W complex binds to DNA in a sequence-independent manner by its histone fold domains (24–27). The N-terminal CENP-T has been postulated to serve as a structural platform to interact with the NDC80 complex in a CDK1-regulated fashion (28, 29). More recently, biophotonic study illustrated a temporal profile of CENP-T/W/S/X assembly onto the centromere during S and G2 phases, suggesting that CENP-T exhibits a temporal loading pattern distinct from that of CENP-A. However, the mechanism underlying CENP-T recruitment to the centromere is still unclear.
Several elegant studies have demonstrated that HJURP is a unique chaperone for CENP-A deposition to the centromere during early G1 phase (30–33). In addition, the highly conserved scm3 domain of HJURP is required for stabilizing the CENP-A–H4 complex and depositing CENP-A (34–39). Our previous work demonstrated that Mis18β binds and specifies the localization of HJURP to the centromere, which is negatively regulated by CDK1 activity (40).
HJURP contains a DNA-binding domain and a dimerization domain, and both domains are required for CENP-A deposition (41–43). Interestingly, both subcellular fractionation studies and previous immunofluorescence observations indicate that HJURP remains in the nucleus after G1 phase, when nascent CENP-A assembly has completed. We were curious about the function of HJURP in S/G2 phase and whether HJURP has a context-dependent function during mitosis.
In this study, we report that HJURP is a novel loading factor responsible for histone fold protein CENP-T assembly to the centromere during G2 phase. A minimal domain of HJURP was mapped to residues 527–532, which are required for its direct interaction with CENP-T and responsible for recruitment of CENP-T to the centromere. Mutation of this domain generated a HJURP binding–deficient mutant, CENP-T6L, which failed to localize at the centromere. Importantly, overexpression of the HJURP binding–defective mutant CENP-T6L promoted chromosome instability, as judged by aberrant segregation of chromosomes 15 and 18 (44, 45). Our study proposes that HJURP orchestrates the centromeric assembly of CENP-A and CENP-T in a temporally controlled manner.
Results
HJURP interacts with CENP-T and specifies its centromere localization
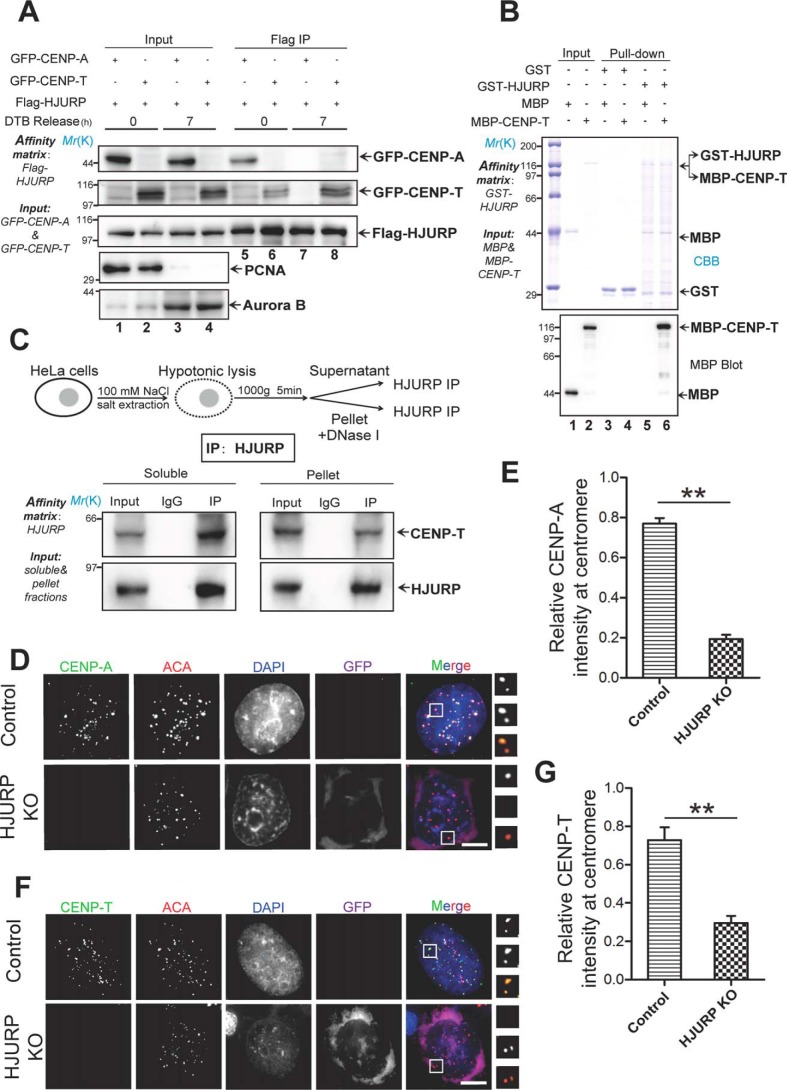
Our trial experiments suggest that HJURP forms a complex with CENP-A, which also contains a proportion of CENP-T (40). To directly assess whether HJURP interacts with and regulates CENP-T distribution at centromeres, we carried out an immunoprecipitation experiment using thymidine-synchronized cells from G1/S and G2, respectively. The synchronized cells were confirmed using Western blot analyses of proliferating cell nuclear antigen (G1/S; Fig. 1A, lanes 1 and 2) and Aurora B (G2; Fig. 1A, lanes 3 and 4). Examination of the temporal pattern of the aforementioned interactions using synchronized cells demonstrated that HJURP association with CENP-T was first detected at early G1 phase (Fig. 1A, lane 6), and the amount of CENP-T association with HJURP reached a peak at early mitosis (Fig. 1A, lane 8). To determine the subcellular distribution of CENP-A and CENP-T relative to HJURP in G1/S phase, we carried out a subcellular fractionation experiment in which nuclear and cytosolic fractions were collected for Western blot analyses. As shown in Fig. S1A, CENP-A is mainly located in the nuclear fraction, whereas the majority of HJURP and CENP-T is distributed in the cytosolic fraction. Next, we performed quantitative immunofluorescence analyses and measured the subcellular distribution profile of CENP-T (Fig. S1B). The distribution of CENP-T in the nucleus in G1/S phase cells prompted us to examine whether CENP-T interacts directly with HJURP. To this end, we employed GST-HJURP as an affinity matrix to isolate MBP–CENP-T from solution. Western blot analyses of MBP confirmed that CENP-T physically binds to HJURP (Fig. 1B, lane 6).
Figure 1.
HJURP interacts with and specifies CENP-T localization to the centromere. A, aliquots of HeLa cells were transiently transfected to express FLAG-HJURP with GFP–CENP-T or GFP–CENP-A. 4 h after transfection, the cells were synchronized by a double thymidine block (DTB) protocol, and synchronized cells were released into G1/S phase (0 h) or G2 phase (7 h). Then the synchronized cells were lysed, and clarified cell lysates were incubated with FLAG-agarose. The beads were washed and boiled in 1× SDS-PAGE sample buffer prior to electrophoresis. Results were analyzed by Western blotting with the indicated antibodies. Each blot was cut around the expected band and is presented with molecular weight information. PCNA, proliferating cell nuclear antigen. B, HJURP interacts directly with CENP-T in vitro. GST-tagged HJURP recombinant protein bound on GSH-agarose beads was used as an affinity matrix to isolate MBP–CENP-T purified from bacteria. Results were analyzed by anti-MBP blot. C, top panel, schematic of the experimental preparation of cell extract. Synchronized HeLa cells were lysed at various hypotonic NaCl concentrations, and the supernatant was separated by centrifugation (1,000 × g, 5 min). The remaining fraction was further treated with DNase I. Bottom panel, HJURP antibody and IgG were incubated with supernatant and pellet fractions for 1 h, and then protein A/G was added to the incubation for another 1 h. Western blotting showed that HJURP interacts with CENP-T in both the supernatant and pellet in vivo. IP, immunoprecipitation. D, HJURP KO plasmid pools were transfected into HeLa cells. Seventy-two hours after transfection, HeLa cells were fixed, followed by a standardized immunofluorescence staining protocol for CENP-A (green), ACA (red), GFP (purple), and DNA (blue). Representative images were collected from three independent experiments and are presented. Scale bar = 5 μm. E, statistical analyses of CENP-A centromere localization level under efficient HJURP knockout. Over 25 cells were tested in each category from three independent preparations. Data represent mean ± S.E. Statistical significance was tested by two-sided t test. **, p < 0.01. F, HJURP KO plasmids were transfected into HeLa cells, and centromere localization of CENP-T was analyzed using the indicated antibodies. As indicated, HJURP has a critical role in CENP-T centromere localization. CENP-T, green; ACA, red; GFP, purple; DNA, blue. Scale bar = 5 μm. G, statistical analysis of CENP-T centromere localization level upon HJURP knockout. Over 25 cells were tested in each category from three independent preparations. Data represent mean ± S.E. Statistical significance was tested by two-sided t test. **, p < 0.01.
To assess whether HJURP plays a role in the CENP-T loading process, aliquots of HeLa cells were transfected with CRISPR knockout (KO) plasmids to suppress the expression of HJURP. As shown in Fig. S1C, this CRISPR-mediated KO resulted in >90% suppression (Fig. S1C). We next examined whether depletion of HJURP interferes with the localization of CENP-T to the centromere. Because KO plasmids fused with the GFP reporter, green fluorescence provided a positive readout for transfection. Using immunofluorescence analyses, we found that GFP-positive cells contain virtually no HJURP expression (Fig. S1D). Our statistical analyses confirmed that more than 90% of HJURP was suppressed by CRISPR-mediated KO (Fig. S1E). Given the efficient suppression of HJURP using CRISPR-mediated KO, we next examined whether suppression of HJURP altered the localization of CENP-T to the centromere. As shown in Fig. 1D, suppression of HJURP abolished the localization of CENP-A to the centromere, which is consistent with earlier reports (37–40). Quantitation of relative intensity (CENP-A/ACA) showed that the level of CENP-A at the centromere was reduced to 25% ± 3% of the control (Fig. 1E, p < 0.01). As shown in Fig. 1F, suppression of HJURP also abolished the centromere localization of CENP-T. Quantitative analyses of relative intensity (CENP-T/ACA) in HJURP-depleted cells showed that the level of CENP-T at the centromere was reduced to 37% ± 5% of the control (Fig. 1G, p < 0.01). These data demonstrate that HJURP is required for stable localization of both CENP-A and CENP-T to the centromere.
HJURP co-localizes with CENP-T from G1 to G2 phase
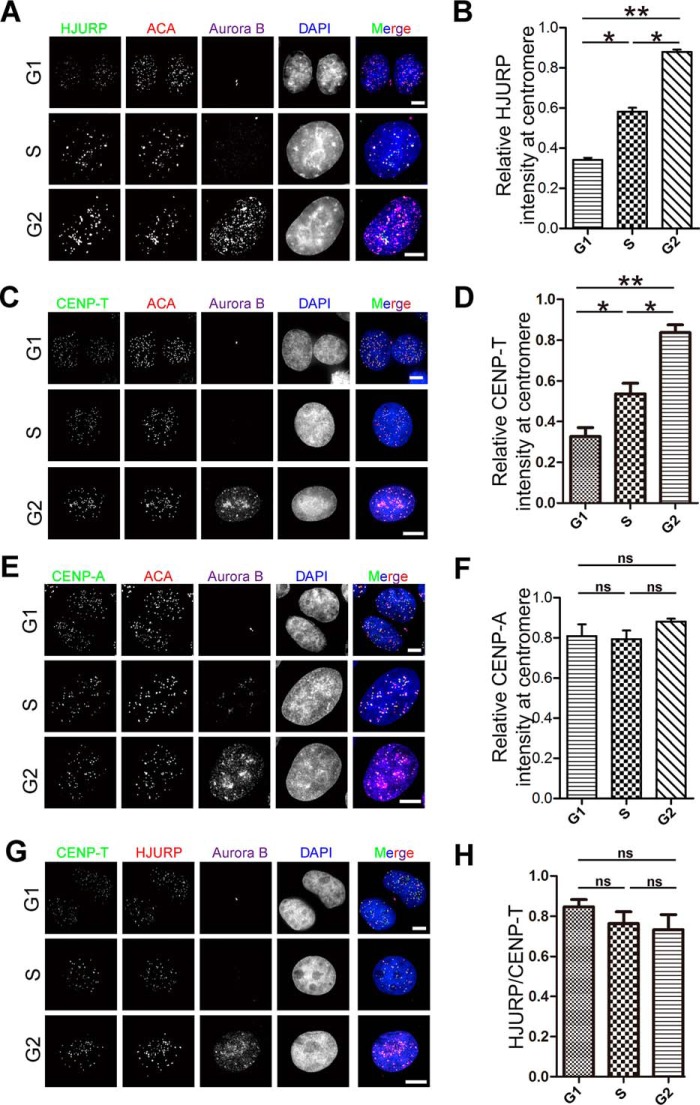
HJURP is critical for loading CENP-A to the centromere. The requirement of HJURP for stable CENP-T localization to the centromere prompted us to determine whether HJURP is a loading factor for CENP-T. To this end, aliquots of synchronized HeLa cells were fixed and immunocytochemically stained for ACA, Aurora B and HJURP, or CENP-T. Quantitative analyses of relative intensity (HJURP/ACA) showed that the intensity of HJURP at the centromere increases from early G1 to G2 phase (Fig. 2, A and B; p < 0.05). Interestingly, quantification of relative intensity (CENP-T/ACA) demonstrated that the intensity of total CENP-A at total centromere CENP-T was also increased from G1 to G2 phase (p < 0.05). However, the intensity level of CENP-A at the centromere showed no significant change from G1 to G2 phase (Fig. 2, E and F; p > 0.05). In contrast, the total protein level of CENP-T increased from G1 to G2 phase (Fig. S2A), which is consistent with quench-chase-pulse assays reported previously (46, 47). To compare the spatiotemporal pattern of CENP-A with that of CENP-T, we carried out another set of immunofluorescence staining of CENP-T and HJURP (Fig. 2G). As shown in Fig. 2H, quantitative analyses of the relative intensity of CENP-T and HJURP from three separate experiments demonstrated that centromere-associated CENP-T and HJURP are stable from S to G2 phase, with a slight reduction. Thus, we conclude that CENP-T is recruited to centromeres by HJURP in S/G2 phase of the cell division cycle.
Figure 2.
HJURP and CENP-T co-localized at centromere from G1 to G2 phase. A, HeLa cells were co-stained for HJURP (green), ACA (red), Aurora B (purple), and DNA (blue). Endogenous staining indicates centromere localization of HJURP increasing from G1 phase to G2 phase. Scale bars = 5 μm. B, statistical analyses of HJURP centromere localization level at different phases. Over 40 cells were tested in each category from three independent preparations. Data represent mean ± S.E. Statistical significance was tested by two-sided t test. *, p < 0.05; **, p < 0.01. C, HeLa cells were co-stained for CENP-T (green), ACA (red), Aurora B (purple), and DNA (blue). Note that centromere localization of CENP-T increased from G1 phase to G2 phase. Scale bars = 5 μm. D, statistical analysis of CENP-T centromere localization level at different phases. Over 55 cells were tested in each category from three independent preparations. Data represent mean ± S.E. Statistical significance was tested by two-sided t test. *, p < 0.05; **, p < 0.01. E, HeLa cells were co-stained for CENP-A (green), ACA (red), Aurora B (purple), and DNA (blue). Note that centromere localization of CENP-A did not show a significant difference from G1 phase to G2 phase. Scale bars = 5 μm. F, statistical analysis of CENP-A centromere localization level at different phases. Over 45 cells were tested in each category from three independent preparations. Data represent mean ± S.E. from three independent experiments. Statistical significance was tested by two-sided t test. ns, no significant difference. G, HeLa cells were co-stained for CENP-T (green), HJURP (red), Aurora-B (purple), and DNA (blue). Note that the ratio of the centromere localization level of HJURP to CENP-T exhibits no significant difference from G1 phase to G2 phase. Scale bars = 5 μm. H, statistical analysis of the ratio of centromere localization level of HJURP to CENP-T at different phases. Over 50 cells were tested in each category from three independent preparations. Data represent mean ± S.E. Statistical significance was tested by two-sided t test.
CENP-T physically binds to C-terminal HJURP
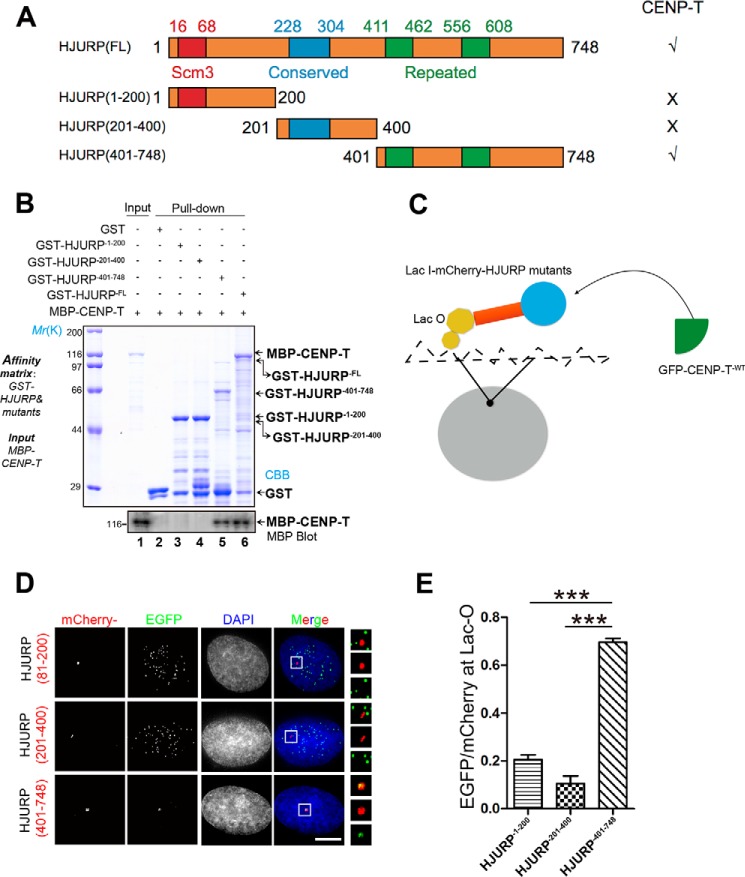
The function of HJURP is conserved from yeast to humans, and the scm3 domain of HJURP is required for direct physical interaction with CENP-A (39, 48). To delineate the structure–function relationship of the HJURP–CENP-T interaction, we next pinpointed the precise region involved in the HJURP–CENP-T interaction. To this end, we designed and generated three truncations of HJURP: GST-HJURP1–200, GST-HJURP201–400, and GST-HJURP401–748 (Fig. 3A). GST-tagged HJURP proteins (full-length and deletion mutants) were purified and used as an affinity matrix to absorb MBP–CENP-T. As shown in Fig. 3B, lanes 1, 5, and 6, both full-length and C-terminal HJURP brought down MBP–CENP-T, whereas other regions of HJURP exhibited no detectable binding activity on CENP-T. To examine whether the C-terminal HJURP also interacts with CENP-T in cells, full-length and three deletion mutants of HJURP were tagged with an ectopic chromosome locus (Lac-I) in U2OS cells (Fig. 3C), as established previously (e.g., Ref. 17). As shown in Fig. 3D, overexpressed Lac-I–HJURP401–748 exhibits the highest co-localization efficiency with GFP–CENP-T and weak localization at the centromere because of competition pressure from endogenous CENP-T (bottom panel, inset), which is consistent with our biochemical characterization using a pulldown assay. Quantification of the relative intensity of CENP-T/HJURP demonstrated that CENP-T is efficiently recruited to Lac-O site–bearing C-terminal HJURP (Fig. 3E).
Figure 3.
CENP-T physically interacts with the C-terminal HJURP. A, schematic of HJURP deletion mutant constructs. B, C-terminal HJURP interacts with CENP-T. GST-tagged HJURP1–200, HJURP201–400, and HJURP401–748 recombinant proteins were used as an affinity matrix to isolate MBP–CENP-T, respectively. GSH beads were subjected to extensive washes, followed by SDS-PAGE and Western blot analysis by anti-MBP antibody. C, schematic of the de novo recruitment system and design. D, U2OS cells stably expressing Lac-O were co-transfected with Lac-I–mCherry–HJURP81–200, Lac-I–mCherry–HJURP201–400, Lac-I–mCherry–HJURP401–748, and GFP–CENP-T. Twenty-four hours after transfection, cells were fixed and stained with DAPI. Scale bar = 5 μm. E, statistical analysis of CENP-T recruitment efficiency of various HJURP truncation mutants. More than 25 cells from three independent experiments were examined in each category. Data represent mean ± S.E. Statistical significance was tested by two-sided t test. ***, p < 0.001.
Because dimerization of HJURP is essential for loading CENP-A to the centromere, we then evaluated whether the dimerization domain of HJURP influences its physical interaction with CENP-T. Consequently, we constructed a dimerization-deficient HJURP plasmid by removing amino acids 554–614 from the C-terminal HJURP, as reported previously (42). The construct was designated GST-HJURP401–748-DE-Di, and purified protein was used as an affinity matrix (Fig. S3A). As shown in Fig. S3B, lanes 3 and 4, the pulldown assay demonstrated that GST-HJURP401–748, but not GST-HJURP401–748-DE-Di, brought down the MBP–CENP-T protein. Thus, we conclude that dimerization of HJURP is essential for its binding to CENP-T.
HJURP binding–deficient mutant CENP-T6L failed to localize correctly
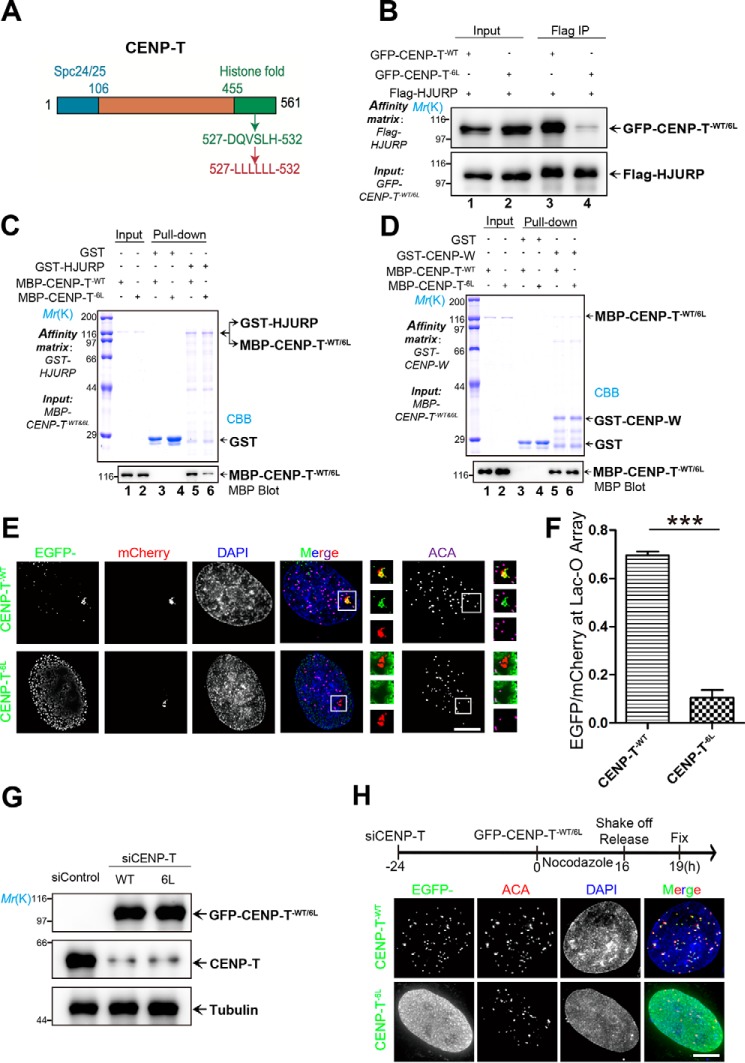
Because HJURP is essential for CENP-A loading, and its dimerization domain plays an important role in centromere loading (42), we sought to evaluate whether CENP-T–HJURP interaction is essential for loading CENP-T to the centromere. To manipulate the interaction between CENP-T and HJURP, we scanned for mutations in the histone fold domain of CENP-T, which compromises CENP-T binding to HJURP, using molecular modeling. Our preliminary analyses suggest that mutation of DQVSLH to six Leu residues (CENP-T6L) in the histone fold domain perturbed the interaction of CENP-T with HJURP (Fig. 4A). To assess whether the aforementioned mutation alters the interaction of CENP-T with HJURP in cells, aliquots of 293T cells were transiently co-transfected to express FLAG-HJURP and GFP–CENP-TWT or FLAG-HJURP plus GFP–CENP-T6L. Transfected cells were subjected to immunoprecipitation using anti-FLAG M2 affinity beads. Western blot analyses show that FLAG-HJURP efficiently brought down WT GFP-CENP-T (Fig. 4B, lane 3) but not the GFP-CENP-T6L mutant (Fig. 4B, lane 4).
Figure 4.
The HJURP binding–deficient mutant CENP-T6L failed to localize at the centromere. A, schematic of the CENP-T histone fold mutant. B, aliquots of HEK293T cells were transiently transfected to express FLAG-HJURP CENP-TWT and CENP-T6L. Thirty-six hours after transfection, the cells were harvested, pelleted, lysed, and clarified. The clarified cell lysates were then incubated with FLAG-agarose. The beads were extensively washed and boiled in 1× SDS-PAGE sample buffer. The samples were resolved for SDS-PAGE, followed by Western blot analyses with the indicated antibodies. IP, immunoprecipitation. C, GST-HJURP pulldown of MBP-CENP-TWT and MBP-CENP-T6L, analyzed by Western blots using anti-MBP antibody. The gel was stained with CBB. The CENP-T6L mutant exhibited much reduced physical interaction with HJURP. D, GST–CENP-W pulldown of MBP–CENP-TWT and MBP–CENP-T6L. GST or GST–CENP-W affinity matrices were incubated with MBP–CENP-TWT or MBP–CENP-T6L. After incubation, the beads were extensively washed and boiled in sample buffer. The samples were resolved by SDS-PAGE and analyzed by Western blotting using anti-MBP antibody. The gel was stained with CBB (top panel). Note that the CENP-T6L mutant interacted with CENP-W as CENP-TWT did. E, U2OS cells stably expressing Lac-O were co-transfected with Lac-I–mCherry–HJURP81–748 and GFP–CENP-TWT or GFP–CENP-T6L. Twenty-four hours after transfection, cells were fixed and stained for DNA (blue), tethered HJURP (red), CENP-T (green), and ACA (purple). Scale bar = 5 μm. F, statistical analysis of CENP-TWT and CENP-T6L recruitment efficiency. More than 25 cells were tested in each category from three independent preparations. Data represent mean ± S.E. Statistical significance was tested by two-sided t test. ***, p < 0.001. G, GFP–CENP-TWT and GFP–CENP-T6L were transfected into HeLa cells depleted of endogenous CENP-T. Cells were collected 48 h after transfection and analyzed by Western blotting using the indicated antibodies. A tubulin blot served as a loading control. H, GFP–CENP-TWT and GFP–CENP-T6L were transfected into HeLa cells depleted of endogenous CENP-T, followed by treatment as indicated in the flow chart. The centromere was labeled red using ACA, whereas exogenously expressed CENP-T (WT and mutant) were labeled green. DNA was stained with DAPI (blue). Scale bar = 5 μm.
To evaluate the binding activity of the CENP-T6L mutant to HJURP, aliquots of GST-HJURP were used as an affinity matrix to absorb recombinant CENP-T WT and mutants. MBP–CENP-T was fully retained on the GST-HJURP beads (Fig. 4C, lane 5), whereas the MBP–CENP-T6L mutant failed to be retained on the GST-HJURP affinity matrix (Fig. 4C, lane 6). Because CENP-T forms a functional dimer with CENP-W, we sought to examine whether the mutant CENP-T6L alters its interaction with CENP-W. CENP-W binding to CENP-T was not altered by the histone fold mutation (Fig. 4D, lanes 5 and 6).
Next, we examined whether HJURP binding–deficient CENP-T is capable of targeting to the centromere engineered by Lac-I–HJURP. To this end, GFP–CENP-T6L or GFP–CENP-TWT was co-transfected with Lac-I–mCherry-HJURP81–748 into U2OS cells stably expressing Lac-O. Our immunofluorescence analyses indicated that WT GFP–CENP-T efficiently targeted to the Lac-O site expressing Lac-I–mCherry-HJURP81–748 (Fig. 4E, top panels). However, GFP–CENP-T6L was absent from Lac-I–mCherry-HJURP81–748 (Fig. 4E, bottom panels). Quantification of the relative intensity (enhanced GFP/mCherry) showed that the level of CENP-T6L at the centromere is about 15.3% ± 0.3% of CENP-TWT (Fig. 4F). Thus, we conclude that the histone fold domain of CENP-T is essential for its interaction with HJUTP, as the CENP-T6L mutant failed to localize to the HJURP foci in U2OS cells.
If HJURP specifies CENP-T to the centromere via its interaction with the CENP-T histone fold domain, then suppression of the endogenous CENP-T level would enable us to test whether GFP-CENP-T6L remains localized to the centromere or other structures. To this end, we carried out siRNA-mediated CENP-T knockdown, followed by transfection of WT and GFP–CENP-T6L. As shown in Fig. 4G, the endogenous CENP-T protein level was minimized, whereas exogenously expressed WT and mutant GFP-CENP-T6L were at the same level of endogenous CENP-T. We next sought to examine whether WT and mutant GFP–CENP-T6L localize to the centromere. As illustrated in Fig. 4H, HeLa cells were first transfected with siRNA, followed by a second transfection of WT and mutant GFP–CENP-T6L. Transfected cells were then exposed to nocodazole for 16 h to synchronize cells at mitosis. Mitotic cells were then collected by shake-off, followed by relocation into a new plate. These transfected and treated cells were released for 3 h (now G1 phase) before fixation. As shown in Fig. 4H, WT GFP–CENP-T expressed in the nucleus as characteristic double dots where the localization was superimposed onto that of ACA labeling, indicating that exogenously expressed GFP–CENP-T is located at the centromere. However, GFP–CENP-T6L diffused in the nucleus with a high background, indicating that the HJURP binding–deficient mutant failed to load CENP-T onto the centromere in early G1 (Fig. 4H). In addition, GFP–CENP-T6L was largely absent from the centromere in mitotic cells, whereas WT GFP–CENP-T localized to the centromere of mitotic cells (Fig. S4, A and B). Thus, we conclude that HJURP is required and sufficient for loading CENP-T to the centromere and its stable deposition.
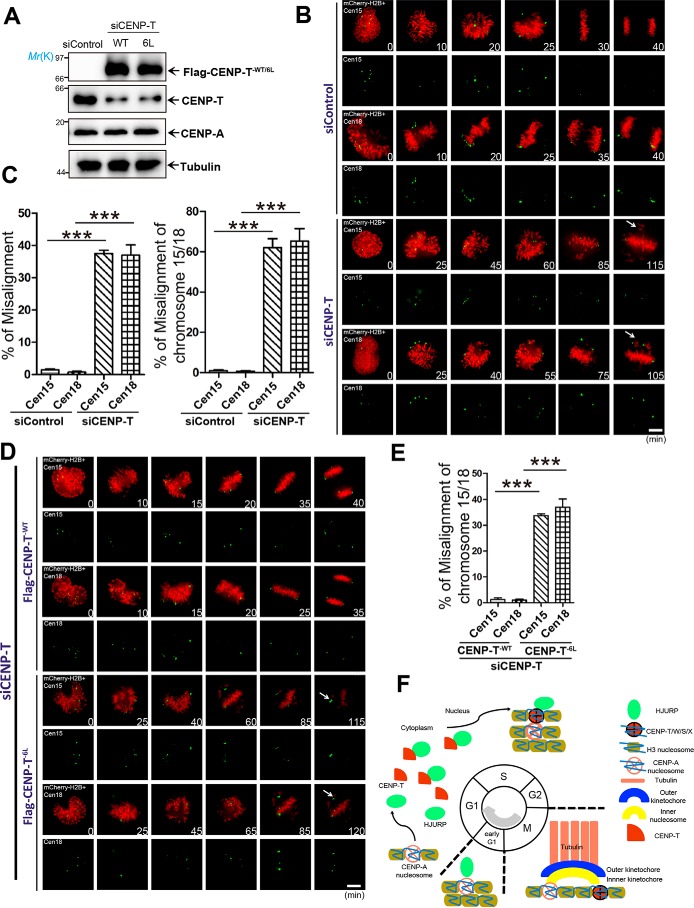
Overexpression of the CENP-T6L mutant perturbed chromosome segregation
Recent studies from The Cell Genome Atlas project indicate that chromosome instability is a common feature of solid tumors and that chromosomes 15 and 18 are more frequently gained and lost during tumorigenesis (49). To test whether perturbation of CENP-T loading alters specific chromosome segregation, we sought to examine the function of CENP-T in chromosome 15 and 18 segregation. Chromosome 18 is highly unstable and often lost in gastrointestinal tumors, whereas chromosome 15 is relatively stable (49). Karyotype analyses indicated that HeLa cells typically contain 82 chromosomes, and our preliminary studies show that centromeres of chromosomes 15 and 18 appeared as four to six points in G1 phase and eight to 12 points in G2 phase (Fig. S5A).
FLAG–CENP-TWT and FLAG–CENP-T6L mutants expressed at comparable levels in HeLa cells depleted of endogenous CENP-T. Western blot analyses showed that the endogenous CENP-T was suppressed to 15% of the control, whereas exogenous FLAG–CENP-TWT expression was comparable with that of the endogenous level.
Next, we tested whether suppression of CENP-T affects chromosome segregation accuracy. To this end, aliquots of CENP-T knock-down HeLa cells were transiently transfected with mCherry-H2B and Cen15 or Cen18, followed by real-time imaging. Surprisingly, suppression of CENP-T resulted in chromosome misalignment, which mainly occurred on chromosome 15 and chromosome 18, as indicated by centromere markers (Fig. 5C).
Figure 5.
Expression of CENP-T6Lperturbed segregation of chromosome 15 and 18 in mitosis. A, assessment of the protein expression level of FLAG–CENP-TWT and its mutants in HeLa cells depleted of endogenous CENP-T. Cells were collected 48 h after transfection and analyzed by Western blot analyses using the indicated antibodies. Tubulin served as a loading control. B, HeLa cells were first transfected with CENP-T siRNA to knock down endogenous CENP-T. Twenty-four hours after siRNA transfection, cells were transfected to express mCherry-H2B and reporters for chromosomes 15 and 18 (Cen15/Cen18, respectively). After another 24 h, live-cell imaging was carried out to follow chromosome segregation into anaphase. Images were acquired at the indicated time points (minutes). Note that control siRNA–treated cells progress into anaphase in 30 ± 5 min, whereas depletion of CENP-T resulted in chronic arrest of mitosis, hallmarked by misaligned chromosomes over 115 min. Scale bar = 5 μm. C, left panel, statistical analysis of HeLa cells with chromosome misalignment in four groups. Right panel, statistical analysis of HeLa cells with chromosome 15 and chromosome 18 misalignment. Over 25 cells were tested in each category from three independent preparations. Data represent mean ± S.E. Statistical significance was tested by two-sided t test. ***, p < 0.001. D, HeLa cells were first transfected with CENP-T siRNA to knock down endogenous CENP-T. Twenty-four hours after siRNA transfection, cells were transfected to express reporters for Cen15/Cen18, mCherry-H2B, and FLAG-CENP-T (WT and mutant CENP-T6L). After another 24 h, live-cell imaging was carried out to follow chromosome segregation into anaphase. Images were acquired at the indicated time points (minutes). Note that control siRNA–treated cells progress into anaphase in 30 ± 5 min, whereas depletion of CENP-T resulted in chronic arrest of mitosis, hallmarked by misaligned chromosomes 15 and 18. Scale bar = 5 μm. E, statistical analysis of misalignment for chromosomes 15 and 18 in four groups. More than 25 cells were tested in each category from three independent preparations. Data represent mean ± S.E. Statistical significance was tested by two-sided t test. ***, p < 0.001. F, hypothetic model to account for the mechanism underlying CENP-T deposition during the cell division cycle.
Because centromeric localization of CENP-T6L was dramatically impaired, we co-transfected mCherry-H2B, Cen15 or Cen18, and FLAG-CENP-TWT or FLAG-CENP-T6L into CENP-T–deficient cells to attempt restoration of CENP-T expression and accurate chromosome segregation. As shown in Fig. 5D, expression of WT but not mutant FLAG-CENP-T6L restored chromosome misalignment induced by CENP-T knockdown. Quantitative analyses showed that chromosome misalignment mainly occurred on chromosomes 15 and 18 (Fig. 5E), suggesting a context-dependent function of CENP-T at different chromosomes.
Discussion
Faithful segregation of the genome depends on the functional centromere, which is built on the CENP-A nucleosome and connects with spindle microtubules via CENP-N/L, CENP-S/X-T/W, and NDC80 complexes. Given the crucial role of CENP-A in centromere specification, several lines of evidence have demonstrated that accurate Mis18β–HJURP interaction provides temporal regulation of CENP-A incorporation into the centromere (16, 36–43). Recent studies have delineated the structural basis underlying CENP-N recognition of the CENP-A nucleosome (19, 20, 50). However, how CENP-S/X-T/W is recruited and assembled onto the centromere remains elusive. Here we uncover the molecular mechanism by which HJURP recruits CENP-T to the centromere.
Although exciting progress has been made toward a better understanding of centromere architecture and interdependence of centromere protein complexes (16, 18, 34, 36, 38), the molecular mechanisms underlying CCAN assembly and their regulation during cell division cycle remain elusive. In particular, very little is known about the respective mechanisms underlying the assembly and disassembly characteristics of the CCAN. In this study, we characterized and established HJURP as a novel assembly factor responsible for accurate CENP-T loading and chromosome segregation. Early studies have established HJURP as CENP-A chaperone and its assembly in early G1 (36–37). The N-terminal region of the CenH3 α2 helix is sufficient for specific recognition by the chaperone in both budding yeast and humans (50–53). However, there is no evidence that shows how HJURP interacts with CENP-A in the cytoplasm. During the preparation of this revision, Foltz and co-workers (54) reported that HJURP is required for centromeric CENP-A nucleosome inheritance. Using auxin-induced degradation of HJURP, they also revealed the requirement of HJURP for CENP-T stable localization at the centromere, which supports our finding.
It is worth noting that the relative level of CENP-T at the centromere was increased from G1 to G2 phase, whereas the intensity level of CENP-A at the centromere exhibited no significant change from G1 to G2 phase. The cellular reservoir of CENP-T, CENP-A, and HJURP is composed of cytoplasmic and nuclear pools, and earlier studies have reported potential dilution of CENP-A intensity at the centromere in S phase (37, 41). However, a follow-up characterization from the same group indicated that the CENP-A level is not quantitatively diluted in S phase (55). Our biochemical characterization and immunofluorescence analyses are consistent with this study (55). Future spectral imaging analyses will provide quantitative and precise measures of spatiotemporal interactions between CENP-A–HJURP and CENP-T–HJURP and dynamic partition of CENP-A/CENP-T in the nucleus, cytoplasm, and centromere during the cell division cycle. Despite our demonstration of a physical interaction between CENP-T and HJURP, the current study does not delineate how HJURP distinguishes CENP-T from CENP-A. Therefore, in the future, it would be of great interest to illustrate how HJURP recognition of CENP-T and CENP-A is determined and regulated by the cell cycle machinery. Because CENP-T forms a functional complex with CENP-W, it would be of great importance to ascertain whether the CENP-T/CENP-W heterodimer is formed at the centromere after individual loading or assembled onto the centromere as a functional heterodimer during the loading process.
Based on our characterization, we propose that HJURP binds to CENP-T both in the cytoplasm and in the nucleus in interphase cells. Recent work from Cheeseman and co-workers (29) demonstrated that the function of CENP-T is regulated by CDK1 kinase activity in mitosis for outer kinetochore connection and spindle plasticity control, which is consistent with our observation of CENP-T activity dynamics during the cell division cycle. Future work will be directed to the structural basis underlying HJURP recognition of CENP-T and how the molecular dynamics of HJURP are regulated by the cell cycle machinery.
Cleveland and co-workers (56) have revealed that mis-segregation of chromatin into micronuclei results in DNA damage and chromosome rearrangement. However, the underlying molecular mechanism remains poorly understood, partially because chromosome mis-segregation into micronuclei appears to be random. A recent study showed that induced CENP-A deficiency resulted in Y centromere inactivation and mis-segregation of the Y chromosome (56). Our study demonstrated that gross suppression of CENP-T resulted in perturbation of chromosome 15 and 18 alignment, which supports the notion that chromosome stability of individual chromosomes in cell division varies. Although in this study we did not carry out an extensive characterization of chromosome stability of all chromosomes, we demonstrated a tight link between chromosome stability and centromere integrity. It will be of great interest to see how chromosome instability phenotypes such as chromothripsis and anaphase chromatin bridges differ and whether CENP-T depletion will result in chromothripsis of chromosome 15 and 18.
In conclusion, we demonstrated that HJURP governs both CENP-A and CENP-T centromeric deposition in a spatiotemporally regulated manner and provided a new mechanism of how HJURP orchestrates CENP-T and CENP-A during S and early G1 phase, respectively. Precise assembly of functional CCAN complex and centromere machinery is critical to ensure robust kinetochore–microtubule attachment for accurate chromosome segregation in mitosis. Thus, the evolutionarily conserved HJURP–CENP-T interactive network may serve as a novel sensor of kinetochore–spindle attachment for accurate mitosis from yeast to humans.
Experimental procedures
Cell culture
HEK293T cells and HeLa cells from the ATCC (Manassas, VA) were maintained as subconfluent monolayers in advanced Dulbecco's modified Eagle's medium (Gibco) with 10% fetal bovine serum (Hyclone) and 100 units/ml of penicillin plus 100 μg/ml of streptomycin (Gibco).
Plasmid construction
GFP, FLAG, and GST-tagged HJURP, full-length and truncations, were constructed as described previously (40). Bacterially expressed constructs of CENP-T, full-length and truncations, were ligated into the pMal-C2 vector (New England Biolabs). Bacterially expressed constructs of CENP-W were cloned into pGEX-6P-1 (GE Healthcare). All plasmid constructs were sequenced for verification.
siRNA treatment
The siRNA sequence (5′-CGGAGAGCCCUGCUUGAAA-3′) used for silencing CENP-T was synthesized by Qiagen. As a control, a scramble sequence was used. All siRNAs were transfected into cells using Lipofectamine 3000 for 48 h, and the knockdown efficiency was assessed by Western blot analysis.
Antibodies
The following antibodies were obtained from commercial sources: anti-CENP-T antibody (Abcam and gifts from Dr. Iain Cheeseman), anti-HJURP antibody (CST), anti-proliferating cell nuclear antigen antibody (CST), anti-CENP-A antibody (CST), anti-Cyclin B antibody (CST), anti-H3 antibody (CST), anti-Aurora-B antibody (CST), anti-GFP antibody (BD Biosciences), anti-MBP antibody (Sigma), and anti-tubulin antibody DM1A (Sigma). FITC-conjugated secondary antibodies (Pierce) and rhodamine-phalloidin (Invitrogen) were obtained commercially.
HJURP Knockout
The HJURP CRISPR knockout plasmids (Santa Cruz) were transiently transfected into HeLa cells using the Lipofectamine protocol, and efficient knockdown was achieved 72 h after transfection. The efficacy and knockout specificity were judged by Western blot analyses.
Expression and purification of recombinant proteins
Purification of recombinant proteins was carried out as described previously (10). Briefly, the soluble GST fusion proteins from the bacterial cell lysates were purified using GSH-agarose beads (Sigma), whereas MBP-tagged proteins were purified using amylose beads (New England Biolabs).
Pulldown assay
Pulldown assays were carried out as described previously (40). Briefly, the GST fusion proteins in the soluble fraction were purified from bacteria by GSH-agarose chromatography, whereas MBP-tagged proteins were purified using amylose beads and then eluted by MBP elution buffer (10 mm maltose in PBS). Then GST fusion protein–bound GSH-agarose beads were incubated with purified MBP-tagged fusion proteins for 1 h at 4 °C. After incubation, the beads were washed three times with PBS containing 0.2% Triton X-100 and once with PBS without Triton X-100 and then boiled in SDS-PAGE sample buffer. The bound proteins were separated on a 10% SDS-PAGE gel for Coomassie Brilliant Blue (CBB) staining or transferred onto a nitrocellulose membrane for Western blot analysis using anti-MBP antibody.
Immunoprecipitation
GFP-C1 vector– or GFP-tagged protein-expressing 293T cells were lysed in lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, and 0.1% Triton X-100) on ice. Lysates were clarified by centrifugation (12,000 rpm for 20 min at 4 °C) and then incubated with GST fusion protein–bound GSH-agarose beads at 4 °C for 3 h. After an extensive wash, the beads were boiled in SDS-PAGE sample buffer for 5 min, and the bound proteins were separated on a 10% SDS-polyacrylamide gel for transfer onto a nitrocellulose membrane for Western blot analysis using anti-GFP antibody as the primary antibody.
Immunofluorescence microscopy
Cells were seeded onto sterile, acid-treated, 12-mm coverslips in 24-well plates (Corning Glass Works) for transfection or drug treatment. Cells were rinsed for 1 min with PHEM buffer (100 mm PIPES, 20 mm HEPES, 5 mm EGTA, 2 mm MgCl2, and 4 m glycerol (pH 6.9)) and permeabilized for 1 min with PHEM plus 0.1% Triton X-100. Extracted cells were then fixed in freshly made 4% paraformaldehyde plus 0.05% glutaraldehyde in PHEM for 10 min and washed three times in PBS. Cells were blocked with 1% BSA (Sigma) for 50 min at room temperature and subsequently incubated with the indicated primary antibodies in a humidified chamber for 1 h at room temperature, followed by secondary antibodies for another 1 h. The DNA was stained with DAPI (Sigma). Images were captured by DeltaVision softWoRx software (Applied Precision) and processed by deconvolution and z-stack projection.
Deconvolution microscopy
Deconvolution images were collected using a Delta Vision wide-field deconvolution microscope system built on an Olympus IX-71 inverted microscope base. For imaging, a ×60/numerical aperture 1.42 PlanApo oil immersion lens was used, and optical sections were taken at intervals of 0.2 μm. Images for display were generated by projecting single optical sections as described previously (40).
Live-cell imaging
For live-cell imaging, HeLa cells were cultured in glass-bottomed culture dishes (MatTek). During imaging, cells were maintained in CO2-independent medium (Invitrogen) containing 10% fetal bovine serum and 2 mm glutamine in a sealed chamber at 37 °C. Images of living cells were taken with a Delta Vision microscopy system and prepared for publication using Photoshop (Adobe).
Fluorescence intensity quantification
Quantification of the level of centromere-associated protein was conducted as described previously (8). In brief, the average pixel intensities from at least 50 kinetochore pairs from five cells were measured, and background pixel intensities were subtracted. The pixel intensities at each kinetochore pair were then normalized against ACA pixel values to account for any variations in staining or image acquisition. The values of specific siRNA-treated cells were then plotted as a percentage of the values obtained from cells transfected with a control siRNA duplex.
Data analyses
Measurements and statistical analyses were calculated using Image J software (National Institutes of Health) and GraphPad Prism (GraphPad Software Inc.). Statistical significance was determined by Student's t test. Differences were considered significant when p < 0.05.
Author contributions
M. D., J. J., F. Y., W. W. Y., Xu Liu, X. D., and J. H. formal analysis; M. D. and J. J. investigation; M. D., J. J., F. Z., Q. W., and C. R. visualization; M. D., J. J., J. H., and X. Y. writing-original draft; J. J., F. Y., F. Z., Q. W., C. R., X. D., J. H., and C. F. validation; F. Y., J. W., and X. D. data curation; J. F., J. W., Xu Liu, P. H., C. F., and X. Y. resources; J. F. and C. F. methodology; W. W. Y., X. D., J. H., C. F., Xing Liu, and X. Y. writing-review and editing; X. G. and M. M. software; P. H., C. F., and X. Y. supervision; P. H., X. D., and J. H. project administration; Xing Liu and X. Y. conceptualization; Xing Liu and X. Y. funding acquisition.
Supplementary Material
Acknowledgments
We thank Drs. Zhen Dou and Wenwen Wang for critical reading, Wei Liu and Gangyin Zhao for technical support, and Dr. Iain Cheeseman for reagents.
This work was supported in part by National Natural Science Foundation of China Grants 31320103904, 31621002, 31430054, and 31671405; National Key Research and Development Program of China Grants 2017YFA0503600 and 2016YFA0100500; Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB19000000; 973 Project Grants 2012CB917204, 2014CB964803, and 2002CB713703; Chinese Academy of Sciences Center for Excellence in Molecular Cell Sciences Grant 2015HSC-UE010; MOE Innovative Team Grant IRT_17R102; NIDDK, National Institutes of Health Grants DK56292 and DK115812; and NCI, National Institutes of Health Grant CA164133. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5.
- CCAN
- centromere-associated network
- HJURP
- Holliday junction recognition protein
- GST
- glutathione S-transferase
- KO
- knockout
- CBB
- Coomassie Brilliant Blue
- DAPI
- 4′,6-diamidino-2-phenylindole
- ACA
- anti-centromere antibodies
- MBP
- maltose-binding protein.
References
- 1. McKinley K. L., and Cheeseman I. M. (2016) The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17, 16–29 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Black B. E., Jansen L. E., Foltz D. R., and Cleveland D. W. (2010) Centromere identity, function, and epigenetic propagation across cell divisions. Cold Spring Harb. Symp. Quant. Biol. 75, 403–418 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cleveland D. W., Mao Y., and Sullivan K. F. (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 4. Yao X., and Fang G. (2009) Visualization and orchestration of the dynamic molecular society in cells. Cell Res. 19, 152–155 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 5. Alabert C., and Groth A. (2012) Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13, 153–167 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 6. Yang Y., and Yu H. (2018) Partner switching for Ran during the mitosis dance. J. Mol. Cell Biol. 10, 89–90 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 7. Zhu L., Wang Z., Wang W., Wang C., Hua S., Su Z., Brako L., Garcia-Barrio M., Ye M., Wei X., Zou H., Ding X., Liu L., Liu X., and Yao X. (2015) Mitotic protein CSPP1 interacts with CENP-H protein to coordinate accurate chromosome oscillation in mitosis. J. Biol. Chem. 290, 27053–27066 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu X., Song Z., Huo Y., Zhang J., Zhu T., Wang J., Zhao X., Aikhionbare F., Zhang J., Duan H., Wu J., Dou Z., Shi Y., and Yao X. (2014) Chromatin protein HP1 interacts with the mitotic regulator borealin protein and specifies the centromere localization of the chromosomal passenger complex. J. Biol. Chem. 289, 20638–20649 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hua S., Wang Z., Jiang K., Huang Y., Ward T., Zhao L., Dou Z., and Yao X. (2011) CENP-U cooperates with Hec1 to orchestrate kinetochore-microtubule attachment. J. Biol. Chem. 286, 1627–1638 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yao X., Abrieu A., Zheng Y., Sullivan K. F., and Cleveland D. W. (2000) CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484–491 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 11. Sarangapani K. K., and Asbury C. L. (2014) Catch and release: how do kinetochores hook the right microtubules during mitosis? Trends Genet. 30, 150–159 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dou Z., Liu X., Wang W., Zhu T., Wang X., Xu L., Abrieu A., Fu C., Hill D. L., and Yao X. (2015) Dynamic localization of Mps1 kinase to kinetochores is essential for accurate spindle microtubule attachment. Proc. Natl. Acad. Sci. U.S.A. 112, E4546–E4555 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mo F., Zhuang X., Liu X., Yao P. Y., Qin B., Su Z., Zang J., Wang Z., Zhang J., Dou Z., Tian C., Teng M., Niu L., Hill D. L., Fang G., et al. (2016) Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat. Chem. Biol. 12, 226–232 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chu L., Zhu T., Liu X., Yu R., Bacanamwo M., Dou Z., Chu Y., Zou H., Gibbons G. H., Wang D., Ding X., and Yao X. (2012) SUV39H1 orchestrates temporal dynamics of centromeric methylation essential for faithful chromosome segregation in mitosis. J. Mol. Cell Biol. 4, 331–340 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu D., Vader G., Vromans M. J., Lampson M. A., and Lens S. M. (2009) Sensing chromosome bi-orientation by spatial separation of Aurora B kinase from kinetochore substrates. Science 323, 1350–1353 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R. 3rd, and Cleveland D. W. (2006) The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 17. McKinley K. L., Sekulic N., Guo L. Y., Tsinman T., Black B. E., and Cheeseman I. M. (2015) The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol. Cell 60, 886–898 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carroll C. W., Silva M. C., Godek K. M., Jansen L. E., and Straight A. F. (2009) Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11, 896–902 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chittori S., Hong J., Saunders H., Feng H., Ghirlando R., Kelly A. E., Bai Y., and Subramaniam S. (2018) Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science 359, 339–343 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tian T., Li X., Liu Y., Wang C., Liu X., Bi G., Zhang X., Yao X., Zhou Z. H., and Zang J. (2018) Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res. 28, 374–378 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takeuchi K., Nishino T., Mayanagi K., Horikoshi N., Osakabe A., Tachiwana H., Hori T., Kurumizaka H., and Fukagawa T. (2014) The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res. 42, 1644–1655 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu D., Liu X., Zhou T., Yao W., Zhao J., Zheng Z., Jiang W., Wang F., Aikhionbare F. O., Hill D. L., Emmett N., Guo Z., Wang D., Yao X., and Chen Y. (2016) IRE1-RACK1 axis orchestrates ER stress preconditioning-elicited cytoprotection from ischemia/reperfusion injury in liver. J Mol. Cell Biol. 8, 144–156 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nishino T., Takeuchi K., Gascoigne K. E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I. M., and Fukagawa T. (2012) CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148, 487–501 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K., and Westermann S. (2012) CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14, 604–613 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 25. Huis In 't Veld P. J., Jeganathan S., Petrovic A., Singh P., John J., Krenn V., Weissmann F., Bange T., and Musacchio A. (2016) Molecular basis of outer kinetochore assembly on CENP-T. eLife 5, e21007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krizaic I., Williams S. J., Sánchez P., Rodríguez-Corsino M., Stukenberg P. T., and Losada A. (2015) The distinct functions of CENP-C and CENP-T/W in centromere propagation and function in Xenopus egg extracts. Nucleus 6, 133–143 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fang C., Guo X., Lv X., Yin R., Lv X., Wang F., Zhao J., Bai Q., Yao X., and Chen Y. (2017) Dysbindin promotes progression of pancreatic ductal adenocarcinoma via direct activation of PI3K. J. Mol. Cell Biol. 9, 504–515 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishino T., Rago F., Hori T., Tomii K., Cheeseman I. M., and Fukagawa T. (2013) CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32, 424–436 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rago F., Gascoigne K. E., and Cheeseman I. M. (2015) Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 25, 671–677 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith L., and Maddox P. S. (2017) Reading the centromere epigenetic mark. Dev. Cell 42, 110–112 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 31. Black B. E., Brock M. A., Bédard S., Woods V. L. Jr., and Cleveland D. W. (2007) An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 104, 5008–5013 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., and Baker R. E. (2007) Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. U.S.A. 104, 10571–10576 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pidoux A. L., Choi E. S., Abbott J. K., Liu X., Kagansky A., Castillo A. G., Hamilton G. L., Richardson W., Rappsilber J., He X., and Allshire R. C. (2009) Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33, 299–311 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanchez-Pulido L., Pidoux A. L., Ponting C. P., and Allshire R. C. (2009) Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell 137, 1173–1174 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barnhart M. C., Kuich P. H., Stellfox M. E., Ward J. A., Bassett E. A., Black B. E., and Foltz D. R. (2011) HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194, 229–243 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dunleavy E. M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., and Almouzni-Pettinotti G. (2009) HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 37. Foltz D. R., Jansen L. E., Bailey A. O., Yates J. R. 3rd, Bassett E. A., Wood S., Black B. E., and Cleveland D. W. (2009) Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137, 472–484 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bassett E. A., DeNizio J., Barnhart-Dailey M. C., Panchenko T., Sekulic N., Rogers D. J., Foltz D. R., and Black B. E. (2012) HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev. Cell 22, 749–762 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shuaib M., Ouararhni K., Dimitrov S., and Hamiche A. (2010) HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. U.S.A. 107, 1349–1354 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang J., Liu X., Dou Z., Chen L., Jiang H., Fu C., Fu G., Liu D., Zhang J., Zhu T., Fang J., Zang J., Cheng J., Teng M., Ding X., and Yao X. (2014) Mitotic regulator Mis18β interacts with and specifies the centromeric assembly of molecular chaperone Holliday junction recognition protein (HJURP). J. Biol. Chem. 289, 8326–8336 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Müller S., Montes de Oca R., Lacoste N., Dingli F., Loew D., and Almouzni G. (2014) Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading. Cell Rep. 8, 190–203 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 42. Zasadzińska E., Barnhart-Dailey M. C., Kuich P. H., and Foltz D. R. (2013) Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 32, 2113–2124 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bodor D. L., and Jansen L. E. (2013) How two become one: HJURP dimerization drives CENP-A assembly. EMBO J. 32, 2090–2092 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma H., Reyes-Gutierrez P., and Pederson T. (2013) Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors. Proc. Natl. Acad. Sci. U.S.A. 110, 21048–21053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bogdanove A. J., and Voytas D. F. (2011) TAL effectors: customizable proteins for DNA targeting. Science 333, 1843–1846 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 46. Prendergast L., van Vuuren C., Kaczmarczyk A., Doering V., Hellwig D., Quinn N., Hoischen C., Diekmann S., and Sullivan K. F. (2011) Premitotic assembly of human CENPs-T and -W switches centromeric chromatin to a mitotic state. PLoS Biol. 9, e1001082 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dornblut C., Quinn N., Monajambashi S., Prendergast L., van Vuuren C., Münch S., Deng W., Leonhardt H., Cardoso M. C., Hoischen C., Diekmann S., and Sullivan K. F. (2014) A CENP-S/X complex assembles at the centromere in S and G2 phases of the human cell cycle. Open Biol. 4, 130229 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu H., Liu Y., Wang M., Fang J., Huang H., Yang N., Li Y., Wang J., Yao X., Shi Y., Li G., and Xu R. M. (2011) Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25, 901–906 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cancer Genome Atlas Research Network (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cho U. S., and Harrison S. C. (2011) Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc. Natl. Acad. Sci. U.S.A. 108, 9367–9371 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hong J., Feng H., Zhou Z., Ghirlando R., and Bai Y. (2013) Identification of functionally conserved regions in the structure of the chaperone/CenH3/H4 complex. J. Mol. Biol. 425, 536–545 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou Z., Feng H., Zhou B. R., Ghirlando R., Hu K., Zwolak A., Miller Jenkins L. M., Xiao H., Tjandra N., Wu C., and Bai Y. (2011) Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472, 234–237 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pentakota S., Zhou K., Smith C., Maffini S., Petrovic A., Morgan G. P., Weir J. R., Vetter I. R., Musacchio A., and Luger K. (2017) Decoding the centromeric nucleosome through CENP-N. eLife 6, e33442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zasadzińska E., Huang J., Bailey A. O., Guo L. Y., Lee N. S., Srivastava S., Wong K. A., French B. T., Black B. E., and Foltz D. R. (2018) Inheritance of CENP-A nucleosomes during DNA replication requires HJURP. Dev. Cell 47, 348–362.e7 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bodor D. L., Valente L. P., Mata J. F., Black B. E., and Jansen L. E. (2013) Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell 24, 923–932 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ly P., Teitz L. S., Kim D. H., Shoshani O., Skaletsky H., Fachinetti D., Page D. C., and Cleveland D. W. (2017) Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 19, 68–75 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.