Abstract
The liver and pancreas are two major digestive organs, and among the different cell types in them, hepatocytes and the insulin-producing β cells have roles in both health and diseases. Accordingly, clinicians and researchers are very interested in the mechanisms underlying the development and regeneration of liver and pancreatic β cells. Gene and enhancer traps such as the Tol2 transposon-based system are useful for identifying genes potentially involved in developmental processes in the zebrafish model. Here, we developed a strategy that combines a Tol2-mediated enhancer trap and the Cre/loxP system by using loxP-flanked reporters driven by β cell– or hepatocyte-specific promoters and the upstream activating sequence (UAS)-driving Cre. Two double-transgenic reporter lines, Tg(ins:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) and Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus), were established to label pancreatic β cells and hepatocytes, respectively. These two double-transgenic lines were each crossed with the Tol2-enhancer trap founder lines to screen for and identify genes expressed in the β cell and hepatocytes during development. This trap system coupled with application of nitroreductase (NTR)/metronidazole (Mtz)–mediated cell ablation could identify genes expressed during regeneration. Of note, pilot enhancer traps captured transiently and weakly expressed genes such as rab3da and ensab with higher efficiencies than traditional enhancer trap systems. In conclusion, through permanent genetic labeling by Cre/loxP, this improved Tol2-mediated enhancer trap system provides a promising method to identify transiently or weakly expressed, but potentially important, genes during development and regeneration.
Keywords: zebrafish, liver, pancreas, development, regeneration, Cre/loxP, endoderm, enhancer trap, gene expression profiling, transposon, pancreatic β cell
Introduction
Diabetes and liver diseases cause global health problems (1). Loss or dysfunction of insulin-producing β cells and hepatocytes is characteristic of diabetes and liver diseases, respectively (2, 3). Although many genes have been reported to regulate pancreas and liver development (4–6), approaches to identify weakly or transiently expressed genes that are potentially important for organogenesis and regeneration remain to be developed.
69% of zebrafish genes have human orthologs (7). High genetic conservation and larval transparency make zebrafish an ideal model to study development and regeneration of liver and β cells (4, 6, 8–12). In addition to the previous work in other vertebrates (13, 14), genetic screens, including N-ethyl-N-nitrosourea mutagenesis, in zebrafish have identified a number of factors and signaling molecules that govern differentiation and morphogenesis of pancreas and liver (15–18). However, because many genes reiteratively instruct multiple development processes, early embryonic lethality or malformations caused by gene mutation will conceal its roles in β cell or liver development and regeneration at later stages (19). Thus, considerable work will still be required to achieve a thorough understanding of the temporal sequences of signaling events underlying pancreatic β cell and liver induction during embryonic development and regeneration, which will in turn benefit therapies for diabetes and liver diseases by replenishing damaged cells in vivo and generating a new supply of β cells and hepatocytes in vitro (20, 21).
Gene and enhancer traps are useful tools to identify genes that potentially regulate developmental processes in zebrafish (22). Previous studies have used Tol2 transposon-mediated Gal4 to target neural circuits (23, 24) and heart (25). Although gene traps have been widely used to study organ development, this system has little been used to explore organ regeneration. The traditional Gal4-based enhancer trap system requires improvements to overcome two limitations. First, the UAS2-driven GFP or other fluorescent proteins can hardly identify weakly or transiently expressed genes involved in organ development and regeneration. Second, traditional enhancer trap lines can hardly be used to trace the cell origins of organogenesis and regeneration.
To improve the traditional gene trap system to overcome the limitations mentioned above, we combined the Tol2-mediate enhancer trap with Cre/loxP and nitroreductase (NTR)/metronidazole (Mtz) systems to screen pancreatic β cell– and hepatocyte-specific genes involved in development or regeneration. Using this strategy, we constructed two transgenic lines, Tg(ins:loxP-CFPNTR-loxP-DsRed)cq67 and Tg(fabp10:loxP-CFPNTR-loxP-DsRed)cq66, which were further crossed with the Tg(10×UAS:Cre, cryaa:Venus)cq64 line to generate double-transgenic reporter lines for β cells and hepatocytes, respectively (26, 27). Pilot screens by crossing these reporter lines with the Tol2-based green fluorescent protein fused to Gal4FF (GGFF)-enhancer trap founders identified six genes with specific expression patterns in β cells or liver during development or regeneration. A traditional enhancer trap strategy using the Tg(10×UAS:Kaeda, cryaa:Venus)cq65 line was performed for comparison. We conclude that this improved Tol2-mediated enhancer trap strategy combining tissue-specific Cre/loxP obtains higher efficiency for identification of weakly or transiently expressed genes.
Results
Constructions of Cre/loxP–based double-transgenic reporter lines
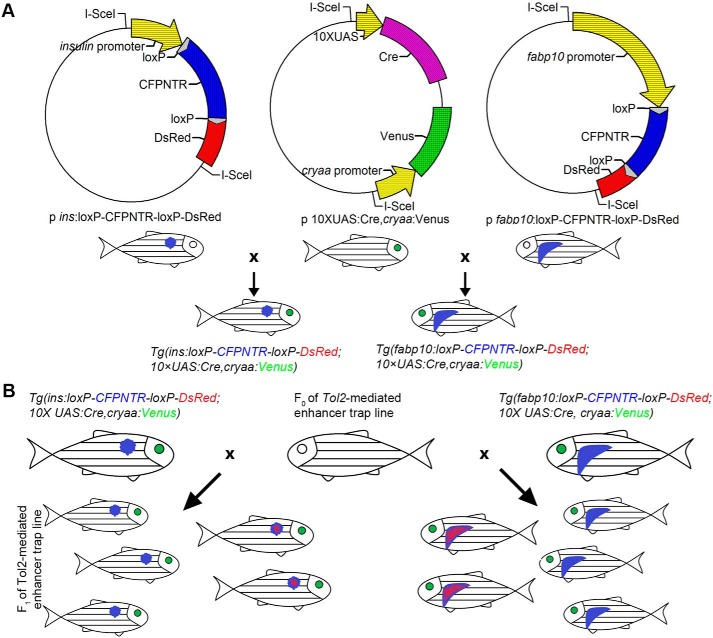
To introduce Cre/loxP into the enhancer trap system for pancreatic β cells and liver, we constructed two double-transgenic reporter lines (Fig. 1A). Under the control of β cell– and hepatocyte-specific promoters, β cells of the Tg(ins:loxP-CFPNTR-loxP-DsRed)cq67 and hepatocytes of the Tg(fabp10:loxP-CFPNTR-loxP-DsRed)cq66 lines were labeled with CFP fluorescence, respectively. In the Tg(10×UAS:Cre, cryaa:Venus)cq64 line, the expression of Cre recombinase was under the control of the UAS. cryaa:Venus was engineered in the same plasmid with 10×UAS:Cre to ensure that the existence of Cre recombinase was visible by the Venus fluorescence in the eyes (Fig. 1A). Then, the Tg(10×UAS:Cre, cryaa:Venus)cq64 was crossed with Tg(ins:loxP-CFPNTR-loxP-DsRed)cq67 and Tg(fabp10:loxP-CFPNTR-loxP-DsRed)cq66 to generate Tg(ins:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) and Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) double-transgenic reporter lines, respectively. Theoretically, by means of crossing these two transgenic reporter lines with Tol2-based enhancer trap founders (F0), F1 larvae with red fluorescence appearing in the β cells or liver will be selected as candidates for further genomic identification (Fig. 1B). Taking advantage of NTR/Mtz–mediated cell ablation (28), F1 individuals can be further subjected to screening for genes activated during β cell and liver regeneration.
Figure 1.
Tol2-mediated enhancer trap is combined with Cre/loxP for the screen of pancreatic β cell– and liver-specific genes. A, Cre/loxP–based transgenic lines Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) and Tg(ins:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) were constructed as schematically illustrated. The expression of Venus in the eyes indicates the 10×UAS:Cre transgene in the genome. B, schema showing the procedure for the screen. Cre/loxP–based transgenic reporter lines for pancreatic β cells (left) and hepatocytes (right) were crossed with Tol2-mediated F0 enhancer trap line, respectively. The F1 larvae with red fluorescence in the β cells or hepatocytes were selected for genomic identification. F1 larvae without red color in targeted organs were subjected to further regeneration studies.
Validation of the Gal4-UAS system in the double-transgenic reporter lines
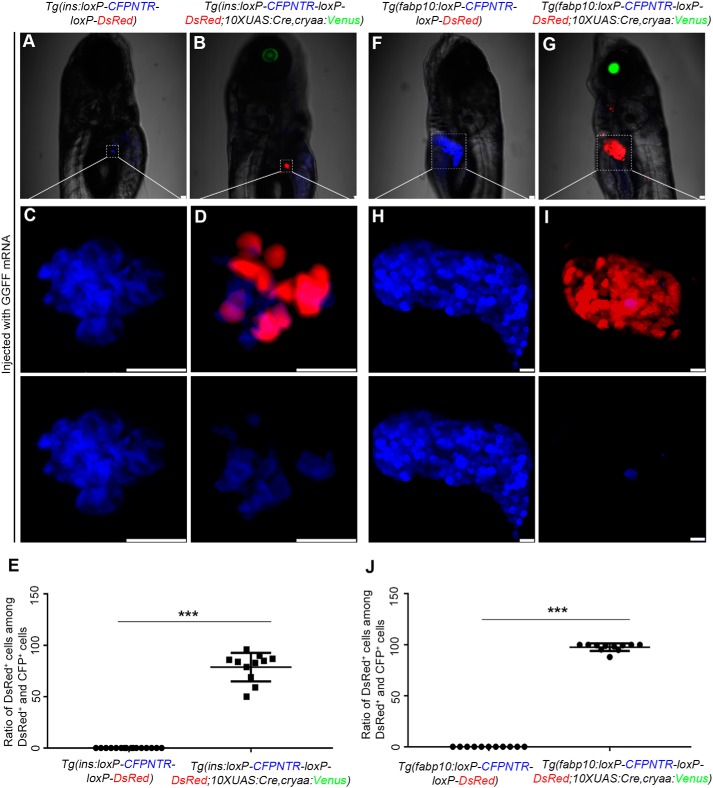
In the Tg(ins:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) and Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) lines, β cells and hepatocytes were respectively labeled by CFP. The expression of Cre recombinase is turned on only when UAS is activated by the Gal4 transcriptional activator protein. In this study, we used GGFF, the optimal version of Gal4 with less toxicity to zebrafish cells (24), to evaluate the effectiveness of GGFF-UAS in the double-transgenic reporter lines. After injection of GGFF mRNA into one-cell-stage embryos of the reporter lines, the UAS activated the expression of Cre recombinase, which in turn excised the CFPNTR cassette flanked by the loxP sites specifically in the pancreatic β cells (Fig. 2, A–E) and hepatocytes (Fig. 2, F–J). Expressions of DsRed were activated in more than 80% of the β cells (Fig. 2, D and E) and in nearly all of the hepatocytes (Fig. 2, I and J). The CFP elimination in hepatocytes was more efficient than in β cells (Fig. 2, D and I). Thus, working efficiencies of GGFF-UAS in these two double-transgenic reporter lines were validated and guaranteed.
Figure 2.
The validation of the Cre/loxP–based transgenic reporter lines. A–D, the fluorescence of the pancreatic β cells in Tg(ins:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) was converted from blue to red by GGFF mRNA. E, quantification of the percentage of the DsRed+ cells among the DsRed+ and CFP+ cells in C and D. F–I, the color of the hepatocytes of the Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) shifts from blue to red when injected with GGFF mRNA at one-cell stage. J, quantification of the percentage of the DsRed+ cells among the DsRed+ and CFP+ cells in H and I. Asterisks indicate statistical significance: ***, p < 0.001. Scale bars, 20 μm. Error bars, ±S.D.
Enhancer trap for genes expressed in pancreatic β cells and hepatocytes during development
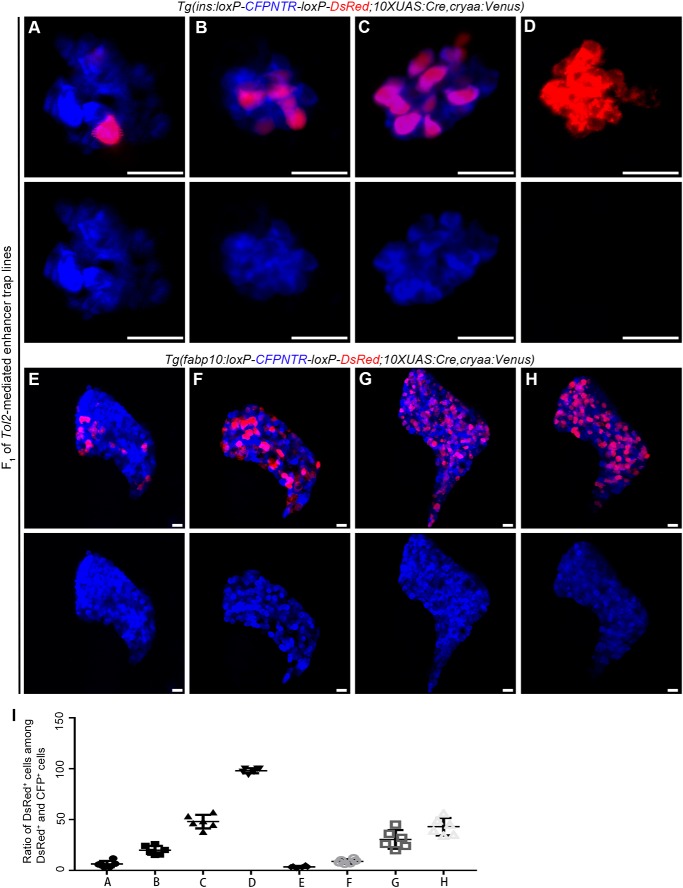
In a pilot screen, over 600 F0 enhancer trap lines were generated using the enhancer trap plasmid T2KhspGGFF. Then, the F0 lines were crossed with two double-transgenic reporter lines, Tg(ins:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) and Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus). F1 larvae with DsRed expression in the β cells or in the liver were raised to adults. Six and 11 lines were identified with DsRed expression in the β cells and liver, respectively. The expression patterns and mosaicity of DsRed were diverse among the different F1 enhancer trap lines (Fig. 3, A–I). In the F1 of β cell enhancer trap lines (Fig. 3, A–D), some lines exhibited only one or two β cells in red (Fig. 3, A and I), whereas one line exhibited DsRed over the entire organ and complete elimination of CFP (Fig. 3, D and I). F1 of hepatocyte enhancer trap lines exhibited similar phenomena (Fig. 3, E–I).
Figure 3.
Improved enhancer trap combined with Cre/loxP is used to screen genes expressed in the β cells and hepatocytes during development. A–D, F1 Tol2-mediated enhancer trap larvae of Tg(ins:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) possess β cells marked by red fluorescence in various degrees. E–H, F1 Tol2-mediated enhancer trap larvae of Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) possess hepatocytes marked by red fluorescence in various degrees. I, quantification of the percentage of the DsRed+ cells among the DsRed+ and CFP+ cells in A–H. Scale bars, 20 μm. Error bars, ±S.D.
Five of the F1 trap lines were raised to the F2 generation, among which two genes with specific expression patterns during development were identified through the reverse PCR method (24). According to the sequencing results of reverse PCR (see Fig. S1A) and BLAST readout from Ensembl (see Fig. S1B), rab3da was found to be highly expressed in the endocrine pancreas in addition to spinal cord at 24 hpf (Fig. 4, A and C). However, its expression in the pancreas was significantly reduced at 48 hpf (Fig. 4, B and D) and became nondetectable at 96 hpf. Although Rab3d has been reported to play roles in maintaining normal-size secretory granules of pancreatic acini in mammals (29, 30) and be critical for secretory granule maturation in PC12 cells (31), its expression in the developing pancreas has not yet been identified. We found high, but transient, expression of its zebrafish ortholog, rab3da, in the pancreas during development. When the F1 of rab3da enhancer trap line was crossed with the traditional reporter line Tg(10×UAS:Kaede, cryaa:Venus)cq65, expression of Kaede in the β cells that represents trap of rab3da could not be detected at 96 hpf (Fig. 4E). By contrast, although expression of rab3da was transient, our Cre/loxP–combined enhancer trap strategy could detect the trap more efficiently at 96 hpf (Fig. 4F).
Figure 4.
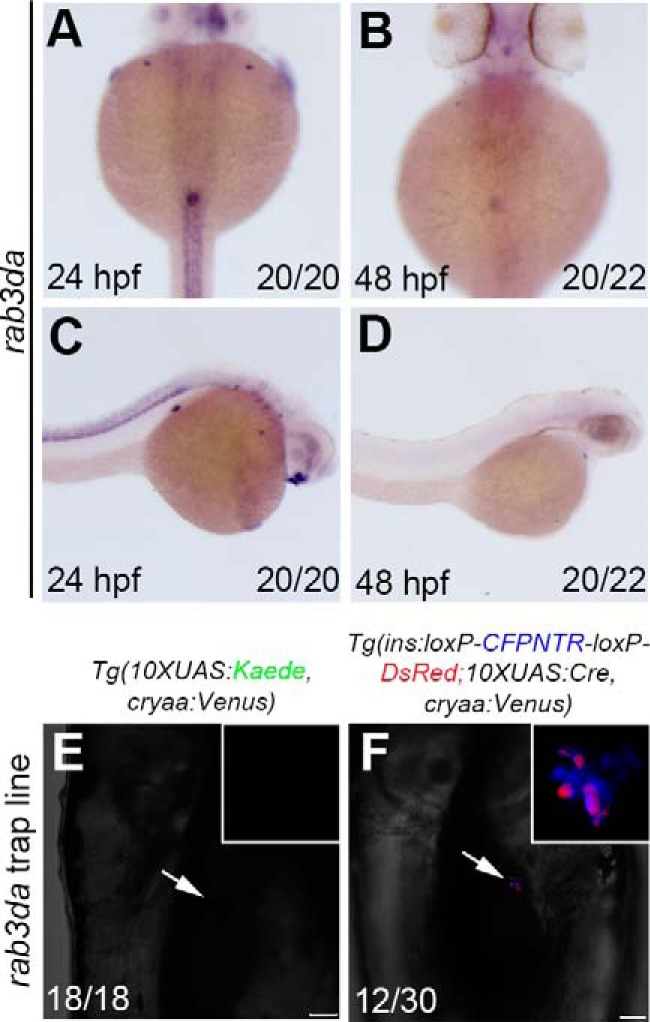
Identification of rab3da expressed in the pancreatic β cells using the improved enhancer trap system. A–D, in situ results show rab3da expressed in pancreatic endocrine cells. E and F, the double-transgenic reporter line Tg(ins:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) expresses the Tol2-mediated GGFF insertion in pancreatic β cells with higher efficiency. Numbers indicate the proportion of larvae exhibiting the expression shown. Arrows indicate the region of pancreatic β cells. Scale bars, 50 μm.
The second gene, rnd2, identified to be highly expressed in the liver and brain at 34 hpf and 58 hpf (Fig. 5, A–D) encodes the Rho family GTPase 2 (see Fig. S2, A and B). In mammals, Rnd2 controls neuron migration in the cerebral cortex (32–34). In addition, the Rnd family is linked to tumorigenesis and metastasis, including lung cancer (35), breast cancer (36), and the most common type of liver cancer, hepatocellular carcinoma (37). We found expression of zebrafish ortholog rnd2 in the developing liver and brain during embryogenesis. The rnd2 insertion could also be present under the background of traditional enhancer trap reporter Tg(10×UAS:Kaede, cryaa:Venus)cq65 (Fig. 5E), but its mosaicity of positive cells was obviously less than the Cre/loxP reporter (Fig. 5F) and became more difficult to be identified. These data demonstrate that the improved enhancer trap system obtains higher screening efficiencies, thus facilitating identification of genes.
Figure 5.
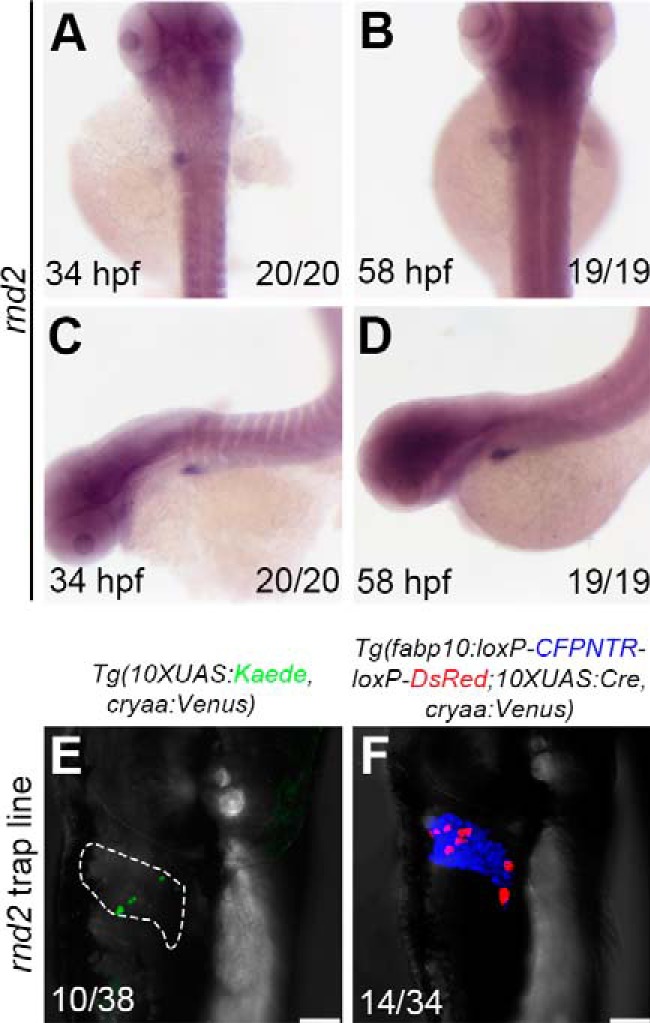
Identification of rnd2 expressed in the liver using the improved enhancer trap system. A–D, in situ results show rnd2 expressed in the liver. E and F, the double-transgenic reporter line Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) expresses the Tol2-mediated GGFF insertion in hepatocytes with higher efficiency. Numbers indicate the proportion of larvae exhibiting the expression shown. Scale bars, 100 μm.
Enhancer trap for genes activated in the regenerating liver
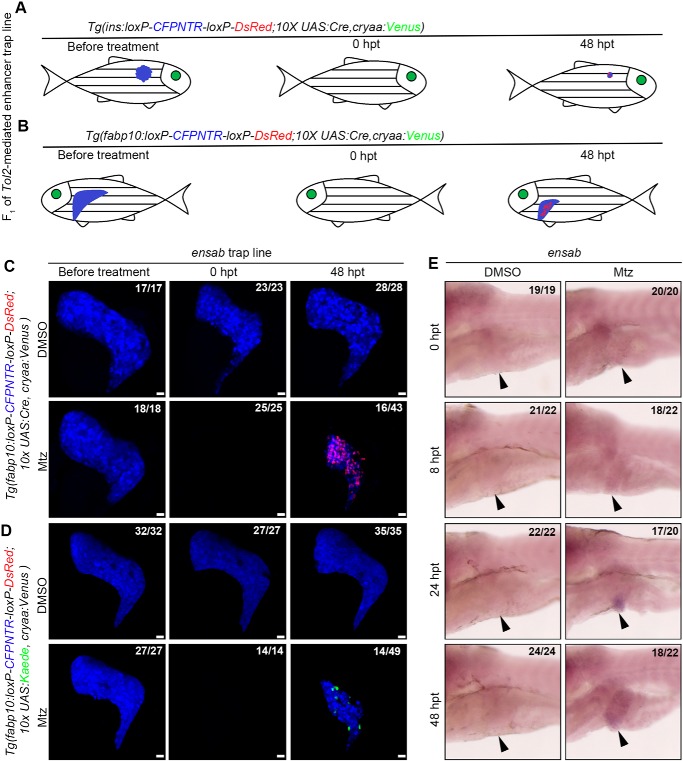
To evaluate the feasibility of this improved enhancer trap system in the identification of genes activated during regeneration (Fig. 6, A and B), we crossed the enhancer trap founder lines with the Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) reporter line followed by Mtz treatment to induce liver injury in F1 larvae (9, 10). DsRed expression in the regenerating liver was found in the F1 of one line at 48 h post-treatment (hpt) (Fig. 6C). The sequencing result of reverse PCR (see Fig. S3A) and BLAST readout from Ensembl (see Fig. S3B) identified the trapped gene ensab. Ensa, the ortholog of zebrafish Ensab, inhibits the activity of protein phosphatase 2A and prompts mitosis (38, 39). After liver injury, expression of ensab in the regenerating liver was initiated at 8 hpt and became moderately up-regulated at 24 and 48 hpt (Fig. 6E, arrowheads), validating the gene trap results (Fig. 6C). When the F0 was crossed with the traditional gene trap reporter line Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Kaede, cryaa:Venus), only a few Kaede-positive cells were present at 48 hpt (Fig. 6D), making it more difficult to be identified from the screen compared with the improved gene trap strategy (Fig. 6C). These data demonstrate that our improved gene trap system provides a useful tool to identify genes activated during regeneration.
Figure 6.
Identification of ensab expressed in the regenerating livers using the improved enhancer trap system. A, the pancreatic β cells marked by red fluorescence would appear exclusively during the regeneration when regeneration-specific genes or genes regulating trans-differentiation have been captured with the improved enhancer trap system. B, the red colored hepatocytes would appear only during the recovery of liver when regeneration-specific genes or genes regulating trans-differentiation have been captured with the improved enhancer trap system. C and D, the double-transgenic reporter line Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) expresses the Tol2-mediated GGFF insertion in regenerating livers with higher efficiency. E, in situ results show ensab expressed in the regenerating livers after Mtz treatment at 24 and 48 hpt (arrowheads). Numbers indicate the proportion of larvae exhibiting the expression shown. Scale bars, 20 μm.
Discussion
Although the process of organ regeneration shares many common molecular pathways with organogenesis, it cannot be ruled out that some molecules play roles only in organ regeneration (40). Moreover, other cell types could convert to regenerating cells through trans-differentiation under certain injury circumstances (9, 10, 41, 42). In addition to the well-established NTR/Mtz injury models in zebrafish (28, 43), this improved enhancer trap system should also be applicable to other injury models to identify transient and weak genes. For example, a previous study has revealed that macrophages repair ruptures of brain blood vessels through direct physical adhesion and mechanical traction forces (44). Generation of double-transgenic reporter lines to label macrophages or blood vessel endothelial cells will enable identifications of genes important for this repair process.
A study of genome-wide enhancer–promoter interactions revealed that the interaction between the enhancer and promoter decreases with increasing distances (45). The transcriptional efficiency of Cre depends on the distance from the insertion site to the candidate enhancer. It accounts in part for the mosaic patterns of DsRed fluorescence embedded on the CFP background in pancreatic β cells and hepatocytes (Fig. 3, A–H). However, this does not overshadow the power of the Cre/loxP–combined enhancer trap to screen the gene of interest. A portion of DsRed-positive pancreatic β cells and hepatocytes retained the CFP fluorescence (Figs. 2 and 3), which could be caused by the activation of Cre at different time points dependent on the insertion sites, and therefore the residual CFP protein has not been degraded yet.
Benefiting from the characteristics of permanent labeling, introduction of the Cre/loxP into the enhancer trap system improves the efficiency to screen genes of interest, in particular those weakly or transiently expressed. For genes with strong expression, this improved enhancer trap system shows no significant difference compared with the traditional reporter. For example, insulin was trapped using both Cre/loxP–combined and traditional reporter lines (see Fig. S4, A–C). Taken together, the Cre/loxP–combined, improved enhancer trap provides an approach to study gene expression in the organ of interest and could be genetically engineered to match the organ injury model for regeneration studies.
Experimental procedures
Ethics statement
All experimental protocols were approved by the School of Life Sciences, Southwest University (Chongqing, China), and the methods were carried out in accordance with the approved guidelines. The zebrafish facility and study were approved by the Institutional Review Board of Southwest University (Chongqing, China). Zebrafish were maintained in accordance with the Guidelines of Experimental Animal Welfare from Ministry of Science and Technology of People's Republic of China (2006) and the Institutional Animal Care and Use Committee protocols from Southwest University (2007).
Plasmid constructs
The 10×UAS fragment was amplified from P5EUAS with PCR and then cloned upstream of Cre coding sequence in the backbone of modified pBluescript, which harbors the meganuclease I-SceI site. On this base, the whole cryaa:Venus was also cloned in the 10×UAS:Cre construct flanked by the I-SceI site. 10×UAS:Kaede was constructed by replacing Cre coding sequence with Kaede coding sequence. fabp10:loxP-CFPNTR-loxP-DsRed was constructed by insertion of the CFPNTR fused sequence into the previously reported fabp10:loxP-stop-loxP-DsRed (9). ins:loxP-CFPNTR-loxP-DsRed was made by replacing the fabp10 promoter of fabp10:loxP-CFPNTR-loxP-DsRed with the insulin promoter. Enhancer trap vector pT2KhspGGFF was a kind gift from the Kawakami Lab.
Zebrafish strains
Transgenic lines Tg(10×UAS:Cre, cryaa:Venus)cq64, Tg(10×UAS:Kaede, cryaa:Venus)cq65, Tg(fabp10:loxP-CFPNTR-loxP-DsRed)cq66, and Tg(ins:loxP-CFPNTR-loxP-DsRed)cq67 were all generated based on the standard I-SceI meganuclease transgenesis technique from the AB genetic background. The Tg(fabp10:loxP-CFPNTR-loxP-DsRed; 10×UAS:Cre, cryaa:Venus) double-transgenic line was generated from the cross of Tg(fabp10:loxP-CFPNTR-loxP-DsRed)cq66 with Tg(10×UAS:Cre, cryaa:Venus)cq64, and the Tg(ins:loxP-CFPNTR-loxp-DsRed; 10×UAS:Cre, cryaa:Venus) double-transgenic line was generated from the cross of Tg(10×UAS:Cre, cryaa:Venus)cq64 with Tg(ins:loxP-CFPNTR-loxP-DsRed)cq67. Enhancer trap F0 was made by injecting Tol2-mediated enhancer trap vector pT2KhspGGFF with transposase mRNA into zebrafish embryos at one-cell stage. All zebrafish lines were brought up and maintained under standard laboratory conditions according to institutional animal care and use committee protocols.
Mtz treatment
The Tg(fabp10:loxP-CFPNTR-loxP-DsRed)cq66 transgenic larvae at 5 days postfertilization was incubated with 10 mm Mtz (Sigma-Aldrich) in 0.2% DMSO for 24 h. Then, larvae were washed three times and recovered in egg water, marking the regeneration 0 hpt.
Microinjection of mRNA
Transposase mRNA was synthesized from the linearized pCS-zTP according to the protocol in the mMESSAGE mMACHINE SP6 kit (Ambion Inc., Austin, TX). GGFF coding sequence was cloned into pCS2(+) plasmid and linearized by XbaI to use as a template to synthesize the GGFF mRNA according to the protocol in the mMESSAGE SP6 kit.
Microscopic analysis
A fluorescence stereomicroscope (M165FC, Leica) was used to observe and screen embryos that express DsRed in their hepatocytes or pancreatic β cells. The selected embryos were mounted with 1.2% low-melting-point agarose and subjected to confocal microscopy using a Zeiss LSM 780 META laser confocal microscope. Images of embryo were acquired as serial sections along the z axis at 1.0-μm intervals and processed using Zeiss LSM 780 Image Browser and Adobe Photoshop CS2.
Whole-mount in situ hybridization
In situ hybridization was performed as described previously (36). The primers used for synthetic probes were as follows: rab3da primers, 5′-AGAGCCGGATAAGATGGCGT-3′ and 5′-ATCAGGGGGCGTGTCTTGAA-3′; rnd2 primers, 5′-CCGTCCACTCACAGTCACAG-3′ and 5′-GTCCCGTAGGCCTCAGTATG-3′; and ensab primers, 5′-CACCGTGGGTGGATCAGATCGG-3′ and 5′-ACCAGTCCTGGTGAAGCTGG-3′.
Quantification and statistical analysis
All statistical tests were performed with GraphPad Prism version 7.0 for Windows (GraphPad Software). Data were analyzed with Student's t test, and multiple comparisons performed with analysis of variance tests were used to determine statistical significance. Statistical significance was defined as follows: *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Author contributions
L. L. writing-review and editing; Y. Z. and L. L. designed the experimental strategy, analyzed data, and wrote the manuscript; W. H. performed plasmid construction; J. D. and Z. W. joined the screen process; J. H. analyzed data and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Koichi Kawakami for the Tol2 plasmid and Li Li for discussions.
This work was supported by National Key Basic Research Program of China Grant 2015CB942800; National Natural Science Foundation of China Grants 31730060, 31801214, and 91539201; National Key Research and Development Program of China Grant 2017YFA0106600; and 111 Program Grant B14037. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
- UAS
- upstream activating sequence
- hpf
- hours postfertilization
- hpt
- hours post-treatment
- Mtz
- metronidazole
- NTR
- nitroreductase
- CFP
- cyan fluorescent protein
- GGFF
- green fluorescent protein fused to Gal4FF.
References
- 1. Picardi A., D'Avola D., Gentilucci U. V., Galati G., Fiori E., Spataro S., and Afeltra A. (2006) Diabetes in chronic liver disease: from old concepts to new evidence. Diabetes Metab. Res. Rev. 22, 274–283 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 2. Malhi H., Gores G. J., and Lemasters J. J. (2006) Apoptosis and necrosis in the liver: a tale of two deaths? Hepatology 43, S31–S44 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 3. Gale E. A. (2001) The discovery of type 1 diabetes. Diabetes 50, 217–226 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 4. Tehrani Z., and Lin S. (2011) Endocrine pancreas development in zebrafish. Cell Cycle 10, 3466–3472 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 5. Tao T., and Peng J. (2009) Liver development in zebrafish (Danio rerio). J. Genet. Genomics 36, 325–334 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 6. Chu J., and Sadler K. C. (2009) New school in liver development: lessons from zebrafish. Hepatology 50, 1656–1663 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., Muffato M., Collins J. E., Humphray S., McLaren K., Matthews L., McLaren S., Sealy I., Caccamo M., Churcher C., Scott C., et al. (2013) The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goessling W., and Sadler K. C. (2015) Zebrafish: an important tool for liver disease research. Gastroenterology 149, 1361–1377 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He J., Lu H., Zou Q., and Luo L. (2014) Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 146, 789–800.e8 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 10. Choi T. Y., Ninov N., Stainier D. Y., and Shin D. (2014) Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 146, 776–788 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kimmel R. A., and Meyer D. (2016) Zebrafish pancreas as a model for development and disease. Methods Cell Biol. 134, 431–461 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 12. Shi W., Fang Z., Li L., and Luo L. (2015) Using zebrafish as the model organism to understand organ regeneration. Sci. China Life Sci. 58, 343–351 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 13. Zorn A. M., and Wells J. M. (2009) Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 25, 221–251 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shih H. P., Wang A., and Sander M. (2013) Pancreas organogenesis: from lineage determination to morphogenesis. Annu. Rev. Cell Dev. Biol. 29, 81–105 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 15. Dong P. D., Munson C. A., Norton W., Crosnier C., Pan X., Gong Z., Neumann C. J., and Stainier D. Y. (2007) Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat. Genet. 39, 397–402 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 16. Ober E. A., Verkade H., Field H. A., and Stainier D. Y. (2006) Mesodermal Wnt2b signalling positively regulates liver specification. Nature 442, 688–691 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 17. Lu H., Ma J., Yang Y., Shi W., and Luo L. (2013) EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev. Cell 24, 543–553 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 18. Kim H. J., Sumanas S., Palencia-Desai S., Dong Y., Chen J. N., and Lin S. (2006) Genetic analysis of early endocrine pancreas formation in zebrafish. Mol. Endocrinol. 20, 194–203 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 19. Andreeva V., Connolly M. H., Stewart-Swift C., Fraher D., Burt J., Cardarelli J., and Yelick P. C. (2011) Identification of adult mineralized tissue zebrafish mutants. Genesis 49, 360–366 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dhawan A., Puppi J., Hughes R. D., and Mitry R. R. (2010) Human hepatocyte transplantation: current experience and future challenges. Nat. Rev. Gastroenterol. Hepatol. 7, 288–298 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 21. Edlund H. (2002) Pancreatic organogenesis—developmental mechanisms and implications for therapy. Nat. Rev. Genet. 3, 524–532 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 22. Kawakami K., Takeda H., Kawakami N., Kobayashi M., Matsuda N., and Mishina M. (2004) A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133–144 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 23. Scott E. K., Mason L., Arrenberg A. B., Ziv L., Gosse N. J., Xiao T., Chi N. C., Asakawa K., Kawakami K., and Baier H. (2007) Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat. Methods 4, 323–326 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 24. Asakawa K., Suster M. L., Mizusawa K., Nagayoshi S., Kotani T., Urasaki A., Kishimoto Y., Hibi M., and Kawakami K. (2008) Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 105, 1255–1260 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poon K. L., Liebling M., Kondrychyn I., Garcia-Lecea M., and Korzh V. (2010) Zebrafish cardiac enhancer trap lines: new tools for in vivo studies of cardiovascular development and disease. Dev. Dyn. 239, 914–926 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 26. Her G. M., Chiang C. C., Chen W. Y., and Wu J. L. (2003) In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett. 538, 125–133 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 27. Huang H., Vogel S. S., Liu N., Melton D. A., and Lin S. (2001) Analysis of pancreatic development in living transgenic zebrafish embryos. Mol. Cell. Endocrinol. 177, 117–124 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 28. Curado S., Anderson R. M., Jungblut B., Mumm J., Schroeter E., and Stainier D. Y. (2007) Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev. Dyn. 236, 1025–1035 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 29. Riedel D., Antonin W., Fernandez-Chacon R., Alvarez de Toledo G., Jo T., Geppert M., Valentijn J. A., Valentijn K., Jamieson J. D., Südhof T. C., and Jahn R. (2002) Rab3D is not required for exocrine exocytosis but for maintenance of normally sized secretory granules. Mol. Cell. Biol. 22, 6487–6497 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen X., Ernst S. A., and Williams J. A. (2003) Dominant negative Rab3D mutants reduce GTP-bound endogenous Rab3D in pancreatic acini. J. Biol. Chem. 278, 50053–50060 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 31. Kögel T., Rudolf R., Hodneland E., Copier J., Regazzi R., Tooze S. A., and Gerdes H. H. (2013) Rab3D is critical for secretory granule maturation in PC12 cells. PLoS One 8, e57321 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fujita H., Katoh H., Ishikawa Y., Mori K., and Negishi M. (2002) Rapostlin is a novel effector of Rnd2 GTPase inducing neurite branching. J. Biol. Chem. 277, 45428–45434 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 33. Li J., and Anton E. S. (2011) Rnd-ing up RhoA activity to link neurogenesis with steps in neuronal migration. Dev. Cell 20, 409–410 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 34. Heng J. I., Nguyen L., Castro D. S., Zimmer C., Wildner H., Armant O., Skowronska-Krawczyk D., Bedogni F., Matter J. M., Hevner R., and Guillemot F. (2008) Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 455, 114–118 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 35. Tang Y., Hu C., Yang H., Cao L., Li Y., Deng P., and Huang L. (2014) Rnd3 regulates lung cancer cell proliferation through notch signaling. PLoS One 9, e111897 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okada T., Sinha S., Esposito I., Schiavon G., López-Lago M. A., Su W., Pratilas C. A., Abele C., Hernandez J. M., Ohara M., Okada M., Viale A., Heguy A., Socci N. D., Sapino A., et al. (2015) The Rho GTPase Rnd1 suppresses mammary tumorigenesis and EMT by restraining Ras-MAPK signalling. Nat. Cell Biol. 17, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grise F., Sena S., Bidaud-Meynard A., Baud J., Hiriart J. B., Makki K., Dugot-Senant N., Staedel C., Bioulac-Sage P., Zucman-Rossi J., Rosenbaum J., and Moreau V. (2012) Rnd3/RhoE Is down-regulated in hepatocellular carcinoma and controls cellular invasion. Hepatology 55, 1766–1775 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 38. Gharbi-Ayachi A., Labbé J. C., Burgess A., Vigneron S., Strub J. M., Brioudes E., Van-Dorsselaer A., Castro A., and Lorca T. (2010) The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science 330, 1673–1677 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 39. Mochida S., Maslen S. L., Skehel M., and Hunt T. (2010) Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330, 1670–1673 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 40. Huch M., Dorrell C., Boj S. F., van Es J. H., Li V. S., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M. J., Haft A., Vries R. G., Grompe M., and Clevers H. (2013) In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thorel F., Népote V., Avril I., Kohno K., Desgraz R., Chera S., and Herrera P. L. (2010) Conversion of adult pancreatic α-cells to beta-cells after extreme β-cell loss. Nature 464, 1149–1154 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chera S., Baronnier D., Ghila L., Cigliola V., Jensen J. N., Gu G., Furuyama K., Thorel F., Gribble F. M., Reimann F., and Herrera P. L. (2014) Diabetes recovery by age-dependent conversion of pancreatic delta-cells into insulin producers. Nature 514, 503–507 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Curado S., Stainier D. Y., and Anderson R. M. (2008) Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat. Protoc. 3, 948–954 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu C., Wu C., Yang Q., Gao J., Li L., Yang D., and Luo L. (2016) Macrophages mediate the repair of brain vascular rupture through direct physical adhesion and mechanical traction. Immunity 44, 1162–1176 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 45. Chepelev I., Wei G., Wangsa D., Tang Q., and Zhao K. (2012) Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 22, 490–503 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.