Abstract
The highly conserved zinc finger CCCTC-binding factor (CTCF) regulates genomic imprinting and gene expression by acting as a transcriptional activator or repressor of promoters and insulator of enhancers. The multiple functions of CTCF are accomplished by co-association with other protein partners and are dependent on genomic context and tissue specificity. Despite the critical role of CTCF in the organization of genome structure, to date, only a subset of CTCF interaction partners have been identified. Here we present a large-scale identification of CTCF-binding partners using affinity purification and high-resolution LC-MS/MS analysis. In addition to functional enrichment of specific protein families such as the ribosomal proteins and the DEAD box helicases, we identified novel high-confidence CTCF interactors that provide a still unexplored biochemical context for CTCF's multiple functions. One of the newly validated CTCF interactors is BRG1, the major ATPase subunit of the chromatin remodeling complex SWI/SNF, establishing a relationship between two master regulators of genome organization. This work significantly expands the current knowledge of the human CTCF interactome and represents an important resource to direct future studies aimed at uncovering molecular mechanisms modulating CTCF pleiotropic functions throughout the genome.
Keywords: mass spectrometry (MS), protein–protein interaction, proteomics, chromatin, transcription factor, BRG1, CTCF, Interactomics, SWI/SNF, transcriptional regulation
Introduction
CTCF4 is a ubiquitously expressed transcription factor with pleiotropic functions driven by recognition and binding of a preferentially unmethylated CpG-rich consensus sequence within several genomic sites. Regulatory functions of CTCF as an enhancer-blocking insulator were first discovered at the β-globin (1) and the imprinted H19-Igf2 loci (2–4). These pioneering studies revealed the ability of CTCF to act as an insulator, thereby preventing the interaction between enhancers and promoters and regulating transcription at selected gene loci. A transcriptional repressor role for CTCF was also reported to be mediated by its binding to different sequences in mouse, human, and chicken MYC promoters (5, 6). Later, the genome-wide mapping of CTCF-binding sites revealed that it can recognize a wide variety of DNA target sequences (7–9), being the high occupancy sites conserved across cell types (10).
Over the past years, a broader view of CTCF as a unique versatile zinc finger protein has emerged, adding knowledge on CTCF functions in transcriptional activation/repression, enhancer-blocking and/or chromatin barrier insulation, hormone-responsive silencing, genomic imprinting, transcription pausing, alternative mRNA splicing, and, more recently, as an architectural protein regulating higher-order chromatin structure and genome topology (11–17). Indeed, recent advances in chromosome conformation capture and high-throughput chromosome conformation capture methods significantly increase our understanding of CTCF roles in mediating long-range interactions as the basis of the genome partitioning into topologically associating domains (TADs), defined as units of chromosomes exhibiting a high frequency of interaction within domains compared with the adjacent domains. Interestingly, CTCF-binding sites have been found to be enriched at TAD boundaries along with transcription start sites (TSSs), further supporting its role as a chromatin organizer (18). On the contrary, Ramírez et al. (19), in contrast to earlier studies, recently found that in flies the CTCF DNA-binding motif is rarely associated with TAD boundaries and that specific DNA motifs can allocate different boundary proteins, thus guiding genome architecture. In particular, a DNA-guided chromatin assembly model has been proposed based on the recognition of boundary elements by specific proteins, which help loading TADs assembly factors onto chromatin (19).
Several efforts have been made to unravel mechanisms at the basis of CTCF pleiotropic functions to achieve a deep understanding of how this unique transcription factor can execute diverse functions in different contexts and cell types. Nakahashi et al. (20) demonstrated that CTCF associates with a wide array of DNA modules via combinatorial clustering of its 11 zinc fingers. An additional strategy widely recognized to modulate CTCF recruitment at various genomic loci is the interaction with other proteins that affects its functional specificity in a genomic context- and tissue-specific manner (12–16, 21, 22). Indeed, an array of classical biochemical techniques has been used to identify binding partners of CTCF including traditional co-immunoprecipitation strategies and binding to CTCF bait in yeast two-hybrid assays; for other proteins, co-localization with CTCF genome-wide by conventional ChIP, ChIP-on-ChIP, or ChIP-Seq experiments have also been reported (12–16, 21, 22). By these approaches, it has been demonstrated that CTCF exerts its function by specific co-associations with a plethora of other proteins belonging to distinct functional groups such as DNA-binding proteins (e.g. Ying yang YY-1, YB1, and Kaiso), DNA and RNA helicases (e.g. CHD8 and DEAD box RNA helicases p68), histones (e.g. H2A and H2A.Z), and other regulatory proteins including poly(ADP-ribose) polymerase, nucleophosmin, topoisomerase II, RNA polymerase II, and transcription factor II-I (15, 21). In addition, cooperation of CTCF with cohesin has emerged to be crucial in determining genomes spatial organization into chromatin loops (23, 24) and TADs (18, 25–27).
It is therefore clear that the identification of novel CTCF-binding partners is of central interest to shed light on mechanisms driving well-known CTCF functions and to open novel perspectives on still unexplored roles of this multivalent transcription factor. Indeed, despite the biological importance of CTCF, our general knowledge of the human CTCF interaction network is limited to selected CTCF protein partners, mainly involved in specific functions such as binding and modification of DNA or chromatin. Recent advances in MS instrumentation and computational tools resulted in the identification of high-confidence interaction proteomes of several biologically relevant protein groups by large-scale affinity purification of proteins coupled to MS (AP-MS) approaches, markedly improving our knowledge of protein interaction networks and functions (28–35).
Here we present a global interaction study of human CTCF by high-resolution nano-LC– electrospray ionization–MS/MS. We identified 90 high confidence protein–protein interactions that constitute a network of proteins with specific functions in chromatin binding, promoter-specific chromatin binding, transcription, and more. In addition to confirming a number of well-known CTCF interactors, our study reveals co-associations of CTCF with still uncharacterized protein partners that are important for genome organization such as BRG1, the major ATPase subunit of the chromatin remodeling complex SWI/SNF. This work significantly expands the current knowledge of the human CTCF interactome and represents an important resource to direct future studies aimed at uncovering molecular mechanisms modulating CTCF pleiotropic functions throughout the genome.
Results
Purification and identification of CTCF-interacting complexes by high-resolution MS
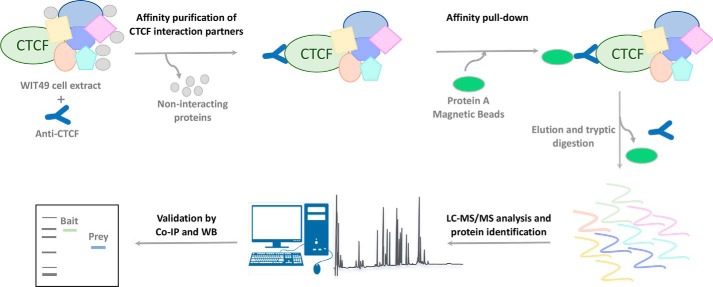
Despite the master role of CTCF in regulating gene expression and genome structure, a large-scale study to identify CTCF interaction partners by high-resolution LC-MS/MS analysis has not been previously reported. Here, we applied an AP-MS approach to characterize the human CTCF interactome in WiT49 cell lines overexpressing CTCF. A schematic outline of the AP-MS procedure used in this study is shown in Fig. 1. Following the transfection of the WiT49 cell line with the pcDNA3 bearing the full-length CTCF DNA encoding sequence, CTCF overexpression was verified by quantitative RT-PCR (Fig. S1). Protein complexes were purified by immunoprecipitation on whole cell lysates and protein A affinity pulldown. Then, after tryptic digestion, peptides were subjected to MS/MS in technical replicates by using a nano-LC Orbitrap system. By applying very stringent filtering criteria including the presence in replicate injections and/or identification with more than one unique peptide, 90 high-confidence proteins, putatively belonging to the CTCF interactome, were identified (Table 1). Details of the identifications are reported in Table S1.
Figure 1.
Schematic workflow of the immunoprecipitation-MS approach to identify the CTCF-interacting proteins. Protein complexes were purified by WiT49 whole cell extract by a two-step affinity purification with an anti-CTCF followed by protein A/G pulldown. Following elution and tryptic digestion, the resulting peptides were subjected to nano-LC–MS/MS in technical replicates for protein identification. Selected preys were then validated by co-immunoprecipitation and Western blotting (WB).
Table 1.
List of high-confidence proteins identified by nano-LC–MS/MS
| Accession | Description | Coverage | No. of peptides | Gene name |
|---|---|---|---|---|
| P49711-1 | Transcriptional repressor CTCF | 9.5 | 5 | CTCF |
| P35579-1 | Myosin-9 | 54.5 | 91 | MYH9 |
| P35580-3 | Isoform 3 of myosin-10 | 48.6 | 75 | MYH10 |
| P78527 | DNA-dependent protein kinase catalytic subunit | 17.2 | 53 | PRKDC |
| O00571 | ATP-dependent RNA helicase DDX3X | 41.7 | 21 | DDX3X |
| Q08211 | Atp-dependent RNA helicase A | 20.1 | 20 | DHX9 |
| P17844 | Probable ATP-dependent RNA helicase DDX5 | 37.6 | 18 | DDX5 |
| P46940 | Ras GTPase-activating-like protein IQGAP1 | 15.6 | 17 | IQGAP1 |
| P51532-1 | Transcription activator BRG1 | 11.2 | 15 | SMARCA4 |
| Q00839 | Heterogeneous nuclear ribonucleoprotein U | 23.2 | 14 | HNRNPU |
| P05783 | Keratin, type I cytoskeletal 18 | 53.5 | 13 | KRT18 |
| P52272 | Heterogeneous nuclear ribonucleoprotein M | 22.5 | 13 | HNRNPM |
| Q92841 | Probable ATP-dependent RNA helicase DDX17 | 28.4 | 11 | DDX17 |
| P23396-1 | 40S ribosomal protein S3 | 46.5 | 11 | RPS3 |
| P49411 | Elongation factor Tu, mitochondrial | 37.6 | 11 | TUFM |
| P63244 | Guanine nucleotide-binding protein subunit β2-like 1 | 39.7 | 10 | RACK1 |
| P25705-1 | ATP synthase subunit α, mitochondrial | 20.6 | 9 | ATP5A1 |
| P15880 | 40S ribosomal protein S2 | 39.2 | 9 | RPS2 |
| P61247 | 40S ribosomal protein S3a | 39.8 | 9 | RPS3A |
| P46781 | 40S ribosomal protein S9 | 37.1 | 9 | RPS9 |
| P54886 | δ1-Pyrroline-5-carboxylate synthase | 15.2 | 9 | ALDH18A1 |
| P11021 | 78-kDa glucose-regulated protein | 19.6 | 9 | HSPA5 |
| P61313-1 | 60S ribosomal protein L15 | 38.7 | 8 | RPL15 |
| P22087 | rRNA 2′-O-methyltransferase fibrillarin | 23.7 | 8 | FBL |
| P62701 | 40S ribosomal protein S4, X isoform | 27.8 | 8 | RPS4X |
| Q00325-2 | Isoform B of phosphate carrier protein, mitochondrial | 25.8 | 8 | SLC25A3 |
| P11142-1 | Heat shock cognate 71-kDa protein | 18.7 | 8 | HSPA8 |
| P06576 | ATP synthase subunit β, mitochondrial | 25.1 | 8 | ATP5B |
| P31943 | Heterogeneous nuclear ribonucleoprotein H | 29.6 | 7 | HNRNPH1 |
| P40429 | 60S ribosomal protein L13a | 30.5 | 7 | RPL13A |
| P62280 | 40S ribosomal protein S11 | 45.6 | 7 | RPS11 |
| P62263 | 40S ribosomal protein S14 | 39.1 | 7 | RPS14 |
| P10809 | 60-kDa heat shock protein, mitochondrial | 19.9 | 7 | HSPD1 |
| Q02543 | 60S ribosomal protein L18a | 31.8 | 6 | RPL18A |
| P61353 | 60S ribosomal protein L27 | 39.7 | 6 | RPL27 |
| P62266 | 40S ribosomal protein S23 | 49.0 | 6 | RPS23 |
| P27635 | 60S ribosomal protein L10 | 27.6 | 6 | RPL10 |
| P36542-1 | ATP synthase subunit γ, mitochondrial | 29.2 | 6 | ATP5C1 |
| O14497 | AT-rich interactive domain-containing protein 1A | 4.5 | 6 | ARID1A |
| P46777 | 60S ribosomal protein L5 | 23.9 | 6 | RPL5 |
| P84098 | 60S ribosomal protein L19 | 18.9 | 5 | RPL19 |
| P18621-3 | Isoform 3 of 60S ribosomal protein L17 | 22.8 | 5 | RPL17 |
| P62829 | 60S ribosomal protein L23 | 39.3 | 5 | RPL23 |
| P46779-3 | Isoform 3 of 60S ribosomal protein L28 | 19.5 | 5 | RPL28 |
| P62826 | GTP-binding nuclear protein RAN | 24.5 | 5 | RAN |
| Q53GQ0 | Very-long-chain 3-oxoacyl-CoA reductase | 22.4 | 5 | HSD17B12 |
| Q15233 | Non-POU domain-containing octamer-binding protein | 14.4 | 5 | NONO |
| P50454 | Serpin H1 | 20.8 | 5 | SERPINH1 |
| P49207 | 60S ribosomal protein L34 | 23.9 | 5 | RPL34 |
| P34897-1 | Serine hydroxymethyltransferase, mitochondrial | 11.5 | 5 | SHMT2 |
| P42677 | 40S ribosomal protein S27 | 40.5 | 5 | RPS27 |
| Q07666 | GAP-associated tyrosine phosphoprotein p62 | 18.3 | 5 | KHDRBS1 |
| P60660 | Myosin light polypeptide 6 | 42.4 | 5 | MYL6 |
| P62277 | 40S ribosomal protein S13 | 37.1 | 5 | RPS13 |
| P05141 | ADP/ATP translocase 2 | 37.6 | 4 | SLC25A5 |
| P50914 | 60S ribosomal protein L14 | 19.5 | 4 | RPL14 |
| P06748 | Nucleophosmin | 26.9 | 4 | NPM1 |
| P62249 | 40S ribosomal protein S16 | 28.1 | 4 | RPS16 |
| P62910 | 60S ribosomal protein L32 | 29.6 | 4 | RPL32 |
| Q9NZ01-1 | Very-long-chain enoyl-CoA reductase | 12.7 | 4 | TECR |
| Q9H9B4 | Sideroflexin-1 | 14.3 | 4 | SFXN1 |
| P62244 | 40S ribosomal protein S15a | 30.8 | 4 | RPS15A |
| P18077 | 60S ribosomal protein L35a | 22.7 | 4 | RPL35A |
| O95864 | Fatty acid desaturase 2 | 13.3 | 4 | FADS2 |
| P61619 | Protein transport protein Sec61 subunit alpha isoform 1 | 8.8 | 4 | SEC61A1 |
| Q16643-3 | Isoform 3 of Drebrin | 12.1 | 4 | DBN1 |
| P60842 | Eukaryotic initiation factor 4A-I | 13.5 | 4 | EIF4A1 |
| Q99623 | Prohibitin-2 | 16.1 | 4 | PHB2 |
| P57088 | Transmembrane protein 33 | 14.6 | 3 | TMEM33 |
| P61254 | 60S ribosomal protein L26 | 15.9 | 3 | RPL26 |
| P04844-1 | Ribophorin II | 7.8 | 3 | RPN2 |
| P62913 | 60S ribosomal protein L11 | 18.5 | 3 | RPL11 |
| P51148-2 | Isoform 2 of Ras-related protein Rab-5C | 14.1 | 3 | RAB5C |
| P60866-2 | Isoform 2 of 40S ribosomal protein S20 | 23.9 | 3 | RPS20 |
| P62899-2 | Isoform 2 of 60S ribosomal protein L31 | 25.0 | 3 | RPL31 |
| Q9UNF1 | Melanoma-associated antigen D2 | 13.4 | 3 | MAGED2 |
| Q96CS3 | FAS-associated factor 2 | 12.6 | 3 | FAF2 |
| P16615 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 5.0 | 3 | ATP2A2 |
| P18085 | ADP-ribosylation factor 4 | 25.0 | 3 | ARF4 |
| Q9UBM7 | 7-Dehydrocholesterol reductase | 6.3 | 3 | DHCR7 |
| P46782 | 40S ribosomal protein S5 | 16.7 | 3 | RPS5 |
| P45880-1 | Voltage-dependent anion-selective channel protein 2_1 | 12.9 | 3 | VDAC2 |
| O15260-1 | Surfeit locus protein 4 | 11.5 | 3 | SURF4 |
| P62854 | 40S ribosomal protein S26 | 18.3 | 2 | RPS26 |
| P62847-4 | Isoform 4 of 40S ribosomal protein S24 | 4.2 | 2 | RPS24 |
| Q14739 | Lamin-B receptor | 4.4 | 2 | LBR |
| P04843 | Ribophorin I | 3.8 | 2 | RPN1 |
| P08238 | Heat shock protein HSP 90β | 3.5 | 2 | HSP90AB1 |
| P14618 | Pyruvate kinase PKM | 6.8 | 2 | PKM |
| Q15555 | Microtubule-associated protein RP/EB family member 2 | 8.6 | 2 | MAPRE2 |
Two proteins of our selected CTCF-interacting proteins (i.e. Nucleophosmin (NPM1) and rRNA 2′-O-methyltransferase fibrillarin (FBL)) are among the CTCF interactors included in the curated BioGRID interaction repository database, as well as other proteins that have also been previously associated with CTCF such as the DEAD box proteins 5 and 17 (DDX5 and DDX17) also known as RNA helicases p68 and p72 (36). We also identified several other known CTCF interactors such as DNA topoisomerase 2 (TOP2B), poly(ADP-ribose) polymerase 1, and Cullin-associated NEDD8-dissociated protein 1 (CAND1) that were not further considered lacking the filtering criteria used in this study. Other previously known CTCF-binding partners were not identified in our screen, likely because of their low abundance that could have prevented their detection by MS. However, we cannot exclude that the failure to identify these proteins could be due to the association of CTCF with different binding partners more relevant in our cell model.
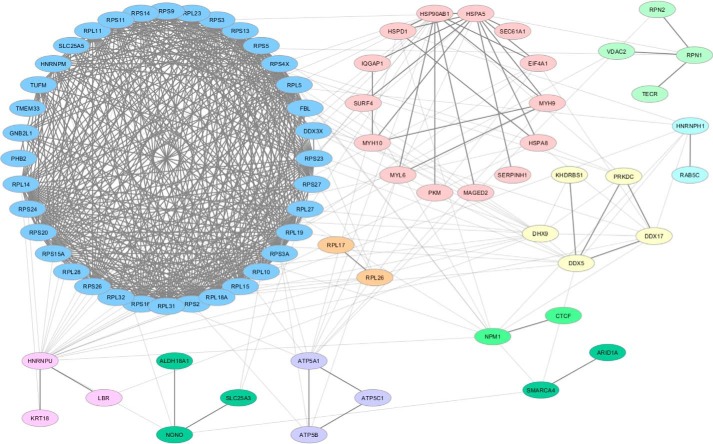
Clustering of CTCF-binding partners based on known protein interactions and functions
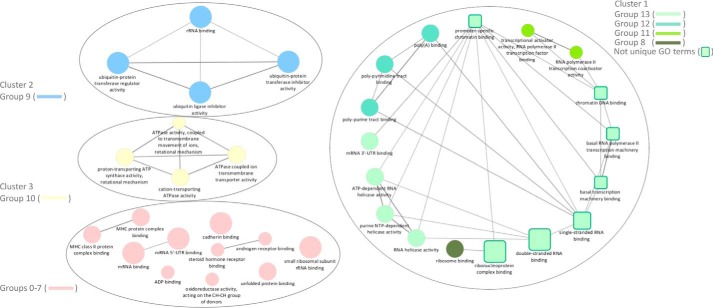
The clustering and visualization of protein–protein interaction networks is critical for the functional interpretation of MS data and for targeting validation on novel binding partners of biological relevance. To this aim, candidate CTCF-interacting proteins were mapped on a single interconnected network by the NetworkAnalyst software using the literature-curated IMEx Interactome database. The ClusterMaker2 Cytoscape plug-in was used for clustering and visualizing network nodes into modules for the detection of previously annotated complexes. By this approach, 76 of the 91 candidate proteins were mapped on a network including a large cluster containing several ribosomal proteins (Fig. 2, blue) connected to several smaller ones including a cluster of several ATP-dependent RNA helicases (Fig. 2, yellow). An unbiased gene ontology-based classification was then applied to investigate functions of proteins associated with CTCF in the pulldown experiment. To this end, the ClueGO cytoscape plug-in was used to generate a functionally grouped gene ontology (GO)/pathway term network of enriched molecular function categories for the identified proteins based on kappa statistics. Identified proteins were assigned to 13 groups that were mapped on a functionally clustered network (Fig. 3 and Table S2). Not surprisingly, the larger cluster of the output network for enriched categories revealed that a subset of identified proteins was involved in several specific functions related to the RNA transcription process, as well as to chromatin DNA binding and promoter-specific chromatin binding. An additional enriched molecular function ontology group in the CTCF interactome is that related to ATP-dependent helicase activity. To provide further insights into proteins associated with GO terms, we visualized these terms with their associated proteins in a heat-map layout showing the individual proteins, resulting in the identification of enriched molecular functions (Fig. S2). This analysis points out the overlapping presence of several DEAD or DEAH box helicases in several groups. Among these, we identified EIF4A1, DDX3X, DHX9, DDX5, DDX17, of which the last two already known CTCF-interacting proteins (36). We also revealed the presence of the transcription activator BRG1, also known as ATP-dependent chromatin remodeler SMARCA4, which is together with BRM (also known as SMARCA2), one of the two mutually exclusive core ATPase subunits of the switch/sucrose non-fermentable (SNF/SWI) chromatin remodeling complex. Interestingly, we also detected the AT-rich interactive domain–containing protein 1 (ARID1A), another component present in only some variants of the SWI/SNF complex. Because a potential interaction of BRG1 and CTCF has been long postulated but still not experimentally demonstrated (37–39), we focused our attention on this interaction and select BRG1 for further investigations. To exclude that the interaction between BRG1 and CTCF was mediated by nucleic acids, we performed pulldown assays in the presence or absence of Benzonase nuclease to degrade DNA/RNA followed by targeted LC-MS/MS analyses. The presence of BRG1 in the CTCF IP was not changed following Benzonase digestion, suggesting that the interaction between the proteins is DNA/RNA-independent (Figs. S3–S20).
Figure 2.
High-confidence interaction partners of CTCF. 76 of the 91 candidate CTCF-interacting proteins were mapped on a single interconnected network constructed by NetworkAnalyst using the literature-curated IMEx Interactome database and visualized as clusters identified by using the ClusterMaker2 plug-in of Cytoscape 3.6.0. The Markov clustering algorithm was used for network clustering.
Figure 3.
Functionally grouped network of enriched molecular function categories for the identified proteins generated by using the ClueGO cytoscape plug-in. The proportion of shared proteins between terms was evaluated using kappa statistics. GO terms are represented as nodes whose size represents the term enrichment significance. Partially overlapping functionally related groups are represented as squares, whereas nonoverlapping terms are represented as circles. Clusters including more than two terms are numbered as clusters 1 (green), 2 (blue), and 3 (yellow). The group number resulting from ClueGO associations of GO terms is indicated for each cluster (Table S2). Nonclustered terms including groups 0–7 are colored pink.
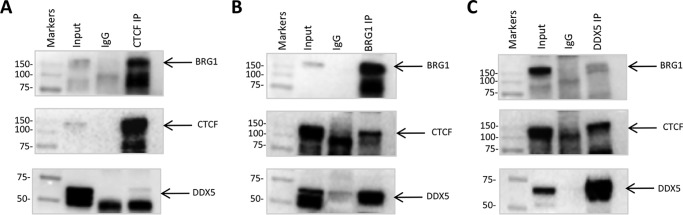
Validation by co-immunoprecipitation of interactions of CTCF with BRG1 and DDX5
Co-immunoprecipitation (co-IP) followed by immunoblot was performed to further validate the interaction of CTCF with BRG1. DDX5 was also selected to validate the specificity of interaction based on previous evidence that report DDX5 as a common interaction partner of both BRG1 and CTCF (36, 40). Both BRG1 and DDX5 were co-immunoprecipitated with anti-CTCF but not with anti-IgG (Fig. 4A). By CTCF IP followed by Western blotting, we also confirmed that Benzonase treatment did not affect the interaction of CTCF with BRG1 (Fig. S21). Moreover, we performed reciprocal co-IP assays using whole cell lysate from WiT49 cells and antibodies against BRG1 (Fig. 4B) and DDX5 (Fig. 4C). The reciprocal co-IP analysis demonstrated that CTCF co-purified with endogenous BRG1 and DDX5 proteins. These results further support AP-MS data and confirm specific interactions of the selected candidate proteins with CTCF.
Figure 4.
Co-immunoprecipitations between CTCF and endogenous DDX5 and BRG1. A, anti-CTCF-immunoprecipitated samples were blotted with anti-BRG1 and anti-DDX5 antibodies. Anti-CTCF was used as positive control. Input, 2% of the cell lysate used for immunoprecipitation. B, anti-BRG1-immunoprecipitated samples were blotted with anti-CTCF and anti-DDX5 antibodies. Anti-BRG1 was used as positive control. Input, 2% of the cell lysate used for immunoprecipitation. C, anti-DDX5-immunoprecipitated samples were blotted with anti-CTCF and anti-BRG1 antibodies. Anti-DDX5 was used as positive control. Input, 4% of the cell lysate used for immunoprecipitation.
Genomic co-occupancy by CTCF, BRG1, and DDX5
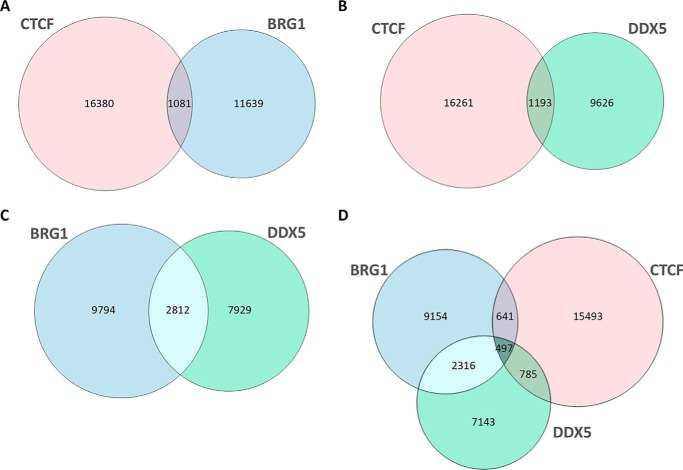
To provide further insights into the functional interaction of CTCF with BRG1 and DDX5, we wondered whether common DNA-binding sites were shared by these proteins. We then reanalyzed ChIP-Seq data on genome-wide binding profiles of the three proteins in HeLa cells (Table S4) to assess their chromatin co-occupancy and to determine whether they co-localized to the same genomic regions. At first, we performed pair-wise comparisons of the genomic sites occupied by each protein (Fig. 5, A–C).
Figure 5.
ChIP-Seq co-localizations between CTCF, BRG1, and DDX5. The Venn diagrams show the overlap of CTCF and BRG1 (A), CTCF and DDX5 (B), BRG1 and DDX5 (C), and CTCF, BRG1, and DDX5 (D) ChIP-seq peaks.
Consistent with the physical interaction revealed by AP-MS, we found significant co-localizations for all the analyzed comparisons (p value < 0.005). In particular, when CTCF and BRG1 sites were compared, we found that ∼9% of BRG1 sites were shared with 6% of CTCF sites (Fig. 5A). Similarly, comparison between CTCF and DDX5 revealed that 11% of DDX5 sites were co-occupied by ∼7% of CTCF sites (Fig. 5B). A higher number of overlapping sites were shared by BRG1 and DDX5, with 26% of DDX5 sites also bound by 22% of BRG1 sites (Fig. 5C). We also identified a set of 497 sites simultaneously co-occupied by CTCF, DDX5, and BRG1 with ∼44% of sites co-occupied by BRG1 and CTCF also bound by DDX5 (Fig. 5D).
To further characterize localizations of the three proteins, we investigated the distribution of both co-occupied sites (Fig. 6, blue bars) and sites occupied by CTCF alone (Fig. 6, green bars) with respect to distances from TSSs. Co-localized regions for CTCF–BRG1, CTCF–DDX5, and CTCF–BRG1–DDX5 were enriched in a window of 0–2 kb around TSSs with respect to sites occupied by CTCF alone (p value < 2.2e−16). Interestingly, the CTCF–BRG1–DDX5 intersection was significantly enriched around TSSs even with respect to both CTCF–BRG1 (p value < 2.2e−16) and CTCF–DDX5 (p value < 6.4e−08), thus suggesting a higher enrichment at promoter regions of sites co-occupied by all the three proteins with respect to overlapping sites of pair-wise comparisons.
Figure 6.
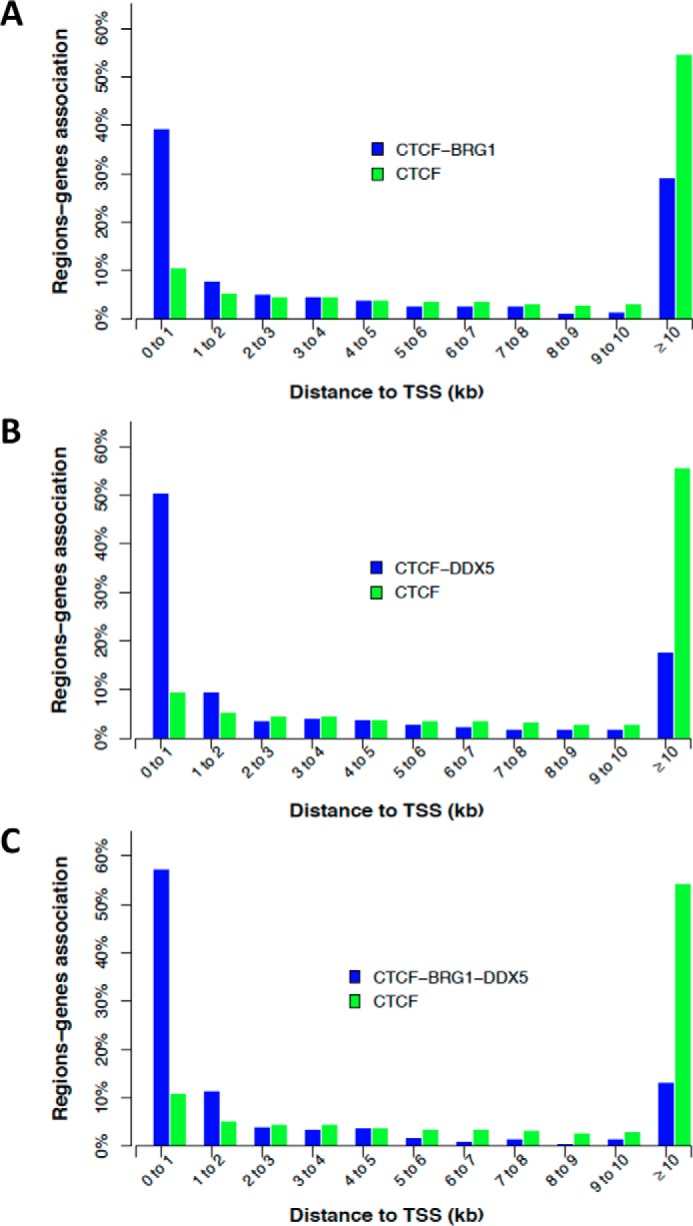
Associations between co-localized regions: genes as a function of the distance to TSS (±2 kb). Co-occupied sites are reported as blue for CTCF–BRG1 (A), CTCF–DDX5 (B), and CTCF–BRG1–DDX5 (C). In all graphs, sites occupied by CTCF alone are reported as green bars.
Moreover, we also found an over-representation (p value < 2.2e−16) with respect to CTCF sites alone, of CTCF–BRG1, CTCF–DDX5, and CTCF–BRG1–DDX5 co-localized regions with trimethylation of histone H3 at lysine 4 (H3K4me3) and trimethylation of histone H3 at lysine 36 (H3K36me3) that are associated with active transcription. Accordingly, for the same co-localized regions, we observed an under-representation (p value < 2.2e−16) of the histone mark of gene repression H3K27me3. Overall, these results suggest the co-occupancy of CTCF, BRG1, and DDX5 on transcriptionally active chromatin regions (Table S3).
Discussion
The transcription factor CTCF plays a pivotal role in a myriad of genomic processes, including transcription, imprinting, and long-range chromatin interactions. It is widely recognized that the versatility of this multitasking master regulator is at least in part determined by co-association with genomic context-specific binding partners (14, 21). Protein–protein interaction maps have proven to be very useful for understanding the protein molecular functions.
Here, we present the first global CTCF-associated protein interactome map performed by high-resolution MS. Our study confirms previously reported interactions and reveals novel potential CTCF-binding partners, suggesting that the CTCF annotated interaction proteome is far from being complete.
Consistent with other studies, we identified several ribosomal proteins together with the nucleolar protein Nucleophosmin, a molecular chaperone involved in the transport of ribosome subunits and histones from the cytoplasm to the nucleus and nucleoli (41). It has been demonstrated that Nucleophosmin interacts with CTCF at the insulator sites in vivo (41). CTCF/Nucleophosmin association has been also hypothesized to be responsible for the co-purification with ribosomal proteins (41). We also confirmed by both MS identification and by co-IP experiments the interaction of CTCF with the DEAD box RNA helicase p68 (DDX5). This complex has been also reported to include the steroid receptor RNA activator and is essential for CTCF function as an enhancer-blocking insulator in vivo (36). Interestingly, together with DDX5 we also identified the highly homologous protein DDX17 (p72), previously reported to be associated with CTCF (36) and several additional members of the DEX(D/H) box family, such as DHX9 (RNA helicase A) and DDX3X. These proteins are engaged in multiple processes of RNA biology including pre-mRNA processing (i.e. cap formation, splicing/alternative splicing, and polyadenylation), ribosome biogenesis, RNA turnover, export, and translation (as reviewed in Refs. 42 and 43). In addition, a growing body of evidence suggests the involvement of several DEX(D/H) box proteins as transcriptional regulators (42, 43). Intriguingly, these roles in the transcriptional machinery appear to be independent from their RNA helicase or unwindase activity. Indeed, it has been reported that they may either stabilize the transcriptional initiation complex or act as bridging factors that facilitate the recruitment of other transcription factors/co-activators such as CBP, p300, and RNA polymerase (Pol) II to responsive promoters (44). RNA helicases p68/p72 and the noncoding steroid receptor RNA activator have been also found associated with MyoD and are directly involved in its co-activation by promoting the assembly of a transcription initiation complex including the TATA-binding protein TBP and the RNA Pol II (40). The catalytic subunit of the ATPase SWI/SNF chromatin remodeling complex, BRG1, that physically interacts with p68/p72 (40) also takes part in this mechanism.
Our AP-MS analysis, for the first time, reveals that BRG1 is co-associated with CTCF. The high-confidence interaction was verified in replicate independent experiments and was further validated by co-IP and reverse co-IP by performing a BRG1 immunoprecipitation and probing the immunoblot with a CTCF antibody. Consistent with the fundamental role of chromatin remodeling complexes in regulating chromatin accessibility for gene expression, a genome-wide screen of SWI/SNF component (i.e. Ini1, BAF155, BAF170, and BRG1) binding sites demonstrated an extensive overlap with promoters, enhancers, and many regions occupied by Pol II and CTCF sites (37). More recently, in addition to the transcriptional role of BRG1 at gene promoters, a more complex scenario is emerging identifying BRG1 as a dynamic component of higher-order chromatin organization enriched at TAD boundaries (38, 39). BRG1 has also been involved in the maintenance of nuclear structure integrity and in mediating specific long-range chromatin interactions through interactions with transcription factors and other co-factors (37, 45). In this context, it was suggested that BRG1 plays a role at TAD boundaries by regulating nucleosome occupancy and possibly CTCF localization. Indeed, an intersection of CTCF ChIP-seq data set carried out using MCF-10A cells (46) with BRG1 peaks revealed that ∼10% of all BRG1 peaks and 12% of BRG1 peaks specifically located at TAD boundaries directly overlapped with CTCF (38). Moreover, a relationship between BRG1 knockdown and the reduction of nucleosome occupancy around the CTCF sites was also observed in mouse fibroblast cells (38). Similar effects were noticed for BRG1 knockdown around TSS of known genes (47). Despite the fact that cross-talk between BRG1 and CTCF has long been hypothesized and supported by genome-wide approaches, attempts to co-purify these factors by AP-MS were to date unsuccessful. In eukaryotic cells, a balance between tight packaging and accessibility of the chromatin is usually achieved by specific proteins that dynamically modify chromatin structure. BRG1 and CTCF are regarded as master regulators of chromatin architecture. Indeed, BRG1 is involved in the fine tuning of DNA accessibility in an ATP-dependent manner, whereas CTCF is widely recognized as a global genome organizer able to coordinate high-order chromatin structures and to regulate gene expression (12, 13, 15). Our data point toward a cooperation between the two proteins that may be crucial in determining their functional specificity. Interestingly, our data suggest an unanticipated interplay in transcriptional regulation between CTCF, BRG1, and DDX5 because we found that regions simultaneously co-occupied by the three proteins are significantly enriched at promoter regions.
The high-resolution map of CTCF-binding sites in human genome revealed that only ∼20% of CTCF sites are near transcription start sites (8). Unlike general transcription factors, the localization of CTCF sites distal to TSS has been suggested to be consistent with its putative role as an insulator-binding protein (8). Nevertheless, much evidence for a direct role of CTCF in transcription regulation on individual genes has been demonstrated (48, 49). Moreover, Peña-Hernández et al. (50) reported that the interaction between CTCF and transcription factor II-I was essential in directing CTCF to the promoter regions of genes involved in metabolism. We also noticed, a significant over-representation, with respect to CTCF sites, of CTCF, BRG1, and DDX5 co-localized regions with H3K4me3 and H3K36me3. These histone marks are usually enriched at TSS/promoter regions with open chromatin structure and known to be positively correlated with gene transcriptional activation. Accordingly, we observed an under-representation of CTCF, BRG1, and DDX5 co-localized regions with the repressive histone modification H3K27me3 associated with silent genes. Taken together, our findings suggest that the CTCF sites where the transcription factor co-localizes with BRG1 and DDX5 mostly include a subset of genome-wide CTCF sites located around the TSS and associated with histone marks of transcriptionally active chromatin. Overall, it can be supposed that, whatever the effect of CTCF on transcription (e.g. repression, activation/transactivation, or pausing), these different outputs can only occur through cooperation with other proteins involved in remodeling chromatin architecture such as BRG1. Additional proteins of the transcriptional machinery such as DDX5 may contribute to the diversification of CTCF functions by means of alternative complexes formation possibly involved in the recruitment of other transcription factors/co-activators to promoters.
Although the roles of several identified proteins are still undefined, our study highlights the capability of AP-MS to fill the gaps in our knowledge about novel CTCF interactors contributing to fine-tuning of its multiple functions. The presented CTCF interaction proteome represents a knowledge base for further elucidating individual protein interaction with CTCF and for instructing future functional experiments to uncover molecular bases responsible for the high versatility of this unique transcription factor.
Materials and methods
Cell culture, cloning, and transfections
The WiT49 cell line derived from a Wilms tumor primary lung metastasis (51) was cultured in Iscove's modified Dulbecco's medium, supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 mg/ml streptomycin at 37 °C in a humidified 5% CO2 atmosphere. The cDNA encoding the full-length CTCF gene was cloned into the pcDNA3 expression vector, under the control of the constitutively expressed cytomegalovirus promoter. For plasmid transfection, WiT49 cells were transfected with the pcDNA3-CTCF plasmid or with the empty control vector using Lipofectamine 3000 according to the manufacturer's protocol (Thermo Fisher Scientific). Stably transfected cells were selected with 1 mg/ml G418 (Life Technologies) and maintained in 0.6 mg/ml G418.
RNA extraction and RT-PCR analysis
Total RNA was isolated from stably transfected WiT49 cell lines using the TRI reagent (Sigma) according to the manufacturer's protocol. Total RNA (1 μg) was reverse-transcribed by using the QuantiTect reverse transcription kit (Qiagen). Real-time PCR was performed using the SYBR Green I DNA-binding dye technology (Bio-Rad) on a C1000 thermal cycler (Bio-Rad). Primer sequences were as follows: 5′-GCAGAGGGAGAGGAAGAGGA-3′ (forward) and 5′-TATGGGTATCCGGCGTAGTC-3′ (reverse) for the CTCF gene and 5′-CAATTCCCCATCTCAGTCGT-3′ (forward) and 5′-GCAGCAGGACACTAGGGAGT-3′ (reverse) for the glyceraldehyde-3-phosphate dehydrogenase gene. The results were expressed relative to the glyceraldehyde-3-phosphate dehydrogenase internal control gene.
Sample preparation for MS analysis
WiT49 cells overexpressing CTCF from ten 150-mm plates were harvested by trypsinization and washed with PBS. The cells were lysed for 45 min at 4 °C in lysis buffer (200 μl of lysis buffer/plate) containing 10 mm Tris-HCl, pH 7.4, 350 mm NaCl, 1 mm EDTA, 1% Triton X-100, 10% glycerol and then clarified at 15,000 × g for 15 min at 4 °C. For Benzonase digestion, the cells were lysed in 10 mm Tris-HCl, pH 7.4, 350 mm NaCl, 1 mm MgCl2, 1% Triton X-100, 10% glycerol and incubated for 30 min at room temperature in the presence or absence of 250 units of Benzonase (Sigma–Aldrich). Aliquots of lysates (10 μl) were analyzed by 1% agarose gel electrophoresis and ethidium bromide staining to verify DNA/RNA degradation (data not shown). Protein concentration was determined by Bradford assay. For IP, protein lysates (1–2 mg for benzonase-treated/untreated samples) were diluted in IP buffer up to 1 ml (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.25% sodium deoxycholate) and incubated for 1 h at 4 °C with DiaMag protein A–coated magnetic beads (40 μl, Diagenode). After the preclearing step, the samples were incubated overnight at 4 °C with polyclonal anti-CTCF (Diagenode C15010210, 10 μg) and polyclonal rabbit anti-IgG (Diagenode C15410206, 10 μg) as negative control. Immunoprecipitated proteins were then incubated for 3 h under rotation at 4 °C with the DiaMag protein A–coated magnetic beads (40 μl, Diagenode) prewashed in the IP buffer. The beads were collected on a magnetic stand, washed three times with 100 μl of 100 mm NH4HCO3, pH 8.0, and resuspended in 100 μl of the same buffer. The proteins were reduced with 10 mm DTT (final concentration) at 55 °C for 1 h and, following a wash step with 100 μl of NH4HCO3, carbamidomethylated with 7.5 mm iodoacetamide (final concentration) at room temperature in the dark for 15 min. Following a further wash step with 100 μl of NH4HCO3, enzymatic hydrolyzes were performed by the addition of 0.2 μg of tosyl phenylalanyl chloromethyl ketone–treated trypsin to the reduced and alkylated mixture. Digestions were performed by incubation at 37 °C for 16 h. After digestions, the samples were centrifuged at 10,000 × g for 15 min, and supernatants were dried under vacuum in a SpeedVac vacuum (Savant Instruments, Holbrook, NY). The samples were then resuspended in 40 μl of H2O, 0.1% TFA and centrifuged at 10,000 × g for 15 min. Aliquots of the supernatant (3 μl) were analyzed by high resolution nano-LC–tandem mass spectrometry.
High resolution nano-LC–tandem mass spectrometry
Mass spectrometry analysis was performed on a Q Exactive Orbitrap mass spectrometer equipped with an EASY-Spray nano-electrospray ion source (Thermo Fisher Scientific) and coupled to a Dionex UltiMate 3000RSLC nano system (Thermo Fisher Scientific). Solvent composition was 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). Peptides were loaded on a trapping PepMapTM100 μ Cartridge Column C18 (300 μm × 0.5 cm, 5 μm, 100 Å) and desalted with solvent A for 3 min with at a flow rate of 10 μl/min. After trapping, eluted peptides were separated on an EASY-Spray analytical column (15 cm × 75 μm inner diameter PepMap RSLC C18, 3 μm, 100 Å), heated to 35 °C, at a flow rate of 300 nl/min by using the following gradient: 4% B for 3 min, from 4 to 22% B in 50 min, from 22 to 35% B in 10 min, and from 35 to 90% B in 5 min. A washing (90% B for 5 min) and a re-equilibration (4% B for 15 min) step was always included at the end of the gradient. Eluting peptides were analyzed on the Q-Exactive mass spectrometer operating in positive polarity mode with capillary temperature of 280 °C and a potential of 1.9 kV applied to the capillary probe. Full MS survey scan resolution was set to 70,000 with an automatic gain control target value of 3 × 106 for a scan range of 375–1500 m/z and maximum ion injection time of 100 ms. The mass (m/z) 445.12003 was used as lock mass. A data-dependent top five method was operated during which higher-energy collisional dissociation (HCD) spectra were obtained at 17,500 MS2 resolution with an automatic gain control target of 1 × 105 for a scan range of 200–2000 m/z, maximum injection time of 55 ms, 2 m/z isolation width, and a normalized collisional energy of 27. Precursor ions targeted for HCD were dynamically excluded for 15 s. Full scans and Orbitrap MS/MS scans were acquired in profile mode, whereas ion trap mass spectra were acquired in centroid mode. Charge state recognition was enabled by excluding unassigned and singly charge states.
MS data processing
The acquired raw files were analyzed with the Proteome Discoverer 2.1 software (Thermo Fisher Scientific) using the SEQUEST HT search engine. The HCD MS/MS spectra were searched against the Homo sapiens Uniprot_sprot database (release 2015_11_11, 42,084 entries) assuming trypsin (full) as digestion enzyme and two allowed number of missed cleavage sites. The mass tolerances were set to 10 ppm and 0.02 Da for precursor and fragment ions, respectively. Oxidation of methionine (+15.995 Da) and N-terminal acetylation (+42.011 Da) were set as dynamic modifications and carbamidomethylation of cysteine (+57.021 Da) as static modification. False discovery rates (FDRs) for peptide spectral matches (PSMs) were calculated and filtered using the target decoy PSM validator node in Proteome Discoverer. The target decoy PSM validator node specifies the PSM confidences on the basis of dynamic score-based thresholds. It calculates the node-dependent score thresholds needed to determine the FDRs, which are given as input parameters of the node. Target decoy PSM validator was run with the following settings: maximum delta Cn 0.05, a strict target FDR of 0.01, a relaxed target FDR of 0.05, and validation based on q value. The protein FDR validator node in Proteome Discoverer was used to classify protein identifications based on q value. Proteins with a q value of <0.01 were classified as high-confidence identifications, and proteins with a q value of 0.01–0.05 were classified as medium-confidence identifications. Only proteins identified with high confidence were retained with an FDR of 1%. The resulting list of CTCF-interacting proteins was finally uploaded into the Contaminant Repository for Affinity Purification (CRAPome, www.crapome.org) (63)5 database to further investigate the presence of potential contaminants within the identified protein list. The obtained results were not used as an exclusion criterion but as an estimate of probability and significance for each interacting protein. The criteria used for inclusion as potential CTCF-interacting protein were the presence in replicate injections with more than one unique peptide and the absence in control IgG IP sample. Proteins identified by searching MS/MS spectra against a custom common contaminant database were also not considered.
Bioinformatic analyses
The list of CTCF interactors identified by LC-MS/MS was imported into the NetworkAnalyst software for integrative analysis of protein data through statistical, visual, and network-based approaches (52). The literature-curated IMEx Interactome database from InnateDB (53) was selected for the protein–protein interaction analysis. The resulting zero-order network was visualized and further analyzed using Cytoscape 3.6.0 (54). The Markov clustering algorithm implemented in the Cytoscape plug-in clusterMaker2 was used for network clustering (55). Molecular function enrichment analysis was performed by using the ClueGO cytoscape plug-in to generate a functionally grouped GO/pathway term network of enriched molecular function categories for the identified proteins based on kappa statistics (56).
ChIP-seq data analysis
The ChIP-seq data used in this study are from previous publications and are listed in Table S4 (36, 37, 57). The numbers of consensus peaks for CTCF, BRG1, DDX5, H3K4me3, H3K36me3, and H3K27me3 is summarized in Table S4. The freely available LiftOver tool (https://genome.ucsc.edu/)5 was used when necessary to convert the genome coordinates from NCBI36/hg18 to GRCh37/hg19. The analyses were carried out using the GRCh37/hg19 coordinates. The consensus regions for CTCF, BRG1, DDX5, H3K4me3, H3K36me3, and H3K27me3 were defined in terms of co-localizations (i.e. overlaps with distance equal to zero) between replicate tracks when available. The CTCF consensus peaks were considered as reference. The Bioconductor package ChIPpeakAnno was used to quantify the co-localizations by computing the number of overlapping/not overlapping regions and the corresponding lists for each comparison (58, 59). The significance of the co-localizations was assessed by a permutation test using the shuffle function of the Bioconductor package ChIPseeker (60). Co-localized regions were annotated with respect to gene positions, and the gene annotation was performed using the package ChIPseeker. The parameters were set up to annotate the regions with the closest gene (in terms of TSS) within a window of 3 kbp. The Ensembl release GRCh37.p13 was considered as a reference database and imported in R using the Bioconductor package biomaRt (https://bioconductor.org/packages/releasebioc/html/biomaRt.html) (64, 65).5 The Fisher's exact test implemented in R was used to evaluate the statistical significance of associations (true odds ratio > 1 to test for over-representation and true odds ratio < 1 to test for under-representation). Statistical significance was reported in terms of p values.
Co-IP and Western blotting analyses
Stably transfected WiT49 cell line overexpressing CTCF grown in 150-mm plates were harvested by trypsinization and washed with PBS. The cells were lysed for 45 min at 4 °C in 200 μl of lysis buffer (10 mm Tris-HCl, pH 7.4, 350 mm NaCl, 1 mm EDTA, 1% Triton X-100, 10% glycerol) and clarified at 15,000 × g for 15 min at 4 °C. Following determination of protein concentration by Bradford assay, 2 mg of total protein lysates was diluted in the IP buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 0.25% sodium deoxycholate) at 2 mg/ml final concentration and incubated overnight at 4 °C (61) with the following antibodies: rabbit polyclonal anti-CTCF (Diagenode C15010210, 10 μg), monoclonal rabbit anti-BRG1 (Abcam ab110641, 0.5 μg), monoclonal rabbit anti-DDX5 (Abcam ab126730, 1.7 μg), and polyclonal rabbit anti-IgG (Diagenode C15410206, 10 μg). 40 μl of DiaMag protein A–coated magnetic beads (Diagenode C03010020–150) were added to samples, and after further incubation for 3 h under rotation at 4 °C, the beads were collected and washed with IP buffer, and the bound proteins were eluted by boiling samples in 2× Laemmli buffer for 10 min. The samples were then resolved by Mini Protean TGX gels (Any-kDa, Bio-Rad catalog no. 4569033) and transferred onto polyvinylidene difluoride membranes (Bio-Rad catalog no. 170-4156) for immunoblotting detection (62) with anti-CTCF (1:1000), anti-BRG1 (1:5000), and anti-DDX5 (1:5000). Following incubation with the anti-rabbit HRP-conjugated antibody (Bio-Rad catalog no. 170-6515, 1:3000), protein bands were revealed by adding the ClarityTM Western ECL substrate (Bio-Rad catalog no. 170-5061) and acquired by using the ChemiDoc XRS System (Bio-Rad).
Author contributions
M. M. M., C. R., R. R., M. V., I. B., I. D. F., C. A., T. X., and A. C. data curation; M. M. M., C. R., R. R., and S. E. investigation; M. M. M., C. R., R. R., M. V., M. T. G., S. E., and I. B. methodology; I. D. F., C. A., T. X., and A. C. formal analysis; G. F. funding acquisition; G. F. writing-review and editing; A. C. and P. V. P. conceptualization; A. C. and P. V. P. supervision; A. C. and P. V. P. writing-original draft.
Supplementary Material
This work was supported by University of Campania “Luigi Vanvitelli” through the VALERE program and the intramural research program of the NIDDK, National Institutes of Health. The National Institutes of Health also provided support to P. V. P. as a visitor. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S4 and Figs. S1–S21.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- CTCF
- CCCTC-binding factor
- TAD
- topologically associating domain
- TSS
- transcription start site
- AP-MS
- affinity purification of proteins coupled to MS
- GO
- gene ontology
- IP
- immunoprecipitation
- Pol
- polymerase
- HCD
- higher-energy collisional dissociation
- FDR
- false discovery rate
- PSM
- peptide spectral match.
References
- 1. Chung J. H., Bell A. C., and Felsenfeld G. (1997) Characterization of the chicken β-globin insulator. Proc. Natl. Acad. Sci. U.S.A. 94, 575–580 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bell A. C., and Felsenfeld G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 3. Hark A. T., Schoenherr C. J., Katz D. J., Ingram R. S., Levorse J. M., and Tilghman S. M. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 4. Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C. F., Wolffe A., Ohlsson R., and Lobanenkov V. V. (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10, 853–856 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 5. Filippova G. N., Fagerlie S., Klenova E. M., Myers C., Dehner Y., Goodwin G., Neiman P. E., Collins S. J., and Lobanenkov V. V. (1996) An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16, 2802–2813 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lobanenkov V. V., Nicolas R. H., Adler V. V., Paterson H., Klenova E. M., Polotskaja A. V., and Goodwin G. H. (1990) A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5, 1743–1753 [PubMed] [Google Scholar]
- 7. Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V. V., and Ren B. (2012) A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim T. H., Abdullaev Z. K., Smith A. D., Ching K. A., Loukinov D. I., Green R. D., Zhang M. Q., Lobanenkov V. V., and Ren B. (2007) Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Renda M., Baglivo I., Burgess-Beusse B., Esposito S., Fattorusso R., Felsenfeld G., and Pedone P. V. (2007) Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J. Biol. Chem. 282, 33336–33345 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 10. Wang H., Maurano M. T., Qu H., Varley K. E., Gertz J., Pauli F., Lee K., Canfield T., Weaver M., Sandstrom R., Thurman R. E., Kaul R., Myers R. M., and Stamatoyannopoulos J. A. (2012) Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 22, 1680–1688 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohlsson R., Renkawitz R., and Lobanenkov V. (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17, 520–527 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 12. Phillips J. E., and Corces V. G. (2009) CTCF: master weaver of the genome. Cell 137, 1194–1211 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ong C. T., and Corces V. G. (2014) CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holwerda S. J., and de Laat W. (2013) CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120369 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wallace J. A., and Felsenfeld G. (2007) We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17, 400–407 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghirlando R., and Felsenfeld G. (2016) CTCF: making the right connections. Genes Dev. 30, 881–891 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zlatanova J., and Caiafa P. (2009) CCCTC-binding factor: to loop or to bridge. Cell Mol. Life Sci. 66, 1647–1660 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., and Ren B. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramírez F., Bhardwaj V., Arrigoni L., Lam K. C., Grüning B. A., Villaveces J., Habermann B., Akhtar A., and Manke T. (2018) High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9, 189 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakahashi H., Kieffer Kwon K. R., Resch W., Vian L., Dose M., Stavreva D., Hakim O., Pruett N., Nelson S., Yamane A., Qian J., Dubois W., Welsh S., Phair R. D., Pugh B. F., et al. (2013) A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 3, 1678–1689 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zlatanova J., and Caiafa P. (2009) CTCF and its protein partners: divide and rule? J. Cell Sci. 122, 1275–1284 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 22. Jabbari K., Heger P., Sharma R., and Wiehe T. (2018) The diverging routes of BORIS and CTCF: an interactomic and phylogenomic analysis. Life (Basel) 8, E4 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hadjur S., Williams L. M., Ryan N. K., Cobb B. S., Sexton T., Fraser P., Fisher A. G., and Merkenschlager M. (2009) Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nativio R., Wendt K. S., Ito Y., Huddleston J. E., Uribe-Lewis S., Woodfine K., Krueger C., Reik W., Peters J. M., and Murrell A. (2009) Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 5, e1000739 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zuin J., Dixon J. R., van der Reijden M. I., Ye Z., Kolovos P., Brouwer R. W., van de Corput M. P., van de Werken H. J., Knoch T. A., van Ijcken W. F., Grosveld F. G., Ren B., and Wendt K. S. (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U.S.A. 111, 996–1001 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sofueva S., Yaffe E., Chan W. C., Georgopoulou D., Vietri Rudan M., Mira-Bontenbal H., Pollard S. M., Schroth G. P., Tanay A., and Hadjur S. (2013) Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 32, 3119–3129 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rao S. S., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., Robinson J. T., Sanborn A. L., Machol I., Omer A. D., Lander E. S., and Aiden E. L. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Köcher T., and Superti-Furga G. (2007) Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods 4, 807–815 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 29. Giambruno R., Grebien F., Stukalov A., Knoll C., Planyavsky M., Rudashevskaya E. L., Colinge J., Superti-Furga G., and Bennett K. L. (2013) Affinity purification strategies for proteomic analysis of transcription factor complexes. J. Proteome Res. 12, 4018–4027 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gingras A. C., Gstaiger M., Raught B., and Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 31. Sardiu M. E., Cai Y., Jin J., Swanson S. K., Conaway R. C., Conaway J. W., Florens L., and Washburn M. P. (2008) Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci. U.S.A. 105, 1454–1459 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sowa M. E., Bennett E. J., Gygi S. P., and Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Varjosalo M., Keskitalo S., Van Drogen A., Nurkkala H., Vichalkovski A., Aebersold R., and Gstaiger M. (2013) The protein interaction landscape of the human CMGC kinase group. Cell Rep. 3, 1306–1320 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 34. Choi H., Larsen B., Lin Z. Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A. C., and Nesvizhskii A. I. (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., Larsen B., Lin Z. Y., Breitkreutz B. J., Stark C., Liu G., Ahn J., Dewar-Darch D., Reguly T., Tang X., Almeida R., et al. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yao H., Brick K., Evrard Y., Xiao T., Camerini-Otero R. D., and Felsenfeld G. (2010) Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 24, 2543–2555 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Euskirchen G. M., Auerbach R. K., Davidov E., Gianoulis T. A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M. B., and Snyder M. (2011) Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7, e1002008 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barutcu A. R., Lajoie B. R., Fritz A. J., McCord R. P., Nickerson J. A., van Wijnen A. J., Lian J. B., Stein J. L., Dekker J., Stein G. S., and Imbalzano A. N. (2016) SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 26, 1188–1201 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barutcu A. R., Lian J. B., Stein J. L., Stein G. S., and Imbalzano A. N. (2017) The connection between BRG1, CTCF and topoisomerases at TAD boundaries. Nucleus 8, 150–155 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caretti G., Schiltz R. L., Dilworth F. J., Di Padova M., Zhao P., Ogryzko V., Fuller-Pace F. V., Hoffman E. P., Tapscott S. J., and Sartorelli V. (2006) The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell 11, 547–560 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 41. Yusufzai T. M., Tagami H., Nakatani Y., and Felsenfeld G. (2004) CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13, 291–298 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 42. Fuller-Pace F. V. (2006) DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 34, 4206–4215 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fuller-Pace F. V. (2013) The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim. Biophys. Acta 1829, 756–763 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 44. Rossow K. L., and Janknecht R. (2003) Synergism between p68 RNA helicase and the transcriptional coactivators CBP and p300. Oncogene 22, 151–156 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 45. Imbalzano A. N., Imbalzano K. M., and Nickerson J. A. (2013) BRG1, a SWI/SNF chromatin remodeling enzyme ATPase, is required for maintenance of nuclear shape and integrity. Commun. Integr. Biol. 6, e25153 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ross-Innes C. S., Brown G. D., and Carroll J. S. (2011) A co-ordinated interaction between CTCF and ER in breast cancer cells. BMC Genomics 12, 593 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tolstorukov M. Y., Sansam C. G., Lu P., Koellhoffer E. C., Helming K. C., Alver B. H., Tillman E. J., Evans J. A., Wilson B. G., Park P. J., and Roberts C. W. (2013) SWI/SNF chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc. Natl. Acad. Sci. U.S.A. 110, 10165–10170 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klenova E. M., Nicolas R. H., Paterson H. F., Carne A. F., Heath C. M., Goodwin G. H., Neiman P. E., and Lobanenkov V. V. (1993) CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 13, 7612–7624 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vostrov A. A., and Quitschke W. W. (1997) The zinc finger protein CTCF binds to the APBβ domain of the amyloid β-protein precursor promoter: evidence for a role in transcriptional activation. J. Biol. Chem. 272, 33353–33359 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 50. Peña-Hernández R., Marques M., Hilmi K., Zhao T., Saad A., Alaoui-Jamali M. A., del Rincon S. V., Ashworth T., Roy A. L., Emerson B. M., and Witcher M. (2015) Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-I. Proc. Natl. Acad. Sci. U.S.A. 112, E677–E686 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alami J., Williams B. R., and Yeger H. (2003) Derivation and characterization of a Wilms' tumour cell line, WiT 49. Int. J. Cancer 107, 365–374 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 52. Xia J., Benner M. J., and Hancock R. E. (2014) NetworkAnalyst: integrative approaches for protein–protein interaction network analysis and visual exploration. Nucleic Acids Res. 42, W167–W174 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Breuer K., Foroushani A. K., Laird M. R., Chen C., Sribnaia A., Lo R., Winsor G. L., Hancock R. E., Brinkman F. S., and Lynn D. J. (2013) InnateDB: systems biology of innate immunity and beyond: recent updates and continuing curation. Nucleic Acids Res. 41, D1228–D1233 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Morris J. H., Apeltsin L., Newman A. M., Baumbach J., Wittkop T., Su G., Bader G. D., and Ferrin T. E. (2011) clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 12, 436 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W. H., Pagès F., Trajanoski Z., and Galon J. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cuddapah S., Jothi R., Schones D. E., Roh T. Y., Cui K., and Zhao K. (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhu L. J. (2013) Integrative analysis of ChIP-chip and ChIP-seq dataset. Methods Mol. Biol. 1067, 105–124 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 59. Zhu L. J., Gazin C., Lawson N. D., Pagès H., Lin S. M., Lapointe D. S., and Green M. R. (2010) ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu G., Wang L. G., and He Q. Y. (2015) ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 61. Gentile M. T., Vecchione C., Marino G., Aretini A., Di Pardo A., Antenucci G., Maffei A., Cifelli G., Iorio L., Landolfi A., Frati G., and Lembo G. (2008) Resistin impairs insulin-evoked vasodilation. Diabetes 57, 577–583 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 62. Gentile M. T., Nawa Y., Lunardi G., Florio T., Matsui H., and Colucci-D'Amato L. (2012) Tryptophan hydroxylase 2 (TPH2) in a neuronal cell line: modulation by cell differentiation and NRSF/rest activity. J. Neurochem. 123, 963–970 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 63. Mellacheruvu D., Wright Z., Couzens A., Lambert J., St-Denis N., Li T., Miteva Y., Hauri S., Sardiu M., Low T., Halim V., Bagshaw R., Hubner N., Hakim A., Bouchard A., et al. (2013) The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 8, 730–736 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Durinck B4S., Spellman P. T., Birney E., and Huber W. (2009) Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., and Huber W. (2005) BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.