Abstract
Celiac disease (CeD) provides an opportunity to study the specificity underlying human T-cell responses to an array of similar epitopes presented by the same human leukocyte antigen II (HLA-II) molecule. Here, we investigated T-cell responses to the two immunodominant and highly homologous HLA-DQ2.5–restricted gluten epitopes, DQ2.5-glia-α1a (PFPQPELPY) and DQ2.5-glia-ω1 (PFPQPEQPF). Using HLA-DQ2.5–DQ2.5-glia-α1a and HLA-DQ2.5–DQ2.5-glia-ω1 tetramers and single-cell αβ T-cell receptor (TCR) sequencing, we observed that despite similarity in biased variable-gene usage in the TCR repertoire responding to these nearly identical peptide–HLA-II complexes, most of the T cells are specific for either of the two epitopes. To understand the molecular basis of this exquisite fine specificity, we undertook Ala substitution assays revealing that the p7 residue (Leu/Gln) is critical for specific epitope recognition by both DQ2.5-glia-α1a– and DQ2.5-glia-ω1–reactive T-cell clones. We determined high-resolution binary crystal structures of HLA-DQ2.5 bound to DQ2.5-glia-α1a (2.0 Å) and DQ2.5-glia-ω1 (2.6 Å). These structures disclosed that differences around the p7 residue subtly alter the neighboring substructure and electrostatic properties of the HLA-DQ2.5–peptide complex, providing the fine specificity underlying the responses against these two highly homologous gluten epitopes. This study underscores the ability of TCRs to recognize subtle differences in the peptide–HLA-II landscape in a human disease setting.
Keywords: T-cell, T-cell receptor (TCR), major histocompatibility complex (MHC), crystal structure, surface plasmon resonance (SPR), human, flow cytometry, Celiac Disease, Gluten, gluten intolerance, Immune response, immunodominant epitope
Introduction
The αβ T-cell antigen receptor (TCR)5 recognizes peptide when presented by a given major histocompatibility complex (MHC) molecule (1). In celiac disease (CeD), an autoimmune-like and HLA-associated disorder, disease-driving T cells recognize deamidated cereal gluten peptides when presented by HLA-DQ2.5, HLA-DQ8, or HLA-DQ2.2 (2). Over 90% of the CeD patients express HLA-DQ2.5, whereas the remainder express HLA-DQ8 or HLA-DQ2.2. The gluten proteome is extremely complex, comprising hundreds of proteins (termed α-, γ-, and ω-gliadins and high- and low-molecular weight glutenins) (3). This system is remarkable in that there is natural immune exposure to a vast array of similar peptide sequences, and strikingly, the subjects who develop CeD make T-cell responses to a limited number of epitopes, many of which share substantial homology (4). In HLA-DQ2.5 associated CeD, most patients respond to DQ2.5-glia-α1a, DQ2.5-glia-α2, DQ2.5-glia-ω1, and DQ2.5-glia-ω2 epitopes (5), whereas most patients expressing HLA-DQ8 respond to DQ8-glia-α1 (6). Hence, these epitopes are termed immunodominant epitopes. T cells reactive with immunodominant epitopes are persistent for decades in the patients (7). Such T cells, expressing gut homing markers, can be isolated from gut biopsies and from peripheral blood, albeit at much lower frequency in blood (7).
Studies of TCRs recognizing immunodominant HLA-DQ2.5 and HLA-DQ8 restricted gluten epitopes have demonstrated biased usage of variable (V)-gene segments (6, 8–13). DQ2.5-glia-α2–specific TCRs exhibit repeated gene usage of TRAV26-1/TRBV7-2 with a conserved CDR3β motif harboring a non-germ line–encoded Arg residue (8, 9, 12). The TCRs specific for DQ2.5-glia-ω2 also display biased usage of TRAV4 and TRBV4 gene segments (12). DQ2.5-glia-α1a–specific TCRs demonstrate biased expression of TRAV4 and TRBV20-1 or TRBV29-1 genes (10). Similarly, the majority of DQ8-glia-α1–specific T cells express TRBV9 preferentially paired with TRBV26-2, expressing a non-germ line–encoded Arg residue in the CDR3β loop (6, 11, 13). The TRBV9-negative T cells specific to DQ8-glia-α1 also show biased usage of TRAV8-3/TRBV6-1+ TCRs, also with a conserved non-germ line–encoded Arg residue in the CDR3β loop (11).
Recently, structural studies have provided molecular insight into TCR recognition of gluten epitopes when presented by HLA-DQ8 or HLA-DQ2.5 (6, 10, 11, 13). Three crystal structures of TRAV26-1/TRBV7-2-TCR–HLA-DQ2.5–DQ2.5-glia-α2 complexes revealed that the TRBV7-2 bias is mostly attributed to the interactions mediated by the CDR3β loop and that the conserved non-germ line–encoded Arg in this loop serves as a lynchpin in the peptide–HLA-II interaction (10). A crystal structure of TRAV4/TRBV20-1 TCR–HLA-DQ2.5–DQ2.5-glia-α1a complex uncovered that the TRAV4 bias is a consequence of interactions between the germ line–encoded TCR residues and HLA-DQ2.5 (10). Crystal structures of TRAV26-2/TRBV9-TCR–HLA-DQ8–DQ8-α1-gliadin and TRAV20/TRBV9-TCR–HLA-DQ8–DQ8-α1-gliadin ternary complexes revealed that germ line–encoded residues interacting with the HLA-DQ8 molecule and gliadin peptide residues formed the basis for the TRBV9 bias (6, 11, 13). These two crystal structures together with a crystal structures of a complex of TRAV8-3/TRBV6-1-TCR–HLA-DQ8–DQ8-α1-gliadin (11) revealed that diverse TCR gene usage by DQ8-restricted gluten-specific T cells converges into a consensus binding that is mediated by germ-line- or non-germ line–encoded Arg residue. However, it remains unclear whether highly homologous gluten epitopes would generate cross-reactive or highly specific T-cell responses.
Here, we have studied the T-cell response toward the epitopes DQ2.5-glia-α1a (PFPQPELPY) and DQ2.5-glia-ω1 (PFPQPEQPF). These immunodominant gluten epitopes in CeD are highly homologous, with only two amino acid differences in the 9-mer core region. Despite the epitope similarity, we found that the majority of patient-derived T cells, both isolated from blood or the celiac gut lesion, are specific for either of the two epitopes with limited cross-reactivity. This suggests that there are important differences in how T cells recognize these peptide–HLA-II complexes, because the immune systems of the patients are exposed simultaneously to both epitopes when they consume gluten-containing food. To explore the basis for specific recognition of the homologous epitopes, we performed T-cell proliferation assays with amino acid–substituted epitopes, undertook single-cell TCRαβ-gene sequencing, and solved the crystal structures of HLA-DQ2.5 complexed with either DQ2.5-glia-α1a or DQ2.5-glia-ω1. Despite biased usage of common V-genes, the majority of the T cells were specific to only one epitope. The p7 residue in both epitopes was critical for the specific TCR recognition by the discriminatory T-cell clones (TCCs). The crystal structures of the two peptide–HLA-DQ2.5 complexes, although similar, exhibited local structural perturbations around the p7 residue. Hence, this study demonstrates the ability of TCR to distinguish subtle differences in peptide–HLA-II topology. Consequently, in human T-cell–mediated diseases like CeD, discrete alterations in the peptide–HLA-II landscape can profoundly shape the disease-driven immune response.
Results
Majority of T cells in blood and gut of CeD patients specific for DQ2.5-glia-α1a or DQ2.5-glia-ω1 show exquisite fine specificity
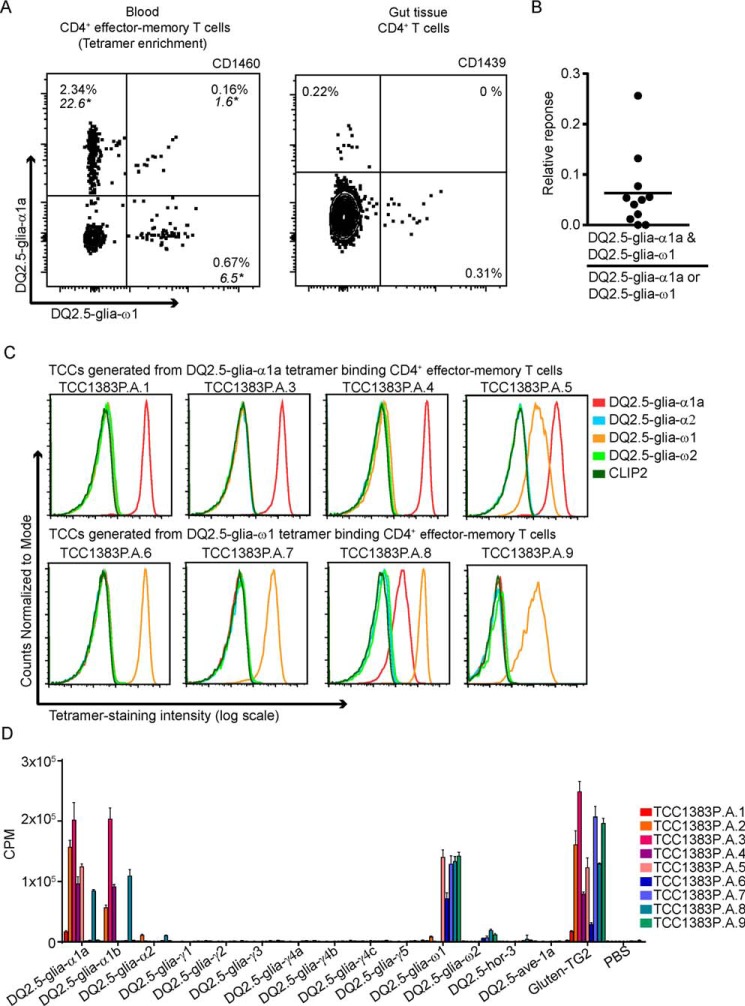
We identified distinct populations of CD4+ T cells that bound the HLA-DQ2.5–DQ2.5-glia-α1a or HLA-DQ2.5–DQ2.5-glia-ω1 tetramers in blood and gut of 12 CeD patients. (Fig. 1A). The relative ratio of CD4+ T cells positive to both DQ2.5–DQ2.5-glia-α1a and DQ2.5–DQ2.5-glia-ω1 tetramers with CD4+ T cells that are positive to either of the two tetramers in samples obtained from 11 CeD patients was 0.06 (mean). This suggests that the majority (∼95%) of these CD4+ T cells recognize either of the two tetramers (Fig. 1B and Table S1), indicating that despite only two amino acids difference in the 9-mer core region of the two gliadin peptides, most of the in vivo primed T cells in the blood and gut of CeD patients discriminate between the two epitopes.
Figure 1.
DQ2.5-glia-α1a- and DQ2. 5-glia-ω1–specific CD4+ T cells in peripheral blood and gut of CeD patients. A, representative plot showing tetramer staining of CD4+ T cells with HLA–DQ gluten tetramers DQ2.5–DQ2.5-glia-α1a and DQ2.5–DQ2.5-glia-ω1 in blood and gut of CeD patients. The PBMCs were stained with tetramers followed by bead enrichment of the tetramer-positive cells. The flow plot shows tetramer-binding CD4+ effector-memory T-cells. *, frequency of tetramer-positive CD4+ T-cells per million total CD4+ T-cells. The gut biopsies were stained with HLA–DQ gluten tetramers; however, bead enrichment was not performed. The plot in the figure shows tetramer-binding CD4+ T cells. B, relative ratio of CD4+ T cells positive to both DQ2.5–DQ2.5-glia-α1a and DQ2.5–DQ2.5-glia-ω1 tetramers with CD4+ T cells that are positive to either of the two tetramers in samples obtained from 11 CeD patients. The horizontal line represents the mean value. C, staining of TCCs generated from HLA-DQ2.5–DQ2.5-glia-α1a or HLA-DQ2.5–DQ2.5-glia-ω1 tetramer binding CD4+ T cells isolated from peripheral blood with HLA-DQ2.5–gluten tetramers (DQ2.5-glia-α1a, DQ2.5-glia-α2, DQ2.5-glia-ω1, DQ2.5-glia-ω2) and CLIP2 tetramer. TCC1383P.A.2 is not included in the figure because of a lack of TCC to carry out this staining (all tetramers attached with PE). However, TCC1383P.A.2 was tested in another staining experiment (tetramers attached with APC or PE) where it stained distinctly only with DQ2.5-glia-α1a (data not shown). D, T-cell proliferation of the TCCs generated from HLA-DQ2.5–DQ2.5-glia-α1a or HLA-DQ2.5–DQ2.5-glia-ω1 tetramer binding CD4+ T cells with panel peptides representing the known HLA-DQ2.5 restricted epitopes: DQ2.5-glia-α1a (QLQPFPQPELPY, underlined 9-mer core amino acid sequence), DQ2.5-glia-α1b (PQPELPYPQPELPY), DQ2.5-glia-α2 (PQPELPYPQPQL), DQ2.5-glia-γ1 (PEQPQQSFPEQERP), DQ2.5-glia-γ2 (GIIQPEQPAQL), DQ2.5-glia-γ3 (FPQQPEQPYPQQP), DQ2.5-glia-γ4a (FSQPEQEFPQPQ), Q2.5-glia-γ4b (FPQPEQEFPQPQ), DQ2.5-glia-γ4c (TEQPEQPFPQP), DQ2.5-glia-γ5 (PEQPFPEQPEQ), DQ2.5-glia-ω1 (PQQPFPQPEQPFP), DQ2.5-glia-ω2 (FPQPEQPFPWQP), DQ2.5-hor3 (PIPEQPQPYPQ), and DQ2.5-ave1a (YQPYPEQEEPFV). Two independent experiments were performed with measurements performed either in duplicate or triplicate. The data show a representative plot in which error bars represent means ± S.D. for the triplicates.
The HLA-DQ–gluten tetramers (including HLA-DQ2.5–DQ2.5-glia-α1a and HLA-DQ2.5–DQ2.5-glia-ω1) give staining of effector memory CD4+ T cells of CeD patients but not of healthy subjects (14, 15). We validated the specific staining of HLA-DQ2.5–DQ2.5-glia-α1a versus HLA-DQ2.5–DQ2.5-glia-ω1 tetramers with TCCs that were generated from tetramer-sorted cells by antigen-free expansion and cloning by limited dilution. On retesting of antigen specificity, all (nine) TCCs generated from the PBMCs of patient CD1383, stained with the tetramer used for sorting and showed a proliferative response toward the respective peptide (Fig. 1, C and D). One of the five TCCs generated from HLA-DQ2.5–DQ2.5-glia-α1a tetramer binding CD4+ T cells and one of four of the TCCs generated from HLA-DQ2.5–DQ2.5-glia-ω1 tetramer binding CD4+ T cells stained with both the tetramers and gave a response to both peptides in T-cell proliferation assays. The cross-reactive T-cell clones displayed a higher fluorescence intensity to the tetramer originally used for their isolation. None of the nine TCCs stained with the other HLA-DQ2.5–gluten tetramers (HLA-DQ2.5–DQ2.5-glia-α2, HLA-DQ2.5–DQ2.5-glia-ω2, and CLIP2), indicating the antigen specificity of the tetramer staining (Fig. 1C). In addition, none of these TCCs showed response to any other gluten epitopes when tested in a T-cell proliferation assay against a panel of known gluten peptides at a high concentration of 10 μm (Fig. 1D). Taken together, the results of the HLA-DQ–gluten tetramer staining of T cells in blood and gut, as well as the proliferation assay and HLA-DQ–gluten tetramer staining of established T-cell clones (the low number here prevents an exact estimate of cross-reactivity), strongly indicate that T cells of CeD patients generally discriminate between the two epitopes.
The p7 residue of the DQ2.5-glia-α1a and DQ2.5-glia-ω1 epitopes is critical for T-cell recognition
To explore the differences in recognition of these two highly homologous epitopes, we measured the effect of single Ala substitutions at each position in the two epitopes (Table 1) in a T-cell proliferation assay. The sequences of the peptides used in these T-cell proliferation assays were identical to the epitope sequences that were part of the HLA-DQ–gluten tetramers. In this analysis TCCs that stained with the HLA-DQ2.5–DQ2.5-glia-α1a or HLA-DQ2.5–DQ2.5-glia-ω1 tetramers and that had reactivity with only one of the two epitopes, were tested (Table 2). Reactivity pattern of the DQ2.5-glia-α1a–specific and DQ2.5-glia-ω1–specific TCCs to the single Ala-substituted DQ2.5-glia-α1a and DQ2.5-glia-ω1 peptides were normalized to the response to the WT peptide.
Table 1.
Peptide-binding register of DQ2.5-glia-α1a and DQ2.5-glia-ω1
The 9-mer core amino acids are in bold type, and the identical residues are shown by dashes.
| Peptide-binding register | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Epitopes | −3 | −2 | −1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| DQ2.5-glia-α1a | Gln | Leu | Gln | Pro | Phe | Pro | Gln | Pro | Glu | Leu | Pro | Tyr | |
| DQ2.5-glia-ω1 | Pro | Gln | — | — | — | — | — | — | — | Gln | — | Phe | Pro |
Table 2.
TCR sequences of DQ2.5-glia-α1a– and DQ2.5-glia-ω1–specific TCCs used in the alanine-substitution assay
| TCC | TRAV | CDR3α | TRAJ | TRBV | CDR3β | TRBJ |
|---|---|---|---|---|---|---|
| DQ2.5-glia-α1a–specific TCCs | ||||||
| 442D.A.45 | 29/DV5*01 | CAASEGDSGTYKYIF | 40*01 | 19*01 | CASSINALVGEQFF | 2-1*01 |
| 1383P.A.3 | 12–2*01 | CAVKFASGTYKYIF | 40*01 | 7-3*01 | CASSPAFSTDTQYF | 2-3*01 |
| 1383P.A.4 | 14/DV4*03 | CAMREGWQAGNTLIF | 15*01 | 5-4*01 | CASSLDGLTNTEAFF | 1-1*01 |
| DQ2.5-glia-ω1–specific TCCs | ||||||
| 442D.A.2 | 8–3*02 | CAVVDASSKLIF | 12*01 | 20-1*01 | CSATLGGDYGYTF | 1-2*01 |
| 442P.C.3/9/23 | 4*01 | CLVGGYNNNDMRF | 43*01 | 20-1*02 | CSAQLAGGGGDTQYF | 2-3*01 |
| 1383P.A.6 | 5*01 | CAESRYSGYSTLTF | 11*01 | 20-1*01 | CSAFPGGDTEAFF | 1-1*01 |
| 1383P.A.7/9 | 19*01 | CALSEYGNKLVF | 47*02 | 7-3*01 | CASSYVGGDTDTQYF | 2-3*01 |
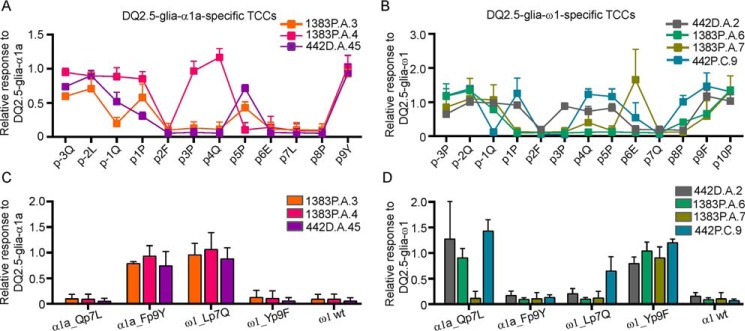
Both DQ2.5-glia-α1a– (Fig. 2A) and DQ2.5-glia-ω1–specific (Fig. 2B) TCCs lost reactivity on Ala substitution at p2 and p7. However, substitution at p9, a position that differs between the two epitopes, had no effect on the reactivity of the TCCs. This is in line with the observations from a previous study where p7 was critical, p9 was dispensable, and the p2 had variable effects on T-cell recognition of the DQ2.5-glia-α1a epitopes by the DQ2.5-glia-α1a–specific TCCs (10). Unique to all DQ2.5-glia-α1a–specific TCCs was the loss of reactivity on Ala substitution of residues p6 and p8. Two of the three TCCs lost reactivity on substitution at p3 and p4, whereas the third TCC was sensitive to substitution at p5. DQ2.5-glia-ω1–specific TCCs exhibited varying responses to Ala substitutions at positions other than p2 and p7. Substitution at p3 and p8 resulted in loss of reactivity in three of four TCCs. In brief, the p2 and p7 amino acids were important for T-cell recognition of both the DQ2.5-glia-α1a and DQ2.5-glia-ω1 epitopes.
Figure 2.
Effect of amino acid substitutions in the DQ2.5-glia-α1a and DQ2.5-glia-ω1 epitopes on T-cell recognition. A and B, reactivity patterns of DQ2.5-glia-α1a–specific- (A) and DQ2.5-glia-ω1–specific (B) TCCs to the single Ala-substituted DQ2.5-glia-α1a and DQ2.5-glia-ω1 peptides normalized to the WT response (1) are shown. C and D, the reactivity of DQ2.5-glia-α1a– (C) and DQ2.5-glia-ω1–specific (D) TCCs against chimeras. α1a_Qp7L, α1a_Fp9Y, ω1_Lp7Q, and ω1_Yp9F peptides normalized to the WT response (1) are shown. The data shown are averages of three independent experiments each with measurements performed in triplicate. The error bars represent means ± S.D. for three experiments.
To further investigate the basis of TCR specificity, T-cell proliferative assays were used to analyze the reactivity of DQ2.5-glia-α1a– and DQ2.5-glia-ω1–specific (Fig. 2, C and D) TCCs against chimeric peptide with a single amino acid exchange at position p7 and p9 of DQ2.5-glia-α1a (α1_Qp7L, α1_Fp9Y) and DQ2.5-glia-ω1 (ω1_Lp7Q, ω1_Yp9F) (Table 3). Because the 9-mer core sequences of the DQ2.5-glia-α1a and DQ2.5-glia-ω1 differ only at p7 and p9, the pair of chimeras α1_Qp7L and ω1_Yp9F, as well as the pair of α1_Fp9Y and ω1_Lp7Q, will have identical 9-mer core sequence but different flanking sequences. We observed that the responses of DQ2.5-glia-α1a– or DQ2.5-glia-ω1–specific TCCs toward these two pairs of chimeras with identical 9-mer core sequence were similar, suggesting that the flanking region is not contributing to the differential reactivity. We observed that both DQ2.5-glia-α1a–specific- (Fig. 2C) and DQ2.5-glia-ω1–specific TCCs (Fig. 2D) recognized chimeric p9 peptides, suggesting that this residue is dispensable. The majority of specific TCCs lost their reactivity when the p7 residue was exchanged. However, these specific TCCs showed reactivity with chimeric peptides that had unchanged p7, i.e. DQ2.5-glia-α1a– and DQ2.5-glia-ω1–specific TCCs reacted with ω1_Lp7Q and α1_Qp7L, respectively. This observation highlights the importance of p7 in the specific T-cell recognition of DQ2.5-glia-α1a or DQ2.5-glia-ω1.
Table 3.
Peptide sequences
| Peptides | −3 | −2 | −1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α1a-wt | Gln | Leu | Gln | Pro | Phe | Pro | Gln | Pro | Glu | Leu | Pro | Tyr | |
| α1a_Qp7L | Gln | Leu | Gln | — | — | — | — | — | — | Gln | — | Tyr | |
| α1a_Fp9Y | Gln | Leu | Gln | — | — | — | — | — | — | Leu | — | Phe | |
| ω1-wt | Pro | Gln | Gln | — | — | — | — | — | — | Gln | — | Phe | Pro |
| ω1_Lp7Q | Pro | Gln | Gln | — | — | — | — | — | — | Leu | — | Phe | Pro |
| ω1_Yp9F | Pro | Gln | Gln | — | — | — | — | — | — | Gln | — | Tyr | Pro |
High similarity in V-gene usage in TCRs specific for DQ2.5-glia-α1a and DQ2.5-glia-ω1
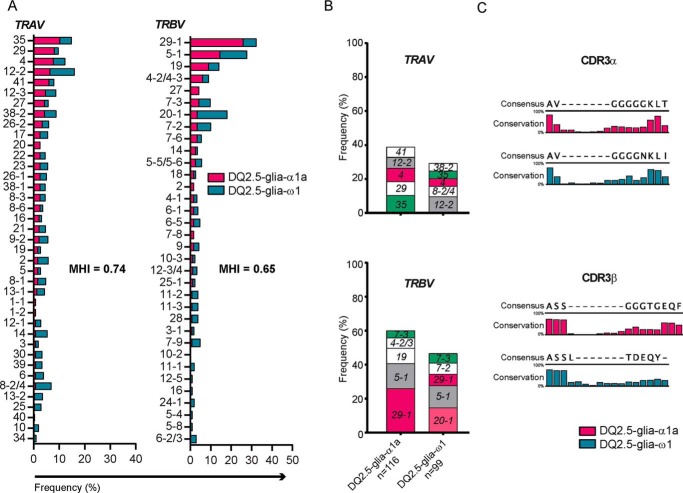
To examine the CeD patient T-cell repertoire reactive to HLA-DQ2.5–DQ2.5-glia-α1a and HLA-DQ2.5–DQ2.5-glia-ω1, we isolated single CD4+ T cells directly from blood or gut biopsies of CeD patients using HLA-DQ2.5 tetramers and determined the sequences of rearranged TCRα and TCRβ gene pairs (Table 4). We looked at the overall usage of TRAV or TRBV gene segments. In cases of dual V-gene usage, each V-gene was given a half score. For DQ2.5-glia-α1a, 165 single T cells isolated from 11 CeD patients produced 116 unique clonotypes expressing 37 TRAV and 26 TRBV genes (Fig. 3A and Fig. S1). Similar analysis of 113 DQ2.5-glia-ω1–specific T cells isolated from 11 CeD patients produced 99 unique clonotypes expressing 35 TRAV and 30 TRBV genes (Fig. 3A and Fig. S1). We then calculated the pair-wise Morisita–Horn index to analyze the similarity in TRAV and TRBV usage, where a higher index value indicates higher similarity. We found high similarity in both TRAV and TRBV usage (0.74 and 0.65, respectively) between DQ2.5-glia-α1a– and DQ2.5-glia-ω1–specific TCRs (Fig. 3A and Fig. S2A). The similarity between DQ2.5-glia-α1a– and DQ2.5-glia-ω1–specific TCRs was highest when compared with the previously published (12) V-gene usage in T cells specific for DQ2.5-glia-α2 and DQ2.5-glia-ω2. (Fig. S2A).
Table 4.
Summary of TCR sequences analyzed in the study
TCD, treated celiac disease; UCD, untreated celiac disease; TCL, T-cell lines generated from gut biopsy.
| Patient | Sample source | DQ2.5-glia-α1a |
DQ2.5-glia-ω1 |
DQ2.5-glia-α2 |
DQ2.5-glia-ω2 |
Total cells | Total clonotypes | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cells | Clonotypes | Cells | Clonotypes | Cells | Clonotypes | Cells | Clonotypes | ||||
| TCD1306 | Gut | 1 | 1 | 0 | 0 | — | — | — | — | 1 | 1 |
| UCD1391 | Gut | 8 | 6 | 1 | 1 | — | — | — | — | 9 | 7 |
| UCD1392 | Blood | 1 | 1 | 5 | 4 | — | — | — | — | 6 | 5 |
| UCD1396 | Gut | 3 | 3 | 0 | 0 | — | — | — | — | 3 | 3 |
| TCD1404 | Gut | 20 | 17 | 7 | 5 | — | — | — | — | 27 | 22 |
| TCD1411 | Gut | 0 | 0 | 14 | 14 | — | — | — | — | 14 | 14 |
| UCD1412 | Gut | 0 | 0 | 3 | 3 | — | — | — | — | 3 | 3 |
| UCD1414 | Gut | 11 | 8 | 1 | 1 | — | — | — | — | 12 | 9 |
| UCD1416 | Gut | 15 | 13 | 10 | 8 | — | — | — | — | 25 | 21 |
| UCD1421 | Gut | 26 | 17 | 7 | 7 | — | — | — | — | 33 | 24 |
| UCD1435 | Gut | 27 | 15 | 21 | 18 | — | — | — | — | 48 | 33 |
| UCD1436 | Gut | 36 | 22 | 30 | 25 | — | — | — | — | 96 | 47 |
| UCD1439 | Gut | 17 | 13 | 14 | 13 | — | — | — | — | 31 | 26 |
| Total | 165 | 116 | 113 | 99 | — | — | — | — | 308 | 215 | |
| TCD364 | TCL | 9 | 5 | 15 | 4 | 20 | 9 | 20 | 5 | 64 | 23 |
| TCD370 | TCL | 39 | 7 | 2 | 2 | 14 | 9 | 22 | 20 | 77 | 38 |
| TCD373 | TCL | 25 | 5 | 0 | 0 | 31 | 6 | — | — | 56 | 11 |
| UCD410 | TCL | 13 | 12 | 2 | 2 | 7 | 3 | 11 | 6 | 33 | 23 |
| UCD411 | TCL | — | — | — | — | 16 | 8 | 20 | 10 | 36 | 18 |
| UCD412 | TCL | 9 | 6 | 10 | 4 | 15 | 8 | 16 | 6 | 50 | 24 |
| TCD436 | TCL | 5 | 4 | 8 | 4 | 15 | 8 | 19 | 8 | 47 | 24 |
| UCD1174 | TCL | 14 | 6 | 16 | 4 | 7 | 4 | 7 | 7 | 44 | 21 |
| UCD1178 | TCL | 12 | 5 | 0 | 0 | 18 | 9 | 4 | 3 | 34 | 17 |
| Total | 126 | 50 | 53 | 20 | 143 | 64 | 119 | 65 | 441 | 199 | |
Figure 3.
DQ2.5-glia-α1a- and DQ2. 5-glia-ω1–specific TCRs. Unique T-cell clonotypes obtained from sequencing of TCRs (pairs of rearranged TCRα and TCRβ genes) of DQ2.5-glia-α1a– and DQ2.5-glia-ω1–specific T cells isolated directly from blood or gut biopsies of CeD patients using corresponding HLA–DQ tetramers were analyzed (A and B). A shows frequencies of TRAV and TRBV usage and Morisita–Horn index for pair-wise comparison of similarity in TRAV and TRBV usage. B shows frequency of the five most dominant TRAVs (upper panel) and TRBVs (lower panel) among the unique clonotypes observed in the respective TCR repertoire. The total number of clonotypes analyzed in each sample is denoted in the x axis. C, CDR3α and CDR3β amino acid sequences of the DQ2.5-glia-α1a and DQ2.5-glia-ω1–specific T cells isolated either directly from blood or gut biopsies or from in vitro cultured TCLs from gut biopsy were aligned and analyzed for the usage of amino acids at different positions.
We examined the five most expressed TRAVs and TRBVs in TCRs specific for DQ2.5-glia-α1a- and DQ2.5-glia-ω1 and observed similar genes being preferentially expressed (Fig. 3B). Of note, TRBV29-1 and TRBV20-1, which were expressed as the most dominant TRBV in each of the two repertoires, are phylogenetically closely related (10). The TRAV35, TRAV4, TRAV12-2, TRBV29-1, or TRBV20-1 and TRBV5-1 were the most frequently expressed TRAV and TRBV gene segments in both TCR repertoires. Further, for both DQ2.5-glia-α1a– and DQ2.5-glia-ω1–specific TCRs, we did not observe preferential pairing of TRAV and TRBV (Fig. S3).
We also performed similar analysis of unique clonotypes obtained by paired TCRα and TCRβ sequencing of single CD4+ T cells isolated from an in vitro cultured T-cell line (TCL) generated from gut biopsies of CeD patients using the HLA-DQ2.5–DQ2.5-glia-α1a (n = 50) or HLA-DQ2.5–DQ2.5-glia-ω1 tetramers (n = 20) (Table 4 and Figs. S1 and S2B). The TRAV and TRBV usage for both epitopes in data derived from T cells that were directly isolated from blood or gut biopsies was generally reproduced in the data obtained from TCLs.
We then aligned the CDR3α and CDR3β sequences of the TCRs specific for DQ2.5-glia-α1a and DQ2.5-glia-ω1 (Fig. 3C) to analyze the CDR3 amino acid usage and positioning. The T cells were isolated using HLA-DQ2.5–DQ2.5-glia-α1a or HLA-DQ2.5–DQ2.5-glia-ω1 tetramers, either directly from blood or gut biopsies or from in vitro cultured TCLs from gut biopsy. We did not observe any significant selection pattern in the amino acid usage and positioning in the CDR3 sequences.
We then examined the most frequently expressed V-genes in TCRs specific for the four immunodominant gluten epitopes (DQ2.5-glia-α1a, DQ2.5-glia-ω1, DQ2.5-glia-α2, and DQ2.5-glia-ω2). We analyzed unique clonotypes obtained by paired TCRαβ sequencing of single T cells isolated either from TCLs or directly from blood or gut biopsies of CeD patients using HLA-DQ2.5 tetramers (Fig. S2B). TRAV4 was among three most frequently used V-gene segments in the TCR repertoires specific for all the four epitopes. Because all of these gluten epitopes are restricted by HLA-DQ2.5, this feature of TRAV4 bias could be dependent on HLA interactions.
Crystal structures of the DQ2.5-glia-α1a and DQ2.5-glia-ω1 complexes
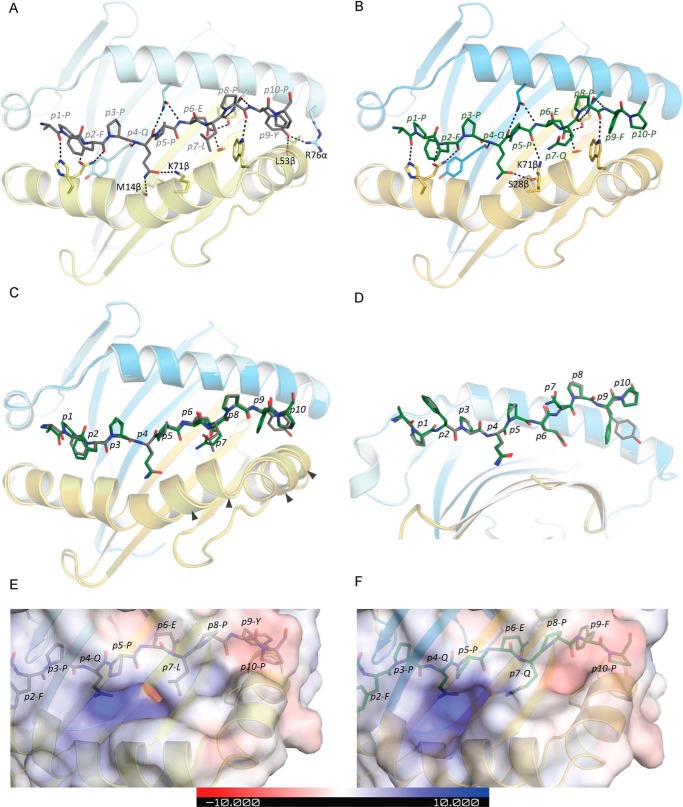
To gain an understanding of how two highly homologous epitopes give rise to two separate populations of T cells, we examined the corresponding peptide–HLA-II landscapes. Given the limited extent of cross-reactivity between the two gliadin determinants, it was unclear whether they would sit differently or within the same register in the HLA-DQ2.5 molecule. The observations made from the Ala substitution experiment performed on the TCCs is also based on the assumption that the substituted peptides bind in the identical binding registers. To establish this, we determined, to high resolution, the crystal structures of HLA-DQ2–DQ2.5-glia-α1a (2.0 Å) and HLA-DQ2–DQ2.5-glia-ω1 (2.6 Å) (Fig. 4, A and B, and Table 5) for data collection and refinement statistics). These complexes were solved with same linker peptide for both epitopes, and accordingly any structural variation between these two binary complexes could be attributed to differences in the two epitopes bound within the antigen-binding cleft. Although the crystal symmetry for the two crystals were different (P222 for DQ2.5-glia-α1 versus C121 for DQ2-glia-ω1), the crystal packing of the two structures was overall similar and, notably, with no crystal contacts involving the peptide positions p7 and p9. The HLA-DQ2.5–DQ2.5-glia-α1a structure aligned well with previously determined binary HLA-DQ2.5–DQ2.5-glia-α1a structure that was solved at lower resolution (16). The higher resolution structure (reported here) will be compared with the HLA-DQ2.5–DQ2.5-glia-ω1 structure. The electron densities corresponding to the two peptides were clear (Fig. S4).
Figure 4.
Structures of HLA-DQ2.5–HLA-DQ2.5-glia-α1a and HLA-DQ2.5–HLA-DQ2-glia-ω1 complexes. HLA-DQ2.5 is shown as cartoon, and peptides are shown as sticks, with HLA-DQ2.5 α-chain in light blue, β-chain in yellow, and DQ2.5-glia-α1a and DQ2.5-glia-ω1 peptides in grayish green, respectively. A, interactions between HLA-DQ2.5 and the DQ2.5-glia-α1a peptide. B, interactions between HLA-DQ2.5 and the DQ2.5-glia-ω1 peptide. C, overlay of HLA-DQ2.5–DQ2.5-glia-α1a and HLA-DQ2.5–DQ2.5-glia-ω1 (top view) showing differences in the HLA-DQ2.5 β-chain helix. The arrowheads show the regions where the helix is shifted by >0.5 Å. D, overlay of HLA-DQ2.5–DQ2.5-glia-α1a and HLA-DQ2.5–DQ2.5-glia-ω1 (side view) showing differences in side-chain conformation in p9. E and F, electrostatic surface potential of HLA-DQ2.5–DQ2.5-glia-α1a (E) and of HLA-DQ2.5–DQ2.5-glia-ω1 (F).
Table 5.
Data collection and refinement statistics
Statistics for the highest-resolution shell are shown in parentheses. TLS = Translation/Libration/Screw.
| DQ2-glia-ω1 | DQ2-glia-α1a | |
|---|---|---|
| Wavelength (Å) | 0.9537 | 0.9537 |
| Resolution range (Å) | 46.94–2.6 (2.74–2.6) | 70.49–2.00 (2.11–2.00) |
| Space group | C 2 2 21 | P 1 21 1 |
| Unit cell | ||
| a, b, c | 93.87, 96.61, 107.05 | 59.23, 101.97, 70.53 |
| α, β, γ | 90.00, 90.00, 90.00 | 90.00, 91.78, 90.00 |
| Total reflections | 191,813 (27,956) | 212,313 (30,339) |
| Unique reflections | 15,331 (2200) | 56,391 (8189) |
| Multiplicity | 12.5 (12.7) | 3.8 (3.7) |
| Completeness (%) | 100 (100) | 99.7 (99.8) |
| Mean I/σ(I) | 13.4 (2.3) | 13.5 (2.0) |
| Rmerge | 0.150 (1.252) | 0.073 (0.649) |
| Rmeas | 0.157 (1.305) | 0.085 (0.761) |
| CC½ | 0.998 (0.719) | 0.999 (0.751) |
| Rpim | 0.044 (0.366) | 0.044 (0.393) |
| Rwork | 0.2086 (0.3656) | 0.1867 (0.2821) |
| Rfree | 0.2503 (0.4207) | 0.222 (0.3378) |
| No. of non-hydrogen atoms | 3052 | 6518 |
| Macromolecules | 2999 | 6011 |
| Ligands | 28 | 14 |
| Protein residues | 372 | 745 |
| Root-mean-square deviation | ||
| Bonds | 0.003 | 0.002 |
| Angles | 0.575 | 0.568 |
| Ramachandran | ||
| Favored (%) | 97 | 97 |
| Allowed (%) | 3.3 | 2.6 |
| Outliers (%) | 0 | 0 |
| Rotamer outliers (%) | 3.9 | 0.89 |
| Clashscore | 6.05 | 3.20 |
| Average B-factor | 64.85 | 40.65 |
| Macromolecules | 64.85 | 40.51 |
| Ligands | 86.08 | 56.36 |
| Solvent | 41.15 | 41.87 |
| Number of TLS groups | 5 | 10 |
Comparison of the HLA-DQ2.5–DQ2.5-glia-α1a and the HLA-DQ2.5–DQ2.5-glia-ω1 structures revealed minor differences (root-mean-square deviation = 0.32 Å for Cα atoms of the peptide-binding domain), revealing that the two epitopes did not bind in a different register within the HLA-DQ2.5 molecule (Fig. 4, C and D). The p7 residue was an anchor residue in both complexes (solvent accessible surface areas of p7L and p7Q were 29.5 and 20.7 Å2, respectively, corresponding to 17 and 11% of the values for Gly–Gln/Leu–Gly). The interactions between the DQ2.5-glia-α1a and DQ2.5-glia-ω1 peptides and HLA-DQ2.5 were essentially conserved (Fig. 4, A and B). Nevertheless, differences were apparent in the local environment of the peptide residues p7 and p9 (p7-Leu and p9-Tyr in HLA-DQ2.5–DQ2.5-glia-α1a; p7-Gln and p9-Phe in HLA-DQ2.5–DQ2.5-glia-ω1). These differences coincided with an adjustment (up to 0.9 Å) in the positioning of the HLA-DQ2.5 β chain helix directly adjacent to p7 (Fig. 4C). The p7 difference between the peptides altered how each peptide was H-bonded to HLA-DQ2.5. Namely, in HLA-DQ2.5–DQ2.5-glia-α1a, Lys-71β formed a H-bond to the side chain of p4-Gln, whereas in HLA-DQ2.5–DQ2.5-glia-ω1, it H-bonded to the backbone of p5-Pro, thus freeing p4-Gln to form an additional H-bond to Ser-28β at the floor of the peptide-binding cleft (Fig. 4, A and B). These structural perturbations also led to electrostatic alterations around the P7 residue (Fig. 4, E and F). These differences are clearly sufficient to be discerned by T cells being specific for either of the two homologous epitopes.
To confirm that TCRs can discriminate between these two epitopes, we undertook surface plasmon resonance–based experiments using the HLA-DQ2.5-glia-α1a–responsive TCRs (S2 and L6) (10) run over HLA-DQ2.5-glia-α1a and HLA-DQ2.5-glia-ω1 (Fig. 5). We found that the S2 TCR specifically recognized HLA-DQ2.5-glia-α1a only and hence discriminated between HLA-DQ2.5-glia-α1 and HLA-DQ2.5-glia-ω1. The L6 TCR, however, bound HLA-DQ2.5-glia-α1a and HLA-DQ2.5-glia-ω1 with similar affinity. Taken together, the functional data and affinity measurements suggest that although a small population of TCRs is able to cross-react with the two highly homologous gliadin epitopes, the majority of TCRs lack cross-reactivity.
Figure 5.
Surface plasmon resonance analysis of TCR interactions with HLA-DQ2.5-glia-α1a and HLA-DQ2.5-glia-ω1. Sensorgram and binding curve fit of S2 (A and B) and L6 (C and D) affinity data into one-site specific binding model toward (A and C) HLA-DQ2.5-glia-α1a and (B and D) HLA-DQ2.5-glia-ω1. For KD determination, all surface plasmon resonance (SPR) data were derived from six (n = 6) and four (n = 4) independent experiments for HLA-DQ2.5-glia-α1a and HLA-DQ2.5-glia-ω1, respectively. Req, response at equilibrium; RU, response units. Error bars indicate S.D.
Discussion
The CD4+ T-cell response to gluten epitopes presented by the disease-predisposing HLA-DQ molecules is an essential part of the pathogenesis of CeD. Here we have studied two homologous and immunodominant gluten epitopes, DQ2.5-glia-α1a (PFPQPELPY) and DQ2.5-glia-ω1 (PFPQPEQPF). Despite only two amino acids difference in the 9-mer core region of the epitopes, the majority (∼ 95%) of T cells in blood and gut of CeD patients discriminate between the two epitopes with only a proportion of the T cells being cross-reactive. We found that the p7 residue was uniformly critical in both epitopes for discriminatory TCR recognition by all TCCs. Accordingly, we addressed the molecular basis underpinning the fine specificity of this response.
The p7 residues served as anchor residues in both HLA-DQ2.5–DQ2.5-glia-α1a and HLA-DQ2.5–DQ2.5-glia-ω1, and their side chains were only partially exposed in the binary crystal structures. Notwithstanding, it is striking that the p7 residue is critical for specific T-cell recognition of both DQ2.5-glia-α1a and DQ2.5-glia-ω1, as well as DQ2.5-glia-α2 and DQ2.5-glia-ω2. In ternary crystal structures of three unique complexes of TCR–HLA-DQ2.5–DQ2.5-glia-α2, the large side chain of p7-Tyr forms critical interactions with the TCR, explaining its vital role for T-cell recognition of this epitope. However, the ternary structure of the S2 TCR (TRAV4*01–TRBV20-1*01) specific for the DQ2.5-glia-α1a epitope in complex with HLA-DQ2.5 (10) revealed that this TCR does not make a direct contact with the less accessible p7-Leu residue. Despite this, the TCC from which the S2 TCR was derived, along with 10 other DQ2.5-glia-α1a–specific TCCs, uniformly lost reactivity toward DQ2.5-glia-α1a on p7 substitution in T-cell proliferation assays (10). The S2 TCR does discriminate between DQ2.5-glia-α1a and DQ2.5-glia-ω1, suggesting that even though the p7-Leu is not making direct contact to the TCR it is, in some way, influencing antigen recognition. To address this conundrum, we determined the crystal structures of HLA-DQ2.5 complexed with the DQ2.5-glia-α1a and DQ2.5-glia-ω1 epitopes, which revealed that the two epitopes were accommodated by HLA-DQ2.5 using the same register. Although the two peptide–HLA-DQ2.5 binary complexes were similar, differences around the p7 residue altered the neighboring substructure of the HLA-DQ2.5 molecule and associated electrostatic properties. Despite these relatively subtle differences in peptide–HLA-II topologies observed for the two immunodominant gluten epitopes, the S2 TCR must be able to detect them in a discriminatory manner. In the S2 TCR ternary complex, these p7 neighboring residues (specifically, HLA-DQ2.5 Asp-66β, Glu-69β, and Arg-70β) form an extensive interface with CDR1α, CDR3β, and α-framework residues of the S2 TCR, suggesting that the nature of this local environment is involved in epitope discrimination. Similar to the previous study, we found that for DQ2.5-glia-α1a–specific TCCs the p7-Leu was uniformly critical in specific TCR recognition. The DQ2.5-glia-α1a–specific TCCs analyzed in the two studies use many different TRAV/TRBV pairings, suggesting that this critical role of p7 is not contingent on a particular TCR gene usage. Interestingly, the p7-Gln of the DQ2.5-glia-ω1 epitope was also critical in specific TCR recognition for the DQ2.5-glia-ω1–specific TCCs that also express fairly diverse TCRs. These observations suggest that the different p7 amino acids in DQ2.5-glia-α1a and DQ2.5-glia-ω1 epitopes induce subtle differences that are sensed by the TCRs giving discriminatory recognition of the two peptide–HLA-II complexes. Similar features of impact on TCR recognition by a MHC buried anchor residues were observed for I-Ek and a hemoglobin epitope where substitution at the p6 residue affected T-cell recognition (17). Hence, these results indicate that anchor residues that are buried in the MHC II and make no direct contact with the TCR can indirectly influence the specific TCR recognition.
The single-cell sequencing of TCRs specific for DQ2.5-glia-α1a and DQ2.5-glia-ω1 epitopes demonstrate the same V-gene bias. Although V-gene bias is not a new phenomenon among TCRs specific for immunodominant gluten epitopes (6, 8–12), the high similarity in V-gene usage and biased expression of common V-genes between DQ2.5-glia-α1a and DQ2.5-glia-ω1 epitopes is noteworthy. However, the TCR bias is not as striking as in TCRs specific to DQ2.5-glia-α2 where we observe biased expression of TRBV7-2 paired with TRAV26-1 and a conserved Arg in CDR3β (8–10, 12) or in TCRs specific for DQ8-glia-α1 with biased expression of TRAV26-2/TRBV9 and a conserved Arg in CDR3α (6, 11). In our hands, the TCR bias toward DQ2.5-glia-α1a and DQ2.5-glia-ω1 is neither characterized by preferential TRAV/TRBV pairing nor conservation of CDR3 motifs that are common for both epitopes. This variability in TRAV/TRBV pairing and CDR3 region could possibly result in variable TCR docking patterns.
Examining the wealth of TCR gene sequence data now accumulated for the four immunodominant HLA-DQ2.5-restricted gluten epitopes (DQ2.5-glia-α1a, DQ2.5-glia-ω1, DQ2.5-glia-α2, and DQ2.5-glia-ω2) in CeD (12), we observe that TRAV4 is among the most frequently expressed TRAVs in T cells specific for all of the four epitopes. This TRAV4 bias does not appear to be associated with a common TRBV usage or conservation of CDR3 sequences. The crystal structure of DQ2.5-glia-α1a–specific TCR using TRAV4 revealed that the TRAV4 bias against DQ2.5-glia-α1a is an effect of interactions between germ line–encoded TCR residues, most prominently Tyr-38α, with residues of the β-chain of HLA-DQ2.5 (Arg-70β and Arg-77β) (10). Similarly, HLA-DQ2.5–DQ2.5-glia-α2–specific TCRs encoded by TRAV26-1, a phylogenetically close V-gene with high sequence relatedness with TRAV4, formed analogous sets of interactions in ternary crystal structures. Therefore, we suggest that the common TRAV4 bias in TCRs specific for immunodominant gluten epitopes restricted by HLA-DQ2.5 could be an outcome of conserved interaction between germ-line residues encoded by TRAV4 and HLA-DQ2.5.
CeD provides an opportunity to study the natural immune response in humans toward a natural antigenic system that comprises a vast array of similar peptide sequences. Here we show that despite minor differences in peptide–HLA-II topologies and high similarity in TCR V-gene usage, the majority of the T cells discriminate between the two homologous and immunodominant gluten epitopes. For CeD, this implicates that highly homologous peptides have the potential to engage separate T-cell populations, thereby resulting in broader and more robust T-cell responses to gluten. Of general implication, the study underscores the exquisite sensitivity of TCRs to detect subtle differences in peptide–HLA-II complexes.
Materials and methods
Patient material
We obtained ∼60 ml of citrated full blood and six gut biopsies taken as part of a gastroduodenoscopy, using an Olympus H-180 endoscope and a regular biopsy forceps (Olympus), from both untreated and treated CeD patients. PBMCs were isolated by Ficoll-based density gradient centrifugation from the blood samples and cryopreserved for later use. Gut biopsies were processed to obtain single-cell suspension prior to cryopreservation. In brief, freshly collected biopsies were washed twice with 2 mm EDTA in 2% fetal calf serum at 37 °C for 10 min (to remove epithelial layer) followed by collagenase digestion (1 mg/ml) at 37 °C for 30–60 min, homogenization by syringe, and filtration. We used previously established and cryopreserved TCLs that had been generated from single gut biopsy, of which some are published (18, 19). Two new TCLs (CD1174 and CD1178) were generated by incubating gut biopsy with native chymotrypsin-digested gluten and deamidated chymotrypsin-digested gluten (both at 20 μg/ml) for 3–5 days followed by addition of interleukin-2/interleukin-15.
Tetramer staining, cell enrichment, and FACS
Tetramers of recombinant HLA-DQ2.5 covalently linked with gluten-derived peptides containing the T-cell epitope DQ2.5-glia-α1a peptide (QLQPFPQPELPY, underlined 9-mer core amino acid sequence) and DQ2.5-glia-α2 peptide (PQPELPYPQPE) were produced and conjugated with phycoerythrin-labeled streptavidin (Invitrogen) or allophycocyanin-labeled streptavidin (ProZyme) as described (20). Likewise, the HLA-DQ2.5–DQ2.5-glia-ω1 (PQQPFPQPEQPFP) and HLA-DQ2.5–DQ2.5-glia-ω2 (FPQPEQPFPWQP) tetramers were generated with the same protocol. Tetramer staining was performed on cryopreserved PBMCs, on single-cell suspension prepared from gut biopsies and on TCLs. In brief, the cryopreserved PBMCs were thawed and stained with the fluorochrome-conjugated tetramers (10 μg/ml each) for 30–45 min at room temperature followed by bead enrichment of tetramer-binding cells before adding surface antibody mix. Cryopreserved single-cell suspensions of gut biopsies were thawed and stained with fluorochrome-conjugated tetramers (10 μg/ml) for 30–45 min at room temperature before adding surface antibody mix. Cryopreserved TCLs were thawed and stained directly with fluorochrome-conjugated tetramers (10 μg/ml) for 2 h at 37 °C before adding surface antibody mix. For PBMCs, cells within the singlet lymphocyte population were further gated to isolate tetramer-binding CD4+ effector-memory T cells that were CD3+, CD11c−, CD14−, CD15−, CD19−, CD56−, CD45RA−, CD62L−, and CD4+. For single-cell suspensions of gut biopsies, live cells within the singlet lymphocyte population were further gated to obtain tetramer-binding CD4+ T cells that were CD3+, CD11c−, CD14−, CD15−, CD19−, CD56−, CD8−, and CD4+. For T-cell lines, live cells within the singlet lymphocyte population were further gated to obtain tetramer-binding CD4+ T cells that were CD3+, CD8−, and CD4+. Finally, the populations of tetramer-binding CD4+ T cells were index sorted for single-cell TCR sequencing or for generation of TCCs by in vitro expansion. The sorting was performed with a FACS Aria II instrument (BD Biosciences) at the Flow Cytometry Core Facility of Oslo University Hospital, and the flow-cytometry data were analyzed with FlowJo software (FlowJo LLC). The antibodies used in the study were CD62L-PerCP/Cy5.5, CD14-Pacific Blue, CD15-Pacific Blue, CD19-Pacific Blue, and CD56-Pacific Blue; CD3-FITC, CD11c-Horizon V450, CD4-APC, or CD4-APC-H7 (BD Biosciences); and CD45RA-PE-Cy7, CD3-eVolve605, CD8-PE-Cy7, or CD8-PerCP (eBioscience). LIVE/DEAD marker fixable violet stain (Thermo Fisher Invitrogen) was used to exclude the dead cells.
Generation and TCR sequencing of TCCs
The TCCs were generated from tetramer-sorted CD4+ T cells and sequenced as described in previous publication (12). In brief, TCCs were generated using cloning by limited dilution and expanded without antigens followed by mRNA isolation, switching mechanism at the 5′-terminus of the RNA transcript based cDNA synthesis, PCR amplification, and finally Sanger sequencing. TCCs from patient CD442 and CD1340 used for Ala scans in the current study were generated in the course of another project (7).
T-cell proliferation assay
T-cell proliferation assays were carried out as described previously (21). In brief, 75,000 antigen-presenting cells (HLA-DQ2.5 homozygous Epstein–Barr virus–transformed cells) were irradiated at 75 grays and incubated with 10 μm peptides at 37 °C for 24 h before adding 50,000 T cells. 48 h later, the cultures were pulsed with 1 μCi of [3H]thymidine followed by harvesting and scintillation counting after 16–20 h. The TCCs were identified as peptide-specific if the stimulation index (ratio of cpm after antigen stimulation and cpm after medium stimulation) was higher than 3.
Single-cell TCR gene sequencing
We used a previously published protocol, based on nested PCR amplification using multiplex TRAV and TRBV primers, for single-cell TCR sequencing (22). The protocol was slightly modified by performing cDNA synthesis and the first PCR in two separate steps. In short, single cells were sorted directly in 96-well plates containing 5 μl of capture buffer (20 mm Tris-HCl, pH 8.0, 1% Nonidet P-40 and 1 unit/μl RNase Inhibitor (optional)). The cDNA mix (5 μl of 1× SSII RT buffer, 1 mm dNTP, 2.5 mm DDT, 1 μm oligo(dT), 1 μm reverse TRAC (5′-AGTCAGATTTGTTGCTCCAGGCC-3′), and TRBC (5′-TTCACCCACCAGCTCAGCTCC-3′) primers, 1.5 units/μl RNase inhibitor, 2.5 units/μl Superscript II in final 10 μl of reaction volume) was added, and cDNA synthesis was carried out at 42 °C for 50 min followed by an inactivation step at 72 °C for 10 min. The original protocol was followed to obtain a purified PCR product, which was then sequenced with 250-bp paired-end sequencing using Illumina MiSeq platform at the Norwegian Sequencing Centre (Oslo University Hospital). All the raw data generated in this study have been uploaded to the European Genome-phenome Archive (EGAS00001003245).
Processing of TCR gene sequences
Processing of the raw sequencing data obtained from Illumina NGS was carried out in a multistep pipeline. The pipeline was composed of quality filtering of low-quality reads (Q < 30), annotation of the header with barcodes (row, plate, and column) and gene (TRA/TRB) information, pairing, and assembly of the annotated forward and reverse reads. These assembled reads were then collapsed to give up to three highest ranking reads (based on dupcount) to remove duplicates. In the next step V-, D-, and J-genes and CDR3 junction sequences were identified using IMGT-HighV-Quest online tool (23). A processing workflow implemented as an in-house Java program together with a custom MySQL database was used to further extract the sequences. In brief, only productive sequences with a dupcount of >100 were collapsed based on identical V-gene, J-gene, and CDR3 (nucleotide level). These sequences were then refiltered, so that only T cells with at least one TCRα and TCRβ chain and dual TCRα or TCRβ chains (but not both dual TCRα and TCRβ, maximum three chains) were considered for downstream analysis. We then categorized T cells as a clonotype if they have identical V and J gene (subgroup level), together with identical CDR3 region allowing for one nucleotide mismatch. These data were then used for further analysis of clonal expansion and V-gene usage. The paired TCRα and TCRβ sequencing of single T cells isolated from TCLs of CeD patients CD364, CD373, CD412, and CD436 used in V-gene usage analysis were generated in the course of another project for tracking clonotypes (7).
CDR3 amino acid sequence alignment
CDR3 amino acid sequence alignment was performed using CLC sequence viewer v8.0 (CLC bio, Aarhus, Denmark) set to accurate with gap open penalty set to 20 and gap extension set to 4.
HLA-DQ2.5 production, crystallization and structure determination
HLA-DQ2.5–DQ2.5-glia-α1a-NC containing the epitope (QPFPQPELPYP) and HLA-DQ2.5–DQ2.5-glia-ω1 containing the epitope (QPFPQPEQPFP) were expressed and purified as described previously (10). Briefly, Fos/Jun zipper fusion constructs of the HLA-DQ2.5 α and β chains with each peptide linked to the N terminus of the HLA-DQ2.5 β chain were co-expressed in Hi5 insect cells and purified via immobilized metal affinity and Superdex S200 gel filtration chromatography. Prior to crystallization, the Fos/Jun zippers were removed by enterokinase cleavage followed by HiTrap-Q ion exchange chromatography. HLA-DQ2.5–DQ2.5-glia-α1a and HLA-DQ2.5–DQ2.5-glia-ω1 were concentrated to 8 mg/ml and crystallized via hanging-drop vapor diffusion at room temperature using mother liquor containing 23% PEG3350 and 0.1 m NaH2PO4 (pH 6). The crystals were cryoprotected in mother liquor supplemented with 20% glycerol and were flash-frozen in liquid N2. X-ray diffraction data were collected at the MX1 and MX2 Beamlines of the Australian Synchrotron and processed using the programs XDS (24) and Scala (25) from the CCP4 package (26). The structures of HLA-DQ2.5–DQ2.5-glia-α1a and HLA-DQ2.5–DQ2.5-glia-ω1 were solved via molecular replacement using Phaser (27), and contained two and one protomers in the asymmetric unit, respectively. Structure models were generated by iterative rounds of model building in Coot (28) and restrained refinement in Phenix. The coordinates and structure factors have been deposited in the Protein Data Bank under the following accession codes: HLA-DQ2.5–DQ2.5-glia-α1a, 6MFG; and HLA-DQ2.5–DQ2.5-glia-ω1, 6MFF.
Surface plasmon resonance
The surface plasmon resonance experiments were conducted using Biacore 3000 (GE Healthcare) with Biacore CAPture sensor chip. This analysis was performed to obtain equilibrium affinity constants of TCRs toward HLA-DQ2.5–DQ2.5-glia-α1a and HLA-DQ2.5–DQ2.5-glia-ω1 constructs. TCRs were produced as described previously (10). The biotinylated ligands HLA-DQ2.5–DQ2.5-glia-α1a, HLA-DQ2.5–DQ2.5-glia-ω1, and HLA-DQ2.5–DQ2.5-CLIP2 (ATPLLMQALPM, negative control for nonspecific binding) were immobilized at 1000–1500 response units as previously shown (6, 10). Flow cell surfaces were activated using 1:1 mixture of Biotin CAPture reagent (streptavidin and complementary oligonucleotide to the one on CAP surface) and HBS buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm EDTA, and 0.005% P20 detergent supplied by the manufacturer) at 25 °C using flow rate of 5 μl/min. Following injection of 1 mm biotin, decreasing concentrations of TCR in HBS buffer were passed over the flow cells for a 1.5-min time interval. The maximum concentration of TCR for dilution series was 200 μm. The affinities of each TCR for HLA-DQ2.5–DQ2.5-glia-α1a (LQPFPQPELPY), HLA-DQ2.5–DQ2.5-glia-α1a-NC (QPFPQPELPYP) HLA-DQ2.5–DQ2.5-glia-ω1 (QPFPQPEQPFP) constructs were tested by conducting four, two, and four independent experiments, respectively. The data for both HLA-DQ2.5-glia-α1a constructs were combined because there was no measurable difference. The equilibrium dissociation constant, KD, was determined by fitting the data into nonlinear one-site specific binding model using GraphPad Prism version 7.0.
Statistical analysis
The Morisita–Horn index for analyzing similarity in TRAV and TRBV usage across TCRs specific for the four immunodominant epitopes was generated in R (3.3.2) with the use of the Vegan (2.4-2) package. We calculated pairwise similarities using sim.table function in the Vegan package, setting 2 as the order of diversity measure (q) to generate the Morisita–Horn indices.
Study approval
The study was approved by Regional Committee for Medical and Health Research Ethics South-East Norway (project 2010/2720). The patients participating in the study gave informed, written consent.
Author contributions
S. D.-K., L. F. R., A. C., S.-W. Q., J. R., and L. M. S. conceptualization; S. D.-K., L. C., and J. P. data curation; S. D.-K. and R. S. N. software; S. D.-K., L. C., J. P., R. S. N., and H. H. R. formal analysis; S. D.-K., L. C., J. P., L. F. R., and A. C. investigation; S. D.-K., L. C., J. P., and R. S. N. methodology; S. D.-K., J. R., and L. M. S. writing-original draft; S. D.-K., L. C., J. P., L. F. R., R. S. N., A. C., K. E. A. L., H. H. R., S.-W. Q., J. R., and L. M. S. writing-review and editing; J. P., H. H. R., S.-W. Q., J. R., and L. M. S. supervision; R. S. N., K. E. A. L., J. R., and L. M. S. resources; H. H. R., J. R., and L. M. S. project administration; J. R. and L. M. S. funding acquisition.
Supplementary Material
Acknowledgments
We are grateful to the patients who participated in this study. We thank S. Furholm, M. H. Bakke, and C. Hinrichs for collecting biological material from patients. We express our gratitude to B. Simonsen and S. R. Lund for producing the biotinylated HLA-DQ–gluten molecules and M. K. Johannesen for technical assistance.
This work was supported by grants from Stiftelsen Kristian Gerhard Jebsen Project SKGJ-MED-017, by the Research Council of Norway Project 179573/V40 through the Centre of Excellence funding scheme and Project 233885, and by South-Eastern Norway Regional Health Authority Projects 2011050, 2013046, and 2015009. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S4.
The atomic coordinates and structure factors (codes 6MFF and 6MFG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
All the raw data generated in this study have been uploaded to the European Genome-phenome Archive (EGAS00001003245).
- TCR
- T-cell receptor
- MHC
- major histocompatibility complex
- CeD
- celiac disease
- HLA
- human leukocyte antigen
- V-gene
- variable gene
- D-gene
- diversity gene
- J-gene
- joining gene
- TCC
- T-cell clone
- PBMC
- peripheral blood mononuclear cell
- TCL
- T-cell line
- PE
- phycoerythrin
- APC
- allophycocyanin.
References
- 1. Rossjohn J., Gras S., Miles J. J., Turner S. J., Godfrey D. I., and McCluskey J. (2015) T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 [DOI] [PubMed] [Google Scholar]
- 2. Sollid L. M., and Jabri B. (2013) Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat. Rev. Immunol. 13, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bromilow S., Gethings L. A., Buckley M., Bromley M., Shewry P. R., Langridge J. I., and Clare Mills E. N. (2017) A curated gluten protein sequence database to support development of proteomics methods for determination of gluten in gluten-free foods. J. Proteomics 163, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sollid L. M., Qiao S. W., Anderson R. P., Gianfrani C., and Koning F. (2012) Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 64, 455–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tye-Din J. A., Stewart J. A., Dromey J. A., Beissbarth T., van Heel D. A., Tatham A., Henderson K., Mannering S. I., Gianfrani C., Jewell D. P., Hill A. V., McCluskey J., Rossjohn J., and Anderson R. P. (2010) Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci. Transl. Med. 2, 41ra51. [DOI] [PubMed] [Google Scholar]
- 6. Broughton S. E., Petersen J., Theodossis A., Scally S. W., Loh K. L., Thompson A., van B. J., Kooy-Winkelaar Y., Henderson K. N., Beddoe T., Tye-Din J. A., Mannering S. I., Purcell A. W., McCluskey J., Anderson R. P., Koning F., Reid H. H., and Rossjohn J. (2012) Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity. 37, 611–621 [DOI] [PubMed] [Google Scholar]
- 7. Risnes L. F., Christophersen A., Dahal-Koirala S., Neumann R. S., Sandve G. K., Sarna V. K., Lundin K. E., Qiao S. W., and Sollid L. M. (2018) Disease-driving CD4+ T cell clonotypes persist for decades in celiac disease. J. Clin. Invest. 128, 2642–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qiao S. W., Raki M., Gunnarsen K. S., Loset G. A., Lundin K. E., Sandlie I., and Sollid L. M. (2011) Posttranslational modification of gluten shapes TCR usage in celiac disease. J. Immunol. 187, 3064–3071 [DOI] [PubMed] [Google Scholar]
- 9. Qiao S. W., Christophersen A., Lundin K. E., and Sollid L. M. (2014) Biased usage and preferred pairing of α- and β-chains of TCRs specific for an immunodominant gluten epitope in coeliac disease. Int. Immunol. 26, 13–19 [DOI] [PubMed] [Google Scholar]
- 10. Petersen J., Montserrat V., Mujico J. R., Loh K. L., Beringer D. X., van L. M., Thompson A., Mearin M. L., Schweizer J., Kooy-Winkelaar Y., van B. J., Drijfhout J. W., Kan W. T., La Gruta N. L., Anderson R. P., Reid H. H., Koning F., and Rossjohn J. (2014) T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease. Nat. Struct. Mol. Biol. 21, 480–488 [DOI] [PubMed] [Google Scholar]
- 11. Petersen J., van Bergen J., Loh K. L., Kooy-Winkelaar Y., Beringer D. X., Thompson A., Bakker S. F., Mulder C. J., Ladell K., McLaren J. E., Price D. A., Rossjohn J., Reid H. H., and Koning F. (2015) Determinants of gliadin-specific T cell selection in celiac disease. J. Immunol. 194, 6112–6122 [DOI] [PubMed] [Google Scholar]
- 12. Dahal-Koirala S., Risnes L. F., Christophersen A., Sarna V. K., Lundin K. E., Sollid L. M., and Qiao S. W. (2016) TCR sequencing of single cells reactive to DQ2.5-glia-α2 and DQ2.5-glia-ω2 reveals clonal expansion and epitope-specific V-gene usage. Mucosal Immunol. 9, 587–596 [DOI] [PubMed] [Google Scholar]
- 13. Petersen J., Kooy-Winkelaar Y., Loh K. L., Tran M., van Bergen J., Koning F., Rossjohn J., and Reid H. H. (2016) Diverse T cell receptor gene usage in HLA-DQ8-associated celiac disease converges into a consensus binding solution. Structure 24, 1643–1657 [DOI] [PubMed] [Google Scholar]
- 14. Christophersen A., Risnes L. F., Bergseng E., Lundin K. E., Sollid L. M., and Qiao S. W. (2016) Healthy HLA-DQ2.5+ Subjects Lack Regulatory and Memory T Cells Specific for Immunodominant Gluten Epitopes of Celiac Disease. J. Immunol. 196, 2819–2826 [DOI] [PubMed] [Google Scholar]
- 15. Sarna V. K., Lundin K. E. A., Morkrid L., Qiao S. W., Sollid L. M., and Christophersen A. (2018) HLA-DQ-Gluten Tetramer Blood Test Accurately Identifies Patients With and Without Celiac Disease in Absence of Gluten Consumption. Gastroenterology 154, 886–896.e886 [DOI] [PubMed] [Google Scholar]
- 16. Kim C. Y., Quarsten H., Bergseng E., Khosla C., and Sollid L. M. (2004) Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl. Acad. Sci. U.S.A. 101, 4175–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kersh G. J., Miley M. J., Nelson C. A., Grakoui A., Horvath S., Donermeyer D. L., Kappler J., Allen P. M., and Fremont D. H. (2001) Structural and functional consequences of altering a peptide MHC anchor residue. J. Immunol. 166, 3345–3354 [DOI] [PubMed] [Google Scholar]
- 18. Molberg O., McAdam S. N., Korner R., Quarsten H., Kristiansen C., Madsen L., Fugger L., Scott H., Noren O., Roepstorff P., Lundin K. E., Sjostrom H., and Sollid L. M. (1998) Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 4, 713–717 [DOI] [PubMed] [Google Scholar]
- 19. Arentz-Hansen H., Korner R., Molberg O., Quarsten H., Vader W., Kooy Y. M., Lundin K. E., Koning F., Roepstorff P., Sollid L. M., and McAdam S. N. (2000) The intestinal T cell response to α-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J. Exp. Med. 191, 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quarsten H., McAdam S. N., Jensen T., Arentz-Hansen H., Molberg O., Lundin K. E., and Sollid L. M. (2001) Staining of celiac disease-relevant T cells by peptide-DQ2 multimers. J. Immunol. 167, 4861–4868 [DOI] [PubMed] [Google Scholar]
- 21. Christophersen A., Raki M., Bergseng E., Lundin K. E., Jahnsen J., Sollid L. M., and Qiao S. W. (2014) Tetramer-visualized gluten-specific CD4+ T cells in blood as a potential diagnostic marker for coeliac disease without oral gluten challenge. United European Gastroenterol J 2, 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han A., Glanville J., Hansmann L., and Davis M. M. (2014) Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lefranc M. P., Giudicelli V., Ginestoux C., Bodmer J., Muller W., Bontrop R., Lemaitre M., Malik A., Barbie V., and Chaume D. (1999) IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 27, 209–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 26. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J Appl Crystallogr 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.