Abstract
Inhibitors of bromodomain and extra-terminal proteins (BETi) suppress oncogenic gene expression and have been shown to be efficacious in many in vitro and murine models of cancer, including triple-negative breast cancer (TNBC), a highly aggressive disease. However, in most cancer models, responses to BETi can be highly variable. We previously reported that TNBC cells either undergo senescence or apoptosis in response to BETi, but the specific mechanisms dictating these two cell fates remain unknown. Using six human TNBC cell lines, we show that the terminal response of TNBC cells to BETi is dictated by the intrinsic expression levels of the anti-apoptotic protein B-cell lymphoma-extra large (BCL-xL). BCL-xL levels were higher in cell lines that senesce in response to BETi compared with lines that primarily die in response to these drugs. Moreover, BCL-xL expression was further reduced in cells that undergo BETi-mediated apoptosis. Forced BCL-xL overexpression in cells that normally undergo apoptosis following BETi treatment shifted them to senescence without affecting the reported mechanism of action of BETi in TNBC, that is, mitotic catastrophe. Most importantly, pharmacological or genetic inhibition of BCL-xL induced apoptosis in response to BETi, and inhibiting BCL-xL, even after BETi-induced senescence had already occurred, still induced cell death. These results indicate that BCL-xL provides a senescent cell death–inducing or senolytic target that may be exploited to improve therapeutic outcomes of TNBC in response to BETi. They also suggest that the basal levels of BCL-xL should be predictive of tumor responses to BETi in current clinical trials.
Keywords: apoptosis, breast cancer, bromodomain-containing protein 4 (BRD4), senescence, B-cell lymphoma 2 (Bcl-2) family, B-cell lymphoma-extra large, BCL-xL, BCL2L1, BET inhibitor, senolytic agent
Introduction
Breast cancers can be categorized based on their gene expression profiles into distinct groups, with one classification including luminal A, luminal B, HER2-overexpressing, claudin-low, and basal (1). The luminal A and B subtypes comprise about 70% of breast cancers and express estrogen and/or progesterone receptors. The HER2 subtype overexpresses the ERBB2 gene and encompasses about 10–15% of breast cancers (2). The basal and claudin-low subtypes are generally triple negative, meaning they lack HER2 overexpression as well as expression of estrogen and progesterone receptors (1). Triple-negative breast cancers (TNBCs)4 are highly proliferative, comprise 15–20% of breast cancers, and are thought to have a larger tumor-initiating, or cancer stem cell, population (2–9). TNBC patients have a poor prognosis because of rapid metastatic recurrence and a lack of effective targeted therapies (3, 5, 10). The gene expression profiles, or transcriptomes, defining the TNBC subtype are well-characterized and control the phenotypes of these tumors (11). It is expected that targeting these transcriptomes, including genes that promote cell cycle progression, may be a viable therapeutic approach to treat this disease.
The bromodomain and extra-terminal (BET) family of proteins is comprised of four family members: BRD2, BRD3, BRD4, and BRDT. BET proteins bind acetylated lysines on histone tails to activate gene transcription. At least one family member, BRD4, is enriched at enhancers in numerous oncogenes and sustains their expression (12). BET family inhibitors (BETi), such as the cell-permeable small molecule JQ1, target the epigenome by competitively binding to the bromodomain regions of BET proteins (3). This blocks the ability of these proteins to interact with acetylated histones and suppresses oncogene transcription. BETi impede tumor growth in multiple cancer models with the only reported adverse effects in mice being reversible male infertility and inhibition of long-term memory formation (13, 14).
BRD4 is amplified or overexpressed in breast cancers of numerous subtypes, including TNBC, supporting its potential importance in disease progression (2). Multiple studies have assessed the efficacy of BETi in TNBC and have found that BETi suppress breast cancer cell growth and invasion. Shi and colleagues found that BRD4 binds to diacetylated Twist to promote migration (15). Inhibition of the BRD4/Twist interaction reduced invasiveness, in vitro, and tumorigenic potential in an in vivo model (15). We and others have reported that BETi suppress the growth of TNBC cells by inducing senescence or apoptosis and are efficacious in numerous xenograft models of this disease, albeit to varying extents (16, 17). Shu et al. (17) further showed that acquired resistance to BETi can occur through hyperphosphorylation of BRD4, resulting from down-regulation of PP2A activity (18). However, the mechanisms underlying de novo BETi resistance in TNBC have not yet been determined.
We recently reported that BETi disrupt proper mitotic progression, thereby inducing mitotic catastrophe in TNBC (19). Mitotic catastrophe is an oncosuppressive mechanism that senses aberrant mitosis, driving cells irreversibly to apoptotic, necrotic, or senescent cell fates (20). TNBC cell lines respond to BETi-induced mitotic catastrophe by undergoing either senescence or apoptosis depending on the cell line (16). However, it is still unclear what dictates the choice between these cell fates in response to BETi. Given the potential for cancer cells to reenter the cell cycle following long-term senescence (21, 22) and the ability of senescent cells to secrete oncogenic growth-stimulatory factors (23), it is essential to elucidate the mechanisms by which the senescent/apoptotic choice is made to provide a potential therapeutic path for preventing the senescent cell fate. This should diminish intrinsic resistance to BETi and provide a more durable response in TNBC patients.
The BCL-2 protein family represents one possible regulator of cell fates in response to BETi. These proteins can modulate cell fates following mitotic catastrophe induced by other, unrelated agents (24–26). The BCL-2 family can be divided into three groups: pro-apoptotic effectors (e.g. BAX and BAK), anti-apoptotic guardian proteins (e.g. BCL-2 and BCL-xL), and BH3-only sensor proteins (e.g. BIM and BID). In normal cells as well as in cancer cells, the decision between survival and death in response to an intrinsic apoptotic signal depends on the ratio of the pro-apoptotic and sensor proteins to the anti-apoptotic BCL-2 family members. Overexpression of anti-apoptotic BCL-2 proteins is a mechanism often used by cancer cells to avoid apoptosis (27–30). In breast cancer, overexpression of BCL-xL is associated with metastasis and higher tumor grade (31). An evaluation of the individual contributions of BCL-2 family members to the viability of senescent cells revealed that inhibition of BCL-W and BCL-XL induces the death of senescent cells, making this treatment approach senolytic (32). Further, the expression of BCL-2 family members has been reported to contribute to BETi response (17, 33), yet their specific impact on BETi-induced senescence or apoptosis has not been addressed.
Here, we show that BCL-xL expression levels dictate the choice of TNBC cells to undergo senescence versus apoptosis in response to BETi-induced mitotic catastrophe. Cells that express high levels of BCL-xL senesce whereas cells that have low BCL-xL expression undergo apoptosis. In addition, BETi further reduce expression of BCL-xL in cells that die whereas BCL-xL expression in cell lines that senesce remains high following BETi treatment. Manipulation of BCL-xL levels was sufficient to promote death, or senolysis, of cells that typically senesce in response to BETi, without altering the mitotic effect itself. Overexpression of BCL-xL further desensitized TNBC cells to BETi-induced apoptosis, indicating that BCL-xL levels can confer intrinsic resistance to BETi. Our data provide evidence for the potential benefit of combining BCL-xL inhibitors with BETi in TNBC patients, particularly those whose tumors express high levels of BCL-xL to avoid intrinsic resistance to BETi. In addition, high BCL-xL levels may provide a useful biomarker for predicting therapeutic response to BETi.
Results
TNBC cells have varied apoptotic/senescent responses to sustained BET inhibition
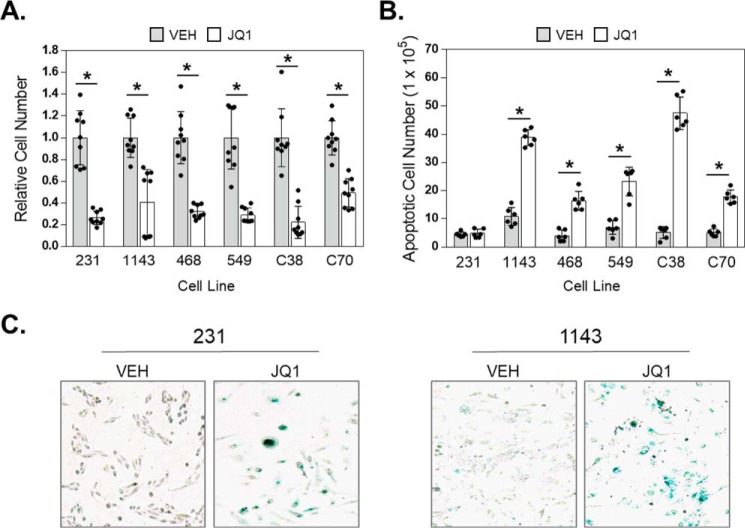
To confirm that BETi induce cell line–dependent apoptotic and/or senescent responses, we treated six TNBC cell lines (MDA-MB-231, HCC38, BT549, HCC1143, HCC70, and MDA-MB-468) with 500 nm of the prototypical BETi JQ1. At 96 h of JQ1 treatment, the growth of all six lines was inhibited to varying degrees (Fig. 1A). The extent of BETi growth inhibition did not correlate with a particular proclivity for apoptosis, as MDA-MB-231 cells have the lowest IC50 for JQ1 but responded to BETi by undergoing senescence, as we demonstrated previously (16). JQ1 induced apoptosis to differing extents, with MDA-MB-231 cells being completely resistant to cell death at this dose and time whereas HCC38 cells demonstrated a 10-fold induction of apoptosis (Fig. 1B). To assess the response of TNBC to extended BETi exposure, all six cell lines were treated with 1 μm JQ1 for 8 days. Only the MDA-MB-231 and HCC1143 cell lines had any surviving cells at this time point. Staining for senescence-associated β-gal revealed that these remaining cells had undergone senescence rather than dying (Fig. 1C). Thus, in response to BETi, TNBC cells can either undergo apoptosis or survive through the activation of senescence. Because senescent cells can contribute to cancer recurrence (34), determining why this fate is chosen by some cells in response to BETi should reveal approaches to improve the clinical efficacy of these drugs.
Figure 1.
TNBC cells have varied apoptotic/senescent responses to sustained BET inhibition. A, TNBC cell line panel: MDA-MB-231, HCC1143, MDA-MB-468, BT549, HCC38, and HCC70 cells were exposed to vehicle (VEH) or 500 nm JQ1 for 96 h and the number of viable cells counted using trypan blue exclusion. B, TNBC cell line panel was treated with VEH or 500 nm JQ1 and apoptosis measured by annexin V–FITC plus PI staining and FACS analysis. Bars show the number of cells that were apoptotic. C, representative images (10×) of MDA-MB-231 and HCC1143 cells that were treated with vehicle or 1 μm JQ1 for 8 days and then stained for senescence-associated β-gal activity. For graphs, data are expressed as mean ± S.D. (* = p < 0.05 compared with vehicle), n = 3 independent experiments.
Cell lines that senesce and survive following BETi exposure express higher basal levels of BCL-xL
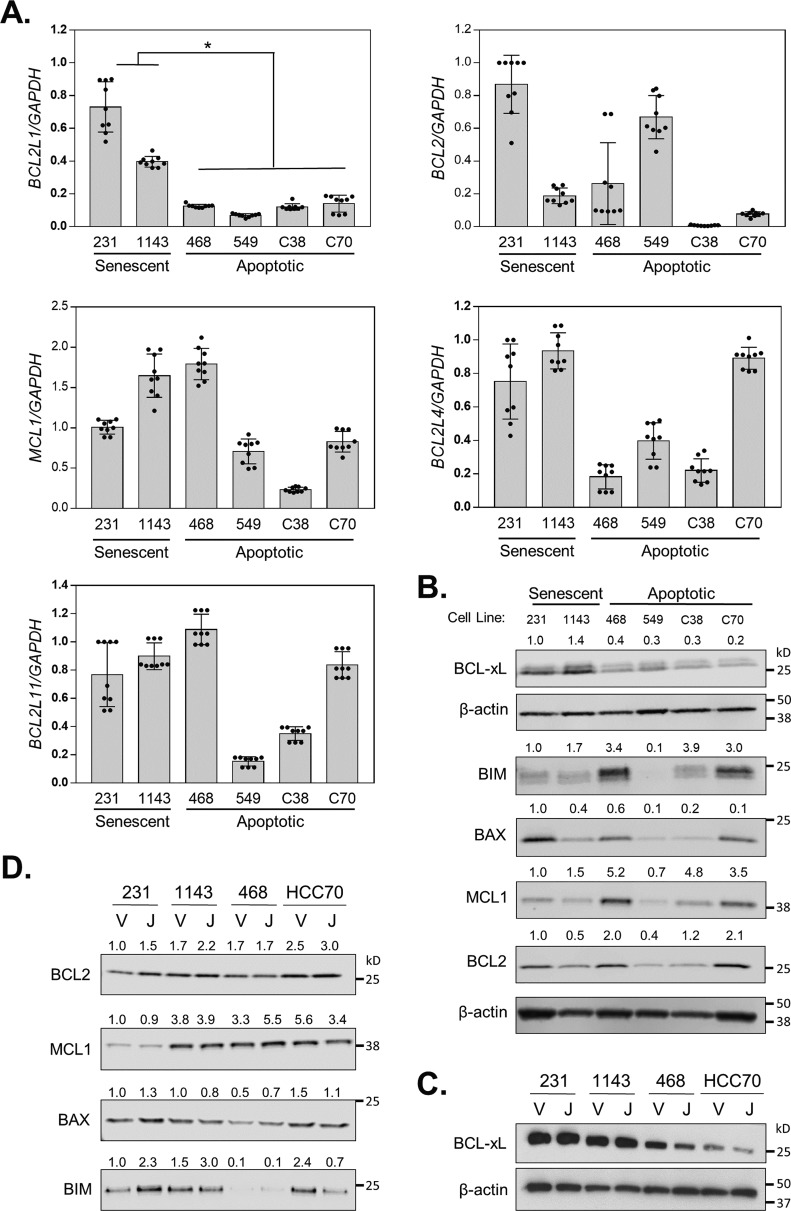
The combined expression patterns of several BCL-2 family members (BCL-2, BCL-xL, and BAD) have been reported to predict the efficacy of BETi in diverse cancer models (33). In addition, a single, experimentally induced TNBC model of acquired BETi resistance displayed increased BCL-xL mRNA and protein expression (17). To determine whether the expression levels of specific BCL-2 family members may correspond with the ability of TNBC cells to undergo senescence in response to BETi, we evaluated basal mRNA (Fig. 2A) and protein (Fig. 2B) levels of pro- and anti-apoptotic members of this family including BCL2, BCL2L1 (BCL-xL), BCL2L11 (BIM), BCL2L4 (BAX), and MCL1 in diverse TNBC cell lines. We anticipated that expression of the anti-apoptotic members, BCL2, BCL2L1, and MCL1 may be elevated, whereas expression of the pro-apoptotic family members BCL2L11 and BCL2L4 would be decreased in the two cell lines (MDA-MB-231 and HCC1143) capable of senescing in response to extended BETi exposure compared with the TNBC lines that completely apoptose. Our results revealed that MCL1, BCL-2, BIM, and BAX were highly variable in all TNBC cell lines and did not correlate with therapeutic response. In contrast, BCL-xL mRNA and protein were more highly expressed in MDA-MB-231 and HCC1143 cells compared with the cell lines that do not survive BETi exposure (HCC38, BT549, HCC70, and MDA-MB-468). Thus, of the BCL-2 family members investigated, only basal expression of BCL-xL was associated with the cell line–specific activation of senescence and survival following BETi.
Figure 2.
Cell lines that senesce and survive following BETi exposure express higher basal levels of BCL-xL. A, RT-qPCR of basal level mRNA expression of BCL2 family members in TNBC panel: MDA-MB-231, HCC1143, MDA-MB-468, BT549, HCC38, and HCC70. Data are presented as mean ± S.D., n = 3 independent experiments. B, Western blot showing protein expression of various BCL-2 family members or β-actin loading control in a panel of TNBC cell lines. C, MDA-MB-231, HCC1143, MDA-MB-468, and HCC70 cells were treated with vehicle (V) or 500 nm JQ1 (J) for 72 h and cell lysates evaluated by Western blotting for BCL-xL and β-actin. D, MDA-MB-231, HCC1143, MDA-MB-468, and HCC70 cells were treated with vehicle (V) or 500 nm JQ1 (J) for 72 h, and cell lysates were evaluated by Western blotting for BCL2, MCL1, BAX, and BIM. Each protein was normalized to total protein.
To further assess the impact of BCL-2 family members on the response of TNBC cells to BETi, we evaluated how the expression of these proteins changed following BETi treatment. Notably, within 72 h JQ1 suppressed BCL-xL expression only in cell lines that die in response to BETi (Fig. 2C). In contrast, the effect of JQ1 on other BCL-2 family member proteins varied and did not correlate with drug response (Fig. 2D). These data indicate that in addition to having low basal BCL-xL levels, certain TNBC cells can be additionally compromised by BETi-induced suppression of BCL-xL expression, leading to a greater pro-apoptotic drive.
BCL-xL inhibition changes cell fate in response to BETi from senescence to apoptosis
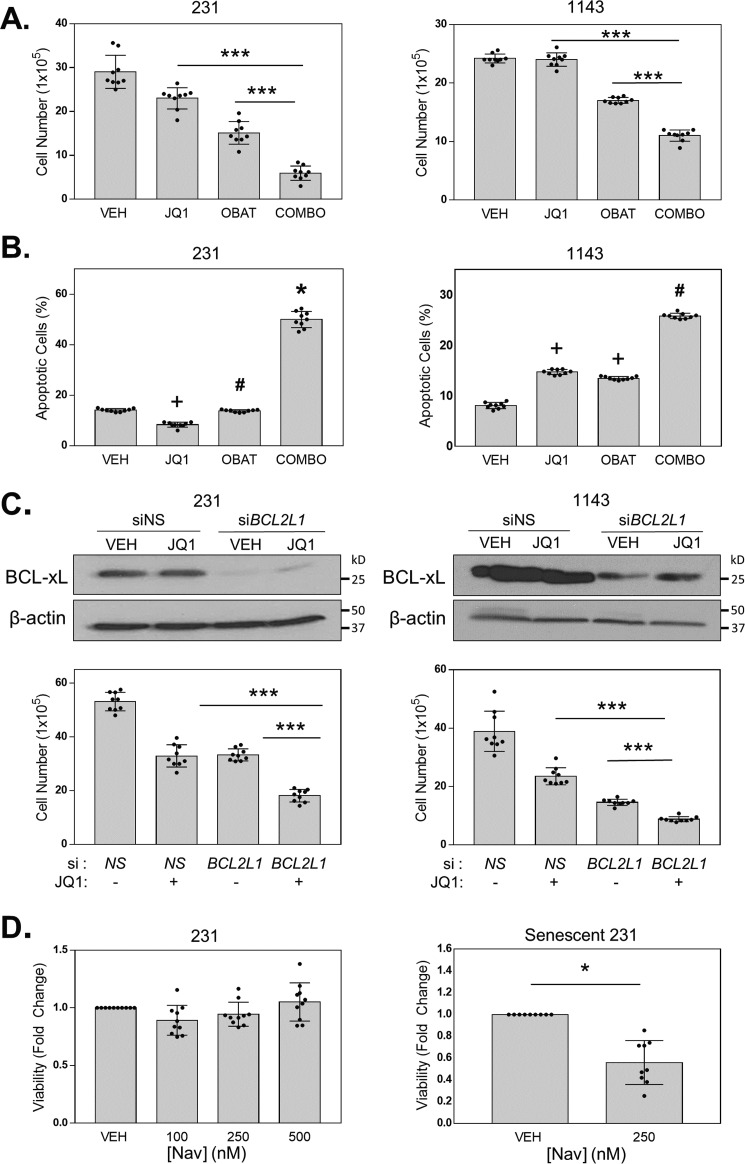
The elevated expression of BCL-xL in cells capable of undergoing senescence in response to BETi suggested that this protein may prevent cells from engaging in a complete apoptotic response. To determine whether inhibiting anti-apoptotic BCL-2 family members could evoke an apoptotic response to BETi rather than senescence, we used Obatoclax, a small molecule that binds to BCL-2 and related proteins and prevents their interactions with pro-apoptotic family members. The two TNBC cell lines that senesce in response to BETi (MDA-MB-231 and HCC1143) were treated with vehicle, a low dose of JQ1 (50 nm) to promote senescence, Obatoclax, or the combination of JQ1 and Obatoclax for 72 h. The number of viable cells were then counted. The combination of JQ1 and Obatoclax treatment resulted in a lower percentage of viable cells when compared with either single agent (Fig. 3A). This decrease could be because of a change in proliferation or cell death. To determine whether the reduction in viable cells was due to an increase in apoptosis, cells were treated as above for 5 days and apoptotic cells quantified by flow cytometry for annexin V–stained cells (Fig. 3B). The combination of the two drugs induced a greater number of apoptotic cells than either drug alone, indicating that blocking BCL-2 anti-apoptotic proteins can promote apoptosis and inhibit the senescence response to BETi.
Figure 3.
BCL-xL inhibition shifts cell fate in response to BETi from senescence to apoptosis. A, MDA-MB-231 cells were treated with DMSO (VEH), 50 nm JQ1, 5 nm Obatoclax, or the combination (COMBO), and HCC1143 cells were treated with DMSO (VEH), 200 nm JQ1, 40 nm Obatoclax, or the combination (COMBO). Viable cells were counted using trypan blue exclusion. Error bars are S.D. *** = p < 0.05. B, MDA-MB-231 and HCC1143 cells were treated with DMSO (VEH), 125 nm JQ1, 40 nm Obatoclax, or the combination (COMBO). Apoptotic cells were quantified using annexin V–FITC plus PI staining and FACS analysis. Error bars are S.D. + = p < 0.05 compared with VEH and COMBO, # = p < 0.05 compared with VEH and COMBO, and * = p < 0.05 compared with VEH, JQ1, and OBAT. C, MDA-MB-231 and HCC1143 cells were transfected with either siRNA targeting BCL2L1 (BCL-xL) or NS siRNA. Reduced BCL-xL expression was confirmed by Western blotting with β-actin serving as a loading control. Cell viability was quantified using the trypan blue exclusion assay after a 72-h treatment of JQ1 (250 nm). D, left, MDA-MB-231 cells were treated with either DMSO (VEH) or 100, 250, or 500 nm Navitoclax for 72 h. Viable cells were counted using the trypan blue exclusion assay. Right, senescence was induced in MDA-MB-231 cells by pretreatment with 1 μm JQ1 for 8 days. Cells were then treated with Navitoclax (250 nm) for 72 h. Error bars are S.D. * = p < 0.05, *** = p < 0.001.
To assess whether BCL-xL is the principal anti-apoptotic protein that dictates the ability to invoke a senescence response to BETi in TNBC cells, we directly silenced its expression with RNAi. MDA-MB-231 and HCC1143 cells were transfected with siRNA directed against BCL-xL or nonsilencing (NS) control siRNA and subsequently treated with either vehicle or JQ1. Similar to the drug combination results, BCL-xL silencing followed by JQ1 treatment reduced the percentage of viable cells compared with either JQ1-treated cells transfected with NS siRNA or vehicle-treated cells with BCL-xL silencing (si-BCL2L1) (Fig. 3C). Together, these results indicate that inhibiting BCL-xL, either pharmacologically or genetically, stimulates a greater cell death response in TNBC cells that are capable of undergoing senescence and surviving BETi treatment.
To determine whether combined inhibition of BET proteins and anti-apoptotic BCL-2 family members can initiate apoptosis in cells that have already senesced in response to BETi, we treated MDA-MB-231 cells with vehicle or three doses of the BH3 mimetic Navitoclax (100, 250, 500 nm). Treatment with Navitoclax lasted for 72 h and began after the cells had already been treated with JQ1 for 96 h to induce senescence. Without JQ1, these doses of Navitoclax did not alter viability (Fig. 3D, right panel). However, when we treated MDA-MB-231 cells that had previously undergone BETi-induced senescence (Senescent 231) with 250 nm Navitoclax, this caused a significant decrease in viability (Fig. 3D, left panel) compared with cells that had previously been treated with vehicle. Thus, while co-inhibition of BET and anti-apoptotic BCL-2 family proteins induces apoptosis in cells that can senesce in response to BETi, sequential treatment with BETi followed by BCL-2 family inhibition also induces greater cell death. These data indicate that Navitoclax is senolytic following BETi treatment.
BCL-xL overexpression suppresses BETi-induced apoptosis
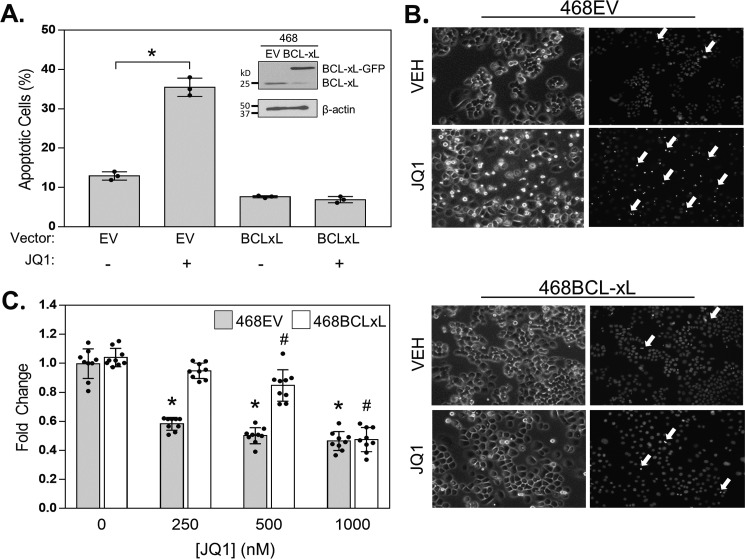
Pharmacologic or genetic inhibition of BCL-xL revealed that sustained activity of this protein is necessary to avoid complete apoptosis in response to BETi. To determine whether enforced BCL-xL expression is sufficient to suppress BETi-induced apoptosis, we overexpressed BCL-xL in cells that have low expression of this protein and are highly apoptotic in response to BETi. MDA-MB-468 cells were transfected with a BCL-xL expression vector (468BCLxL) or an empty vector (468EV) control. They were then treated with vehicle or JQ1 (500 nm) for 72 h, and pyknotic nuclei were assessed with Hoechst. JQ1 increased the number of apoptotic cells in 468EV cells but not in 468BCLxL cells (Fig. 4, A and B). In addition, 468EV and 468BCLxL cells were treated with increasing concentrations of JQ1, and cell viability was assessed using trypan blue exclusion assays. As expected, there was a dose-dependent inhibition of viable cells in the 468EV controls. In contrast, only 1000 nm of JQ1 induced a significant decrease in cell number in BCL-xL overexpressing cells (Fig. 4C). These results demonstrate that increasing BCL-xL expression levels is sufficient to blunt the growth inhibitory effects of BETi and suppress apoptosis in cells that normally die in response to BETi. These data suggest that not only could increased expression of BCL-xL contribute to acquired BETi resistance (17) but also could confer de novo resistance by preventing the induction of apoptosis.
Figure 4.
BCL-xL overexpression suppresses BETi-induced apoptosis. A and B, 468EV and 468BCLxL cells were treated with vehicle or 500 nm JQ1 for 72 h. A, apoptotic cells were stained with Hoechst, and pyknotic nuclei (cells undergoing apoptosis) were counted. Inset, Western blot confirming overexpression of BCL-xL in MDA-MB-468 cells (468BCLxL) compared with MDA-MB-468 cells expressing the empty vector (468EV). B, representative images of Hoechst staining. White arrows indicate pyknotic nuclei. C, 468EV and 468BCLxL cells were treated with increasing concentrations of JQ1, and viable cells were counted after 72 h. For graphs, data are presented as mean ± S.D., n = 3 independent experiments. * = p < 0.05 compared with vehicle for 468EV cell line. # = p < 0.05 compared with vehicle for 468BCLxL cell line.
BCL-xL regulation of apoptosis occurs downstream of BETi-induced mitotic catastrophe
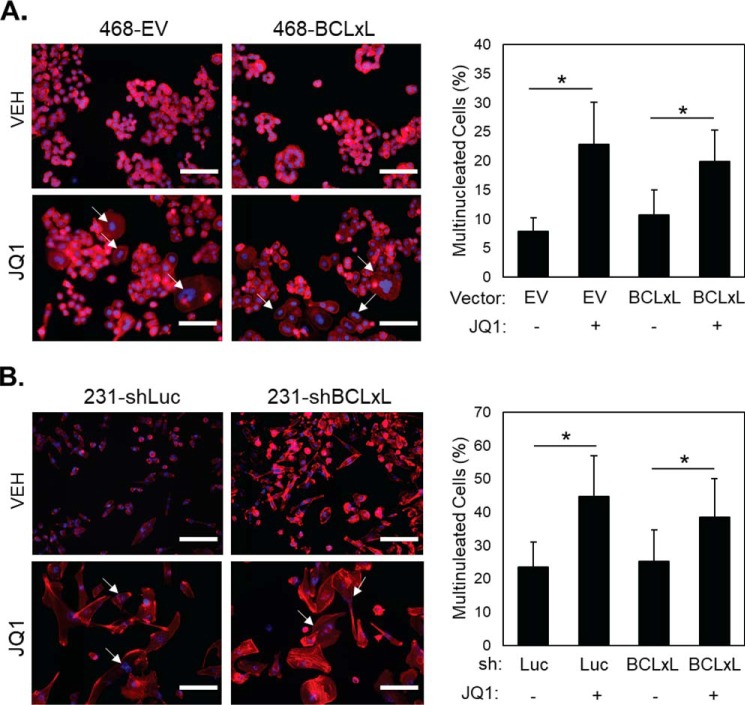
Multiple BETi cause apoptosis or senescence in TNBC cells as a terminal response to the more proximal induction of mitotic catastrophe (16, 19). BETi-treated TNBC cells first become multi-nucleated which leads to varying degrees of apoptosis or senescence, depending on the cell line. Manipulation of BCL-xL levels could either alter the BETi-induced mitotic defect or the terminal cell fate that occurs in response to disrupted mitosis. To determine whether modulating BCL-xL expression levels impacts the accumulation of multi-nucleated cells in response to BETi, 468EV and 468BCLxL cells were treated with 1000 nm JQ1 for 4 days. Multi-nucleated cells were visualized using DAPI (DNA stain) to observe the nucleus and phalloidin (F-actin/cytoskeleton stain) to identify cell borders. There was an accumulation of multi-nucleated cells following JQ1 treatment regardless of BCL-xL overexpression (Fig. 5A). Moreover, silencing BCL-xL expression in cells that normally express high basal levels of BCL-xL (MDA-MB-231) had no impact on the accumulation of multi-nucleated cells in response to BETi (Fig. 5B). Thus, BCL-xL expression levels do not impact the ability of BETi to induce multi-nucleation in TNBC cells.
Figure 5.
BCL-xL modulation does not affect BETi-induced multi-nucleation. A, left, representative images (20×) of 468EV and 468BCLxL cells treated with vehicle (VEH) or 500 nm JQ1 for 4 days and stained with DAPI (blue, nucleus) and phalloidin (red, F-actin). White arrows indicate multi-nucleated cells; scale bars are 120 μm. Right, quantitation of multi-nucleated cells. Data are mean ± S.D., * = p < 0.05 compared with vehicle. B, left, representative images (20×) of MDA-231-shLuc and MDA-231-shBCLxL cells treated with vehicle (VEH) or 500 nm JQ1 for 8 days and stained with DAPI (blue, nucleus) and phalloidin (red, F-actin). White arrows indicate multi-nucleated cells. Right, quantitation of multi-nucleated cells. Data are mean ± S.D., * = p < 0.05 compared with vehicle.
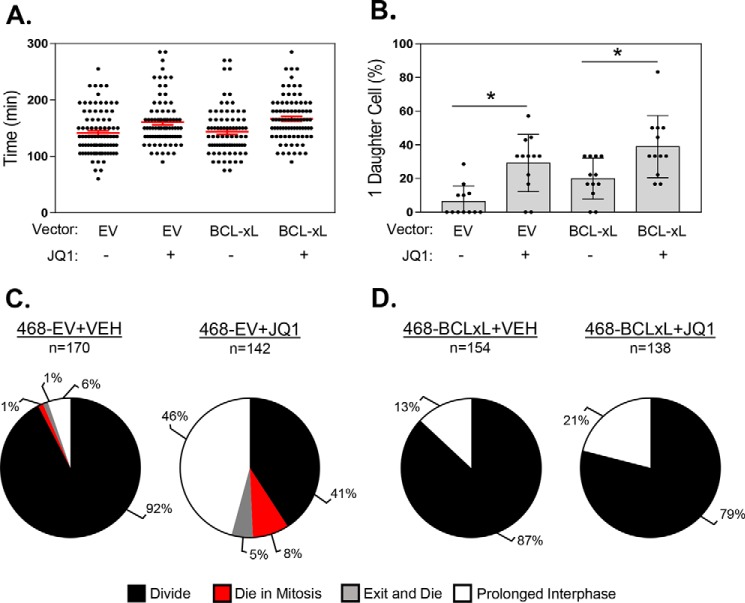
To directly assess the impact of modulating BCL-xL levels on BETi-induced mitotic cell fate, 468EV and 468BCLxL cells were treated with vehicle or 1000 nm JQ1, and live cell imaging was used to follow individual cells through mitosis and evaluate their post-mitotic cell fate. Treatment with JQ1 significantly increased the amount of time needed for cells to complete mitosis compared with vehicle-treated cells (Fig. 6A). This effect was independent of BCL-xL expression, because 468EV and 468BCLxL cells did not demonstrate any statistically significant differences in time to mitotic completion. JQ1 treatment also more frequently led to the generation of a single daughter cell instead of two following mitosis compared with vehicle-treated cells (Fig. 6B). Yet again, there was no difference in the frequency of this aberrant event associated with BCL-xL expression. These data indicate that JQ1 disrupts proper mitotic progression independent of BCL-xL expression levels.
Figure 6.
BCL-xL regulation of apoptosis occurs downstream of BETi-induced mitotic catastrophe. A, bee swarm plot of the length of time required by individual 468EV and 468BCLxL cells to complete mitosis starting 6 h after the addition of vehicle or 1000 nm JQ1. Red lines are mean ± S.E. p < 0.05 between 1) 468EV+VEH and 468EV+JQ1 and 2) 468BCLxL+VEH and 468BCLxL+JQ1 according to Tukey's multiple comparison test. B, quantitation of the percent of single daughter cells produced following mitosis in 468EV and 468BCLxL cells. * = p < 0.05 compared with vehicle. C and D, pie charts showing the percentage of vehicle- and JQ1-treated 468EV (C) and 468BCLxL (D) cells that underwent different mitosis-associated cell fates: divide (black), die in mitosis (red), exit and die (gray), or prolonged interphase (white).
We then determined if overexpression of BCL-xL alters the terminal outcome of cells following BETi-induced mitotic dysfunction. We visually tracked cells from the time they exited mitosis until they died or entered a second round of mitosis. The vast majority of vehicle-treated 468EV cells (92%) were able to undergo a second mitosis, with only a small percentage (2%) of cells dying either during mitosis or immediately following mitotic exit (Fig. 6C, left panel). The remaining (6%) cells survived but did not divide again, hence they entered a prolonged interphase. As expected, when 468EV cells were treated with JQ1, there was a large shift in post-mitotic outcome, with less than 50% of cells entering a second mitosis (Fig. 6C, right panel). A greater proportion of cells died during mitosis or after mitotic exit compared with vehicle-treated cells, and there was a large (∼8-fold) increase in the percentage of cells that entered a prolonged interphase. In contrast to these results, BCL-xL overexpression completely protected cells against mitosis-associated cell death, with none of the vehicle- or JQ1-treated 468BCLxL cells dying during this experiment (Fig. 6D). BCL-xL overexpression also reduced the percentage of cells that entered a prolonged interphase in response to JQ1. These data indicate that although BCL-xL overexpression does not inhibit BETi-induced mitotic catastrophe, it does alter the terminal fate of TNBC cells in response to this defect.
Discussion
Patients diagnosed with TNBC have a poor prognosis because of the scarcity of targeted therapies available for this disease. TNBCs initially respond to cytotoxic therapies; however, they have a high rate of metastatic recurrence and a median overall survival of 13 months following detection of metastasis (35). BETi have previously been shown to reduce TNBC growth in both in vitro and in vivo models (15, 17, 36) and are currently being evaluated in clinical trials for this disease. We recently reported that BETi suppress growth of TNBCs by inducing mitotic catastrophe, resulting in either senescence or apoptosis (16, 19), yet the mechanisms underlying the choice between these two terminal responses remained unknown. Given the ability of senescent cells to promote recurrence in various cancer models (34), identifying pathways that prevent senescence in response to BETi will be critical for maximizing their efficacy. Here, we show for the first time that intrinsic levels of BCL-xL expression dictate TNBC cell fate decisions in response to BETi: high levels empower senescence whereas low levels promote apoptosis. In addition, BCL-xL expression is further reduced by JQ1 in cells that eventually die in response to JQ1 (MDA-MB-468 and HCC70) but is unaffected in cells that senesce in response to BETi (MDA-MB-231 and HCC1143). This is unique to BCL-xL, as other BCL-2 family member proteins have variable responses to JQ1 that do not correspond with terminal outcome. When BCL-xL is overexpressed, cells that typically undergo BETi-induced apoptosis are less likely to die in response to the drug. Moreover, suppression of BCL-xL in cells that are capable of senescing in response to BETi causes a shift in cell fate to apoptosis. Notably, the senolytic action of BCL-xL inhibition has previously been observed in other settings, with the BH3 mimetic Navitoclax (ABT-263) efficiently eliminating DNA damage–induced senescent cells in the lungs of mice (32) and senescent hematopoietic bone marrow stem cells in aged animals (37). In cancer models, Bernards and colleagues have also shown that senescence induced by etoposide or Alisertib, an Aurora A kinase inhibitor, can be precluded by co-treating with senolytic agents such as Navitoclax (38). Our observation that Navitoclax stimulates cell death in BETi-induced senescent cells provides supporting evidence for combining BCL-xL and BET inhibition in TNBC.
The results presented herein indicate that BCL-xL may be a major factor in defining the long-term response of TNBC patients to BETi. Although patients whose tumors have low intrinsic BCL-xL expression may have a durable response to BETi, those with higher levels may suffer from intrinsic resistance or recurrence. Our findings underscore the need to conduct further studies evaluating BCL-xL levels in tumors prior to BETi treatment to determine whether they correlate with patient outcomes. Moreover, blocking the induction of senescence in tumors that have high BCL-xL may convey improved outcomes in response to BETi. As proof of principle, we used both pharmacological and genetic approaches to combat the anti-apoptotic effects of BCL-xL in TNBC cells treated with BETi. These studies not only revealed that suppression of BCL-xL in cells that are capable of senescing in response to BETi results in a shift to maximal apoptosis, but they also demonstrated that BCL-xL inhibition eliminates cells that have already undergone BETi-induced senescence. Thus, BH3 mimetic drugs provide a senolytic approach that may increase the efficacy of BETi in TNBC. BETi are currently in clinical trials for various cancer types, and we expect our findings to be applicable beyond breast cancer.
Although BCL-xL ultimately regulates cell fates in response to BETi, modulating BCL-xL levels did not impact the ability of BETi to induce mitotic dysfunction. Cells with silenced or overexpressed BCL-xL still become multi-nucleated following BETi treatment. However, elevated BCL-xL expression prevents apoptosis as well as decreases the number of cells entering a prolonged interphase, even though these cells are multi-nucleated. The increased viability of multi-nucleated cells in the context of high BCL-xL levels raises the intriguing possibility that BETi may actually increase the rate of chromosomal instability in tumor cells that do not die in response to these drugs. It is possible that tumors that recur following treatment with BETi could have greater genomic disruption as it is well-established that polyploidy can induce numerical and structural chromosome aberrations (39).
Overexpression of BCL-xL has been shown to confer resistance to several currently used chemotherapies including doxorubicin, methotrexate, and 5-fluorouracil (40–42). Our study is the first to demonstrate that although BCL-xL can control cell fates, it does not alter the initial mitosis-associated defect that is conveyed by BETi. A recent report describing the design of a degrader of BET proteins, BETd-246, revealed that BCL-xL can also confer resistance to BETd-246–induced apoptosis (43). Moreover, this work also found that inhibitors of BCL-xL can synergize with BETd-246 in TNBC. However, these studies did not evaluate the impact of this novel BET degrader on mitotic events or the role of BCL-xL in dictating cell fate in response to these events. Combined with the findings reported herein, these data underscore the role of BCL-xL as a key modulator of therapeutics targeting the BET family of proteins and support the development of clinical trials that combine BET-targeting agents and BCL-xL inhibitors for the treatment of TNBC.
In summary, we have found that the intrinsic ability of BETi to induce apoptosis in TNBC cells is associated with basal expression of BCL-xL. Although BCL-xL levels do not impact the ability of BETi to induce mitotic catastrophe in TNBC cells, they do determine how these cells ultimately respond to mitotic dysfunction. As a result, BCL-xL expression may prove useful in predicting patient response to BETi as well as serve as a useful target for improving BETi efficacy. Moreover, these studies suggest that the induction of senescence in tumors with elevated BCL-xL expression may ultimately result in intrinsic resistance or more aggressive recurrences because of cell survival in the presence polyploidization and subsequent chromosomal defects.
Experimental procedures
Cell culture and reagents
All cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cells were maintained at 37 °C with 5% CO2. All lines were cultured in RPMI 1640, supplemented with 10% FBS and 1% penicillin-streptomycin (Invitrogen; 15140122). Doses of Navitoclax (Selleckchem, Houston, TX; Z258143) and Obatoclax (LC Laboratories, Woburn, MA; O-3077) were experimentally derived, and JQ1 (Cayman Chemical, Ann Arbor, MI; 11187) doses were based on our previously reported growth inhibitory values (16). Vectors encoding BCL-xL-targeting shRNAs (HS0000182676, HS0000182677, HS0000182678, HS0000182679, HS0000182680) and a control vector encoding an shRNA to Luciferase, shLuc (SHC007), were purchased from Sigma-Aldrich. Cells were transduced in the presence of polybrene (5 μg/ml) for 24 h, allowed to recover in normal growth medium for 48 h, and then selected for stable shRNA expression with puromycin.
An expression vector for WT BCL-xL or an empty vector (Clark Distelhorst) were used to generate stable populations of MDA-MB-468 cells as described previously (44). To quantify the number of viable cells, cells were trypsinized and counted using trypan blue exclusion on a Countess II FL (Thermo Fisher; AMQAF1000).
siRNA transfections
HCC1143 and MDA-MB-231 cells were seeded in 6-well plates to be 50% confluent upon transfection. siRNA targeting BCL-xL (Dharmacon, Lafayette, CO; L-003458-00-0005) or nontargeting siRNA (Dharmacon; D-001810-02-20) was transfected in Opti-MEM media (Invitrogen; 31985088) to a final concentration of 100 nm. Culture media were changed to complete growth medium after 6 h.
RNA analysis
RNA was isolated using TRIzol Reagent (Ambion, Waltham, MA; 15596018), treated with DNase (Ambion; AM1906), and cDNA produced using SuperScript II Reverse Transcriptase (Invitrogen; 18064-014). Quantitative real-time PCR was performed using Applied Biosystems (Foster City, CA) TaqMan Gene Expression Assays (Assay IDs = BCL2, Hs00608023_m1; BCL2L1 (BCL-xL), Hs00236329_m1; BCL2L4 (BAX), Hs00180269_m; BCL2L11 (BIM), Hs00708019_s1; MCL1, Hs01050896_m1; GAPDH, Hs02758991_g1).
Western blot analysis
Cells were lysed in radioimmune precipitation assay buffer (20 mm Tris-HCl, 150 mm NaCl, 1 mm Na2, EDTA, 1 mm EGTA, 1% Nonidet P-40, 1% sodium deoxycholate, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 μg/ml leupeptin) supplemented with 1 mm Na3VO4 (Sigma-Aldrich; 13721396) and protease inhibitors (Sigma-Aldrich; P8340). Immunoblots were probed with antibodies from Cell Signaling Technology that recognized BIM (2933), BAX (5023), BCL-xL (2764), BCL-2 (15071), MCL1 (94296), and β-actin (Sigma-Aldrich; A1978). Pierce ECL Western Blotting Substrate (Thermo Scientific; 32106) was used to develop blots and ImageJ was used for protein band densitometry.
Flow cytometric detection of apoptotic death
Annexin V staining for apoptotic cells was performed according to the manufacturer's protocol. Cells were incubated with FITC-conjugated annexin V (Molecular Probes, Eugene, OR; V13245) and propidium iodide (Invitrogen; P3566). Stained cells were analyzed by flow cytometry using the GUAVA-PCA flow cytometry system (Guava Technologies, Burlington, MA). Each experiment represents a minimum of 2 × 103 gated events from two wells. Percent apoptotic cells (annexin V–positive, PI-negative) and percent dead cells (annexin V–positive, PI-positive) were determined using the Guava software.
Hoechst staining
Hoechst staining was performed as described previously (16). Briefly, treated cells were stained with 10 μm Hoechst 33342 (Thermo Scientific; 62249) and the percentage of apoptotic cells with pyknotic nuclei was determined.
Immunocytochemistry
Cells grown on coverslips were fixed with 3.7% formaldehyde for 10 min and immunostained with Texas Red-X phalloidin (Invitrogen; T7471) for F-actin according to the manufacturer's instructions. Cells were mounted using Vectashield with DAPI (Vector Laboratories, Burlingame, CA; H-1500) and visualized at 20× magnification on a Leica inverted microscope.
Senescence-associated β-gal activity stain
Cells were fixed in 2% formaldehyde and 0.2% glutaraldehyde for 5 min followed by staining for 12–16 h at 37 °C in 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 2 mm MgCl2, 150 mm NaCl, 30 mm citric acid/phosphate buffer, pH 6, 1 mg/ml X-gal (Invitrogen; 15520).
Live cell imaging
MDA-MB-468-EV and MDA-MB-468–BCL-xL cells were treated with vehicle or 1000 nm JQ1. The IncuCyte Zoom System (Essen BioScience, Ann Arbor, MI) was used to collect images every 20 min at 20× for a total of 48 h. After the first 6 h of treatment, cells were tracked through mitosis and the time it took for cells to complete mitosis (mitotic timing) was recorded. Subsequent daughter cells were also tracked to determine whether they died, divided, or entered a prolonged interphase.
Statistical methods
Statistical analyses were performed as described for each experiment. Unless otherwise specified, all experiments in cell culture were independently performed three times in triplicate, with the results expressed as the mean of the nine values. For comparisons between two groups of values, statistical analysis of the results was performed by the Student's t test. For comparisons between more than two groups of values, one-way analysis of variance (ANOVA) and Tukey's multiple corrections tests were used. p values less than 0.05 were considered statistically significant.
Author contributions
S. S. G., J. M. S., and R. A. K. conceptualization; S. S. G., J. M. S., B. M. W., K. L. W.-B., M. S. S., R. S., and M. K. S. data collection; S. S. G., J. M. S., B. M. W., K. L. W.-B., M. S. S., R. S., M. K. S., and R. A. K. data analysis; S. S. G. and J. M. S. writing-original draft; S. S. G., J. M. S., B. M. W., K. L. W.-B., M. S. S., R. S., E. E. B., M. K. S., and R. A. K. writing-review and editing; J. M. S. validation; E. E. B. and M. K. S. resources; R. A. K. supervision; R. A. K. funding acquisition.
Acknowledgments
We thank Clark Distelhorst (Case Western Reserve University) for providing the BCL-xL expression vector.
This work was supported by National Institutes of Health Grants T32CA059366 (to S. S. G. and J. M. S.), F32CA210426 (to S. S. G.), F99CA212460 (to J. M. S.), F31CA224809 (to B. M. W.), RO1CA187780 (to E. E. B.), R01GM112895 (to M. K. S.), R01GM108743 (to M. K. S.), and R01CA206505 (to R. A. K.). The authors declare that they have no conflicts of interest with the contents of this article.The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- TNBC
- triple-negative breast cancer
- BETi
- BET family inhibitors
- NS
- nonsilencing.
References
- 1. Lovén J., Hoke H. A., Lin C. Y., Lau A., Orlando D. A., Vakoc C. R., Bradner J. E., Lee T. I., and Young R. A. (2013) Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carey L. A., Perou C. M., Livasy C. A., Dressler L. G., Cowan D., Conway K., Karaca G., Troester M. A., Tse C. K., Edmiston S., Deming S. L., Geradts J., Cheang M. C., Nielsen T. O., Moorman P. G., Earp H. S., and Millikan R. C. (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295, 2492–2502 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 3. Badve S., Dabbs D. J., Schnitt S. J., Baehner F. L., Decker T., Eusebi V., Fox S. B., Ichihara S., Jacquemier J., Lakhani S. R., Palacios J., Rakha E. A., Richardson A. L., Schmitt F. C., Tan P. H., Tse G. M., Weigelt B., Ellis I. O., and Reis-Filho J. S. (2011) Basal-like and triple-negative breast cancers: A critical review with an emphasis on the implications for pathologists and oncologists. Mod. Pathol. 24, 157–167 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 4. Bauer K. R., Brown M., Cress R. D., Parise C. A., and Caggiano V. (2007) Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the California Cancer Registry. Cancer 109, 1721–1728 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 5. Chen X., Li J., Gray W. H., Lehmann B. D., Bauer J. A., Shyr Y., and Pietenpol J. A. (2012) TNBCtype: A subtyping tool for triple-negative breast cancer. Cancer Inform. 11, 147–156 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dent R., Trudeau M., Pritchard K. I., Hanna W. M., Kahn H. K., Sawka C. A., Lickley L. A., Rawlinson E., Sun P., and Narod S. A. (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 13, 4429–4434 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 7. Giatromanolaki A., Sivridis E., Fiska A., and Koukourakis M. I. (2011) The CD44+/CD24- phenotype relates to 'triple-negative' state and unfavorable prognosis in breast cancer patients. Med. Oncol. 28, 745–752 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 8. Ma F., Li H., Wang H., Shi X., Fan Y., Ding X., Lin C., Zhan Q., Qian H., and Xu B. (2014) Enriched CD44+/CD24− population drives the aggressive phenotypes presented in triple-negative breast cancer (TNBC). Cancer Lett. 353, 153–159 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 9. Reuben J. M., Lee B. N., Gao H., Cohen E. N., Mego M., Giordano A., Wang X., Lodhi A., Krishnamurthy S., Hortobagyi G. N., Cristofanilli M., Lucci A., and Woodward W. A. (2011) Primary breast cancer patients with high risk clinicopathologic features have high percentages of bone marrow epithelial cells with ALDH activity and CD44+CD24lo cancer stem cell phenotype. Eur. J. Cancer 47, 1527–1536 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Irvin W. J. Jr., and Carey L. A. (2008) What is triple-negative breast cancer? Eur. J. Cancer 44, 2799–2805 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 11. Craig D. W., O'Shaughnessy J. A., Kiefer J. A., Aldrich J., Sinari S., Moses T. M., Wong S., Dinh J., Christoforides A., Blum J. L., Aitelli C. L., Osborne C. R., Izatt T., Kurdoglu A., Baker A., et al. (2013) Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol. Cancer Ther. 12, 104–116 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 12. Khongkow P., Gomes A. R., Gong C., Man E. P., Tsang J. W., Zhao F., Monteiro L. J., Coombes R. C., Medema R. H., Khoo U. S., and Lam E. W. (2015) Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene 35, 990–1002 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belkina A. C., Nikolajczyk B. S., and Denis G. V. (2013) BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J. Immunol. 190, 3670–3678 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Korb E., Herre M., Zucker-Scharff I., Darnell R. B., and Allis C. D. (2015) BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat. Neurosci. 18, 1464–1473 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi J., Wang Y., Zeng L., Wu Y., Deng J., Zhang Q., Lin Y., Li J., Kang T., Tao M., Rusinova E., Zhang G., Wang C., Zhu H., Yao J., Zeng Y. X., Evers B. M., Zhou M. M., and Zhou B. P. (2014) Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 25, 210–225 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sahni J. M., Gayle S. S., Bonk K. L., Vite L. C., Yori J. L., Webb B., Ramos E. K., Seachrist D. D., Landis M. D., Chang J. C., Bradner J. E., and Keri R. A. (2016) Bromodomain and extraterminal protein inhibition blocks growth of triple-negative breast cancers through the suppression of Aurora kinases. J. Biol. Chem. 291, 23756–23768 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shu S., Lin C. Y., He H. H., Witwicki R. M., Tabassum D. P., Roberts J. M., Janiszewska M., Huh S. J., Liang Y., Ryan J., Doherty E., Mohammed H., Guo H., Stover D. G., Ekram M. B., et al. (2016) Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529, 413–417 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. da Motta L. L., Ledaki I., Purshouse K., Haider S., De Bastiani M. A., Baban D., Morotti M., Steers G., Wigfield S., Bridges E., Li J. L., Knapp S., Ebner D., Klamt F., Harris A. L., and McIntyre A. (2017) The BET inhibitor JQ1 selectively impairs tumour response to hypoxia and downregulates CA9 and angiogenesis in triple negative breast cancer. Oncogene 36, 122–132 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sahni J. M., Gayle S. S., Webb B. M., Weber-Bonk K. L., Seachrist D. D., Singh S., Sizemore S. T., Restrepo N. A., Bebek G., Scacheri P. C., Varadan V., Summers M. K., and Keri R. A. (2017) Mitotic vulnerability in triple-negative breast cancer associated with LIN9 is targetable with BET inhibitors. Cancer Res. 77, 5395–5408 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vitale I., Galluzzi L., Castedo M., and Kroemer G. (2011) Mitotic catastrophe: A mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 12, 385–392 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 21. Roberson R. S., Kussick S. J., Vallieres E., Chen S. Y., and Wu D. Y. (2005) Escape from therapy-induced accelerated cellular senescence in p53-null lung cancer cells and in human lung cancers. Cancer Res. 65, 2795–2803 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 22. Wang Q., Wu P. C., Roberson R. S., Luk B. V., Ivanova I., Chu E., and Wu D. Y. (2011) Survivin and escaping in therapy-induced cellular senescence. Int. J. Cancer 128, 1546–1558 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coppé J. P., Patil C. K., Rodier F., Sun Y., Muñoz D. P., Goldstein J., Nelson P. S., Desprez P. Y., and Campisi J. (2008) Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fernández Y., España L., Mañas S., Fabra A., and Sierra A. (2000) Bcl-xL promotes metastasis of breast cancer cells by induction of cytokines resistance. Cell Death Differ. 7, 350–359 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 25. Harley M. E., Allan L. A., Sanderson H. S., and Clarke P. R. (2010) Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 29, 2407–2420 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Terrano D. T., Upreti M., and Chambers T. C. (2010) Cyclin-dependent kinase 1-mediated Bcl-xL/Bcl-2 phosphorylation acts as a functional link coupling mitotic arrest and apoptosis. Mol. Cell. Biol. 30, 640–656 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ghaneh P., Kawesha A., Evans J. D., and Neoptolemos J. P. (2002) Molecular prognostic markers in pancreatic cancer. J. Hepatobiliary Pancreat. Surg. 9, 1–11 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 28. Gobé G., Rubin M., Williams G., Sawczuk I., and Buttyan R. (2002) Apoptosis and expression of Bcl-2, Bcl-XL, and Bax in renal cell carcinomas. Cancer Invest. 20, 324–332 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 29. Krajewska M., Krajewski S., Epstein J. I., Shabaik A., Sauvageot J., Song K., Kitada S., and Reed J. C. (1996) Immunohistochemical analysis of bcl-2, bax, bcl-X, and mcl-1 expression in prostate cancers. Am. J. Pathol. 148, 1567–1576 [PMC free article] [PubMed] [Google Scholar]
- 30. Watanabe J., Kushihata F., Honda K., Mominoki K., Matsuda S., and Kobayashi N. (2002) Bcl-xL overexpression in human hepatocellular carcinoma. Int. J. Oncol. 21, 515–519 [PubMed] [Google Scholar]
- 31. Olopade O. I., Adeyanju M. O., Safa A. R., Hagos F., Mick R., Thompson C. B., and Recant W. M. (1997) Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J. Sci. Am. 3, 230–237 [PubMed] [Google Scholar]
- 32. Yosef R., Pilpel N., Tokarsky-Amiel R., Biran A., Ovadya Y., Cohen S., Vadai E., Dassa L., Shahar E., Condiotti R., Ben-Porath I., and Krizhanovsky V. (2016) Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 7, 11190 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Conery A. R., Centore R. C., Spillane K. L., Follmer N. E., Bommi-Reddy A., Hatton C., Bryant B. M., Greninger P., Amzallag A., Benes C. H., Mertz J. A., and Sims R. J. 3rd (2016) Preclinical anticancer efficacy of BET bromodomain inhibitors is determined by the apoptotic response. Cancer Res. 76, 1313–1319 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 34. Demaria M., O'Leary M. N., Chang J., Shao L., Liu S., Alimirah F., Koenig K., Le C., Mitin N., Deal A. M., Alston S., Academia E. C., Kilmarx S., Valdovinos A., Wang B., et al. (2017) Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 7, 165–176 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kassam F., Enright K., Dent R., Dranitsaris G., Myers J., Flynn C., Fralick M., Kumar R., and Clemons M. (2009) Survival outcomes for patients with metastatic triple-negative breast cancer: Implications for clinical practice and trial design. Clin. Breast Cancer 9, 29–33 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 36. Stratikopoulos E. E., Dendy M., Szabolcs M., Khaykin A. J., Lefebvre C., Zhou M. M., and Parsons R. (2015) Kinase and BET inhibitors together clamp inhibition of PI3K signaling and overcome resistance to therapy. Cancer Cell 27, 837–851 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang J., Wang Y., Shao L., Laberge R. M., Demaria M., Campisi J., Janakiraman K., Sharpless N. E., Ding S., Feng W., Luo Y., Wang X., Aykin-Burns N., Krager K., Ponnappan U., Hauer-Jensen M., Meng A., and Zhou D. (2016) Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22, 78–83 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang L., Leite de Oliveira R., Wang C., Fernandes Neto J. M., Mainardi S., Evers B., Lieftink C., Morris B., Jochems F., Willemsen L., Beijersbergen R. L., and Bernards R. (2017) High-throughput functional genetic and compound screens identify targets for senescence induction in cancer. Cell Rep. 21, 773–783 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 39. Storchova Z., and Kuffer C. (2008) The consequences of tetraploidy and aneuploidy. J. Cell Sci. 121, 3859–3866 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 40. Liu R., Page C., Beidler D. R., Wicha M. S., and Núñez G. (1999) Overexpression of Bcl-xL promotes chemotherapy resistance of mammary tumors in a syngeneic mouse model. Am. J. Pathol. 155, 1861–1867 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Minn A. J., Boise L. H., and Thompson C. B. (1996) Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 10, 2621–2631 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 42. Park S. S., Kim M. A., Eom Y. W., and Choi K. S. (2007) Bcl-xL blocks high dose doxorubicin-induced apoptosis but not low dose doxorubicin-induced cell death through mitotic catastrophe. Biochem. Biophys. Res. Commun. 363, 1044–1049 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 43. Bai L., Zhou B., Yang C. Y., Ji J., McEachern D., Przybranowski S., Jiang H., Hu J., Xu F., Zhao Y., Liu L., Fernandez-Salas E., Xu J., Dou Y., Wen B., et al. (2017) Targeted degradation of BET proteins in triple-negative breast cancer. Cancer Res. 77, 2476–2487 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang N. S., Unkila M. T., Reineks E. Z., and Distelhorst C. W. (2001) Transient expression of wild-type or mitochondrially targeted Bcl-2 induces apoptosis, whereas transient expression of endoplasmic reticulum-targeted Bcl-2 is protective against Bax-induced cell death. J. Biol. Chem. 276, 44117–44128 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]